Abstract
Norwalk virus (NV) is the prototype strain of a group of human caliciviruses responsible for epidemic outbreaks of acute gastroenteritis. While these viruses do not grow in tissue culture cells or animal models, expression of the capsid protein in insect cells results in the self-assembly of recombinant NV virus-like particles (rNV VLPs) that are morphologically and antigenically similar to native NV. The X-ray structure of the rNV VLPs has revealed that the capsid protein folds into two principal domains: a shell (S) domain and a protruding (P) domain (B. V. V. Prasad, M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes, Science 286:287-290, 1999). To investigate the structural requirements for the assembly of rNV VLPs, we performed mutational analyses of the capsid protein. We examined the ability of 10 deletion mutants of the capsid protein to assemble into VLPs in insect cell cultures. Deletion of the N-terminal 20 residues, suggested by the X-ray structure to be involved in a switching mechanism during assembly, did not affect the ability of the mutant capsid protein to self-assemble into 38-nm VLPs with a T=3 icosahedral symmetry. Further deletions in the N-terminal region affected particle assembly. Deletions in the C-terminal regions of the P domain, involved in the interactions between the P and S domains, did not block the assembly process, but they affected the size and stability of the particles. Mutants carrying three internal deletion mutations in the P domain, involved in maintaining dimeric interactions, produced significantly larger 45-nm particles, albeit in low yields. The complete removal of the protruding domain resulted in the formation of smooth particles with a diameter that is slightly smaller than the 30-nm diameter expected from the rNV structure. These studies indicate that the shell domain of the NV capsid protein contains everything required to initiate the assembly of the capsid, whereas the entire protruding domain contributes to the increased stability of the capsid by adding intermolecular contacts between the dimeric subunits and may control the size of the capsid.
Norwalk virus (NV) is the prototype strain of a group of noncultivable human caliciviruses responsible for epidemic outbreaks of acute gastroenteritis (11, 14). NV is a small, icosahedral, single-stranded, positive-stranded RNA virus whose capsid is formed by multiple copies of a single major structural protein (7, 38). The NV genome is composed of three open reading frames (ORFs). The NV capsid protein is encoded by ORF2. Expression of the capsid protein in insect cells results in the self-assembly of recombinant NV empty virus-like particles (rNV VLPs) that are morphologically and antigenically similar to native NV (12, 27, 39).
The three-dimensional structure of rNV VLPs was first determined by electron cryomicroscopy and computer image-processing techniques to a resolution of 22 Å (40). That study revealed that the rNV capsid has a diameter of 38.0 nm and exhibits T=3 icosahedral symmetry, with a defined surface structure that resembles typical animal and human caliciviruses, in which cup-like depressions or hollows are evident at the three- and fivefold axes of symmetry. Each virus particle is composed of 180 molecules of the capsid protein, which form 90 arch-like capsomers at all the local and strict twofold axes surrounding the hollows.
A high-resolution (3.4 Å) structure of the NV capsid has also been determined by using X-ray crystallography techniques (38). The capsid protein folds into two principal domains, a shell (S) domain and a protruding (P) domain, which contains two subdomains, P1 and P2. The S domain is formed by the N-terminal 225 residues, and residues 50 to 225 fold into an eight-stranded antiparallel β sandwich structure, a folding commonly seen in many viral capsids (20). The P domains of neighboring subunits, related by the local and icosahedral twofold axes, interact with each other to form a prominent protrusion (38). The residues beyond 225 form the P1 and P2 domains.
Similar to the NV capsid structure, the shell and protruding domains are also found in tombusviruses such as tomato bushy stunt virus and turnip crinkle virus (21, 25). These plant viruses have an additional internal basic region, the R domain, absent in NV, formed by the 60 N-terminal residues in tomato bushy stunt virus and 40 N-terminal residues in turnip crinkle virus. The R domain in these plant viruses is involved in RNA binding (20, 25).
To form a T=3 icosahedral structure, the capsid protein has to adapt to three quasiequivalent positions, and the subunits at these positions are conventionally referred to as A, B, and C (20). In the modular structure of the NV capsid protein, the S domain is involved in the icosahedral contacts, and the P domains are involved in the dimeric contacts (38). NV and other caliciviruses are unique among the animal viruses because they possess a capsid composed of a single major structural protein. Consequently, all the functional entities required for calicivirus structural integrity, immunogenicity, and infectivity are encoded in one structural protein. Understanding the structure and functions of this unique viral capsid protein should facilitate the development of antiviral strategies for caliciviruses.
To identify functional domains of the NV capsid protein involved in assembly, we produced and analyzed the ability of a set of deletion mutants of the capsid protein to assemble into VLPs after expression in insect cell cultures. Our results demonstrate that most of the NV assembly determinants reside in the shell domain of the capsid protein.
MATERIALS AND METHODS
Cloning of NV full-length and mutant capsid genes into baculovirus transfer vectors.
The baculovirus transfer vectors chosen to express the NV capsid protein were the nonfusion plasmids pVL1393 and pVL1392, kindly provided by M. Summers (Texas A&M University, College Station, Tex.). In addition, pFastBAC (Gibco-BRL, Gaithersburg, Md.) was used for the construction of some of the mutants (Fig. 1). The primers used for this study, with the nucleotide (nt) positions, polarities, restriction enzyme sites, and sequences, are shown in Table 1.
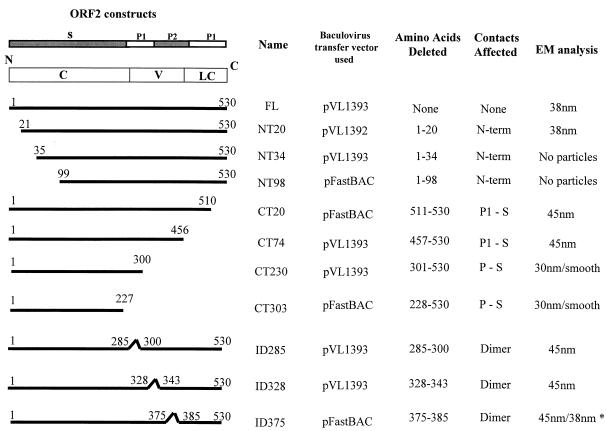
FIG. 1.
Schematic representation of the constructs used in this study to produce NV capsid mutants using the baculovirus expression system. The left column shows a schematic representation of the NV capsid protein and each mutant. The conserved (C), variable (V), and less-conserved (LC) regions based on sequence identity among Norwalk-like viruses are shown within the schematic of the full-length (FL) capsid protein. Above this schematic representation, the amino acid sequence that relates to each domain of the capsid structure is indicated. The figure also shows the transfer vector used for expression, which amino acids were deleted, and what contacts were predicted to be affected. The right column shows the size of particles observed by electron microscopy when the particles were purified over iodixanol or sucrose gradients. ∗ indicates that the 38-nm particles were observed only when particles were purified over iodixanol gradients. Mutant NT34 was incorrectly called NT35 when first described (47).
TABLE 1.
Names, sequences, nucleotide positions on ORF2, and polarities of primers used in this study
| Primer | Sequence (5′ to 3′) | Positionsa (nt) | Sense | Restriction enzyme site | Mutant(s) generated |
|---|---|---|---|---|---|
| NV110a | GCGGCGAGATCTAATTCGTAAATGATGATGGCG | 5349-5371 | Plus | BglII | CT303, ID285, ID328, CT74, CT230 |
| NV111a | GCGGCGAGATCTAATTGCACCAATTATGGCTT | 6957-6976 | Minus | BglII | NT20, ID285, ID328 |
| NV115a | GCGGCGAGATCTATGGTACCGGAGGTTAATGCTT | 5419-5439 | Plus | BglII | NT20 |
| NV102 | CGGGATCCATGAAGATGGCGTCGAATGAC | 5358-5378 | Plus | BamHI | NT34 |
| NV109 | CCCGGGGGATCCAATTGCACCAATTATGGCTT | 6957-6977 | Minus | BamHI | NT34 |
| NV-NT98-AUG | CCGGGGATCCGAAATGATGATGGCGTATAATGGTTGGGTTGG | 5655-5671 | Plus | BamHI | NT98 |
| NV-3′VCR-Xho | CCCGCTCGAGAACATCAAATTAAACCTAAT | 7635-7654 | Minus | XhoI | NT98 |
| NV-CT20 | GCGGCGCTCGAGAAATCTGGACACCCATGAAAC | 6857-6888 | Minus | XhoI | CT20 |
| NV112a | GCGCGAGATCTTCAAGCCTCACCTACAGTAGGG | 6707-6725 | Minus | BglII | CT74 |
| NV114a | GCGGCGAGATCTTCAGTTGATTACAGTGCCATT | 6240-6257 | Minus | BglII | CT230 |
| NV123 | GGCACCACCCCAGTTTCATTGCTTACTGAATTGGATGGCACA | 6189-6209/6257-6278 | Plus | None | ID285 |
| NV125 | TGTGCCATCCAATTCAGTAAGCAATGAAACTGGGGTGGTGCC | 6189-6209/6257-6278 | Minus | None | ID285 |
| NV124 | CCAGACCTCGGTGGTTGTGATGATGTAGACACCACCCCTGAC | 6318-6338/6387-6408 | Plus | None | ID328 |
| NV126 | GTCAGGGGTGGTGTCTACATCATCACAACCACCGAGGTCTGG | 6318-6338/6387-6408 | Minus | None | ID328 |
| NV132 | GGCCTGATCAATGATGATGGCGTCTAAGGAC | 5357-5378 | Plus | BclI | CT20, ID375 |
| NV133 | CCCGCTCGAGCATCGCCTATTATTTGCGAAT | 7567-7588 | Minus | XhoI | ID375 |
| NV134 | TATGTTGGTGTTCTTAGCTCCCAAGTTGACCTTTGGAAGATC | 6462-6478/6508-6533 | Plus | None | ID375 |
| NV135 | GATCTTCCAAAGGTCAACTTGGGACCAGCTAAGAACACCAACATA | 6462-6482/6510-6533 | Minus | None | ID375 |
| NV-CT303 | CCGCTCGAGAACATCAAATTAAACCTAATTAAACCTAATTAAATGAA TATGATGCCCACATTTCATATTA CAACAATAGGTTTTCTGCTCCAC CGTAGGAG | 6319-6341/7589-7654 | Minus | XhoI | CT303 |
NV accession number M87661 in GenBank.
Recombinant baculovirus transfer plasmids containing the full-length gene of ORF2 that codes for the NV capsid protein and N-terminally and C-terminally truncated mutants were generated by inserting PCR-amplified fragments flanked by BglII or BamHI restriction enzyme sites into those restriction enzyme sites in the multiple cloning site of the transfer vector pVL1393 or pVL1392, as indicated in Table 1 and Fig. 1. The templates used for PCR were plasmids pGNV2 and pG4145, which contain the region of NV ORF2 (26, 27). The generation of recombinant baculoviruses expressing the full-length capsid protein and the mutants NT34, CT74, and CT230 was described previously (47).
The N-terminally truncated gene NT20, which spans from nt 5419 to 6976, was amplified using primers NV-115a and NV-111a. The PCR products were phenol-chloroform extracted, ethanol precipitated, and suspended in Milli-Q water. The fragments were digested with BglII (New England Biolabs, Beverly, Mass.) for 2 h at 37°C. The digested fragments were purified by agarose gel electrophoresis and QIAquick extraction (Qiagen Inc., Chatsworth, Calif.). The digested PCR products were ligated to the pVL1393 transfer vector previously linearized with BglII, purified by agarose gel electrophoresis, treated with calf intestinal alkaline phosphatase, phenol-chloroform extracted, and ethanol precipitated. A similar procedure was used for the generation of the NT98 mutant, which spans from nt 5655 to 7454.
The primers used for the generation of the truncated NT98 gene were NV-NT98-AUG and NV-3′NCR-XhoI. The NT98 PCR-generated fragment was ligated into a previously BamHI- and XhoI-digested pFastBAC. The 3′-terminally truncated gene CT20, which spans from nt 5357 to 6888, was amplified using primers NV-132 and NV-CT20. The PCR product was digested using BclI and XhoI and ligated into a pFastBAC plasmid that was digested with BamHI and XhoI. The same procedure was followed for generation of the CT303 mutant, which spans from nt 5349 to 6341. The primers used to generate the truncated CT303 PCR product were NV-CT303 and the forward primer 110a (47).
The recombinant transfer plasmids containing the internal deletion mutants (ID) of the NV capsid protein were generated by splicing by overlap extension PCR (24), using two sequential PCRs. For the ID285 mutant (containing nt 5349 to 6209 and 6258 to 6976), two separate primary PCRs were performed, using the primer pairs 123/111a and 125/110a (47). Primers NV123 and NV125 are internal primers, complementary to each other, that contain in continuous sequence the regions flanking the one to be deleted. The two PCR products generated by these primary PCRs overlap in the region flanking the deletion. After gel purification (QIAquick gel extraction kit) of the PCR products, the DNA fragments were denatured and annealed together in a secondary PCR in which the extended template was amplified using the external primers NV110a and NV111a.
To make the ID328 mutant (spanning nt 5349 to 6338 and 6387 to 6976), internal primers for the first PCR were NV124 and NV126, while external primers were the same (NV110a and NV111a) as the ones used to generate the ID285 mutant. The PCR products from the secondary PCRs were purified by agarose gel electrophoresis, followed by extraction with the QIAquick gel extraction kit, digestion with BglII, and ligation to the BglII-linearized, calf intestine alkaline phosphatase-treated pVL1393 transfer vector. The same procedure was followed to produce ID375 (spanning nt 5349 to 6482 and 6510 to 7654).
The deletion was generated in two steps. First, two fragments of the ORF2 gene that overlap in the region that flanks the deletion were generated by PCR using primers NV135 and NV132 or NV134 and NV133. These two fragments were used as templates in a second PCR using only the external primers NV133 and NV132. The PCR product was digested using BclI and XhoI for cloning into the pFastBAC plasmid that was previously digested with BamHI and XhoI. The sequence in all the recombinant plasmids was confirmed to be correct by sequencing using the T7 Sequenase version 2.0 DNA sequencing kit (USB, Cleveland, Ohio).
Generation of recombinant baculoviruses containing full-length and mutant capsid protein.
Baculovirus recombinants containing the full-length or mutant forms of NV ORF2 in a pVL vector (pVL1392 or pVL1393, Table 1) were obtained by homologous recombination between the respective transfer vector and linearized baculovirus DNA containing a lethal deletion (Baculogold; PharMingen, San Diego, Calif.) that is complemented by sequences in the transfer vector. Spodoptera frugiperda Sf9 cells were cotransfected by liposome-mediated transfection with the recombinant baculovirus transfer vector plasmid DNA and the Baculogold DNA as described previously (47).
In the case of mutants CT20, NT98, CT303, and ID375, which were cloned into pFastBAC, the generation of recombinant baculoviruses was performed using the Bac-to-Bac baculovirus expression system (Gibco-BRL, Gaithersburg, Md.) according to the manufacturer's instructions. Four purified clones for each recombinant baculovirus were confirmed for the presence of specific NV DNA sequences by PCR using the same primers used for the cloning procedure. The recombinant baculovirus stocks were diluted 10−2 to avoid inhibition of the PCR by the cell medium, and 1 μl of the dilution was loaded in a 100-μl PCR.
Expression in insect cells.
Expression of full-length and mutant capsid proteins was evaluated in cell lysates as well as from the medium of Sf9 cultures. The Sf9 cells grown in monolayers were infected at a multiplicity of infection (MOI) of 10 with the appropriate recombinant baculovirus, as indicated above, and the cell lysates were collected 34 h postinfection (hpi). The expression of the recombinant NV capsid proteins in the medium was analyzed by infection of a 200-ml insect cell culture at a density of 3 × 106 cells/ml with an MOI of 5. The supernatant was collected 5 days postinfection (dpi). Then, the supernatant was clarified by sedimentation for 15 min at 1,500 rpm (Beckman JS-4.2 rotor) followed by sedimentation for 30 min at 12,000 rpm (Beckman JA-14 rotor). Next, the culture medium was examined after concentration by sedimentation for 2 h at 26,000 rpm in an SW28 rotor (Beckman).
Metabolically radiolabeled VLPs were prepared by incubating the monolayers of infected Sf9 cells in methionine-free Grace's insect medium at 28 hpi for 30 min, followed by addition of 25 to 30 μCi of [35S]methionine (Trans35S-label; ICN, Irvine, Calif.) per ml. After 4 h of labeling, cell monolayers were scraped, pelleted, and lysed in lysis buffer (100 mM phosphate buffer [pH 6], 150 mM NaCl, 1% NP-40, 1.5 mM MgCl2, 0.5 μg/ml of each of the protease inhibitors aprotinin and leupeptin [Sigma, St. Louis, Mo.]). Cell lysates were kept on ice for 20 min and then pelleted at 10,000 × g for 10 min at 4°C. The supernatant (S10 fraction) was used immediately for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
For the analysis of radiolabeled capsid proteins in the medium, cultures received 50 μg of cold methionine (Gibco-BRL) per ml at 32 hpi and protease inhibitors daily (0.5 μg of aprotinin [Sigma], 0.7 μg of pepstatin [Calbiochem, San Diego, Calif.], and 0.5 μg of leupeptin [Sigma] per ml) and were harvested at 6 to 7 dpi. The culture medium was examined after concentration by sedimentation for 2 h at 26,000 rpm in an SW28 rotor (Beckman).
Pulse-chase labeling of baculovirus-infected insect cells.
Pulse-chase experiments to test the protein stability of some of the mutants were performed following a protocol described for recombinant proteins expressed in baculovirus (35, 36). Briefly, 1.2 × 107 Sf9 cells growing in suspension were infected in a 15-ml tube with an MOI of 20. At 36 hpi, the medium was removed and the cells were suspended in methionine-free Grace's medium. The cells were starved for 1 h before the medium was changed and 200 μCi of [35S]-methionine was added to the mix. After 15 min, the medium was replaced with fresh methionine-free Grace's medium with a 20-fold excess of cold methionine and 100 μg of cycloheximide per ml. Aliquots (1 ml) equivalent to 3 × 106 cells were collected every 12 h postlabeling for 48 h. The cell pellet was suspended in loading buffer and analyzed by SDS-12% PAGE.
Antigen ELISAs.
To quantitate the antigen titer of the different mutant proteins, an antigen enzyme-linked immunoassay (ELISA) specific for NV capsid protein was used (27). Briefly, 96-well polyvinyl chloride microtiter plates were coated overnight at room temperature with 100 μl of guinea pig hyperimmune serum (27) per well prepared against rNV diluted 1:5,000 in phosphate-buffered saline (PBS). Guinea pig preimmune serum and uncoated wells were used to control for background binding. The plates were blocked with 5% Blotto (Carnation nonfat dry milk in 0.01 M PBS) for 2 h at 37°C and then washed five times with 0.05% (vol/vol) Tween 20 (Sigma Chemical Co., St. Louis, Mo.) in PBS (PBS-T).
The antigen was prepared as a 400-μl cell lysate of 3 × 106 insect cells infected with the different mutants or the full-length rNV capsid protein recombinant baculoviruses. The antigen was serially diluted in 10% Blotto, added to the coated and blocked plates (100 μl), and incubated at 37°C for 2 h. Purified rNV was added as a positive antigen control at concentrations ranging from 62 to 0.5 ng. The plates were washed five times with PBS-T and reacted with rabbit anti-rNV hyperimmune serum (27) at a final dilution of 1:5,000 in 2% Blotto. After 2 h of incubation at 37°C, the plates were again washed five times with PBS-T and reacted with 100 μl of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (HyClone laboratories, Logan, Utah) diluted 1:5,000 in 2% Blotto and containing 2% normal guinea pig serum. The plates were incubated for 2 h at 37°C, and following a final five washes, TMB-peroxidase substrate (Kirkegaard and Perry, Gaithersburg, Md.) was added, and the color was allowed to develop for 10 min at room temperature. The reaction was stopped by addition of 1 M phosphoric acid, and the optical density at 450 nm was determined with a Titerket Multiskan plate reader (Flow laboratories, Inc., McLean, Va.).
SDS-PAGE and immunoblotting analysis.
Analysis of proteins by SDS-PAGE was done according to the method of Laemmli with modifications (28). Polyacrylamide gels with 12% acrylamide in the separating gel and 4% in the stacking gel were used. Samples to be analyzed were boiled for 2 min in sample buffer containing 1% SDS, 10% 2-mercaptoethanol, 0.05 M Tris-HCl (pH 6.8), 10% glycerol, and 0.0025% phenol red.
The proteins separated by SDS-PAGE were transferred onto a nitrocellulose membrane (Hybond-C; Amersham, Life Sciences, Buckinghamshire, England) as described previously (47). The full-length or mutant capsid protein was detected using a rabbit hyperimmune anti-rNV serum (27) at a dilution of 1:5,000 in PBS. All the secondary antibodies used were conjugated to horseradish peroxidase (HyClone Laboratories Inc., Logan, Utah). The membranes were developed colorimetrically and by chemiluminescence using the ECL detection reagent (Amersham, Life Science, Buckinghamshire, England) following the manufacturer's protocol.
Rate zonal centrifugation in sucrose gradients.
Sucrose gradients were used to separate and analyze the physical state of the capsid protein. A six-step gradient of 10 to 60% (wt/vol) sucrose was prepared in PBS (pH 7.2) or in phosphate buffer (0.2 M NaH2PO4, 0.2 M Na2HPO4·7H2O, pH 6.0) and used to separate soluble capsid protein from protein assembled in VLPs. The gradients were prepared in 2-ml tubes by loading 0.3 ml/step and incubated for 2 h at 4°C to allow diffusion into a continuous gradient. The sample (0.1 ml) was loaded on the gradients and centrifuged for 1 to 1.5 h at 35,000 rpm (Beckman TLS-55 rotor) at 4°C. Alternatively, the gradients were prepared in polyallomer (1 by 3.5 in. [ca. 2.5 by 9 cm]) centrifuge tubes (Seton Scientific, Los Gatos, Calif.), loading 5 ml/step and using a sample volume of approximately 0.5 ml. These gradients were centrifuged for 1.5 h at 25,000 rpm (Beckman SW28 rotor). All gradients were fractionated by bottom puncture, and aliquots of each fraction were analyzed by Western blotting as described above.
Centrifugation in iodixanol gradients.
As an alternative to sucrose gradients, iodixanol was used for the separation of soluble protein from VLPs. This density gradient medium forms self-generating iso-osmotic gradients (31). Opti-prep, a 60% solution of iodixanol in water (Gibco-BRL, Gaithersburg, Md.) was mixed with the sample and phosphate buffer in the right proportions to obtain a 30% iodixanol solution. These gradients were centrifuged for 3 h at 63,000 rpm in a Beckman NVT-65.2 rotor. All gradients were fractionated by bottom puncture, and aliquots of each fraction were analyzed by Western blotting as described above.
Isopycnic separations in self-generating CsCl gradients.
In some experiments, particles were purified in CsCl gradients. The pellets obtained after centrifugation of the supernatant of infected cells were suspended in a solution of CsCl (1.362 g/cm3 in phosphate buffer) and centrifuged for 24 h at 35,000 rpm in a Beckman SW50.1 rotor. The gradients were fractionated by bottom puncture, and the rNV peak fractions, determined by Western blot, were pooled. The CsCl was diluted by the addition of phosphate buffer, and particles were sedimented for 2 h at 26,000 rpm in a Beckman SW28 rotor.
Electron microscopy.
Negative-stain electron microscopy was used to analyze the presence, integrity, and morphology of the rNV VLPs. The stain used throughout this work was 1% ammonium molybdate prepared in water at pH 5. A 5-μl drop of the sample was applied to a parafilm sheet, and a carbon-coated and glow-discharged 400-mesh electron microscopy grid (Ted Pella, Inc, Redding, Calif.) was set on the drop for 10 min at room temperature in a humid chamber. Excess liquid was absorbed from the side of the grid with a filter paper; the grid was washed for a few seconds on a drop of water if necessary (if sucrose or high salts were present in the sample), and finally the grid was placed on a drop of the ammonium molybdate stain for 15 to 30 s and excess liquid was blotted as before, followed by air drying for 5 min. After the excess fluid had been blotted and the grid had been air dried, the grid was examined in a Phillips CM10 electron microscope operating at 80 kV.
Electron cryomicroscopy.
Mutant NV VLPs for electron cryomicroscopy were embedded in a thin layer of vitreous ice on holey carbon films using standard procedures (5, 29). Frozen hydrated specimens were imaged in a Jeol 1200 electron cryomicroscope, using a 100-kV electron beam with a dose of ≈10 e−/Å2 at a magnification of 40,000×. For each specimen area, focal pairs were recorded, the first one at a defocus of ≈1 μm and the subsequent one at a defocus of ≈2 μm. Images were recorded on Kodak SO-163 electron films with a 1-s exposure time. Micrographs were developed for 12 min in a Kodak D-19 developer at 21°C and fixed for 10 min in Kodak fixer.
Three-dimensional structural analysis.
Micrographs were chosen for structural analysis based on the criteria of ice quality, particle concentration, and optimum defocus. Images were digitized on a Zeiss SCA1 microdensitometer (Carl Zeiss, Inc., Englewood, Colo.) using a 7-μm step size. Pixels were averaged to give a 14-μm step size, which corresponded to ≈3.5 Å per pixel in the object. Particles from both images of digitized focal pairs were boxed into individual particle images and masked with a suitable radius. While the closer-to-focus images were used for the three-dimensional reconstruction, the farther-from-focus images were used to confirm the orientations of the particles. Orientations of the particles were determined using common lines (1) and refined using the cross-common lines procedures as described previously (30). The three-dimensional reconstructions were computed using cylindrical expansion methods (1).
The distributions of the particle orientations were assessed by plotting them on the icosahedral asymmetric unit represented in terms of φ and θ and estimating mean inverse eigenvalues (2). The defocus value of each micrograph was estimated from the positions of the contrast transfer function rings in the sum of individual particle image Fourier transforms. The final reconstructions were computed to a resolution that contained information within the first zero of the contrast transfer function. Corrections for contrast transfer function were carried out using a Wiener filter, assuming an amplitude contrast factor of 0.14 and a signal-to-noise ratio of 0.3, as described previously (50). Effective resolution of each reconstruction was estimated using Fourier cross-correlation coefficients and using equation 3 from van Heel et al. (46), and phase residuals were estimated using equation 6 between independent reconstructions of the same specimen or between two independent reconstructions obtained by randomly dividing the data from a single micrograph into two sets. The reconstructions were visualized using IRIS Explorer (NAG, Inc.) with several customized modules (30).
RESULTS
Design of capsid protein deletion mutants and rationale.
Based mainly on structural information, 10 NV capsid protein deletion mutants were designed (Fig. 1). Some of these deletions were in the N-terminal S domain, and others were in the P domain. The N-terminal domain of the NV capsid protein is located toward the interior of the particle (38), and its amino acid sequence is highly conserved among Norwalk-like viruses (15, 34). In the crystal structure of the NV capsid, the N-terminal 28 residues of the A and C subunits are disordered. However, in the B subunit, only the first 9 N-terminal residues are disordered. The residues from 9 to 15 in this subunit are involved in hydrogen-bonding interactions with the S domain of the neighboring C subunit around the threefold axis (38). To better understand the role of this region in the assembly of the capsid, we designed three N-terminal deletions of 20, 34, and 98 amino acids (NT20, NT34, and NT98, respectively).
The P domain is the most exposed part of the protein in the particle (38), and sequence comparisons between capsid proteins of Norwalk-like viruses indicate that this domain contains regions that are most variable (15, 34). Interactions between the P domains of the opposing subunits form the prominent protrusions, and there are several contact regions between them, which may be relevant for the assembly or stability of the capsid (38). In addition, particularly in the A and B subunits, the C-terminal residues interact with the S domain residues by forming hydrogen bonds.
Do these interactions play a role in capsid assembly or stability? In order to map the regions in the protruding domain that may be involved in the assembly of the particle, we designed three deletion mutants lacking 20, 75, or 230 amino acids at the C terminus (CT20, CT74, and CT230, respectively), which are in the P1 subdomain, and three internal deletions of 10 to 15 amino acids, each located in the P2 domain (ID285, ID328, and ID375) (Fig. 1). The CT20 and CT74 mutants were constructed to examine the effect of interfering with the interactions between the P and S domains on capsid assembly. The CT230 deletion mutant was designed to analyze the function of four dimeric interaction regions between amino acids 441 and 448, 426 and 428, 374 and 376, and 332 and 340 in the assembly of the NV capsid. Other mutations in the P domain, such as ID285, ID328, and ID375, were constructed to evaluate the role of individual dimeric contacts in the assembly.
Finally, we completed the set of mutants by designing a deletion lacking the entire protruding domain. This CT303 deletion consists only of the shell domain of the capsid protein. The shell domain is involved in forming the icosahedral shell and has a classical eight-stranded antiparallel β sandwich motif, a common fold seen in many viral capsid proteins (20, 38). The shell domain is the most conserved region of the capsid, an indication of the possible importance of this region for the integrity of the capsid.
Comparative analysis of expression levels of full-length and mutant forms of NV capsid protein in Sf9 cells.
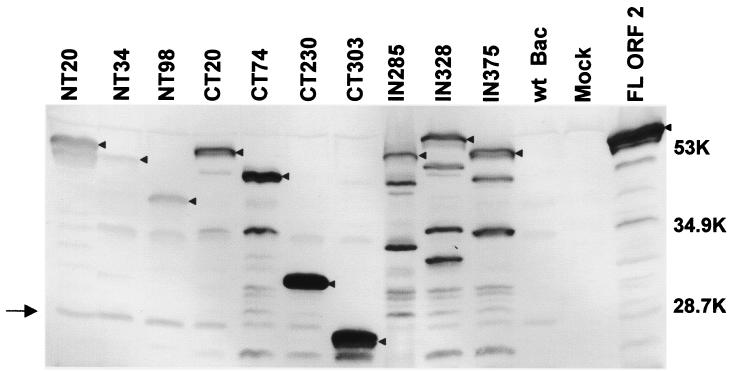
To ensure that the inability of a particular mutant to form particles was not because of low protein expression levels, we first analyzed and compared the levels of expression of the mutant proteins with that of the full-length capsid protein both in cell lysates and in supernatants of infected cells. In such comparative analyses with cell lysates, the same number (3 × 106) of confluent Sf9 cells were infected with the different recombinant baculoviruses at the same MOI of 10. The infected cells were lysed at 36 hpi in the same volume of PBS by freezing and thawing. The full-length and truncated capsid proteins were detected using rabbit hyperimmune antiserum to rNV VLPs by Western blotting (Fig. 2). The truncated capsid proteins were detected at the expected different molecular weights by Western blotting (Fig. 2).
FIG. 2.
Expression of NV mutant capsid proteins. Infected cell lysates (3 × 106 cells) were harvested at 36 hpi in 300 μl of 5× SDS-PAGE sample buffer (3 parts disruption buffer [10% SDS, 50% 2-mercaptoethanol, 5 M urea, 1:1:1]; 2 parts F/2 [1:1; stacking gel buffer {0.5 M Tris-HCl, 0.46% TEMED, pH 6.6 to 6.8}), 80% glycerol; 0.02 g of phenol red]) and analyzed by SDS-12% PAGE and Western blotting. In order to compare the relative levels of protein expression of the mutant proteins, the same volume of lysate was loaded for each mutant. Western blotting was performed using a rabbit hyperimmune serum to rNV VLPs (27). At the dilution used, this antiserum also recognizes one nonspecific band in the wild-type-baculovirus-infected cell lysates at around 20 kDa (→). Although protease inhibitors were added to the infected cells, some degradation products were observed for all the constructs. Arrowheads indicate the bands that correspond to the uncleaved mutant or full-length capsid proteins. The levels of expression of each mutant were quantitated by ELISA (see text). Sizes are shown in kilodaltons.
The levels of capsid protein expression were quantitated by using an ELISA and a standard curve of known amounts of full-length capsid protein (data not shown). Comparison of the levels of expressed protein in the cell lysates from 3 × 106 cells at 36 hpi, prepared as described above for the Western blot, showed the highest expression level for the full-length capsid protein (509 μg) followed by the mutant NT20 (263 μg). The other mutants showed intermediate levels of expression (0.3 to 2.6 μg). The relative yields of purified VLPs for the full-length, NT20, and CT303 proteins (20 mg, 10 mg, and 1 mg, respectively) from the same number of infected cells (6 × 108) correlated with the values observed for the ELISA analysis. Expression of the NT34 mutant protein was the lowest, almost undetectable (0.01 μg).
To attempt to understand if the reason for the reduced yields of the NT34 mutant was because of lower stability of the protein, we performed pulse-chase experiments in insect cells infected with the recombinant baculovirus expressing the mutated protein. The cells were pulsed at 36 hpi with [35S]methionine for 15 min, and an excess of unlabeled methionine was added during the chase period, when samples were collected every 12 h for 48 h. Analysis of these samples showed that the stability of the NT34 protein was comparable to that of the full-length capsid protein, but the expression levels were very low (data not shown).
The expression levels of the mutant proteins were also compared in the medium of the infected Sf9 cell cultures 6 days postinfection. In these experiments, Sf9 cells were infected with individual recombinant baculoviruses at an MOI of 5. Protease inhibitors were added daily to the cultures to minimize protein degradation. The culture medium was examined after concentration by sedimentation for 2 h at 26,000 rpm in an SW28 rotor (Beckman), which were the same conditions used to pellet full-length rNV VLPs. Western blot analysis showed that all mutant capsid proteins were present in the pelleted medium, suggesting that particles or aggregates had formed (data not shown).
Biophysical analysis of full-length and mutant proteins by sucrose and iodixanol gradient sedimentation.
To determine if the different capsid protein mutants were in soluble or VLP form, we analyzed the proteins by sucrose and iodixanol gradients. We used two different gradient purification procedures to minimize the possibility that the lack of detecting particle assembly with certain mutants was due to the purification protocol. Sucrose gradients have been used successfully before for the isolation of rNV VLPs (27, 47). However, the high osmolarity and viscosity of sucrose solutions are known to affect weak electrostatic interactions between proteins (31). Iodixanol gradients are reported to preserve protein-protein interactions better than concentrated high-osmotic-strength sucrose solutions (3, 31). Iodixanol gradients have been used with excellent results to purify viral particles whose integrity is sensitive to other separation systems (22, 31, 51).
The purification procedure using iodixanol gradients was standardized by using full-length rNV VLPs. The supernatants of Sf9 cells expressing full-length capsid protein were analyzed at 5 dpi by sedimentation on 30% iodixanol solution followed by fractionation and analysis of the proteins in each fraction by SDS-10% PAGE and Western blotting. The full-length capsid protein was detected in fractions 2 to 6, with a peak in fraction 3, corresponding to the top half of a 30% iodixanol gradient (Fig. 3A). Analysis of the peak fraction with the highest protein concentration by electron microscopy showed the presence of rNV VLPs (data not shown). With each of the mutant proteins, both sucrose and iodixanol gradient purification procedures were used. Both purification schemes worked well for separating full-length rNV VLPs; however, with some of the mutants, particles were only detected when purified over iodixanol gradients.
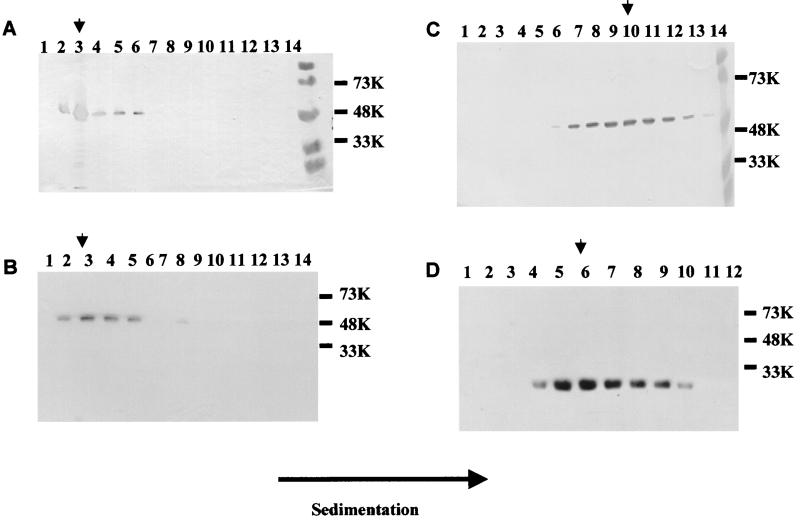
FIG. 3.
Analysis of the presence of capsid protein in fractions of iodixanol and sucrose gradients. The supernatant of infected cells (3 × 106 cells/ml in 200 ml) was sedimented and separated through sucrose or iodixanol gradients. The figure shows Western blotting analysis of fractions from iodixanol (A and B) and sucrose (C and D) gradients. The fractions shown to contain capsid protein were pooled, and the sucrose or iodixanol was removed by dilution in phosphate buffer and sedimentation. (A) Full-length NV capsid, (B) NT20, (C) ID375, and (D) CT230. Sizes are shown in kilodaltons. The arrowhead shows the peak fraction in each gradient.
As for the full-length capsid protein, all the mutants were purified from the medium of infected Sf9 cells. The NT20 VLPs sedimented in sucrose gradients in the central region of the gradient, similar to the full-length capsid protein (data not shown). Over iodixanol gradients, the mutant protein NT20 behaved similarly to the full-length capsid protein and was localized in the top half of the gradient at fractions 3 and 4 (Fig. 3B), suggesting that, as for the full-length capsid protein, NT20 was assembled into VLPs. Although the NT20 protein sedimented similarly to rNV VLPs, the NT98 or NT34 protein sedimented in the top regions of sucrose (fractions 2 and 3) and iodixanol (fraction 1) gradients, suggesting that the protein was in a soluble form (data not shown).
We next analyzed the two C-terminal deletion mutants, CT20 and CT74, and the three internal deletion mutants, ID285, ID328, and ID375, generated in this study. When the mutant proteins obtained from the sedimented supernatants of the infected insect cells were purified over 10 to 60% sucrose gradients, the CT20 and CT74 proteins were found in the middle fractions of the sucrose gradients, suggesting the presence of particles. However, the CT20 and CT74 protein peaks of the gradients were slightly shifted toward the bottom fractions (fractions 9, 10, and 11) with respect to the full-length NV capsid protein (fractions 7 and 8) (data not shown). The faster sedimentation of the CT20 and CT74 proteins suggested that the particles might be larger than full-length NV VLPs. Similarly, the ID285, ID328, and ID375 proteins were also found shifted toward the bottom of the sucrose gradient with respect to the full-length NV capsid protein (fractions 9, 10, and 11) (Fig. 3C). In addition, these mutant proteins (CT20, CT74, ID285, ID328, and ID375) were analyzed by iodixanol gradients and were found at the top half of the gradients, suggesting the presence of particles. The density of these CT20, CT74, and internal deletion particles was determined in sucrose gradients to be >1.4 g/cm3, which is higher than that of the full-length NV VLPs (1.31 g/cm3 in sucrose).
The CT230 protein was located in the middle fractions (fractions 5 and 6) of the sucrose gradient (Fig. 3D) and the top half of the fractions of an iodixanol gradient, suggesting the presence of particles. The CT230 particles were not stable in CsCl. The CT303 VLPs migrated similarly to the CT230 particles when purified over sucrose and iodixanol gradients, but they were stable in CsCl. The density of the CT303 particles in CsCl gradients was 1.32 g/cm3, the same as for full-length NV VLPs.
Electron microscopy and cryoelectron microscopy analysis of the deletion mutants of NV capsid protein.
Next, we analyzed the iodixanol and sucrose gradient peak fractions that contained the capsid protein by electron microscopy to confirm the presence or absence of particles and the morphological characteristics of the particles. Whenever the yield of the particles was sufficiently high, some of the particles were also examined by cryoelectron microscopy.
(i) N-terminal deletions.
Negative-stain electron microscopy analysis of the peak fractions from sucrose and iodixanol gradients of the NT20 mutant showed that the protein was assembled into VLPs. These VLPs were similar in size and overall surface structure to the full-length capsid particles (Fig. 4A and Fig. 5A, B, D, and 5E). The yield of the NT20 particles was comparable to that of the full-length capsid particles and permitted cryoelectron microscopic analysis. The cryoimages of NT20 particles showed that these particles were morphologically identical to the full-length VLPs, with a diameter of ≈380 Å.
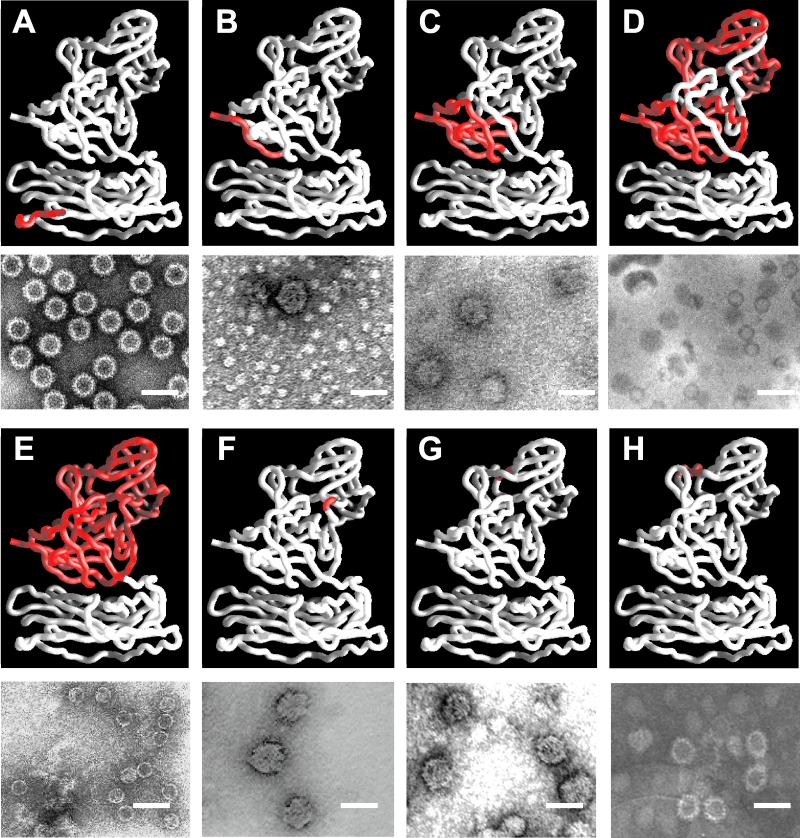
FIG. 4.
Electron microscopic analysis of NV capsid mutants. For each mutant, the top panel shows the region mutated (red) on a rope representation (white) of the full-length NV capsid protein crystal structure (38), and the lower panel shows an electron micrograph of the different mutant particles purified from undiluted supernatant material from the Sf9 cell cultures. The electron micrographs show a representative area of grids prepared with gradient fractions. Ammonium molybdate (1%) was used for staining. (A) NT20 forms VLPs that resemble full-length capsid protein VLPs. (B and C) CT20 and CT74 form VLPs that are 45 nm in diameter. (D and E) CT230 and CT303 form VLPs that lack the characteristic arches formed by the protruding domains and resemble smooth particles. (F and G) ID285 and ID328 form VLPs that are 45 nm in diameter. (H) ID375 forms VLPs that are 45 nm in diameter, as well as VLPs that are morphologically similar to full-length capsid protein VLPs when purified over iodixanol gradients. Bar, 50 nm.
FIG. 5.
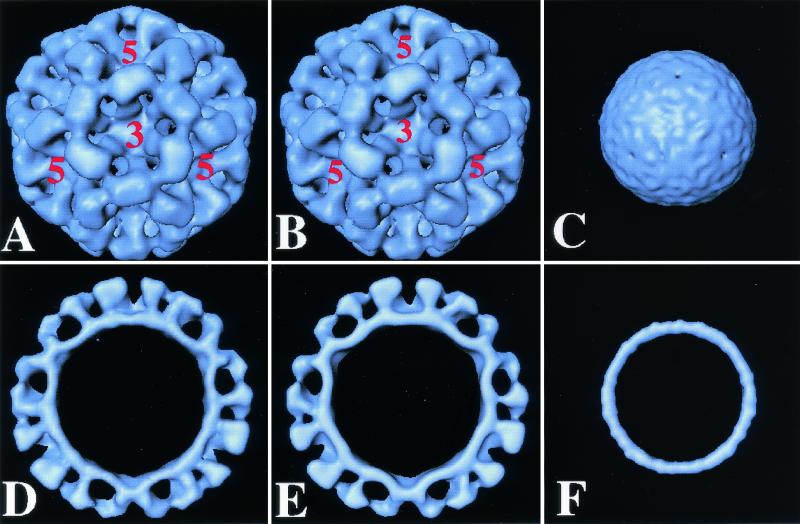
Comparison of the structure of full-length and mutant NT20 and CT303 rNV VLPs. Surface representation of the cryoelectron microscopic reconstruction of full-length rNV VLPs (A) at 22 Å and its cross-section (D), the NT20 mutant structure (B) and its cross-section (E), and the CT303 mutant structure (C) and its cross-section (F). The five- and threefold axes are indicated.
Three-dimensional reconstruction of the NT20 VLPs was carried out by combining 76 particle images which adequately sampled the icosahedral asymmetric unit to a resolution of 23 Å (Fig. 5B and 5E). The reconstruction showed T=3 icosahedral symmetry with a contiguous shell and distinctive arch-like capsomers. A comparative analysis of this reconstruction with that of the full-length rNV VLPs showed no discernible differences at this resolution. The radial dispositions of the shell and the protruding portions in the two reconstructions were identical. The resolution of the cryostructure of the NT20 and full-length proteins is not high enough to allow us to see differences between the N termini.
In contrast to the NT20 construct, particles were not detected by electron microscopy in the preparations of NT34 which were purified over sucrose gradients or iodixanol (data not shown). Due to the low levels of expression of this mutant, it is difficult to assess the involvement of amino acids 20 to 34 in assembly. Next, we analyzed the NT98 deletion, which lacks 98 amino acids, 48 of which are located in the β-barrel motif. We predicted that this deletion would have a dramatic effect on the assembly of the VLPs. As expected from the sedimentation results, assembled particles were not detected by electron microscopy in NT98 preparations from the sucrose or iodixanol gradient peak protein fractions (data not shown).
(ii) C-terminal deletions.
VLPs were visualized in preparations containing the sedimented C-terminal CT20 and CT74 mutant capsid proteins (Fig. 4B and 4C), and as predicted by the sedimentation on sucrose and iodixanol gradients, the particles had a larger diameter (approximately 45 nm) than the VLPs made from the full-length capsid protein. The surface morphological features of these particles appeared different from those in the full-length capsid protein VLPs: the arches are less evident than in the full-length rNV VLPs. A better characterization of structural differences will require cryoelectron microscopy studies. However, such an analysis has not been feasible because of low concentrations of particles.
The CT230 mutant assembled into archless smooth particles that had a diameter of approximately 30 nm (Fig. 4D). The morphology of the particles seen after purification over iodixanol gradients was the same as that of particles purified over sucrose gradients, but a greater number of particles were seen when iodixanol gradients were used, suggesting a low stability of these particles when exposed to high osmotic pressure. Moreover, the 45-nm particles and the CT230 smooth particles were not stable when purified over CsCl gradients (data not shown) as opposed to the full-length capsid VLPs.
The CT303 deletion mutant lacks the 303 amino acids corresponding to the entire P domain. Conventional negative-stain electron microscopy analysis of the VLPs assembled from the CT303 mutant revealed that these VLPs lacked the characteristic arches formed by the protruding domain (Fig. 4E), similar to the CT230 particles. This observation was further confirmed by cryoelectron microscopy of these particles (Fig. 5C and 5F). The cryoimaging of the CT303 smooth particles revealed that they have a diameter of 27 nm and the core has a thickness of 4 nm. The diameter was smaller than expected (30 nm) for the icosahedral shell of the rNV particles. The shrinkage was confirmed by imaging a mixture of smooth particles and the native rNV particles, eliminating the possibility that the difference was due to magnification differences during the cryoimaging procedure. The total protein mass inside the shell calculated from the volume of the shell in the smooth particle reconstruction, assuming a protein density value of 1.30 g/ml, is consistent with 180 molecules of the CT303 protein of 20K. This strongly suggests that the shell in the CT303 mutant has a T=3 icosahedral organization.
(iii) Internal deletions.
The particles containing the internal deletions ID285 and ID328 had a larger diameter (approximately 45 nm) than the VLPs made from the full-length capsid protein (Fig. 4F and 4G), and they were similar to the particles observed with the C-terminal mutants CT20 (Fig. 4B) and CT74 (Fig. 4C). Forty-five-nanometer particles were obtained for ID285 and ID328 independently of the type of gradient (iodixanol or sucrose) used for their purification (Fig. 4F and 4G). In contrast, for the ID375 mutant, which lacks a region that forms a loop on top of the P2 domain, 38-nm VLPs were observed when purified over iodixanol gradients (Fig. 4H), but 45-nm particles were observed when the particles were purified over sucrose gradients (data not shown). Furthermore, none of the 45-nm particles formed by the internal deletions or the CT20 and CT74 mutant proteins remained assembled when purified over CsCl gradients. Therefore, deletions in the protruding domains affected the stability of the particles.
DISCUSSION
NV is a prototypical T=3 icosahedral virus. Near-atomic resolution structures of several T=3 viruses (20, 42), including that of NV capsid (38), have been determined by X-ray crystallography. These studies have provided valuable information about the structural organization in these viruses as well as important insights into possible mechanisms of capsid assembly. The simple architecture of the capsid made from a single gene product and the ability of the expressed protein to spontaneously assemble into a T=3 capsid make NV an excellent model system to further our understanding of the structural requirements for the assembly of a T=3 capsid. Close structural relatives of NV, a human virus, are the tombusviruses, which are plant viruses. The capsid protein in these viruses, including NV, has two distinct domains. The S domain, which forms an eight-stranded β-barrel, is involved exclusively in icosahedral contacts, and the other, more external P domain is involved in dimeric contacts. In contrast, capsid proteins of other T=3 viruses, like sobemoviruses and nodaviruses, such as southern bean mosaic virus and flock house virus, respectively, have only one domain which is structurally homologous to the S domain (20). An interesting question is whether the S domain alone in NV can form an icosahedral capsid. This study reports the design and characterization of several mutants to begin to map the role of different domains of the capsid protein in the assembly and structural integrity of the NV capsid.
Role of N-terminal residues in the assembly of NV VLPs.
The effect of deleting N-terminal residues has been studied in a number of other T=3 viruses. The N-terminal 60 residues in tomato bushy stunt virus (17), 52 residues in turnip crinkle virus (25, 45), and 25 residues in cowpea chlorotic mottle virus, a bromovirus (49), which are not part of the eight-stranded β-barrel domain are oriented toward the interior of the particle and have been shown to be required for the encapsidation of RNA but not for assembly of empty particles (25, 49). Proteolytic digestion of the N-terminal portions of the capsid protein of southern bean mosaic virus, a sobemovirus, leads to the assembly of T=1 particles instead of T=3 particles (6, 44). Likewise, deletions of 31 amino acid residues from the N terminus of flock house virus, a T=3 virus member of the family Nodaviridae, resulted in the formation of variously shaped particles (4).
Our study shows that the N-terminal 20 residues are dispensable for T=3 particle assembly. This finding is similar to that reported for the capsid protein of the Physalis mottle virus, a T=3 plant tymovirus, which does not switch the assembly of particles to T=1 when the N-terminal 30 amino acid are deleted, suggesting that the determinants for the T=3 capsid assembly for some of the T=3 viruses may lie outside of the N terminus (43). While we were studying the assembly of N-terminal mutants of NV capsid protein, it was reported that the N-terminal 30 amino acid residues of VP60, the rabbit hemorrhagic disease virus capsid protein, can be deleted without affecting the assembly of the particles (32, 33); however, the capsid protein of rabbit hemorrhagic disease virus, although similar to the NV capsid protein, is larger (49 extra amino acids) and has an extended N-terminal domain (37, 48).
A switching region that is postulated to control the variations in the conformation of the coat protein of the T=3 viruses involves either the N-terminal arm of the capsid protein or the genomic RNA (18, 19, 42). Elimination of this “molecular switch” is expected to result in a loss of precision in viral assembly. In tombusviruses and sobemoviruses, the N-terminal arm of the S domain is ordered in the C subunits and disordered in the A and B subunits, allowing the required switch for the establishment of the T=3 icosahedral symmetry (19, 21, 41). The ordered portions of the C subunits prevent C/C dimers from assuming a bent conformation that is seen in the A/B dimers. In the case of nodaviruses, ordered RNA interacting with C/C dimers has been suggested to play an important role in modulating the curvature by constraining the C/C dimers to a flat conformation (8, 9).
The rNV particles are different from either of these classes of T=3 capsids because neither the interactions involving ordered N-terminal residues of the C subunit nor the interactions with RNA are observed. The rNV capsid protein readily forms T=3 capsids without RNA. In the native NV VLPs, the N-terminal residues (amino acids 10 to 14) of the B subunit interact with the F-strand of the eight-stranded β-barrel of the neighboring C subunit in the T=3 lattice (38). Similar (quasi-equivalent) interactions between the neighboring A subunits around the fivefold axis or the C subunits around the threefold axis are lacking because the first 29 residues of the A and C subunits are disordered.
Our studies presented here with the NT20 mutant, in which the N-terminal 20 residues are deleted, clearly indicate that this interaction between the B and the C subunits is not obligatory for the formation of the T=3 capsid. In the NT20 mutant, a compensatory conformational change, such as further ordering of the C subunit that interacts with the B subunit, may retain the requirements for icosahedral assembly. The N termini of the neighboring B and C subunits are spatially close to each other in the native NV structure. In the native structure, it appears that ordering of the N-terminal residues of the B subunit forces the N-terminal residues of the C subunit to adapt different conformations. In the case of NT20, it is possible that instead of the B subunit's interacting with the C subunit, the N-terminal residues of the C subunit may interact with the B subunit to provide similar interactions as in the native structure.
Contrary to the NT20 mutant, NT98 did not assemble into detectable particles. This deleted region of the capsid protein consists of three of the eight β-strands in the eight-stranded β-barrel motif and a pivotal α-helix. Some of the residues in this region are well conserved across calicivirus sequences and are involved in interactions between the dimeric subunits. It is therefore not unexpected that the integrity of the particle is compromised in the NT98 mutant. The level of protein expression of the NT34 mutant was significantly lower than for the other mutants. If a minimal amount of protein is required to trigger particle assembly, it is possible that the low yields of this mutant may be the reason for the lack of assembly. Therefore, we cannot make conclusions about the involvement of the first 21 to 34 residues in particle formation.
Role of the P domain in the assembly of NV VLPs.
Our studies clearly indicate that the role of the P domain is not only to confer increased stability to the icosahedral capsid, but also to provide a control for the size of the particle. Deletion of all the dimeric contacts, as in CT303, or individual contacts, as in the internal deletion mutants, produces particles which lack native dimensions. The larger 45-nm particles, which are produced in low yields, appeared to be less stable and structurally heterogeneous when examined by electron microscopy. It remains unclear if these particles are icosahedral or if they are aggregates of the mutated capsid proteins. It is possible that elimination of the dimeric contacts in some of these mutants may force the P domain to adopt alternate conformations, which directly influence the packing of the S domains and result in increased size and lower stability. In the ID375 mutant, which lacks 10 amino acids (375 to 385), some particles purified over iodixanol gradients looked similar to full-length NV capsid VLPs, in addition to 45-nm particles. It is likely that the region composed of amino acids 375 to 385 will tolerate the insertion of heterologous peptides better than a deletion. If this proves to be true, rNV VLPs could be used as carriers of heterologous epitopes for the induction of mucosal immunity using oral or intranasal vaccine protocols in mice or humans (13, 32).
Our results also clearly indicate that interaction between the P and S domains plays an important role in capsid assembly. Small deletions in the protruding domain, as in the CT20 and CT74 mutants, affect only the interactions between the P1 and S domains and result in the production of particles that are 45 nm in diameter. The same was observed when the C-terminal 48 amino acids were removed (data not shown). Notice that all these mutants retain a significant portion of the P domain. In the deletion mutants in which either the entire P domain (CT303) or a significant portion of the P domain (CT230) was deleted, eliminating the interactions between the P and S domains, the effect is different. The size of the formed particle is compromised, but it is in the opposite direction. These mutants produce well-formed archless smooth particles. Cryoelectron microscopic analysis of CT303 indicated that the particles formed are indeed icosahedral but with a noticeably smaller diameter than the diameter expected from the rNV shell structure.
Thus, in all these particles, an absence of interactions between the P and the S domains compromised the size of the particles. It is plausible that the hydrogen bond interactions between the C-terminal residues of the P domain with the S domain residues, as observed in the crystal structure of the rNV capsid, control the size of the icosahedral capsid, and the lack of such interactions may be responsible for the observed alterations in size. Another interesting feature is that the CT303 particles appear to be more stable than the CT230 particles, as they maintain their integrity in CsCl gradients. One reason may be that the 73 residues of the P domain still remaining in the CT230 may adversely affect the S domain interactions and make these particles less stable than the CT303 particles, which contain only the S domain portion of the capsid protein. Taken together these results indicate that while the S domain by itself has all the attributes to assemble a stable icosahedron, the interactions between the entire P domain and the S domain enhance the stability of the assembled particles and perhaps help in conferring the appropriate size on the particle.
Biological significance of smooth particles.
The ability of the S domain alone to form an icosahedral structure may have some biological implications. In the liver of infected rabbits, the calicivirus rabbit hemorrhagic disease virus forms archless particles, which are believed to be composed of the N terminus of the capsid protein (10), which resemble CT230 or CT303 smooth particles. Furthermore, Hillman et al., in 1982 (23), reported that chymotrypsin digestion of calicivirus-like viruses that infect worms transformed the particles into smooth particles.
Although smooth particles have thus far not been described for NV, NV may be similar to rabbit hemorrhagic disease virus and the virus described by Hillman et al. For example, the 32K soluble protein present in high concentrations in stool samples of NV-infected patients represents the C terminus of the capsid protein that forms the protruding domains. The trypsin cleavage site in the protein is at amino acid 227 and is highly conserved in NV-like caliciviruses (16). Although trypsin cleavage of the capsid in vitro is not possible when the particles are intact, there may be factors during natural infection that modify the conformation of the capsid and expose the trypsin cleavage site and result in production of smooth particles. In addition, since the shell domain that forms the smooth particles is highly conserved among the Norwalk-like viruses, the smooth particles may represent a valuable tool for the production of cross-reactive antibodies for use in diagnostic assays that will allow the capture and detection of different strains of Norwalk-like viruses.
Acknowledgments
We thank Sue E. Crawford and Pamela J. Glass for helpful suggestions and discussions and Carl Q.-Y. Zeng for assistance with insect cell cultures and electron microscopy.
This work was supported by Public Health Service grants AI38036 and AI46581, DK-56338, which supports the Texas Gulf Coast Digestive Diseases Center, and training grant T32 DK-07664, from the National Institutes of Health. B. V. V. Prasad acknowledges support from the R. Welch Foundation.
REFERENCES
- 1.Crowther, R. A. 1971. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Phil. Trans. R. Scot. Lond. B Biol. Sci. 261:221-230. [DOI] [PubMed] [Google Scholar]
- 2.Crowther, R. A., D. J. DeRosier, and A. Klug. 1970. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proc. R. Soc. Lond. 317:319.. [DOI] [PubMed] [Google Scholar]
- 3.Delgadillo, R. A., R. M. Smets, Q. R. Yang, D. Vanden Berghe, and A. Neetens. 1988. The use of iodinated density gradient medium for the isolation of rod outer segments. Experientia 44:702-704. [DOI] [PubMed] [Google Scholar]
- 4.Dong, X. F., P. Natarajan, M. Tihova, J. E. Johnson, and A. Schneemann. 1998. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J. Virol 72:6024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21:129-228. [DOI] [PubMed] [Google Scholar]
- 6.Erickson, J. W., A. M. Silva, M. R. Murthy, I. Fita, and M. G. Rossmann. 1985. The structure of a T=1 icosahedral empty particle from southern bean mosaic virus. Science 229:625-629. [DOI] [PubMed] [Google Scholar]
- 7.Estes, M. K., and M. E. Hardy. 1995. Norwalk virus and other enteric caliciviruses, p. 1009-1034. In M. Blaser, P. Smith, J. Ravdin, H. B. Greenberg, and R. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, New York, N.Y.
- 8.Fisher, A. J., and J. E. Johnson. 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361:176-179. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, A. J., B. R. McKinney, A. Schneemann, R. R. Rueckert, and J. E. Johnson. 1993. Crystallization of viruslike particles assembled from flock house virus coat protein expressed in a baculovirus system. J. Virol. 67:2950-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granzow, H., F. Weiland, H.-G. Strebelow, C. M. Liu, and H. Schirrmeier. 1996. Rabbit hemorrhagic disease virus (RHDV): ultrastructure and biochemical studies of typical and core-like particles present in liver homogenates. Virus Res. 41:163-172. [DOI] [PubMed] [Google Scholar]
- 11.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadephia, Pa. [Google Scholar]
- 12.Green, K. Y., J. F. Lew, X. Jiang, A. Z. Kapikian, and M. K. Estes. 1993. Comparison of the reactivities of baculovirus-expressed recombinant Norwalk virus capsid antigen with those of the native Norwalk virus antigen in serologic assays and some epidemiologic observations. J. Clin. Microbiol. 31:2185-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrero, R. A., J. M. Ball, S. S. Krater, S. E. Pacheco, J. D. Clements, and M. K. Estes. 2001. Recombinant Norwalk virus-like particles administered intranasally to mice induce systemic and mucosal (fecal and vaginal) immune responses. J. Virol. 75:9713-9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale, A., K. Mattick, D. Lewis, M. Estes, X. Jiang, J. Green, R. Eglin, and D. Brown. 2000. Distinct epidemiological patterns of Norwalk-like virus infection. J. Med. Virol. 62:99-103. [DOI] [PubMed] [Google Scholar]
- 15.Hardy, M. E., S. F. Kramer, J. J. Treanor, and M. K. Estes. 1997. Human calicivirus genogroup II capsid sequence diversity revealed by analyses of the prototype Snow Mountain agent. Arch. Virol. 142:1469-1479. [DOI] [PubMed] [Google Scholar]
- 16.Hardy, M. E., L. J. White, J. M. Ball, and M. K. Estes. 1995. Specific proteolytic cleavage of recombinant Norwalk virus capsid protein. J. Virol. 69:1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison, S. C. 1980. Protein interfaces and intersubunit bonding. The case of tomato bushy stunt virus. Biophys. J. 32:139-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison, S. C. 1984. Multiple modes of subunit association in the structures of simple spherical viruses. Trends Biochem. Sci. 9:345-351. [Google Scholar]
- 19.Harrison, S. C. 1989. Common features in the design of small RNA viruses, p. 3-19. In M. B. A. Oldstone and A. Notkins (ed.), Concepts in viral pathogenesis, vol. 3. Springer-Verlag, New York, N.Y. [Google Scholar]
- 20.Harrison, S. C. 2001. Principles of virus structure, p. 53-85. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 21.Harrison, S. C., A. Olson, C. E. Schutt, F. K. Winkler, and G. Bricogne. 1978. Tomato bushy stunt virus at 2.9 angstrom resolution. Nature 276:368-373. [DOI] [PubMed] [Google Scholar]
- 22.Hermens, W. T., O. ter Brake, P. A. Dijkhuizen, M. A. Sonnemans, D. Grimm, J. A. Kleinschmidt, and J. Verhaagen. 1999. Purification of recombinant adeno-associated virus by iodixanol gradient ultracentrifugation allows rapid and reproducible preparation of vector stocks for gene transfer in the nervous system. Hum. Gene Ther. 10:1885-1891. [DOI] [PubMed] [Google Scholar]
- 23.Hillman, B., T. J. Morris, W. R. Kellen, D. Hoffman, and D. E. Schlegl. 1982. An invertebrate calici-like virus. Evidence for partial virion disintegration in host excreta. J. Gen. Virol. 60:115-123. [Google Scholar]
- 24.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 25.Hogle, J. M., A. Maeda, and S. C. Harrison. 1986. Structure and assembly of turnip crinkle virus. I. X-ray crystallographic structure analysis at 3.2 angstrom resolution. J. Mol. Biol. 191:625-638. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, X., D. Y. Graham, K. N. Wang, and M. K. Estes. 1990. Norwalk virus genome cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 27.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli, U. K., F. Beguin, and G. Gujer-Kellenberger. 1970. A factor preventing the major head protein of bacteriophage T4 from random aggregation. J. Mol. Biol. 47:69-85. [DOI] [PubMed] [Google Scholar]
- 29.Lawton, J. A., M. K. Estes, and B. V. V. Prasad. 1997. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 4:118-121. [DOI] [PubMed] [Google Scholar]
- 30.Lawton, J. A., and B. V. V. Prasad. 1996. Automated software package for icosahedral virus reconstruction. J. Struct. Biol. 116:209-215. [DOI] [PubMed] [Google Scholar]
- 31.Moller-Larsen, A., and T. Christensen. 1998. Isolation of a retrovirus from multiple sclerosis patients in self-generated iodixanol gradients. J. Virol. Methods 73:151-161. [DOI] [PubMed] [Google Scholar]
- 32.Nagesha, H. S., L. F. Wang, and A. D. Hyatt. 1999. Virus-like particles of calicivirus as epitope carriers. Arch. Virol. 144:2429-2439. [DOI] [PubMed] [Google Scholar]
- 33.Nagesha, H. S., L. F. Wang, A. D. Hyatt, C. J. Morrissy, C. Lenghaus, and H. A. Westbury. 1995. Self-assembly, antigenicity, and immunogenicity of the rabbit haemorrhagic disease virus (Czechoslovakian strain V-351) capsid protein expressed in baculovirus. Arch. Virol. 140:1095-1108. [DOI] [PubMed] [Google Scholar]
- 34.Neill, J. D. 1992. Nucleotide sequence of the capsid protein gene of two serotypes of San Miguel sea lion virus: identification of conserved and non-conserved amino acid sequences among calicivirus capsid proteins. Virus Res. 24:211-222. [DOI] [PubMed] [Google Scholar]
- 35.Oker-Blom, C., R. F. Pettersson, and M. D. Summers. 1989. Baculovirus polyhedrin promoter-directed expression of rubella virus envelope glycoproteins, E1 and E2, in Spodoptera frugiperda cells. Virology 172:82-91. [DOI] [PubMed] [Google Scholar]
- 36.Oker-Blom, C., and M. D. Summers. 1989. Expression of Sindbis virus 26S cDNA in Spodoptera frugiperda (Sf9) cells, using a baculovirus expression vector. J. Virol. 63:1256-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parra, F., J. A. Boga, M. S. Marin, and R. Casais. 1993. The amino terminal sequence of VP60 from rabbit hemorrhagic disease virus supports its putative subgenomic origin. Virus Res. 27:219-228. [DOI] [PubMed] [Google Scholar]
- 38.Prasad, B. V. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 39.Prasad, B. V. V., M. E. Hardy, and M. K. Estes. 2000. Structural studies of recombinant Norwalk capsids. J. Infect. Dis. 181:S317-S321. [DOI] [PubMed] [Google Scholar]
- 40.Prasad, B. V. V., R. Rothnagel, X. Jiang, and M. K. Estes. 1994. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J. Virol. 68:5117-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossmann, M. G., C. Abad-Zapatero, M. A. Hermodson, and J. W. Erickson. 1983. Subunit interactions in southern bean mosaic virus. J. Mol. Biol. 166:37-73. [DOI] [PubMed] [Google Scholar]
- 42.Rossmann, M. G., and J. E. Johnson. 1989. Icosahedral RNA virus structure. Annu. Rev. Biochem. 58:533-573. [DOI] [PubMed] [Google Scholar]
- 43.Sastri, M., D. S. Reddy, S. S. Krishna, M. R. Murthy, and H. S. Savithri. 1999. Identification of a discrete intermediate in the assembly/disassembly of physalis mottle tymovirus through mutational analysis. J. Mol. Biol. 289:905-918. [DOI] [PubMed] [Google Scholar]
- 44.Savithri, H. S., and J. W. Erickson. 1983. The self-assembly of the cowpea strain of southern bean mosaic virus: formation of T=1 and T=3 nucleoprotein particles. Virology 126:328-335. [DOI] [PubMed] [Google Scholar]
- 45.Sorger, P. K., P. G. Stockley, and S. C. Harrison. 1986. Structure and assembly of turnip crinkle virus. II. Mechanism of reassembly in vitro. J. Mol. Biol. 191:639-658. [DOI] [PubMed] [Google Scholar]
- 46.van Heel, M., G. Harauz, E. V. Orlova, R. Schmidt, and M. Schatz. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 47.White, L. J., J. M. Ball, M. E. Hardy, T. N. Tanaka, N. Kitamoto, and M. K. Estes. 1996. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 70:6589-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wirblich, C., H.-J. Theil, and G. Meyers. 1996. Genetic map of the calicivirus hemorrhagic disease virus as deduced from in vitro translation studies. J. Virol. 70:7974-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao, X., J. M. Fox, N. H. Olson, T. S. Baker, and M. J. Young. 1995. In vitro assembly of cowpea chlorotic mottle virus from coat protein expressed in Escherichia coli and in vitro-transcribed viral cDNA. Virology 207:486-494. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, Z. H., B. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]
- 51.Zolotukhin, S., B. J. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. J. Samulski, and N. Muzyczka. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973-985. [DOI] [PubMed] [Google Scholar]