Abstract
The regulatory proteins of human immunodeficiency virus may represent important vaccine targets. Here we assessed the role of Tat-specific cytotoxic T lymphocytes (CTL) in controlling pathogenic simian immunodeficiency virus SIVmac239 replication after using a DNA-prime, vaccinia virus Ankara-boost vaccine regimen. Despite the induction of Tat-specific CTL, there was no significant reduction in either peak or viral set point compared to that of controls.
Recent reports have suggested that immune responses directed against the smaller regulatory proteins might be able to control human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) replication (7, 8, 10, 17, 19, 21, 22). We have recently shown that Mamu-A*01-positive rhesus macaques mount an immunodominant cytotoxic T-lymphocyte (CTL) response to an epitope in Tat (Tat28-35SL8) (1). This CTL response exerts significant selective pressure by eliminating wild-type virus replication by 4 weeks postinfection (p.i.) in the majority of Mamu-A*01-positive macaques. Additionally, the HIV Tat protein is highly variable, suggesting that CTL exert selective pressure on this region of the virus (12, 13). We therefore assessed whether vaccine-induced Tat-specific CTL might prove effective in controlling SIVmac239 replication.
Induction of Tat28-35SL8-specific CTL by DNA/MVA.
We employed a DNA-prime, attenuated vaccinia virus Ankara (MVA)-boost vaccination regimen to induce Tat-specific CTL in three Mamu-A*01-positive (animal 96016, 1975, and 97085) and one Mamu-A*01-negative (animal 97007) rhesus macaques. Four Mamu-A*01-positive macaques (animal 96111, 95086, 93057, and 85013) served as controls. Vaccinees were immunized three times with DNA at 6-week intervals as previously described (2). Two DNA vectors were employed. One vector encoded full-length SIVmac239 Tat, and the second vector encoded a single Mamu-A*01-restricted CTL epitope Tat28-35SL8 (STPESANL) inserted within the immunodominant region of hepatitis B core antigen. A gene gun was used to deliver gold particles coated with plasmid DNA into the epidermis. Each immunization consisted of a total of 32 μg of DNA administered at eight skin sites over the abdominal and inguinal lymph nodes, as previously described (2)
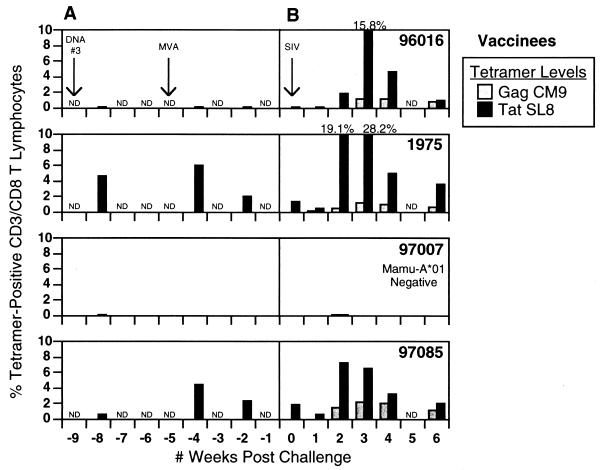
Following the final DNA immunization, tetramer analysis detected Tat28-35SL8-specific CTL in fresh peripheral blood mononuclear cells (PBMC) of two of the three Mamu-A*01-positive vaccinees (4.7 and 0.64% of all CD3/CD8 T lymphocytes) (Fig. 1A). Background tetramer staining in control animals was less than 0.08%. Surprisingly, in one of these animals (macaque 1975), the Tat28-35SL8 response reached levels in excess of 4%, a magnitude of CTL response not previously observed in rhesus macaques following DNA vaccination alone (2-5, 11).
FIG. 1.
Tat28-35SL8 and Gag181-189CM9 tetramer levels in vaccinees. PBMC were stained for CD3, CD8, and both the Mamu-A*01-restricted Gag181-189CM9 and Tat28-35SL8 tetramers.
One month after receiving the last DNA immunization, rhesus macaques were then boosted intradermally with 5 × 108 PFU of MVA encoding full-length Tat, based on the tat gene sequences derived from SIVmac251 32H (molecular clone pJ5), as previously described (2). Animals also received the same dose of MVA-Tat intrarectally (i.r.), delivered atraumatically by using a needle-free 1-ml syringe, following dilution of the MVA in 500 μl of 1× phosphate-buffered saline. One week after being boosted with MVA-Tat, levels of Tat28-35SL8-specific CTL in all three Mamu-A*01-positive macaques were elevated to 0.14, 6.0, and 4.5% (of all CD8+/CD3+ T lymphocytes) in animals 96016, 1975, and 97085, respectively. These levels of CD8+ T cells were confirmed by enzyme-linked immunospot assay (ELISPOT), which yielded values of 430, >2,000, and >2,000 spot-forming cells (SFC)/106 PBMC, respectively (data not shown). While tetramers could not be used to assess Tat-specific CTL in the one Mamu-A*01-negative vaccinee (macaque 97007), responses were detected by ELISPOT to two other regions of Tat (amino acids 37 to 59 and 61 to 83) (data not shown). Responses to these two regions, which reached levels of 1,000 and 2,100 SFC/106 PBMC, respectively, were comparable in magnitude to the Tat28-35SL8 responses observed in the Mamu-A*01-positive vaccinees.
Vaccine-induced proliferative responses.
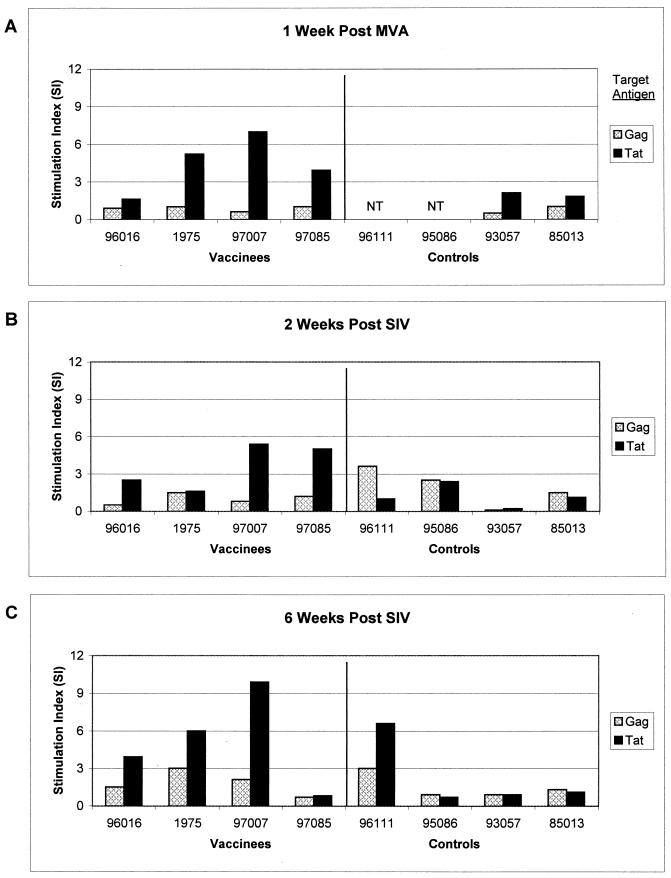
CD4 T-cell proliferative responses against Tat were also measured using recombinant Tat protein. PBMC taken 1 week following administration of the Tat MVA were depleted of CD8-positive T lymphocytes by anti-CD8 antibody conjugated to magnetic beads (Dynal, Lake Success, N.Y.). CD8-depleted PBMC (100,000/well) were then plated in quadruplicate in 150 μl of AB+ medium into 96-well round-bottomed plates. Antigens were added in 50 μl of AB+ medium at 1 μg/well. Plates were incubated for 4 days before adding 1 μCi of tritiated thymidine per well. After a further 16 to 18 h of incubation, cells were harvested onto glass fiber mats and counted via scintillation. Tat-specific proliferative responses were detected in three of four vaccinees (Fig. 2A; stimulation index [SI] of >3.0 considered significant).
FIG. 2.
CD4 proliferative responses after MVA vaccination and SIV infection. PBMC from both vaccinated and control macaques were tested for proliferative responses to Gag and Tat recombinant proteins, as well as to concanavalin A (positive control; data not shown). Animals were tested 1 week after MVA inoculation (A), 2 weeks after SIV infection (B), and 6 weeks after SIV infection (C). Responses are reported as SI, with SIs greater than 3.0 considered significant.
Challenge of Tat-vaccinated macaques with SIVmac239.
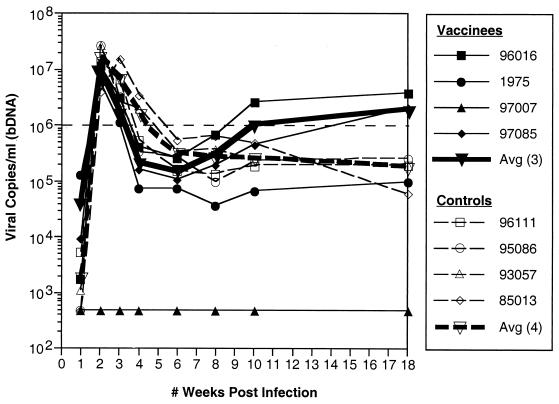
The eight vaccinated and control macaques were then challenged i.r. with 3.16 × 103 50% tissue culture infectious doses of SIVmac239/nef-open (14) 6 weeks after receiving the MVA booster immunization. Mamu-A*01-positive animals were selected as controls, since Mamu-A*01-positive animals have demonstrated a lower viral set point following mucosal challenges with SIVmac251 (18). Viral loads were quantitated with the SIVmac branched DNA (bDNA) RNA assay by the Bayer Reference Testing Laboratory (Emeryville, Calif.). Since we could find no evidence of viral replication in the Mamu-A*01-negative vaccinee (macaque 97007), it was omitted from further analysis. It is unlikely that this represents an example of sterilizing immunity against SIVmac239 challenge because occasional technical challenge failures have been observed after an i.r. challenge (although this is the first such failure in our hands using this stock of virus). Comparison of viral loads in the three remaining vaccinees to those of the controls revealed a slight, but not statistically significant, reduction in peak viremia (P = 0.954; t-test after log-transforming data to improve normality and homoscedasticity) and a slightly more pronounced diminution in viral loads at week 4 in the vaccinees. However, by week 10, viral loads in the vaccinees and controls were indistinguishable (P = 0.954; t-test) (Fig. 3). This suggested that there might have been some short-term control of infection in the early weeks following challenge. Interestingly, however, there was a 1.5-log difference in viral loads of vaccinees verses controls at 1 week p.i., with the three vaccinees demonstrating higher viral loads at this early point in the infection.
FIG. 3.
Viral loads of vaccinated and control macaques. Viral loads were quantitated from plasma by the Bayer Reference Testing Laboratory's SIVmac bDNA RNA Assay. Viral loads are indicated for each animal in the study, with the average viral loads of the vaccinees and controls plotted separately.
Following challenge with SIVmac239, there was a massive Tat28-35SL8-specific CTL recall response. In vaccinees, peak levels of 15.8, 28.8, and 7.3% were detected between 2 and 3 weeks p.i. (Fig. 1B). In comparison, in the control animals peak CTL levels were not achieved until 3 to 4 weeks p.i. Furthermore, these CTL responses in the controls (data not shown) were lower (between 2.0 and 8.5%) than those seen in the vaccinees (between 7.3 and 28.8%). Gag181-189CM9-specific acute-phase Mamu-A*01-restricted CTL responses were also measured. With the exception of control animal 85013, which exhibited levels of Gag181-189CM9-specific CTL of >6.0%, there was no difference in the peak levels of the Gag181-189CM9-specific CTL between vaccinees and controls. Interestingly, however, Gag181-189CM9-specific CTL appeared to peak earlier in the vaccinees, occurring at 3 weeks p.i. compared to 4 weeks p.i. in the controls.
In light of our previous discovery that the Tat28-35SL8 CTL response selects for escape variants during early infection, we reasoned that strong anamnestic Tat28-35SL8-specific CTL responses, coupled with high viral loads, would select for CTL escape variants in the vaccinees. Indeed, Tat28-35SL8 CTL escape mutants largely replaced wild-type virus in the plasma within 4 weeks p.i. in both the vaccinated and control animals (data not shown). This finding suggests that reduction of peak viremia to a level that does not support the emergence of escape variants may be critically important for vaccine regimens that include CTL epitopes that escape rapidly during natural infection.
Tat-specific proliferative responses were again measured at 2 and 6 weeks post-SIV infection (Fig. 2B and C). Robust proliferative responses were now detectable in the majority of vaccinees compared to only one of four controls at these time points tested. Interestingly, vaccinee 97007, the animal that did not become infected, continued to exhibit the highest levels of Tat-specific proliferative responses. This could represent either a boosting of the proliferative immune response from some low-level exposure to the virus during challenge or simply maintenance of the initial DNA/MVA-induced proliferative response.
These rather disappointing results should be interpreted with caution. We challenged vaccinated animals with a highly pathogenic molecular clone. With the exception of live-attenuated SIV (9, 16) or prior exposure to simian-human immunodeficiency virus clone 89.6 (15), no vaccine regimen has been able to effectively control SIVmac239 replication. However, the SIVmac239 clone represents a good challenge virus to evaluate potential HIV vaccines since, like most primary HIV type 1 isolates, this virus is highly resistant to antibody-mediated neutralization (6, 20) and causes a gradual depletion of CD4 T lymphocytes. Furthermore, challenge of rhesus macaques with SIVmac239 yields reproducible viral set points of approximately 106 viral copies/ml, thus facilitating clear identification of vaccine efficacy. Additionally, despite our induction of high levels of Tat-specific CTL, we may not have induced these CTL at important mucosal sites. Furthermore, induction of a CTL response against a single CTL epitope may be similar to the use of single antiretroviral drug therapy, and induction of immune responses against multiple CTL epitopes may prove more effective, possibly analogous to the situation with combination drug therapy. Finally, while this vaccine induced strong CTL responses, the regimen was not designed to similarly induce strong CD4 T-cell helper responses, which might play an important role in the containment of HIV and SIV infections.
Acknowledgments
We thank Jenny Booth for performing the bDNA assays in a timely fashion. We also thank the Immunology/Virology Core Laboratory at WRPRC for infection and monitoring of macaques. We also acknowledge Marian Ohlmann for expert MVA vaccine preparation. We thank Deb Fuller (Powderject Vaccines, Inc.) for assistance with immunizations.
This work was supported by grants from the National Institutes of Health (AI49120 and AI46366 to D.I.W.; and RR00167 to the Wisconsin Regional Primate Research Center) and the European Community (QLK2-2000-1040 to G.S.). D.I.W. is an Elizabeth Glaser Scientist.
REFERENCES
- 1.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 2.Allen, T. M., T. U. Vogel, D. H. Fuller, B. R. Mothe, S. Steffen, J. E. Boyson, T. Shipley, J. Fuller, T. Hanke, A. Sette, J. D. Altman, B. Moss, A. J. McMichael, and D. I. Watkins. 2000. Induction of AIDS virus-specific CTL activity in fresh, unstimulated PBL from rhesus macaques vaccinated with a DNA prime/MVA boost regimen. J. Immunol. 164:4968-4978. [DOI] [PubMed] [Google Scholar]
- 3.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H.-L. Ma, B. D. Grimm, M. L. Hulsey, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS in rhesus macaques by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., A. Craiu, S. Santra, M. A. Egan, J. E. Schmitz, M. J. Kuroda, T. M. Fu, J. H. Nam, L. S. Wyatt, M. A. Lifton, G. R. Krivulka, C. E. Nickerson, C. I. Lord, B. Moss, M. G. Lewis, V. M. Hirsch, J. W. Shiver, and N. L. Letvin. 2001. Elicitation of high-frequency cytotoxic T-lymphocyte responses against both dominant and subdominant simian-human immunodeficiency virus epitopes by DNA vaccination of rhesus monkeys. J. Virol. 75:2462-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barouch, D. H., S. Santra, T. D. Steenbeke, X. X. Zheng, H. C. Perry, M. E. Davies, D. C. Freed, A. Craiu, T. B. Strom, J. W. Shiver, and N. L. Letvin. 1998. Augmentation and suppression of immune responses to an HIV-1 DNA vaccine by plasmid cytokine/Ig administration. J. Immunol. 161:1875-1882. [PubMed] [Google Scholar]
- 6.Burns, D. P., C. Collignon, and R. C. Desrosiers. 1993. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J. Virol. 67:4104-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cafaro, A., A. Caputo, C. Fracasso, M. T. Maggiorella, D. Goletti, S. Baroncelli, M. Pace, L. Sernicola, M. L. Koanga-Mogtomo, M. Betti, A. Borsetti, R. Belli, L. Akerblom, F. Corrias, S. Butto, J. Heeney, P. Verani, F. Titti, and B. Ensoli. 1999. Control of SHIV-89.6P-infection of cynomolgus monkeys by HIV-1 Tat protein vaccine. Nat. Med. 5:643-650. [DOI] [PubMed] [Google Scholar]
- 8.Cafaro, A., F. Titti, C. Fracasso, M. T. Maggiorella, S. Baroncelli, A. Caputo, D. Goletti, A. Borsetti, M. Pace, E. Fanales-Belasio, B. Ridolfi, D. R. Negri, L. Sernicola, R. Belli, F. Corrias, I. Macchia, P. Leone, Z. Michelini, P. ten Haaft, S. Butto, P. Verani, and B. Ensoli. 2001. Vaccination with DNA containing tat coding sequences and unmethylated CpG motifs protects cynomolgus monkeys upon infection with simian/human immunodeficiency virus (SHIV89.6P). Vaccine 19:2862-2877. [DOI] [PubMed] [Google Scholar]
- 9.Daniel, M. D., F. Kirchhoff, S. C. Czajak, P. K. Sehgal, and R. C. Desrosiers. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938-1941. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein, G., K. Manson, G. Tribbick, and R. Smith. 2000. Minimization of chronic plasma viremia in rhesus macaques immunized with synthetic HIV-1 Tat peptides and infected with a chimeric simian/human immunodeficiency virus (SHIV33). Vaccine 18:2789-2795. [DOI] [PubMed] [Google Scholar]
- 11.Hanke, T., T. J. Blanchard, V. C. Neumann, J. E. Boyson, P. Sweeney, T. M. Allen, D. I. Watkins, G. L. Smith, and A. McMichael. 1999. Effective induction of simian immunodeficiency virus-specific cytotoxic T lymphocytes in macaques using a DNA prime-measles virus Ankara boost vaccination regimen. J. Virol. 73:7524-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, A. L., K. Westover, J. da Silva, D. H. O'Connor, and D. I. Watkins. 2001. Simultaneous positive and purifying selection on overlapping reading frames of the tat and vpr genes of simian immunodeficiency virus. J. Virol. 75:7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korber, B., B. Gaschen, K. Yusim, R. Thakallapally, C. Kesmir, and V. Detours. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19-42. [DOI] [PubMed] [Google Scholar]
- 14.Lewis, M. G., S. Bellah, K. McKinnon, J. Yalley-Ogunro, P. M. Zack, W. R. Elkins, R. C. Desrosiers, and G. A. Eddy. 1994. Titration and characterization of two rhesus-derived SIVmac challenge stocks. AIDS Res. Hum. Retrovir. 10:213-220. [DOI] [PubMed] [Google Scholar]
- 15.Miller, C. J., M. B. McChesney, X. Lu, P. J. Dailey, C. Chutkowski, D. Lu, P. Brosio, B. Roberts, and Y. Lu. 1997. Rhesus macaques previously infected with simian/human immunodeficiency virus are protected from vaginal challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 71:1911-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori, K., Y. Yasutomi, S. Ohgimoto, T. Nakasone, S. Takamura, T. Shioda, and Y. Nagai. 2001. Quintuple deglycosylation mutant of simian immunodeficiency virus SIVmac239 in rhesus macaques: robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild-type strain. J. Virol. 75:4023-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osterhaus, A. D., C. A. van Baalen, R. A. Gruters, M. Schutten, C. H. Siebelink, E. G. Hulskotte, E. J. Tijhaar, R. E. Randall, G. van Amerongen, A. Fleuchaus, V. Erfle, and G. Sutter. 1999. Vaccination with Rev and Tat against AIDS. Vaccine 17:2713-2714. [DOI] [PubMed] [Google Scholar]
- 18.Pal, R., D. Venzon, N. L. Letvin, S. Santra, D. C. Montefiori, N. R. Miller, E. Tryniszewska, M. G. Lews, T. C. Vancott, V. Hirsch, R. Woodward, A. Gibson, M. Grace, E. Dobratz, P. D. Markham, Z. Hel, J. Nacsa, M. Klein, J. Tartaglia, and G. Franchini. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus-induced immunodeficiency. J. Virol. 76:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pauza, C. D., P. Trivedi, M. Wallace, T. J. Ruckwardt, H. Le Buanec, W. Lu, B. Bizzini, A. Burny, D. Zagury, and R. C. Gallo. 2000. Vaccination with Tat toxoid attenuates disease in simian/HIV-challenged macaques. Proc. Natl. Acad. Sci. USA 97:3515-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 21.van Baalen, C. A., O. Pontesilli, R. C. Huisman, A. M. Geretti, M. R. Klein, F. de Wolf, F. Miedema, R. A. Gruters, and A. D. Osterhaus. 1997. Human immunodeficiency virus type 1 Rev- and Tat-specific cytotoxic T lymphocyte frequencies inversely correlate with rapid progression to AIDS. J. Gen. Virol. 78:1913-1918. [DOI] [PubMed] [Google Scholar]
- 22.Van Baalen, C. A., M. Schutten, R. C. Huisman, P. H. Boers, R. A. Gruters, and A. D. Osterhaus. 1998. Kinetics of antiviral activity by human immunodeficiency virus type 1-specific cytotoxic T lymphocytes (CTL) and rapid selection of CTL escape virus in vitro. J. Virol. 72:6851-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]