Abstract
Bovine herpesvirus 5 (BHV-5) is a neurovirulent alphaherpesvirus that causes fatal encephalitis in calves. In a rabbit model, the virus invades the central nervous system (CNS) anterogradely from the olfactory mucosa following intranasal infection. In addition to glycoproteins E and I (gE and gI, respectively), Us9 and its homologue in alphaherpesviruses are necessary for the viral anterograde spread from the presynaptic to postsynaptic neurons. The BHV-5 Us9 gene sequence was determined, and the predicted amino acid sequence of BHV-5 Us9 was compared with the corresponding Us9 sequences of BHV-1.1. Alignment results showed that they share 77% identity and 83% similarity. BHV-5 Us9 peptide-specific antibody recognized a doublet of 17- and 19-kDa protein bands in BHV-5-infected cell lysates and in purified virions. To determine the role of the BHV-5 Us9 gene in BHV-5 neuropathogenesis, a BHV-5 Us9 deletion recombinant was generated and its neurovirulence and neuroinvasive properties were compared with those of a Us9 rescue mutant of BHV-5 in a rabbit model. Following intranasal infection, the Us9 rescue mutant of BHV-5 displayed a wild-type level of neurovirulence and neural spread in the olfactory pathway, but the Us9 deletion mutant of BHV-5 was virtually avirulent and failed to invade the CNS. In the olfactory mucosa containing the olfactory receptor neurons, the Us9 deletion mutant virus replicated with an efficiency similar to that of the Us9 rescue mutant of BHV-5. However, the Us9 deletion mutant virus was not transported to the bulb. Confocal microscopy of the olfactory epithelium detected similar amounts of virus-specific antigens in the cell bodies of olfactory receptor neuron for both the viruses, but only the Us9 rescue mutant viral proteins were detected in the processes of the olfactory receptor neurons. When injected directly into the bulb, both viruses were equally neurovirulent, and they were transported retrogradely to areas connected to the bulb. Taken together, these results indicate that Us9 is essential for the anterograde spread of the virus from the olfactory mucosa to the bulb.
Bovine herpesvirus 5 (BHV-5) is a neurovirulent alphaherpesvirus that causes fatal encephalitis in calves (4, 19). BHV-1 is associated with abortions, respiratory infections (subtype 1.1), and genital infections (subtype 1.2) in cattle (42) but does not cause encephalitis. Both BHV-1 and BHV-5 are neurotropic viruses, and they establish latency in the trigeminal ganglion (TG) following intranasal and conjunctival inoculation (1, 35). The viruses show 85% DNA homology but differ in their ability to cause neurological disease in calves (4). In a rabbit seizure model, BHV-1.1 and BHV-5 infections are distinguished by their differential neuropathogenesis (14). For example, following intranasal inoculation nasal swabs yielded higher quantities of BHV-1 than BHV-5; however, only BHV-5 invades the central nervous system (CNS) via the olfactory pathway (28). When inoculated intranasally, BHV-5 invades the brain via the olfactory pathway and produces acute neurological signs that are comparable to those seen in calves (28). The neural spread and neuronal damage are located in the olfactory bulb and the areas connected to the olfactory bulb (the anterior olfactory nucleus, piriform or entorhinal cortex, frontal or cingulate cortex, hippocampus or dentate gyrus, amygdala, dorsal raphe, and locus coeruleus) (28). In addition, virus can be found within the TG, but further invasion of the virus to the second-order neurons in the trigeminal pathway of the pons and medulla does not occur (28). In BHV-1-inoculated rabbits, the virus does not invade the CNS, and no neurological signs develop. However, like BHV-5, BHV-1-infected rabbits also have infected neurons in the TG (14, 28). When inoculated intracerebrally, both BHV-1 and BHV-5 are neurovirulent (S. I. Chowdhury, unpublished data).
In pseudorabies virus (PRV), three gene products in the unique short (Us) region of the viral genome are important for anterograde transneural spread. These genes encode the envelope glycoproteins E and I (gE and gI, respectively) and an envelope-tegument protein, Us9 (8, 10, 25, 26, 31). The gE/gI homologues in alphaherpesviruses are expressed on the infected-cell membrane. They complex together to form a functional unit that is required for cell-to-cell spread in vitro and virulence in vivo (16, 21). Both in herpes simplex virus (HSV) and in PRV, either gE or gI deletion affects the anterograde transport of the virus to visual centers of the rat brain after intraocular inoculation (10, 17, 20, 40). In BHV-5, gE deletion affected the anterograde transport of the virus from the olfactory receptor neurons to the CNS and neurovirulence in the brain (14).
In PRV, Us9 is an unglycosylated envelope protein (6). While the same is likely true for all alphaherpesviruses, this has not been demonstrated. In HSV type 1 (HSV-1), Us9 protein is ubiquitinated, and the ubiquitinated form of the protein is incorporated in the virion (5). In the virus-infected cells, it is associated with proteasome (5). Since the ubiquitinated Us9 remains stable and the Us9 coding sequence is well conserved in alphaherpesviruses, it has been suggested that one of its functions may be to perturb ubiquitin-mediated protein degradation (5). Unlike the gE/gI deletion mutant viruses, which show a small-plaque phenotype, the Us9 deletion mutant of PRV forms wild-type-size plaques in cell cultures, indicating that Us9 is not involved in cell-to-cell spread of the virus (7, 8). In PRV, Us9 does not form complexes with either gE or gI in infected cells or in the virus particle (8). Interestingly, the Us9 deletion mutant of PRV displays a defect in the anterograde spread of the virus in vivo in the neurons (8, 9). Therefore, the in vivo phenotype of the Us9 deletion mutant virus is very similar to the in vivo phenotype of the gE/gI deletion mutant viruses (9). In PRV, Us9 is predicted to be involved in the transport of the newly enveloped viral particles and/or vesicles containing the enveloped glycoproteins (37, 38). The gE-gI complex in HSV-1 is predicted to interact with a putative cellular receptor on the postsynaptic cell (16). Together, these functions of the three envelope proteins are critical for the transneural spread of the virus.
In this study, we investigated the role of Us9 in the neuropathogenesis of BHV-5. We determined the nucleotide sequence of the BHV-5 Us9 open reading frame (ORF) and compared it with the published coding sequence for the BHV-1 Us9 ORF. To determine the role of Us9 in BHV-5 neuropathogenesis, we generated Us9 deletion mutant and Us9 rescue mutant BHV-5 viruses and analyzed their neuropathogenicity in a rabbit seizure model (14). Using immunofluorescence-confocal microscopy and immunohistochemistry, we compared the ability of Us9 deletion mutant and wild-type BHV-5 to infect the olfactory receptor neurons and its spread to the olfactory bulb and the areas connected to the olfactory bulb. Additionally, spread of wild-type and Us9 deletion mutant BHV-5 within the CNS was evaluated after direct olfactory bulb inoculation.
MATERIALS AND METHODS
Virus strains and cell lines.
The BHV-1 Cooper (Colorado-1) strain, obtained from the American Type Culture Collection (Manassas, Va.), and the BHV-5 TX-89 strain (19) were used in this study. The viruses were propagated and titrated in Madin-Darby bovine kidney (MDBK) cells as previously described (11).
Mapping, cloning, and sequencing of the BHV-5 Us9 gene.
The locations of the BHV-5 Us9 gene coding region on the virus genome and of the pertinent restriction sites for the subcloning and the sequencing strategy are illustrated in Fig. 1. A pUC19- and pBR322-based plasmid library containing the BamHI genomic fragments of BHV-5 DNA was developed (11). To determine the location of the BHV-5 Us9 gene, the BamHI C fragment, which includes the entire Us region of the BHV-5 genome (map units 0.827 and 0.944), was digested with several restriction endonucleases, separated on an agarose gel, and analyzed by hybridization with BHV-1 Us9 coding sequences. Based on the hybridization results, a 2.9-kb PstI/PstI fragment was identified and cloned (pKS99-23) (Fig. 1). A restriction map of the cloned 2.9-kb fragment was constructed, and subclones spanning the entire fragment were generated (Fig. 1B). The DNA sequence was determined by the Iowa State University sequencing facility.
FIG. 1.
BHV-5 genomic structure, the cloning and sequencing strategies of BHV-5 Us9, and schematic map of Us9 deletion plasmid. The genomic organization of BHV-5 depicted at the top consists of unique long (UL) and Us regions and two repeat regions (IR and TR). Localization of the Us9 gene and its flanking gE and bICP22 genes are shown. (A) A restriction site map of the 2.9-kb PstI/PstI fragment contained in plasmid pKS99-23 is shown. (B) Various clones used for the sequencing are shown. (C) The regions of interest are shown in the schematic structure of plasmid pBHV5Us9Δ.
Sequence analysis.
The sequences obtained from clones pKS99-49, pKS99-36, pKS99-47, and pKS99-26 spanning the entire Us9 coding region (Fig. 1) were assembled using the seqaid II sequence analysis software (D. D. Rhodes and D. J. Roufa, Center for Basic Cancer Research, Kansas State University [KSU], Manhattan). The predicted amino acid sequences of BHV-5 Us9 (this study) and BHV-1 Us9 (GenBank accession no. 298199) were aligned using the CLUSTALW alignment program with a ktuple value of 1 and a window of 5. Hydropathicity analysis was performed (27) using a nine-amino-acid (9-aa) window, and the antigenicity profile of the predicted amino acid sequence was analyzed (39) using a 7-aa window. The prediction of signal sequence was determined using the Signal PV1.1 program from the World Wide Web (32).
Production of anti-BHV-5 Us9 peptide-specific polyclonal rabbit sera.
Based on the predicted, regional hydropathicity and antigenicity, the predicted aa 9 to 22 ([H]-NENYRGADAADAAS-[OH]) from BHV-5 Us9 (Fig. 2B) were selected. The peptides were synthesized at the Biotechnology Center of KSU using the 9-fluorenyl-methyloxycarbonyl chemistry on an ABI model 431A automated peptide synthesizer (Applied Biosystems, Inc., Foster City, Calif.) as described earlier (11). After conjugation with polyethylene glycol, the peptides were used to immunize New Zealand White rabbits as described earlier (13).
FIG. 2.
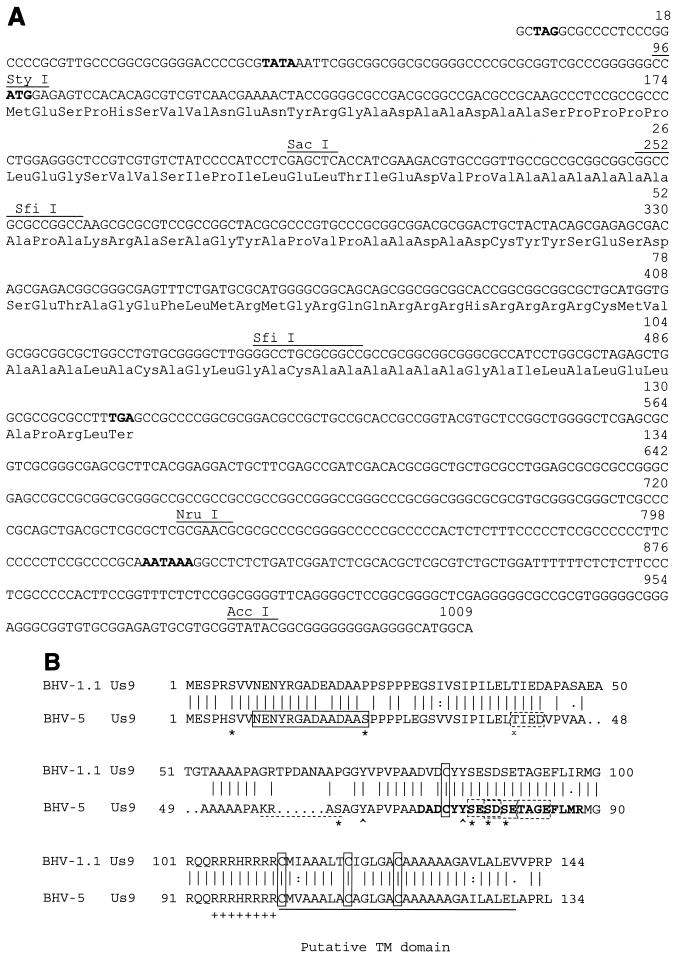
(A) The nucleotide sequence of the BHV-5 Us9 gene. The nucleotide sequence of the ORF is depicted. Putative termination of the neighboring gE gene (TAG) is shown in boldface type. The amino acid sequence of the predicted polypeptide is presented. The putative start codon (ATG), the TATA box, and the consensus polyadenylation signal (AATAAA) associated with the ORF are shown in boldface type. The locations of the StyI and SfiI sites are shown. (B) Comparison of the predicted amino acid sequence of the BHV-5 Us9 with the predicted amino acid sequence of BHV-1.1 (GenBank accession no. 298199). The sequences were aligned by using the Genetics Computer Group Gap program. Conserved cysteine residues are boxed. The transmembrane (TM) anchor sequences are underlined with a solid line. The peptide sequence selected for raising antibody is indicated by a rectangular box (solid lines). The acidic domain is shown in boldface type, and the cluster of positively charged amino acids preceding the putative transmembrane domain is indicated by a string of plus signs. Four casein kinase II phosphorylation sites are indicated (dashed rectangular boxes). A potential cAMP- and cGMP-dependent protein kinase phosphorylation site (KRAS) present only in BHV-5 sequence is indicated (dashed underline). Putative tyrosine kinase-mediated phosphorylation sites (Y62 and Y74) are indicated (∧). In addition six serine residues (S6, S22, S59, S75, S77, S79) (∗) and one threonine (T42) (x) are indicated.
Western blot analysis.
Cellular extracts and purified virion lysates were electrophoresed, unless otherwise mentioned in the figure legend, on a 10%-to-20% gradient sodium dodecyl sulfate (SDS) polyacrylamide gel and transferred to nitrocellulose membrane. Proteins were visualized by using rabbit polyclonal antibodies as described previously (13).
PAA treatment.
To determine the kinetic class of BHV-5 Us9 gene expression, we tested whether its expression is dependent on ongoing DNA synthesis (true γ2 protein) and sensitive to phosphonoacetic acid (PAA). MDBK cells were treated with PAA at a concentration of 400 μg/ml of culture medium 4 h before infection, and this level was maintained throughout infection. As a control, the expression of gC, a known γ2 protein, was tested in a parallel experiment.
Construction of BHV-5 Us9 deletion, β-Gal insertion plasmid.
The BHV-5 Us9 gene is flanked upstream by the gE gene and downstream by bICP22, the gene homologue of the HSV-1 ICP22 gene (Fig. 1A). To delete the Us9 coding region, the plasmid clone pKS99-23, which carries the Us9 gene and its flanking sequences (Fig. 1A), was digested with restriction enzymes SfiI and StyI and blunt ended with T4 polymerase and Klenow enzymes, respectively. Double digestion of the plasmid released a 162-bp StyI/SfiI fragment and a 189-bp SfiI/SfiI fragment containing the majority of the Us9 coding region (Fig. 1A). The blunt-ended larger StyI/SfiI fragment was gel purified and ligated to the blunt-ended 4.5-kb PstI/PstI fragment of plasmid pCMVβ (Clontech, Palo Alto, Calif.) containing the bacterial β-galactosidase (β-Gal) gene. In the resulting plasmid (pBHV5Us9Δβ), the β-Gal gene is flanked by virus-specific 0.72-kb Us9 upstream and 1.8-kb Us9 downstream sequence (Fig. 1C).
Generation of recombinant viruses. (i) Us9 deletion mutant of β-Gal-expressing BHV-5 (BHV-5Us9Δ).
To generate a BHV-5 Us9 deletion mutant recombinant virus, linearized plasmid pBHV5Us9Δβ DNA (Fig. 1) and full-length wild-type BHV-5 DNA were cotransfected in MDBK cells using Lipofectamine (Gibco BRL, Life Technologies, Inc., Grand Island, N.Y.) following the manufacturer's protocol. Recombinant viruses expressing β-Gal were plaque purified three times by screening for blue plaques under a Bluo-Gal (Gibco BRL, Life Technologies, Inc.) overlay as described previously (12). The recombinant viruses were characterized further by Southern blot analysis using the Us9 ORF coding and β-Gal sequences as probes and were confirmed by Western blot analysis with anti-BHV-5 Us9 peptide-specific rabbit polyclonal serum.
(ii) Us9 rescue mutant of BHV-5 (BHV-5Us9R).
BHV-5Us9R was created by cotransfection of BHV-5Us9Δ viral DNA and linearized pKS99-23 plasmid DNA containing the entire Us9 gene and its flanking gE and bICP22 sequences. Recombinant viruses with colorless plaques were picked, plaque purified, and analyzed by Western blotting for the expression of wild-type Us9.
Virus growth curve experiment.
One-step virus growth experiments (12) were conducted to compare the growth kinetics of BHV-5Us9Δ and its parent wild-type BHV-5. A series of replicate cultures of MDBK cells were infected separately with 5 PFU per cell. Infected cultures were harvested at successive intervals postinfection, and their titers were determined by plaque titration assays (12).
Animal experiments.
Four-week-old New Zealand White rabbits weighing 500 to 600 g (Myrtles Rabbitry, Thomson Station, Tenn.) were used. Rabbits were maintained in laboratory isolation cages in our vivarium throughout the experiments, with food and water freely available. All procedures were approved by the KSU Animal Care and Use Committee.
(i) Intranasal infection and immunohistochemical processing.
To compare the neuropathogenic properties of BHV-5Us9Δ and BHV-5Us9R, the rabbit seizure model described previously (14, 28) was used. Ten rabbits were inoculated with BHV-5Us9Δ and five were inoculated with BHV-5Us9R (experiment 1). In each case rabbits were infected intranasally with 2 × 107 PFU of the respective virus (per nostril) into the paranasal sinuses as described earlier (14, 28). Following infection, the rabbits were observed four times a day for the appearance of neurological symptoms. For the group infected with BHV-5Us9Δ, five rabbits were euthanatized at 10 days postinfection (dpi) and the remaining five were euthanatized at 13 dpi. For the group infected with BHV-5Us9R, rabbits were euthanatized at 10 dpi or earlier when they showed severe neurological signs. In experiment 2, nine rabbits were similarly infected with BHV-5Us9Δ, and on days 4, 6, and 8 dpi three rabbits were euthanatized. The rabbits were deeply anesthetized by intramuscular injection of ketamine (35 mg/kg of body weight) and xylazine (5 mg/kg) prior to all experimental manipulations. To determine the neural spread of the virus, the animals were anesthetized and perfused transcardially, and the brain and the TG were processed for immunohistochemistry as described earlier (28).
(ii) Nasal swabs.
To test whether Us9 deletion affected virus replication in the naso-olfactory epithelium, the amount of virus shed in the nasal epithelium was quantitated. Six animals were infected with BHV-5Us9Δ virus and six animals were infected with BHV-5Us9R virus as mentioned above. Two animals from each group were euthanatized at 2, 3, and 5 dpi. Nasal swabs were obtained and the virus was quantitated by plaque assay.
(iii) Immunofluorescence labeling and confocal microscopy.
To determine the ability of BHV-5Us9Δ and BHV-5Us9R viruses to infect the olfactory receptor neurons and to spread anterogradely to the bulbs, four rabbits were similarly infected by either of these viruses. Two rabbits from each group were euthanatized at 3 dpi, and the remaining two were euthanatized at 5 dpi. Following euthanasia, the head was removed and the section of the skull containing the midmaxillary region to the middle of the orbit (including the bulb) was dissected and en bloc fixed in 10% buffered neutral formalin (2 to 3 days). For the whole-mount immunofluorescence-confocal microscopy, the sensory epithelium and underneath connective layers of the nasal cavity including the nasal turbinates were separated from the nostril bone structures under a stereo microscope. After two washes in phosphate-buffered saline (PBS), they were further treated for 10 min with 0.1% sodium borohydrate in PBS, followed by two rinses in PBS. They were treated with 100% cold methanol (5 min at −20°C), followed by a rinse in plain PBS (15 min) and a wash in PBS (15 min) containing 0.1% Triton X-100. All incubations and washes for the immunofluorescence labeling were carried out at room temperature on a rocker. After blocking in 3% immunoglobulin G (IgG)-free bovine serum albumin (Jackson ImmunoResearch, Inc.) in PBS for 1 h, samples were incubated for 2 h with a cocktail of the following primary antibodies: monoclonal antibodies to neurofilament 160- and 200-kDa proteins (Sigma) and bovine anti-BHV-5 polyclonal antibodies (14, 28). Unless otherwise mentioned antibody dilutions for the immunofluorescence experiment were done in PBS containing 1% IgG-free bovine serum albumin. Following three washes in PBS (∼30 min each), the tissues were incubated with secondary antibodies, cyanin 2-conjugated donkey anti-bovine IgGs and cyanin 5-conjugated donkey anti-mouse IgGs, for 1 h. The double-labeled samples were washed three times in PBS (30 min each) and treated with propidium iodide (2 μg/ml) for 5 min for nuclear staining. After a 5-min rinse in PBS, samples were placed in PBS in a 60-mm-diameter petri dish and examined using 20× or 40× water-immersion objective lenses with a Bio-Rad MRC1024Es confocal laser scanning microscope. Series of z-axis optical sections of the triple-labeled samples for viral antigens, DNA in the cell nucleus, and neurofilaments were collected simultaneously with a three-line laser excitation mode (488, 560, and 640 nm) and triple emission (522, 598, and 680 nm) filter set using the Bio-Rad LaserSharp imaging program.
For immunofluorescence labeling of the olfactory bulb neurons, frozen sections were cut. The immunofluorescence labeling procedures were the same as those for the whole-mount tissues described above.
(iv) Olfactory bulb injections.
Four rabbits were anesthetized as described above. The heads of anesthetized animals were mounted in a stereotaxic frame and leveled. A burr hole was created in the skull between the lambda and Bregma sutures (20 to 22 mm rostral to the Bregma suture and 5 mm lateral to the midsagittal suture). A Hamilton 10-μl syringe with an attached 30-gauge needle was used for injection; a separate syringe was used for each virus. The tip of the needle was lowered 1.5 to 2 mm below the surface of the bulb. Virus was injected slowly over a period of 2 min, and the needle was left in the brain for another 2 min following completion of the injection to reduce the reflux of the inoculum along the injection tract. Two rabbits were inoculated with Us9 deletion mutant BHV-5, and two were inoculated with BHV-5. After survival times of 5 dpi or when the animals showed severe neurological signs, rabbits were euthanatized as before and processed for immunohistochemical staining of the brain sections (28).
Preparation of brain tissues for BHV-5-specific PCR.
Two rabbits were infected with BHV-5, and two were infected with the Us9 deletion mutant of BHV-5. To detect the BHV-5-specific DNA sequence in the different brain sections of these animals, the animals were euthanatized at 6 dpi. Brains of these infected animals along with one from an uninfected rabbit were dissected into olfactory bulb, anterior and posterior cortex, cerebellum, and pons-medulla blocks. They were kept at −70°C until DNA was extracted.
DNA extraction.
Brain tissues were homogenized at 20% (wt/vol) with 10 mM Tris-1 mM EDTA, pH 7.5 (TE). Samples were frozen and thawed twice and incubated at 37°C for 30 min with 5 μl of RNase (10 mg/ml) in the presence of 1% SDS. Proteinase K (14.4 mg/ml) digestion was done at 60°C for 30 min. Following phenol-chloroform-isoamyl alcohol (25:24:1) and ether extractions, DNA was ethanol precipitated and resuspended in 15 μl of TE.
PCR.
Primer pairs gI+ (5′-GTG CTC TTC TCC ATC GCC-3′; sense) and gI− (5′ CGG GAG GAGGAG TTG TCG G-3′; antisense) were derived from BHV-5 gI sequence (Al-Mubarak and S. I. Chowdhury, unpublished data). These primers amplify a 435-bp product in BHV-5. PCR was carried out in a 50-μl volume containing 1× PCR buffer, 2.0 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, a 0.5 mM concentration of each primer, 5% dimethyl sulfoxide, and 1.5 U of Taq polymerase. Briefly, 44.5 μl of master mix lacking Taq and template was aliquoted in reaction tubes. A 0.5-μl aliquot of the appropriate template was added to each reaction tube. Following an 8-min incubation at 99°C, the temperature was decreased to 90°C, while 5 μl of 1× PCR buffer containing 1.5 U of Taq was added to each reaction (hot start). Cycling consisted of four cycles of denaturation at 97°C for 1 min, annealing and extension at 68°C for 1 min, and 36 cycles of denaturation at 97°C for 1 min and annealing and extension at 66°C for 1 min. Upon completion of the last cycle, the reaction mixtures were further incubated at 72°C for 5 min to ensure complete extension of the amplified products.
Nucleotide sequence accession number.
The nucleotide sequence of the BHV-5 Us9 gene has been submitted to the GenBank with the accession no. AY064172.
RESULTS
Analysis of BHV-5 Us9 ORF and comparison of predicted amino acid sequences of BHV-5 and BHV-1 Us9.
The nucleotide sequence analysis spanning the 1.47-kb SacI (located in pKS99-49)/AccI (located in pKS99-47) fragment (Fig. 1C) revealed that the BHV-5 Us9 ORF is 405 nucleotides long and encodes a 134-aa polypeptide which is large enough to encode the BHV-5 Us9 gene (Fig. 2A). The ATG start codon is located 91 bp downstream of the stop codon of the gE gene. The putative TATA box is located 43 nucleotides upstream of the start codon. The polyadenylation site is located 313 nucleotides downstream of the stop codon TGA. The nucleotide composition of the BHV-5 Us9 ORF showed 75.5% G+C content, which is higher than those of the BHV-1.1 (GenBank accession no. 298199) and BHV-1.2 (not shown) (29) Us9 coding regions (70.4%). Hydropathic analysis of the predicted protein revealed the presence of a highly hydrophobic domain of 27 aa at the C terminus, the length and relative hydrophobicity of which are characteristic of a transmembrane anchor sequence (data not shown). The analysis for the potential signal sequence did not reveal an obvious signal sequence. The predicted 134 aa encoded by BHV-5 Us9 ORF are predicted to yield a 13.7-kDa protein, whereas the BHV-1.1 Us9 is predicted to be 144 aa long and have a corresponding molecular mass of 16.3 kDa, respectively. In BHV-5, the total number of negatively charged residues (Asp and Glu) is 15 and the total number of positively charged amino acid residues (Arg and Lys) is 13, whereas in BHV-1 Us9 the number of negatively and positively charged aa is 18 and 13, respectively. Based on the predicted amino acid sequence the BHV-1.1 Us9 protein has a net charge of −4, and the BHV-5 Us9 protein has a net charge of −8 at pH 7.0 (Antheprot protein analysis software; Institut de Biologie et Chimie des Proteins, Lyon, France). When aligned using the CLUSTALW alignment program, the predicted protein product of the BHV-5 Us9 gene shows 77% identity and 83% similarity to that of BHV-1.1 (Fig. 2B) and 32% identity and 46% similarity to that of PRV (data not shown).
Analysis using the NetPhos prediction program (version 2.0; http://www.cbs.dtu.dk/cgi-bin) showed that six serine (6S, 22S, 59S, 75S, 77S, and 79S), one threonine (40T), and two tyrosine (62Y and 74Y) residues were potential phosphorylation sites (Fig. 2B). Analysis using the PROSITE database (2) showed that the protein had a potential cyclic AMP (cAMP)- and cGMP-dependent protein kinase phosphorylation site (KRAS) at aa 56 to 59, which does not exist in BHV-1 Us9. In addition, there are four potential casein kinase II phosphorylation sites at aa 40 to 43 (TIED), 75 to 78 (SESD), 77 to 80 (SDSE), and 81 to 84 (TAGE) which are conserved in BHV-1 Us9 (Fig. 2B). Potential N-linked glycosylation sites are not present in both the Us9 sequences. BHV-5 Us9 protein contains an acidic motif between aa 68A and 85F, which is highly conserved in BHV-1 (GenBank accession no. 298199 [29]) and PRV Us9 (6, 7). This domain contains three serine residues (S75, S77, and S79), two tyrosine residues (Y62, Y74) which are potential phosphorylation sites, and three potential casein kinase II phosphorylation sites (Fig. 2B). These phosphorylation sites are conserved in both BHV-1 and BHV-5 sequences. The sequence alignment also showed that a region in BHV-5 Us9 between aa 46 and 60 differs significantly when compared with the corresponding region of BHV-1 Us9. For example, there are cAMP- and cGMP-dependent protein kinase phosphorylation sites (KRAS) in BHV-5 Us9 which are not present in BHV-1 Us9 (Fig. 2B). In both the viruses, the 27-aa hydrophobic putative transmembrane domain of Us9 is preceded by a cluster of eight positively charged amino acids. The cytoplasmic domain of BHV-5 Us9 is 101 aa long, while in BHV-1 it is 111 aa long. They both are predicted to be located upstream of the transmembrane domain.
Identification and analysis of the BHV-5 Us9 protein in infected-cell lysates and virions. (i) Anti-BHV-5 Us9 peptide antibody recognizes a 17- to 19k-Da protein.
Based on predicted regional hydropathicity and antigenicity, the predicted aa 9 through 22 ([H]-NENYRGADAADAAS-[OH]) (14-mer) and 90 through 101 ([H]-GRQQRRRHRRRR-[OH]) (12-mer) were synthesized, and rabbit polyclonal sera were generated.
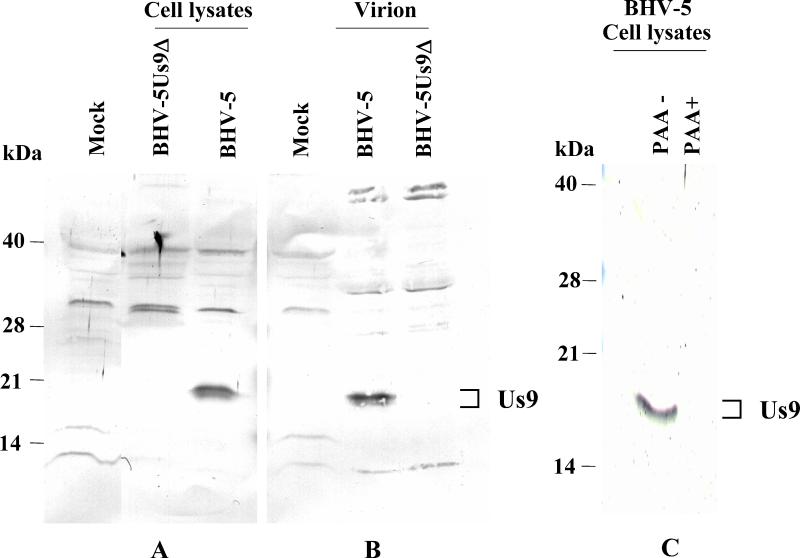
To identify the BHV-5 Us9 protein, virus- and mock-infected cell lysates were analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting with the peptide-specific antibodies. The antipeptide-specific antibody generated against the 14-mer peptide [H]-NENYRGADAADAAS-[OH]) reacted strongly with a doublet of protein bands in BHV-5-infected cell lysates (Fig. 3). The apparent molecular masses of these polypeptides were estimated to be 17 and 19 kDa. The antipeptide-specific antibody generated against the 12-mer peptide failed to react specifically to any virus-specific protein (data not shown). To determine whether Us9 is present in the virions, partially purified virion lysates were similarly analyzed by immunoblotting. As shown in Fig. 3B, the peptide-specific serum reacted with a doublet of 17- and 19-kDa protein bands of purified virions.
FIG. 3.
Identification and characterization of BHV-5 Us9 ORF-specific proteins with a rabbit antibody to 14-mer BHV-5 Us9-specific polypeptides. Immunoblot analysis of mock-infected, BHV-5Us9Δ-, and BHV-5-infected cell lysates (A); mock-infected cell lysates and partially purified BHV-5 and BHV-5Us9Δ virion lysates (B); and BHV-5 polypeptides produced in untreated (PAA−) and PAA-treated (PAA+) BHV-5-infected cell lysates (C). (C) Western blot of an SDS-12% polyacrylamide gel.
(ii) BHV-5 Us9 is a γ2 protein.
To determine the kinetic class of BHV-5 Us9 gene expression, we tested whether its expression is dependent on ongoing DNA synthesis and sensitive to PAA (true γ2 protein). In the absence of PAA both Us9 and gC (data not shown) are expressed; however, in the presence of PAA the expression of both Us9 (Fig. 3C) and gC (data not shown) was inhibited. Thus, the BHV-5 Us9 gene is a γ2 protein.
Construction and analysis of BHV-5Us9Δ and BHV-5Us9R recombinants.
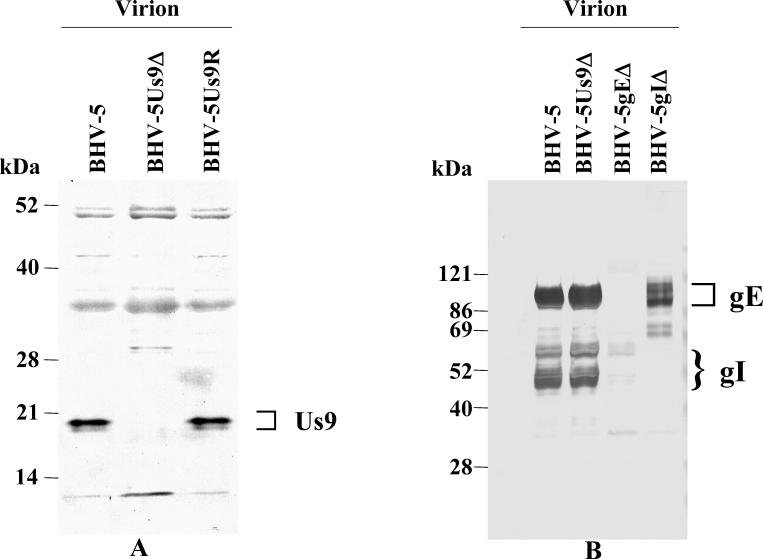
Viral DNA from BHV-5Us9Δ recombinant and wild-type BHV-5 was analyzed by Southern blot hybridization to confirm the intended deletion and insertion of β-Gal sequences at the Us9 locus (data not shown). The absence of a StyI/SfiI fragment sequence and the presence of a β-Gal sequence in the BHV-5Us9Δ recombinant isolate but not in the parental BHV-5 demonstrated that recombination in these isolates had taken place in a site-specific manner (data not shown). Consistent with this finding, the 17- and 19-kDa BHV-5 Us9 proteins were absent in the BHV-5Us9Δ but were detected in both the parent and BHV-5Us9R viruses (Fig. 4A).
FIG. 4.
Characterization of BHV-5Us9Δ virus. (A) Identification of Us9 protein in wild-type BHV-5, BHV-5Us9Δ, and BHV-5Us9R viruses by immunoblotting with anti-BHV-5 Us9 peptide-specific antibody. (B) Characterization of gE and gI in BHV-5 and BHV-5Us9Δ purified virion lysates by a cocktail of rabbit anti-gE/gI polyclonal antibodies (15, 41). As controls, BHV-5gEΔ (15) and BHV-5gIΔ (A. Al-Mubarak and S. I. Chowdhury, unpublished) virion lysates are shown.
In alphaherpesviruses the transcription at the gI/gE and Us9 sites is complex, and the manipulation of the Us9 gene could potentially affect the gE/gI gene expression. To verify the gE/gI expression in the BHV-5Us9Δ, we tested the expression of these glycoproteins or their incorporation into the virion by immunoblotting. The results presented in Fig. 4B show that both gE and gI are present in BHV-5Us9Δ virion at wild-type levels.
One-step growth experiments were conducted to analyze the growth kinetics of the BHV-5Us9Δ recombinant virus in MDBK cells in comparison to the parental strain BHV-5 TX-89. The results (data not shown) demonstrated that the time course and yield of infectious progeny of BHV-5Us9Δ virus were similar to those of the parental virus. In addition we have observed that BHV 5Us9Δ virus produces wild-type-size plaques (data not shown).
Pathogenicity of Us9 deletion mutant virus in rabbits. (i) Us9 deletion mutant BHV-5 is nonneurovirulent.
In the first experiment, 10 rabbits were infected with BHV-5Us9Δ and 5 rabbits were infected with BHV-5Us9R. Five of the animals infected with BHV-5Us9Δ recombinant virus were sacrificed at 10 dpi, and the remaining animals were sacrificed at 13 dpi. These animals did not display any noticeable neurological signs until the time of their sacrifice. Of the five rabbits infected with BHV-5Us9R, three showed severe neurological signs which consisted of circular movement, upright sitting posture, seizures, convulsions, and opisthotonus. The BHV-5Us9R-infected animals were euthanatized at 10 dpi or 3 to 4 h after the onset of seizures. The brains were processed for immunohistochemistry to localize the virus-specific antigens in the brain (28). As seen in Fig. 5, no virus-specific antigen was detected in the representative brain sections of animals infected with Us9 deletion mutant BHV-5. In contrast, rabbits infected with BHV-5Us9R showed a wild-type level of viral spread in the olfactory pathway (28).
FIG. 5.
Localization of viral antigen in brain sections. Animals were inoculated intranasally with either BHV-5Us9R or BHV-5Us9Δ as described earlier (14, 28). The animals were euthanatized on 4, 6, 8, 10, and 13 dpi or when they showed neurological signs, and their brains were processed for immunohistochemical analysis as described earlier (28). On the left is shown a low-power montage of Nissl-stained sections that indicate the locations of the micrographs shown on the right (indicated by boxes). Representative sections of the anterior olfactory nucleus (A), piriform cortex (B), and amygdala (C) are pictured. In this assay, BHV-5Us9R spread to the anterior olfactory nucleus, piriform cortex, amygdala, and hippocampus (not shown) at 8 dpi; no labeling was observed in BHV-5Us9Δ virus-infected animals at 4 to 13 dpi.
In experiment 2, nine rabbits were intranasally infected with BHV-5Us9Δ. On days 4, 6, and 8 dpi, three rabbits were euthanatized. Their brains and the TG were processed for immunohistochemistry. No virus-specific antigens were detectable in the brain sections at any of the survival times tested. However, two to four infected neurons were detected in the TG at 6 to 8 dpi (data not shown).
Since immunohistochemistry did not detect the presence of the Us9 deletion mutant virus within the CNS, the brain tissue of infected rabbits was analyzed by PCR for the presence of virus-specific DNA (BHV-5 gI-specific sequences). At 6 dpi no BHV-5 DNA sequences could be amplified from the olfactory bulbs of four rabbits infected with BHV-5US9Δ. In contrast BHV-5 DNA sequences were amplified from two rabbits infected with BHV-5Us9R (Fig. 6).
FIG. 6.
PCR amplification of BHV-5Us9Δ and BHV-5Us9R virus DNA from infected rabbit brain. Lane 1, PCR product from BHV-5 control DNA; lane 2, PCR product from olfactory bulb of normal rabbit brain; lanes 3 and 4, PCR product from olfactory bulb of rabbits 1 and 2, respectively, infected with BHV-5 Us9R; lanes 5 to 8, PCR product from olfactory bulb of rabbits 1 to 4, respectively, infected with BHV-5Us9Δ; lane M, DNA molecular size marker.
(ii) BHV-5Us9Δ infects the olfactory receptor neurons but failed to get transported to the bulb.
To test whether the deletion of Us9 affects the virus growth in the naso-olfactory epithelium, we quantitated the amount of virus shed in the nasal epithelium. Nasal swabs were obtained at 3 and 5 dpi, and the virus was quantitated by plaque assay. Our results from two independent experiments indicated that both BHV-5Us9Δ and BHV-5Us9R are shed with a comparable yield. At 3 and 5 dpi, 1.8 × 103 PFU (range, 0.7 × 103 to 2.5 × 103 PFU) and 0.8 × 103 PFU (range, 0.5 × 103 to 1.1 × 103 PFU) of BHV-5Us9R were detected, respectively. At 3 and 5 dpi, 1.4 × 103 PFU (range, 0.7 × 103 to 2.0 × 103 PFU) and 0.6 × 103 PFU (range, 0.4 × 103 to 1.0 × 103 PFU) of BHV-5Us9Δ were detected, respectively.
To determine whether the deletion of Us9 affects the ability of the virus to infect the olfactory receptor neurons and/or to be transported from the olfactory receptor neurons to the bulb, four rabbits were infected with Us9 deletion mutant BHV-5 and four were infected with wild-type BHV-5. Immunofluorescence-confocal microscopy of the olfactory epithelium was performed at 3 and 5 dpi using the anti-BHV-5 and antineurofilament-specific (neurofilament 200- and 160-Da proteins) antibodies. As shown in Fig. 7 the olfactory receptor cells (bipolar neurons) within the olfactory epithelium were infected by both types of virus. Optical images of connective tissues underneath the respiratory layer revealed viral proteins in olfactory nerve fibers of rabbits infected with wild-type BHV-5 but not for rabbits infected with BHV-5Us9Δ. Viral proteins were also detected at 5 dpi in the olfactory bulbs of the rabbits infected with wild-type virus but not in the rabbits infected with BHV-5Us9Δ (Fig. 8).
FIG. 7.
Localization of virus-specific antigens by immunofluorescence-confocal microscopy in the olfactory receptor neurons of rabbits infected with BHV-5Us9Δ and BHV-5Us9R viruses. Confocal images were taken at the nasal-olfactory epithelial tissues of rabbits infected with BHV-5Us9Δ (A to C) and BHV-5Us9R (D to F). (A and D) Viral antigens (green fluorescence) as detected by BHV-5-specific bovine polyclonal antibody are seen in the cell bodies of both the epithelial and olfactory receptor neuronal cells. (B and E) Olfactory receptor neurons and their processes are labeled by neurofilament-specific antibodies (red fluorescence). (C and F) Merged images show colocalization of viral and neurofilament antigens. Note that BHV-5Us9R viral antigens but not BHV-5Us9Δ viral antigens are detectable in neuronal fibers as indicated by arrows. Scale bar = 30 μm.
FIG. 8.
Confocal images of immunofluorescence-labeled viral antigens in the neurons of the olfactory bulb. Frozen sections of the olfactory bulb of rabbits infected with BHV-5Us9Δ (A to C) and BHV-5Us9R (D to F) were labeled for viral antigens (green fluorescence) as well as for neurofilaments (red fluorescence). Note that viral antigens were detected in neuronal cell bodies and fibers (arrow) in sections from BHV-5-infected rabbit (D) but were not detected in those infected with Us9 deletion mutant virus (A). (C and F) The merged images of viral antigens and neuronal markers are shown. Scale bar = 30 μm (for all images).
(iii) Us9 deletion mutant BHV-5 is equally virulent when injected into the olfactory bulb.
When injected into the bulb directly, both the BHV-5Us9R and BHV-5Us9Δ viruses were equally neurovirulent. These rabbits showed severe neurological signs at 3 to 5 dpi. Based on the immunohistochemistry results, the neural spread of BHV-5Us9Δ and BHV-5Us9R viruses within the olfactory pathway is indistinguishable (Fig. 9).
FIG. 9.
Localization of viral antigen in brain sections of animals that received inoculations directly into the olfactory bulb. Animals were inoculated with either BHV-5Us9R or BHV-5Us9Δ as described in Materials and Methods. The animals were euthanatized on 5 dpi or when they showed neurological signs. Their brains were processed for immunohistochemical analysis (28). On the left is shown a low-power montage of Nissl-stained sections that indicate the locations of the micrographs shown on the right (indicated by boxes). Representative sections of the olfactory bulb (A), anterior olfactory nucleus (B), and piriform cortex (C) are pictured. In this assay, both BHV-5Us9R and BHV-5Us9Δ spread to the structures connected to the bulb.
DISCUSSION
This study was initiated to determine the role of the Us9 gene in BHV-5 neuroinvasion in a rabbit seizure model. First, the BHV-5 Us9 gene sequence was determined, and the predicted amino acid sequence of BHV-5 Us9 was compared with that of BHV-1.1. Second, we generated BHV-5 Us9 deletion recombinant and BHV-5 Us9 rescue mutant viruses. Third, we characterized the in vitro and in vivo properties of the BHV-5Us9Δ and BHV-5Us9R viruses in cell culture and in a rabbit seizure model. The study demonstrated that BHV-5 Us9Δ virus is virtually avirulent in the rabbit seizure model. Even though BHV-5Us9Δ infects the olfactory receptor neurons (first-order neurons in the olfactory pathway), it is defective in the anterograde transport to the bulb.
Us9 gene homologs are found in most of the alphaherpesviruses, including BHV-1 and BHV-5 (5, 6, 34, 38). Alignment of the predicted amino acid sequence of the BHV-5 Us9 gene with the BHV-1.1 Us9 showed that they have a short (6-aa) ectodomain at the carboxy end of the protein; there is no signal sequence, and their cytoplasmic domain is located upstream of the putative transmembrane domain. In addition, a cluster of positively charged amino acids precedes the transmembrane domain. Based on studies from other Us9 homologs, the alignment is consistent with a type II membrane protein (6, 22, 34).
BHV-5 Us9 ORF is 10 aa shorter than the BHV-1 Us9 ORF (the BHV-5 Us9 ORF contains 134 aa, whereas that of BHV-1 Us9 contains 144 aa). The predicted molecular mass of BHV-5 Us9 is 13.7 kDa. Yet, BHV-5 Us9 specific antibody recognized two distinct polypeptides with molecular masses of 17 and 19 kDa. This electrophoretic mobility implies that it is modified after translation by covalent addition of sugar, phosphates, or other small molecules. The BHV-5 Us9 sequence analysis revealed that there are several potential phosphorylation sites. In HSV-1, Us9 is ubiquitinated and the ubiquitinated form of the protein is packaged in the virion (5). These modifications in BHV-5 Us9 need to be examined in the future. Nevertheless, these modifications would account for the size difference between the predicted and the actual Us9 size. This is consistent with the findings in PRV (6) and HSV-1 Us9 proteins (5, 21).
Following infection of rabbits, our Us9 deletion mutant BHV-5 is not neurovirulent and failed to invade the CNS. Intranasal inoculation of BHV-5 Us9 deletion and BHV-5 Us9-rescue mutant viruses revealed that the Us9 rescue mutant of BHV-5 displayed the wild-type level of neurovirulence and neural spread in olfactory pathway. In contrast, the Us9 deletion mutant of BHV-5 is virtually avirulent in similarly infected rabbits, and spread within the CNS was not detected by immunostaining. In the nasal mucosa the Us9 deletion mutant of BHV-5 replicated with equal efficiency when compared with the Us9 rescue mutant of BHV-5. To spread via the olfactory pathway to the deeper tissues of the CNS, the virus must infect the cell bodies of first-order olfactory receptor neurons located within the olfactory epithelium and spread anterogradely to the second-order neurons in the bulb. Confocal microscopy data indicated that BHV-5Us9Δ infects the olfactory receptor neurons (first-order neurons), but it is not transported to the bulb. It is possible that animals infected with BHV-5Us9Δ virus have fewer viruses that invade the olfactory bulb and thus are not detected by immunohistochemistry. Alternatively, the virus may be clarified more easily by the immune response following infection. To exclude this possibility, we conducted PCR experiments for the presence of virus-specific nucleic acid in the bulb. Interestingly, rabbits infected with BHV-5Us9Δ did not have viral DNA in the bulb, indicating that the virus did not invade the bulb.
Subsequent invasion from the olfactory bulb to the third-, fourth-, and/or fifth-order neurons involves retrograde transneuronal spread (28). The results of direct olfactory bulb injection showed that both BHV-5Us9Δ and the wild-type BHV-5 are equally neurovirulent and that the viral spread in the olfactory pathway is very similar. Taken together, the results of immunofluorescence-confocal microscopy after intranasal infection and direct olfactory bulb injections suggest that the BHV-5Us9Δ infects the olfactory receptor neurons equally efficiently, like the wild-type BHV-5, but failed to spread anterogradely to the bulb. Since the virus spreads efficiently after direct olfactory bulb injection, we conclude that the deletion of the Us9 gene does not affect the retrograde neuronal transport. Consistent with this finding, we have determined that, like the wild-type BHV-5, BHV-5Us9Δ virus can be transported retrogradely to the TG. This finding is in agreement with what has been reported for PRV (8, 9).
Besides Us9, gE and gI (3, 15, 17, 18, 20, 31, 40) are known to play an important role in the anterograde transport of BHV-5, PRV and HSV-1. Both gE and gI of alphaherpesviruses are required for efficient cell-to-cell spread in a variety of tissue culture cells, and gE/gI deletion mutants produce small plaques (17, 23, 24, 43, 44). Interestingly, Us9 deletion mutants of PRV and BHV-5 produce wild-type-size plaques (reference 8 and this study). Yet, in vivo, deletion of Us9 in PRV (8) and BHV-5 (this study) results in a phenotype with a very similar (PRV) or more pronounced (BHV-5) defect in the anterograde transport than that observed with their respective gE deletion (8, 15). The gE-gI complex promotes the spread of virus from the presynaptic axon terminals to the postsynaptic cell (20). As reported by Tirabassi et al. (36), these glycoproteins act at the site of egress and need not be in viral envelopes to promote viral spread between the synaptically connected neurons. Brideau et al. (8) predicted that the gE-gI complex of PRV mediates spread through the interaction of the gE-gI ectodomains with a putative cellular receptor on the postsynaptic cell. Dingwell and Johnson (16) proposed a similar model for the interaction of gE-gI with a cellular ligand for the spread of HSV-1 between epithelial cells. Since the expression of both gE and gI in the BHV-5Us9Δ was not affected, we conclude that the anterograde spread defect of Us9 deletion mutant virus was due to the loss of Us9 and not due to changes in gE or gI expression.
Earlier studies with HSV-1 and PRV indicated that glycoproteins, capsids, and/or capsid-associated tegument proteins are transported separately from cell bodies to axon terminals (30, 33, 37, 38). Infection of primary cultures of fetal sympathetic ganglia neurons with the PRV-Us9 null mutant showed markedly reduced immunostaining for gE, gB, and gC in axons when compared to PRV wild-type infection (37). Additionally, PRV data suggested that Us9 plays a role in delivering vesicles containing envelope proteins, such as gE and gI, to the axonal terminals (37, 38). These observations indicated that some viral envelope proteins require Us9 for entry and transport in axons (37, 38). Our confocal microscopy data showed that anterograde axonal transport of the Us9 deletion mutant BHV-5 does not occur in vivo in the olfactory pathway. Rabbits infected intranasally with the Us9 deletion mutant of BHV-5 did not contain virus-specific antigens in the nerve processes of olfactory receptor neurons. While BHV-5 Us9 deletion mutant virus did enter and replicate efficiently in the olfactory receptor neurons and shed at the wild-type level, it failed to infect the olfactory bulb. In contrast, direct inoculation of Us9 deletion mutant virus into the bulb resulted in wild-type-level spread within the olfactory pathway. Taking together PRV in vitro data and our in vivo data, we believe that the lack of detectable viral antigens within the nerve processes of olfactory receptor neurons in rabbits infected with Us9 deletion mutant virus is due to the lack of viral glycoprotein transport from the cell body to the axon termini.
In an eye model in rats, PRV Us9 is involved in the anterograde neuronal transport of the virus (8, 9). It is hypothesized that, in the absence of Us9, PRV glycoproteins are not transported to the synapse, where they are required for the spread of the virus across the synapse (37, 38). The PRV in vivo studies revealed that Us9 deletion mutant viruses show defect in anterograde spread to some subsets of the visual pathway but not all (8, 9). However, the PRV data do not provide direct evidence demonstrating that the Us9 deletion mutant PRV infects and replicates equally in all retinal cell types in vivo (8). We have shown that BHV-5 Us9 is not required for the entry and replication in the first-order olfactory receptor neuron in vivo, but it is essential for the transport of the virus anterogradely from the olfactory receptor neurons to the second-order neurons within the bulb in a rabbit model. In the PRV eye model, the Us9 mutant could infect and spread anterogradely in functional subsets of visual pathway neurons. Presumably, in these instances, Us9 was not required for anterograde spread. This phenomenon observed in PRV neuronal transport is not well understood. Therefore, our olfactory neuroepithelium studies provide complementary data suggesting that Us9 is essential for anterograde axonal transport of BHV-5 glycoproteins but it is not essential for the entry and replication in the neuronal cell body.
BHV-1, like Us9 deletion mutant BHV-5, can infect and replicate within olfactory receptor neurons but fails to infect the olfactory bulb. However, when injected into the bulb BHV-1 is highly neurovirulent and neuroinvasive (Chowdhury and Weiss, unpublished data). It is possible that BHV-1 and BHV-5 Us9 differ with respect to axonal transport of virion glycoprotein.
Acknowledgments
We thank Lynn Enquist, Princeton University, for the rabbit anti-BHV-1 gE- and gI-specific antibodies and Lois Morales for immunocytochemistry.
This work was supported by USDA grants 97-35204-4700 and 00-02103 to S. I. Chowdhury.
Footnotes
Published as contribution 02-99-J of the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Ashbaugh, S. E., K. E. Thompson, E. B. Belknap, P. C. Schultheiss, S. I. Chowdhury, and J. K. Collins. 1997. Specific detection of shedding and latency of bovine herpesvirus 1 and 5 using a nested polymerase chain reaction. J. Vet. Diagn. Investig. 9:387-394. [DOI] [PubMed] [Google Scholar]
- 2.Bairoch, A., P. Bucher, and K. Hoffman. 1997. The PROSITE data base, its status in 1997. Nucleic Acids Res. 25:217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balan, P., N. Davis-Poynter, S. Bell, H. Atkinson, H. Browne, and T. Minson. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 75:1245-1258. [DOI] [PubMed] [Google Scholar]
- 4.Belknap, E. B., J. K. Collins, V. K. Ayers, and P. C. Schulteiss. 1994. Experimental infection of neonatal calves with neurovirulent bovine herpes virus type 1.3. Vet. Pathol. 31:358-365. [DOI] [PubMed] [Google Scholar]
- 5.Brandimarti, R., and B. Roizman. 1997. Us9, a stable lysine-less herpes simplex virus 1 protein, is ubiquitinated before packaging into virions and associates with proteasomes. Proc. Natl. Acad. Sci. USA 94:13973-13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brideau, A. D., B. W. Banfield, and L. W. Enquist. 1998. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J. Virol. 72:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brideau, A. D., T. del Rio, E. J. Wolffe, and L. W. Enquist. 1999. Intracellular trafficking and localization of the pseudorabies virus Us9 type II envelope protein to host and viral membranes. J. Virol. 73:4372-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brideau, A. D., J. P. Card, and L. W. Enquist. 2000. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J. Virol. 74:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brideau, A. D., G. Eldridge, and L. W. Enquist. 2000. Directional transneuronal infection by pseudorabiesvirus is dependent on an acidic internalization motif in the Us9 cytoplasmic tail. J. Virol. 74:4549-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Card, J. P., and L. W. Enquist. 1995. Neurovirulence of pseudorabies virus. Crit. Rev. Neurobiol. 9:137-162. [PubMed] [Google Scholar]
- 11.Chowdhury, S. I. 1995. Molecular basis of antigenic variation between the glycoprotein C of respiratory bovine herpesvirus 1 (BHV-1) and neurovirulent BHV-5. Virology 213:558-568. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury, S. I. 1996. Construction and characterization of an attenuated bovine herpesvirus type 1 (BHV-1) recombinant virus. Vet. Microbiol. 52:13-23. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhury, S. I., C. S. D. Ross, B. J. Lee, V. Hall, and H.-J. Chu. 1999. Construction and characterization of a glycoprotein E gene-deleted bovine herpesvirus type 1 recombinant. Am. J. Vet. Res. 60:227-232. [PubMed] [Google Scholar]
- 14.Chowdhury, S. I., B. J. Lee, D. Mosier, J.-H. Sur, F. A. Osorio, G. Kennedy, and M. L. Weiss. 1997. Neuropathology of bovine herpesvirus type 5 (BHV-5) meningo-encephalitis in a rabbit seizure model. J. Comp. Pathol. 117:295-310. [DOI] [PubMed] [Google Scholar]
- 15.Chowdhury, S. I., B. J. Lee, A. Ozkul, and M. L. Weiss. 2000. Bovine herpesvirus 5 glycoprotein E is important for the neuroinvasiveness and neurovirulence in the olfactory pathway of the rabbit. J. Virol. 75:2094-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dingwell, K. S., and D. C. Johnson. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dingwell, K. S., L. C. Doering, and D. C. Johnson. 1995. Glycoprotein E and I facilitate neuron-to-neuron spread of herpes simplex virus. J. Virol. 69:7087-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Offay, J. M., R. E. Mock, and R. W. Fulton. 1993. Isolation and characterization of encephalitic bovine herpesvirus type 1 isolates from cattle in North America. Am. J. Vet. Res. 54:534-539. [PubMed] [Google Scholar]
- 20.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1999. Spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 21.Frame, M. C., D. J. McGeoch, F. J. Rixon, A. C. Orr, and H. S. Marsden. 1986. The 10K virion protein encoded by gene Us9 from herpes simplex virus type 1. Virology 150:321-332. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman, K., and W. Stoffel. 1993. Tmbase--a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 347:166. [Google Scholar]
- 23.Jacobs, L. 1994. Glycoprotein E of pseudorabies virus and homologous proteins in other alphaherpesvirinae. Arch. Virol. 137:209-228. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, D. C., M. C. Frame, M. W. Ligas, A. M. Cross, and N. D. Stow. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 62:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kritas, S. K., H. J. Nauwynck, and M. B. Pensaert. 1995. Dissemination of wild-type and gC-, gE- and gI-deleted mutants of Aujeszky's disease virus in the maxillary nerve and trigeminal ganglion of pigs after intranasal inoculation. J. Gen. Virol. 76:2063-2066. [DOI] [PubMed] [Google Scholar]
- 26.Kritas, S. K., M. B. Pensaert, and T. C. Mettenleiter. 1994. Role of envelope glycoproteins gI, gp63 and gIII in the invasion and spread of Aujeszky's disease virus in the olfactory nervous pathway of the pig. J. Gen. Virol. 75:2319-2327. [DOI] [PubMed] [Google Scholar]
- 27.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 28.Lee, B. J., M. L. Weiss, D. Mosier, and S. I. Chowdhury. 1999. Spread of bovine herpesvirus type 5 (BHV-5) in the rabbit brain after intranasal inoculation. J. Neurovirol. 5:473-483. [DOI] [PubMed] [Google Scholar]
- 29.Leung-Tack, P., J. C. Audonnet, and M. Rivière. 1994. The complete DNA sequence and genetic organization of the short unique region (US) of the bovine herpes virus type 1 (ST strain). Virology 199:409-421. [DOI] [PubMed] [Google Scholar]
- 30.Miranda-Saksena, M., P. Armati, R. A. Boadle, D. J. Holland, and A. L. Cunningham. 2000. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J. Virol. 74:1827-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulder, W., L. Jacobs, J. Priem, G. L. Kok, F. Wagenaar, T. G. Kimman, and J. M. Pol. 1994. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and trigeminal routes in the porcine central nervous system. J. Gen. Virol. 75:3095-3106. [DOI] [PubMed] [Google Scholar]
- 32.Nielson, H., J. Engelbrecht, S. Brunak, and G. V. Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 33.Penfold, M. E. T., P. Armati, and A. L. Cunningham. 1994. Axonal transport of herpes simplex virions to epidermal cells: evidence for a specialized mode of virus transport and assembly. Proc. Natl. Acad. Sci. USA 91:6529-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrovskis, E. A., and L. E. Post. 1987. A small open reading frame in pseudorabies virus and implications for evolutionary relationships between herpesviruses. Virology 159:193-195. [DOI] [PubMed] [Google Scholar]
- 35.Rock, D. L., W. A. Hagemoser, F. A. Osorio, and D. E. Reed. 1986. Detection of bovine herpesvirus type 1 RNA in trigeminal ganglia of latently infected rabbits by in situ hybridization. J. Gen. Virol. 67:2515-2520. [DOI] [PubMed] [Google Scholar]
- 36.Tirabassi, R. S., R. A. Townley, M. G. Eldridge, and L. W. Enquist. 1997. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J. Virol. 71:6455-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomishima, M. J., and L. W. Enquist. 2001. A conserved α-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomishima, M. J., G. A. Smith, and L. W. Enquist. 2001. Sorting and transport of alpha herpesviruses in axons. Traffic 2:429-436. [DOI] [PubMed] [Google Scholar]
- 39.Welling, G. W., W. J. Weijer, R. van der Zee, and S. Welling-Wester. 1985. Prediction of sequential antigenic regions in proteins. FEBS Lett. 188:215-218. [DOI] [PubMed] [Google Scholar]
- 40.Whealy, M. E., J. P. Card, A. K. Robbins, J. R. Dubin, H. J. Rhiza, and L. W. Enquist. 1993. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J. Virol. 67:3786-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitbeck, J. C., A. C. Knapp, L. W. Enquist, W. C. Lawrence, and L. J. Bello. 1996. Synthesis, processing, and oligomerization of the bovine herpes virus 1 gE and gI membrane proteins. J. Virol. 70:7878-7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyler, R., M. Engels, and M. Schwyzer. 1989. Infectious bovine rhinotracheitis/vulvovaginitis (BHV-1), p. 1-72. In G. Wittman (ed.), Herpesvirus diseases of cattle, horses and pigs. Kluwer Academic Publishers, Hingham, Mass.
- 43.Zsak, L., F. Zuckermann, N. Sugg, and T. Ben-Porat. 1992. Glycoprotein gI of pseudorabies virus promotes cell fusion and virus spread via direct cell-to-cell transmission. J. Virol. 66:2316-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuckermann, F. A., T. C. Mettenleiter, C. Schreurs, N. Sugg, and T. Ben-Porat. 1988. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J. Virol. 62:4622-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]