Abstract
Members of the Snf1/AMPK family of protein kinases are activated by distinct upstream kinases that phosphorylate a conserved threonine residue in the Snf1/AMPK activation loop. Recently, the identities of the Snf1- and AMPK-activating kinases have been determined. Here we describe the purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. The identities of proteins associated with the Snf1-activating kinases were determined by peptide mass fingerprinting. These kinases, Sak1, Tos3 and Elm2 do not appear to require the presence of additional subunits for activity. Sak1 and Snf1 co-purify and co-elute in size exclusion chromatography, demonstrating that these two proteins form a stable complex. The Snf1-activating kinases phosphorylate the activation loop threonine of Snf1 in vitro with great specificity and are able to do so in the absence of β and γ subunits of the Snf1 heterotrimer. Finally, we showed that the Snf1 kinase domain isolated from bacteria as a GST fusion protein can be activated in vitro and shows substrate specificity in the absence of its β and γ subunits.
Keywords: Elm1, Hym1, Sak1, Snf1, Snf1-activating kinase, Tos3
Abbreviations: CkII, casein kinase II; GST, glutathione S-transferase; HA, haemagglutinin; TAP, tandem affinity purification
INTRODUCTION
The Snf1 kinase of Saccharomyces cerevisiae and its mammalian orthologue, AMPK belong to a highly conserved family of serine/threonine protein kinases that are found in all eukaryotes. In general, Snf1 and AMPK are active under conditions of energy limitation and inactive when energy supplies are abundant. In S. cerevisiae, Snf1 plays an important role as cells deplete glucose supplies and are forced to either ferment alternative carbon sources or grow by aerobic respiration [1,2]. In mammals, AMPK is activated by an elevated AMP:ATP ratio such as that observed in skeletal muscles during exercise [3]. Interest in AMPK has been intensified by the finding that activation of AMPK may be the molecular mechanism of action for two classes of drugs that are widely used to treat type 2 diabetes [4].
The Snf1/AMPK kinase complexes are heterotrimers composed of a catalytic α subunit and two regulatory subunits, β and γ. Co-transfection experiments in mammalian cells and genetic experiments in yeast have shown that all three subunits are required for kinase function in vivo [5–7]. The γ subunit is required for efficient activation of Snf1/AMPK by counteracting an inhibitory domain present in the C-terminus of the α subunit [8,9]. In addition, the γ subunit contains two Bateman domains that directly bind to AMP and contribute to the activation of AMPK by AMP [10]. A second activation step involves the phosphorylation of a conserved threonine in the activation loop of the α subunit's kinase domain. This mechanism for kinase regulation is widespread and examples can be found for both auto-phosphorylation and trans-phosphorylation carried out by a distinct upstream kinase [11]. The activation loops of the Snf1 and AMPK enzymes are phosphorylated by distinct upstream kinases [12,13]. The identities of the Snf1- and AMPK-activating kinases have been revealed only recently. We and others have found that yeast expresses three distinct Snf1-activating kinases [14–16] that are each capable of activating Snf1. In mammals, the primary activating kinase for AMPK is LKB1 (also known as STK11) [17,18], while CaMKKβ appears to activate AMPK under certain stress conditions [19].
The three Snf1-activating kinases present in yeast are Pak1, Tos3 and Elm1. Pak1 (also known as Sak1 or YER129W) was originally identified as a high-copy-number suppressor of a mutation in DNA polymerase α [20]. The acronym Pak1 stood for polymerase A kinase. However, in the intervening years, the acronym Pak has become widely used to refer to p21-activated kinases, a distinct sub-family of serine/threonine protein kinases that are activated by the binding of small G-proteins. The p21-activated kinases of S. cerevisiae are Ste20, Cla4 and Skm1 but not Pak1. In order to avoid confusion, we will henceforth use the acronym Sak1 to refer to YER129W. The Saccharomyces genome database has agreed to this name change.
The focus of this paper is the purification and characterization of the three Snf1-activating kinases from S. cerevisiae. In particular, we sought to address whether or not the Snf1-activating kinases require additional subunits for activity. The mammalian AMPK-activating kinase, LKB1, requires the binding of two auxiliary subunits, STRAD and MO25, for activity [17]. STRAD is a pseudokinase related to the yeast Ste20, whereas MO25 appears to act as a scaffolding protein for LKB1 and STRAD [21]. Yeast do not encode any pseudokinases and there is no evidence to link the Ste20 kinase to the glucose signalling pathway. However, yeast do encode a homologue of MO25 called Hym1. The S. cerevisiae HYM1 gene was thus named due to its homology to the Aspergillus nidulans hymA gene. Mutations in hymA cause aberrant cell morphology [22]. In S. cerevisiae, the Hym1 protein is also thought to function in cellular morphogenesis, cell cycle progression and cell separation [23–25]. Since Hym1 is a homologue of MO25, we address in this study whether or not the Hym1 protein is associated with any of the three Snf1-activating kinases. Of the three Snf1-activating kinases, Elm1 was an intriguing candidate for interaction with Hym1 since mutations in ELM1 also cause defects in the regulation of cell morphology [26,27]. To address the subunit composition of the three Snf1-activating kinases, each kinase was affinity purified, and associated proteins were identified by mass spectrometry.
MATERIALS AND METHODS
Strains and media
The S. cerevisiae strains used in this study were MSY182 (MATa ura3–52 leu2Δ1 trp1Δ63 his3Δ200), MSY912 (MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 sak1Δ::KAN), MSY913 (MATa ura3Δ0 leu2Δ0 his3Δ1 sak1Δ::KAN snf1Δ10), MSY915 (MATα ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 elm1Δ::KAN), MSY917 (MATa ura3 leu2 his3 trp1Δ63 lys2Δ0 met15Δ0 elm1Δ::KAN snf1Δ10), MSY916 (MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 tos3Δ::KAN), MSY918 (MATα ura3 leu2 his3 tos3Δ::KAN snf1Δ10), MSY923(MATα ura3 leu2 his3 sak1Δ::KAN elm1Δ::KAN tos3Δ::KAN snf1Δ10) and MSY904 (MATa ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 met15Δ0 hym1Δ::KAN+ pHYM1–3HA). For studies of CKII (casein kinase II), the strains YPH250 (MATa ade2–101 his3Δ200 leu2Δ1 lys2–801 trp1Δ1 ura3–52), YDH13 and YAR109 were used. YDH13 and YAR109 are isogenic to YPH250 except they also contain cka1Δ1::HIS3 cka2Δ1::TRP1. YDH13 and YAR109 are viable due to the presence of low-copy-number plasmids expressing temperature-sensitive alleles of CKA1 and CKA2 respectively [28,29]. Yeast strain JWY7128 (MATα ura3–52 lys2–801 ade2–101 trp1Δ63 his3Δ200 leu2Δ1 ytm1–1) contains a temperature-sensitive allele of YTM1 [30]. Strains were grown at 30 °C in synthetic complete media lacking the appropriate nutrient for plasmid selection unless otherwise noted. Glucose or sucrose was used as carbon source at 2% (w/v), while a mixture of glycerol and ethanol was used at 3% and 2% (v/v) respectively.
Plasmid constructions
Plasmids used in this study were generated using gap repair and standard subcloning protocols. The plasmids used for TAP (tandem affinity purification) used the TAP cassette from plasmid pBS1479 [31] amplified by PCR, such that the tag integrated at the C-terminus of the targeted open reading frame. The SNF1–TAP plasmid has been described in [32]. The SAK1–TAP fusion is expressed from the endogenous SAK1 promoter in pRS426 (2μ URA3). The kinase dead allele of SAK1–TAP was created such that the catalytic aspartate (Asp277) was changed to alanine. The ELM1–TAP and TOS3–TAP fusions were expressed from the ADH1 promoter also in a pRS426 backbone. The Snf1 kinase domain (amino acids 1–392) as well as the Thr210 to Ala210 (T210A) mutant were expressed in bacteria as GST (glutathione S-transferase) fusions using pGEX2T (Pharmacia). The plasmid expressing Snf1 with three copies of the HA (haemagglutinin) epitope at the C-terminus has been described [13]. Construction of V5-tagged Sak1, Elm1 and Tos3 behind their endogenous promoters has previously been described [33]. The pHYM1-HA plasmid was constructed by gap repair such that the Hym1 gene was expressed from its endogenous promoter with three copies of the HA epitope at its C-terminus.
TAP purification
TAP-tagged Snf1, Sak1, Elm1 and Tos3 were purified as described by Rigaut et al. [31]. Protein extracts from 2 litres of cell culture were prepared by grinding in liquid nitrogen. Cells were grown in media containing sucrose (Snf+) or glucose (Snf−). Proteins were eluted from the final calmodulin affinity resin (Stratagene) in five 200 μl fractions. Fractions two, three and four were pooled, glycerol added to a final concentration of 5% (v/v), and stored in aliquots at −80 °C. Approx. 5–10 μg of protein was recovered in a typical TAP purification from 2 litres of cells. Protein complexes were analysed by SDS/5% PAGE and visualized using silver staining (Bio-Rad Laboratories).
Immunoprecipitation
Immunoprecipitation was carried out using extracts from cells co-transformed with 2μ plasmids expressing Hym1–HA [13] and Sak1–V5, Elm1–V5 or Tos3–V5 [33]. Cells were grown in selective media containing 2% glucose and shifted to 0.05% glucose for 3 h. Hym1–HA was collected by incubation of 400–800 μg of extract with 20 μl of agarose-conjugated monoclonal anti-HA antibody (Santa Cruz Biotechnology) for 2 h at 4 °C. Beads were washed three times with RIPA buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 0.1% (w/v) SDS, 1% Nonidet P40, 0.5% sodium deoxycholate and phosphatase inhibitors (50 mM NaF, 5 mM sodium pyrophosphate)] and protein complexes were eluted by boiling beads for 5 min at 95 °C in 2× SDS sample buffer.
Western blotting
Epitope-tagged proteins were detected by Western blotting using HRP (horseradish peroxidase)-conjugated mouse monoclonal antibody (1:1000) directed against the HA epitope (Santa Cruz Biotechnology) and HRP-conjugated mouse monoclonal antibody (1:5000) against the V5 epitope (Invitrogen). Snf1 was also detected, when indicated (Figure 2), by the use of an HRP-conjugated polyhistidine antibody (1:1000) (Santa Cruz Biotechnology) which recognizes the 13 consecutive histidine residues naturally occurring in Snf1. TAP-tagged proteins were detected using rabbit polyclonal antibody (1:2000) directed against the calmodulin binding peptide (Upstate).
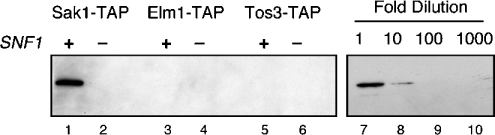
Figure 2. Association of Snf1 with the Snf1-activating kinases.
Sak1, Elm1 and Tos3 kinases were TAP-purified from SNF1 (+) or snf1Δ10 (−) strains. Samples were analysed by Western blotting using an antibody directed against polyhistidine (lanes 1–6). A dilution series of the GST–Snf1 kinase domain was analysed with the same antibody (lanes 7–10).
GST-fusion protein purification
GST–Mig1 containing a fragment of Mig1 (residues 202–414) including the four serines phosphorylated by Snf1 was purified as previously described by Nath et al. [32]. The GST–Snf1 kinase domains (wild-type and T210A; residues 1–392) were purified from 1 litre of Escherichia coli cells after induction by 0.1 mM IPTG (isopropyl β-D-thiogalactoside) for 3 h. Eluates were dialysed against the kinase assay buffer [20 mM Hepes (pH 7.0), 0.5 mM EDTA, 0.5 mM dithiothreitol, 5 mM magnesium acetate]. Glycerol was added to a final concentration of 5% (v/v), and aliquots were stored at −80 °C.
Enzyme assays
Kinase assays were conducted for 1 h at 30 °C in reactions (20 μl) containing 0.2 mM [γ-32P]ATP (1000 c.p.m. pmol), kinase assay buffer and protein substrates at approx. 50 μg/ml. Snf1-activating kinases were assayed at a concentration of approx. 1 μg/ml. For invertase assays of temperature-sensitive strains, cells were grown in rich media with 2% (w/v) glucose at 28 °C to a D600 of 0.6–0.8. An aliquot was removed, and the remaining cells were shifted to 0.05% YPD medium [1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose] for 2 h. For non-permissive conditions, cells were grown in rich media with 2% (v/v) glucose at 37 °C for 2 h. An aliquot was removed, and the remaining cells were shifted to 0.05% (w/v) glucose for an additional 2 h at 37 °C. Samples were taken during the four different growth conditions and assayed as described previously in [34].
Protein identification
TAP-purified protein complexes were precipitated using trichloroacetic acid, separated on SDS/7.5% PAGE gels and visualized with Simply Blue Colloid Coomassie staining (Invitrogen). Individual protein bands were excised and digested with trypsin. Peptides were concentrated with a Zip Tip (Millipore) and analysed using a PerSeptive Biosystems MALDI Voyager Elite STR DE reflectron TOF (time-of-flight) mass spectrometer. Proteins were identified by peptide mass fingerprinting using the Mascot search engine (Matrix Science). For the identification of proteins associated with Snf1, samples were precipitated with trichloroacetic acid and analysed by multidimensional protein identification technology [35].
Gel filtration chromatography
Yeast cell protein extracts were prepared from cells disrupted using glass beads in NHTG buffer [350 mM NaCl, 40 mM Hepes (pH 7.3), 0.1% (v/v) Tween-20 and 10% (v/v) glycerol]. Extracts were clarified by centrifugation at 12000 g for 20 min. The supernatant fraction (200 μl, containing approx. 600 μg of protein) was subjected to chromatography on a Superose 12 column at a flow rate of 0.5 ml/min in NHTG buffer. Fractions (0.5 ml) were collected and examined by Western blotting. The size standards used to calibrate the column were bovine thyroglobulin (670 kDa), rabbit muscle aldolase (158 kDa) and chicken ovalbumin (43 kDa).
RESULTS AND DISCUSSION
Identification of Snf1-interacting proteins
In order to determine the identity of proteins that associate with the Snf1 kinase complex, we first used the TAP strategy to isolate Snf1 complexes [31,32]. Associated proteins were identified using multidimensional protein identification technology [35] and 50 such proteins were identified in the Snf1 sample. In Table 1, we show only those proteins for which at least ten peptides were detected. The complete list of proteins found associated with Snf1 is presented in a supplementary table at http://www.BiochemJ.org/bj/393/bj3930797add.htm. Our results are very similar to those reported earlier in a systematic analysis of yeast protein complexes [36]. Five of the Snf1 kinase subunits (Snf1, Snf4, Sip2 and Gal83) were detected, while one of the alternative β subunits, Sip1, was not detected. We can only speculate as to whether the failure to detect Sip1 is due to its lower abundance, its poor extraction from the cells or its recalcitrance to mass spectrometry. In addition to the Snf1 subunits themselves, we also detected abundant Reg1, a regulatory subunit for the PP1 Glc7 known to regulate Snf1 activation loop phosphorylation [37], and two 14-3-3 proteins, Bmh1 and Bmh2, that associate with Reg1 [38]. Also abundant in the Snf1 preparation are members of the Hsp70 family of chaperone proteins. Finally, we also detected the Snf1-activating kinase, Sak1. Although not abundant, the identification of Sak1 was unambiguous. Absent even from the full list of 50 proteins of Snf1-associated proteins were the other Snf1-activating kinases, Tos3 and Elm1.
Table 1. Snf1 complex components.
| Peptides | |||||
|---|---|---|---|---|---|
| Protein | ORF | Total | Unique | Coverage | Protein function |
| Snf1 | YDR477W | 953 | 221 | 80.10% | α Subunit of Snf1 kinase complex |
| Gal83 | YER027C | 861 | 169 | 86.6% | β Subunit of Snf1 kinase complex |
| Reg1 | YDR028C | 483 | 122 | 63.50% | Regulatory subunit of Glc7 phosphatase |
| Snf4 | YGL115W | 433 | 86 | 82.90% | γ Subunit of Snf1 kinase complex |
| Ssb1 | YDL229W | 119 | 54 | 60.70% | Hsp70 protein |
| Ssb2 | YNL209W | 119 | 56 | 60.70% | Hsp70 protein |
| Sip2 | YGL208W | 133 | 43 | 58.80% | β Subunit of Snf1 kinase complex |
| Bmh1 | YER177W | 52 | 22 | 58.10% | 14-3-3 protein |
| Bmh2 | YDR099W | 51 | 22 | 56.00% | 14-3-3 protein |
| Ubi4 | YLL039C | 40 | 3 | 5.80% | Ubiquitin polyprotein |
| Sak1 | YER129W | 14 | 11 | 12.30% | Snf1-activating kinase |
| Ssa1 | YAL005C | 10 | 9 | 14.00% | Hsp70 protein |
| Ssa2 | YLL024C | 10 | 9 | 13.80% | Hsp70 protein |
Purification of the Snf1-activating kinases
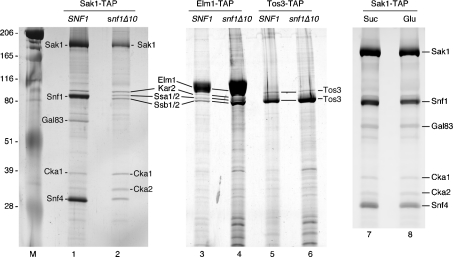
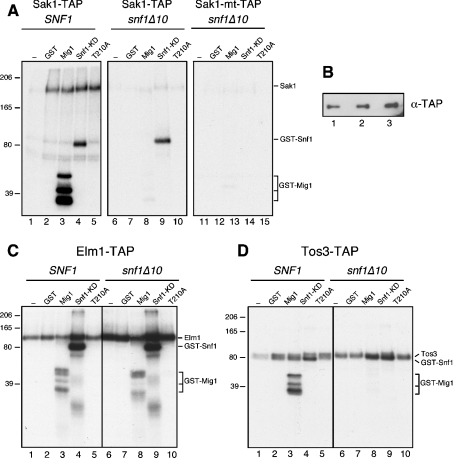
Earlier systematic analyses of yeast protein complexes did not include the three Snf1-activating kinases. In order to characterize these proteins and to identify associated proteins, we used the TAP purification strategy for each of the Snf1-activating kinases, Sak1, Tos3 and Elm1. In all cases, the TAP tag was added to the C-terminus of the full-length open reading frames present on high-copy plasmids. The TAP-tagged genes were functional in vivo as they were able to complement the Snf-phenotypes of a sak1Δ tos3Δ elm1Δ strain (results not shown). In each case, the Snf1-activating kinase, expressed from the TAP-tagged gene, was purified from strains that lacked the specific Snf1-activating kinase being purified. The purifications were also performed from strains that additionally lacked the SNF1 gene. The proteins present in these preparations were analysed on SDS gels and visualized by silver staining (Figure 1). In all cases, the most abundant protein had a gel mobility consistent with the predicted molecular mass of the Snf1-activating kinase with the residual TAP tag (132 kDa for Sak1, 78 kDa for Elm1 and 67 kDa for Tos3).
Figure 1. TAP purification of the Snf1-activating kinases.
Sak1, Elm1 and Tos3 were TAP-purified from SNF1 and snf1Δ10 strains. Protein complexes were separated by SDS/PAGE and silver stained. Proteins identified by tryptic peptide mass fingerprinting are indicated. The mobilities of molecular mass standards (kDa) are shown on the right. Sak1 was also purified from SNF1 cells grown in glucose (Glu) or sucrose (Suc) media (lanes 7 and 8).
Identification of proteins associated with the Snf1-activating kinases
A goal of this study was to determine whether the Snf1-activating kinases required additional subunits for activity. Purified Sak1 protein contained significant quantities of proteins other than Sak1 itself. In contrast, the purified Tos3 and Elm1 enzymes (Figure 1, lanes 3 and 5) appeared to be largely homogeneous without significant quantities of other proteins. The proteins that associated with Sak1 were excised from gels and identified by tryptic peptide mass fingerprinting. All the proteins identified are shown in Figure 1. The major protein that co-purified with Sak1 is the Snf1 protein. The Snf1 subunits, Snf4 and Gal83, were also detected in association with Sak1. The β subunits Sip2 and Sip1 were not detected in this analysis and the relative abundance of the Gal83 subunit appeared sub-stoichiometric, relative to the α and γ subunits. This finding may reflect a propensity of the β subunits to dissociate from the complex during purification or a difference in silver staining. To confirm that Snf1 associates with Sak1, we also purified Sak1 from snf1Δ10 cells and found that all three Snf1 subunits disappeared (Figure 1, lane 2). Snf1 was not detected by a similar analysis with Elm1 or Tos3. However, Elm1, Tos3, Snf1 and the Hsp70 proteins all have very similar gel mobilities, making clear conclusions difficult. Therefore, we analysed the purified kinase preparations by Western blotting using an antibody that recognizes polyhistidine tracts. Snf1 has 13 consecutive histidine residues near its N-terminus and is readily detected by this antibody in the Sak1 preparation (Figure 2). Snf1, if present, is below the level of detection in the Elm1 and Tos3 preparations. We also detected proteins of the Hsp70 class of chaperones (Ssa1, Ssa2, Ssb1, Ssb2 and Kar2) in association with Sak1 and Elm1. The functional significance of these proteins is uncertain. A minor role for the Ssb proteins in glucose repression has been noted [38]. However, the fact that some of the Hsp70 proteins (Ssa and Ssb) are cytoplasmic while one (Kar2) is found in the lumen of the endoplasmic reticulum suggest that these proteins may associate after cell lysis. Finally, both catalytic subunits of CKII were present in the Sak1 preparation. The regulatory subunits for CKII were not detected. The functional significance of CKII for glucose signalling is addressed below. Other polypeptides visible in the stained gel, in particular a protein migrating slightly more slowly than Snf4, were not successfully identified. Finally, we have shown previously by co-immunoprecipitation that the association of Snf1 and Sak1 is increased when cells are grown under conditions that activate Snf1 signalling [16]. In contrast, TAP purification of Sak1 demonstrated that Snf1 association was independent of carbon source (Figure 1, lanes 7 and 8). This discrepancy might be explained by previous observations that the process of centrifugation promotes activation of Snf1 [39]. Alternatively, it is possible that Sak1 and Snf1 are constitutively associated, but the stability of the interaction varies with carbon source and extraction method.
Sak1 and Snf1 form a complex
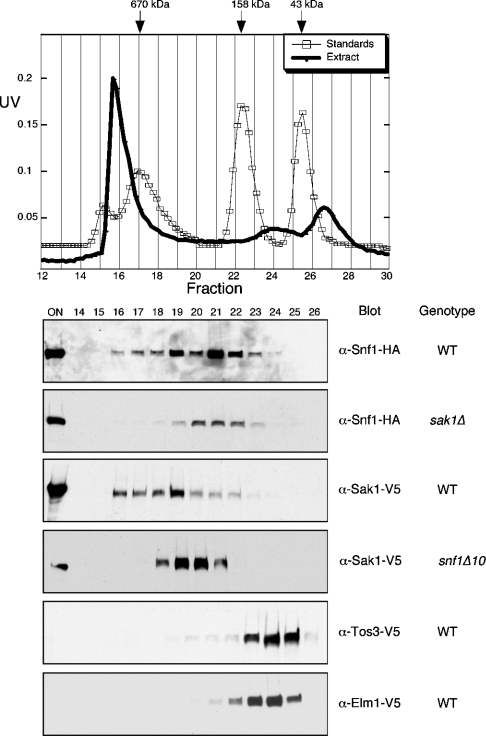
Purified Sak1 contains significant quantities of Snf1, at levels that may approach a stoichiometry of 1:1. However, the TAP-tagged Snf1-activating kinases were expressed from high-copy-number plasmids, complicating the interpretation of the stoichiometry of complex components. To further investigate the association of Snf1 with its activating kinases under more natural conditions, the elution profiles of these proteins were determined by size exclusion chromatography. Protein extracts were prepared from cells expressing epitope-tagged proteins from low-copy-number plasmids under control of their cognate promoters. Extracts were applied to a Superose 12 column and the presence of the Snf1-activating kinases and of Snf1 in fractions was determined by Western blotting (Figure 3). Snf1 elutes from the Superose 12 column with a major peak in fraction 21, with higher-molecular-mass species eluting in fractions 16–19. Size standards suggest that the peak in fraction 21 is consistent with the Snf1 kinase complex heterotrimer (156 kDa). The larger Snf1 complexes eluting in fraction 19 and earlier could be due to the association of a subset of the Snf1 molecules with Sak1. Indeed, when the same experiment is performed with cells lacking Sak1, the larger Snf1 complexes are much less abundant. The Sak1 protein elutes with a peak in fraction 19, with larger species eluting in fractions 16–18. Deletion of Snf1 greatly impacts on the elution profile of Sak1, shifting its elution to a smaller size with a peak in fractions 20 and 21, consistent with a monomer of Sak1 (132 kDa). The elution profile of Snf1 is shifted to smaller complexes when Sak1 is absent. Simlarly, the elution profile of Sak1 is shifted to smaller complexes when Snf1 is absent. Taken together with the abundance of Snf1 in the purified Sak1 preparation, we conclude that these proteins form a stable complex. The overall distribution of these proteins in the size exclusion chromatogram suggests that most of the Sak1 exists in a complex with Snf1 while only a fraction of the total Snf1 is associated with Sak1. This interpretation is consistent with the finding that purified Sak1 contains abundant Snf1 (Figure 1), while the purified Snf1 contains only trace amounts of Sak1 (Table 1 and [32]). Neither Elm1 nor Tos3 co-elute with Snf1. Both of these Snf1-activating kinases peak in fraction 24, a position that is consistent with the enzymes existing as monomers. Similarly, the TAP-purified Tos3 and Elm1 do not show any evidence for additional subunits present in stoichiometric amounts. Taken together, our results indicate that the Sak1 kinase forms a stable complex with Snf1 in vivo and that the Tos3 and Elm1 kinases, while capable of activating Snf1, do not stably associate with Snf1.
Figure 3. Gel filtration of Snf1 and the Snf1-activating kinases.
Protein extracts were size-fractionated on a 20 ml Superose 12 column (Pharmacia). The UV traces for a representative protein extract (closed circles) and molecular mass size standards (open squares) are shown. Proteins of interest were detected by Western blotting. The onput sample (ON) and fraction numbers of the samples are indicated. The relevant genotypes of the cells are shown on the right: wt, wild-type.
Hym1 is not associated with the Snf1-activating kinases
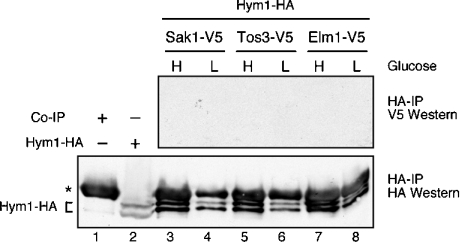
The AMPK-activating kinase in mammalian cells, LKB1, requires additional subunits STRAD and MO25 for full activity [17]. We sought to determine whether the yeast homologue of MO25, the Hym1 protein, associates with the Snf1-activating kinases. Hym1 has a predicted molecular mass of 46 kDa. Purified Sak1, Tos3 and Elm1 do not contain significant amounts of a 46 kDa protein (Figure 1), nor was Hym1 identified by peptide mass fingerprinting. To further investigate the association of Hym1 with the Snf1-activating kinases, we performed co-immunoprecipitation experiments using epitope-tagged versions of Hym1 and the Snf1-activating kinases. Hym1–HA was functional, since it complemented a hym1Δ strain. Hym1–HA was pulled down with anti-HA agarose beads, and the presence of V5-tagged Snf1-activating kinases was tested by Western blotting. We were completely unable to detect any association of Hym1 with any of the Snf1-activating kinases (Figure 4), Hym1 is expressed in these cells (lower panel) as are the Snf1-activating kinases (results not shown). Similar experiments performed with Sak1 and Snf1 readily detect association of these proteins [16]. Our data suggest that S. cerevisiae Hym1 does not associate with the Snf1-activating kinases and does not play role in the Snf1 activation pathway.
Figure 4. Hym1 does not associate with the Snf1-activating kinases.
Extracts were prepared from cells grown in high (H) or low (L) glucose. Cells were transformed with plasmids expressing Hym1–HA and one of the three V5-tagged Snf1-activating kinases. Proteins were immunoprecipitated (IP) with anti-HA beads and analysed with anti-V5 (upper panel) or anti-HA (lower panel) antibodies. A non-specific band from the immunoprecipitation is indicated (*) as is the mobility of a doublet of bands representing the Hym1–HA protein.
CKII is not required for glucose signalling
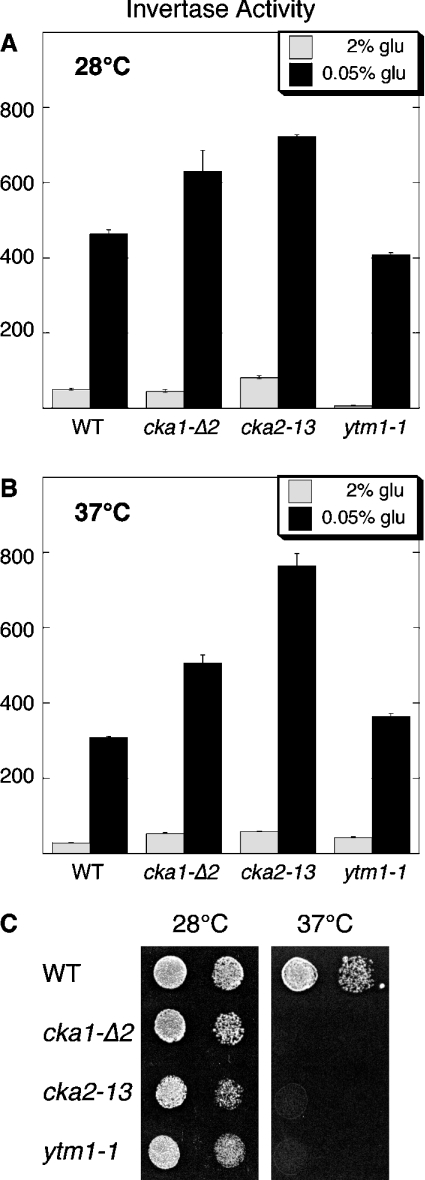
Earlier studies also identified CKII subunits associated with Snf4, the γ subunit of Snf1 kinase [36]. We found the catalytic subunits of CKII associated with purified Sak1 (Figure 1). These results led us to test whether CKII plays a role in glucose signalling. Deletion of the genes for both CKII catalytic subunits is lethal. Therefore we utilized strains that lack both CKA1 and CKA2 but are viable due to the presence of a plasmid expressing a temperature-sensitive allele of either CKA1 or CKA2 [28,29]. The strains were tested for Snf1 function by assessing their ability to induce expression of invertase in response to a low-glucose signal (Figure 5). When shifted to the non-permissive temperature, these strains lose CKII activity and ultimately their ability to grow (Figure 5C); however, they are still able to induce invertase expression (Figure 5B). An unrelated strain with a temperature-sensitive allele of YTM1 was also able to induce invertase. We conclude that CKII does not play a critical role in the Snf1-activation pathway. The idea that the co-purification of CKII with Sak1 and Snf4 is spurious is supported by the finding that CKII was found associated with 13 distinct TAP complexes [36].
Figure 5. CKII is not required for Snf1 signalling.
Invertase activity was determined in strains grown in high glucose, after they were shifted to low glucose for 2 h. Cells were either grown at the permissive temperature, 28 °C (A), or for 2 h at the non-permissive termperature, 37 °C, prior to shifting to low glucose (B). Relevant genotypes are indicated. (C) Growth phenotypes on rich media for the temperature-sensitive strains were measured by spot dilution assays at 28 °C and 37 °C.
Activity and specificity of the Snf1-activating kinases
Purified Snf1-activating kinases were assayed for their ability to phosphorylate a panel of recombinant proteins fused to GST and purified from E. coli. The substrates used were GST itself, GST–Mig1, GST–Snf1 kinase domain (Snf1–KD) and GST–Snf1 kinase domain with the activation loop threonine changed to alanine (T210A). Analysis of Sak1 kinase activity and specificity is complicated by the fact that the purified Sak1 fraction contains three kinases: Sak1, Snf1 and CKII. Therefore, we compared the activity of three preparations of Sak1. The Sak1 purified from wild-type cells is unable to phosphorylate GST but shows good activity toward GST–Mig1 and GST–Snf1 (Figure 6A, lanes 2–4). Mutation of the activation loop threonine of Snf1 completely abolishes all incorporation of radioactivity (lane 5), thereby confirming that the site of GST–Snf1 phosphorylation is limited to the activation loop threonine. In addition, the Sak1 polypeptide itself is phosphorylated in these reactions, as are the Snf1 and Gal83 proteins that co-purify with Sak1. When Sak1 was purified from snf1Δ10 cells, the GST–Mig1 substrate was no longer phosphorylated (lane 8). We conclude that it was the Snf1 kinase that associates with Sak1 that was responsible for the phosphorylation of the GST–Mig1. In addition, the incorporation of radioactivity into the Sak1 protein is greatly reduced by deletion of SNF1 (compare lanes 2 and 7), suggesting that the Snf1 kinase phosphorylates Sak1 in vitro. In order to distinguish the kinase activity of Sak1 from the CKII present in the preparation, a kinase dead Sak1 was purified and assayed (Sak1-mt–TAP). Phosphorylation of the GST–Snf1 substrate was abolished by the active site mutation in Sak1, demonstrating that Sak1, and not CKII, is responsible for the phosphorylation of the Snf1 activation loop. Finally, we detect very little kinase activity that can be attributed to CKII (lanes 11–15). Silver stained protein gels show that CKII is present at comparable levels in all three preparations (results not shown) as is the Sak1 protein as judged by Western blotting (Figure 6B).
Figure 6. Kinase activity assays of Snf1-activating kinases.
(A) Sak1 or kinase-dead Sak1 (Sak1-mt) was purified from SNF1 or snf1Δ10 strains and assayed for incorporation of [γ-32P]ATP using recombinant proteins as substrates. (B) Sak1 enzyme preparations were analysed by Western blotting using an antibody directed against the calmodulin portion of the TAP tag. Samples were the same as those assayed in (A). Sak1–TAP from SNF1 cells (lane 1), Sak1–TAP from snf1Δ10 cells (lane 2) and Sak1-mt–TAP from snf1Δ10 cells (lane 3). (C and D) Elm1 and Tos3 were purified from SNF1 and snf1Δ10 cells and assayed as above. Recombinant substrates were GST, GST–Mig1, GST–Snf1 kinase domain (KD), or GST–Snf1 kinase domain with the T210A mutation as indicated, or no added substrate (−). Proteins were resolved on 7.5% acrylamide (A and B) or SDS/10% PAGE gels (C and D). Incorporation of radioactivity was detected by autoradiography.
The activity and specificity of the Elm1 and Tos3 kinases were measured using the same substrates that were tested with Sak1. The purified Elm1 fraction phosphorylated GST–Mig1 and GST–Snf1, but not GST or the GST–Snf1 lacking the activation loop threonine (Figure 6C). When Elm1 was purified from cells lacking the SNF1 gene, the activity toward GST–Mig1 was slightly reduced and the pattern of labelled Mig1 bands was altered. This result suggests that trace amounts of Snf1 may in fact co-purify with Elm1. However, Elm1 itself appears to recognize Mig1 (lane 8), which complicates interpretation. Elm1 phosphorylation of GST–Snf1 is specific and limited to the activation loop since the T210A construct is not phosphorylated (lanes 5 and 10). We also detect significant incorporation of radioactivity into the Elm1 protein itself and this activity appears to be enhanced in the enzyme purified from snf1Δ10 cells. Measuring the activity of the Tos3 enzyme is complicated by the fact that the GST–Snf1 kinase domain substrate co-migrates with the Tos3 polypeptide. Despite this complication, it is apparent that Tos3 readily phosphorylates itself and GST–Snf1 (Figure 6D). The phosphorylation of Snf1 is specific since it is eliminated by the T210A mutation (lanes 5 and 10). We also detect incorporation into GST–Mig1 that is completely abrogated when Tos3 is purified from snf1Δ10 cells (Figure 6D, compare lanes 3 and 8). Thus, it seems likely that trace amounts of Snf1 are also present in the Tos3 preparation. The level of Snf1 in the Elm1 and Tos3 preparations was below the level of detection by silver staining (Figure 1) and Western blotting (Figure 2); neither Elm1 nor Tos3 was detected in a complex with Snf1 by size exclusion chromatography (Figure 3). Nonetheless, the phosphorylation of Mig1 appears to be a more sensitive assay, and our results are most consistent with trace amounts of Snf1 co-purifying with Elm1 and Tos3. We conclude that that all three Snf1-activating kinases are active and show a high degree of specificity for the Snf1 activation loop threonine. As previously noted for Tos3 and Elm1 [14], none of the Snf1-activating kinases require the presence of the γ or β subunits for the recognition of the Snf1 activation loop.
Catalytic activity of the Snf1 kinase domain
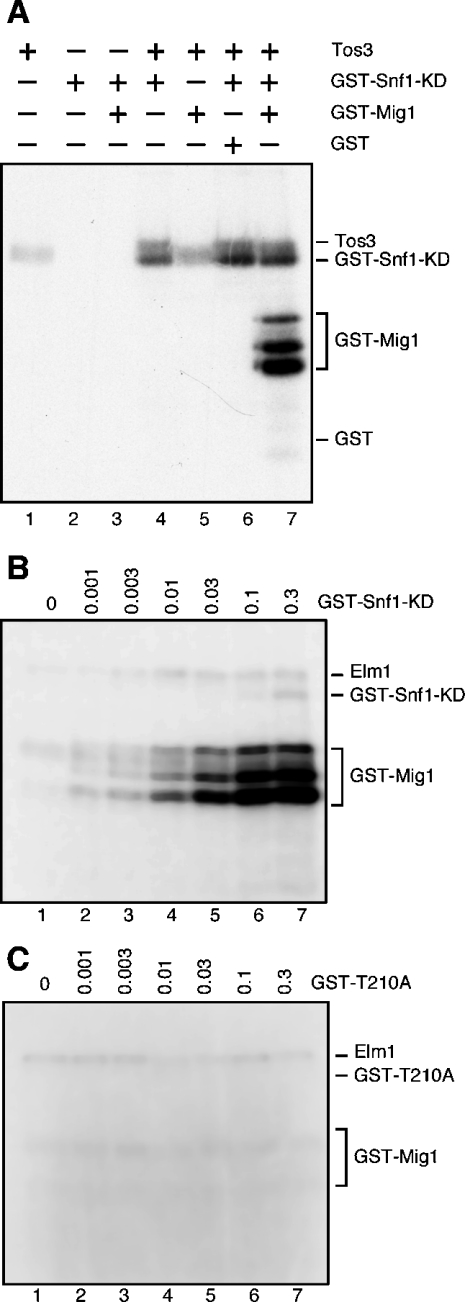
During the course of these studies, certain experiments suggested that the GST–Snf1 kinase domain was catalytically active. However, this idea was not easily reconciled with genetic data that demonstrated that the β and γ subunits were required for Snf1 function in vivo. In order to test directly whether the isolated Snf1 kinase domain was active, kinase reactions were conducted using the Tos3 enzyme purified from snf1Δ10 cells since this enzyme does not phosphorylate GST–Mig1 (Figure 6D, lane 8; Figure 7A, lane 5). The GST–Snf1 kinase domain purified from E. coli did not show any activity when incubated by itself (Figure 7A, lane 2) or with GST–Mig1 (lane 3), a result consistent with the requirement for activation loop phosphorylation. When Tos3 was incubated with GST–Snf1, incorporation into Snf1 could be detected, although the co-migration of Tos3 and GST–Snf1 partially obscures this result. Only when Tos3, GST–Snf1 kinase domain and GST–Mig1 were incubated together is robust incorporation into GST–Mig1 detected. The GST–Snf1 kinase domain shows substrate specificity, since the GST–Mig1 protein is recognized while GST alone is not (lanes 6 and 7). The simplest interpretation of these data is that Tos3 activates the GST–Snf1 kinase domain which phosphorylates the GST–Mig1. To further confirm the presence of catalytic activity in the GST–Snf1 kinase domain, the Elm1 enzyme, purified from snf1Δ10 cells, was incubated with GST–Mig1 and increasing concentrations of GST–Snf1 (Figure 7B). The Elm1 enzyme was present at one tenth of the level present in previous assays and at that concentration its ability to phosphorylate GST–Mig1 was limited (lane 1). As increasing amounts of GST–Snf1 were added to the reactions, the incorporation of radioactivity into the GST–Mig1 protein greatly increased. When the same reactions were performed in parallel with the GST–Snf1 containing the activation loop mutation (T210A), no increase in GST–Mig1 labelling was observed (Figure 7C). We conclude from these experiments that the Snf1 kinase domain purified from bacteria can be phosphorylated and activated by the Snf1-activating kinases. The Snf1 kinase domain by itself does not require the β or γ subunits for activity or for substrate definition, since the Mig1 protein is readily and specifically phosphorylated by the activated kinase domain. The idea that the Snf1 kinase domain can recognize substrates, without the need for β or γ subunits, may help to explain earlier studies showing that the Snf1 kinase domain was partially functional in vivo [40].
Figure 7. Recombinant Snf1 kinase domain is catalytically active in vitro.
(A) Kinase assays containing [γ-32P]ATP, purified Tos3, GST–Snf1 kinase domain (KD), GST-Mig1 or GST proteins as indicated. (B) GST–Snf1 kinase domain was titrated into reactions containing constant amounts of [γ-32P]ATP, purified Elm1 and GST–Mig1. (C) GST–Snf1 kinase domain with a T210A mutation was titrated into reactions as above. Incorporation of radioactivity was detected by autoradiography.
Concluding remarks
The study of the Snf1/AMPK family of protein kinases has greatly benefited from the fact that these enzymes are being studied both in mammalian cells and in a model genetic organism. Insights from one system have led to rapid progress in the other. In this study, we have found both parallels and differences between the yeast and mammalian Snf1/AMPK signalling pathways. First, both Snf1 and AMPK are activated by an upstream kinase. However, the mammalian LKB1 kinase requires additional subunits for full activity. The Snf1-activating kinases of S. cerevisiae do not require any additional subunits. The MO25 protein is required for full activation of LKB1, yet its yeast homologue, the Hym1 protein, does not associate with the Snf1-activating kinases. The finding that yeast encodes three Snf1-activating kinases leads to the question of why there are three. In this study, we show that only one of the Snf1-activating kinases, Sak1, forms a stable complex with Snf1. These findings, as well as studies reported elsewhere [33,41], suggest that Sak1 may well be the primary activator of Snf1 in vivo, with Tos3 and Elm1 acting only under certain stress conditions [42]. In mammalian cells, a similar story is developing whereby LKB1 appears to be the primary activator of AMPK, although CAMKKβ and possibly other kinases [17–19] activate AMPK in restricted cell types or under certain stress conditions.
Online Data
Acknowledgments
We thank Mr Scott Anderson and Dr John Yates III at the Scripps Institute Proteomic Mass Spectrometry Facility (Department of Cell Biology, The Scripps Research Institute, La Jolla, CA, U.S.A.), and Dr Mark Bier (Department of Chemistry), Dr Susan Dowd (Department of Chemistry) and Dr John Woolford (Department of Biological Sciences) at Carnegie Mellon University Center for Molecular Analysis (Carnegie Mellon University, Pittsburgh, PA, U.S.A.) for performing mass spectrometry. We thank Ms Anna Leech for construction of the pHYM1-HA plasmid as well as Dr Claiborne Glover (Department of Biochemistry and Molecular Biology, The University of Georgia, GA, U.S.A.) and John Woolford for providing yeast strains. This work was supported by grant GM46443 from National Institutes of Health.
References
- 1.Carlson M. Glucose repression in yeast. Curr. Opin. Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 2.Gancedo J. M. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aschenbach W. G., Sakamoto K., Goodyear L. J. 5′ adenosine monophosphate-activated protein kinase, metabolism and exercise. Sports Med. 2004;34:91–103. doi: 10.2165/00007256-200434020-00003. [DOI] [PubMed] [Google Scholar]
- 4.Fryer L. G., Parbu-Patel A., Carling D. The anti-diabetic drugs rosiglitazone and metformin stimulate AMP-activated protein kinase through distinct signalling pathways. J. Biol. Chem. 2002;277:25226–25232. doi: 10.1074/jbc.M202489200. [DOI] [PubMed] [Google Scholar]
- 5.Dyck J. R. B., Gao G., Widmer J., Stapleton D., Fernandez C. S., Kemp B. E., Witters L. A. Regulation of 5′-AMP-activated protein kinase activity by the noncatalytic β and γ subunits. J. Biol. Chem. 1996;271:17798–17803. doi: 10.1074/jbc.271.30.17798. [DOI] [PubMed] [Google Scholar]
- 6.Celenza J. L., Eng F. J., Carlson M. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the Snf4 protein with the Snf1 protein kinase. Mol. Cell. Biol. 1989;9:5045–5054. doi: 10.1128/mcb.9.11.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt M. C., McCartney R. R. β-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 2000;19:4936–4943. doi: 10.1093/emboj/19.18.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leech A., Nath N., McCartney R. R., Schmidt M. C. Isolation of mutations in the catalytic domain of the Snf1 kinase that render its activity independent of the Snf4 subunit. Eukaryot. Cell. 2003;2:265–273. doi: 10.1128/EC.2.2.265-273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang R., Carlson M. Glucose regulates protein interactions within the yeast Snf1 protein kinase complex. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 10.Scott J. W., Hawley S. A., Green K. A., Anis M., Stewart G., Scullion G. A., Norman D. G., Hardie D. G. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolen B., Taylor S., Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 12.Stein S. C., Woods A., Jones N. A., Davison M. D., Carling D. The regulation of AMP-activated protein kinase by phosphorylation. Biochem. J. 2000;345:437–443. [PMC free article] [PubMed] [Google Scholar]
- 13.McCartney R. R., Schmidt M. C. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 2001;276:36460–36466. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- 14.Hong S. P., Leiper F. C., Woods A., Carling D., Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutherland C. M., Hawley S. A., McCartney R. R., Leech A., Stark M. J., Schmidt M. C., Hardie D. G. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae Snf1 complex. Curr. Biol. 2003;13:1299–1305. doi: 10.1016/s0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- 16.Nath N., McCartney R. R., Schmidt M. C. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 2003;23:3909–3917. doi: 10.1128/MCB.23.11.3909-3917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawley S. A., Boudeau J., Reid J. L., Mustard K. J., Udd L., Makela T. P., Alessi D. R., Hardie D. G. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2003;2:28–44. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woods A., Johnstone S. R., Dickerson K., Leiper F. C., Fryer L. G., Neumann D., Schlattner U., Wallimann T., Carlson M., Carling D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 19.Hurley R. L., Anderson K. A., Franzone J. M., Kemp B. E., Means A. R., Witters L. A. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J. Biol. Chem. 2005;280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- 20.Hovland P. G., Tecklenberg M., Sclafani R. A. Overexpression of the protein kinase Pak1 suppresses yeast DNA polymerase mutations. Mol. Gen. Genet. 1997;256:45–53. doi: 10.1007/s004380050544. [DOI] [PubMed] [Google Scholar]
- 21.Milburn C. C., Boudeau J., Deak M., Alessi D. R., van Aalten D. M. Crystal structure of MO25 α in complex with the C terminus of the pseudo kinase STE20-related adaptor. Nat. Struct. Mol. Biol. 2004;11:193–200. doi: 10.1038/nsmb716. [DOI] [PubMed] [Google Scholar]
- 22.Karos M., Fischer R. Molecular characterization of HymA, an evolutionarily highly conserved and highly expressed protein of Aspergillus nidulans. Mol. Gen. Genet. 1999;260:510–521. doi: 10.1007/s004380050924. [DOI] [PubMed] [Google Scholar]
- 23.Bidlingmaier S., Weiss E. L., Seidel C., Drubin D. G., Snyder M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:2449–2462. doi: 10.1128/MCB.21.7.2449-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson B., Kurischko C., Horecka J., Mody M., Nair P., Pratt L., Zougman A., McBroom L. D., Hughes T. R., Boone C., Luca F. C. RAM: a conserved signalling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell. 2003;14:3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogomolnaya L. M., Pathak R., Guo J., Cham R., Aramayo R., Polymenis M. Hym1p affects cell cycle progression in Saccharomyces cerevisiae. Curr. Genet. 2004;46:183–192. doi: 10.1007/s00294-004-0527-3. [DOI] [PubMed] [Google Scholar]
- 26.Sreenivasan A., Kellogg D. The Elm1 kinase functions in a mitotic signalling network in budding yeast. Mol. Cell. Biol. 1999;19:7983–7994. doi: 10.1128/mcb.19.12.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouquin N., Barral Y., Courbeyrette R., Blondel M., Snyder M., Mann C. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J. Cell Sci. 2000;113:1435–1445. doi: 10.1242/jcs.113.8.1435. [DOI] [PubMed] [Google Scholar]
- 28.Hanna D. E., Rethinaswamy A., Glover C. V. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:25905–25914. doi: 10.1074/jbc.270.43.25905. [DOI] [PubMed] [Google Scholar]
- 29.Rethinaswamy A., Birnbaum M. J., Glover C. V. Temperature-sensitive mutations of the CKA1 gene reveal a role for casein kinase II in maintenance of cell polarity in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:5869–5877. doi: 10.1074/jbc.273.10.5869. [DOI] [PubMed] [Google Scholar]
- 30.Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 31.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 32.Nath N., McCartney R. R., Schmidt M. C. Purification and characterization of Snf1 kinase complexes containing a defined B subunit composition. J. Biol. Chem. 2002;277:50403–50408. doi: 10.1074/jbc.M207058200. [DOI] [PubMed] [Google Scholar]
- 33.McCartney R. R., Rubenstein E. M., Schmidt M. C. Snf1 kinase complexes with different β subunits display stress-dependent preferences for the three Snf1-activating kinases. Curr. Genet. 2005;47:335–344. doi: 10.1007/s00294-005-0576-2. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt M. C., McCartney R. R., Zhang X., Tillman T. S., Solimeo H., Wolfl S., Almonte C., Watkins S. C. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Washburn M. P., Wolters D., Yates J. R., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 36.Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature (London) 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 37.Sanz P., Alms G. R., Haystead T. A., Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 2000;20:1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dombek K. M., Kacherovsky N., Young E. T. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:39165–39174. doi: 10.1074/jbc.M400433200. [DOI] [PubMed] [Google Scholar]
- 39.Wilson W. A., Hawley S. A., Hardie D. G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 40.Ludin K., Jiang R., Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedbacker K., Hong S. P., Carlson M. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell. Biol. 2004;24:8255–8263. doi: 10.1128/MCB.24.18.8255-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim M. D., Hong S. P., Carlson M. Role of Tos3, a Snf1 protein kinase kinase, during growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Eukaryot. Cell. 2005;4:861–866. doi: 10.1128/EC.4.5.861-866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.