Abstract
We have examined the influence of RNA upon the interaction of Gag-Pol with Gag during human immunodeficiency virus type 1 (HIV-1) assembly. COS7 cells were transfected with protease-negative HIV-1 proviral DNA, and Gag/Gag-Pol complexes were detected by coimmunoprecipitation with anti-integrase. In COS7 cells, Gag/Gag-Pol is found almost entirely in pelletable, membrane-bound complexes. Exposure of cells to 1% Triton X-100 releases Gag/Gag-Pol from bulk membrane, but the complexes remain pelletable. The role of RNA in facilitating the interaction between Gag and Gag-Pol was examined in these bulk membrane-free, pelletable complexes. The specific presence of viral genomic RNA is not required to maintain the Gag/Gag-Pol interaction, but some type of RNA is, since exposure to RNase destabilized the Gag/Gag-Pol complex. When present only in Gag, the nucleocapsid mutation R7R10K11S, which inhibits Gag binding to RNA, inhibits the formation of both Gag and Gag/Gag-Pol complexes. When present only in Gag-Pol, this mutation has no effect upon complex formation. This result indicates that Gag-Pol may not interact directly with RNA but rather requires RNA-facilitated Gag multimerization for its interaction with Gag.
The Gag precursor in human immunodeficiency virus type 1 (HIV-1) is alone sufficient to produce viral particles (14, 21, 28, 45). Several putative regions of interactions between Gag molecules have been delineated, mainly in the C-terminal half of Gag, and include the C-terminal half of the capsid (CA) (3, 12, 31, 35), p2 (1, 34, 46), the nucleocapsid (NC) (6, 7, 10, 43), and p6 (13). It is generally assumed that in order to obtain the interactions required for assembly, the Gag molecules must first be concentrated at a cellular site. In addition to membrane, RNA has been proposed to act as a scaffold for aligning Gag molecules and facilitating their interaction with each other (2). In vitro studies using truncated Gag molecules have indicated that RNA is important in facilitating a membrane-independent interaction between Gag molecules and that this RNA need not be viral. For example, it was found that when Rous sarcoma virus or HIV-1 peptides containing only CA-NC sequences were expressed in Escherichia coli, they assembled into hollow cylinders in vitro, but only in the presence of added RNA, with the cylinder length dependent upon the length of the RNA (7). The RNA was isolated from E. coli, i.e., assembly did not depend upon the presence of virus-specific RNA. In another study (19), similar cylindrical structures were also reported to form in vitro in the absence of RNA when HIV-1 CA alone was used, but RNA greatly facilitated the speed of the reaction. In vivo findings have also demonstrated that Gag particles are formed even when unable to package viral genomic RNA either because of the absence of the viral RNA packaging signal (30, 33, 40, 42, 47) or because of the mutations in the Cys-His boxes in viral NC (15-17, 40). Most mutations in the Cys-His boxes, while affecting specific genomic RNA incorporation, do not inhibit the packaging of cellular RNA into virions and do not affect viral assembly or viral density (5, 16). More recently it was shown that RNA plays a structural role in retrovirus particles (37). Murine leukemia virus particles that lack genomic RNA were found to contain cellular mRNA in place of genomic RNA, and the retroviral cores were disrupted by treatment with RNase.
During HIV-1 assembly, Gag-Pol is also incorporated into the virus. The factors facilitating the interaction between Gag and Gag-Pol have been less studied, although it has been assumed that these molecules interact with each other through similar sequences involved in Gag/Gag interactions. Evidence for this includes the fact that unmyristylated Gag or Gag-Pol molecules can also be rescued into assembly complexes by myristylated Gag (36, 39, 45). An apparent exception to this assumption is the fact that mutations in the major homology region in the C-terminal half of CAp24 can still allow formation of Gag particles but inhibit the packaging of Gag-Pol into these particles (24, 47). In this work, we show that an RNA requirement for Gag/Gag-Pol interaction probably reflects a requirement for an RNA-facilitated Gag polymerization, and not a direct interaction of Gag-Pol with RNA.
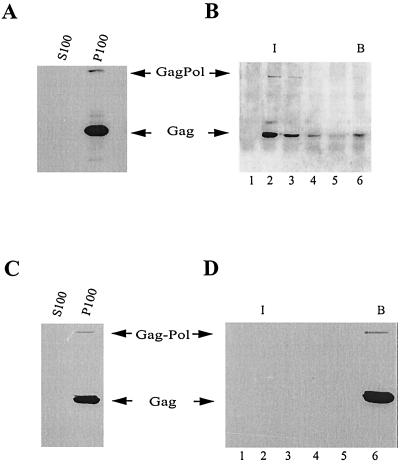
Table 1 provides an overview of the expression constructs used in the study, including appropriate references to their origin. In Fig. 1, we have transfected COS7 cells with pSVC21 BH10.P−, a plasmid coding for protease-negative HIV-1. We have examined the distribution of Gag/Gag-Pol complexes in three cellular fractions: nonpelletable, membrane free; pelletable, membrane free; and pelletable, membrane bound. Resh and colleagues reported that nearly all steady-state Gag in HIV-1-transfected COS1 cells is pelletable and membrane bound (22, 48). The data in Fig. 1 suggest a similar conclusion for Gag/Gag-Pol complexes. In the experiments represented in panels A and B, cells were swollen in hypotonic buffer without detergent and lysed by Dounce homogenization (25 to 30 strokes). The lysate was first centrifuged at low speed (1,500 × g) for 30 min, and the resulting supernatant (S1) was then centrifuged at 100,000 × g for 1 h, resulting in a pellet (P100) and supernatant (S100). Nonpelletable, membrane-free Gag and Gag-Pol were defined as the molecules remaining in the S100 supernatant, while the pelletable material (P100) was further resolved by discontinuous sucrose gradient centrifugation into membrane-free components remaining at the bottom of the gradient and membrane-bound components located at the interface between the 65 and 10% sucrose layers (flotation assay).
TABLE 1.
List of HIV-1 proviral DNA constructs
| Constructa | Viral sequencesb | Mutations in coding sequencec | Major viral protein(s) | Reference(s) |
|---|---|---|---|---|
| PSVC21 BH10 | A | None | All | 18 |
| PSVC21 BH10.P− | B | None | All | 18 |
| R7R10K11S | A | R7R10K11S | All | 26, 38 |
| C15S/C18S | A | C15S/C18S | All | 17, 26 |
| S3(32-34) | A | R32K33K34S | All | 26, 38 |
| C36S/C39S | A | C36S/C39S | All | 17, 26 |
| pSVGAG | C | None | Pr55gag | 45 |
| pSVGAG/GAGPOL.P− | D | None | Pr55gag and Pr160gag-pol | 45 |
| pSVGAGPOL.P− | E | None | Pr160gag-pol | 45, 46 |
| PCMV-rev | F | None | Rev protein | 45 |
| PSVR7-GAG | C | R7R10K11S | Pr55gag | 26 |
| PSVS3-GAG | C | R32K33K34S | Pr55gag | 26 |
| PSVR7-GAGPOL.P− | E | R7R10K11S | Pr160gag-pol | 26 |
| PSVS3-GAGPOL.P− | E | R32K33K34S | Pr160gag-pol | 26 |
pSVGAG, pSVGAG/GAGPOL.P−, pSVGAGPOL.P−, and pCMV-rev were kind gifts of David Rekosh (45, 46). pSVC21 BH10 contains wild-type HIV-1 proviral DNA sequence. pSVC21 BH10.P− differs from pSVC21 BH10 by a single point mutation at position 25 of the protease region, converting Asp25 to Arg25. Transfection of pSVC21 BH10.P− produces noninfectious viral particles containing wild-type genomic RNA and unprocessed precursor proteins Gag and Gag-Pol (18). pSVC21 BH10 and pSVC21 BH10.P− are gifts from E. Cohen, University of Montreal. Plasmids containing the NC mutations R7R10K11S and S3(32-34) were obtained from J. L. Darlix (38), while those containing the NC mutations C15S/C18S and C36S/C39S were obtained from A Rein and R. Gorelick (17). Their cloning into full-length HIV-1 sequences was previously described (26). The R7R10K11S and S3(32-34) mutants were subcloned into vectors expressing only Gag (pSVR7-GAG and pSVS3-GAG, respectively) or only Gag-Pol (pSVR7-GAG-POL.P− and pSVS3-GAG-POL.P−, respectively), as previously described (8).
Some constructs contain mutations that render them protease deficient or frameshift positive, viral sequence types are as follows. A, full-length HIV-1 proviral DNA. B, full-length HIV-1 proviral DNA, but protease negative (D25R substitution). C, proviral DNA starting at HXB2 nucleotide sequence 679 (SacI site) and terminating immediately after-the Gag open reading frame (ORF), HXB2 nucleotide sequence 2428 (BclI site). Proviral DNA is thus missing all the 5′ long termine repeat (LTR) and leader sequences, as well as all coding regions downstream of Gag, but includes the RRE sequence. This construct requires cotransfection with pCMV-rev for protein expression. D, proviral DNA starting at HXB2 nucleotide sequence 679 (SacI site) and terminating immediately after the Vif ORF, HXB2 nucleotide sequence 5785 (SalI site). Proviral DNA is thus missing all the 5′ LTR and leader sequences, as well as all coding regions downstream of Vif, but includes the RRE sequence. Contains an inactive protease (D25G substitution), which results in the synthesis of unprocessed Pr55gag and Pr160gag-pol only. This construct requires cotransfection with pCMV-rev for protein expression. E, proviral DNA starting at HXB2 nucleotide sequence 679 (SacI site) and terminating immediately after the Vif ORF, HXB2 nucleotide sequence 5785 (SalI site). Proviral DNA is thus missing all the 5′ LTR and leader sequences, as well as all coding regions downstream of Vif, but includes the RRE sequence. Contains an inactive protease (D25G) and a deletion of five Ts (nucleotides 2082 to 2086), which results in only Pr160gag-pol, and not Pr55gag, being synthesized. This construct requires cotransfection with pCMV-rev for protein expression. F, proviral DNA starting at HXB2 nucleotide sequence 5954 and terminating immediately after the rev ORF.
Substitutions indicated are in NC.
FIG. 1.
Distribution of Gag/Gag-Pol complexes in COS7 cells. Culture and transfection of COS7 cells by the calcium phosphate method and viral isolation were as previously described (8, 27). COS7 cells were lysed 48 h posttransfection at 4°C in two ways. (i) With a hypotonic medium, lysis was done by Dounce homogenization in hypotonic Tris-EDTA buffer (20 mM Tris-HCl [pH 7.4], 1 mM EDTA, 0.01% β-mercaptoethanol) supplemented with a protease inhibitor cocktail (Complete; Boehringer Mannheim). (ii) With a nonionic detergent, cells were lysed in TNT buffer (20 mM Tris-HCl [pH 7.5], 200 mM NaCl, 1% Triton X-100) supplemented with a protease inhibitor cocktail (Complete; Boehringer Mannheim). For either method, the cell homogenate was then centrifuged at 1,500 × g for 30 min to remove nuclei and unbroken cells. The supernatant (S1) was then centrifuged for 1 h at 100,000 × g in an SW 55Ti rotor (Beckman, Columbia, Md.) at 4°C, yielding the supernatant (S100) and the pellet (P100). Further fractionation of the P100 into membrane-free and membrane-bound protein was done by the membrane flotation assay (44). P100 was resuspended in 1 ml of 73% sucrose. Two milliliters of 65% sucrose in TNE (20 mM Tris [pH 7.8], 100 mM NaCl, 1 mM EDTA) was layered on top of the 73% sucrose, and 2 ml of 10% sucrose was layered on top of the 65% sucrose. The gradients were then centrifuged at 100,000 × g in a Beckman SW55 Ti rotor overnight at 4°C. Fractions (0.8 ml) were collected and diluted with an equal volume of 2× TNT, and each fraction was immunoprecipitated at 4°C with anti-IN. A polyclonal antibody to integrase protein (anti-IN), directed against either the first 16 amino acids (NIH AIDS Research and Reference Reagent Program) or amino acids 276 to 288 (a gift from Mark Muesing, Aaron Diamond AIDS Research Center), was used to immunoprecipitate the Gag/Gag-Pol complexes. Equal amounts of protein, 200 to 500 μg (Bio-Rad assay), were incubated with 30 μl of antibody cross-linked to protein A-Sepharose (Pharmacia Amersham Biotech, Quebec) for 1 h at 4°C. The immunoprecipitate was then washed three times with TNT and twice with phosphate-buffered saline. After the final supernatant was removed, 30 μl of 2× sample buffer (120 mM Tris HCl [pH 6.8], 20% glycerol, 4% sodium dodecyl sulfate [SDS], and 0.02% bromophenol blue) was added and the precipitate was then boiled for 5 min to release the precipitated proteins. After microcentrifugation, the resulting supernatant was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Western blots were probed with mouse anti-CA antibody (Cellular Products, Inc., Buffalo, N.Y.), at a dilution of 1:2,000. It was used as the primary antibody, and horseradish peroxidase-linked goat anti-mouse antibody was used as a secondary antibody. Antibody binding was detected by enhanced chemiluminescence (ECL kit; Pharmacia Amersham Biotech). (A and C) Western blots of anti-IN immunoprecipitates of the S100 and P100 fractions from hypotonic-lysed cells (A) or Triton X-100-lysed cells (C). (B and D) The P100 fraction from hypotonic-lysed cells (B) or Triton X-100-lysed cells (D) was resolved by discontinuous sucrose gradient centrifugation into membrane-bound (I, interface) and membrane-free (B, bottom) protein. Each fraction was immunoprecipitated with anti-IN and analyzed by Western blotting using anti-CA.
For immunoprecipitation experiments with anti-integrase (anti-IN), the P100 and S100 fractions were in the same volumes of TNT buffer. Western blot analysis of the immunoprecipitates using anti-CA as a primary antibody shows that all Gag/Gag-Pol in transfected COS cells are found in the P100 fraction (Fig. 1A), and that almost all Gag/Gag-Pol complexes are membrane bound (Fig. 1B). It is not known if the small amount of membrane-free Gag/Gag-Pol detected in the P100 fraction dissociated from the membrane or was never associated with it.
Although Gag and Gag/Gag-Pol complexes are found in the pelletable, membrane-bound fraction when COS7 cells are lysed in hypotonic medium with Dounce homogenization, lysing cells in 1% Triton X-100 alters this distribution, i.e., Gag/Gag-Pol complexes are released from the membrane but remain pelletable. This is shown in Fig. 1C and D, depicting an assay in which Gag/Gag-Pol complexes are immunoprecipitated from the detergent lysate using anti-IN and a Western blot analysis of the immunoprecipitates uses anti-CA to detect Gag and Gag-Pol. Gag/Gag-Pol complexes remain in the P100 fraction (Fig. 1C) but are released from bulk membrane (Fig. 1D). Although discontinuous sucrose gradient analysis indicates that the P100 fraction contains Gag-Pol that has become membrane free, it may be more appropriate to refer to this as bulk membrane free, since this type of analysis may not distinguish membrane-free molecules from those bound to detergent-resistant, high-density membrane subdomains. For example, it has been reported that in HIV-1-transfected COS1 cells, a fraction of Gag determined to be membrane free by discontinuous sucrose gradient analysis is in fact present in such membrane subdomains. These domains were termed barges because their buoyant densities were greater than those found for lipid raft membrane subdomains, perhaps due to the large sizes of the multimeric Gag complexes (32).
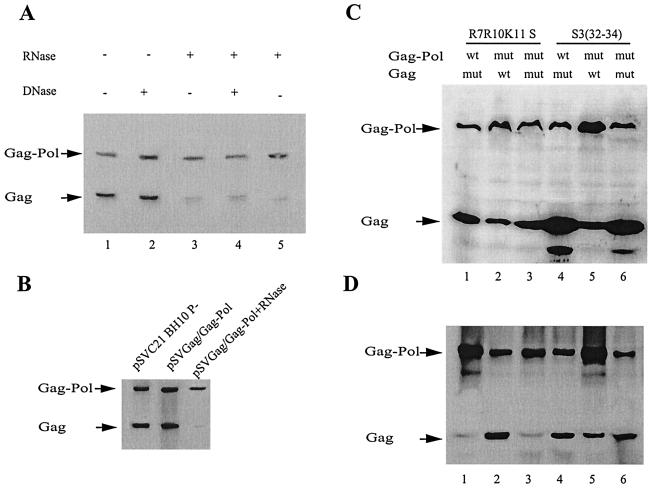
Since RNA (genomic or cellular) plays an important role in the formation of the Gag complex (2, 37), we have examined the role of RNA in the Gag/Gag-Pol interaction. Studies were done with the P100 fraction isolated from cells lysed in 1% Triton X-100. We first examined the ability of the Gag/Gag-Pol complex to be immunoprecipitated in the presence of nucleases. COS7 cells were transfected with BH10 P-, and the results (Western blots of anti-IN immunoprecipitates of the P100 fraction, stained with anti-CA and anti-reverse transcriptase [RT]) are shown in Fig. 2A. Lane 1 shows the coimmunoprecipitation of Gag with Gag-Pol in the absence of nuclease, while lane 2 shows that this interaction is unaffected by the presence of DNase. The addition of RNase (lane 3) or DNase and RNase (lane 4) before immunoprecipitation results in the inhibition of coimmunoprecipitation of Gag with Gag-Pol. Similar results are obtained if RNase is added after immunoprecipitation but before washing and release of the anti-IN/Gag-Pol complex from the beads (lane 5). These results indicate that RNA is required for maintaining the stability of the Gag/Gag-Pol complex. It will be noted regarding this figure that immunoprecipitation with anti-IN always results in a small amount of precipitation of a species migrating in a manner similar to that of Gag. This species, which may be the heavy chain of anti-IN detected by the secondary antibody, is considered background since it appears even in anti-IN immunoprecipitates from lysates of nontransfected COS7 cells (data not shown).
FIG. 2.
Role of RNA in the formation and/or stability of Gag/Gag-Pol complexes. COS7 cells were transfected with mutant HIV-1 proviral DNA. Forty-eight hours posttransfection, cells were lysed on ice in TNT buffer containing 1% Triton X-100 and after clarification by low-speed centrifugation were centrifuged at 100,000 × g for 1 h at 4°C to produce the S100 and P100 fractions. The P100 fraction resolved from the lysates was immunoprecipitated with anti-IN, and Western blots were probed with mouse anti-RT antibodies (38) and mouse anti-CA antibodies (Cellular Products, Inc.). (A) Effects of RNase A upon Gag/Gag-Pol complex formation. The P100 fractions from COS7 cells transfected with pSVC21 BH10.P− proviral DNA were pretreated with no DNase or RNase (lane 1), 20 μg of DNase (lane 2), 20 μg of RNase (lane 3), or 20 μg of both DNase and RNase (lane 4) and then immunoprecipitated with anti-IN. In lane 5, the pSVC21 BH10.P− P100 fraction was first immunoprecipitated and then treated with 20 μg of RNase, followed by washing with TNT buffer and phosphate-buffered saline and analysis by Western blotting. (B) Genomic RNA packaging is not required for Gag/Gag-Pol complex formation, but RNA is. COS7 cells were transfected with pSVC21 BH10.P− proviral DNA (lane 1) or with a construct expressing both Gag and Gag-Pol, pSVGAG/GAGPOL.P−, which lacks the five-leader region of the genomic RNA, including the Ψ packaging signal (lanes 2 and 3). The P100 fractions were immunoprecipitated with anti-IN and analyzed by Western blotting. In lane 3, the sample was treated with RNase before immunoprecipitation. (C and D) Effects of mutations in the basic amino acid regions flanking the first Cys-His box in NC upon Gag/Gag-Pol complex formation. COS7 cells were cotransfected with different combinations of plasmids coding for either Gag or Gag-Pol, which either were wild type or contained mutations in the basic amino acid regions flanking the first Cys-His box. All plasmids were inactive for the viral protease. (C) Western blot analysis of viral proteins in the P100 fraction of COS7 cells cotransfected with plasmids (described in Table 1) coding for the following: wild-type Gag-Pol, pSVGAGPOL.P−, and mutant Gag, pSVR7-GAG (lane 1); wild-type Gag, pSVGAG, and mutant Gag-Pol, pSVR7-GAGPOL.P− (lane 2); mutant Gag, pSVR7-GAG, and mutant Gag-Pol, pSVR7-GAGPOL.P− (lane 3); wild-type Gag-Pol, pSVGAGPOL.P−, and mutant Gag, pSVS3-GAG (lane 4); wild-type Gag, pSVGAG, and mutant Gag-Pol, pSVS3-GAGPOL.P− (lane 5); and mutant Gag, pSVS3-GAG, and mutant Gag-Pol, pSVS3-GAGPOL.P− (lane 6). (D) The corresponding P100 fractions were immunoprecipitated with anti-IN and analyzed by Western blotting.
Panel B in Fig. 2 further indicates that this RNA need not be genomic RNA since the Gag/Gag-Pol complex is formed when COS7 cells are transfected with pSVGAG/GAG-POL.P−, a construct missing the HIV-1 leader sequence and which produces Gag/Gag-Pol particles which are defective in viral RNA packaging (34, 46). However, the formation of this Gag/Gag-Pol complex still depends upon cellular RNA, since the complex is still destroyed by RNase, as shown in lane 3.
Since RNA has been shown to be important for Gag multimerization, we investigated whether the dependence upon RNA for stability of the Gag/Gag-Pol complex reflected a direct interaction of Gag-Pol with RNA or if the interaction of Gag-Pol with Gag depends primarily upon the ability of RNA to facilitate Gag multimerization. NC sequences are required for the interaction of Gag with genomic RNA and have been shown to be important for Gag multimerization in vitro. It is currently thought that specific interactions of Gag with viral genomic RNA involve the interaction of Cys-His boxes in NC (15-17) with specific stem-loop structures in the 5′ leader sequence of the RNA (1, 9, 11, 29). On the other hand, the basic amino acids flanking the first (or only) Cys-His box have been proposed to interact via ionic bonds nonspecifically with viral or cellular RNA, a process which may serve to concentrate Gag molecules for intermolecular interactions during assembly (5, 7, 20). Mutations in the basic amino acid regions flanking the first Cys-His box in HIV-1 NC reduce by approximately 80% both viral RNA packaging into virus and the in vivo annealing of primer tRNA3Lys onto the genomic RNA that is packaged (8, 25). We have studied the effects of these mutations upon the formation of the Gag/Gag-Pol complex by cotransfecting COS7 cells with pSVGAG and pSVGAG-POL vectors, which code for either Gag or Gag-Pol, respectively. Mutations were made in these vectors, which flank the first, Cys-His box in NC, using either the upstream mutation, R7R10K11S, or the downstream mutation, S3(32-34). The presence of both Gag and Gag-Pol in lysates of cells cotransfected with various combinations of wild-type and mutant pSVGAG and pSVGAG-POL vectors is shown in the Western blots in Fig. 2C, while Western blots of the anti-IN immunoprecipitates from these lysates are shown in Fig. 2D. Gag/Gag-Pol complex formation is inhibited by the R7R10K11S mutation, but not by the S3(32-34) mutation. Furthermore, the R7R10K11S mutation is inhibitory only when present in Gag (Fig. 2D, lanes 1 and 3). This mutation has no effect upon complex formation when present only in the Gag-Pol precursor (Fig. 2D, lane 2) and indicates that the interaction of Gag-Pol with Gag does not require Gag-Pol to first interact with RNA. The requirement of RNA for Gag-Pol to interact with Gag may therefore reflect a requirement for Gag multimerization.
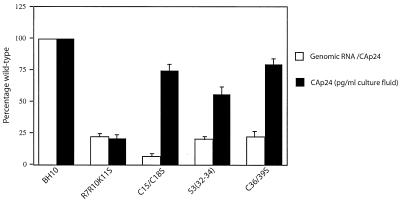
The difference in the abilities of the R7R10K11S and S3(32-34) mutations to disrupt Gag/Gag-Pol complex formation is reflected in their different abilities to disrupt extracellular viral particles and cytoplasmic Gag particle formation, as shown in Fig. 3 and 4, respectively. Both R7R10K11S and S3(32-34) reduce packaging of viral genomic RNA in virions equally well (about 75 to 80% in Fig. 3), but it may be that the ability to bind to nonviral RNA is more severely reduced by the R7R10K11S mutation than by the S3(32-34) mutation. This should affect extracellular viral and cytoplasmic Gag complex production, and in fact, this is shown to be the case by the results given in Fig. 3 and 4, respectively. In the assay presented in Fig. 3, we measured both the relative viral production in culture (sedimentable CAp24) and the relative amount of genomic RNA found per sedimentable CAp24 in virus-like particles produced from COS7 cells transfected with wild-type and mutant HIV-1 proviral DNA. As previously reported, genomic RNA packaging is maximally affected (94% reduction) by mutations in the first Cys-His box (C15S/C18S), while mutations in the second Cys-His box (C36S/C39S) or in the regions flanking the first Cys-His box [R7R10K11S and S3(32-34)] reduce genomic RNA packaging by 75 to 80%. But viral particle production is not correlated with the lack of genomic RNA packaging, and the R7R10K11S mutation clearly produces the greatest inhibition of viral particle production. The particles produced could have resulted from a weaker, but maintained, ability of plasma membrane to organize Gag molecules into particles ready for budding, i.e., the virus may be capable of using less efficient alternative assembly pathways (23, 41). Although the R7R10K11S particles are capable of packaging both Gag-Pol and tRNA3Lys (26), Western blots detecting the RT protein in these particles show that RT protein is present at only 25 to 50% of the levels found in the wild-type virions (4, 26), so that the total particle-associated Gag-Pol seen in the R7R10K11S mutant virions is probably only 5 to 10% of that seen in the wild type.
FIG. 3.
Genomic RNA packaging and CAp24 production in extracellular wild-type and mutant viruses. Culture and transfection of COS7 cells by the calcium phosphate method and viral isolation were as previously described (8, 27). Viral particle release into cell culture medium was measured by CAp24 production. CAp24 was determined by using the commercial kit available for CAp24 antigen capture (Abbott Laboratories). Viral RNA isolation and quantification by dot blot hybridization was performed as previously described (8, 25).
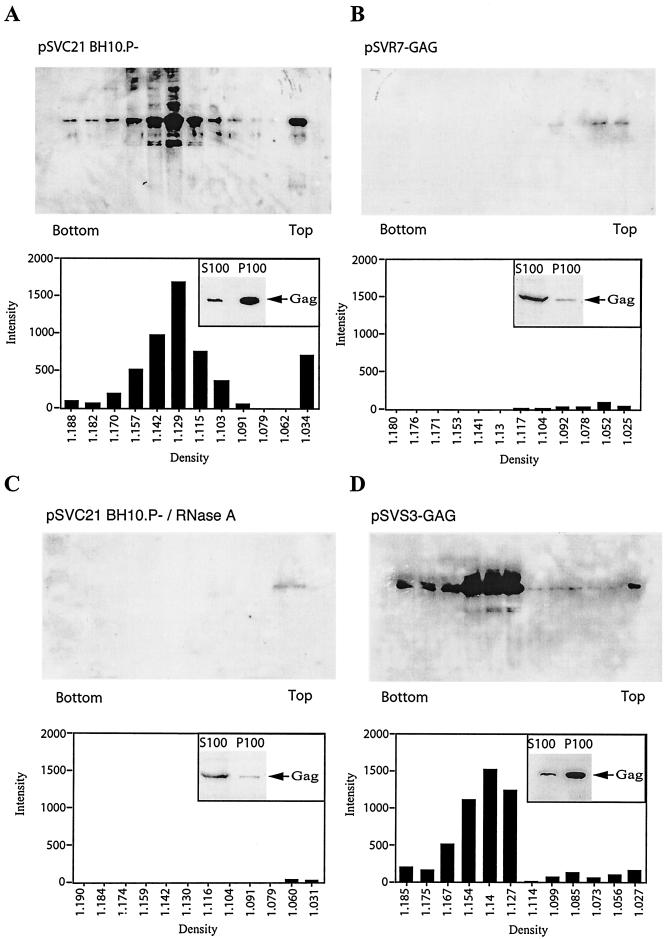
FIG. 4.
Formation of cytoplasmic wild-type and mutant Gag complexes. COS7 cells were transfected with pSVC21 BH10.P−, pSVR7-GAG, and pSVS3-GAG. Forty-eight hours posttransfection, cells were lysed on ice in TNT buffer containing 1% Triton X-100, and after clarification by low speed centrifugation, the resulting S1 supernatant was analyzed by equilibrium density centrifugation. Prior to centrifugation, one-half of the S1 fraction from cells transfected with pSVC21 BH10.P− was treated with RNase A, and equal amounts of the four lysate preparations were resolved by ultracentrifugation. Twelve fractions were collected, each fraction was centrifuged at 100,000 × g for 1 h at 4°C, and the resulting P100 pellet was analyzed by Western blotting. The small inserts in each panel represent analysis of the S100 and P100 fractions before gradient fractionation, in which Gag was immunoprecipitated by anti-CA and analyzed by Western blotting.
Figure 4 indicates that R7R10K11S has a greater ability than the S3(32-34) mutation to reduce the amount of pelletable, cytoplasmic Gag. COS7 cells were transfected with either pSVC21 BH10.P− or pSVGAG proviral DNA containing the NC mutations R7R10K11S or S3(32-34) (pSVR7-GAG or pSVS3-GAG, respectively). Transfected cells were lysed in Triton X-100 as described above, and one-half of the lysate of cells transfected with pSVC21 BH10.P− was exposed to RNase A. Equal amounts of the four lysate preparations were then resolved by equilibrium density centrifugation. Gradient fractions were collected, and each fraction was centrifuged at 100,000 × g to pellet any Gag complexes. The pellet was resuspended and analyzed by using Western blots probed with anti-CA. The results are shown in Fig. 4. Lysates of cells transfected with either pSVC21 BH10.P− or pSVS3-GAG contain similar amounts of Gag complexes. However, the presence of pelletable Gag complexes is severely reduced either in RNase A-treated lysates of cells transfected with pSVC21 BH10.P− or in lysates of cells transfected with the pSVR7-GAG mutant. The low Gag signal seen for pSVR7-GAG or pSVC21 BH10P−/RNase A results from most of the Gag being in the S100 fraction as opposed to most of the Gag being in the P100 fraction for pSVC21 BH10P− and pSVS3-Gag (see inserts in these panels). Although almost all of the Gag in COS cells appears in the P100 fraction when cells are lysed in hypotonic medium with Dounce homogenization (our unpublished data and reference 32), lysis of cells in 1% Triton X-100 does release 20 to 25% of Gag into the S100.
Thus, although RNA is part of the Gag/Gag-Pol complexes (Fig. 2A and B) and need not be genomic RNA (Fig. 2B), the influence of RNA upon the Gag/Gag-Pol interaction appears to act through RNA's ability to facilitate Gag/Gag interactions. Gag-Pol does not seem to interact directly with RNA. This conclusion is supported by the fact that the R7R10K11S mutation inhibits the Gag/Gag-Pol interaction only when present in Gag, which disrupts Gag polymerization but does not inhibit the Gag/Gag-Pol interaction when present only in Gag-Pol (Fig. 2D, lanes 1 to 3). This observation makes it unlikely that the disruption of the Gag/Gag-Pol complex is due to either (i) the disruption of an RNA bridge to which both precursors bind or (ii) disruption of a protein-protein interaction at the NC sequences containing R7R10K11. The tertiary structure of Gag-Pol is not known, but it is possible that the RNA binding sequence within the Gag NC is hidden by the large Pol sequence during the early stages of assembly.
Acknowledgments
A.K. and R.H. contributed equally to this work.
REFERENCES
- 1.Aldovini, A., and R. A. Young. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 64:1920-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging, p. 177-218. In H. G. Krausslich (ed.), Morphogenesis and maturation of retroviruses, vol. 214. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 3.Berthet-Colominas, C., S. Monaco, A. Novelli, G. Sibai, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (CAp24) complexed with a monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthoux, L., C. Pechoux, M. Ottmann, G. Morel, and J.-L. Darlix. 1997. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J. Virol. 71:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cen, S., Y. Huang, A. Khorchid, J.-L. Darlix, M. A. Wainberg, and L. Kleiman. 1999. The role of Pr55gag in the annealing of tRNA3Lys to human immunodeficiency virus type 1 genomic RNA. J. Virol. 73:4485-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clavel, F., and J. M. Orenstein. 1990. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J. Virol. 64:5230-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craven, R. C., and L. J. Parent. 1996. Dynamic interactions of the Gag polyprotein. Curr. Top. Microbiol. Immunol. 214:65-94. [DOI] [PubMed] [Google Scholar]
- 11.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 12.Gamble, T. R., S. Yoo, F. Vajdos, U. K. Von Schwedler, J. McCutcheon, and W. I. Sundquist. 1997. Structure of the carboxy-terminal dimerization domain of HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 13.Garnier, L., L. Ratner, B. Rovinski, S. X. Cao, and J. W. Wills. 1998. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J. Virol. 72:4667-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 15.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. O. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorelick, R. J., J. S. M. Nigida, J. J. W. Bess, L. O. Arthur, L. E. Henderson, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross, I., H. Hohenberg, C. Huckhagel, and H. G. Krausslich. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross, I., H. Hohenberg, and H. G. Krausslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 21.Haffar, O., J. Garrigues, B. Travis, P. Moran, J. Zarling, and S. L. Hu. 1990. Human immunodeficiency virus-like, nonreplicating, gag-env particles assemble in a recombinant vaccinia virus expression system. J. Virol. 64:2653-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hockley, D. J., M. V. Nermut, C. Grief, J. B. M. Jowett, and I. M. Jones. 1994. Comparative morphology of Gag protein structures produced by mutants of the Gag gene of human immunodeficiency virus type 1. J. Gen. Virol. 75:2985-2997. [DOI] [PubMed] [Google Scholar]
- 24.Huang, M., and M. A. Martin. 1997. Incorporation of Pr160gag-pol into virus particles requires the presence of both the major homology region and adjacent C-terminal capsid sequences within the Gag-Pol polyprotein. J. Virol. 71:4472-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, Y., A. Khorchid, J. Gabor, J. Wang, X. Li, J.-L. Darlix, M. A. Wainberg, and L. Kleiman. 1998. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA3Lys genomic placement and initiation of reverse transcription in HIV-1. J. Virol. 72:3907-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, Y., A. Khorchid, J. Wang, M. A. Parniak, J. Darlix, M. A. Wainberg, and L. Kleiman. 1997. Effect of mutations in nucleocapsid protein (NCp7) upon Pr160gag-pol and tRNALys incorporation into HIV-1. J. Virol. 71:4378-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, Y., J. Mak, Q. Cao, Z. Li, M. A. Wainberg, and L. Kleiman. 1994. Incorporation of excess wild-type and mutant tRNA3Lys into HIV-1. J. Virol. 68:7676-7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karacostas, V., K. Nagashima, M. A. Gonda, and B. Moss. 1989. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc. Natl. Acad. Sci. USA 86:8964-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lever, A., H. Göttlinger, W. Haseltine, and J. Sodroski. 1989. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 63:4085-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin, J. G., P. M. Grimley, J. M. Ramseur, and I. K. Berezesky. 1974. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 14:152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 32.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linial, M. L., and A. D. Miller. 1990. Retroviral RNA packaging: sequence requirements and implications. Curr. Top. Microbiol. Immunol. 157:125-152. [DOI] [PubMed] [Google Scholar]
- 34.Mak, J., M. Jiang, M. A. Wainberg, M.-L. Hammarskjold, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Momany, C., L. Kovari, A. Prongay, W. Keller, R. Gitti, B. Lee, A. Gorbalenya, L. Tong, J. McClure, L. Ehrlich, M. Summers, C. Carter, and M. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 36.Morikawa, Y., S. Hinata, H. Tomoda, T. Goto, M. Nakai, C. Aizawa, H. Tanaka, and S. Omura. 1996. Complete inhibition of human immunodeficiency virus Gag myristoylation is necessary for inhibition of particle budding. J. Biol. Chem. 271:2868-2873. [DOI] [PubMed] [Google Scholar]
- 37.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ottmann, M., C. Gabus, and J.-L. Darlix. 1995. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J. Virol. 69:1778-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park, J., and C. D. Morrow. 1992. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into virus-like particles. J. Virol. 66:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rein, A., D. P. Harvin, J. Mirro, S. M. Ernst, and R. J. Gorelick. 1994. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J. Virol. 68:6124-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Royer, M., S. S. Hong, B. Gay, M. Cerutti, and P. Boulanger. 1992. Expression and extracellular release of human immunodeficiency virus type 1 Gag precursors by recombinant baculovirus-infected cells. J. Virol. 66:3230-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakalian, M., J. W. Wills, and V. M. Vogt. 1994. Efficiency and selectivity of RNA packaging by Rous sarcoma virus Gag deletion mutants. J. Virol. 68:5969-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon, J. H. M., E. A. Carpenter, R. A. M. Fouchier, and M. H. Malim. 1999. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 73:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, A. J., M. I. Cho, M. L. Hammarskjöld, and D. Rekosh. 1990. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into virus-like particles. J. Virol. 64:2743-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, A. J., N. Srinivasakumar, M.-L. Hammarskjöld, and D. Rekosh. 1993. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J. Virol. 67:2266-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srinivasakumar, N., M.-L. Hammarskjöld, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of HIV-1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]