Abstract
Hepatitis delta virus (HDV) is a subviral human pathogen that uses specific RNA editing activity of the host to produce two essential forms of the sole viral protein, hepatitis delta antigen (HDAg). Editing at the amber/W site of HDV antigenomic RNA leads to the production of the longer form (HDAg-L), which is required for RNA packaging but which is a potent trans-dominant inhibitor of HDV RNA replication. Editing in infected cells is thought to be catalyzed by one or more of the cellular enzymes known as adenosine deaminases that act on RNA (ADARs). We examined the effects of increased ADAR1 and ADAR2 expression on HDV RNA editing and replication in transfected Huh7 cells. We found that both ADARs dramatically increased RNA editing, which was correlated with strong inhibition of HDV RNA replication. While increased HDAg-L production was the primary mechanism of inhibition, we observed at least two additional means by which ADARs can suppress HDV replication. High-level expression of both ADAR1 and ADAR2 led to extensive hyperediting at non-amber/W sites and subsequent production of HDAg variants that acted as trans-dominant inhibitors of HDV RNA replication. Moreover, we also observed weak inhibition of HDV RNA replication by mutated forms of ADARs defective for deaminase activity. Our results indicate that HDV requires highly regulated and selective editing and that the level of ADAR expression can play an important role: overexpression of ADARs inhibits HDV RNA replication and compromises virus viability.
Hepatitis delta virus (HDV) is a subviral satellite of hepatitis B virus (HBV) that increases the severity of HBV-related disease (33). The HDV particle has three components: the HDV RNA genome, hepatitis delta antigen (HDAg), which is the sole HDV protein, and the hepatitis B surface antigen (HBsAg), which is the sole helper function provided by the helper virus, HBV (4, 20, 33). The RNA genome of HDV is a single-stranded circular molecule in which about 70% of the nucleotides can form Watson-Crick base pairs in an unbranched rod structure (18, 40). The RNA may resemble an imperfect double-stranded RNA (dsRNA) with short (<15 bp) double-stranded regions interspersed with numerous mismatches, bulges, and internal loops.
HDV produces two forms of HDAg (2), HDAg-S and HDAg-L, which have distinct roles in the viral replication cycle. The shorter form, HDAg-S, is required for viral RNA replication, whereas the longer form, HDAg-L, is required for viral particle formation but is a potent trans-dominant inhibitor of replication (reviewed in reference 20). The HDV replication cycle begins with the production of HDAg-S, which is encoded in the infectious viral genome and supports viral RNA replication. The large form is subsequently produced by the process of RNA editing (5, 24), the specific deamination of adenosine 1012 in the antigenome (numbering is for the genomic strand, according to Wang et al. [40]) by a host activity known as adenosine deaminase that acts on RNA (ADAR). This adenosine is within the amber stop codon (UAG) that terminates HDAg-S synthesis. After replication and transcription, deamination of this adenosine to inosine, which base pairs preferentially with C, leads to the production of an HDAg-encoding mRNA in which the UAG amber termination codon has been changed to UGG (tryptophan), thereby extending the open reading frame (ORF) to encode HDAg-L, which is 19 amino acids longer at the carboxyl terminus (38, 41). Because of the codon change produced by this editing event, the editing site is called the amber/W site (31).
Two genes, ADAR1 and ADAR2, have been identified in mammalian cells that encode proteins capable of editing adenosines in double-stranded RNA (dsRNA) (26, 28, 29, 45). Both ADAR1 and ADAR2 contain a catalytic deaminase domain along with three or two, respectively, copies of dsRNA-binding motifs (reviewed in reference 1). In vitro studies using dsRNA as the substrate have shown that as many as 50% of the adenosines in a single dsRNA can be deaminated by these proteins (1). A third member of this gene family, ADAR3, has been identified by genetic analysis but has not demonstrated any catalytic activity (10, 25).
It is not yet clear to what extent dsRNAs are substrates for editing by ADAR1 and ADAR2 in vivo, but several cellular and viral RNAs have been identified that are substrates in vitro and in vivo (reviewed in references 1, 15, and 37). Significantly, for all of these substrates, editing is highly specific for certain adenosines that are located within particularly structured segments of the RNA (1, 15, 37). The HDV amber/W site in the HDV antigenome is one of these substrates, and it is likely that the HDV amber/W site is edited by one or both of these gene products in infected hepatocytes. The Xenopus laevis homologue of ADAR1 edits the amber/W site in HDV RNA with high specificity in vitro, and the effects of site-directed mutations on editing activity in vivo closely match those on editing by the purified enzyme in vitro (31). Moreover, overexpression of human ADAR1 and ADAR2 can increase editing of an HDV amber/W site in an editing reporter mRNA in transfected cells (35, 42).
Both the extent of editing at the amber/W site and the specificity of editing that occurs on the HDV antigenome are likely to have important consequences for HDV viability. Excessive editing at the amber/W site could result in reduced levels of RNA replication and reduced production of viable virions because edited antigenomes encode HDAg-L, which is a trans-dominant inhibitor of HDV RNA replication (9, 14). The dominant negative phenotype of HDAg-L is due to the fact that HDAg forms dimers and higher-order complexes (11, 21, 39, 44). Analysis of monoclonal antibody epitopes has led to the suggestion that HDAg-L assumes a different conformation than HDAg-S and may alter the conformation of the larger complex as well, thus inhibiting replication (16).
Because of the sensitivity of HDV replication to dominant negative effects, excessive promiscuous editing at non-amber/W sites could also produce HDAg variants that form alternative conformations and act as trans-dominant inhibitors. It is therefore likely that the virus also requires that editing be highly specific for the amber/W site. In fact, previous analysis of HDV RNA editing in transfected cells has shown that editing is highly specific for the amber/W site; 13 days following transfection, over 25% of genomes were edited at the amber/W site, while an average of only 0.16 of the 337 non-amber/W adenosines were edited per genome (32).
As noted above, it has recently been shown that overexpression of ADAR1 and ADAR2 can increase the amount of amber/W editing in an HDV editing reporter mRNA in transfected cells (35, 42). In this study, we analyzed the effects of such overexpression on the extent and specificity of editing on replicating HDV RNA and on HDV RNA replication. We found that increased expression of ADARs is correlated with strong inhibition of HDV RNA replication. This inhibition is due to several effects, including increased editing at the amber/W site and the production of dominant negative HDAg variants that are edited at numerous additional non-amber/W sites.
MATERIALS AND METHODS
Plasmids.
Construct pHDV, used for expressing the replicating HDV RNA in transfected cells, was described previously as pCMV3DC1X1.2 (7). The construct pM6, which expresses the site-directed HDV mutant G580A, has also been reported before (7, 31). In this construct, guanosine 580 (numbering is according to the genomic RNA) was altered to adenosine, so that it expresses replicating HDV RNA that is poorly edited at the amber/W site (5, 7). Both pHDV and pM6 produce a 1.2-mer antigenomic HDV RNA that replicates in transfected Huh7 cells. Plasmid pHDVI(+)Ag(−) is identical to pCMV3DC1X1.2 except that the reading frame for HDAg has been disrupted by the insertion of a stop codon/frameshift at codon 7 (6); replication of HDV RNA produced by this plasmid can be rescued by cotransfection with an expression construct for HDAg-S (6).
The HDAg-S expression plasmid (pHDS) and the HDAg-L expression plasmid (pHDL) were previously reported as pCMVAg-S and pHDAg-L, respectively (6). The ADAR2-edited, mutant HDAg-S expression constructs pAgSM11 and pAgSM25 were generated by replacing the HDAg-S coding regions in pHDS with the HDAg-S coding regions in the ADAR2-edited clones M11 and M25, respectively. M11 and M25 are clones from reverse transcription-PCR (RT-PCR) products of HDV RNA coexpressed with ADAR2 generated by the cloning procedure described under generation of PCR clones from ADAR-edited RNA (see below).
Plasmids pAD1 and pAD2 were generated to express rat ADAR1a and rat ADAR2b, respectively, in transfected cells. Briefly, the entire coding regions of rat ADAR1 and ADAR2 were amplified by RT-PCR from rat brain cDNA (Clontech) and cloned. The fidelity of the ADAR1 and ADAR2 clones obtained was confirmed by sequence analysis. For ADAR1 expression, sequences 658 to 3547 (GenBank accession no. U18942) were inserted into the mammalian expression vector pCI (Clontech), which contains the cytomegalovirus immediate-early promoter, to yield the ADAR1 expression construct pAD1. This construct directs synthesis of the p110 isoform of ADAR1, which is the major isoform found in Huh7 cells. All ADAR2 clones initially contained a 47-nucleotide intronic segment, which disrupts the ADAR2 reading frame (34); this segment was removed by PCR mutagenesis, and the sequences between 88 and 2255 (GenBank accession no. U43534) were inserted into pCI to yield the expression construct pAD2.
Constructs for expression of site-directed mutants of ADAR1 (pQA1) and ADAR2 (pQA2) defective for deaminase activity were created by PCR amplification with primers containing the desired mutant sequences. In these mutants, the highly conserved CHAE amino acid sequence in the catalytic deaminase domain of the proteins was altered to CQAA (23). For both ADAR1 and ADAR2, the sequence CACGCAGAG was changed to CAAGCAGCG. The mutation C → A corresponds to positions 2588 and 1301, respectively, and the mutation A → C corresponds to positions 2593 and 1306, respectively, on ADAR1 and ADAR2. The sequences of the final clones obtained verified the presence of the desired mutations and the absence of any extraneous mutations.
Cell culture and transfections.
Huh7 human hepatoma cells were cultured in six-well dishes using Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% fetal bovine serum and 1 mM glutamine. Cells were transfected using the Lipofectamine Plus reagent (Life Technologies), following the manufacturer's protocol. Transfections were done in duplicate and were repeated at least once. When appropriate, the total amount of DNA in the transfection mix was adjusted to 2 μg by adding the plasmid vector pCI (InVitrogen). In order to normalize for efficiency of transfection, we included 0.1 μg of the construct pSEAP2Control (Clontech) in all transfections (13).
Preparation of RNA samples.
RNA samples were prepared from cells 4 days posttransfection using the RN-Easy mini kit (Qiagen), following the manufacturer's protocol.
Analysis of amber/W editing on HDV RNA.
Editing assays were performed as described previously (7, 31). Briefly, DNase-treated RNA samples were subjected to RT-PCR using PCR primers 5414 and 5415 (27). The effectiveness of DNase treatment was verified by the absence of PCR products after PCR amplification without prior reverse transcription. PCR products were labeled with [α-32P]dCTP and analyzed for amber/W editing by restriction digestion with StyI. PCR products obtained without prior DNase treatment and without prior reverse transcription did not yield StyI digestion fragments, and undigested PCR products did not yield bands that could interfere with those produced as a result of restriction digestion.
Editing was quantified by polyacrylamide gel electrophoresis followed by radioanalytic imaging (InstantImager; Packard Instruments, Meriden, Conn.). Because editing creates a StyI restriction site that is not present in unedited RNA (5, 31), the percent editing is determined by dividing the sum of the band intensities due to the StyI digestion products by the sum of these bands plus the intensity of the PCR product band not digested by StyI. We have shown that the results of such analyses are highly reproducible and agree with sequence analysis of clones from PCR products (31, 32).
Generation of PCR clones from ADAR-edited HDV RNA and sequencing of the clones.
DNase-treated RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (100 U; Life Technologies) in 10-μl reaction mixtures containing 1× forward reaction buffer (supplied by the manufacturer), 2 nmol of random hexamer, 1 mM deoxynucleoside triphosphate mix, 10 mM dithiothreitol, and 10 U of RNasin (Promega). Reaction mixtures were incubated at 25°C for 10 min followed by 42°C for 30 min. The reverse transcription products were then amplified with Pfu polymerase (Stratagene, La Jolla, Calif.). Forty microliters of PCR master mix, containing 1.25 U of Pfu DNA polymerase, 1× Pfu polymerase buffer (supplied by the manufacturer), and 25 pmol of primers 5414 (27) and 6657 (6), was added to the 10-μl reverse transcription mixture. The cDNA was amplified for 30 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C. PCR products were cloned into the vector pCR-Blunt (Invitrogen) following the manufacturer's protocol. Barrier pipette tips were used to set up all reactions, and standard precautions were taken to minimize potential contamination of samples prior to PCR (19). Control reactions lacking reverse transcriptase or RNA were performed to ensure that the reactions were not contaminated.
Multiple clones were obtained from independent amplification reactions from three different aliquots of DNase-treated RNA. Both strands of cloned PCR products were sequenced by the dye terminator sequencing system on an ABI platform at MWG Biotech, High Point, N.C. Sequence changes were considered bona fide only if observed on both strands.
Northern blot analyses for HDV RNA replication.
RNA was electrophoresed through 1.5% agarose gels containing 2.2 M formaldehyde, transferred to positively charged nylon membranes, and hybridized with an antigenome sense 32P-labeled probe, as described previously (7). The hybridization temperature was 60°C and the posthybridization wash temperature was 70°C. The integrity of the RNA samples and equivalency of loading were assessed by visualization of rRNA bands after staining gels with ethidium bromide. Relative levels of HDV RNA were determined by radioanalytic scanning of blots with a Packard InstantImager. Expression levels were corrected for transfection efficiency by monitoring expression of the cotransfected secreted alkaline phosphatase reporter (13); duplicate values within an experiment varied by 35% or less (difference between duplicates divided by their average).
RESULTS
Overexpression of ADARs increases RNA editing at the HDV amber/W site and inhibits HDV RNA replication.
Amber/W editing is increased in vitro by increasing amounts of added Xenopus ADAR (31). Recently it was shown that overexpression of ADAR1 or ADAR2 in transfected cells could increase editing of an amber/W site located in an mRNA reporter construct (35, 42). We sought to examine whether high-level expression of ADAR1 and ADAR2 in cells could increase amber/W editing in replicating HDV RNA and, if so, analyze the effects of increased RNA editing on HDV RNA replication. Huh7 cells were transfected with pHDV, an expression construct for replicating HDV antigenomic RNA, and expression construct pAD1 or pAD2 for ADARs. Construct pAD1 expresses the p110 form of ADAR1, which is the predominant endogenous form present in Huh7 cells (29, 30); pAD2 expresses ADAR2. Northern blot analysis indicated that expression levels for ADAR1 and ADAR2 were increased more than 20-fold over endogenous levels following transfection.
RNA was harvested 4 days posttransfection and analyzed for editing at the amber/W site by restriction enzyme digestion of PCR-amplified cDNA with StyI (Fig. 1). Because editing at amber/W creates a StyI site that is not present in unedited RNA, StyI digestion of RT-PCR products can be used to quantify edited and unedited RNA templates (7, 31). In previous studies (31, 32) and in this one, we have found that values for amber/W editing obtained with this StyI digestion assay agree well with those obtained by sequencing of numerous (>50) clones of PCR products. We found that overexpression of both ADAR1 and ADAR2 led to hyperediting at the amber/W site; 54 and 68% of antigenomic RNAs were edited at the amber/W site 4 days after cotransfection of pHDV with pAD1 and pAD2, respectively, compared with 4 to 6% edited by endogenous levels of ADARs (Fig. 1). This increase is comparable to that observed for an HDV amber/W site located in an mRNA reporter construct (35, 42).
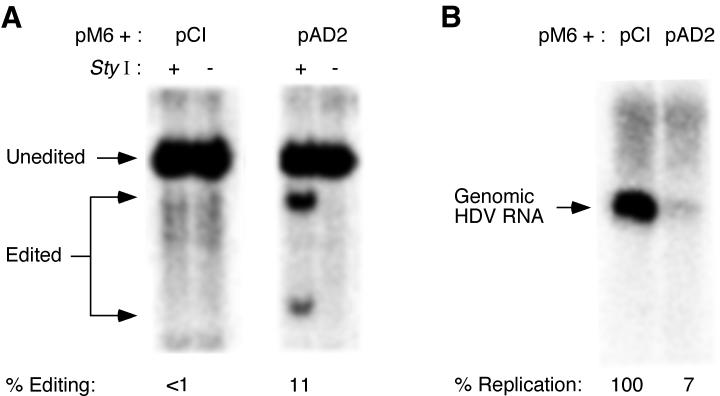
FIG. 1.
Effects of ADAR1 and ADAR2 overexpression on HDV amber/W RNA editing and replication. Huh7 human hepatoma cells were cotransfected with 1 μg each of the replicating HDV RNA expression construct pHDV, designed to synthesize antigenomic HDV RNA, and either the control expression vector pCI or an ADAR1 (pAD1) or ADAR2 (pAD2) expression construct. RNAs were harvested 4 days posttransfection. (A) Editing of the HDV RNA was analyzed by StyI digestion of RT-PCR products, as described in Materials and Methods. Smaller fragments produced by restriction digestion with StyI indicate editing of the RNA at the amber/W site. The autoradiogram shows 32P-labeled RT-PCR products, uncut (−) or cut with StyI (+). The percent editing is obtained by dividing the sum of the bands due to editing by the sum of all three bands (edited plus unedited). (B) Northern blot analysis of HDV genomic RNA. Results for pCI are shown twice because the data shown are from two independent transfections.
Editing at the amber/W site of HDV RNA yields production of HDAg-L, which functions as a trans-dominant inhibitor of HDV RNA replication (9, 14). During HDV replication, HDAg-L is normally present only in the later stages, when sufficient viral RNA has been made for packaging. As HDV replication proceeds, amber/W editing levels increase gradually and reach a maximum of about 30%; in cell culture transfection experiments, this level is achieved 10 to 14 days posttransfection (5, 8, 43). Early onset of high levels of amber/W editing could cause excessive HDAg-L synthesis that could inhibit HDV RNA replication.
To determine the effects of ADAR overexpression on HDV RNA replication, we analyzed the level of HDV genomic RNA, which is produced from the expressed antigenome via RNA replication in the cotransfected cells (Fig. 1B). Both ADAR1 and ADAR2 strongly suppressed HDV RNA replication; ADAR1 inhibited replication by about 10-fold, and ADAR2 inhibited replication 100-fold. This inhibition was observed without any apparent cytotoxicity; expression of a cotransfected secreted alkaline phosphatase transfection reporter gene was affected by neither ADAR1 nor ADAR2.
Increased amber/W editing is not the only mechanism by which ADAR overexpression inhibits HDV RNA replication.
As noted above, excessive premature production of HDAg-L could well account for the observed inhibition of HDV RNA replication by ADAR1 and ADAR2. To determine the extent to which HDAg-L overproduction was responsible for the inhibition, we analyzed the effects of ADAR2 expression on replication of a site-directed HDV mutant (G580A) that is edited much less efficiently than the wild type. The mutation G580A (nucleotide identities and numbering are in the genome sense) changes the A-C mismatch pair in the amber/W editing site to an A-U pair; this single base change decreases amber/W editing by about 10-fold without affecting HDV RNA replication (5, 31, 35).
Cotransfection of the ADAR2 expression construct pAD2 with the G580A HDV mutant led to increased editing at the amber/W site (11%; Fig. 2A); however, the level of editing attained was significantly less than that of the wild type (Fig. 1) and was not substantially greater than that observed for the wild type without cotransfected ADAR expression constructs. Despite the markedly lower amber/W editing levels, replication of the G580A mutant was strongly inhibited by cotransfection of ADAR2 (Fig. 2B). Similar results were obtained for the effect of ADAR1 overexpression on G580A RNA editing and replication (data not shown). These results suggested that increased amber/W editing might not be the sole explanation for the observed inhibition of HDV replication by ADAR overexpression.
FIG. 2.
Effect of ADAR2 overexpression on amber/W editing and replication of the HDV mutant G580A. Huh7 cells were cotransfected with 1 μg each of an ADAR2 expression construct (pAD2) and pM6, which produces replication-competent antigenomic HDV RNA containing the G580A mutation, which strongly reduces amber/W editing. RNAs were harvested 4 days posttransfection and analyzed for both amber/W editing and HDV genomic RNA as for Fig. 1. (A) Amber/W editing. (B) Northern blot analysis of HDV genomic RNA.
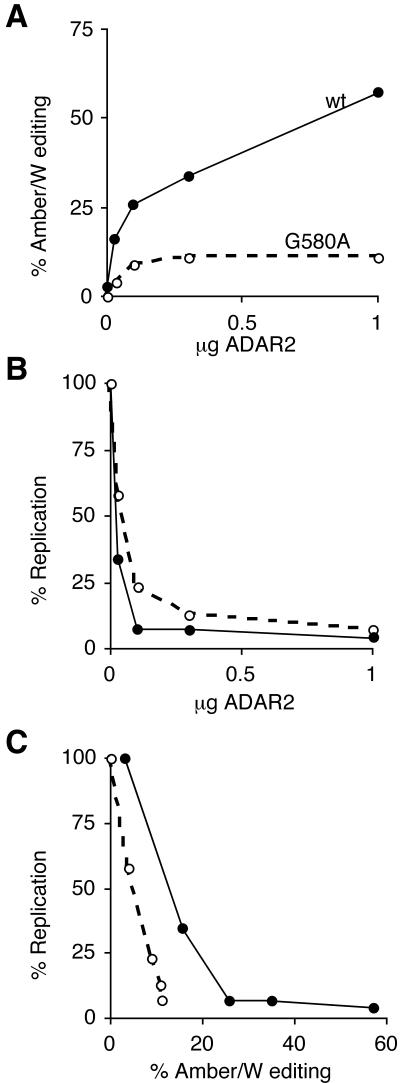
To further analyze the role of amber/W editing in the inhibition of replication by high-level ADAR expression, we cotransfected various amounts of the ADAR2 expression construct pAD2 with constructs expressing either wild-type HDV or the G580A mutant and assessed both amber/W editing and HDV RNA replication (Fig. 3). Replication of both wild-type and G580A mutant HDV was progressively inhibited by increasing amounts of the cotransfected ADAR2 expression construct (Fig. 3B). Notably, comparison of wild-type and G580A mutant activities indicated that both amber/W editing and RNA replication of wild-type HDV were more sensitive to ADAR2 overexpression (Fig. 3A and 3B). For wild-type HDV, greater than 95% inhibition was observed with 0.1 μg of pAD2 cotransfected, which produced 25% amber/W editing. Neither this level of editing nor inhibition of replication was achieved with the G580A mutant for any amount of pAD2 transfected. These results indicate that increased amber/W editing is the primary mechanism by which ADAR overexpression inhibits HDV RNA replication.
FIG. 3.
Effect of ADAR2 expression on both amber/W editing and replication of wild-type and G580A mutant HDV. Huh7 cells were cotransfected with 1 μg of either pHDV (wild type) or pM6 (G580A mutant) and either 0, 0.03, 0.1, 0.3, or 1 μg of pAD2. The total amountof transfected DNA was made up to 2 μg with the control expression vector pCI. RNAs were harvested 4 days posttransfection and analyzed for amber/W editing and replication as in Fig. 1. For all plots, values for wild-type HDV are indicated by solid circles and solid lines; values for the G580A mutant are indicated by open circles and dashed lines. (A) Amber/W editing versus amount of pAD2 transfected. (B) HDV RNA replication versus amount of pAD2 transfected. Percent replication is the amount of genomic RNA detected relative to the amount detected when pCI alone was cotransfected with either pHDV or pM6. All values were first normalized to levels of secreted alkaline phosphatase, which was expressed from a cotransfected plasmid included as a control for transfection variations. (C) Values from A and B replotted as replication versus amber/W editing.
To more clearly distinguish the effects of amber/W editing from other effects on replication, we took the data from Fig. 3A and 3B and plotted the level of replication versus the amount of amber/W editing for both the wild type and the G580A mutant (Fig 3C). For both, increased editing (and ADAR expression) was negatively correlated with RNA replication, as was expected given the effect of HDAg-L on HDV RNA replication. If amber/W editing were the sole factor responsible for the inhibition of replication by ADAR2, the two curves in Fig. 3C would be superimposed. The observed displacement between the two curves (Fig. 3C) indicates that ADAR overexpression produces additional effects that can also inhibit HDV RNA replication.
Hyperediting at non-amber/W sites produces trans-dominant inhibitors of HDV replication.
Several factors led us to consider deamination of non-amber/W sites as a possible additional mechanism by which ADAR expression could inhibit HDV replication. Although editing of HDV RNA by Xenopus ADAR is highly specific for the amber/W site in vitro, a significant number of non-amber/W sites were modified (31). Moreover, high-level expression of ADARs can induce readily detectable amber/W site editing in RNAs containing mutations that virtually eliminate editing by endogenous ADAR activity (Fig. 2) (35, 42). Similarly, high-level ADAR expression may induce significant editing at non-amber/W sites that are not edited at detectable levels by endogenous ADARs. Because HDAg functions as a multimer (21, 39, 44), some such HDAg mutations could act as dominant inhibitors of replication, as observed for HDAg-L.
To analyze non-amber/W editing, we reverse transcribed and amplified the HDAg coding region from HDV RNAs expressed with ADAR1 and ADAR2 (Fig. 1) and obtained 50 cDNA clones of each. As described in Materials and Methods, these clones were derived from three separate PCR amplifications starting from three different aliquots of RNA collected from Huh7 cells 4 days after cotransfection with 1 μg each of pHDV and pAD1 or pAD2. By the same protocol, we also obtained 20 clones from RNA collected 4 days after cotransfection of Huh7 cells with 1 μg each of pHDV and the control vector plasmid pCI, which does not produce ADAR. The sequence of the HDAg coding region in these clones was determined.
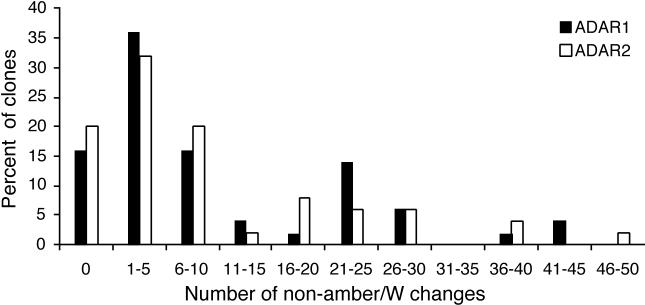
Sequence analysis of the clones obtained indicated amber/W editing in 46 and 74% of those derived from HDV RNA cotransfected with ADAR1 and ADAR2, respectively. These values are in good agreement with those obtained by the RT-PCR/StyI digestion assay (Fig. 1). While the amber/W site was the preferred site for editing, ADAR overexpression induced extensive editing at multiple non-amber/W sites (Fig 4). Both ADAR1 and ADAR2 edited 5.5% of the non-amber/W adenosines in the clones analyzed. This level of non-amber/W editing is substantially higher than that observed previously for replicating HDV RNA in transfected cells (32) or in cells transfected with the control expression construct pCI in this study, for which less than 0.1% of non-amber/W adenosines were deaminated.
FIG. 4.
Percentage of HDV RNA cDNA clones with non-amber/W A → G changes in the HDAg coding region (nucleotides 949 to 1598). The percentage of clones (represented on the y axis) with the indicated number of non-amber/W changes (represented on the x axis) is shown. As described in Materials and Methods, the cDNA populations were derived, by RT-PCR, from replicating HDV RNAs harvested from Huh7 human hepatoma cells 4 days after cotransfection with 1 μg each of the replicating HDV RNA expression construct (pHDV), designed to synthesize antigenomic HDV RNA, and either an ADAR1 (pAD1) or ADAR2 (pAD2) expression construct. Fifty cDNA clones each were analyzed for ADAR1 and ADAR2.
Analysis of the distribution of editing among the clones showed that over 25% of the clones were obtained from RNAs modified at 16 or more of the 187 adenosines (8.6%) in the HDAg coding region, and more than 80% of the clones contained at least one edited non-amber/W adenosine (Fig. 4). On the other hand, 15 to 20% of clones contained no edited adenosines, and nearly half contained two or fewer modifications.
While 5.5% of the adenosines in these clones were changed to guanosine for both ADAR1 and ADAR2, consistent with A→I editing on antigenomic RNA, the number of U→C changes, which are consistent with A→I editing on the genomic RNA, was much smaller (0.27% for ADAR1 and 0.19% for ADAR2). (Other changes occurred at frequencies of <0.05%.) The difference is likely due to the fact that we transfected a construct that generated antigenomic RNA, which then served as a substrate for editing. The genome RNA is apparently equally susceptible to deleterious hyperediting, because ADAR cotransfection with an HDV construct that generated genomic RNA also strongly inhibited HDV RNA replication (not shown).
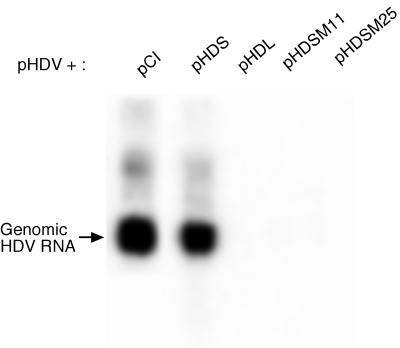
Many of the modifications found changed the coding sequence of HDAg. To determine whether the hyperediting observed at non-amber/W sites produced HDAg that would act as a trans-dominant inhibitor of replication, we selected two highly edited HDAg clones from those obtained from HDV RNA coexpressed with ADAR2. These clones, M11 and M25, were edited at 21 and 16 non-amber/W adenosines, respectively, and contained 13 and 12 amino acid substitutions, respectively. Expression constructs for HDAg-S derived from these clones were cotransfected into Huh7 cells along with the replicating HDV RNA expression construct pHDV; control transfections included wild-type HDAg-S, which has little effect on replication, and wild-type HDAg-L, which strongly inhibits HDV RNA replication (9, 14). RNA was collected 4 days posttransfection, and replication of HDV was assayed by Northern blot analysis of HDV genomic RNA, as described in Materials and Methods. We observed that expression of HDAg-S containing the mutations found in the M11 and M25 clones led to substantial inhibition of HDV replication (Fig. 5). Indeed, the inhibition due to M11 and M25 was at least as great as that due to HDAg-L, the prototypical trans-dominant inhibitor of HDV RNA replication.
FIG. 5.
Inhibition of HDV RNA replication by ADAR-edited HDAg-S mutants. Huh7 cells were cotransfected with 1 μg of pHDV and 0.04 μg of either pCI or the following HDAg expression constructs: pHDS, expressing HDAg-S; pHDL, expressing HDAg-L; or pHDSM11 or pHDSM25, expressing HDAg-S containing mutations induced by ADAR2 overexpression. RNA was extracted and Northern blot analyses were performed as described in Materials and Methods.
The above results suggest that high levels of ADAR expression inhibit HDV RNA replication by a combination of effects on the HDAg coding region—excessive production of HDAg-L caused by amber/W site editing and production of dominant negative HDAg variants caused by extensive non-amber/W editing. Both of these effects are related to increased deamination of HDV RNA in the presence of high levels of ADAR expression. To determine whether hyperediting could also directly inhibit the ability of the RNA to replicate, for example, by interfering with essential RNA structures such as the ribozymes, we analyzed the ability of ADAR2 to inhibit replication of a site-directed HDV RNA mutant that was deficient for HDAg synthesis (6); replication of HDV RNA produced by this construct is supported by cotransfection of an HDAg-S expression construct (6). We found that ADAR2 overexpression did not inhibit replication of the HDV RNA defective for HDAg synthesis (not shown). This result is consistent with the interpretation that the effects of ADAR overexpression on HDV replication are primarily mediated by sequence changes in HDAg rather than by mutations that affect RNA structures required for replication.
Inhibition of HDV RNA replication by ADARs deficient in deaminase activity.
We created site-directed mutations in the catalytic deaminase domains of ADAR1 and ADAR2 to determine the extent to which ADAR inhibition of HDV RNA replication might be due to effects other than RNA editing, such as by binding HDV RNA. In these mutants, the highly conserved CHAE sequence in the deaminase domain was altered to CQAA; this mutation has been shown to eliminate the deaminase activity of ADAR1 without affecting its ability to bind dsRNA (23). Expression constructs for the resultant mutant proteins were transfected with pHDV; cells were harvested 4 days posttransfection, and levels of amber/W editing and HDV RNA replication were assessed.
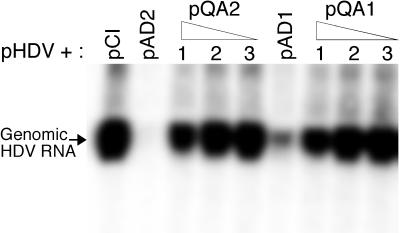
We observed that overexpression of both ADAR1 and ADAR2 deficient in deaminase activity was nevertheless able to partially inhibit HDV RNA replication (Fig. 6). This residual inhibition, which was not due to increased amber/W editing (not shown), does not appear to be a dominant mechanism by which ADAR overexpression inhibits HDV replication. The inhibition by the deaminase-deficient ADARs was much less than that observed for the wild-type ADARs cotransfected with either wild-type HDV or the G580A mutant (Fig. 2, 3, and 4).
FIG. 6.
Effect of deaminase-deficient forms of ADAR1 and ADAR2 on HDV RNA replication. Huh7 cells were cotransfected with pHDV and either pCI, pAD1, different amounts of the deaminase-deficient ADAR1 expression construct pQA1, pAD2, or different amounts of the deaminase-deficient ADAR2 expression construct pQA2. The amounts of pQA1 or pQA2 cotransfected were as follows: lanes 1, 1.0 μg; lanes 2, 0.3 μg; and lanes 3, 0.1 μg. RNAs were harvested 4 days posttransfection and analyzed for HDV genomic RNA as described in Materials and Methods. Percent replication is the amount of genomic RNA detected relative to the amount detected when pCI alone was cotransfected with pHDV. The values are the averages for two independent transfection experiments. All values were first normalized to levels of secreted alkaline phosphatase, which was expressed from a cotransfected plasmid included as a control for transfection variations.
DISCUSSION
We have found that HDV RNA editing and replication are sensitive to levels of ADAR expression. Both ADAR1 and ADAR2 were able to increase amber/W site editing in full-length replicating HDV RNA. This observation is consistent with recent findings that editing of an HDV amber/W site in an mRNA editing reporter construct is increased by high levels of both ADAR1 and ADAR2 expression (35, 42). These observations do not directly address which ADAR is responsible for editing HDV RNA in infected liver but are consistent with either ADAR1 or ADAR2 playing a role. The higher level of ADAR1 expression in uninfected liver (17, 26) suggests that ADAR1 may be the primary source of activity in infected cells, but further analysis will likely be necessary to determine whether levels of either enzyme are influenced by HDV infection.
Inhibition of HDV RNA replication by overexpression of ADAR1 or ADAR2 was primarily due to increased editing at the amber/W site (Fig. 1 to 3), which yields excessive premature production of HDAg-L, a potent trans-dominant inhibitor of HDV RNA replication. Inhibition was quite dramatic at the highest expression levels, which increased amber/W editing from about 4% to well over 50%, but was also evident at lower expression levels. ADAR2 expression levels which induced 18% amber/W editing caused a two-thirds reduction in HDV RNA replication (Fig. 3). The mechanisms by which HDV regulates amber/W editing during replication have yet to be fully determined, but our data suggest that increased expression of ADARs is a potential mechanism. Moreover, the substantial negative effect of increased editing on replication underscores the significance of regulating amber/W editing in the HDV life cycle.
The endogenous editing activity in Huh7 cells, which is likely due to some combination of ADAR1 and ADAR2 activities, has been shown to be highly selective for the amber/W site (32), and similar results were obtained in this study. When overexpressed, both ADAR1 and ADAR2 edited HDV RNA extensively at multiple non-amber/W adenosines. Although the amber/W site was still the most preferred site under these conditions, some of the other edited sites were nearly as active (not shown). The decrease in selectivity compared to the endogenous editing activity was likely due to the high level of ADAR expression. The effects of ADAR levels on editing selectivity have not been extensively analyzed in other substrates for highly specific editing; Melcher et al. (26) reported that editing of the glutamate receptor subunit B Q/R site by ADAR2 (RED1) in vitro was more specific at low enzyme levels; as more ADAR2 was added to the reaction mixture, editing at an intron hot spot increased. The presence of multiple potential editing sites is more likely to be important for HDV, because the size of the extensively base-paired HDV RNA is substantially greater than that of other editing substrates.
Editing at many of the additional sites produced coding changes in HDAg that could yield HDAg variants with dominant negative phenotypes. The production of such variants is a likely secondary means by which ADAR overexpression inhibits HDV RNA replication. Analysis of two HDAg-S clones that were derived from those found in HDV RNA coexpressed with ADAR2 and contained 13 or 12 amino acid changes demonstrated that these ADAR-induced variants do indeed act as trans-dominant inhibitors of replication, similar to HDAg-L. While we do not know which amino acid change(s) is responsible for the dominant negative effect, over 25% of the RNAs analyzed by cloning showed non-amber/W editing at levels similar to or greater than that of the two clones tested. It is likely that many if not all of these RNAs would produce similarly effective dominant negative HDAg variants. A population of genomes in which 25% of the HDAg produced exhibited the dominant negative phenotype would be severely impaired for replication.
The dominant negative phenotype likely results from the disruption of an HDAg-S complex by interaction with a variant form of HDAg, similar to the proposed mechanism by which HDAg-L inhibits HDV RNA replication (16, 44). Previously identified dominant negative variants have included HDAg-L, HDAg-S with an N-terminal addition, and genotype III HDAg-S; however, none of a series of deletion mutations exhibited this phenotype (21, 22), and no site-directed mutations of HDAg-S have been shown to act in this manner. It has been suggested that, at least for HDAg-L, the dominant negative phenotype is due to an alternative protein fold that may induce a conformational change (16). In this regard, it is worth noting that in both of the dominant negative HDAg variants reported here, the region between amino acids 120 and 145 (which is predicted to form an alpha helix) contains multiple glycine substitutions that could alter the conformation of this region.
The weak inhibition of HDV RNA replication by overexpression of deaminase-deficient ADARs is likely due to binding of ADARs to HDV RNA via interactions between the dsRNA-binding motifs and the unbranched rod structure of the RNA, which contains short dsRNA segments interspersed with mismatches, bulges, and internal loops. Some such imperfect dsRNAs have been shown to interact with dsRNA-binding motifs (3). Indeed, editing at the amber/W site, which neither contains nor neighbors a significant dsRNA region, likely requires interactions between HDV RNA and ADAR dsRNA-binding motifs, as does editing at the less selective sites found when ADARs were expressed at high levels. In a study that may be related in this regard, Circle et al. (12) showed that HDV RNA can bind and activate dsRNA-activated protein kinase in vitro. These observations raise interesting questions about the nature of interactions between HDV RNA and dsRNA-binding motifs. Such interactions may play a role in HDV RNA replication; moreover, the avoidance or subversion of these interactions by HDV may permit the virus to replicate without triggering cellular responses to dsRNA.
In addition to the three mechanisms we have identified by which ADAR overexpression inhibits HDV RNA replication, at least two may merit further investigation in future studies. First, extensive editing may yield RNAs that are incapable of replication because of the disruption of important functional structures, such as the ribozymes or as yet undefined elements required for RNA replication. The analysis of the replication competence of extensively modified RNAs could yield valuable information about the role of different sequences and structures in HDV RNA replication. Second, extensive deamination may expose HDV RNA to cleavage by a recently identified cellular RNase activity that specifically cleaves RNAs containing inosine (36).
While interesting, neither of these potential effects, which would operate in cis, seems likely to be as great as those observed for amber/W editing or extensive editing at non-amber/W sites, because the latter effects operate via trans-dominant inhibitors. Mutations that only affect RNA structural features, such as ribozymes or transcription control elements, are only likely to affect replication of the RNA on which they occur, but will not inhibit the replication of other RNAs. On the other hand, HDAg mutations that create dominant negative inhibitors can inhibit the replication of even wild-type RNAs, as shown in Fig. 5. Thus, for HDV, inhibitors that operate via trans effects can be much more potent than those that operate in cis.
Our sequence analysis showed that about half of the clones analyzed were edited at two sites or fewer (15 to 20% were not edited at all; Fig. 4) and that the locations of these sites were heterogeneous (and therefore not likely to be uniformly detrimental to replication). Thus, even if all RNAs edited at three or more sites were completely compromised for further replication (which seems unlikely), we would see no more than a 50% reduction in RNA replication if inhibition were limited to cis effects. On the contrary, ADAR overexpression results in greater than 90% inhibition of RNA replication; this strong inhibition is entirely consistent with and explained by the ability of HDAg mutants to act as trans-dominant negative inhibitors.
The demonstration that HDV editing and replication are sensitive to the levels of ADAR expression raises questions for both HDV and other viruses. First, regarding the role of ADARs in HDV replication, it will be interesting to determine whether HDV replication regulates ADAR expression, perhaps by inducing higher expression levels after sufficient amounts of viral RNA have accumulated.
ADAR1 was initially cloned by Patterson et al. (29) in a screen of interferon-responsive genes, and its expression has been shown to be upregulated following interferon treatment (29, 30). This interferon responsiveness and the inhibition of HDV replication that we have observed raise the possibility that ADARs may function as part of the cellular response to viral infection. The strong inhibition of HDV replication in the absence of apparent toxicity also raises the possibility that overexpression of ADARs may be a useful tool for combating not only HDV replication but that of other RNA viruses as well.
Acknowledgments
This work was supported by grant R01-AI42324 from the National Institutes of Health.
REFERENCES
- 1.Bass, B. L. 1997. RNA editing and hypermutation by adenosine deamination. Trends Biochem. Sci. 22:157-162. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann, K. F., and J. L. Gerin. 1986. Antigens of hepatitis delta virus in the liver and serum of humans and animals. J. Infect. Dis. 154:702-706. [DOI] [PubMed] [Google Scholar]
- 3.Bevilacqua, P. C., C. X. George, C. E. Samuel, and T. R. Cech. 1998. Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry 37:6303-6316. [DOI] [PubMed] [Google Scholar]
- 4.Bonino, F., B. Hoyer, J. W. Shih, M. Rizzetto, R. H. Purcell, and J. L. Gerin. 1984. Delta hepatitis agent: structural and antigenic properties of the delta-associated particle. Infect. Immun. 43:1000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casey, J. L., K. F. Bergmann, T. L. Brown, and J. L. Gerin. 1992. Structural requirements for RNA editing in hepatitis delta virus: evidence for a uridine-to-cytidine editing mechanism. Proc. Natl. Acad. Sci. USA 89:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casey, J. L., and J. L. Gerin. 1998. Genotype-specific complementation of hepatitis delta virus RNA replication by hepatitis delta antigen. J. Virol. 72:2806-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casey, J. L., and J. L. Gerin. 1995. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J. Virol. 69:7593-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casey, J. L., A. G. Polson, B. L. Bass, and J. L. Gerin. 1997. Hepatitis delta virus genetic variations and RNA editing, p. 320-326. In M. Rizzetto, R. H. Purcell, J. L. Gerin, and G. Verme (ed.), Viral hepatitis and liver disease. Edizioni Minerva Medica, Turin, Italy.
- 9.Chao, M., S. Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C. X., D. S. Cho, Q. Wang, F. Lai, K. C. Carter, and K. Nishikura. 2000. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6:755-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, P. J., F. L. Chang, C. J. Wang, C. J. Lin, S. Y. Sung, and D. S. Chen. 1992. Functional study of hepatitis delta virus large antigen in packaging and replication inhibition: role of the amino-terminal leucine zipper. J. Virol. 66:2853-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Circle, D. A., O. D. Neel, H. D. Robertson, P. A. Clarke, and M. B. Mathews. 1997. Surprising specificity of PKR binding to delta agent genomic RNA. RNA 3:438-448. [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen, B. R., and M. H. Malim. 1992. Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 216:362-368. [DOI] [PubMed] [Google Scholar]
- 14.Glenn, J. S., and J. M. White. 1991. trans-dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 65:2357-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gott, J. M., and R. B. Emeson. 2000. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34:499-531. [DOI] [PubMed] [Google Scholar]
- 16.Hwang, S. B., and M. M. Lai. 1993. A unique conformation at the carboxyl terminus of the small hepatitis delta antigen revealed by a specific monoclonal antibody. Virology 193:924-931. [DOI] [PubMed] [Google Scholar]
- 17.Kim, U., Y. Wang, T. Sanford, Y. Zeng, and K. Nishikura. 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. USA 91:11457-11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo, M. Y., J. Goldberg, L. Coates, W. Mason, J. Gerin, and J. Taylor. 1988. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J. Virol. 62:1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 20.Lai, M. M. 1995. The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 64:259-286. [DOI] [PubMed] [Google Scholar]
- 21.Lazinski, D. W., and J. M. Taylor. 1993. Relating structure to function in the hepatitis delta virus antigen. J. Virol. 67:2672-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazinski, D. W., and J. M. Taylor. 1993. Structure and function of the delta virus antigens. Prog. Clin. Biol. Res. 382:35-44. [PubMed] [Google Scholar]
- 23.Liu, Y., C. X. George, J. B. Patterson, and C. E. Samuel. 1997. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J. Biol. Chem. 272:4419-4428. [DOI] [PubMed] [Google Scholar]
- 24.Luo, G. X., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melcher, T., S. Maas, A. Herb, R. Sprengel, M. Higuchi, and P. H. Seeburg. 1996. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem. 271:31795-31798. [DOI] [PubMed] [Google Scholar]
- 26.Melcher, T., S. Maas, A. Herb, R. Sprengel, P. H. Seeburg, and M. Higuchi. 1996. A mammalian RNA editing enzyme. Nature 379:460-464. [DOI] [PubMed] [Google Scholar]
- 27.Niro, G. A., A. Smedile, A. Andriulli, M. Rizzetto, J. L. Gerin, and J. L. Casey. 1997. The predominance of hepatitis delta virus genotype I among chronically infected Italian patients. Hepatology 25:728-734. [DOI] [PubMed] [Google Scholar]
- 28.O'Connell, M. A., S. Krause, M. Higuchi, J. J. Hsuan, N. F. Totty, A. Jenny, and W. Keller. 1995. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol. Cell. Biol. 15:1389-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson, J. B., and C. E. Samuel. 1995. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 15:5376-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patterson, J. B., D. C. Thomis, S. L. Hans, and C. E. Samuel. 1995. Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 210:508-511. [DOI] [PubMed] [Google Scholar]
- 31.Polson, A. G., B. L. Bass, and J. L. Casey. 1996. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380:454-456. [DOI] [PubMed] [Google Scholar]
- 32.Polson, A. G., H. L. Ley 3rd, B. L. Bass, and J. L. Casey. 1998. Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol. Cell. Biol. 18:1919-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizzetto, M. 1983. The delta agent. Hepatology 3:729-737. [DOI] [PubMed] [Google Scholar]
- 34.Rueter, S. M., T. R. Dawson, and R. B. Emeson. 1999. Regulation of alternative splicing by RNA editing. Nature 399:75-80. [DOI] [PubMed] [Google Scholar]
- 35.Sato, S., S. K. Wong, and D. W. Lazinski. 2001. Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. J. Virol. 75:8547-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scadden, A. D., and C. W. Smith. 2001. Specific cleavage of hyper-edited dsRNAs. EMBO J. 20:4243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seeburg, P. H., M. Higuchi, and R. Sprengel. 1998. RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res. Rev. 26:217-229. [DOI] [PubMed] [Google Scholar]
- 38.Wang, J. G., J. Cullen, and S. M. Lemon. 1992. Immunoblot analysis demonstrates that the large and small forms of hepatitis delta virus antigen have different C-terminal amino acid sequences. J. Gen. Virol. 73:183-188. [DOI] [PubMed] [Google Scholar]
- 39.Wang, J. G., and S. M. Lemon. 1993. Hepatitis delta virus antigen forms dimers and multimeric complexes in vivo. J. Virol. 67:446-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta viral genome. Nature 323:508-514. [DOI] [PubMed] [Google Scholar]
- 41.Weiner, A. J., Q.-L. Choo, K.-S. Wang, S. Govindarajan, A. G. Redeker, J. L. Gerin, and M. Houghton. 1988. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24δ and p27δ. J. Virol. 62:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong, S. K., S. Sato, and D. W. Lazinski. 2001. Substrate recognition by ADAR1 and ADAR2. RNA 7:846-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, T. T., H. J. Netter, D. W. Lazinski, and J. M. Taylor. 1997. Effects of nucleotide changes on the ability of hepatitis delta virus to transcribe, process, and accumulate unit-length, circular RNA. J. Virol. 71:5408-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia, Y. P., and M. M. Lai. 1992. Oligomerization of hepatitis delta antigen is required for both the trans-activating and trans-dominant inhibitory activities of the delta antigen. J. Virol. 66:6641-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, J. H., P. Sklar, R. Axel, and T. Maniatis. 1997. Purification and characterization of a human RNA adenosine deaminase for glutamate receptor B pre-mRNA editing. Proc. Natl. Acad. Sci. USA 94:4354-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]