Abstract
Nucleotide sequences were determined for the complete S genome segments of the six distinct hantavirus genotypes from Argentina and for two cell culture-isolated Andes virus strains from Chile. Phylogenetic analysis indicates that, although divergent from each other, all Argentinian hantavirus genotypes group together and form a novel phylogenetic clade with the Andes virus. The previously characterized South American hantaviruses Laguna Negra virus and Rio Mamore virus make up another clade that originates from the same ancestral node as the Argentinian/Chilean viruses. Within the clade of Argentinian/Chilean viruses, three subclades can be defined, although the branching order is somewhat obscure. These are made of (i) “Lechiguanas-like” virus genotypes, (ii) Maciel virus and Pergamino virus genotypes, and (iii) strains of the Andes virus. Two hantavirus genotypes from Brazil, Araraquara and Castello dos Sonhos, were found to group with Maciel virus and Andes virus, respectively. The nucleocapsid protein amino acid sequence variability among the members of the Argentinian/Chilean clade does not exceed 5.8%. It is especially low (3.5%) among oryzomyine species-associated virus genotypes, suggesting recent divergence from the common ancestor. Interestingly, the Maciel and Pergamino viruses fit well with the rest of the clade although their hosts are akodontine rodents. Taken together, these data suggest that under conditions in which potential hosts display a high level of genetic diversity and are sympatric, host switching may play a prominent role in establishing hantavirus genetic diversity. However, cospeciation still remains the dominant factor in the evolution of hantaviruses.
Hantaviruses, members of the Bunyaviridae family, are enveloped, single-stranded, negative-sense RNA viruses (10). The virus genome consists of three segments, designated large (L), medium (M), and small (S), that are packed into helical nucleocapsids. These segments encode the RNA polymerase (20, 42), a glycoprotein precursor that is cotranslationally processed to yield two envelope glycoproteins (G1 and G2) and a nucleocapsid (N) protein, respectively (38, 42).
Each hantavirus is typically predominantly associated with a specific rodent host indigenous to the geographic area from which the hantavirus was characterized (15, 21, 30, 32, 33), although there appear to be occasional occurrences of spillover infections in related rodent species (3, 5, 15, 27, 28, 41). According to most studies, the rodent host appears to be an asymptomatic carrier with a persistent lifetime infection (21, 51). However, recent data indicate that North American Sin-Nombre-like viruses cause some pathology within their reservoir hosts (25, 31). The genus Hantavirus includes approximately 30 distinct virus serotypes/genotypes (20, 38, 39). As only a small percentage of known rodent species has been examined for the presence of hantavirus genetic material, this number is likely to increase. In North America, hantaviruses are the cause of hantavirus pulmonary syndrome (HPS), a severe respiratory illness with a greater than 40% mortality rate (6, 8, 28, 43, 44). Since the 1993 identification of HPS in the United States (32), many new hantaviruses have been discovered throughout the New World.
During the last decade, epidemiological and serological data have provided evidence of hantavirus-positive rodents and HPS-like human disease in several South American countries (1, 14, 17, 36, 49, 52, 53, 55, 56; S. C. Levis, G. E. Calderon, N. Pini, T. G. Ksiazek, C. J. Peters, and D. A. Enria, 5th Argent. Congr. Virol., abstr. 144, 1996). As a result, several distinct hantavirus genotypes have been described in South America. These include Rio Mamore (RM) virus, amplified and sequenced from the sigmodontine rodent Oligoryzomys microtis collected in Bolivia (1, 14); Andes (AND) virus recovered from a human HPS case autopsy and O. longicaudatus rodents from southern Argentina (7, 24, 35, 48, 53); and LN virus recovered from Calomys laucha rodents and human HPS case samples in Paraguay (17). Three novel HPS-associated hantavirus genotypes, whose rodent hosts are unknown, were identified from human HPS cases that occurred in Brazil. These are Juquitiba virus, the causative agent of the 1993 Brazilian HPS cases (49), and the Castelo dos Sonhos (CAS) and Araraquara (ARA) viruses, which are responsible for three fatal HPS cases occurring in 1995 and 1996 (18). More recently, two distinct hantavirus genotypes, Choclo virus (from O. fulvescens) and Calabazo virus (from Zygodontomys brevicauda), were genetically identified in Panama, and the former virus was associated with human HPS cases (50).
Finally, six novel genetically related virus genotypes from the three major foci of hantavirus infections in Argentina (the northwestern, central, and southwestern regions [22, 23, 36]) were partially genetically characterized in our laboratory (22, 23). The rodent reservoirs of five of the virus genotypes, termed Pergamino (PRG), Maciel (MAC), Lechiguanas (LEC), Bermejo (BMJ), and Oran (ORN), were found to be Akodon azarae, Necromys benefactus (formerly Bolomys obscurus), O. flavescens, O. chacoensis, and O. longicaudatus, respectively. Of the six hantavirus genotypes, LEC, ORN, and Hu39694 (rodent host under investigation) have been associated with human disease (22, 23). Analysis of a 1,654-nucleotide (nt) M segment fragment showed these six Argentinian virus genotypes to be monophyletic and to group together with AND virus (23). The genotypes LEC, BMJ, ORN, and Hu39694 are most closely related and make up a well-supported subclade (23). However, phylogenetic analysis of the M genome segment-derived nucleotide sequences failed to resolve conclusively the interrelationships of the PRG, MAC, and AND viruses within this clade.
In the present study, the complete S segment nucleotide sequence was determined for the six novel Argentinian hantavirus genotypes and for two cell culture-isolated AND virus strains from Chile. The primary aim of the S segment sequencing was to further analyze the phylogenetic relationship among the newly recognized Argentinian genotypes and previously characterized New World hantaviruses since the M segment sequence data are not available for these previously known viruses. Additional objectives were to examine aspects of the molecular epidemiology and evolution of the Argentinian/Chilean hantaviruses.
MATERIALS AND METHODS
Patient and rodent samples.
For this study, one human blood sample and five rodent tissue samples, described previously (23), were used (Table 1.). Briefly, the human sample (Hu39694) was obtained from an HPS patient residing in Pergamino who contracted HPS at an unknown location. The rodent samples were from Oran, in the Salta province of northwestern Argentina (O. longicaudatus /ORN and O. chacoensis/BMJ) and from Pergamino (A. azarae/PRG), the Lechiguanas Islands (O. flavescens/LEC), and Maciel (N. benefactus/MAC) in central Argentina. In addition, two AND virus isolates, AND/Chile/OL58/97 and AND/Chile/OL123/97, referred to as strains AND58 and AND123, respectively, were used. The O. longicaudatus lung tissue samples studied (kindly provided by T. Ksiazek, Centers for Disease Control and Prevention, Atlanta, Ga.) were collected in the Aysen region of Chile (48), and the two strains mentioned above were subsequently isolated and grown in cell culture in our laboratory.
TABLE 1.
Argentinian and Chilean hantaviruses analyzed in the current study
| Virus genotypea | Source of RNA | Place of origin, country | Date |
|---|---|---|---|
| Lechiguanas (LEC) | O. flavescens (lung) | Lechiguanas Islands, Argentina | 1997 |
| Bermejo (BMJ) | O. chacoensis (lung) | Oran, Argentina | 1997 |
| Oran (ORN) | O. longicaudatus (lung) | Oran, Argentina | 1997 |
| Human no. 39694 (Hu39694) | Human HPS case (blood) | Pergamino, Argentina | 1997 |
| Maciel (MAC) | N. benefactus (lung) | Maciel, Argentina | 1997 |
| Pergamino (PRG) | A. azarae (lung) | Pergamino, Argentina | 1997 |
| Andes/Chile/OL58/97 (AND58) | O. longicaudatus (lung) | Aysen, Chile | 1997 |
| Andes/Chile/OL123/97 (AND123) | O. longicaudatus (lung) | Aysen, Chile | 1997 |
Standard abbreviations for the Argentinian/Chilean viruses referred to in this study are given in parentheses.
Extraction and preparation of total RNA.
Total RNA was extracted from human and rodent samples, and the procedures were performed in a laminar-flow hood located inside a biosafety level 3 facility. The RNaid PLUS kit (Bio 101, Inc., La Jolla, Calif.) was used to extract RNA from 100 mg of tissue or 20 μl of blood in accordance with the manufacturer's recommendations. RNA was purified with RNA matrix beads, eluted with 30 μl of RNase-free H2O, and stored at −80°C.
Nested reverse transcription-PCR and sequence analysis.
cDNA syntheses using virus RNAs and subsequent nested PCRs were performed as previously described (23). Amplified products from the nested PCRs were analyzed by gel electrophoresis. Fragments of the expected sizes were excised from the gels and purified with the GeneClean Kit (Bio 101, Inc.) in accordance with the manufacturer's recommendations. Purified PCR products were sequenced directly with the dRhodamine Terminator Cycle Sequencing Kit as specified by the manufacturer (Applied Biosystems, Inc., Foster City, Calif.) and subsequently analyzed with an ABI Prism 310 Genetic Analyzer as previously described (Applied Biosystems, Inc.) (23).
Oligonucleotide primer design.
The first-round primers used, with slight modifications, have been described previously and were based upon conserved regions of characterized hantaviruses to encompass the entire S segment open reading frame (ORF) (9). The nested second-round primers subsequently used have been previously described (11). To amplify the long variable 3′ noncoding region (NCR), first- and second-round reverse primers were designed based on the El Moro Canyon (ELMC) virus sequence (12) at nucleotide positions 1872 and 1850, respectively, of the cRNA. The forward primers were based upon sequence data from the 3′ end of the N gene ORFs of the specific viruses under study as the data became available. All sequence gaps were filled in with virus-specific primers. A previously described primer that contains the generic hantavirus terminal sequence (29) was used to obtain remaining portions of the S segment sequence close to the termini. Specific primer sequences are available upon request.
Sequencing of the 5′ and 3′ termini of virus RNA.
The exact nucleotide sequences of the 5′ and 3′ termini of the LEC and AND (AND58) virus S segments (sequenced from both the 3′ and 5′ sides) were determined by the RNA ligation method described previously (26). In earlier studies, it was determined that viral genomic RNAs are effective substrates for direct ligation of the 5′ and 3′ ends (4, 26). Briefly, ligations were done with 4 μl of viral genome RNA in a reaction mixture containing 5 U of T4 RNA ligase (Promega, Madison, Wis.), 12 U of the RNase inhibitor RNasin (Promega), and ligation buffer (50 mM Tris [pH 7.8], 10 mM MgCl2, 5 mM dithiothreitol, 1 mM ATP). The reaction mixture was incubated at 37oC for 30 min, and then 5 μl of the ligated virus RNA was used in the first-round PCR. All of the primers used were specific for the S segment being analyzed. For the BMJ, ORN, Hu39694, AND123, MAC, and PRG viruses, nucleotide sequences were obtained through the ligation region from the 3′ side.
Nucleotide sequence comparison and phylogenetic analysis.
Sequence overlaps were obtained with the AutoAssembler computer program. Alignments and comparisons of the hantavirus S genome segments were performed with the LINEUP and PILEUP programs of the GCG (Genetics Computer Group, Madison, Wis.) software package. Nucleotide sequence differences were accessed with the GAP program of the GCG software package. Phylogenetic analyses of the nucleotide sequences (the entire S genome segment and the N gene ORF) and the N protein sequences of different hantaviruses were performed with both PAUP version 3.1.1 (45) and PAUP* version 4.0b8 (46) computer program software. Routinely used methods for reconstruction of hantavirus phylogeny included the maximum-parsimony (MP) method (supported by both PAUP version 3.1.1. and PAUP* version 4.0b8), the distance-based neighbor-joining (NJ) method (supported by PAUP* 4.0b8), and quartet puzzling (supported by PAUP* 4.0b8). The more computationally intensive maximum-likelihood (ML) method, supported by PAUP* 4.0b8, was also used to analyze a smaller subset of the sequence data that included two novel hantaviruses from Brazil (18).
In MP analyses, phylogenetic trees were obtained by the heuristic search method using either equal weighting of all changes or weighting of transversions over transitions. Weighting schemes of 3:1, 6:1, and 10:1 (which represent estimated transition/transversion ratios within major virus clades, within small subclades, and among closely related strains of the same virus, respectively) were employed. Gaps were treated alternatively either as missing data or as a fifth character state. Analyses of the deduced N protein amino acid sequences were conducted by using either equal weighting of all changes or the amino acid weighting matrix found in the PROTPARS example file of the PAUP software package. NJ analysis employed several distance corrections supported by PAUP* 4.0b8. However, the Kimura two-parameter algorithm (with or without gamma-distributed rate variation across the site) was used in the majority of the NJ-based phylogenetic reconstructions. Bootstrap confidence limits were obtained by 1,000 heuristic (MP) or Kimura two-parameter (NJ) search repetitions. The previously published sequences of the hantavirus S genome segments used in this study are listed in the legend to Fig. 1.
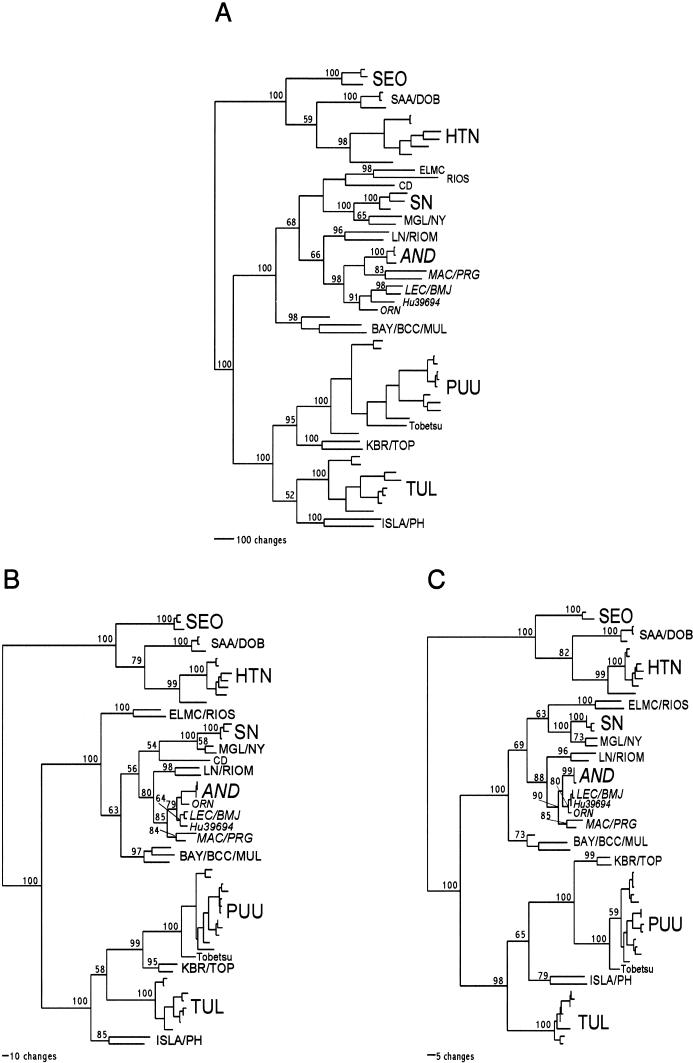
FIG.1.
Phylogenetic relationships among the Argentinian/Chilean virus genotypes examined in the present study. Phylogenetic trees were generated on the basis of nucleotide sequence differences in the 1,129-nt fragment of the N gene ORF (A and B) or amino acid sequence differences in the entire N protein (C). Lengths of the horizontal branches are proportional to the nucleotide (A and B) and amino acid (C) step differences. Vertical branches are for visual clarity only. Bootstrap values of greater than 50%, obtained from 1,000 replicates of the analysis, are shown for the New World hantaviruses and for all other major hantavirus clades at the appropriate branch points. The Argentinian/Chilean virus genotypes under study are in italics. Previously published hantavirus S segment sequences used in the analysis include the following: Hantaan (HTN) virus strains 76118 (GenBank accession number M14626), CUMC-B11 (U37768), Q32 (AB027097), Chen4 (AB027101), Hu (AB027111), Z10 (AF184987), and NC167 (AB027523); Seoul (SEO) virus strains SR-11 (M34881), Z37 (AF187082), and Gou3 (AF184988); Dobrava (DOB) virus strain 3970/87 (L41916); Saaremaa (SAA) virus strains Saa/160V (AJ009773) and Saa/90Aa/97 (AJ009775); Puumala (PUU) virus strains Puu/Vindeln/L20Cg/83 (Z48586), Vranica (U14137), CG1820 (M32750), K27 (L08804), Puu/Kazan (Z84204), Udmurtia/458Cg/88 (Z30707), Udmurtia/894Cg/91 (Z21497), Puu/Virrat/25Cg/95 (Z69985), Evo/12Cg/93 (Z30702), Sotkamo (X61035), Puu/Solleftea/Cg6/95 (AJ223377), CG 13891 (U22423), and Tobetsu-60Cr-93 (AB010731); Khabarovsk (KBR) virus strain MF-43 (U35255); Topografov (TOP) virus strain Ls136V (AJ011646); Tula (TUL) virus strains Tula/76Ma/87 (Z30941), Tula/53Ma/87 (Z30942), Cacak (AF017659), Tula/Kosice667/Ma/95 (Y13980), Tula/Moravia/5293Ma/94 (Z48574), Tula/Koziky/5276Ma/94 (AJ223601), Malacky/Ma370/94 (Z68191), and Lodz-2 (AF063897); PH virus strain PH-1 (M34011); Isla Vista (ISLA) virus strain MC-SB-1 (U31534); ELMC virus strain RM-97 (U11427); Rio Segundo (RIOS) virus strain RMx-Costa-1 (U18100); Cano Delgadito (CD) virus strain VHV-574 (AF000140); SN virus strains Convict Creek 74 (L33816), Convict Creek 107 (L33683), and NM H10 (L25784); Monongahela (MGL) virus strain Monongahela-1 (U32591); New York (NY) virus strain RI-1 (U09488); Bayou (BAY) virus strain Bayou-1 (L36929); Black Creek Canal (BCC) virus strain BCC-1 (L39949); Muleshoe (MUL) virus strain SH-Tx-339 (U54575); Laguna Negra (LN) virus strain 510B (AF005727); RM (RIOM) virus strain OM-556 (U52136); and AND virus strain AH-1 (AF004660). (A) MP analysis of the nucleotide sequence, using the heuristic search option and a 3:1 weighting of transversions over transitions, generated a single most parsimonious tree. (B) MP analysis of the nucleotide sequence without third-position bases of the N gene ORF, using the heuristic search option and a 3:1 weighting of transversions over transitions, generated four equally parsimonious trees. One representative tree is shown here; three other trees differ only in the placement of certain genotypes (although without meaningful bootstrap support) within the clade of Argentinian/Chilean virus genotypes (data not shown). (C) Unweighted MP analysis of the complete amino acid sequence of the N protein generated 48 equally parsimonious trees. One representative tree is shown here. Other trees differ only in the placement of certain minor branches that have no significant bootstrap support.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been submitted to the GenBank database and assigned accession numbers AF291702 and AF482712 through AF482717.
RESULTS
Sequence analysis of the S genome segment of the six Argentinian and Chilean hantavirus genotypes.
The nucleotide sequences of the S genome segments of the six virus genotypes from Argentina and the two virus strains from Chile were determined. The sequences of the S genome segments from the LEC, BMJ, ORN, Hu39694, AND58, MAC, and PRG viruses, respectively, are 1,938, 1,934, 1,919, 1,900, 1,871, 1,869, and 1,860 nt long. The sequences of the S segment termini were determined directly for the LEC and AND58 virus genotypes as described in Materials and Methods. For the rest of the viruses in this study, 3′-terminal virus RNA sequences were derived by sequencing with the generic primer mentioned in Materials and Methods (29). However, since the terminal 8 nt at the 3′ and 5′ ends are invariant in all three genome segments of the eight representatives of the currently known serologic groups of the Hantavirus genus (4), it is most probable that these nucleotides are identical in the viruses in this study as well. Comparison of the 3′ and 5′ termini of the viral RNA shows complementarity between the terminal 22 nt, with two mismatches at positions 9 and 19 and a noncanonical G · U pair in the viral RNA sense at position 10.
The first potential methionine initiation codon for all seven viruses is located at nucleotide position 43 from the 5′ terminus of the cRNA, and this ORF terminates at nucleotide position 1326. The ORF, 1,287 nt long, has the potential to code for the putative 429-amino-acid (aa) N protein. A second small overlapping ORF was found in all seven of the viruses in this study (the LEC, BMJ, ORN, Hu39694, AND, MAC, and PRG viruses). The methionine initiation codon for the putative second ORF starts at nucleotide position 122 (76 nt downstream from the initiation codon of the N protein) and extends to a termination codon at nucleotide position 311. This ORF potentially codes for a 64-aa protein. As seen with previously analyzed viruses (2, 29, 37, 40, 44), a lower third-base substitution frequency in this region provides theoretical evidence that this second ORF codes for a functional protein in the Argentinian viruses. However, there are no experimental data that support the functionality of this second ORF. No additional ORF capable of encoding proteins of more than 50 aa was found on either strand.
The 3′ NCRs of the Argentinian/Chilean LEC, BMJ, ORN, Hu39694, AND (AND58), MAC, and PRG virus cRNA S segments are 609, 605, 590, 571, 542, 540, and 531 nt long, respectively. As with all previously characterized hantaviruses, this region is relatively variable in sequence. A more detailed analysis of the 3′ NCR nucleotide sequence disclosed the presence of a number of precise and imprecise repeats. For instance, a search of the 3′ NCR for precise repeats 8 bases long (Repeat Program; GCG) showed 5 (Hu39694 virus) to 11 (LEC virus) various repeated motifs in the 3′ NCR. While most of the repeats seen were unique to the particular virus genotypes, others appeared to be shared, especially among the genotypes that were most closely related phylogenetically. The most prominent repeats observed in the NCRs of all of the Argentinian/Chilean viruses (cDNA sense) included short GGGT (triple-G-containing) repeats and the CTACCTCA (C-rich) repeats. The former sequence, or the sequence with one base mismatch, appeared several times in the region (approximately between positions 1330 and 1500 in different virus genotypes). The latter repeats are located close to the S segment 5′ end and occur three times in all of the Argentinian/Chilean virus genotypes, with the exception of the MAC and AND123 viruses, where one of the copies has a one-base mismatch. Among the repeats shared by the most closely related viruses, the most prominent appears to be the sequence ATTGCTT, which is present in the LEC, BMJ, ORN, and Hu39694 viruses.
Phylogenetic relationship of the Argentinian and Chile hantavirus genotypes.
Analysis of nucleotide sequence differences in the entire S segment and the N gene ORF (Fig. 1A to C) confirmed the existence of seven distinct Argentina/Chile hantavirus genotypes. As was seen with the M segment of these viruses (23), the genotypes are genetically unique in comparison with previously characterized hantaviruses. All South American hantaviruses form a new, distinct, and well-supported clade that diverges from the same ancestral node as three other groups of New World hantaviruses (i.e., Peromyscus-borne viruses, Reithrodontomys-borne viruses, and Oryzomys/Sigmodon-borne viruses). Within the clade of South American hantaviruses, the seven genotypes described in the present study show a distinct evolutionary divergence from the LN and RM viruses found in Paraguay and Bolivia, respectively (1, 17). Strong support for these nodes is provided by bootstrap analysis (Fig. 1A to C).
Within the clade composed of the Argentinian hantaviruses and the Chilean AND virus, the S segment-based phylogenetic reconstructions reveal three distinct subclades. The first subclade (LEC-like virus genotypes) is made up of the LEC, BMJ, Hu39694, and ORN genotypes, with the LEC and BMJ genotypes being the most closely related. A nucleotide sequence comparison of the ORFs of these two viruses shows 91% similarity between the LEC and BMJ viruses. Other genotypes within this subclade display about 86% nucleotide similarity (Table 2). The akodontine rodent-borne PGM and MAC genotypes are placed together in the second subclade. Finally, strains of the AND virus form the third subclade. Our analysis cannot resolve decisively the branching order of the three subclades within the clade of Argentinian/Chilean hantavirus genotypes; however, some phylogenetic reconstructions (Fig. 1B) show a certain affinity between the AND virus and the LEC-like genotypes.
TABLE 2.
Comparison of the nucleotide (top) and amino acid (bottom) sequences of the N protein ORF among seven Argentinian virus lineages and other known hantaviruses
| Virus | % Similarity
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LEC | BMJ | ORN | Hu | AND | PRG | MAC | RIOM | LN | SN | BAY | BCC | TUL | PUU | SEO | HTN | |
| LEC | 91.1 | 86.6 | 86.7 | 83.4 | 81.7 | 80.7 | 78.4 | 77.2 | 74.7 | 73.9 | 74.6 | 65.1 | 63.4 | 56.4 | 56.2 | |
| BMJ | 99.5 | 86.4 | 86.6 | 82.8 | 80.2 | 80.3 | 77.4 | 76.2 | 74.7 | 73.3 | 74.7 | 64.0 | 63.1 | 55.9 | 56.6 | |
| ORN | 98.4 | 98.6 | 87.3 | 83.0 | 81.3 | 81.0 | 78.9 | 76.8 | 74.3 | 75.3 | 72.8 | 64.7 | 64.1 | 56.3 | 57.0 | |
| Hu39694 | 99.3 | 99.5 | 98.8 | 82.2 | 80.6 | 80.4 | 77.3 | 76.9 | 72.6 | 74.0 | 72.8 | 64.6 | 64.1 | 56.5 | 56.1 | |
| AND123 | 96.3 | 96.5 | 96.5 | 96.7 | 79.5 | 79.3 | 78.7 | 76.6 | 73.4 | 74.3 | 71.7 | 65.4 | 64.4 | 55.4 | 56.6 | |
| PRG | 95.3 | 95.6 | 95.8 | 95.8 | 94.9 | 81.4 | 77.1 | 76.0 | 72.3 | 74.0 | 72.7 | 65.1 | 63.2 | 55.0 | 56.0 | |
| MAC | 93.9 | 94.2 | 94.4 | 94.4 | 93.7 | 95.8 | 75.9 | 75.0 | 75.1 | 73.3 | 73.7 | 65.8 | 64.7 | 56.0 | 56.6 | |
| RIOM | 90.4 | 90.2 | 90.4 | 90.4 | 90.9 | 89.7 | 89.3 | 80.7 | 72.7 | 74.9 | 73.6 | 65.4 | 64.3 | 54.4 | 57.2 | |
| LN | 89.5 | 89.7 | 90.0 | 90.0 | 90.2 | 89.5 | 88.1 | 93.0 | 73.3 | 74.4 | 72.7 | 65.3 | 65.0 | 55.6 | 58.0 | |
| SN NM H10 | 86.9 | 87.2 | 86.7 | 86.9 | 85.8 | 85.8 | 85.1 | 84.4 | 85.1 | 73.8 | 71.1 | 62.0 | 63.6 | 55.8 | 56.3 | |
| BAY | 88.3 | 88.6 | 88.8 | 88.8 | 88.1 | 88.3 | 87.4 | 87.9 | 86.9 | 86.7 | 78.5 | 65.9 | 64.1 | 52.6 | 58.3 | |
| BCC | 85.8 | 86.0 | 86.2 | 86.2 | 86.2 | 85.8 | 86.2 | 86.5 | 85.3 | 83.7 | 92.1 | 66.1 | 66.4 | 57.6 | 57.7 | |
| TUL/53Ma/87 | 74.8 | 74.6 | 74.4 | 74.8 | 74.4 | 74.1 | 73.7 | 75.5 | 74.4 | 73.2 | 75.5 | 75.3 | 69.5 | 56.8 | 55.5 | |
| PUU CG1820 | 72.7 | 73.0 | 73.0 | 73.0 | 73.0 | 72.3 | 73.2 | 73.2 | 73.0 | 71.6 | 73.7 | 73.7 | 80.0 | 56.9 | 53.6 | |
| SEO SR-11 | 62.9 | 62.7 | 62.7 | 62.7 | 63.4 | 62.2 | 62.0 | 62.9 | 62.2 | 60.6 | 61.8 | 62.5 | 61.9 | 61.4 | 68.8 | |
| HTN76118 | 65.3 | 65.0 | 64.8 | 64.8 | 64.3 | 63.6 | 63.4 | 64.1 | 63.9 | 62.0 | 63.4 | 63.6 | 62.6 | 60.7 | 82.1 | |
For the virus genotypes in this study, S segment- and N ORF-based phylogenetic trees display branching orders very similar to that of the tree based on a 1,654-nt fragment from the M coding region (23). Based on this observation, there is no indication that genetic reassortment has occurred among these seven virus genotypes.
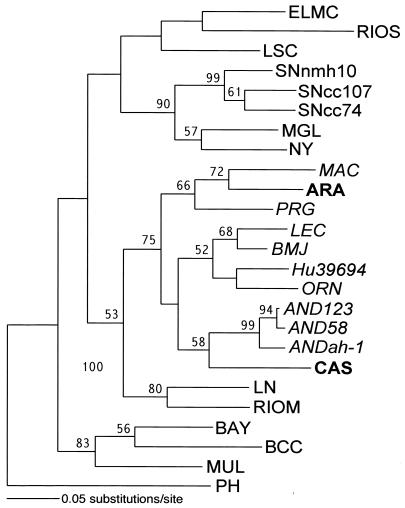
Finally, ML analysis of a 643-nt N gene region (Fig. 2) placed two recently recognized pathogenic Brazilian hantavirus genotypes, ARA and CAS, within the clade of Argentinian/Chilean viruses together with the MAC and AND viruses, respectively.
FIG. 2.
Phylogenetic relationships of the two novel virus genotypes from Brazil, ARA (GenBank accession number AF307325) and CAS (AF307324), to the Argentinian/Chilean viruses examined in the present study. A phylogenetic tree was generated on the basis of nucleotide sequence differences in the 643-nt region of the N gene ORF, which is available for these Brazilian viruses. ML analysis was conducted by using the heuristic search option and the default ML settings of PAUP* version 4b8. The lengths of horizontal branches represent numbers of substitutions per site between corresponding taxa (see scale bar). Vertical branches are for visual clarity only. Bootstrap values of greater than 50%, obtained from 1,000 replicates of the ML analysis, are shown at the appropriate branch points. Argentinian/Chilean virus genotypes are in italics, and two novel Brazilian genotypes are in boldface.
DISCUSSION
The GenBank hantavirus database contains a larger number of complete S genome segment than complete M genome segment nucleotide sequences. Since incorporating the additional genetically related taxa into a typical analysis usually leads to better phylogenetic resolution, we proceeded to amplify and sequence the entire S genome segments of the Argentinian and Chilean hantaviruses mentioned above. Not surprisingly, the S genome segment nucleotide sequences of these virus genotypes appear to be quite conservative, with the nucleotide sequence identity among different genotypes ranging from 79 to 91% (Table 2). This is reminiscent of the situation with the North American SN-like viruses, where genetically distinct genotypes specifically associated with closely related species or distinct subspecies of Peromyscus rodents reach a similar degree of genetic diversity (13, 28, 30, 43). Interestingly, the large S segment-derived ORF of the Argentinian/Chilean virus genotypes is exactly the same length (429 aa) as that of the prototype Hantaan virus (42) but is 1 aa longer than the N protein of other previously described sigmodontine rodent-borne New World hantaviruses, such as the Sin Nombre (SN), ELMC, and Bayou viruses (12, 29, 44). The N protein of the Argentinian/Chilean genotypes displays the same degree of within-group amino acid sequence identity (94 to 96%) as that of the previously investigated North American SN-like viruses (30).
In all previously characterized hantaviruses, the nucleotide sequence of the S genome segment 3′ NCR is quite variable and contains numerous imprecise oligonucleotide repeats, with the 3′ NCR length ranging from 331 nt in Prospect Hill (PH) virus to 728 nt in SN virus (37, 44). Although the Argentinian/Chilean virus genotypes are genetically closely related, they also display a significant degree of 3′ NCR length variability, with lengths ranging from 531 nt in PRG virus to 609 nt in LEC virus. The 3′ NCR length does appear to correlate directly with the genetic relatedness of these genotypes. This can be seen from the positions of the LEC (609 nt) and BMJ (604 nt) virus genotypes, as well as the MAC (540 nt) and PRG (531 nt) virus genotypes, on the phylogenetic tree (Fig. 1A to C). These closely related virus genotypes also share a greater number of the characteristic imprecise oligonucleotide repeats. Others (34) consider AND virus to be the prototype South American hantavirus that encompasses all Oligoryzomys-borne virus strains. Our studies show that it is AND virus that appears to be the most divergent member of the clade of Argentinian/Chilean viruses with respect to S segment 3′ NCR length and nucleotide sequence features (i.e., sequence divergence, as well as characteristic nucleotide repeats and short deletions/insertions). Analysis of the S segment cRNA 3′ NCR nucleotide alignment of Argentinian viruses (data not shown; alignment available upon request) reveals additional interesting features. Genetic diversity appears to vary significantly throughout the entire 3′ NCR, with several conserved “islands” surrounded by hypervariable regions. The most prominent conserved islands are located at approximately alignment positions 1410 to 1445, 1451 to 1471, 1502 to 1522, 1670 to 1700, 1795 to 1825, and 1830 to 1886 and also include the last 5′-terminal 37 nt. Some of these islands are likely to represent functional nucleotide motifs that have been implicated in transcription termination (repeated triple-G-containing motif) (16, 29, 34) or virus replication (repeated CTACCTCA motif) (34, 40) or are involved in some unknown functions. Hypervariable regions seem to evolve very rapidly. Only among the most closely related virus genotypes (for instance, among three strains of AND virus or among LEC and BMJ virus genotypes) can such regions be easily recognized as variations that have originated through the introduction of multiple nucleotide substitutions and small deletions/insertions into the common ancestral sequence. However, such comparisons of the hypervariable regions become more difficult to perform as one compares more divergent members of the clade. The ability of the complementary 3′ and 5′ termini of the genomic RNA to form panhandle-like structures is characteristic of Bunyaviridae (19, 38) and other negative-stranded RNA viruses (47, 54). In a similar manner (47, 54), the 5′/3′-terminal panhandle-like structure of hantaviruses with the mismatch at position 9 could play a role in polymerase binding and regulation of virus RNA transcription and replication.
Despite the fact that hantavirus S genome segments are generally less variable, the present analysis seems to provide additional insights into the phylogenetic relationship of the seven hantaviruses comprising the Argentinian/Chilean clade. Most of the phylogenetic reconstructions (MP with different weighting schemes, NJ, QP, and ML) place the LEC, BMJ, ORN, and Hu39694 virus genotypes together in a well-supported subclade (i.e., a smaller clade within the Argentinian/Chilean clade), with LEC and BMJ being the most closely related (Fig. 1A to C). The only exception is observed in the MP analysis when the third base positions of the N gene ORF are excluded; the ORN genotype switches closer to the AND virus (Fig. 1B). However, due to the nature of this particular analysis (i.e., absence of the most common neutral third-base substitutions), credible phylogenetic resolution (reliable bootstrap support) is not provided for the grouping of the closely related genotypes within the clade of Argentinian viruses (although the data shown in Fig. 1C suggest an ancestral link of the “LEC-like” genotypes and AND virus). In fact, the analysis without third-base positions was conducted to better resolve the phylogenetic relationships among the four major clades of the New World hantaviruses mentioned in Results.
All of the LEC-like virus genotypes are hosted by the Oligoryzomys sp. rodents from both northwestern (BMJ, associated with O. chacoensis, and ORN, associated with O. longicaudatus) and central (LEC, associated with O. flavescens) Argentina. This observation is consistent with the hypothesis that cospeciation of hantaviruses with their specific rodent hosts is a predominant pattern in the evolution of these viruses (15, 30, 33, 38, 39). Very recently, the Hu39694 virus genotype was detected in several O. flavescens rodents in central Argentina (S. Levis and D. A. Enria, personal communication). A study is under way to determine if these particular O. flavescens rodents are genetically distinct from those previously shown to carry the LEC virus genotype.
The second subclade of Argentinian viruses is composed of akodontine rodent-borne PRG and MAC virus genotypes. The present analysis places these two genotypes together, further supporting the cospeciation hypothesis. Finally, Argentinian and Chilean strains of the AND virus together form the third subclade. This is not surprising in light of our recent finding that the southern (carrying AND) and northern (carrying ORN) populations of what are currently known as O. longicaudatus rodents are genetically distinct with respect to their mitochondrial DNA (J. D. Boone and S. P. Morzunov, unpublished data). However, our present study still does not decisively resolve interrelationships among the three subclades within the major clade of Argentinian/Chilean virus genotypes.
Visual comparison of the M (23) and S (present study) genome segment-based phylogenetic trees suggests that genetic reassortment has not occurred between the individual Argentinian/Chilean virus genotypes. However, no definite conclusions can be drawn at this time, as the previously described phylogenetic analysis based on the partial M genome segment sequences (23) does not clearly resolve the phylogenetic relationship of Argentinian virus genotypes.
Phylogenetic analysis shows that the next closest relatives to the Argentinian/Chilean virus genotypes are the RM virus from Bolivia (78 to 80% nucleotide sequence similarity to the genotypes of the Argentinian/Chilean clade) and the LN virus from Paraguay (77 to 79% similarity). The RM and LN viruses are placed together, although their hosts belong to two different tribes of sigmodontine rodents. Therefore, association of the RM virus with the oryzomyine rodent O. microtis is likely to represent a host-switching event (17, 39). However, such a conclusion calls for further confirmatory study of this particular previously described virus-host association (1, 14).
Although with modest bootstrap support, two recently described human HPS case-derived virus genotypes from Brazil (18) consistently fall into two separate subclades of the Argentinian/Chilean viruses (Fig. 2). The rodent hosts of these viruses are still unknown. Based on the present analysis, one can predict that these might be akodontine and oligoryzomyine rodents for the ARA and CAS genotypes, respectively. It is noteworthy that this is the first piece of evidence suggesting, although indirectly, that akodontine rodent-borne viruses can cause HPS. Some caution should be exercised, however, as the emerging picture of the genetic diversity, geographic distribution, and host association of the South American hantaviruses appears to be quite complex (15, 33, 39). Our present data support preliminary conclusions recently drawn by Plyusnin and Morzunov (39). Cospeciation remains a dominant force driving the evolution of South American hantaviruses, as evidenced by the compact phylogenetic grouping of the majority of Oligoryzomys-borne virus genotypes, as well as by the phylogenetic grouping of the akodontine rodent-borne MAC and PRG viruses.
Conversely, factors that are known to occasionally disrupt cospeciation patterns (such as high rodent genetic diversity and sympatric coexistence of closely related rodent species) (15, 30, 33, 39) seem to play a prominent role as well. Two pieces of evidence in this study support this conclusion. A clearly “territorial” phylogenetic grouping of the akodontine rodent-borne virus genotypes in one clade with the Oligoryzomys-borne viruses in Argentina suggests an ancient host-switching event by the ancestor of the PRG and MAC viruses. In addition, the grouping of the Oligoryzomys-borne RM virus with the phyllotine rodent-borne LN virus strongly indicates a host-switching event.
There is no question that an evaluation of more rodent samples from different countries, and possibly the identification and characterization of new hantavirus genotypes, is required to complete the characterization of the phylogenetic relationships among South American hantaviruses, the taxonomic status of the different virus lineages, and their association with particular rodent host species. As mentioned above, the virus genotypes comprising the Argentinian/Chilean clade apparently display a degree of genetic diversity on both the nucleotide and amino acid levels very similar to that previously observed among the North American SN-like virus genotypes (30). However, there is a remarkable difference between the observed host ranges of these two groups of viruses. The SN-like genotypes are associated with closely related species or distinct subspecies of the same species of Peromyscus rodents. On the contrary, Argentinian/Chilean viruses (i.e., LEC-like virus genotypes, AND, MAC, and PRG) are harbored not only by several Oligoryzomys species but also by akodontine rodents. One explanation for this fact could be that rapid adaptive radiation of the ancestral oryzomyine rodents in South America (15, 39) has not yet resulted in significant changes on the cellular level, thus restricting evolution of the hantavirus N protein. Consequently, a 7% cutoff level for protein diversity (one of the hantavirus species recognition criteria established by the International Committee on Taxonomy of Viruses) might not serve as a reliable hantavirus species designation criterion for the South American hantaviruses. In addition, considering the possibility of host switching for the ancestor of the MAC and PRG virus genotypes, one should be cautious not to make any decisions on the taxonomic status of any of the virus genotypes mentioned above until additional data are collected.
Acknowledgments
Marlene C. Bohlman and Sergey P. Morzunov contributed equally to this study.
We thank Tom Ksiazek for providing rodent samples for this study.
This work was supported by National Institutes of Health grants 1R01 AI45059 and T32 CA09563.
REFERENCES
- 1.Bharadwaj, M., J. Botten, N. Torrez-Martinez, and B. Hjelle. 1997. Rio Mamore virus: genetic characterization of a newly recognized hantavirus of the pygmy rice rat, Oligoryzomys microtis, from Bolivia. Am. J. Trop. Med. Hyg. 57:368-374. [DOI] [PubMed] [Google Scholar]
- 2.Bowen, M. D., H. Kariwa, P. E. Rollin, C. J. Peters, and S. T. Nichol. 1995. Genetic characterization of a human isolate of Puumala hantavirus from France. Virus Res. 38:279-289. [DOI] [PubMed] [Google Scholar]
- 3.Childs, J. E., T. G. Ksiazek, C. F. Spiropoulou, J. W. Krebs, S. Morzunov, G. O. Maupin, K. L. Gage, P. E. Rollin, J. Sarisky, and R. E. Enscore. 1994. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J. Infect. Dis. 169:1271-1280. [DOI] [PubMed] [Google Scholar]
- 4.Chizhikov, V. E., C. F. Spiropoulou, S. P. Morzunov, M. C. Monroe, C. J. Peters, and S. T. Nichol. 1995. Complete genetic characterization and analysis of isolation of Sin Nombre virus. J. Virol. 69:8132-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dearing, M. D., A. M. Mangione, W. H. Karasov, S. Morzunov, E. W. Otteson, and S. St Jeor. 1998. Prevalence of hantaviruses in four species of Neotoma from Arizona and Utah. J. Mammal. 79:1254-1259. [Google Scholar]
- 6.Drebot, M. A., I. Gavrilovskaya, E. R. Mackow, Z. Chen, R. Lindsay, A. J. Sanchez, S. T. Nichol, and H. Artsob. 2001. Genetic and serotypic characterization of Sin Nombre-like viruses in Canadian Peromyscus maniculatus mice. Virus Res. 75:75-86. [DOI] [PubMed] [Google Scholar]
- 7.Enria, D., P. Padula, E. L. Segura, N. Pini, A. Edelstein, C. R. Posse, and M. C. Weissenbacher. 1996. Hantavirus pulmonary syndrome in Argentina. Possibility of person to person transmission. Medicina (Buenos Aires) 56:709-711. [PubMed] [Google Scholar]
- 8.Fabbri, M., and M. J. Maslow. 2001. Hantavirus pulmonary syndrome in the United States. Curr. Infect. Dis. Rep. 3:258-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldmann, H., A. Sanchez, S. Morzunov, C. F. Spiropoulou, P. E. Rollin, T. G. Ksiazek, C. J. Peters, and S. T. Nichol. 1993. Utilization of autopsy RNA for the synthesis of the nucleocapsid antigen of a newly recognized virus associated with hantavirus pulmonary syndrome. Virus Res. 30:351-367. [DOI] [PubMed] [Google Scholar]
- 10.Gonzales-Scarano, F., and N. Nathanson. 1990. Bunyaviruses, p. 1195-1238. In B. N. Fields and D. M. Knipe (ed.), Virology. Raven Press, Ltd., New York, N.Y.
- 11.Henderson, W. W., M. C. Monroe, S. C. St Jeor, W. P. Thayer, J. E. Rowe, C. J. Peters, and S. T. Nichol. 1995. Naturally occurring Sin Nombre virus genetic reassortants. Virology 234:602-610. [DOI] [PubMed] [Google Scholar]
- 12.Hjelle, B., F. Chavez-Giles, N. Torrez-Martinez, T. Yates, J. Sarisky, J. Webb, and M. Ascher. 1994. Genetic identification of a novel hantavirus of the harvest mouse, Reithrodontomys megalotis. J. Virol. 68:6751-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hjelle, B., S.-W. Lee, W. Song, N. Torrez-Martinez, J.-W. Song, R. Yanagihara, I. Gavrilovskaya, and E. R. Mackow. 1995. Molecular linkage of hantavirus pulmonary syndrome to the white-footed mouse, Peromyscus leucopus: genetic characterization of the M genome of New York virus. J. Virol. 69:8137-8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hjelle, B., N. Torrez-Martinez, and F. T. Koster. 1996. Hantavirus pulmonary syndrome-related virus from Bolivia. Lancet 347:57.. [DOI] [PubMed] [Google Scholar]
- 15.Hjelle, B., and T. Yates. 2001. Modeling hantavirus maintenance and transmission in rodent communities. Curr. Top. Microbiol. Immunol. 256:77-90. [DOI] [PubMed] [Google Scholar]
- 16.Hutchinson, K. L., C. J. Peters, and S. T. Nichol. 1996. Sin Nombre virus mRNA synthesis. Virology 234:139-149. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, A. M., M. D. Bowen, T. G. Ksiazek, R. J. Williams, R. T. Bryan, J. N. Mills, C. J. Peters, and S. T. Nichol. 1997. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology 238:115-127. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, A. M., L. T. de Souza, I. B. Ferreira, L. E. Pereira, T. G. Ksiazek, P. E. Rollin, C. J. Peters, and S. T. Nichol. 1999. Genetic investigation of novel hantaviruses causing fatal HPS in Brazil. J. Med. Virol. 59:527-535. [DOI] [PubMed] [Google Scholar]
- 19.Jonsson, C. B., and C. S. Schmaljohn. 2001. Replication of hantaviruses. Curr. Top. Microbiol. Immunol. 256:15-32. [DOI] [PubMed] [Google Scholar]
- 20.Kukkonen, S. K., A. Vaheri, and A. Plyusnin. 1998. Completion of the Tula hantavirus genome sequence: properties of the L segment and heterogeneity found in the 3′ termini of S and L genome RNAs. J. Gen. Virol. 79:2615-2623. [DOI] [PubMed] [Google Scholar]
- 21.LeDuc, J. 1987. Epidemiology of Hantaan and related viruses. Lab. Anim. Sci. 37:413-418. [PubMed] [Google Scholar]
- 22.Levis, S., J. E. Rowe, S. Morzunov, D. A. Enria, and S. St Jeor. 1997. New hantaviruses causing hantavirus pulmonary syndrome in central Argentina. Lancet 349:998-999. [DOI] [PubMed] [Google Scholar]
- 23.Levis, S., S. P. Morzunov, J. E. Rowe, D. Enria, N. Pini, G. Calderon, M. Sabattini, and S. C. St Jeor. 1998. Genetic diversity and epidemiology of hantaviruses in Argentina. J. Infect. Dis. 177:529-538. [DOI] [PubMed] [Google Scholar]
- 24.Lopez, N., P. Padula, C. Rossi, M. E. Lazaro, and M. T. Franze-Fernandez. 1996. Genetic identification of a new hantavirus causing severe pulmonary syndrome in Argentina. Virology 220:223-226. [DOI] [PubMed] [Google Scholar]
- 25.Lyubsky, S., I. Gavrilovskaya, B. Luft, and E. Mackow. 1996. Histopathology of Peromyscus leucopus naturally infected with pathogenic NY-1 hantaviruses: pathologic markers of HPS viral infection in mice. Lab. Investig. 74:627-633. [PubMed] [Google Scholar]
- 26.Mandl, C. W., F. X. Heinz, E. Puchhammer-Stockl, and C. Kunz. 1991. Sequencing the termini of capped viral RNA by 5′-3′ ligation and PCR. BioTechniques 10:484-486. [PubMed] [Google Scholar]
- 27.Mills, J. N., J. M. Johnson, T. G. Ksiazek, B. A. Ellis, P. E. Rollin, T. L. Yates, M. O. Mann, M. R. Johnson, M. L. Campbell, J. Miyashiro, M. Patrick, M. Zyzak, D. Lavender, M. G. Novak, K. Schmidt, C. J. Peters, and J. E. Childs. 1998. A survey of hantavirus antibody in small-mammal populations in selected United States national parks. Am. J. Trop. Med. Hyg. 58:525-532. [DOI] [PubMed] [Google Scholar]
- 28.Monroe, M. C., S. P. Morzunov, A. M. Johnson, M. D. Bowen, H. Artsob, T. Yates, C. J. Peters, P. E. Rollin, T. G. Ksiazek, and S. T. Nichol. 1999. Genetic diversity and distribution of Peromyscus-borne hantaviruses in North America. Emerg. Infect. Dis. 5:75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morzunov, S. P., H. Feldmann, C. F. Spiropoulou, V. A. Semenova, P. E. Rollin, T. G. Ksiazek, C. J. Peters, and S. T. Nichol. 1995. A newly recognized virus associated with a fatal case of hantavirus pulmonary syndrome in Louisiana. J. Virol. 69:1980-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morzunov, S. P., J. E. Rowe, T. G. Ksiazek, C. J. Peters, S. C. St Jeor, and S. T. Nichol. 1998. Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America. J. Virol. 72:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Netski, D., B. H. Thran, and S. C. St Jeor. 1999. Sin Nombre virus pathogenesis in Peromyscus maniculatus. J. Virol. 73:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichol, S. T., C. F. Spiropoulou, S. Morzunov, P. E. Rollin, T. G. Ksiazek, H. Feldmann, A. Sanchez, J. Childs, S. Zaki, and C. J. Peters. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262:914-917. [DOI] [PubMed] [Google Scholar]
- 33.Nichol, S. T. 1999. Genetic analysis of hantaviruses and their host relationships, p. 99-109. In J. F. Saluzzo and B. Dodet (ed.), Emergence and control of rodent-borne viral diseases. Elsevier Publishers, Paris, France.
- 34.Padula, P. J., S. B. Colavecchia, V. P. Martinez, D. Gonzalez, V., A. Edelstein, S. D. Miguel, J. Russi, J. M. Riquelme, N. Colucci, M. Almiron, and R. D. Rabinovich. 2000. Genetic diversity, distribution, and serological features of hantavirus infection in five countries in South America. J. Clin. Microbiol. 38:3029-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Padula, P. J., A. Edelstein, S. D. Miguel, N. M. Lopez, C. M. Rossi, and R. D. Rabinovich. 1998. Hantavirus pulmonary syndrome outbreak in Argentina: molecular evidence for person-to-person transmission of Andes virus. Virology 241:323-330. [DOI] [PubMed] [Google Scholar]
- 36.Parisi, M., D. Enria, N. C. Pini, and M. S. Sabattini. 1995. Deteccion retrospectiva de infecciones clinicas por hantavirus en la Argentina. Medicina (Buenos Aires) 56:1-13. [PubMed] [Google Scholar]
- 37.Parrington, M. A., and C. Y. Kang. 1990. Nucleotide sequence analysis of the S genomic segment of Prospect Hill virus: comparison with the prototype hantavirus. Virology 175:167-175. [DOI] [PubMed] [Google Scholar]
- 38.Plyusnin, A., O. Vapalahti, and A. Vaheri. 1996. Hantaviruses: genome structure, expression and evolution. J. Gen. Virol. 77:2677-2687. [DOI] [PubMed] [Google Scholar]
- 39.Plyusnin, A., and S. P. Morzunov. 2001. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr. Top. Microbiol. Immunol. 256:47-75. [DOI] [PubMed] [Google Scholar]
- 40.Ravkov, E. V., P. E. Rollin, T. G. Ksiazek, C. J. Peters, and S. T. Nichol. 1995. Genetic and serologic analysis of Black Creek Canal virus and its association with human disease and Sigmodon hispidus infection. Virology 210:482-489. [DOI] [PubMed] [Google Scholar]
- 41.Rowe, J. E., S. C. St Jeor, J. Riolo, E. W. Otteson, M. C. Monroe, W. W. Henderson, T. G. Ksiazek, P. E. Rollin, and S. T. Nichol. 1995. Coexistence of several novel hantaviruses in rodents indigenous to North America. Virology 213:122-130. [DOI] [PubMed] [Google Scholar]
- 42.Schmaljohn, C. S., S. E. Hasty, S. A. Harrison, and J. M. Dalrymple. 1983. Characterization of Hantaan virions, the prototype virus of hemorrhagic fever with renal syndrome. J. Infect. Dis. 148:1005-1012. [DOI] [PubMed] [Google Scholar]
- 43.Song, J. W., L. J. Baek, I. N. Gavrilovskaya, E. R. Mackow, B. Hjelle, and R. Yanagihara. 1996. Sequence analysis of the complete S genomic segment of a newly identified hantavirus isolated from the white-footed mouse (Peromyscus leucopus): phylogenetic relationship with other sigmodontine rodent-borne hantaviruses. Virus Genes 12:249-256. [DOI] [PubMed] [Google Scholar]
- 44.Spiropoulou, C. F., S. Morzunov, H. Feldmann, A. Sanchez, C. J. Peters, and S. T. Nichol. 1994. Genome structure and variability of a virus causing hantavirus pulmonary syndrome. Virology 200:715-723. [DOI] [PubMed] [Google Scholar]
- 45.Swofford, D. L. 1991. PAUP: phylogenetic analysis using parsimony, version 3.1.1. Illinois Natural History Survey, Champaign.
- 46.Swofford, D. L. 1998. PAUP*, phylogenetic analysis using parsimony (* and other methods), version 4.0b8. Sinauer, Sunderland, Mass.
- 47.Tiley, L. S., M. Hagen, J. T. Matthews, and M. Krystal. 1994. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J. Virol. 68:5108-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toro, J., J. D. Vega, A. S. Khan, J. N. Mills, P. Padula, W. Terry, Z. Yadon, R. Valderrama, B. A. Ellis, C. Pavletic, R. Cerda, S. Zaki, W. J. Shieh, R. Meyer, M. Tapia, C. Mansilla, M. Baro, J. A. Vergara, M. Concha, G. Calderon, D. Enria, C. J. Peters, and T. G. Ksiazek. 1998. An outbreak of hantavirus pulmonary syndrome, Chile, 1997. Emerg. Infect. Dis. 4:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasconcelos, M. I., V. P. Lima, L. B. Iversson, M. D. Rosa, A. P. da Rosa, E. S. da Rosa, L. E. Pereira, E. Nassar, G. Katz, L. H. Matida, M. A. Zaparoli, J. J. Ferreira, and C. J. Peters. 1997. Hantavirus pulmonary syndrome in the rural area of Juquitiba, Sao Paulo metropolitan area, Brazil. Rev. Inst. Med. Trop. Sao Paulo 39:237-238. [DOI] [PubMed] [Google Scholar]
- 50.Vincent, M. J., E. Quiroz, F. Gracia, A. J. Sanchez, T. G. Ksiazek, P. T. Kitsutani, L. A. Ruedas, D. S. Tinnin, L. Caceres, A. Garcia, P. E. Rollin, J. N. Mills, C. J. Peters, and S. T. Nichol. 2000. Hantavirus pulmonary syndrome in Panama: identification of novel hantaviruses and their likely reservoirs. Virology 277:14-19. [DOI] [PubMed] [Google Scholar]
- 51.Weigler, B. J. 1995. Zoonotic hantaviruses: new concerns for the United States. J. Am. Vet. Med. Assoc. 206:979-987. [PubMed] [Google Scholar]
- 52.Weissenbacher, M. C., M. S. Merani, V. L. Hodara, G. de Villafane, D. C. Gajdusek, Y. K. Chu, and H. W. Lee. 1990. Hantavirus infection in laboratory and wild rodents in Argentina. Medicina (Buenos Aires) 50:43-46. [PubMed] [Google Scholar]
- 53.Wells, R. M., E. S. Sosa, Z. E. Yadon, D. Enria, P. Padula, N. Pini, J. N. Mills, C. J. Peters, E. L. Segura, et al. 1997. An unusual hantavirus outbreak in southern Argentina: person-to-person transmission? Emerg. Infect. Dis. 3:171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wertz, G. W., S. Whelan, A. LeGrone, and L. A. Ball. 1994. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc. Natl. Acad. Sci. USA 91:8587-8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams, R. J., R. T. Bryan, J. N. Mills, R. E. Palma, I. Vera, F. De Velasquez, E. Baez, W. E. Schmidt, R. E. Figueroa, C. J. Peters, S. R. Zaki, A. S. Khan, and T. G. Ksiazek. 1997. An outbreak of hantavirus pulmonary syndrome in western Paraguay. Am. J. Trop. Med. Hyg. 57:274-282. [DOI] [PubMed] [Google Scholar]
- 56.Zaparoli, M. A., L. B. Iversson, M. D. Rosa, E. Travassos da Rosa, L. E. Pereira, P. Rollin, and C. J. Peters. 1995. Investigation on case-contacts of human disease caused by hantavirus in Juquitiba, state of Sao Paulo, Brazil. Am. J. Trop. Med. Hyg 53(Suppl.):232-233. [Google Scholar]