Abstract
Hepatitis delta virus (HDV) contains a viroid-like circular RNA that is presumed to replicate via a rolling circle replication mechanism mediated by cellular RNA polymerases. However, the exact mechanism of rolling circle replication for HDV RNA and viroids is not clear. Using our recently described cDNA-free transfection system (L. E. Modahl and M. M. Lai, J. Virol. 72:5449-5456, 1998), we have succeeded in detecting HDV RNA replication by metabolic labeling with [32P]orthophosphate in vivo and obtained direct evidence that HDV RNA replication generates high-molecular-weight multimeric species of HDV RNA, which are processed into monomeric and dimeric forms. Thus, these multimeric RNAs are the true intermediates of HDV RNA replication. We also found that HDV RNA synthesis is highly temperature sensitive, occurring most efficiently at 37 to 40°C and becoming virtually undetectable at temperatures below 30°C. Moreover, genomic HDV RNA synthesis was found to occur at a rate roughly 30-fold higher than that of antigenomic RNA synthesis. Finally, in lysolecithin-permeabilized cells, the synthesis of full-length antigenomic HDV RNA was completely resistant to high concentrations (100 μg/ml) of α-amanitin. In contrast, synthesis of genomic HDV RNA was totally inhibited by α-amanitin at concentrations as low as 2.5 μg/ml. Thus, these results suggest that genomic and antigenomic HDV RNA syntheses are performed by two different host cell enzymes. This observation, combined with our previous finding that hepatitis delta antigen mRNA synthesis is likely performed by RNA polymerase II, suggests that the different HDV RNA species are synthesized by different cellular transcriptional machineries.
Circular RNAs of viroids and virusoids and satellite RNA have been postulated to undergo replication by a rolling circle mechanism (1). This model is supported by the detection of multimeric RNAs in infected cells (1, 18) and, in some cases, by the ability of these RNAs to undergo autocatalytic cleavage into monomer RNAs in vitro (36). However, the exact role of multimer RNAs in viroid replication is not clear, as the nascent intermediates of RNA replication have not, so far, been detected. Moreover, the reverse process of ligation of monomer RNAs into multimer RNAs can also occur in vitro (2, 36), making it difficult to determine whether the multimer RNAs are genuine intermediates of RNA replication. Furthermore, the multimer RNAs often assume a rod-like secondary structure that is incompatible with the ribozyme conformation necessary for the processing of these RNAs (9, 13). Viroid RNA replication is presumably carried out by a cellular RNA-dependent RNA polymerase, which has been detected in some plant species (38). However, neither whole-cell nor in vitro transcription studies have established that this polymerase is capable of carrying out rolling circle replication of viroid RNA. Thus, although the rolling circle replication model most elegantly explains the replication of circular RNAs, it is supported only by circumstantial evidence, and the molecular mechanisms of such an RNA replication remain largely unknown.
Hepatitis delta virus (HDV) is the only animal virus containing a circular RNA. HDV is usually associated with hepatitis B virus and frequently causes severe acute and chronic liver disease in humans (reviewed in reference 31). The HDV genome consists of a small, single-stranded, circular RNA molecule of roughly 1.7 kb that, in the virion, is associated with the only known virus-encoded protein, hepatitis delta antigen (HDAg). Due to a high degree of intramolecular base pairing, HDV RNA usually folds into an unbranched rod-like structure similar to that of viroids (40). HDV RNA contains a ribozyme activity, which requires a unique conformation of five helices formed by a stretch of 85 nucleotides (nt) (7). This ribozyme activity is essential for HDV RNA replication (19, 27). The HDV RNA itself (i.e., the genomic strand) does not encode any protein. However, the complementary strand (antigenomic HDV RNA), which is detected in HDV-infected cells, encodes HDAg. HDAg occurs as two species, small HDAg (S-HDAg or p24) and large HDAg (L-HDAg or p27), which play different roles in HDV replication. S-HDAg is an essential activator of HDV RNA replication (21), whereas L-HDAg has potent inhibitory activity (11) but is essential for virion assembly (3).
HDV RNA replication is thought to occur via a double rolling circle mechanism similar to that proposed for some viroids (1, 22). Most features of this model have not been experimentally proven. Moreover, since it has been shown that the HDV RNA ligase reaction can occur both inter- and intramolecularly (37), it is conceivable that the multimeric HDV RNA species detected in HDV-infected cells are a result of aberrant ligations between monomer species and may represent dead-end products. In addition to the HDV RNA species described above, a much less abundant subgenomic, antigenomic-sense, poly(A)+ species of approximately 800 nt is also detected in HDV-infected cells (4, 30). This serves as the mRNA for the synthesis of HDAg.
HDV RNA is the first and, so far, only example of an RNA species that can be copied by cellular enzymes in mammalian cells. Several reports have suggested the possible presence of an RNA-dependent RNA polymerase activity in mammals; however, this activity has yet to be clearly identified (6, 39). Studies using nuclear run-on assays or in vitro transcription systems have shown that HDV RNA synthesis is sensitive to α-amanitin (8, 10, 26), suggesting the involvement of RNA polymerase II (pol II). However, for a variety of reasons (reviewed in reference 33), these studies may not reflect the natural requirements of HDV RNA replication. Our recent study examining HDV RNA synthesis in cell cultures showed that HDAg mRNA production was very sensitive to α-amanitin (33). In contrast, synthesis of full-length antigenomic HDV RNA was resistant to α-amanitin at concentrations up to 25 μg/ml, implying the involvement of pol I or enzymes other than pol II in the latter reaction (33). However, this study relied on the measurement of steady-state RNA, which complicated the interpretation of the kinetics of HDV RNA synthesis.
In this study, we succeeded in metabolic labeling with [32P]orthophosphate of HDV RNA intermediates in HuH7 cells transfected by using our recently developed HDV cDNA-free transfection method (30). This approach allowed us to determine the likely processing events of rolling circle replication. We further show the different cellular polymerase requirements for genomic and antigenomic HDV RNA synthesis, suggesting that more than one mammalian cellular polymerase is involved in RNA-dependent RNA synthesis.
MATERIALS AND METHODS
Cell culture and transfection.
Human hepatoma cell line HuH7 (35) was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin and incubated at 37°C in 5% CO2. For transfection studies, cell cultures were seeded overnight in six-well plates or 60-mm-diameter petri dishes and transfected with 5 and 10 μg, respectively, of 50:50 mixtures of HDAg mRNA and HDV RNA (1.2 times the genome length) using DMRIE-C reagent (Gibco BRL) according to the manufacturer's directions. Following transfection, the cultures were incubated overnight, the medium was changed, and incubation was continued for an additional 2 to 7 days.
Plasmids and cloning.
Plasmid pBSδG-Basic was prepared by cloning a SmaI-XbaI fragment from the Italian HDV sequence (nt 479 to 781, according to the numbering of Wang et al. [40]) between the KpnI and XbaI sites of pBluescript SK (Stratagene). The 302-bp HDV cDNA fragment in this vector includes the complete sequence for the genomic-strand HDV ribozyme. Similarly, pBSδAG-Basic contains a SmaI-XbaI (nt 1108 to 781) fragment of HDV cDNA that includes the complete antigenomic HDV ribozyme sequence cloned between the KpnI and XbaI sites of pBluescript SK. Plasmids pBSδ1.2G and pBSδ1.2AG (in vitro transcription templates for the genomic and antigenomic HDV RNAs, respectively, whose lengths were 1.2 times the genome length) were obtained by cloning XbaI monomer fragments of HDV cDNA from plasmid pSVLD3 (21) into the XbaI sites of pBSδG-Basic and pBSδAG-Basic, respectively. Plasmids pX9-1/II and pKS/HDV1.9, used as in vitro transcription templates for HDAg mRNA and 1.9-kb antigenomic HDV RNA synthesis, respectively, and plasmids pTMδSalA and pTMδSalB, used for the generation of unlabeled and 32P-labeled monomers of genomic and antigenomic HDV RNA, respectively, have been described elsewhere (25, 30). pBSδHX, used in the generation of 32P-labeled riboprobes for detection of the antigenomic HDV RNA not included in the HDAg mRNA species, was constructed by inserting a HindIII-XbaI (nt 1 to 781) cDNA fragment from the Italian HDV sequence (40) into the same sites of pBluescript SK.
In vitro transcription.
HDV RNAs 1.2 times the genome length were transcribed from plasmids pBSδ1.2G, pBSδ1.2AG, pBSδ1.2G(2xS), and pBSδ1.2AG(2xS) with T7 MEGAscript kits (Ambion) after linearization with restriction enzyme NotI. Capped mRNA for HDAg was transcribed from plasmid pX9-I/II after linearization with HindIII by using a T7 m-Message m-Machine kit (Ambion). Unlabeled monomer genomic and antigenomic HDV RNAs were transcribed from pTMδSalA and pTMδSalB with T7 MEGAscript after linearization by PstI digestion. The method for generation of HDV-specific 32P-labeled riboprobes has been described elsewhere (25). Production of 1.9-kb antigenomic HDV RNA from pKS/HDV1.9 has been described previously (33).
Partitioning of nuclear and cytoplasmic fractions.
Partitioning of cell lysates into nuclear and cytoplasmic fractions was performed by a modification of a previously published protocol (29) as described in the accompanying article (28).
Northern blot and reverse hybridization analyses.
RNA was extracted from intact cells or nuclear and cytoplasmic fractions with Tri-Reagent (Molecular Research Center, Inc.) according to the manufacturer's protocol. For analysis by Northern blotting, the RNA samples were treated with formaldehyde and separated by electrophoresis through MOPS (morpholinepropanesulfonic acid)-formaldehyde-containing 1.2% agarose gels. RNA was then transferred to a BrightStar-Plus nylon membrane (Ambion) according to the method recommended by the manufacturer using a high-salt buffer containing 10 mM NaOH to ensure even transfer of low- and high-molecular-weight RNA species. Hybridizations for the detection of genomic and antigenomic HDV RNA were performed at 68°C using ULTRAhyb reagent (Ambion) and in vitro-transcribed 32P-labeled probes generated from pTMδSalB and pBSδHX, respectively. The membrane was washed at 75°C and exposed to Biomax MR or MS X-ray films (Kodak). Quantitation was performed by phosphorimagery using ImageQuant, version 1.11, software (Molecular Dynamics).
For analysis by reverse hybridization, probes were fixed to the membrane by applying 1 μl (1 μg) of heat-denatured, unlabeled RNA or DNA samples directly to strips of the BrightStar-Plus membrane and immobilizing them by baking at 80°C for 30 min in a vacuum oven. The probes used were as follows. For the detection of genomic and antigenomic HDV RNA, unlabeled in vitro-transcribed RNA was derived from pTMdSalB and pTMdSalA, respectively, using T7 RNA polymerase. For ChoA mRNA detection, a ChoA cDNA fragment was excised from plasmid pChoA (14) by digestion with restriction enzyme PstI. Hybridizations and stringent washes for the detection of HDV RNA were performed as described above. Both hybridization and washes for the detection of ChoA mRNA were performed at 42°C.
[32P]orthophosphate metabolic labeling.
Three to 4 days posttransfection, cultures were washed once with phosphate-free DMEM (Gibco BRL) and the medium was replaced with phosphate-free DMEM containing 10% dialyzed FBS. Following incubation at 37°C for 90 min, the medium was replaced with phosphate-free DMEM containing 10% dialyzed FBS and between 1 and 100 μg of actinomycin D (AMD; Fisher Biotech)/ml. After another 30-min incubation at 37°C, [32P]orthophosphate (3,000 Ci/mM; ICN) was added to a final concentration of 0.25 mCi/ml. Labeling was performed for 30 min to 4 h at 30 to 40°C depending on the individual experiment. RNA was then extracted from whole cells or nuclear and cytoplasmic fractions, treated with formaldehyde, and separated by electrophoresis on 1.2% MOPS-formaldehyde agarose gels. The gels were dried, and labeled RNA was visualized by autoradiography.
Transcription assays with lysolecithin-permeabilized cells.
All steps prior to 37°C incubation were performed at room temperature. Four days posttransfection, cultures of HuH7 cells grown in 60-mm-diameter petri dishes were washed sequentially in TBS (150 mM NaCl, 10 mM Tris-HCl [pH 7.4], 5 mM MgCl2) and glycerol buffer (20 mM Tris-HCl [pH 7.4], 5 mM MgCl2, 25% glycerol, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM EGTA). The cells were then permeabilized by treatment for 90 s with glycerol buffer containing 100 μg of lysolecithin (Sigma)/ml and 10 U of RNasin (Promega)/ml. Cultures were washed in glycerol buffer to remove lysolecithin, and 2 ml of synthesis buffer (100 mM KCl; 50 mM Tris-HCl [pH 7.4]; 10 mM MgCl2; 0.5 mM EGTA; 25% glycerol; 25 μM S-adenosyl-l-methionine [Sigma]; 20 U of RNAsin/ml; 0.5 mM [each] ATP, CTP, and GTP; 0 to 100 μg of α-amanitin [Molecular Probes]/ml; 100 μCi of 32P-labeled UTP [3,000 Ci/mM; ICN]/ml) was added. AMD (50 μg/ml) was also included in assays for the detection of HDV RNA but was excluded in those for detection of ChoA mRNA. Following incubation at 37°C for 30 min, total RNA was extracted and subjected to reverse hybridization analysis as described above.
RESULTS
Detection of HDV RNA synthesis by metabolic labeling.
We have recently developed a cDNA-free HDV RNA transfection system (30) which avoids the artificial requirement for DNA-dependent HDV RNA transcription associated with the expression plasmid transfection methods commonly used in most HDV studies. In this approach, we cotransfect an in vitro-transcribed HDV RNA of ca. 1.2 times the genome length and a capped mRNA for HDAg. This method not only reflects more closely the natural HDV replication cycle but also leads to robust HDV RNA replication at levels much higher than those possible with HDV cDNA transfection. We therefore attempted to perform metabolic labeling of HDV RNA with [32P]orthophosphate to study the mechanism of HDV RNA synthesis.
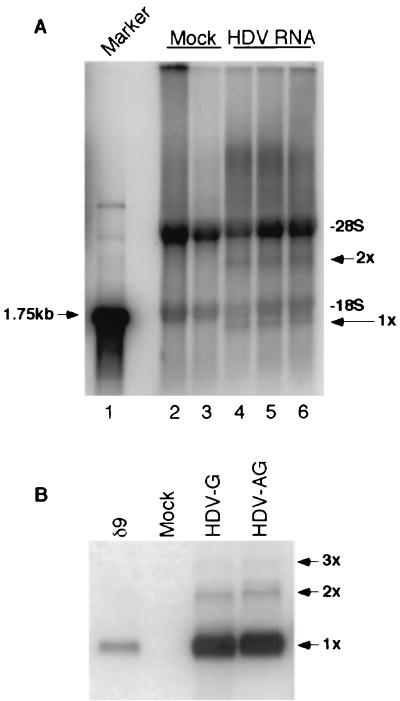
Replicate cultures of HuH7 cells were labeled for 4 h with [32P]orthophosphate in the presence of AMD 4 days after transfection with HDV RNA and HDAg mRNA. Total cellular RNA was then extracted and separated on 1.2% denaturing agarose gels (Fig. 1A). In all samples, two major bands representing the 28S and 18S rRNA species were detected. However, in HDV RNA-transfected cells, two additional bands were observed (Fig. 1A, compare lanes 2 and 3 to lanes 4 to 6). The lower band migrated below the 1.75-kb RNA size marker (Fig. 1A, lane 1) and likely represents monomer HDV RNA. Similarly, the upper band (3.4 kb) most likely represents dimeric HDV RNA (Fig. 1A, 2x). The detection of these two RNA species in HDV-transfected cells was reproducible in three different independent transfections (Fig. 1A, lanes 4 to 6). In addition to these two RNA species, there appeared to be some RNA species even longer than the 28S rRNA. The nature of these RNAs will be examined in more detail below. Thus, this experiment detected, for the first time, newly synthesized HDV RNA species by metabolic labeling.
FIG. 1.
Analysis of HDV RNA-transfected cells by metabolic labeling and Northern blotting. (A) 32P-labeled RNA from antigenomic HDV RNA-transfected HuH7 cells. RNA was separated by electrophoresis on a 1.2% denaturing agarose gel. Lane 1, 1.75-kb RNA marker; lanes 2 and 3, mock-transfected cells; lanes 4 to 6, HDV RNA-transfected cells. Each lane represents independently transfected cell cultures. 1x and 2x, positions of monomer and dimer HDV RNAs, respectively. The 28S and 18S rRNA species are also labeled. (B) Northern blot analysis. HuH7 cells transfected with either genomic (HDV-G) or antigenomic (HDV-AG) HDV RNA were blotted for the detection of genomic HDV RNA at 4 days after transfection. Lane δ9, genomic HDV RNA marker (25). 3x, position of trimer HDV RNA.
When RNA from similarly transfected cells was analyzed by Northern blotting, monomer and dimer HDV RNAs were again detected as well as some trimeric forms (Fig. 1B). Strikingly, by this approach, the monomer RNA was at least 30 times more abundant than the dimer RNA, whereas the two RNA species were detected in almost equimolar amounts by metabolic labeling (compare bands 1x and 2x between Fig. 1A and B). Similar results were obtained regardless of whether genomic (Fig. 1B, lane HDV-G) or antigenomic (Fig. 1B, lane HDV-AG) HDV RNA was used for transfection and whether genomic or antigenomic HDV RNA (data not shown) was detected by Northern blotting. This result suggested that the newly synthesized dimeric HDV RNA might be preferentially synthesized and processed into monomer forms, resulting in the accumulation of monomeric HDV RNA in the steady state.
HDV RNA synthesis was temperature sensitive and resistant to AMD.
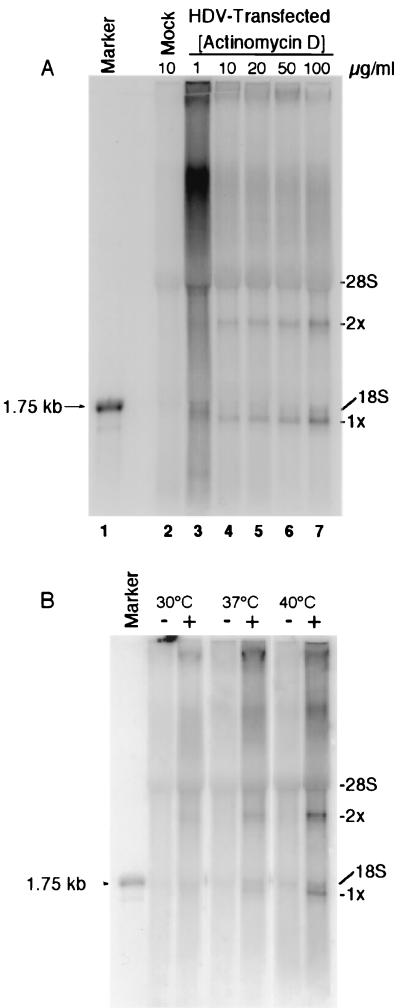
The metabolic labeling experiment shown in Fig. 1A was performed in the presence of 5 μg of AMD/ml, consistent with an earlier study indicating that HDV RNA synthesis is resistant to this inhibitor (26). To determine the extent of this resistance, we performed [32P]orthophosphate metabolic labeling in the presence of different concentrations of AMD on HuH7 cells 4 days after transfection with HDV RNA (Fig. 2A). Monomeric and dimeric HDV RNA species were detected in all HDV-transfected cultures at all concentrations of AMD (Fig. 2A). Moreover, the amounts of HDV-specific transcripts increased with the concentration of AMD (Fig. 2A, compare tracks 3 to 7), suggesting that higher concentrations of AMD liberated more of the cellular transcription machinery for HDV replication. A concentration of 50 μg of AMD/ml was used in all subsequent metabolic labeling experiments.
FIG. 2.
HDV RNA synthesis is resistant to AMD but sensitive to low temperature. Shown is 32P-labeled RNA from HDV-transfected HuH7 cells metabolically labeled in the presence of different concentrations of AMD (A) and at different temperatures (B). − and + (B), mock- and HDV RNA-transfected cells, respectively. 28S and 18S, major cellular rRNA species. 1x and 2x are as defined for Fig. 1.
We also examined the effect of different temperatures on HDV RNA synthesis and found that the level of HDV RNA synthesis at 40°C was approximately eight- and threefold higher than those at 30 and 37°C, respectively (Fig. 2B). In contrast, the levels of background rRNA synthesis were similar at all temperatures. Thus, HDV RNA synthesis appears to be more efficient at high temperatures, a finding consistent with previous studies of the effect of temperature on steady-state levels of HDV RNA (17). This enhanced HDV RNA synthesis at high temperatures may reflect an increase in stability of the HDV RNA rod structure at low temperatures, which, in turn, may impede strand separation and render the HDV RNA a less efficient template for transcription. Similarly, HDV ribozyme activity, which is also assumed to require local destabilization of the HDV rod structure, is likely to be inhibited at low temperatures. Alternatively, a cellular factor required for HDV RNA synthesis may be nonfunctional or not synthesized at low temperatures.
HDV RNA is gradually processed from higher-molecular-weight forms to monomer species.
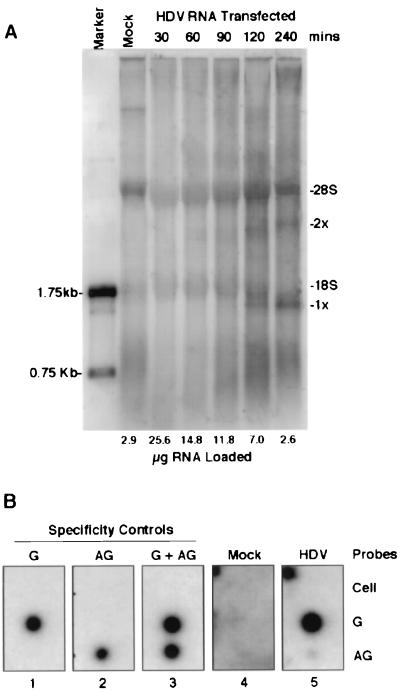
The finding that the ratio of metabolically labeled dimeric to monomeric HDV RNA species was much higher than the steady-state ratio of dimeric to monomeric HDV RNA detected by Northern blotting (compare Fig. 1A and 2A and B to 1B) suggests that at least some of dimeric HDV RNA is a precursor to the monomer. To examine this possibility, HDV RNA-transfected HuH7 cells were labeled with [32P]orthophosphate for various periods of time (Fig. 3A). The same quantity of 32P was loaded into each lane; this resulted in a requirement for about 10 times more RNA from cultures labeled for 30 min than from cells labeled for 240 min. After 30 min of labeling, no HDV RNA species could be identified. However, after 60 min, a slightly diffuse band of dimeric HDV RNA could be detected; this band gradually became more intense and sharper as the labeling period increased. The monomeric HDV RNA was not observed until 90 to 120 min of labeling. The band of this RNA species was always relatively sharp and also became more intense as the labeling period increased. By 240 min, there was more monomer than dimer HDV RNA. The sequential appearance of the dimeric and monomeric HDV RNA species suggests that the monomer RNA was likely derived from the processing of dimeric or larger RNA species. Significantly, some faint and diffuse RNA species larger than the 28S rRNA were also detected, which could represent the precursor to dimeric HDV RNA. These RNA species were much more discrete when nuclear fractions were examined (see below). We attempted to perform pulse-chase labeling experiments; however, these experiments failed, as labeled nucleotides continued to be incorporated into HDV RNA during the chase period. This was likely the result of the large phosphate pool in the cells. We also have been unable to detect the HDAg mRNA species (0.8 kb) by metabolic labeling, reflecting the known low abundance of this transcript in HDV-transfected cells. Nevertheless, the kinetics of the appearance of the dimer and monomer RNAs suggests strongly that the dimer and/or multimer RNAs are precursors to the monomer RNA.
FIG. 3.
Monomeric HDV RNA is derived from the processing of higher-molecular-weight species. (A) 32P-labeled RNA from antigenomic HDV RNA-transfected cells, metabolically labeled for 30 to 240 min. The duration of labeling and amount of RNA loaded in each lane are indicated at the top and bottom of each track, respectively. The mock-transfected HuH7 cells were labeled for 240 min. 1x and 2x, monomer and dimer HDV RNA, respectively. (B) 32P-labeled RNA from mock- (panel 4) and HDV RNA-transfected (panel 5) HuH7 cells was reverse hybridized with membranes containing unlabeled RNA probes specific for genomic (G) and antigenomic (AG) HDV RNA. Cell, probe consisting of total cellular RNA extracted from untransfected cells. The specificity controls shown in panels 1 to 3 are reverse hybridizations with unlabeled HuH7 cell RNA spiked with [32P]orthophosphate-labeled (20,000 cpm) full-length genomic or antigenomic HDV RNA or a 50:50 mixture of both (G + AG) to the membranes as described above.
To determine the polarity of the HDV RNA transcripts synthesized, metabolically labeled RNA extracted from HDV RNA-transfected HuH7 cells was subjected to reverse hybridization to HDV RNA-specific and control probes (Fig. 3B). The three probes used were total HuH7 cell RNA (to assess the cross-reactivity of the various labeled RNA species) and genomic and antigenomic HDV RNA. As a control for specificity of the hybridization conditions, we first performed a reverse hybridization experiment using unlabeled total HuH7 RNA spiked with 32P-labeled, in vitro-transcribed genomic or antigenomic HDV RNA or a combination of both (Fig. 3B, panels 1 to 3). The 32P-labeled HDV RNAs only hybridized with the correct HDV probes and showed no cross hybridization with the cellular RNA. As expected, metabolically labeled RNA from mock-transfected cells did not hybridize to any probe (Fig. 3B, panel 4). In contrast, the sample from the HDV RNA-transfected cells annealed strongly with the probe for detecting genomic HDV RNA and hybridized only weakly (ca. 30-fold-lower efficiency) with the probe for detecting antigenomic HDV RNA (Fig. 3B, panel 5). This finding indicates that the majority of HDV RNA synthesis was of genomic sense. Since Northern blotting of RNA from HDV-infected cells also shows 10- to 20-fold more genomic than antigenomic HDV RNA (4), the newly synthesized genomic and antigenomic HDV RNAs identified by metabolic labeling are likely equally stable.
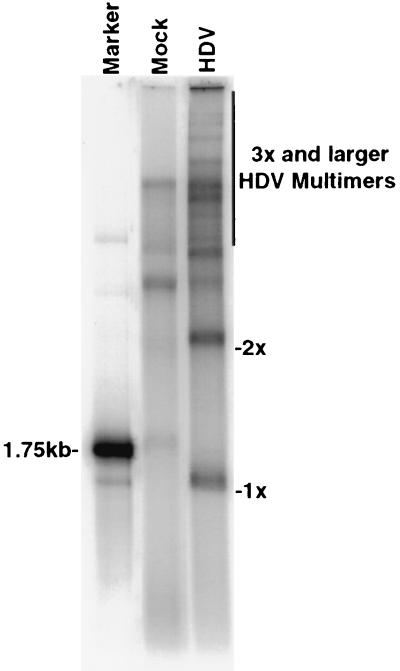
To characterize the higher-molecular-weight HDV RNA species in more detail, we examined the nuclear fraction of metabolically labeled, HDV RNA-transfected cells, as HDV RNA was thought to accumulate in the nucleus (12) and thus the relative proportion of HDV RNA to cellular RNA would be higher in the nucleus than in the whole cell. Indeed, a large number of slowly migrating bands consistent in size with trimeric and longer multimeric species of HDV RNA (up to 10 times the genome length) were detected in the nuclear fraction following 240 min of labeling (Fig. 4). Since the majority of these species are not detected by Northern blotting (Fig. 1B), it seems likely that they represent precursors to the lower-molecular-weight species. Surprisingly, a substantial amount of monomeric and dimeric HDV RNA species were also detected in the cytoplasmic fractions (data not shown). This observation has been examined in more detail in the accompanying study (28).
FIG. 4.
Analysis of metabolically labeled HDV RNA in nuclear fractions. 32P-labeled nuclear RNA from genomic HDV RNA-transfected or mock-transfected HuH7 cells labeled for 240 min was separated on a 1.5% denaturing agarose gel. 1x and 2x are as defined for Fig. 1.
Genomic and antigenomic HDV RNA syntheses are likely carried out by different host cell RNA polymerases.
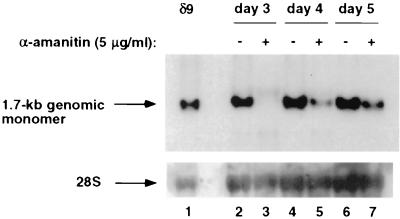
We previously demonstrated that synthesis of the 1.7-kb antigenomic HDV RNA species was resistant to inhibition by α-amanitin while the synthesis of the 0.8-kb HDAg mRNA was sensitive to a low concentration of α-amanitin, suggesting that the synthesis of these two RNA species is carried out by two different polymerases (33). We have also shown that genomic and antigenomic HDV RNA syntheses are differentially sensitive to inhibition by L-HDAg (32), again suggesting that genomic and antigenomic RNA syntheses are carried out by two different mechanisms. Therefore, we explored the possibility that replication of full-length genomic and antigenomic HDV RNA may exhibit different sensitivities to inhibition by α-amanitin. We first tested the sensitivity of genomic HDV RNA synthesis to α-amanitin (Fig. 5) by using the same steady-state conditions previously described for antigenomic HDV RNA synthesis (33). HuH7 cells transfected with antigenomic HDV RNA were treated with 5 μg of α-amanitin/ml at various days posttransfection, and genomic HDV RNA was detected by Northern blotting 2 days later. Such a long-term treatment was necessary, because we found that the uptake of α-amanitin by intact cells was very slow. Under such treatment conditions, cellular DNA-dependent transcription by pol III was not affected (33), indicating that α-amanitin did not cause a general shutdown of RNA transcription in cells. When α-amanitin was added at day 1 posttransfection, synthesis of genomic RNA was completely inhibited and was undetectable at day 3 (Fig. 5, lanes 3 and 4). When α-amanitin was added at day 2 posttransfection, synthesis of genomic RNA was detected at day 4 but the level was much lower than that in untreated cells (Fig. 5, lanes 5 and 6). When α-amanitin was added at day 3 posttransfection, again only a small amount of genomic RNA was detected at day 5 relative to the amount in untreated cells (Fig. 5, lanes 6 and 7). The amount of RNA in α-amanitin-treated cells at day 5 was less than that in untreated cells harvested at day 3, indicating that little or no new HDV RNA was synthesized in the presence of α-amanitin and that the genomic RNA detected at day 5 was likely the remains of genomic RNA present at the time of α-amanitin treatment. This is in contrast to the effect of α-amanitin on antigenome synthesis, where antigenomic HDV RNA continued to increase in the presence of α-amanitin when it was added at day 3 (33). Thus, unlike what was found for antigenomic HDV RNA, where synthesis was shown to be sensitive to α-amanitin only at the beginning of the replication cycle (due to its dependence on the synthesis of HDAg mRNA) (33), synthesis of genomic RNA was sensitive to α-amanitin throughout the replication cycle. These results strongly suggest that synthesis of antigenomic HDV RNA and synthesis of genomic RNA have different sensitivities to inhibition by α-amanitin. However, these observations were made following long-term treatment of transfected cells with α-amanitin. Thus, we could not exclude the possibility that these findings may have been the result of an unusual cytotoxicity reaction caused by α-amanitin.
FIG. 5.
Sensitivity of genomic HDV RNA synthesis to α-amanitin analyzed with steady-state RNA. HuH7 cells were transfected with 1.9-kb antigenomic HDV RNA and mRNA encoding S-HDAg and treated with α-amanitin (5 μg/ml) at various time points posttransfection. Total RNA was extracted 2 days after treatment and subjected to Northern blot analysis using a 32P-labeled antigenomic-sense riboprobe. Lane 1, positive control marking the position of the 1.7-kb genome; lanes 3 to 7, total RNA harvested from treated and untreated transfected cells at the indicated time points. Cells were treated for 48 h prior to harvest. For example, in lane 3, cells were treated with α-amanitin at day 1 and harvested at day 3 posttransfection.
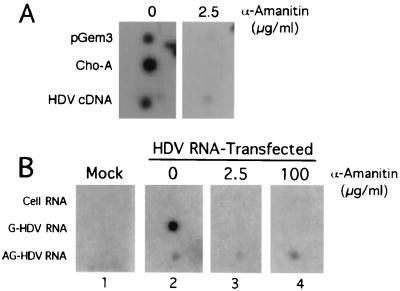
To eliminate this possibility, we performed a modified nuclear run-on experiment, in which HuH7 cells transfected with antigenomic HDV RNA were permeabilized with lysolecithin and incubated in a transcription mixture that included 32P-labeled UTP and various concentrations of α-amanitin for 40 min at 37°C. Total RNA was extracted and subjected to reverse hybridization analysis using strand-specific RNA as probes (Fig. 6). In the absence of α-amanitin, both genomic and antigenomic HDV RNAs were detected (Fig. 6B, panel 2). Moreover, the signal for genomic HDV RNA was approximately 12-fold greater than that for antigenomic HDV RNA, consistent with the ratio of these species normally detected in intact HDV-transfected cells. The signal for genomic HDV RNA was completely eliminated in the presence of low concentrations of α-amanitin (2.5 μg/ml), whereas the signal for antigenomic HDV was unaffected by this or much higher concentrations (100 μg/ml) of the inhibitor (Fig. 6B, panels 3 and 4). Transcription of the pol II-dependent ChoA mRNA (as detected by hybridization to ChoA cDNA) was also eliminated in the presence of 2.5 μg of α-amanitin/ml (Fig. 6A) under the same conditions, confirming the accuracy of our assay (cDNA probes used in Fig. 6A yielded some nonspecific hybridization, which was eliminated by α-amanitin). These data strongly suggest that pol II is the cellular polymerase responsible for genomic HDV RNA synthesis. However, the α-amanitin resistance of antigenomic HDV RNA synthesis (even at concentrations expected to inhibit pol III) implies that pol I is likely involved in antigenomic HDV RNA synthesis. The above observations, together with our previous study showing that synthesis of HDAg mRNA is performed by pol II (33), indicate that at least two different cellular RNA polymerases are likely to be responsible for HDV RNA replication.
FIG. 6.
Genomic and antigenomic HDV RNA synthesis is performed by two different host cell RNA polymerases. (A) Reverse hybridization analysis of runoff transcripts from untransfected HuH7 cells permeabilized with lysolecithin and incubated for 30 min at 37°C in the presence of [32P]UTP and either 0 or 2.5 μg of α-amanitin/ml. Probes, pGem3 (plasmid) DNA and ChoA and HDV cDNA, were fixed on the membrane. Hybridization and washes were performed at 42°C. (B) Reverse hybridization analysis of runoff transcripts from mock- (panel 1) and HDV RNA-transfected Huh7 cells (panels 2 to 4) labeled in the presence of 50 μg of AMD/ml. Cells were permeabilized and incubated with [32P]UTP and either 0 (panels 1 and 2), 2.5 (panel 3), or 100 (panel 4) μg of α-amanitin/ml as for panel A. Reverse hybridization probes (RNA) were as described in the legend of Fig. 3B and were fixed on the membrane. Hybridization and washes were performed at 68 and 75°C, respectively. G, genomic; AG, antigenomic.
DISCUSSION
In this study, we have successfully combined the strength of the HDV cDNA-free transfection system with [32P]orthophosphate metabolic labeling to provide a glimpse of the mechanism of rolling circle replication for a viroid-like RNA. We have obtained evidence that dimeric HDV RNA is likely the replication intermediate in the synthesis of the monomer HDV RNA species and not a dead-end product. Two lines of evidence support this conclusion. First, dimeric HDV RNA appeared before the monomer (Fig. 3A). Second, the ratio of dimeric to monomeric HDV RNA was much higher after metabolic labeling (which identifies HDV RNA synthesized over a short period of time) than that observed by Northern blot hybridization (which shows the accumulated HDV RNA species over a number of days). In addition, we observed multimer RNAs of up to 10 genome lengths, which were not detected by Northern blotting. Although we failed in the true pulse-chase labeling experiments, the accumulated data from this study strongly suggest that the dimer and, very likely, multimer RNA species are precursors to monomer RNAs. It is interesting that, during metabolic labeling, the dimer and multimer RNAs were often heterogeneous in size; they may represent newly synthesized, but unprocessed, RNA molecules. To our knowledge, this is the first successful detection of the nascently synthesized replication intermediates of viroid-like RNAs, thus further supporting the validity of the rolling circle replication model (1). Our previous finding that ribozyme activity is necessary for HDV RNA replication in vivo (19, 27) suggests that these multimer intermediates are cleaved by the ribozyme. However, dimer RNA was found to be more abundant than monomer RNA during short-term labeling, suggesting that the processing is relatively slow compared to HDV RNA replication. Thus, ribozyme cleavage occurs most likely on the dimer and multimer RNA molecules, but not, as previously suggested, on the nascent RNA molecule immediately after its ribozyme domain is synthesized (22, 23). This conclusion raises the following question: how is the ribozyme conformation formed on these multimeric RNA molecules? Since HDAg has been shown to have an RNA chaperone activity (16) and can enhance ribozyme activity in vitro (16) and in vivo (20), HDAg seems a likely facilitator. Another point of interest is that, despite the presence of proportionally more templates for antigenomic HDV RNA production than for genomic RNA synthesis, the latter outpaces the former throughout the viral replication cycle. The molecular basis for the different rates of HDV RNA synthesis is not clear.
Our study demonstrated that genomic and antigenomic HDV RNAs are likely replicated by two different cellular polymerases. This conclusion was first suggested from Northern blot analysis of HDV RNA made in the presence of α-amanitin (33) (Fig. 5). The use of lysolecithin-permeabilized, HDV RNA-transfected cells finally allowed us to study HDV RNA synthesis at an optimal temperature in an almost in vivo condition. Genomic HDV RNA synthesis is most likely carried out by pol II, based on its sensitivity to low concentrations of α-amanitin. In contrast, the polymerase that replicates antigenomic HDV RNA demonstrated a level of resistance to α-amanitin that is normally associated with pol I. These conclusions are in complete agreement with present and previous findings (33) made by long-term treatment of HDV-replicating cells with α-amanitin. The combined results further establish a clear distinction between genomic and antigenomic HDV RNA syntheses in their dependence on different cellular RNA polymerases. The possible synthesis of antigenomic HDV RNA by pol I is consistent with the findings that HDAg interacts with nucleolar proteins nucleolin (24) and B23 (15). However, the possibilities that a novel RNA-dependent RNA polymerase is involved and that synthesis of antigenomic HDV RNA somehow induces pol II to form a structure that is resistant to α-amanitin cannot be rigorously ruled out. In any case, the conclusion that genomic and antigenomic HDV RNAs are synthesized by different polymerases and probably in different subcellular compartments may explain why these two HDV RNA species are synthesized at different rates.
While this paper was undergoing editorial review, another paper reporting the metabolic requirements of HDV RNA synthesis based on some of the methods used in our study appeared (34). It claimed that HDV RNA replication involved only pol II, not pol I or pol III. However, that study (34) did not actually examine antigenomic RNA synthesis, which we found to be resistant to α-amanitin and which is, therefore, likely mediated by pol I. Furthermore, the runoff transcription reported in that study did not reflect the natural metabolic requirements of intact cells, as several inhibitors of cellular transcription, such as DRB and tagetitoxin, were reported to have no effects in that system (34). Therefore, although the major conclusion of that study agrees with our finding that HDV genomic RNA synthesis is sensitive to α-amanitin and is likely mediated by pol II, it neglected to examine antigenomic RNA synthesis, which we found to have a different metabolic requirement.
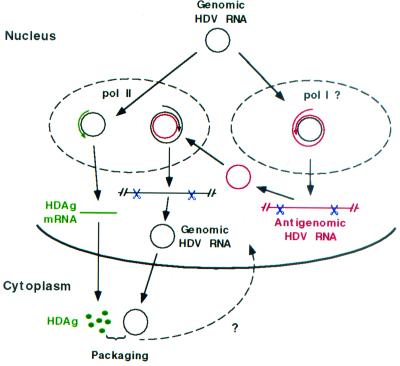
When the findings of this study are combined with our recent findings that pol II is responsible for HDAg mRNA production (33) and that genomic HDV RNA is preferentially exported from the nucleus after synthesis (28), a novel picture of HDV RNA synthesis begins to emerge (Fig. 7). Genomic HDV RNA is used as a template by pol I (or an unknown RNA polymerase) for full-length antigenomic HDV RNA synthesis and by pol II for HDAg mRNA synthesis. The pol II transcription machinery recognizes the poly(A) addition signal on the RNA transcript to generate the HDAg mRNA, whereas this signal will be ignored by pol I, thus allowing rolling circle replication. Therefore, transcription of HDAg mRNA and synthesis of antigenomic RNA from the genomic RNA template are likely carried out in different compartments in the nucleus. On the other hand, antigenomic RNA is used as a template by pol II only, which, because the antigenomic RNA lacks a poly(A) termination signal (5), should allow continuous synthesis of multimeric genomic HDV RNA. This process may be further facilitated by a recently described property of HDAg of promoting the elongation of pol II transcripts (41). Because both HDAg mRNA (antigenomic) and genomic RNA are synthesized by the pol II transcriptional machinery, they are linked to the RNA export mechanisms for cellular RNAs. Therefore, template genomic HDV RNA is likely to be localized in both the nucleoplasm and nucleolus (or another compartment), whereas template antigenomic HDV RNA may stay mainly in the nucleoplasm with the pol II transcription machinery. Such a separation of template and polymerases allows the mRNA and the full-length antigenomic HDV RNA to be differentially synthesized from the same genomic template and explains the dissimilar rates of synthesis and the different fates of export of different HDV RNA species.
FIG. 7.
Proposed model for HDV rolling circle replication incorporating nuclear export (28) and the use of two different cellular RNA polymerases. See text for details.
The ability of cellular polymerases to carry out RNA-dependent RNA synthesis of HDV RNAs reveals an unusual property of mammalian cells. The identification of these enzymes and the mechanisms of RNA synthesis will expand the horizon of understanding of gene expression and amplification.
Acknowledgments
Thanks to John Taylor (Fox Chase Cancer Center, Philadelphia) for provision of plasmid pSVLD3.
M.M.C.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNA's. Science 223:450-455. [DOI] [PubMed] [Google Scholar]
- 2.Buzayan, J. M., W. L. Gerlach, and G. Bruening. 1986. Nonenzymic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature 323:349-353. [Google Scholar]
- 3.Chang, F.-L., P.-J. Chen, S.-J. Tu, C.-J. Wang., and D.-S. Chen. 1991. The large form of hepatitis delta antigen is crucial for the assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, P.-J., G. Kalpana, J. Goldberg, W. Mason, B. Werner, J. Gerin, and J. Taylor. 1986. Structure and replication of the genome of the hepatitis delta virus. Proc. Natl. Acad. Sci. USA 83:8774-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dye, M. J., and N. J. Proudfoot. 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105:669-681. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschel. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 7.Ferre-D'Amare, A. R., K. Zhou, and J. A. Doudna. 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395:567-574. [DOI] [PubMed] [Google Scholar]
- 8.Filipovska, J., and M. M. Konarska. 2000. Specific HDV RNA-templated transcription by pol II in vitro. RNA 6:41-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forster, F. C., and R. H. Symons. 1987. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell 49:211-220. [DOI] [PubMed] [Google Scholar]
- 10.Fu, T. B., and J. Taylor. 1993. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J. Virol. 67:6965-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glenn, J. S., and J. M. White. 1991. trans-Dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 65:2357-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gowans, E. J., B. M. Baroudy, F. Negro, A. Ponzetto, R. H. Purcell, and J. L. Gerin. 1988. Evidence for replication of hepatitis delta virus RNA in hepatocyte nuclei after in vivo infection. Virology 167:274-278. [DOI] [PubMed] [Google Scholar]
- 13.Hampel, A., and R. Tritz. 1989. RNA catalytic properties of the minimum (-)sTRSV sequence. Biochemistry 28:4929-4933. [DOI] [PubMed] [Google Scholar]
- 14.Harpold, M. M., R. M. Evans, M. Salditt-Georgieff, and J. E. Darnell. 1979. Production of mRNA in Chinese hamster cells: relationship of the rate of synthesis to the cytoplasmic concentration of nine specific mRNA sequences. Cell 17:1025-1035. [DOI] [PubMed] [Google Scholar]
- 15.Huang, W. H., B. Y. Yung, W. J. Syu, and Y. H. Lee. 2001. The nucleolar phosphoprotein b23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J. Biol. Chem. 276:25166-25175. [DOI] [PubMed] [Google Scholar]
- 16.Huang, Z. S., and H. N. Wu. 1998. Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides. J. Biol. Chem. 273:26455-26461. [DOI] [PubMed] [Google Scholar]
- 17.Hwang, S. B., K.-S. Jeng, and M. M. C. Lai. 1995. Studies of functional roles of hepatitis delta antigen in delta virus RNA replication, p. 95-109. In G. Dinter-Gottlieb (ed.), The unique hepatitis delta virus. R. G. Landes Company, Austin, Tex.
- 18.Ishikawa, M., T. Meshi, T. Ohno, Y. Okada, T. Sano, I. Ueda, and E. Shikata. 1984. A revised replication cycle for viroids: the role of longer than unit length RNA in viroid replication. Mol. Gen. Genet. 196:421-428. [DOI] [PubMed] [Google Scholar]
- 19.Jeng, K.-S., A. Daniel, and M. M. C. Lai. 1996. A pseudoknot ribozyme structure is active in vivo and required for hepatitis delta virus RNA replication. J. Virol. 70:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeng, K.-S., P.-Y. Su, and M. M. C. Lai. 1996. Hepatitis delta antigens enhance the ribozyme activities of hepatitis delta virus RNA in vivo. J. Virol. 70:4205-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo, M. Y.-P., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai, M. M. C. 1995. The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 64:259-286. [DOI] [PubMed] [Google Scholar]
- 23.Lazinski, D. W., and J. M. Taylor. 1995. Regulation of the hepatitis delta virus ribozymes: to cleave or not to cleave? RNA 3:225-233. [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, C. H., S. C. Chang, C. J. Chen, and M.-F. Chang. 1998. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 273:7650-7656. [DOI] [PubMed] [Google Scholar]
- 25.Macnaughton, T. B., E. J. Gowans, A. R. Jilbert, and C. J. Burrell. 1990. Hepatitis delta virus RNA, protein synthesis and associated cytoxicity in a stably transfected cell line. Virology 177:692-698. [DOI] [PubMed] [Google Scholar]
- 26.Macnaughton, T. B., E. J. Gowans, S. P. McNamara, and C. J. Burrell. 1991. Hepatitis δ antigen is necessary for access of hepatitis virus RNA to the cell transcriptional machinery but is not part of the transcriptional complex. Virology 184:387-390. [DOI] [PubMed] [Google Scholar]
- 27.Macnaughton, T. B., Y.-J. Wang, and M. M. C. Lai. 1993. Replication of hepatitis delta virus RNA: effect of mutations of the autocatalytic cleavage sites. J. Virol. 67:2228-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macnaughton, T. B., and M. M. C. Lai. 2002. Genomic but not antigenomic hepatitis delta virus RNA is preferentially exported from the nucleus immediately after synthesis and processing. J. Virol. 76:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makino, S., F. Taguchi, N. Hirano, and K. Fujiwara. 1984. Analysis of genomic and intracellular viral RNAs of small plaque mutants of mouse hepatitis virus, JHM strain. Virology 139:138-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modahl, L. E., and M. M. C. Lai. 1998. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J. Virol. 72:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modahl, L. E., and M. M. C. Lai. 2000. Hepatitis delta virus: the molecular basis of laboratory diagnosis. Crit. Rev. Clin. Lab. Sci. 37:45-92. [DOI] [PubMed] [Google Scholar]
- 32.Modahl, L. E., and M. C. Lai. 2000. The large delta antigen of hepatitis delta virus potently inhibits genomic but not antigenomic RNA synthesis: a mechanism enabling initiation of viral replication. J. Virol. 74:7375-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modahl, L. E., T. B. Macnaughton, N. Zhu, D. L. Johnson, and M. M. C. Lai. 2000. RNA-dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol. Cell. Biol. 20:6030-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moraleda, G., and J. Taylor. 2001. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J. Virol. 75:10161-10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cell lines with differentiated function in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 36.Prody, G. A., J. T. Bakos, J. M. Buzayan, I. R. Schneider, and G. Bruening. 1986. Autolytic processing of dimeric plant virus satellite RNA. Science 231:1577-1580. [DOI] [PubMed]
- 37.Reid, C. E., and D. W. Lazinski. 2000. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. USA 97:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiebel, W., T. Pelissier. L. Riedel, S. Thalmeir, R. Schiebel, D. Kempe, F. Lottspeich, H. L. Sanger, and M. Wassenegger. 1998. Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 10:2087-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volloch, V., B. Schweitzer, and S. Rits. 1996. Antisense globin RNA in mouse erythroid tissues: structure, origin, and possible function. Proc. Natl. Acad. Sci. USA 93:2476-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, K.-S., Q.-L. Choo, A. J. Weiner, J.-H. Ou, R. C. Najerian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis δ viral genome. Nature 323:508-514. (Erratum, 328:456, 1987.) [DOI] [PubMed]
- 41.Yamaguchi, Y., J. Filipovska, K. Yano, A. Furuya, N. Inukai, T. Narita, T. Wada, S. Sugimoto, M. M. Konarska, and H. Handa. 2001. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 293:124-127. [DOI] [PubMed] [Google Scholar]