Abstract
Early steps of retroviral replication involve reverse transcription of the viral RNA to yield a linear double-stranded cDNA copy and then integration of the viral cDNA into a chromosome of the host cell. A portion of the viral cDNA can also follow nonproductive pathways in which it becomes circularized. In one pathway, the ends of the linear cDNA become joined together by the cellular nonhomologous DNA end-joining system to form two-long terminal repeat (2-LTR) circles. It has been argued that 2-LTR circles are quickly degraded in human immunodeficiency virus (HIV)-infected cells, allowing the presence of 2-LTR circles to be used as a marker for ongoing de novo infection in patients. Following this idea, detection of 2-LTR circles in patients undergoing successful highly active antiretroviral therapy has led to the proposal that viral replication persists despite treatment. We have used fluorescence-monitored PCR (Taqman) to quantitate the metabolism of HIV cDNA early after infection. Contrary to previous work, we find that 2-LTR circles are actually quite stable in experiments where confounding variables are controlled. Thus, studies relying on the lability of 2-LTR circles are open to reinterpretation. We also used the quantitative PCR methods to analyze the effects of MG132, a proteasome inhibitor, which revealed that viral complexes containing mostly completed cDNAs are the primary substrates for proteasome-mediated degradation.
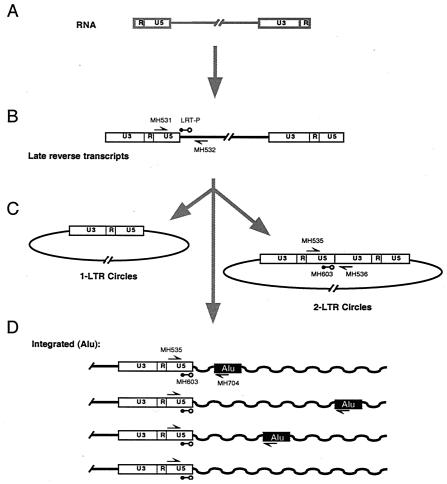
Following introduction of the viral core into the cell cytoplasm, the viral RNA is reverse transcribed to yield a linear cDNA copy (Fig. 1). In productive infections, the viral cDNA is then integrated into a chromosome of the host cell. A fraction of the linear cDNA molecules instead becomes circularized, a reaction that is thought to be a dead end for the virus since circular cDNAs are not substrates for the viral integration machinery (3). Circularization can be accomplished by (i) ligation of the cDNA ends by the host cell nonhomologous DNA end-joining system (9), yielding the two-long terminal repeat (2-LTR) circle; (ii) recombination between the two LTRs, yielding a 1-LTR circle; (iii) stalling of reverse transcription to yield 1-LTR circles; or (iv) integration of the linear cDNA into itself, yielding an internally rearranged form.
FIG. 2.
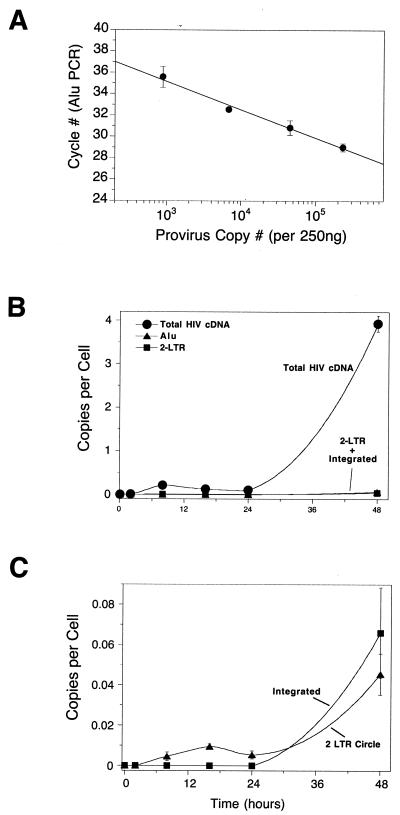
HIV cDNA metabolism in a multicycle HIV infection of SupT1 cells assayed by quantitative PCR. (A) Standard curve relating the number of integrated HIV cDNA copies quantitated by the total cDNA amplicon to the Alu PCR cycle of threshold. Using the standard curve the number of proviruses can be determined from a measurement of the Alu PCR signal. (B) Accumulation of total HIV cDNA (i.e., cDNAs that have completed the second strand transfer step of reverse transcription), 2-LTR circles, and integrated proviruses during multicycle HIV infection. In this range of copies per cell (y axis) the 2-LTR circles and integrated provirus signals are at the baseline. In this and subsequent figures, total HIV cDNA copies are indicated by filled circles, 2-LTR circles are indicated by filled triangles, and integrate proviruses are indicated by filled squares. (C) Accumulation of 2-LTR circles and integrated proviruses replotted against lower levels of copies per cell (y axis). The multiplicity of infection was 0.04 infectious unit per cell as determined by infection of 5.25 indicator cells. Error bars, standard deviations.
We have probed factors that modulate metabolism of the human immunodeficiency virus (HIV) cDNA by using fluorescence-monitored PCR quantitation. Three separate assays can be used to monitor (i) accumulation of total viral cDNA (6), (ii) 2-LTR circle formation (8, 21), and (iii) integration (2). The first of these assays uses primers internal to the HIV cDNA and detects all of the viral cDNA forms that have completed the second template switch of reverse transcription. The assay for 2-LTR circles requires amplification across a circle junction. The assay for integration uses one primer in an HIV LTR and a second in cellular Alu repeats. The latter assay is made quantitative by using a standard curve made with infected-cell DNA as described elsewhere (2).
In some studies we have used HIV-based vectors instead of replication-competent HIV to minimize the toxicity of infection (4, 14, 26). The system used yields particles containing the HIV Gag and Pol proteins and a vector RNA transducing the gene for green fluorescent protein (gfp), all packaged as pseudotypes with the vesicular stomatitis virus G envelope.
Previous quantitative studies revealed that total HIV cDNA accumulated quickly after infection, reaching maximum abundance after 12 to 24 h. Initially this corresponds to linear double-strand viral DNA (2). The 2-LTR circles and integrated forms accumulated more slowly, reaching maximal abundance after 24 h, consistent with the expected precursor-product relationship. The final number of proviruses per cell was considerably lower than the total number of cDNA copies detected initially, indicating that only a fraction of the HIV DNA was converted to proviruses. After 24 to 48 h, all of the viral cDNA detected could be accounted for by proviruses and circular forms. These methods have been validated by demonstrating that reverse transcriptase inhibitors and integrase inhibitors block accumulation of the expected cDNA forms (2) and that a catalytic site mutant of integrase (E152A) selectively blocked integration of an HIV-based vector (9).
In this work, we report the use of quantitative PCR assays to address a controversial aspect of HIV cDNA metabolism--the stability of 2-LTR circles. A critical question in HIV biology is that of whether there is ongoing HIV replication in patients undergoing highly active antiretroviral therapy (HAART). To analyze this, it is critical to develop assays for de novo infection in patients, since this allows latently infected cells to be distinguished from recently infected cells resulting from ongoing replication. It has been proposed that 2-LTR circles provide a marker for ongoing infection, since 2-LTR circles have been proposed to be rapidly degraded once formed (21), so detection of circles is inferred to indicate de novo infection. It was found that 2-LTR circles could still be detected in samples from many patients on HAART, leading to the proposal that “covert” HIV replication persists despite successful therapy (21). Several other studies have also used 2-LTR circles as markers to investigate the dynamics of HIV infection in patients (16, 17, 25).
We have studied the stability of 2-LTR circles under a variety of experimental conditions and obtained contrary data, indicating instead that 2-LTR circles are quite stable when confounding variables are controlled. These 2-LTR circles, once formed, can fall in number per cell by any of three means. (i) They can be degraded by cellular nucleases, (ii) they can be diluted by growth of cells, since they are unable to replicate as episomes, and (iii) they may be lost by death of infected cells. We have developed a method to distinguish among these possibilities. We have carried out infections of cells in culture and analyzed them using the three quantitative PCR assays for cDNA forms. We used cell cycle inhibitors to block cell division, thereby eliminating the complication of dilution of unintegrated viral DNA during cell growth. To eliminate complications due to cell killing by HIV, we compared results with HIV to those obtained using nontoxic HIV-based vectors.
We found that 2-LTR circles, once formed, persisted indefinitely. Similar results have been obtained by Pierson et al. (17a). These data call into question the use of 2-LTR circles as a marker for ongoing viral replication in patients.
We have also used the proteasome inhibitor MG132 to analyze the effects of the proteasome on the viral cDNA generated after infection (19). Addition of the inhibitor does not greatly affect the initial accumulation of unintegrated viral cDNA but greatly promotes its persistence. This indicates that the main substrate for the proteasome is cDNA complexes that have mostly completed reverse transcription.
MATERIALS AND METHODS
Cells and viruses.
SupT1 cells were maintained and infected in RPMI 1640 medium-10% fetal bovine serum (BioWhittaker). 293T cells were maintained in Dulbecco's modified Eagle medium-10% fetal bovine serum.
The SM2 HIV-based vector was produced from the cell line SKSM2 as described (5). The self-inactivating (SIN) HIV vector (p156RRLsinPPTCMVGFPPRE) is described elsewhere (4). The SIN HIV-based vector supernatants were prepared by three-plasmid cotransfection into 293T cells with pVSVG, pdeltaR9 (14), and p156RRLsinPPTCMVGFPPRE. Titers of vector stocks were measured as the number of infectious units forming gfp-positive centers on 293T cells per ml. Typical vector stocks were about 3 × 107 infectious units per ml. The 293T cells are highly permissive for infection, so titers seen on the SupT1 cells used here are lower.
The HIV type 1 (HIV-1) virus used was generated by transfection of a plasmid encoding the R9 strain of HIV; stocks of HIV-1 (R9) (24) were expanded by infecting SupT1 cells in the presence of Polybrene (8 μg/ml). Supernatants were harvested when maximal supernatant p24 levels (240 to 540 ng/ml) were reached, and the resulting virus was used to initiate experimental infections immediately after filtration through 0.45-μm-pore-size filters. For stocks of HIV, viral titers were measured as the number of positive centers generated by infection of 5.25 reporter cells per milliliter of viral supernatant.
Viral infections and DNA isolation.
Infections were started with ∼5 × 106 cells in 25-ml flasks. Supernatants of vector-derived virus were filtered through 0.45-μm-pore-size filters and treated with DNase I (Roche) at 10 to 20 U/ml for 60 min at room temperature to prevent viral DNA carryover. Vector-derived virus was added to cells in minimal volume with DEAE-dextran (20 μg/ml) and incubated at 37°C. Single-round infections with replication-competent HIV were carried out by adding amprenivir and nelfinavir, each at 5 μM. The viral infections in the single-round HIV infection were initiated using a modification of the spinoculation protocol previously reported (15). In a 12-well plate, 7 × 106 cells were resuspended in fresh viral supernatant and any indicated inhibitor. The plates were spun at 1,200 × g for 1 h and incubated for another hour before the supernatant was replaced with fresh medium. DNA time points were collected by removing ∼105 cells (as a constant fraction of the culture volume) and washing them before lysis. For late time points in cytopathic HIV infections, where many cells were in syncytia, samples were harvested as a constant fraction of the culture volume. Titrations revealed that a wide range of aphidicolin concentrations (0.3 to 10 μM) were sufficient to block division of SupT1 cells. Inhibitors—aphidicolin (0.3 to 10 μg/ml; Sigma) or 50 μM Z-Leu-Leu-Leu-CHO (MG132; BioMol)—were added. Dimethyl sulfoxide concentrations, generally 0.1 to 0.2% (final), were normalized in all cultures. To isolate DNA, cells were washed with phosphate-buffered saline, and DNA was harvested using the DNeasy tissue kit (Qiagen).
Quantitative PCR.
DNA samples were quantitated by optical density at 260 nm and brought to the same concentration before use in quantitative PCR. Because similar numbers of cells were harvested at each time point, the adjustments necessary at this stage were minimal except in the cytopathic HIV infections, where yields were lower at the later time points. The primer sets used to detect each sequence (purchased from GenSet or Integrated DNA Technologies) were as follows: total cDNA, MH531 (5′-TGTGTGCCCGTCTGTTGTGT-3′); total cDNA reverse, MH532 (5′-GAGTCCTGCGTCGAGAGAGC-3′); total cDNA probe, LRT-P [5′-(FAM)-CAGTGGCGCCCGAACAGGGA-(TAMRA)-3′]; 2-LTR circle forward, MH535 (5′-AACTAGGGAACCCACTGCTTAAG-3′); 2-LTR reverse, MH536 (5′-TCCACAGATCAAGGATATCTTGTC-3′); 2-LTR probe, MH603 [5′-(FAM)-ACACTACTTGAAGCACTCAAGGCAAGCTTT-(TAMRA)-3′]; Alu forward, MH535 (above); Alu reverse, SB704 (5′-TGCTGGGATTACAGGCGTGAG-3′); Alu probe, MH603 (above). With each experiment, a standard curve of the amplicon being measured was run in duplicate, ranging from 10 to 107 copies plus a negative control lacking template. For the total viral cDNA and 2-LTR circle assays, standard curves were generated by dilution of cloned DNAs with matching sequences. In all cases the DNA standards were diluted into 250 ng of uninfected cellular DNA to match the cellular DNA samples.
The Alu copy number standard DNA was generated as described previously (2). 293T or SupT1 cells were infected with a high-titer HIV-based vector and then grown for several weeks to dilute away all the unintegrated viral cDNA forms. Cellular DNA was harvested, and then a dilution series was prepared by diluting the infected-cell DNA into DNA from uninfected cells. The number of proviruses per weight of total DNA was then determined using the total HIV cDNA primers. Separately, the Alu PCR signal was determined for each point in the titration. This allowed a standard curve to be generated relating the number of integrated proviruses to the Alu PCR signal. DNA samples from infected cells could then be quantitated by measuring the Alu PCR signal and reading the number of proviruses off the standard curve (Fig. 2A).
FIG. 3.
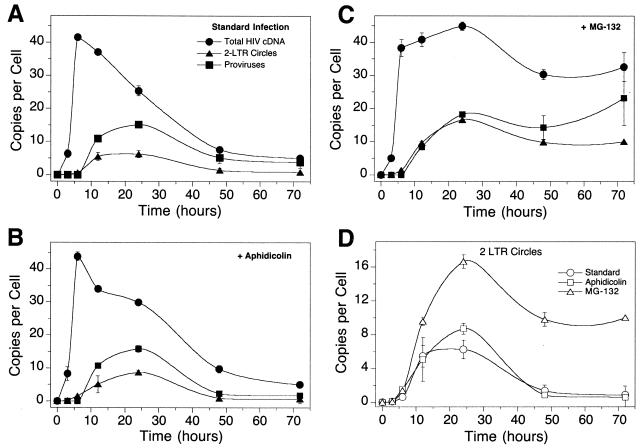
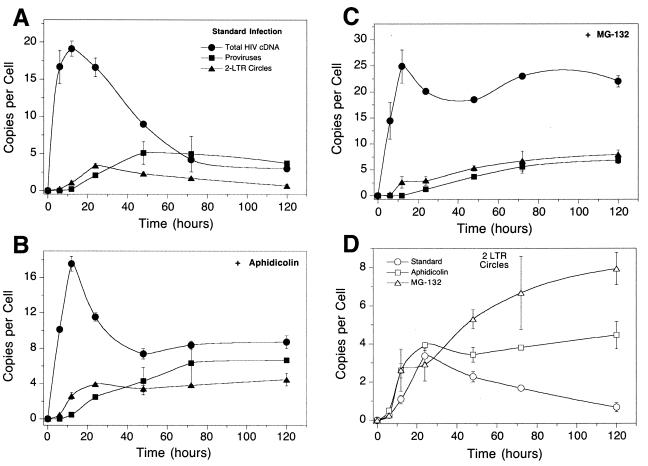
HIV cDNA metabolism in single-cycle infections of SupT1 cells with HIV-1 assayed by quantitative PCR. (A) Infection of SupT1 cells with HIV R9 in the absence of added cell cycle inhibitors. The numbers of copies of each cDNA form per cell are as indicated. Points are labeled as in Fig. 1. (B) Infection as shown in panel A but with aphidicolin (10 μg/ml). (C) Infection as shown in panel A but with 50 μM MG132. (D) Comparison of 2-LTR circle formation in the assays in panels A to C. Error bars, standard deviations.
Reaction mixtures contained 1× Taqman universal master mix (Perkin-Elmer), 300 nM forward primer, 300 nM reverse primer, 100 nM probe primer, and 250 ng of template DNA. After initial incubations at 50°C for 2 min and 95°C for 10 min, 40 cycles of amplification were carried out for 15 s at 95°C followed by 1 min at 60°C (1 min 30 s for Alu PCR). Reactions were carried out in optical-quality 96-tube PCR plates and analyzed using the ABI Prism 7700 sequence detection system (PE-Applied Biosystems). Results were analyzed with ABI Prism SDS software. Detailed protocols can be found at http://www.salk.edu/LABS/idl/idlb/idlbfram.html.
RESULTS
Fluorescence-monitored PCR analysis of low-multiplicity infection with HIV.
Our previous studies with the fluorescence-monitored PCR assays for reverse transcription, 2-LTR circle formation, and integration focused on infections with HIV-based vectors (2, 9). Since the goal of this study was to understand the stability of cDNA forms in cells infected with replication-competent HIV, we began by characterizing infection of SupT1 lymphoid cells with wild-type HIV. Cells were exposed to viral supernatants for 2 h, and then the supernatants were removed and samples were harvested as a function of time after initiating infection. Samples of total DNA, including both integrated and unintegrated forms, were then analyzed at each time using the three quantitative assays.
We first characterized a multicycle infection at a low multiplicity (0.04 infectious units per cell; titers determined by infection of 5.25 reporter cells). The number of total HIV cDNA copies (Fig. 2B) showed a transient increase, peaking at 8 to 12 h after infection, at about 0.25 copy per cell, and then declining. At about 30 h the number of HIV cDNA copies again ascended, reaching about four copies per cell at 48 h. At this point the cultures were terminated due to the extensive cytopathic effect of infection. This second increase in total HIV DNA copies likely represents a second round of infection by HIV produced from the initially infected cells.
Analysis of 2-LTR circles (Fig. 2C) showed an initial peak at 15 h of 0.01 copy per cell, followed by a decline and then a substantial increase by 48 h. Integrated proviruses, analyzed by Alu PCR, were not detectable until the second round, accumulating to 0.06 copy per cell by 48 h. To investigate the mechanism of decrease of 2-LTR circles in a manner more like that of Sharkey et al. (21)—where 2-LTR circles were judged to be labile—we next carried out single-cycle infections with HIV.
Single-cycle infection with HIV-1.
The protease inhibitors amprenivir and nelfinavir were added to infected cultures to restrict HIV replication to a single round. HIV was applied immediately after harvesting to SupT1 cells using spinoculation. These manipulations resulted in a greatly enhanced multiplicity of infection (15). (Similar high levels of infection have been seen in several studies [1, 2, 7, 13, 18; J. Olvera and F. D. Bushman, unpublished data].) Figure 2 presents quantitation of total HIV cDNA, 2-LTR circles, and integrated proviruses present in the cells as a function of time after initial infection. Reverse transcription products peaked in abundance after 12 h at 40 copies per cell (Fig. 3A) and then declined thereafter. The 2-LTR circles reached maximum abundance of about six per cell at 12 to 24 h and then decreased. Integrated proviruses peaked in abundance at about 12 per cell at 24 h and then declined thereafter.
FIG. 1.
Diagram of the early steps of HIV infection, indicating the locations of primers used for the quantitative PCR assays. The RNA genome (A) is converted to a linear double-stranded cDNA form by reverse transcription (B). Some of the viral cDNA is circularized (C), while other HIV cDNA molecules are integrated in to the cellular DNA (D). Different proviruses are integrated at different distances from cellular Alu repeats.
By 48 h many of the cells were in syncytia, and by 72 h most cells were in syncytia and considerable debris had accumulated in the cultures, indicative of cell death. In studies of infections with HIV-based vectors (2; also see below), the abundance of integrated proviruses reached a maximal level at about 48 h that did not change thereafter, and no syncytium formation or cell death was seen. Thus, the loss of integrated proviruses correlates with death of cells in the culture. At late times, rare uninfected cells are expected to survive disproportionately and continue to replicate. Thus, standardizing the amount of DNA in Taqman samples to 250 ng resulted in increasing the representation of uninfected-cell DNA at late times, accounting for the observed decline in the HIV DNA forms.
Effect of aphidicolin on HIV replication.
We next asked whether the decline in abundance of 2-LTR circles might be due to dilution during cell growth. If cells divide and segregate unintegrated cDNA and the quantitative assays are always normalized to the mass of input cellular DNA (250 ng), then dilution during growth will lead to an apparent loss of 2-LTR circles. To examine the possible effects of dilution, aphidicolin (10 μg/ml) was added to inhibit the progression of the cell cycle (Fig. 3B) and DNAs analyzed as in Fig. 3A.
The quantitative data obtained were similar to those seen in the absence of aphidicolin. The number of unintegrated HIV cDNA copies increased to a high level and then declined. The 2-LTR circles and integrated proviruses peaked at about 24 h at the same levels and then declined with similar kinetics. The aphidicolin-treated culture (Fig. 3B) showed a strong cytopathic effect, as was seen with the standard culture in Fig. 3A. Most cells were in syncytia by 72 h, and considerable cell debris was present. HIV DNA forms decreased in abundance over time, again correlating with death of infected cells. We conclude that cell division was not a major factor in the untreated HIV-infected culture shown in Fig. 3A, but rather the cytopathic effect dominated the outcome of the quantitative assays.
Effect of MG132 on HIV replication.
In contrast, arresting the cell cycle by treatment with 50 μM proteasome inhibitor MG132 had a dramatic effect on the accumulation of viral cDNA (Fig. 3C). The number of total cDNA copies rose to the same level as in the standard culture over the first 12 h but then remained at a much higher level for the remainder of the experiment. MG132-treated infections showed a slight decrease in all forms of viral DNA between 24 and 48 h potentially due to cell death, but the overall amount of degradation was greatly reduced. These data confirm that the proteasome is responsible for the degradation of a significant fraction of the viral cDNA, as previously reported (19). The proteasome is not expected itself to degrade the viral cDNA but probably degrades proteins bound to cDNA, thereby exposing the cDNA to nucleases that carry out degradation.
Figure 3D plots the accumulation of 2-LTR circles in the infections depicted in Fig. 3A to C. The time of peak accumulation was the same in all cases, but the peak was about twice as high in the presence of MG132, and the values remained at a high level (at least 10 copies per cell) instead of falling steadily.
Comparison with infections using an HIV-based vector.
The studies in Fig. 3 showed that 2-LTR circles can decline in abundance during a single-cycle infection, but in the example studied this was due mainly to the loss of infected cells. The stability of two LTR circles was therefore studied in an experimental paradigm in which infected cells remained healthy, which was achieved by using lentivirus vectors instead of HIV. The vector particles used were produced by a cell engineered to express (i) HIV Gag-Pol, (ii) a packageable vector portion transducing gfp, and (iii) the vesicular stomatitis virus type G envelope. Infection of SupT1 cells with high titers of the HIV vectors did not lead to any noticeable cytopathic effect (unpublished data). To examine the effects of dilution of viral cDNAs during cell growth, the infections were also carried out in the presence of aphidicolin and MG132.
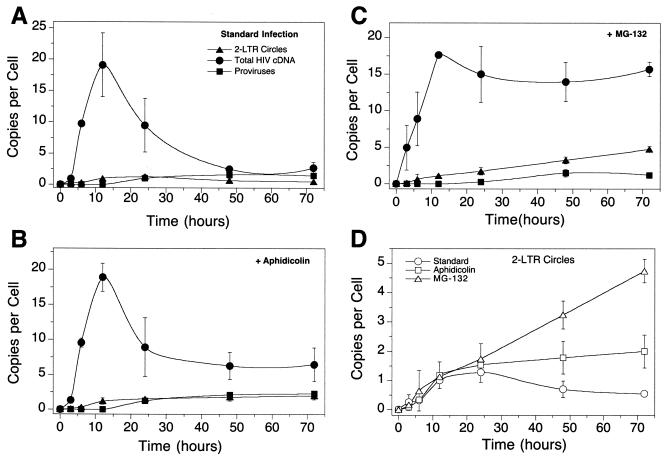
In the absence of any inhibitors (Fig. 4A), total HIV cDNA copies peaked around 12 h postinfection at 20 copies per cell with the vector stock studied. The amount of total cDNA decreased over the next 50 to 60 h. By about 50 h, the 2-LTR circles and integrated proviruses accounted for most of the measured total viral cDNA, and this continued to be the case out to 72 h. This is consistent with a published analysis of a similar infection assayed by Southern blotting, which revealed that all detectable linear cDNA was gone by 72 h and only proviral and 2-LTR circle forms could be observed (2).
FIG. 4.
Metabolism of cDNA synthesized by infection with the SM2 HIV-based vector. (A) Infection of SupT1 cells with the SM2 vector in the absence of added cell cycle inhibitors. (B) Infection as shown in panel A but with aphidicolin (10 μg/ml). (C) Infection as shown in panel A but with 50 μM MG132. (D) Comparison of 2-LTR circle formation in the assays in panels A to C. Error bars, standard deviations.
The 2-LTR circles peaked in abundance around 24 h at about 1.5 copies per cell and decreased slowly afterward. The rate of decline of 2-LTR circles was about that expected due to dilution during growth. Integrated proviruses reached a maximum by 48 h of about one copy per cell and remained constant thereafter (since proviruses are duplicated with each cell division).
Infections with an HIV-based vector in the presence of aphidicolin.
When the cell cycle is arrested with aphidicolin, the contribution of dilution during cell growth is eliminated, allowing the chemical stability of 2-LTR circles to be measured. Figure 4B shows that, in the absence of cell replication, total cDNA copies still peak at 12 h and subsequently decrease rapidly, but that rate of decrease slows to a plateau value of about eight cDNA copies per cell. By 24 to 48 h postinfection, the number of total cDNA copies per cell can be mostly accounted for by the measured proviruses and 2-LTR circles—that is, the linear cDNA has all been transformed to the circular and integrated forms.
The 2-LTR circles peak in abundance after 12 to 24 h but do not decrease in abundance in nondividing cells (Fig. 4B and D). Thus, eliminating (i) dilution during cell growth and (ii) loss of infected cells due to cytopathic effects blocks the decline in the abundance of 2-LTR circles. These data indicate that 2-LTR circles are not degraded by cellular nucleases but are actually quite stable.
Infections with an HIV-based vector in the presence of MG132.
Infections with HIV-based vectors in the presence of the proteasome inhibitor MG132 highlight the role of the proteasome in degradation of the viral cDNA (Fig. 4C). Total HIV cDNA accumulated as in Fig. 4A and B but did not decline significantly in abundance thereafter. Integrated proviruses accumulated to similar levels in the presence of MG132 as in the other infections, but the level of 2-LTR circles went up steadily instead of leveling off with time. The lack of degradation apparently allowed accumulation of circles over a longer time than is possible during normal infection. In the MG132-treated culture, the 2-LTR circles and provirus did not account for the total viral cDNA detected; thus, there does seem to be linear cDNA present that can be slowly converted to 2-LTR circles. This is not the case in the standard infection or in the aphidicolin-treated culture, where proviruses and 2-LTR circles did account for most or all of the DNA detected after 24 to 48 h. Figure 2D compares the 2-LTR circle data for all three infections, indicating that 2-LTR circles did not decline in abundance when cells were not dividing. The decline in abundance seen in dividing cells is about that expected for dilution during cell growth.
Comparison with results using a SIN HIV vector.
To test the generality of the results in Fig. 4, similar studies were carried out with another type of HIV-based vector. The SIN retroviral vector of Follenzi et al. (4), which contains a large deletion in U3 that becomes incorporated into both LTRs during reverse transcription, was studied. Figure 5A presents data from an infection with these vectors. The results are generally similar to those with the SM2 vector with respect to the timing of the appearance of the viral cDNA forms and the maximum number of cDNA copies produced (about 20 copies per cell). The formation of proviruses was more efficient in this experiment, with a maximum of four to five (Fig. 5A) proviruses per cell instead of one to two (Fig. 4A). The reason for the observed differences in the efficiency in provirus formation is unclear and a topic of ongoing study. As with the SM2 vector, 2-LTR circles formed with the SIN vector peak in abundance by about 24 h and then declined roughly as expected by dilution during cell growth.
FIG. 5.
Metabolism of cDNA synthesized by infection with the SIN HIV-based vector. (A) Infection of SupT1 cells with the SIN vector in the absence of added cell cycle inhibitors. (B) Infection as shown in panel A but with aphidicolin (10 μg/ml). (C) Infection as shown in panel A but with 50 μM MG132. (D) Comparison of 2-LTR circle formation in the assays in panels A to C. Error bars, standard deviations.
Addition of aphidicolin to block cell cycle progression changed the outcome substantially (Fig. 5B), yielding data similar to that seen with the SM2 vector in the presence of aphidicolin (Fig. 4B). The maximum number of total HIV cDNA copies was formed by about 12 h but then declined to a plateau by about 40 h (compare Fig. 5A and B). The 2-LTR circles accumulated to about four per cell and then remained at that level for the rest of the time analyzed (out to 120 h), again confirming the stability of 2-LTR circles. Proviruses climbed in abundance to about six per cell and then remained constant. After about 48 h, most of the cDNA detected could be accounted for by the 2-LTR circles and integrated proviruses.
Addition of MG132 (Fig. 5C) yielded data as with the SM2 vector (Fig. 4C). The number of total cDNA copies reached a maximum (around 24 copies per cell) at about 12 h but then remained high. The numbers of proviruses and 2-LTR circles increased gradually for the full 120 h of the experiment. Figure 5D presents the data for 2-LTR circles generated by the SIN vector in the three infections. In summary, studies with the SIN vector generally mirrored those with the SM2 vector, with the exception that the conversion of unintegrated cDNA copies (linear form) to proviruses was somewhat more efficient.
A covert steady state does not account for the apparent stability of 2-LTR circles.
We have argued that in infections with HIV vectors in the presence of aphidicolin, 2-LTR circles were stable once formed (Fig. 4B and D and 5B and D; 48 h and after). One possible alternative was that 2-LTR circles are degraded but replaced at the same rate. That is, perhaps the measured constant number of 2-LTR circles per cell late during infection (Fig. 4B and D and 5B and D) was due to a covert steady state. The data argue against the idea that preexisting linear cDNA was converted to 2-LTR circles in the hypothetical steady state, since after about 48 h most or all of the cDNA was either circular or integrated. This can be seen in Fig. 4B and 5B, where after about 48 h the measured 2-LTR circular DNA and proviral DNA accounted for most or all of the observed total HIV cDNA. Thus, there was not enough linear DNA present to support a covert steady state.
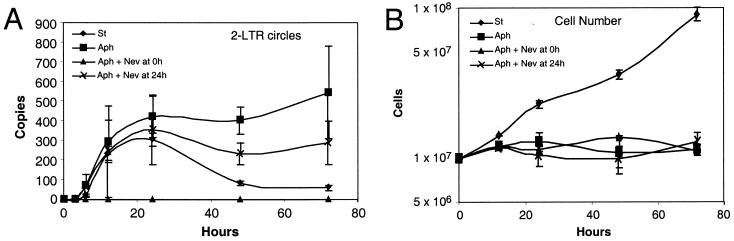
However, a related possibility is that ongoing reverse transcription forms new linear cDNA molecules late during infection that are quickly circularized and that this underlies a covert steady state. To challenge this idea, we infected SupT1 cells with an HIV-based vector in the presence of aphidicolin but added the reverse transcriptase inhibitor nevirapine at different times after infection (Fig. 6). Addition of nevirapine with virus (time zero) blocked accumulation of 2-LTR circles, as expected (Fig. 6A). Addition of nevirapine at 24 h after infection resulted in a slight decrease in reverse transcription products, but importantly the numbers of 2-LTR circles remained constant thereafter (Fig. 6A). Measurement of cell number confirmed that aphidicolin blocked cell replication (Fig. 6B). We conclude that continued reverse transcription cannot support a covert steady state and that 2-LTR circles must indeed be stable in the infections studied.
FIG. 6.
A covert steady-state model does not account for the observed stability of 2-LTR circles. Cells were infected with the SM2 HIV-based vector (spinoculation was not used), and aliquots were analyzed. Cells were treated with 0.3 μM aphidicolin (Aph), aphidicolin plus nevirapine at time zero (Aph + Nev at 0 h), aphidicolin plus nevirapine at 24 h (Aph + Nev at 24 h), or no added drug (St [for “standard”]). (A) Assay of 2-LTR circle DNA. (B) Viable cell number in cultures studied (assayed by trypan blue exclusion), indicating that aphidicolin arrested the cell cycle. Error bars, standard deviations.
DISCUSSION
Stability of 2-LTR circles.
It has been proposed that 2-LTR circles are unstable in cells and so are useable as a marker for ongoing infection in patients undergoing successful HAART (21). However, this work and the accompanying work (17a) report that 2-LTR circles are in fact quite stable in the infections studied, suggesting that caution should be used in interpreting results from studies of 2-LTR circles in patients.
2-LTR circles, once formed, can decline in abundance (copy number per cell) by any of three pathways. They can be degraded by cellular nucleases, they can be diluted by cell division, or they can be lost due to death of infected cells. We found that in infections with replication-competent HIV, 2-LTR circles did indeed decline in abundance, but this correlated with death of infected cells. When noncytotoxic HIV-based vectors were studied, 2-LTR circles also declined in abundance, but in this case the cells were doubling normally and the 2-LTR circles declined in abundance as expected for dilution during cell growth. A careful study by Pierson and coworkers has documented this point particularly clearly (17a). To eliminate the effect of dilution, we arrested the cell cycle with aphidicolin, and to reduce the toxicity of infection, we infected cells with HIV-based vectors. We found that 2-LTR circles reached their maximal abundance after 12 to 24 h and remained constant thereafter, indicating that 2-LTR circles, once formed, are quite stable.
These observations parallel findings in other systems in which circular DNA forms were found to be stable in animal cells. The circular DNAs generated by RAG-mediated recombination at the T-cell receptor locus (T-cell receptor excision circles) are quite stable, allowing them to be used as markers for T-cell development (10-12, 20, 22). Circular parvovirus genomes have also been found to persist for long periods inside cells (see reference 23 and references therein). We have argued elsewhere that circularization of linear HIV DNA by the cellular nonhomologous DNA end-joining pathway diminishes its toxicity, leading to a preservation of infected cells (9). All of these findings support the idea that 2-LTR circles are likely to be stable in cells once formed.
In contrast, a previous report proposed that 2-LTR circles did not persist over time if ongoing replication was blocked. This formed the basis for analyzing samples from patients undergoing successful HAART. The finding that 2-LTR circles were present in patient samples supported the conclusion that infection was ongoing despite successful HAART (21). Several scenarios could explain the observed decline in the number of 2-LTR circles seen in this study. Any toxicity of HIV infection in the cells studied could have caused the 2-LTR circles to fall in number with the death of infected cells. Effects of dilution during cell division could also have caused the fall in number of 2-LTR circles observed. If these conditions hold in vivo, then detection of 2-LTR circles in patients undergoing prolonged HAART would imply ongoing covert HIV replication. However, another explanation is that 2-LTR circles persist in long-lived nondividing cells, also potentially explaining their detection after prolonged HAART. In this case the conclusion of ongoing covert replication would be mistaken. We feel that the results in the present work, and similar studies by Pierson et al. (17a), establish that 2-LTR circle detection is not by itself a reliable marker for ongoing HIV replication.
The use of cell cycle inhibitors raises the question of whether this approach might have influenced the outcome of the experiment. Possibly cell cycle-regulated nucleases were not expressed at G1, the point of arrest by aphidicolin, thereby artifactually stabilizing 2-LTR circles. Arguing against this possibility is the observation that in dividing cells, the abundance of 2-LTR circles falls at about the rate expected for dilution by cell division, also indicating stability.
Antiviral effect of the proteasome.
Schwartz and coworkers reported previously that the proteasome exerts an antiviral effect by suppressing accumulation of viral cDNA (19). They found that HIV infection of cells treated with the proteasome inhibitor MG132 or lactacystin yielded increased titers and that unintegrated cDNA forms (linear cDNA and circles) were increased in abundance in the treated cells compared to untreated controls. They also found that Tat-activated transcription from the HIV LTR was normal in the presence of the inhibitors, allowing them to propose that inhibiting the proteasome resulted in the formation of increased numbers of integrated proviruses.
Data from the quantitative assays reported here strengthen and extend these conclusions. The quantitative Alu PCR method allowed the increase in provirus formation to be measured directly rather than inferred. Moreover, comparison of infections in the presence and absence of MG132 revealed that inhibiting proteasome-mediated degradation does not result in a higher peak of production of total viral cDNA copies. Rather, the viral cDNA reaches roughly the same peak level of abundance but does not decline thereafter, indicating that the proteasome is acting after the completion of reverse transcription (or at least after the second-strand transfer event, which is required for formation of the amplicon used to detect total HIV cDNA). Inhibiting proteasome-mediated degradation apparently increased the level of substrates for the circularization reaction, leading to increased numbers of 2-LTR circles per cell. Increased provirus formation was also seen in some experiments. These findings indicate that the substrates for proteasome-mediated degradation are complexes containing largely completed reverse transcription products or fully formed preintegration complexes. In the future, the ability to stabilize viral replication intermediates by addition of MG132 may aid biochemical efforts to purify cDNA complexes.
Acknowledgments
We thank R. Siliciano, T. Pierson, and members of the Salk Institute Infectious Disease Laboratory for helpful discussions.
This work was supported by NIH grants GM56553 and AI34786 to F.D.B., the James B. Pendleton Charitable Trust, the Berger Foundation, and Cornelia Mackey.
REFERENCES
- 1.Bell, P., L. J. Montaner, and G. G. Maul. 2001. Accumulation and intranuclear distribution of unintegrated human immunodeficiency virus type 1 DNA. J. Virol. 75:7683-7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler, S., M. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV cDNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 3.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 4.Follenzi, A., L. E. Ailes, S. Bakovic, M. Gueuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 5.Hansen, M. S. T., G. J. I. Smith, T. Kafri, V. Molteni, J. S. Siegel, and F. D. Bushman. 1999. Integration complexes derived from HIV vectors for rapid assays in vitro. Nat. Biotechnol. 17:578-582. [DOI] [PubMed] [Google Scholar]
- 6.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 7.Kim, S., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewin, S. R., M. Vesanen, L. Kostrikis, A. Hurley, M. Duran, L. Zhang, D. D. Ho, and M. Markowitz. 1999. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 73:6099-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, L., J. M. Olvera, K. Yoder, R. S. Mitchell, S. L. Butler, M. R. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livak, F., and D. Schatz. 1996. T-cell receptor alpha locus V(D)J recombination by-products in thymocytes and mature T cells. Mol. Cell. Biol. 16:609-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markert, M. L., C. B. Hicks, J. A. Bartlett, J. L. Harmon, L. P. Hale, et al. 2000. Effect of highly active antiretroviral therapy and thymic transplantation on immunoreconstitution in HIV infection. AIDS Res. Hum. Retrovir. 16:403-413. [DOI] [PubMed] [Google Scholar]
- 12.McFarland, R. D., D. C. Douek, R. A. Koup, and L. J. Picker. 2000. Identification of a human recent thymic emigrant phenotype. Proc. Natl. Acad. Sci. USA 97:4215-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muesing, M. A., D. A. Smith, C. D. Cabradilla, C. V. Benton, L. A. Lasky, and D. J. Capon. 1985. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature 313:450-458. [DOI] [PubMed] [Google Scholar]
- 14.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 15.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panther, L. A., R. W. Coombs, J. E. Zeh, A. C. Collier, and L. Corey. 1998. Unintegrated circular HIV-1 DNA in the peripheral mononuclear cells of HIV-1-infected subjects: association with high levels of plasma HIV-1 RNA, rapid decline in CD4 count, and clinical progression to AIDS. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 17:303-313. [DOI] [PubMed] [Google Scholar]
- 17.Pauza, C. D., P. Trivedi, T. S. McKechnie, D. D. Richman, and F. M. Graziano. 1994. 2-LTR circular viral DNA as a marker for human immunodeficiency virus type 1 infection in vivo. Virology 205:470-478. [DOI] [PubMed] [Google Scholar]
- 17a.Pierson, T., T. L. Kieffer, C. T. Ruff, C. Buck, S. J. Gange, and R. F. Siliciano. 2002. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J. Virol. 76:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson, H. L., and D. M. Zinkus. 1990. Accumulation of human immunodeficiency virus type 1 DNA in T cells: results of multiple infection events. J. Virol. 64:4836-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz, O., V. Marechal, B. Friguet, F. Arenzana-Seisdedos, and J.-M. Heard. 1998. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J. Virol. 72:3845-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sempowski, G. D., L. P. Hale, J. S. Sundy, J. M. Massey, R. A. Koup, et al. 2000. Leukemia inhibitory factor, oncostatin M, IL-6, and stem cell factor mRNA expression in human thymus increases with age and is associated with thymic atrophy. J. Immunol. 164:2180-2187. [DOI] [PubMed] [Google Scholar]
- 21.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. I. Ellision, B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sodora, D. L., D. C. Douek, G. Silvestri, L. Montgomery, M. Rosenzweig, T. Igarashi, B. Bernacky, R. P. Johnson, M. B. Feinberg, M. A. Martin, and R. A. Koup. 2000. Quantification of thymic function by measuring T cell receptor excision circles within peripheral blood and lymphoid tissues in monkeys. Eur. J. Immunol. 30:1145-1153. [DOI] [PubMed] [Google Scholar]
- 23.Song, S., P. J. Laipis, K. I. Berns, and T. R. Flotte. 2001. Effect of DNA-dependent protein kinase on the molecular fate of the rAAV2 genome in skeletal muscle. Proc. Natl. Acad. Sci. USA 98:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swingler, S., P. Gallay, D. Camaur, J. Song, A. Abo, and D. Trono. 1997. The Nef protein of human immunodeficiency virus type 1 enhances serine phosphorylation of the viral matrix. J. Virol. 71:4372-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zazzi, M., L. Romano, M. Catucci, G. Venturi, A. De Milito, P. Almi, A. Gonnelli, M. Rubino, U. Occhini, and P. E. Valensin. 1997. Evaluation of the presence of 2-LTR HIV-1 unintegrated DNA as a simple molecular predictor of disease progression. J. Med. Virol. 52:20-25. [DOI] [PubMed] [Google Scholar]
- 26.Zufferey, R., T. Dull, R. Mandel, A. Bukovsky, D. Quiroz, L. Naldini, and D. Trono. 1998. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72:9873-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]