Abstract
The emergence of drug-resistant variants has posed a significant setback against effective antiviral treatment for human immunodeficiency virus (HIV) infections. The choice of a nonmutable region of the viral genome such as the conserved transactivation response element (TAR element) in the 5′ long terminal repeat (LTR) may potentially be an effective target for drug development. We have earlier demonstrated that a polyamide nucleotide analog (PNA) targeted to the TAR hairpin element, when transfected into cells, can effectively inhibit Tat-mediated transactivation of HIV type 1 (HIV-1) LTR (T. Mayhood et al., Biochemistry 39:11532-11539, 2000). Here we show that this anti-TAR PNA (PNATAR), upon conjugation with a membrane-permeating peptide vector (transportan) retained its affinity for TAR in vitro similar to the unconjugated analog. The conjugate was efficiently internalized into the cells when added to the culture medium. Examination of the functional efficacy of the PNATAR-transportan conjugate in cell culture using luciferase reporter gene constructs resulted in a significant inhibition of Tat-mediated transactivation of HIV-1 LTR. Furthermore, PNATAR-transportan conjugate substantially inhibited HIV-1 production in chronically HIV-1-infected H9 cells. The mechanism of this inhibition appeared to be regulated at the level of transcription. These results demonstrate the efficacy of PNATAR-transportan as a potential anti-HIV agent.
In AIDS patients, rapid turnover and high mutation rates result in a diverse population of human immunodeficiency virus type 1 (HIV-1) quasispecies. Efforts to control the AIDS epidemic have relied on interactions between viral proteins or enzymes and new drug candidates. Current combination therapy that utilizes drug cocktails of inhibitors of viral protease and reverse transcriptase can initially reduce the viremia levels (1). However, the rapid emergence of variants due to mutations in the reverse transcriptase and/or protease gene eventually make chemotherapy ineffective.
To overcome the barrier of virus mutability and consequent drug resistance, the viral targets selected for drug intervention should include conserved regulatory regions resistant to mutational changes. The 5′ nontranslated region of the viral genome (5′ long terminal repeat [LTR]) comprises several important domains essential for viral replication and gene expression. These regions individually or collectively could be potential targets for drug design and development.
The HIV-1 LTR contains a unique region known as the transactivation response (TAR) region which is critical for transcriptional activation by the transactivator protein Tat (13). The TAR element extends between nucleotides +1 and +59 and forms a unique, stable stem-loop RNA structure (3, 46). Intensive research over the last decade on the transactivation mechanism governing the Tat-TAR interaction has yielded significant biological and virological insights. It has now been established that the Tat protein of HIV-1 is a potent transactivator of viral gene expression and is essential for viral replication (25).
The primary role of Tat is in regulating productive and processive transcription after binding to its RNA target, the TAR element in the HIV-1 LTR (21). The structure of TAR has been well studied, and the salient features essential for its interaction with Tat, centering on a U-rich bulge near the apex of the loop, have been characterized (10). The structural integrity of TAR is important because natural or induced mutations that destabilize TAR by disrupting base pairing in the stem region abolish Tat-stimulated transcription, resulting in premature transcription termination at random locations downstream of the viral RNA start site (59). Thus, given the functional importance of the Tat-TAR interaction to the viral life cycle, the Tat protein and the TAR element both present attractive targets for drug design.
Agents affecting the interactions of Tat and TAR prevent transcriptional activation of the HIV-1 genome either by steric hindrance, a shear displacement mechanism, or deprivation of the functional molecules. For example, shielding the bulge-loop region of TAR with Tat peptides or analogs was shown to inhibit transactivation of HIV-1 LTR (6, 14, 26). Alteration of the stem-loop conformation of TAR by neomycin induces dissociation of Tat from TAR by an allosteric mechanism (65). In other strategies, either Tat protein or TAR RNA has been captured by a TAR RNA decoy (4, 37, 40, 60), TAR circle (3), anti-Tat monoclonal antibody and single-chain antibody (41, 54), ALX40-C (an oligocationic peptide) (49), or CGP64222 (a 9-residue basic oligomer of Tat) (19) to intervene in their functional interaction, thereby reducing transcription and ultimately the viral load.
The oligodeoxyribonucleotides complementary to the stem-loop and bulge regions of the TAR RNA have been employed successfully to block transcriptional activation of the HIV-1 viral genome. Several chemical modifications have been made in oligodeoxyribonucleotides in order to improve their stability and effectiveness in the cellular milieu. The therapeutic potential of phosphoramides has recently been evaluated against the HIV-1 untranslated region as the target (11). In 1991, Nielsen described the synthesis of a polyamide nucleotide analog (PNA) in which the bases are linked with peptide bonds instead of a sugar phosphate backbone (48). The PNA molecules are resistant to nucleases and proteases (8) and have strong affinity for the cDNA or RNA sequences (31). Its ability to form an energetically favorable structure that can bind to its target in an orientation-independent manner has made it an excellent molecule for gene expression studies, but the low solubility of PNA in aqueous media and its inefficient cellular uptake are major drawbacks.
Recently, methods have been developed for the delivery of exogenous proteins into living cells with the help of membrane-permeating carrier peptides derived from HIV-1 Tat (residues 48 to 60) and antennapedia (residues 43 to 58) (9, 12, 57). The basic nature of these peptides and the locations of aromatic groups on the peptides enable them to penetrate the cell membrane. Using a range of basic peptides, Futaki et al. have demonstrated that a peptide containing eight arginine residues can efficiently translocate across the membrane (16). Recently, a chimeric peptide derived from galparan and transportan has been used as an effective peptide vector for biodelivery of PNA molecules (52).
Previously, we have shown that transfection of an anti-TAR PNA complementary to the stem-loop and bulge regions of HIV-1 TAR can effectively inhibit the Tat-mediated transactivation of HIV-1 LTR in CEM cells (40). In this report, we conjugated the anti-TAR PNA with a membrane-permeating peptide vector, transportan, and examined the functional efficacy and antiviral activity of this chimeric molecule (PNATAR-transportan conjugate) in cell culture. Our results demonstrate that the PNATAR-transportan conjugate retained affinity for its target sequence and was able to effectively block Tat-mediated transactivation and HIV-1 production when added to the culture medium.
MATERIALS AND METHODS
PNA oligomers.
The sequence of PNATAR-transportan conjugate, tetramethylrhodamine (TAMRA)-labeled PNA-transportan conjugate, and scrambled-PNA-transportan conjugate are shown in Fig. 1. PNA oligomers and their conjugated derivatives were obtained from Applied Biosystems. To avoid the problem of precipitation of PNA at high concentrations, the working stocks of PNA oligomers as well as the PNA-transportan conjugates were maintained at a 100 μM concentration and stored at 4°C as recommended. The stocks were prepared by dissolving the PNA in water and heating to 50°C for 10 min, followed by incubation at 37°C for 30 min prior to quantification. Unused portions of the PNA stocks were aliquoted after 2 weeks, lyophilized, and stored at 4°C. The TAMRA-labeled PNA was protected from light to avoid photobleaching during its preparation and during the experiments.
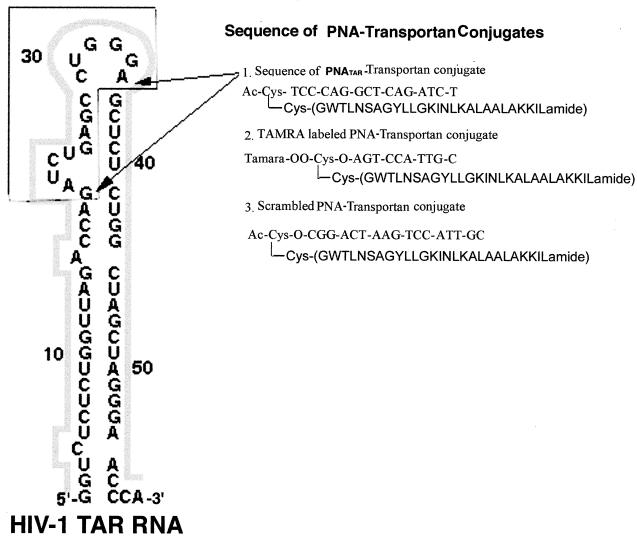
FIG. 1.
Sequence of PNA-transportan conjugates. Secondary structure of the HIV-1 TAR RNA is as shown. The TAR sequences interacting with the PNATAR-transportan conjugate are indicated by arrows. The inset corresponds to the sequences of PNATAR, TAMRA-tagged PNA, and scrambled PNA conjugated with the 27-amino-acid transportan peptide.
Plasmid constructs.
Plasmids for expression in mammalian cells were as follows: pHIV-1 LTR-Luc (a kind gift from M. B. Mathews), containing the firefly luciferase gene cloned downstream of the HIV-1 LTR; pcDNA3-Tat (pCMV-Tat), encoding the 72-amino-acid Tat protein under the control of the cytomegalovirus (CMV) promoter (15); pCMV-R.Luc (Promega, Madison, Wis.), for expressing Renilla luciferase under the control of the CMV promoter; and pcDNA3.1 (Invitrogen Corp., Carlsbad, Calif.), encoding the CMV promoter.
Plasmids used for preparing transcripts for gel shift assays.
pEM7 and pTAR-BS were used for generating the wild-type TAR RNA and its mutant derivative carrying a deletion at the top of the stem (18), and pET-28a-RT was used for transcribing a 427-base-long HIV-1 reverse transcriptase coding fragment (27).
Plasmids used for generating probes for RNase protection assay.
The plasmids used for generating probes for the RNase protection assay were as follows: plasmids pSP-luc+ and pSP-rluc (Promega), encoding the firefly and Renilla luciferase gene cassettes, respectively, and having an opposing T7 promoter located downstream of the luc+ and rluc insert, whereas pGEM23 (34) has an opposing SP6 promoter located downstream of the TAR gene insert.
Cell culture and transfection.
CEM and Jurkat T-cell lymphocytes were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 4 mM l-glutamine, 100 U of penicillin, and 100 μg of streptomycin per ml at 37°C in 5% CO2-containing humidified air. The chronically HIV-1-infected H9 cells were maintained under identical conditions except that they were supplemented with 20% fetal calf serum. Jurkat T cells (5.0 × 106 cells in 300 μl of RPMI 1640 supplemented with 20% fetal calf serum) were transfected with requisite amounts of the experimental plasmids pHIV-1 LTR-Luc and pCMV-Tat by electroporation using a Bio-Rad gene pulser II at 280 V and 975 μF capacitance. In order to monitor the efficiency of transfection, the cells were cotransfected with the reporter plasmid pCMV-R.Luc. The cells were stimulated with phorbol 12-myristate 13-acetate (PMA; 20 ng/ml) and the calcium ionophore A23187 (1 μM) at 24 h posttransfection.
CEM cells were washed with equal volumes of phosphate-buffered saline (PBS) (without Ca2+ or Mg2+) before transfection. After washing, cells were resuspended in unsupplemented RPMI 1640 medium (5.0 × 106 cells in 250 μl) and electroporated with experimental and reporter plasmids at 230 V and 800 μF capacitance. Cells were then grown in 10 ml of complete RPMI 1640 medium.
Cellular uptake of PNA-transportan conjugate.
The uptake of TAMRA-tagged PNA-transportan conjugate was examined in CEM cells and Jurkat cells. Briefly, the log-phase cells were washed with PBS and resuspended in a six-well plate in serum-free RPMI medium at a cell density of 3 × 105 cells/ml. The TAMRA-PNA-transportan conjugate (75 nM) was added to the culture medium, and the cells were incubated at 37°C. At various time intervals, aliquots (700 μl) of the cells were removed, washed with PBS, and examined by fluorescence microscopy. Simultaneously, the effect of PNA-transportan conjugate on cell viability was examined using the calcein AM component from the live-dead viability kit (Molecular Probes) as per the manufacturer's protocol.
Electrophoretic mobility shift assay.
The affinity and specificity of the PNATAR-transportan conjugate for the TAR RNA was examined by gel mobility shift analysis using the wild-type TAR RNA probe, a mutant TAR RNA probe, and the HIV-1 reverse transcriptase RNA probe. The 32P-labeled TAR and TAR BS mutant RNA probes were prepared by in vitro transcription of the linearized plasmid templates as described previously (40). The 427-base HIV-1 reverse transcriptase (RT) RNA probe was generated by digesting pET-28a-RT with EcoRV and transcribing it using the T7 RNA polymerase as per the manufacturer's protocol (Roche Biochemicals). TAR RNA transcript (5 × 103 Cerenkov cpm) was incubated with PNATAR-transportan conjugate, unconjugated PNATAR, or scrambled PNA-transportan conjugate at various molar ratios. In another set, PNATAR-transportan conjugate was incubated with the mutant TAR BS or the HIV-1 RT radiolabeled probe at various molar ratios in order to evaluate its binding specificity.
Incubations were carried out for 3 h at 37°C in a binding buffer containing 50 mM Tris-HCl (pH 7.8), 60 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol (DTT), 10% glycerol, 0.01% bovine serum albumin, 0.01% NP-40, and 500 ng of poly(rI-rC), in a final volume of 15 μl. Then 3 μl of RNA gel loading dye (0.27% bromophenol blue and 20% glycerol) was added to the samples and subjected to gel retardation analysis on native 6% polyacrylamide gel in Tris-borate-EDTA (TBE) buffer, pH 8.2. The gels were routinely run at 100 V for 30 min at 4°C in TBE buffer. The various complexes were resolved at a constant voltage of 150 V at 4°C. The gels were dried, visualized on a PhosphorImager (Molecular Dynamics), and quantified using Image-Quant software (Molecular Dynamics).
Reverse transcription of TAR RNA transcript.
Reverse transcription catalyzed by HIV-1 RT on the TAR RNA transcript in the presence or absence of PNATAR-transportan conjugate, unconjugated PNATAR, or scrambled PNA-transportan conjugate was monitored by extension of 5′-32P-labeled 17-mer DNA primer annealed with the TAR RNA template. The PNATAR-transportan conjugate or PNATAR at 1 μM concentration was incubated with 10 nM annealed template-primer at 37°C for 1 h in a reaction buffer containing 50 mM Tris-HCl (pH 7.8), 10 mM DTT, 100 μg of bovine serum albumin per ml, 60 mM KCl, and 5 mM MgCl2 and used in the extension reaction. Reverse transcription was initiated by the addition of 50 nM HIV-1 RT and 100 μM each of the four deoxynucleoside triphosphates. The reaction mixture was incubated at 25°C, and the reaction was terminated at the indicated time by the addition of an equal volume of Sanger's gel loading solution (56). The products were resolved on an 8% polyacrylamide-urea gel and visualized by PhosphorImager.
Assay for Tat-mediated transactivation of HIV-1 LTR.
The amounts of Tat required for the transactivation of the HIV-1 LTR were optimized in Jurkat and CEM cells. In Jurkat cells, the plasmid pHIV-1 LTR-Luc (4 μg) was cotransfected with pCMV-R.Luc (1 μg) and various amounts of the Tat expression clone (pCMV-Tat) and electroporated under standard conditions as described above. The cells were plated in 10 ml of complete RPMI 1640 medium and stimulated with PMA and the Ca2+ ionophore A23187 at 24 h posttransfection. Following 8 h of stimulation, the cells were harvested and assayed for luciferase activity.
In another set of experiments, CEM cells were transfected with identical amounts of the plasmid cocktail comprising pHIV-1 LTR-Luc and pCMV-R.Luc and various amounts of plasmid pCMV-Tat under standard reaction conditions and assayed for luciferase activity at 16 h posttransfection.
Tat-mediated transactivation of HIV-1 LTR in cell culture in the presence of PNATAR-transportan conjugate.
The effect of the PNATAR-transportan conjugate on Tat-mediated transactivation of HIV-1 LTR was assessed in both CEM and Jurkat cells as described below. CEM and Jurkat cells (5.0 × 106 cells) were electroporated with a plasmid cocktail comprising pHIV-1 LTR-Luc (4 μg) and pCMV-R.Luc (1 μg) in the presence or absence of optimal amounts of plasmid pCMV-Tat as determined from the above experiment. The Jurkat and CEM cells were allowed to recover from the effects of electroporation in complete RPMI medium containing 10% fetal bovine serum for 12 h and 3 h, respectively.
To facilitate the uptake of PNA-transportan conjugate, the cells were washed with PBS and resuspended in RPMI medium containing various concentrations of the PNATAR-transportan conjugate. The Jurkat cells were then stimulated with PMA and Ca2+ ionophore at 20 h posttransfection and harvested 8 h after stimulation. The CEM cells were harvested at 20 h posttransfection. The cells were analyzed for the luciferase reporter activities in order to evaluate the effect of the PNATAR-transportan on Tat-mediated transactivation of the HIV-1 LTR. Identical sets of experiments were carried out to determine the effect of the unconjugated PNATAR, scrambled PNA-transportan conjugate, and transportan on Tat-mediated transactivation of the HIV-1 LTR.
In a similar set of experiments, the CEM and Jurkat cells were electroporated with the plasmid pCMV-R.Luc and incubated in the presence or absence of PNATAR-transportan, unconjugated PNATAR, scrambled PNA-transportan conjugate, or transportan. The cells were harvested at 16 to 24 h posttransfection and analyzed for Renilla luciferase activity as described below.
Luciferase assays.
Luciferase assays were performed by using the Promega dual luciferase assay kit. Briefly, the cells harvested at 1,500 rpm for 7 min were washed once with PBS and lysed by the addition of 50 μl of the reporter lysis buffer (Promega). Following incubation at 25°C for 15 min on a rocking shaker, the lysates were centrifuged at 15,000 rpm for 10 min, and the supernatant was collected in fresh tubes. Luciferase assays were performed by mixing 30 μl of the supernatant with 75 μl of the firefly luciferase substrate in a 96-well Fluorotrac plate, and the light emission was measured using a Packard Top Count luminescence counter. The firefly luciferase activity was quenched by the addition of Stop and Glo reagent, which also served as a substrate for estimating the Renilla luciferase activity. Transfection efficiencies were normalized by the expression levels of the Renilla luciferase reporter gene construct cotransfected along with the experimental plasmid. The results of at least three separate transfections were analyzed for each experiment.
HIV-1 production in H9 cells in the presence of PNATAR-transportan conjugate.
The chronically HIV-1-infected H9 cells were centrifuged and washed extensively in PBS to remove the previously produced virions. The cells were suspended in 1.0 ml of serum-free RPMI 1640 medium at 5 × 106 cells/well and incubated in a six-well culture plate at 37°C for 6 h in the absence or presence of various concentrations of PNATAR-transportan conjugate, unconjugated PNATAR, scrambled PNA-transportan conjugate, or transportan alone. The cells were then reconstituted to a final cell density of 106 cells/ml in complete RPMI 1640 medium and incubated at 37°C. The cells were harvested on the third day at 1,200 rpm for 7 min, and the levels of p24 antigen were analyzed in the supernatants with an enzyme-linked immunosorbent assay (ELISA) p24 antigen kit (Abbott Laboratories). The cells were also analyzed for HIV-1 mRNA levels after total RNA isolation and RNase protection analysis as described below.
Total RNA isolation and RNase protection assay.
The plasmids pSP-luc+, pSP-rluc, and pGEM23 were linearized with the restriction enzymes HincII, BsaAI, and XbaI, respectively. The former two linearized plasmids were used to generate radioactively labeled riboprobes for the firefly and Renilla luciferase corresponding to 390 nucleotides and 245 nucleotides, respectively. Briefly, the digested plasmids were transcribed using T7 RNA polymerase in the presence of [α-32P]UTP (Perkin Elmer Life Sciences, Inc.) as per the MAXIscript in vitro transcription kit (Ambion Inc., Austin, Tex.). A 195-nucleotide-long riboprobe for the TAR RNA was synthesized from linearized pGEM23 with SP6 RNA polymerase (New England Biolabs) and [α-32P]UTP under standard reaction conditions. The DNA templates were removed by DNase I digestion, and the RNA probes were purified by gel electrophoresis.
For the RNase protection assay, transfection and other experimental conditions as described above were maintained in the CEM, Jurkat, and H9 cells. The cells were harvested at the stated time and washed with PBS, and total RNA was isolated from the immortalized cell lines using the RNAqueous kit (Ambion Inc., Austin, Tex.). Seven micrograms of total RNA extracted from the experimental and control samples was hybridized to 7 × 104 to 10 × 104 cpm of the individual riboprobe and analyzed as per the RPA III kit (Ambion Inc., Austin, Tex.). Protected fragments were separated on a 6% polyacrylamide-urea gel and detected by PhosphorImager analysis.
Determination of [3H]thymidine incorporation into cellular DNA in chronically HIV-1-infected H9 cells.
Cellular proliferation of chronically HIV-1-infected H9 cells in the presence of PNATAR-transportan conjugate was determined by estimating the levels of [3H]thymidine incorporated in their nuclei. Briefly, freshly split cells were grown in the presence or absence of PNATAR-transportan conjugate (5 μM concentration) and supplemented with 10 μCi of [methyl-3H]thymidine/ml (83.7 Ci/mmol). The cells were withdrawn at 3, 12, 24, 36, and 48 h, and the cell number was determined using a Coulter counter. The cells were harvested, washed with PBS, and resuspended in 200 μl of lysis buffer containing 1% NP-40 in PBS. The nucleic acids were precipitated by adding cold 10% trichloroacetic acid (TCA). Precipitates were collected on GF/C glass fiber filters (Whatman, Inc., Maidstone, Kent, England) and washed extensively with ice-cold 10% TCA and once in 70% ethanol. Filters were dried and placed in scintillation vials, and radioactivity was counted in the scintillation counter. Protein content in each lysate was estimated by the Bio-Rad protein assay. Results were expressed as counts per minute per milligram of protein.
RESULTS
Synthesis of PNA and PNA-transportan conjugates.
PNA and PNA-transportan conjugates were synthesized at Applied Biosystems. The PNA molecules are linked to the transportan peptide through a disulfide linkage, as shown in Fig. 1. Secondary structure of the HIV-1 TAR RNA is also shown in this figure. The TAR sequences interacting with the PNATAR-transportan conjugate are indicated by arrows. The molecular mass of the PNATAR-transportan conjugate was analyzed by matrix-assisted laser desorption ionization-time of flight-mass spectrometry (MALDI-TOF-MS). The observed mass of the conjugate (7,382.3 Da) corresponded to the calculated mass (7,377.3 Da) with a variance of −0.07%.
Affinity of PNATAR-transportan conjugate to TAR.
In order to ascertain whether PNATAR conjugated with the transportan peptide vector retained its binding affinity for its target sequence on the TAR RNA, we performed gel mobility shift assays with the 32P-labeled 82-base-long TAR RNA transcript and various concentrations of PNATAR-transportan conjugate, scrambled PNA-transportan conjugate, or unconjugated PNA TAR (Fig. 2A to C). Titration of PNATAR-transportan conjugate with TAR RNA at molar ratios of the conjugate to TAR RNA of less than 1 resulted in a stoichiometric band shift of the labeled TAR RNA (Fig. 2A, lanes 2 to 4).
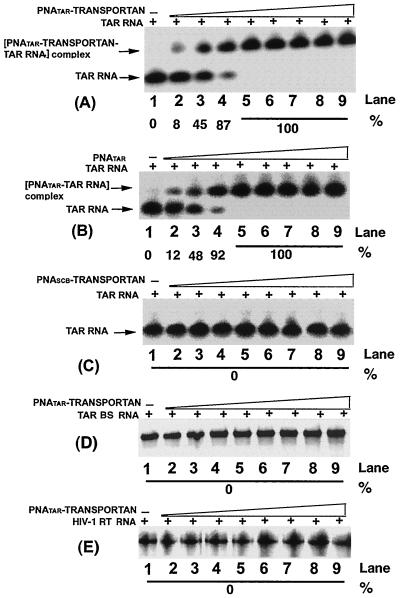
FIG. 2.
Binding specificity of PNATAR-transportan conjugate to its target sequence. The affinity of the PNATAR-transportan conjugate (A) for its target sequence on the TAR RNA was assessed by gel mobility shift analysis as described in Materials and Methods. Unconjugated PNATAR (B) was included as a positive control, and scrambled PNA-transportan conjugate (PNASCB-transportan) (C) was included as a negative control. The specificity of this interaction was determined by analyzing the relative gel mobility shift upon interaction of PNATAR-transportan conjugate with a mutant TAR RNA (TAR BS; panel D) as well as with an unrelated RNA such as the HIV-1 RT RNA (panel E). In panel A, lanes 1 through 9 represent molar ratios of PNATAR-transportan conjugate to TAR RNA of 0, 0.1, 0.5, 0.8, 1.0, 2.5, 5, 7.5, and 10, respectively. Panel B and panel C show gel mobility shifts performed at similar ratios as indicated in panel A except that unconjugated PNATAR and scrambled PNA-transportan conjugate, respectively, were used. Similar ratios are also shown in panels D and E except that the mobility shift of the PNATAR-transportan conjugate was carried out with the TAR BS and HIV-1 RT RNA probes, respectively. The extent of gel shift was determined by quantifying the probe RNA band on the PhosphorImager using Image-Quant software (Molecular Dynamics). The percentage of labeled TAR RNA retarded due to PNA binding is indicated.
As shown in the figure, at molar ratios of 0.1, 0.5, and 0.8 of the conjugate to labeled TAR RNA, the extent of band shift corresponded to approximately 8, 45, and 87%, respectively. A complete shift in the mobility was achieved at an equimolar ratio or a molar excess of the conjugate to TAR RNA (panel A, lanes 5 to 9). A similar titration was carried out in the presence of unconjugated PNATAR (panel B, lanes 2 to 9) and scrambled PNA-transportan conjugate (panel C, lanes 2 to 9). As shown in the figure, the binding of PNATAR to the TAR RNA was similar to that obtained with the PNATAR-transportan conjugate, suggesting that conjugation of PNATAR with the transportan peptide vector had no effect on its binding affinity. Scrambled PNA-transportan conjugate at similar molar ratios did not result in any gel shift, suggesting the specificity of the interaction. Further evidence of this specificity was demonstrated by our observation that interaction of PNATAR-transportan conjugate with TAR BS, a 63-base-long mutant TAR RNA carrying a deletion in the stem and bulge region (panel D), and with HIV-1 RT RNA, an unrelated RNA (panel E), also exhibited no shift in mobility.
Reverse transcription of TAR RNA region is blocked by PNATAR-transportan conjugate.
Since the PNATAR-transportan conjugate displayed affinity for TAR RNA similar to unconjugated PNATAR, it was interesting to examine if the conjugate could effectively block reverse transcription of TAR RNA. Ability to block reverse transcription, in turn, would have multiple impacts on viral replication besides influencing Tat-mediated transactivation. For this purpose, the TAR RNA transcript primed with the labeled 17-mer DNA primer was incubated in the absence or presence of 100 nM PNATAR-transportan conjugate, PNATAR, or scrambled PNA at 37°C, followed by initiation of reverse transcription by HIV-1 RT. The results are presented in Fig. 3. PNATAR and PNATAR-transportan conjugate showed a similar pattern of reverse transcription, with a prominent pause at nucleotide position 42, prior to the loop site targeted by these two PNAs. These results suggest that conjugation of PNATAR with transportan did not alter the ability of PNATAR to block reverse transcription of TAR RNA.
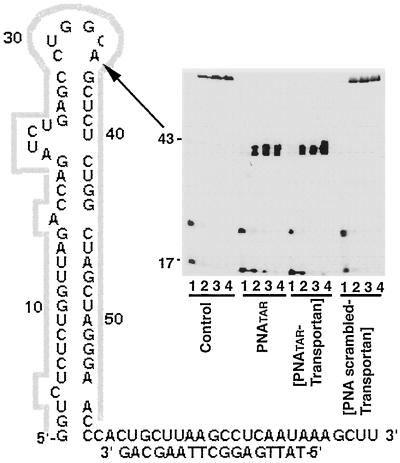
FIG. 3.
Effect of PNATAR-transportan conjugate on reverse transcription on TAR RNA primed with the 17-mer DNA primer. TAR RNA template primed with the 5′-32P-labeled 17-mer DNA primer was incubated in the absence or presence of PNATAR-transportan conjugate, unconjugated PNATAR, or scrambled PNA-transportan conjugate at 37°C for 1 h and used in the gel extension reaction as described under Materials and Methods. The reaction products were analyzed on a denaturing 8% polyacrylamide-urea gel and subjected to PhosphorImager analysis. Lanes 1 through 4 in each set represent the extension reactions at 25°C for 5, 10, 15, and 20 min, respectively. The control set represents reactions carried out in the absence of PNATAR-transportan conjugate, unconjugated PNATAR, or scrambled PNA-transportan conjugate. The arrow indicates the position on the gel corresponding to the sequence targeted by PNATAR-transportan conjugate as well as unconjugated PNATAR on TAR RNA.
Cellular uptake of PNA-transportan and its intracellular localization.
Although PNAs display strong binding affinity to their target sequence and have great potential as antisense agents, their biodelivery in the cell is very poor (47). The biodelivery of PNA into cells has been enhanced by cationic detergents (39) and by linking them with membrane-translocating peptides (51). To evaluate the uptake of the PNA-transportan conjugate in our system, we used a transportan-conjugated 10-mer PNA tagged with a TAMRA fluorophore probe. The entry of the dye-linked conjugate into the cells was then monitored by fluorescence microscopy and analyzed by fluorescence-activated cell sorting (FACS) analysis. The results are shown in Fig. 4 and Table 1.
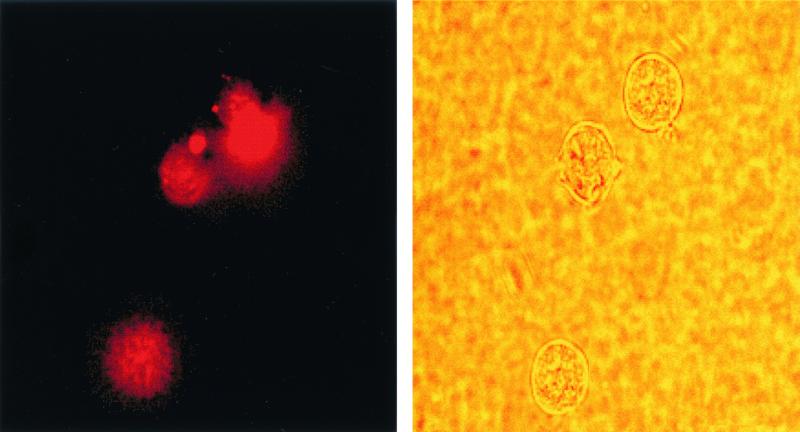
FIG. 4.
Cellular uptake of PNA-transportan conjugate. CEM cells were incubated with 70 nM TAMRA-tagged PNA-transportan conjugate in a six-well plate in RPMI 1640 medium in the absence of serum. At various times, aliquots of the cells were washed, resuspended in PBS, and examined by fluorescence microscopy for monitoring cellular uptake. The panel on the left is the fluorescence image, and that on the right is the phase-contrast image of the same field.
TABLE 1.
Time course of TAMRA-tagged PNA-transportan conjugate uptake in cellsa
| Cells | Uptake of PNA-transportan (%) at time posttransfection:
|
||||
|---|---|---|---|---|---|
| 1 h | 2 h | 4 h | 6 h | 12 h | |
| CEM | 5 | 12 | 35 | 55 | 90 |
| Jurkat | 0 | 3 | 16 | 27 | 52 |
The extent of uptake of TAMRA-tagged PNA-transportan conjugate was monitored in CEM and Jurkat cells as described in Materials and Methods. The values represent averages of three independent experiments.
It was observed that the PNA-transportan conjugate is able to enter the cells in a time-dependent manner, confirming that transportan linked with PNA retained its ability to cross the cell membrane (Fig. 4). At 12 h, approximately 90% of the CEM cells were found to display fluorescence, as against 52% of the Jurkat cells (Table 1). This suggests that the PNA-transportan conjugate can efficiently cross the membrane of both cell types, although the kinetics of cell entry varied. Furthermore, the PNA-transportan conjugate had no detrimental effect on cell viability up to the 10 μM concentration tested in both CEM and Jurkat cells (results not shown). These results demonstrate that membrane-permeating peptide vectors such as transportan may be effective carriers for intracellular delivery of PNA.
Inhibition of Tat-mediated transactivation of HIV-1 LTR by PNATAR-transportan conjugate.
Since the transporter peptide was found to be an efficient vehicle for biodelivery of PNA, we examined the ability of PNATAR-transportan conjugate to block Tat-mediated transactivation of HIV-1 LTR in CEM and Jurkat cells. Using the reporter plasmid, we first established the optimum amounts of Tat required for the expression of the HIV-1 LTR in these cells. For this, we used the highly sensitive assay system involving expression of Renilla luciferase and firefly luciferase. Expression of the firefly luciferase (pHIV-1 LTR-Luc) and Renilla luciferase (pCMV-R.Luc) was under the control of the HIV-1 LTR and CMV promoter, respectively. The cells were transfected with these two plasmids in the presence of various concentrations of the Tat expression vector (pCMV-Tat).
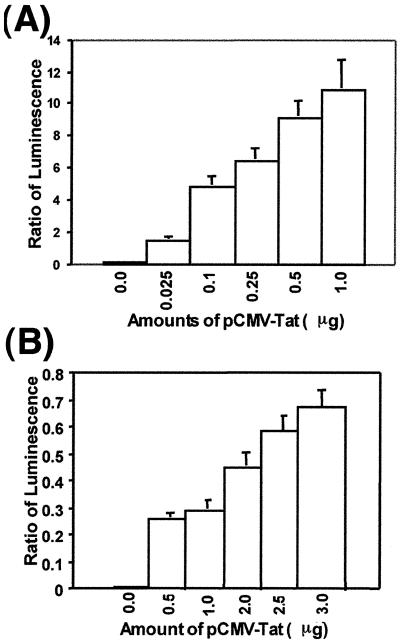
In the Jurkat cells, the transfected cells were activated with PMA and Ca2+ ionophore for expression of the reporter plasmids. When Tat was expressed in trans, the firefly luciferase was transactivated severalfold, while the Renilla luciferase expression, used as an internal control to monitor transfection efficiency, remained constant. Expression of the firefly luciferase in CEM as well as Jurkat cells was found to be proportional to the concentration of pCMV-Tat used in the transfection. The amount of pCMV-Tat for optimum stimulation of HIV-1 LTR in CEM cells (100-fold stimulation) and Jurkat cells (150-fold stimulation) was 1 μg and 3 μg, respectively (Fig. 5).
FIG. 5.
Tat-mediated transactivation of HIV-1 LTR. The indicated amounts of pCMV-Tat were cotransfected along with a plasmid cocktail comprising pHIV-1 LTR-Luc (4 μg) and pCMV-R.Luc (1 μg) in Jurkat and CEM cells (5.0 × 106 cells) in order to establish the extent of Tat-mediated transactivation of the HIV-1 LTR. The highly sensitive luciferase reporter was used for this analysis. The extent of stimulation of the HIV-1 LTR was monitored as a function of the expression of the firefly luciferase gene, with the Renilla luciferase reporter serving as an internal control for normalizing the transfection efficiency. The results are expressed as the ratio of firefly to Renilla luciferase activity. Panels A and B represent the extent of transactivation in CEM and Jurkat cells, respectively, as a function of pCMV-Tat concentration. The results are presented as an average of three independent experiments. The bars represent the standard deviation.
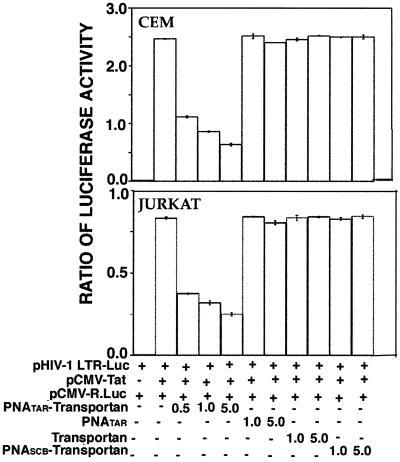
Using this standardized system, we determined the functional ability of the PNATAR-transportan conjugate to inhibit Tat-mediated transactivation of the HIV-1 LTR. The results are shown in Fig. 6. As shown in the figure, the PNATAR-transportan conjugate substantially inhibited Tat-mediated transactivation of the HIV-1 LTR, as judged from the expression levels of luciferase in both CEM and Jurkat cells. Approximately 55% inhibition was achieved at 500 nM PNA-peptide conjugate, which gradually leveled off to 70 to 80% inhibition at higher concentrations. Unconjugated PNATAR, scrambled PNA-transportan, and transportan itself had no effect on the luciferase activity under identical conditions. These results demonstrate that PNATAR-transportan present in the culture medium is able to enter the cells and prevent Tat-mediated transactivation by efficiently sequestering the TAR region of the HIV-1 LTR.
FIG. 6.
Effect of PNATAR-transportan conjugate on Tat-mediated transactivation of HIV-1 LTR. CEM and Jurkat cells (5.0 × 106 cells), cotransfected with the plasmid cocktail comprising pHIV-1 LTR-Luc (4 μg) and pCMV-R.Luc (1 μg) in the presence or absence of the Tat expression clone pCMV-Tat, were incubated with the indicated concentrations of the PNATAR-transportan, unconjugated PNATAR, transportan, or scrambled PNA-transportan (PNASCB-transportan) under the conditions described in Materials and Methods. At 20 h (CEM) or 28 h (Jurkat) posttransfection, cell lysates were assayed for firefly and Renilla luciferase activities. The results are presented as the relative ratio of firefly to Renilla luciferase activity. The amounts of the various plasmids, transportan, and PNA used in the experiment are indicated. The results are presented as an average of three independent experiments. The bars represent the standard deviation.
PNATAR-transportan inhibits at the level of transcription.
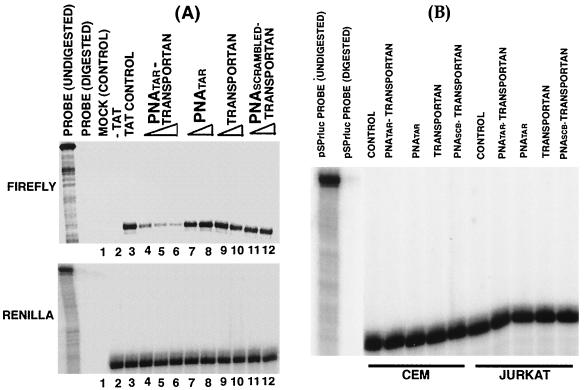
A substantial reduction in the expression of the luciferase reporter in both CEM and Jurkat cells suggested that the PNATAR-transportan inhibits the Tat-mediated transactivation of the HIV-1 LTR. To determine if the inhibition was at the level of transcription, we performed RNase protection assays. CEM and Jurkat cells transfected with an HIV-1 LTR promoter-driven luciferase reporter plasmid in the absence or presence of a Tat expression plasmid were treated with various concentrations of PNATAR-transportan, PNATAR, scrambled PNA-transportan, or transportan and harvested at the specified times for total RNA isolation. In addition, the Renilla luciferase reporter driven by a CMV promoter was included as a control for monitoring transfection efficiency. The RNA samples were hybridized with the 32P-labeled pSPluc+ and pSPrluc probes, digested with the RNase A and T1 mix, and examined by denaturing gel electrophoresis for protected fragments corresponding to the firefly and Renilla transcripts, respectively.
A representation of one such analysis is presented in Fig. 7. Our results indicated that the accumulation of firefly RNA was inversely proportional to the concentration of PNATAR-transportan in CEM and Jurkat cells (Fig. 7A). This provides direct evidence that the PNATAR-transportan interferes with the transcription process, presumably by binding to the TAR region of the HIV-1 LTR, thereby preventing Tat-mediated transactivation. Neither the unconjugated PNATAR, transportan itself, nor scrambled PNA-transportan affected the firefly RNA expression levels, indicating the specificity of the interaction of PNATAR-transportan with its target sequence. Furthermore, the effect of PNATAR-transportan on the HIV-1 LTR appeared to be promoter specific, as noted from our observation that PNATAR-transportan, PNATAR, transportan, and scrambled PNA-transportan did not affect the expression levels of the Renilla RNA driven by the CMV promoter (Fig. 7B).
FIG. 7.
RNase protection assay. (A) Jurkat cells were transfected with 4 μg of pHIV-1 LTR-Luc and 1 μg of pCMV-R.Luc in the absence or presence of pCMV-Tat (2.5 μg). The cells were grown in the presence of PNATAR-transportan (0.5 to 5.0 μM), PNATAR (1.0 and 5.0 μM), transportan (1.0 and 5.0 μM), or scrambled PNA-transportan (1.0 and 5.0 μM). At 28 h posttransfection, total RNA was isolated from the cells and hybridized with RNA probes for the different reporter transcripts (firefly and Renilla luciferase). The pSPrluc probe coding for the Renilla RNA served as a transfection control. The firefly RNA levels were normalized to the Renilla RNA signal. The lanes are as follows: mock-transfected cells (lane 1); luciferase expression in the absence of Tat (lane 2); and luciferase expression in the presence of Tat and the indicated inhibitors (lanes 3 to 12). (B) CEM and Jurkat cells were transfected with 1 μg of pCMV-R.Luc and incubated in the absence or presence of PNATAR-transportan, unconjugated PNATAR, transportan, or scrambled PNA-transportan (PNASCB-transportan) at 5 μM in order to investigate the promoter specificity of PNATAR-transportan. Total RNA was isolated from cells and subjected to the RNase protection assay with the pSPrluc probe. The protected fragments were analyzed on a denaturing polyacrylamide-urea gel and visualized on a PhosphorImager. Renilla RNA was normalized to total RNA in the cell. The lane marked control represents Renilla RNA signal in transfected cells in the absence of the inhibitor.
Effect of PNATAR-transportan on HIV-1 production in chronically HIV-1-infected H9 cells.
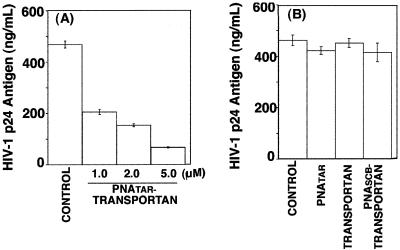
Since PNATAR-transportan was able to enter the cells and interfere with Tat-mediated transactivation, we examined its antiviral efficacy in H9 cells chronically infected with HIV-1. These cells were grown in the absence or presence of PNATAR-transportan (1.0 to 5.0 μM), unconjugated PNATAR (5.0 μM), transportan (5.0 μM), or scrambled PNA-transportan (5.0 μM) for 3 days, and their culture supernatants were analyzed for levels of p24 antigen by ELISA, since the levels of p24 antigen provided a measure of viral titers. The results are shown in Fig. 8.
FIG. 8.
Effect of PNATAR-transportan conjugate on HIV-1 production in chronically HIV-1-infected H9 cells. The indicated concentrations of PNATAR-transportan (A), unconjugated PNATAR, transportan, or scrambled PNA-transportan (PNASCB-transportan) (B) were added to the culture medium of chronically HIV-1-infected H9 cells, and their ability to inhibit HIV-1 production was examined by monitoring the levels of p24 antigen in the culture supernatant. The results are presented as relative amounts of HIV-1 p24 antigen present in the supernatants of H9 cells cultured in the absence (control) or presence of PNATAR-transportan (1.0 to 5.0 μM), unconjugated PNATAR (5.0 μM), transportan (5.0 μM), or scrambled PNA-transportan (5.0 μM). The results are expressed as mean values along with standard deviation for three independent experiments.
The levels of p24 antigen decreased by 60% at 1 μM PNATAR-transportan conjugate. Further increases in the concentration of the PNATAR-transportan conjugate resulted in a substantial decrease (70 to 90%) in the p24 levels, suggesting its potential for blocking HIV-1 production (Fig. 8A). Our observation that unconjugated PNATAR at similar concentrations did not decrease viral production indicated the efficacy of PNATAR-transportan. Furthermore, the effect of PNATAR-transportan appeared to be quite specific, since neither scrambled PNA-transportan nor transportan by itself decreased HIV-1 production under identical conditions (Fig. 8B).
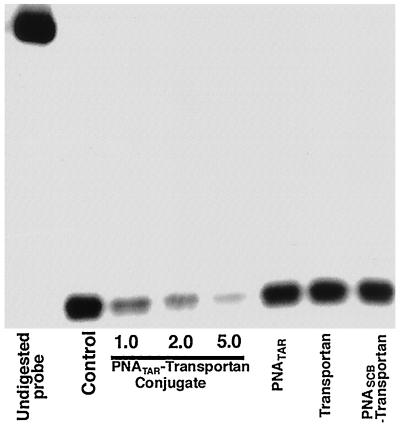
PNATAR-transportan inhibits transcription of HIV-1 mRNA in chronically HIV-1-infected H9 cells.
We also examined the mechanism by which PNATAR-transportan inhibited HIV-1 production in chronically HIV-1-infected H9 cells. Total RNA isolated from the cells treated with PNATAR-transportan, unconjugated PNATAR, transportan, or scrambled transportan were hybridized to the radiolabeled probe synthesized from pGEM23 and subjected to an RNase protection assay. The results are shown in Fig. 9.
FIG. 9.
PNATAR-transportan inhibits transcription of the HIV-1 mRNA in chronically HIV-1-infected H9 cells. Total RNA was isolated from chronically HIV-1-infected H9 cells treated with the indicated concentrations of PNATAR-transportan or 5.0 μM PNATAR, transportan alone, or scrambled PNA-transportan (PNASCB-transportan). The RNA was hybridized with the pGEM23 probe and digested with the RNase A and T1 mix, and the protected fragments were separated on a polyacrylamide-urea gel and detected by PhosphorImager analysis. The undigested probe corresponds to a 195-base fragment, whereas HIV-1 mRNA initiating at +1 of the HIV-1 LTR protected an 83-nucleotide fragment of the riboprobe corresponding to the anti-TAR sequence.
The mRNA initiating at +1 of the HIV-1 LTR protected an 83-nucleotide fragment of the riboprobe corresponding to the anti-TAR sequence. A decrease in levels of HIV-1 mRNA in cells treated with PNATAR-transportan suggested that the inhibition occurred at the transcription step. The specificity of the effect of PNATAR-transportan was seen from the observation that neither transportan itself nor scrambled PNA-transportan altered the HIV-1 mRNA levels. Furthermore, PNATAR alone also did not influence the levels of HIV-1 mRNA synthesis, indicating the efficiency of transportan as a vehicle for delivering PNATAR to its target site.
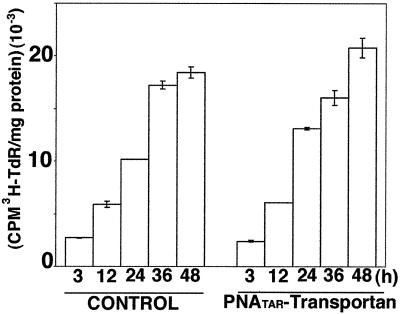
PNATAR-transportan conjugate has no effect on cellular proliferation of chronically HIV-1-infected H9 cells.
We examined the incorporation of [3H]thymidine into cellular DNA in order to determine if the PNATAR-transportan conjugate impacted cellular proliferation of chronically HIV-1-infected H9 cells. The cells were grown in the presence or absence of PNATAR-transportan and supplemented with [3H]thymidine. At the indicated time points, aliquots were withdrawn, and the DNA was precipitated with TCA to determine thymidine incorporation. The results of this analysis are shown in Fig. 10. As shown in the figure, similar amounts of [3H]thymidine were incorporated into cellular DNA in both the treated and untreated cells at all time points analyzed, indicating that cellular proliferation was not altered in the presence of PNATAR-transportan.
FIG. 10.
[3H]thymidine incorporation into cellular DNA in chronically HIV-1-infected H9 cells. Chronically HIV-1-infected H9 cells grown in the presence or absence of 5.0 μM PNATAR-transportan were supplemented with 10 μCi of [methyl-3H]thymidine/ml. At the indicated time points, the cells were harvested and assayed for the amounts of [3H]thymidine (TdR) incorporated into DNA by TCA precipitation of the acid-insoluble material onto glass fiber filters. The total amount of radioactivity was determined by scintillation counting, and the results are represented as counts per minute incorporated per milligram of protein. The results are expressed as average values along with the standard deviation for three independent experiments.
DISCUSSION
In spite of massive efforts for the prevention of AIDS, the AIDS epidemic has reached alarming proportions worldwide. The current antiviral treatment for HIV-1 infection is restricted to drugs that mainly target two key viral enzymes in its life cycle, reverse transcriptase and protease (7, 22, 23, 28, 32, 42-44, 55, 67). Successful HIV-1 therapy, such as the highly active antiretroviral therapy, is hampered by the appearance of drug-resistant strains of HIV-1, which have been directly correlated with mutations in the genes encoding the viral reverse transcriptase and protease (1, 5).
In the present study we have used specific PNA-transportan conjugate to target the TAR RNA, an important regulatory element in the nontranslated 5′ end of the retroviral genome. The rationale for targeting TAR was that it forms a stable nuclease-resistant stem-loop structure which is averse to mutational changes (25, 59). HIV-1 Tat, a nuclear transcriptional activator, interacts with TAR to enhance the processivity of RNA polymerase II complexes (25). The importance of the Tat-TAR interaction in regulation of gene expression has been well established (21, 25). We had earlier demonstrated that a novel inhibitor, PNA, targeted to the TAR sequences of the viral RNA genome blocks the Tat-TAR interaction, resulting in the inhibition of transactivation of the HIV-1 LTR (40). In another study, PNA has been employed to inhibit in vitro protein synthesis at both the transcriptional and translational levels (20, 31).
Effective antisense inhibition in cells requires efficient cell entry. Though PNAs have been shown to inhibit gene expression and have several advantages over conventional DNA or RNA antisense oligomers, their use as an alternative therapy has been limited chiefly by their inability to cross the cell membrane efficiently (47). More recent findings demonstrating that PNAs conjugated to transporter peptides exhibit efficient biodelivery into cells have redefined its potential as a therapeutic agent (52).
For the present studies, we used the transportan peptide vector for biodelivery of PNATAR. Transportan is a 27-amino-acid chimeric peptide consisting of 13 amino acids from the amino terminus of the neuropeptide galanin and 14 amino acids from the amino terminus of the wasp venom toxic peptide mastoparan (33). Transportan penetrates every cell type in a rapid and efficient way and is localized mostly in membranous structure; upon prolonged incubation, it concentrates in the nucleus (50). In moderate concentrations (10 μM), it does not affect the growth of cells in culture (50). These unique properties of the chimeric transportan peptide make it potentially useful as a vector for biodelivery of different proteins or nonprotein drugs into cells. We used this approach to address the issues of (i) uptake of the PNA-transportan conjugate by cells and its effect on cell viability and (ii) functional efficacy of the PNATAR-transportan conjugate in blocking the Tat-TAR interaction, thereby inhibiting viral transcription and replication.
To ascertain the ability of the PNA-transportan conjugate to enter the cells, we tagged it with TAMRA, a fluorescent probe, and monitored its uptake in the cell. A substantial accumulation of the fluorophore in the cell in 12 h (Table 1) indicated efficient uptake, though the overall kinetics of its internalization was slower than with transportan alone (50, 51). In marked contrast to an earlier report demonstrating that cell type did not influence the cell-penetrating ability of transportan (51), the uptake of the PNA-transportan conjugate by CEM and Jurkat cells varied significantly (Table 1). This may be attributed to a difference in the morphology and membrane composition of these two cells, or it is likely that the presence of the PNA moiety influences uptake. Nonetheless, the accumulation of the PNA-transportan conjugate in the cells is encouraging and suggests that the transportan vector may be used effectively for biodelivery of this class of compounds.
PNATAR-transportan substantially inhibited Tat-mediated transactivation of the HIV-1 LTR in both CEM and Jurkat cells (Fig. 5). The PNA-peptide conjugate may have dual functions; it may prevent the TAR-Tat interaction by sequestering the TAR sequence of the nascent RNA, or it may prevent transcription directly by interacting with the TAR sequence on the proviral DNA. One may visualize that the large transportan moiety may interfere with the PNA, targeting its site inside the cell. However, this did not appear to be true in this study, since the PNATAR-transportan conjugate displayed similar binding affinity for its target sequence on the TAR RNA (Fig. 2) and was also able to block reverse transcription to a similar extent as the unconjugated PNATAR (Fig. 3). Moreover, a substantial inhibition of Tat-mediated transactivation was also achieved by PNATAR-transportan conjugate in cell culture (Fig. 6). The mechanism of this inhibition appeared to be at the level of transcription (Fig. 7).
Our results demonstrating that PNATAR-transportan can enter chronically HIV-1-infected H9 cells and inhibit virus production are very promising (Fig. 8). This is the first report on the use of a PNA-transporter peptide conjugate to inhibit the replication of HIV-1. Interestingly, recent studies by Sei et al. (58) have demonstrated that PNA targeted to the Gag-Pol transframe domain can arrest virus production. However, very high concentrations of PNA (100 μM) were required to achieve this effect. In contrast, the PNA-transportan conjugates used in this study were able to inhibit virus replication at significantly lower concentrations. It may also be noted that the effect of the PNATAR-transportan conjugate on transactivation of the HIV-1 LTR appeared to be more prominent in the transfected cells than in H9 cells. This is not surprising, as the events associated with the replication of HIV-1 in H9 cells are far more complex than those associated with the expression of the luciferase gene in cells.
The choice of antisense targets appears quite challenging, since targeting the various regulatory genes of the HIV-1 genome has not always been effective against diverse HIV-1 strains (2, 29, 30, 36-38, 66). The unique 5′ untranslated region (U5) (nucleotides 1 to 333) of the HIV-1 genome, containing several critical domains essential for viral replication, may be an ideal target for drug intervention. These critical domains include (i) the primer-binding site (nucleotides 183 to 201), essential for tRNA3Lys-primed initiation of reverse transcription (45, 53, 62); (ii) the A-loop region located upstream of the primer-binding site (nucleotides 168 to 173), essential for the selection and interaction of tRNA3Lys primer (24, 35, 63); and (iii) the LTR sequences at the 5′ and 3′ ends, essential for viral transcription and integration (61).
Studies in our laboratory are under way to analyze the effect of PNA-transportan conjugates targeted against these sites as effective antiviral agents. A PNA-transportan conjugate directed against galanin receptor mRNA has been shown to inhibit the expression levels of galanin receptor in Bowes melanoma cells in cell culture and also alleviate pain in the rat model (52). Recently, Good et al. have shown that PNA conjugated with a 10-mer peptide vector targeted against rRNA and against mRNA of Acp protein of bacteria can effectively cure HeLa cells infected with Escherichia coli K-12 strains, raising the possibility of anti-infective drug development (17).
In summary, our studies have demonstrated that cellular uptake of anti-TAR PNA is significantly enhanced upon conjugation with a transportan peptide, resulting in effective blockage of the Tat-TAR interaction at relatively low inhibitor concentrations and inhibiting transcription. These studies with transportan peptides as the vector for biodelivery of PNA have created a therapeutic niche for the application of PNA against HIV-1. Thus, our approach using PNA-transportan to target the critical regions of the HIV-1 genome in the quest for novel inhibitors of HIV-1 appears quite promising. Optimal chemical modification of the existing transporter peptides by treatment with polyethylene glycol (64) in order to increase the circulating half-life of PNA-transporter conjugates will further advance the efforts to develop potent antiviral therapeutics targeting viral genes.
Acknowledgments
The first two authors made an equal contribution.
We thank Michael Mathews and M. Peterlin for the kind gift of the plasmids and John Stephens for assistance with the HIV-1 p24 assay.
This research was supported by a grant from the NIAID (AI42520).
REFERENCES
- 1.Aalen, O. O., V. T. Farewell, D. De Angelis, N. E. Day, and O. N. Gill. 1999. New therapy explains the fall in AIDS incidence with a substantial rise in number of persons on treatment expected. AIDS 13:103-108. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, S., T. Ikeuchi, D. Sun, P. S. Sarin, A. Konopka, J. Maizel, and P. C. Zamecnik. 1989. Inhibition of human immunodeficiency virus in early infected and chronically infected cells by antisense oligodeoxynucleotides and their phosphorothioate analogues. Proc. Natl. Acad. Sci. USA 86:7790-7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat transactivates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 4.Bohjanen, P. R., R. A. Colvin, M. Puttaraju, M. D. Been, and M. A. Garcia-Blanco. 1996. A small circular TAR RNA decoy specifically inhibits Tat-activated HIV-1 transcription. Nucleic Acids Res. 24:3733-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1998. HIV/AIDS surveillance report, midyear ed., vol. 10. Centers for Disease Control and Prevention, Public Health Service, Department of Health and Human Services, Atlanta, Ga.
- 6.Cordingley, M. G., R. L. Lafemina, P. L. Callahan, J. H. Condra, V. V. Sardana, D. J., Graham, T. M. Nguyen, K. Legrow, L. Gotlib, A. J. Schlabach, and R. J. Colonno. 1990. Sequence-specific interaction of Tat protein and Tat peptides with the transactivation-responsive sequence element of human immunodeficiency virus type 1 in vitro. Proc. Natl. Acad. Sci. USA 87:8985-8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Clercq, E. 1992. HIV inhibitors targeted at the reverse transcriptase. AIDS Res. Hum. Retrovirol. 8:119-134. [DOI] [PubMed] [Google Scholar]
- 8.Demidov, V. V., V. N. Potaman, M. D. Frank-Kamenetskii, M. Egholm, O. Buchard, S. H. Sonnichsen, and P. E. Nielsen. 1994. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 48:1310-1313. [DOI] [PubMed] [Google Scholar]
- 9.Derossi, D., S. Calvet, A. Trembleau, A. Brunissen, G. Chassaing, and A. Prochiantz. 1996. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem. 271:18188-18193. [DOI] [PubMed] [Google Scholar]
- 10.Dingwall, C., I. Ernberg, M. J. Gait, S. M. Green, S. Heaphy, J. Karn, A. D. Lowe, M. Singh, and M. A. Skinner. 1990. HIV-1 Tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 12:4145-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faria, M., D. G. Spiller, C. Dubertret, J. S. Nelson, M. R. White, D. Scherman, C. Helene, and C. Giovannangeli. 2001. Phosphoramidate oligonucleotides as potent antisense molecules in cells and in vivo. Nat. Biotechnol. 19:40-44. [DOI] [PubMed] [Google Scholar]
- 12.Fawell, S., J. Serry, Y. Daikh, C. Moore, L. L. Chen, B. Pepinsky, and J. Barsoum. 1994. Tat-mediated delivery of heterologous proteins into cells. Proc. Natl. Acad. Sci. USA 91:664-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, S., and E. C. Holland. 1988. HIV-1 Tat trans-activation requires the loop sequence within tar. Nature 334:165-167. [DOI] [PubMed] [Google Scholar]
- 14.Frankel, A. D., S. Blancalana, and D. Hudson. 1989. Activity of synthetic peptides from the Tat protein of HIV-1. Proc. Natl. Acad. Sci. USA 86:7397-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Futaki, H., T. Suzuki, W. Ohashi, T. Yagami, S. Tanaka, K. Ueda, and Y. Sugiura. 2001. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 276:5836-5840. [DOI] [PubMed] [Google Scholar]
- 17.Good, L., S. K. Awasthi, R. Dryselius, O. Larsson, and P. E. Nielsen. 2001. Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 19:360-364. [DOI] [PubMed] [Google Scholar]
- 18.Gunnery, S., S. R., Green, and M. B. Matthews. 1992. Tat-responsive region RNA of human immunodeficiency virus type 1 stimulates protein synthesis in vivo and in vitro: relationship between structure and function. Proc. Natl. Acad. Sci. USA 89:11557-11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamy, F., E. R. Felder, G. Heizmann, J. Lazdins, F. Aboul-ela, G. Varani, J. Karn, and T. Klimkait. 1997. An inhibitor of the Tat/TAR RNA interaction that effectively suppresses HIV-1 replication. Proc. Natl. Acad. Sci. USA 94:3548-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanvey, J. C., N. J. Peffer, J. E. Bisi, S. A. Thomas, R. Cadilla, J. A. Josey, et al. 1992. Antisense and antigene properties of peptide nucleic acids. Science 258:1481-1485. [DOI] [PubMed] [Google Scholar]
- 21.Jeang, K. T., H. Xiao, and E. A. Rich. 1999. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 274:28837-28840. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, V. A., D. P. Merrill, T. C. Chou, and M. S. Hirsch. 1992. Human immunodeficiency virus type 1 (HIV-1) inhibitory interactions between protease inhibitor Ro 31-8959 and zidovudine, 29,39-dideoxycytidine, or recombinant interferon-alpha A against zidovudine-sensitive or -resistant HIV-1 in vitro. J. Infect. Dis. 166:1143-1146. [DOI] [PubMed] [Google Scholar]
- 23.Kageyama, S., J. N. Weinstein, T. Shirasaka, D. J. Kempf, D. W. Norbeck, J. J. Plattner, J. Erickson, and H. Mitsuya. 1992. In vitro inhibition of human immunodeficiency virus (HIV) type 1 replication by C2 symmetry-based HIV protease inhibitors as single agents or in combinations. Antimicrob. Agents Chemother. 36:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang, S. M., Z. Zhang, and C. D. Morrow. 1997. Identification of a sequence within U5 required for human immunodeficiency virus type 1 to stably maintain a primer binding site complementary to tRNAMet. J. Virol. 71:207-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karn, J. 1999. Tackling Tat. J. Mol. Biol. 293:235-254. [DOI] [PubMed] [Google Scholar]
- 26.Kashanchi, F., M. R. Sadaie, and J. N. Brady. 1997. Inhibition of HIV-1 transcription and virus replication using soluble Tat peptide analogs. Virology 227:431-438. [DOI] [PubMed] [Google Scholar]
- 27.Kaushik, N., N. Rege, P. N. S. Yadav, S. G. Sarafianos, M. J. Modak, and V. N. Pandey. 1996. Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Biochemistry 35:11536-11546. [DOI] [PubMed] [Google Scholar]
- 28.Kempf, D. J., K. C. Marsh, J. F. Denissen, E. McDonald, S. Vasavanonda, C. A. Flentge, B. E. Green, L. Fino, C. H. Park, X. P. Kong, et al. 1995. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc. Natl. Acad. Sci. USA 92:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, S. G., T. Hatta, S. Tsukahara, H. Nakashima, N. Yamamoto, Y. Shoji, K. Takai, and H. Takaku. 1995. Antiviral effect of phosphorothioate oligodeoxyribonucleotides complementary to human immunodeficiency virus. Bioorg. Med. Chem. 3:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinchington, D., S. Galpin, J. W. Jaroszewski, K. Ghosh, C. Subasinghe, and J. S. Cohen. 1992. A comparison of gag, pol and rev antisense oligodeoxynucleotides as inhibitors of HIV-1. Antivir. Res. 17:53-62. [DOI] [PubMed] [Google Scholar]
- 31.Knudsen, H., and P. E. Nielsen. 1996. Antisense properties of duplex- and triplex-forming PNAs. Nucleic Acids Res. 24:494-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam, P. Y., P. K. Jadhav, C. J. Eyermann, C. N. Hodge, Y. Ru, L. T. Bacheler, J. L. Meek, M. J. Otto, M. M. Rayner, Y. N. Wong, et al. 1994. Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors. Science 263:380-384. [DOI] [PubMed] [Google Scholar]
- 33.Langel, Ü., M. Pooga, C., Kairane, M., Zilmer, and T. Bartfai. 1996. A galanin-mastoparan chimeric peptide activates the Na+, K+-ATPase and reverses its inhibition by ouabain. Regul. Peptides 62:47-52. [DOI] [PubMed] [Google Scholar]
- 34.Laspia, M. F., A. P. Rice, and M. B. Mathews. 1989. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 59:283-292. [DOI] [PubMed] [Google Scholar]
- 35.Li, Y., S. M. Kang, and C. D. Morrow. 1997. Stability of HIV type 1 proviral genomes that contain two distinct primer-binding sites. AIDS Res. Hum. Retrovir. 13:253-262. [DOI] [PubMed] [Google Scholar]
- 36.Lisziewicz, J., D. Sun, M. Klotman, S. Agrawal, P. Zamecnik, and R. Gallo. 1992. Specific inhibition of human immunodeficiency virus type 1 replication by antisense oligonucleotides: an in vitro model for treatment. Proc. Natl. Acad. Sci. USA 89:11209-11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lisziewicz, J., D. Sun, J. Smythe, P. Lusso, F. Lori, A. Louie, P. Markham, J. Rossi, M. Reitz, and R. C. Gallo. 1993. Inhibition of human immunodeficiency virus type 1 replication by regulated expression of a polymeric Tat activation response RNA decoy as a strategy for gene therapy in AIDS. Proc. Natl. Acad. Sci. USA 90:8000-8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lisziewicz, J., D. Sun, V. Metelev, P. Zamecnik, R. C. Gallo, and S. Agrawal. 1993. Long-term treatment of human immunodeficiency virus-infected cells with antisense oligonucleotide phosphorothioate. Proc. Natl. Acad. Sci. USA 90:3860-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ljungstrom, T., H. Knudsen, and P. E. Nielsen. 1999. Cellular uptake of adamantyl conjugated peptide nucleic acids. Bioconjug. Chem. 10:965-972. [DOI] [PubMed] [Google Scholar]
- 40.Mayhood, T., N. Kaushik, P. K. Pandey, F. Kashanchi, L Deng, and V. N. Pandey. 2000. Inhibition of Tat-mediated transactivation of HIV-1 LTR transcription by polyamide nucleic acid targeted to TAR hairpin element. Biochemistry 39:11532-11539. [DOI] [PubMed] [Google Scholar]
- 41.Mhashilkar, A. M., D. K. Biswas, J. LaVecchio, A. B. Pardee, and W. A. Marasco. 1997. Inhibition of human immunodeficiency virus type 1 replication in vitro by a novel combination of anti-Tat single-chain intrabodies and NF-κB antagonists. J. Virol. 71:6486-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitsuya, H., and S. Broder. 1986. Inhibition of the in vitro infectivity and cytopathic effect of human T-lymphotrophic virus type III/lymphadenopa-thy-associated virus (HTLV-III/LAV) by 29,39-dideoxynucleosides. Proc. Natl. Acad. Sci. USA 83:1911-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitsuya, H., R. F. Jarrett, M. Matsukura, F. Di Marzo Veronese, A. L. DeVico, M. G. Sarngadharan, D. G. Johns, M. S. Reitz, and S. Broder. 1987. Long-term inhibition of human T-lymphotropic virus type III/lymph-adenopathy-associated virus (human immunodeficiency virus) DNA synthesis and RNA expression in T cells protected by 29,39-dideoxynucleosides in vitro. Proc. Natl. Acad. Sci. USA 84:2033-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitsuya, H., K. J. Weinhold, P. A. Furman, M. H. St. Clair, S. N. Lehrman, R. C. Gallo, D. Bolognesi, D. W. Barry, and S. Broder. 1985. 39-Azido-39-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphoid nopathy-associated virus in vitro. Proc. Natl. Acad. Sci. USA 82:7096-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muesing, M. A., D. H. Smith, C. D. Cabradilla, C. V. Benton, L. A. Lasky, and D. J. Capon. 1985. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature 313:450-458. [DOI] [PubMed] [Google Scholar]
- 46.Muesing, M. A., D. H. Smith, and D. J. Capon. 1987. Regulation of mRNA accumulation by a human immunodeficiency virus transactivator protein. Cell 48:691-701. [DOI] [PubMed] [Google Scholar]
- 47.Nielsen, P. E. 1999. Peptide nucleic acids as therapeutic agents. Curr. Opin. Struct. Biol. 9:353-357. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen, P. E., M. Egholm, R. H. Berg, and O. Buchardt. 1991. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 254:1497-1500. [DOI] [PubMed] [Google Scholar]
- 49.O'Brien, W. A., M. Sumner-Smith, S. H. Mao, S. Sadeghi, J. Q. Zhao, and I. S. Chen. 1996. Anti-human immunodeficiency virus type 1 activity of an oligocationic compound mediated via gp120 V3 interactions. J. Virol. 70:2825-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pooga, M., M. Hallbrink, M. Zorko, and U. Langel. 1998. Cell penetration by transportan. FASEB J. 12:67-77. [DOI] [PubMed] [Google Scholar]
- 51.Pooga, M., M. Lindgren, M. Hallbrink, E. Brakenhielm, and U. Langel. 1998. Galanin-based peptides, galparan and transportan, with receptor-dependent and independent activities. Ann. N. Y. Acad. Sci. 863:450-453. [DOI] [PubMed] [Google Scholar]
- 52.Pooga, M., U. Soomets, M. Hallbrink, A. Valkna, K. Saar, K. Rezaei, U. Kahl, J. X. Hao, X. J, Xu, Z Wiesenfeld-Hallin, T. Hokfelt Bartfai, and U. Langel. 1998. Cell penetrating PNA constructs regulate galanin receptor levels and modify pain transmission in vivo. Nat. Biotechnol. 16:857-861. [DOI] [PubMed] [Google Scholar]
- 53.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, L. Ivanoff, J. S. R. Petteway, M. L. Pearson, J. A. Lautenberge, T. S. Papas, J. Ghrayeb, N. T. Chang, R. C. Gallo, and F. Wong-Staal. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature (London) 313:277-284. [DOI] [PubMed] [Google Scholar]
- 54.Re, M. C., G. Furlini, M. Vignoli, E. Ramazzotti, G. Roderigo, V. De Rosa, G. Zauli, S. Lolli, S. Capitani, and M. La Placa. 1995. Effect of antibody to HIV-1 Tat protein on viral replication in vitro and progression of HIV-1 isease in vivo. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 10:408-416. [DOI] [PubMed] [Google Scholar]
- 55.Roberts, N. A., J. A. Martin, D. Kinchington, A. V. Broadhurst, J. C. Craig, I. B. Duncan, S. A. Galpin, B. K. Handa, J. Kay, A. Krohn, et al. 1990. Rational design of peptide-based HIV proteinase inhibitors. Science 248:358-361. [DOI] [PubMed] [Google Scholar]
- 56.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwarze, S. R., A. Ho, A. Vocero-Akbani, and S. F. Dowdy. 1999. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science 285:1569-1572. [DOI] [PubMed] [Google Scholar]
- 58.Sei, S., Q. Yang, D. O'Neill, K. Yoshimura, K. Nagashima, and H. Mitsuya. 2000. Identification of a key target sequence to block human immunodeficiency virus type 1 replication within the gag-pol transframe domain. J. Virol. 74:4621-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selby, M. J., E. S. Bain, P. A. Luciw, and B. M. Peterlin. 1989. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by Tat through the HIV-1 long terminal repeat. Genes Dev. 3:547-558. [DOI] [PubMed] [Google Scholar]
- 60.Smith, C. S., W. Lee, E. Wong, H. Gallardo, K. Page, O. Gaspar, J. Lebkowski, and E. Gilboa. 1996. Transient protection of human T-cells from human immunodeficiency virus type 1 infection by transduction with adeno-associated viral vectors which express RNA decoys. Antivir. Res. 32:99-115. [DOI] [PubMed] [Google Scholar]
- 61.Vink, C., D. C. Van Gent, Y. Elgersma, and R. H. A. Plasterk. 1991. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol. 65:4636-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wakefield, J. K., A. G. Wolf, and C. D. Morrow. 1995. Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNA3Lys. J. Virol. 69:6021-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wakefield, J. K., S.-M. Kang, and C. D. Morrow. 1996. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J. Virol. 70:966-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, J., S. Y. Huang, I. Choudhury, M. J. Leibowitz, and S. Stein. 1995. Use of polyethylene glycol-peptide conjugate in a competition gel shift assay for screening potential antagonists of HIV-1 Tat protein binding to TAR RNA. Anal. Biochem. 232:238-242. [DOI] [PubMed] [Google Scholar]
- 65.Wang, S., P. W. Huber, M. Cui, A. W. Czarnik, and H. Y. Mei. 1998. Binding of neomycin to the TAR element of HIV-1 RNA induces dissociation of Tat protein by an allosteric mechanism. Biochemistry 37:5549-5557. [DOI] [PubMed] [Google Scholar]
- 66.Weichold, F. F., J. Lisziewicz, R. A. Zeman, L. S. Nerurkar, S. Agrawal, M. S. Reitz, Jr., and R. C. Gallo. 1995. Antisense phosphorothioate oligodeoxynucleotides alter HIV type 1 replication in cultured human macrophages and peripheral blood mononuclear cells. AIDS Res. Hum. Retrovirol. 11:863-867. [DOI] [PubMed] [Google Scholar]
- 67.Yarchoan, R., R. W. Klecker, K. J. Weinhold, P. D. Markham, H. K. Lyerly, D. T. Durack, E. Gelmann, S. N. Lehrman, R. M. Blum, D. W. Barry, et al. 1986. Administration of 39-azido-39-deoxythymidine, an inhibitor of HTLV-III/LAV replication, to patients with AIDS or AIDS-related complex. Lancet i:575-580. [DOI] [PubMed] [Google Scholar]