Abstract
BCL-2 family proteins constitute a critical control point for the regulation of apoptosis. Protein interaction between BCL-2 members is a prominent mechanism of control and is mediated through the amphipathic α-helical BH3 segment, an essential death domain. We used a chemical strategy, termed hydrocarbon stapling, to generate BH3 peptides with improved pharmacologic properties. The stapled peptides, called “stabilized alpha-helix of BCL-2 domains” (SAHBs), proved to be helical, protease-resistant, and cell-permeable molecules that bound with increased affinity to multidomain BCL-2 member pockets. A SAHB of the BH3 domain from the BID protein specifically activated the apoptotic pathway to kill leukemia cells. In addition, SAHB effectively inhibited the growth of human leukemia xenografts in vivo. Hydrocarbon stapling of native peptides may provide a useful strategy for experimental and therapeutic modulation of protein-protein interactions in many signaling pathways.
BCL-2 is the founding member of a protein family (1-3) composed of pro- and anti-apoptotic molecules that serve as an essential control point in apoptosis, governing susceptibility to cell death (4-6). The BCL-2 family is defined by the presence of up to four conserved BCL-2 homology (BH) domains, all of which include α-helical segments. Anti-apoptotic proteins (for example, BCL-2 and BCL-XL) display sequence conservation in all BH domains, whereas pro-apoptotic proteins are divided into multidomain members (such as BAX and BAK), and BH3-only members (such as BID and BAD) that display sequence similarity only to the BH3 α-helical domain. The amphipathic α-helical BH3 segment of pro-apoptotic family members is a required death domain (7, 8) that binds to the hydrophobic groove formed by the juxtaposition of BH1, BH2, and BH3 domains of anti-apoptotic multidomain members (9, 10).
The α helix, a major structural motif of proteins, frequently mediates intracellular protein-protein interactions that govern many biological pathways (7, 11). Theoretically, helical peptides, such as the BH3 helix, could be used to selectively interfere with or stabilize protein-protein interactions and thereby manipulate physiological processes. However, biologically active helical motifs within proteins typically have little structure when taken out of context and placed in solution. Although peptides are attractive candidates for stabilizing or disrupting protein-protein interactions, their efficacy as in vivo reagents is severely compromised by their loss of secondary structure, susceptibility to proteolytic degradation, and difficulty in penetrating intact cells.
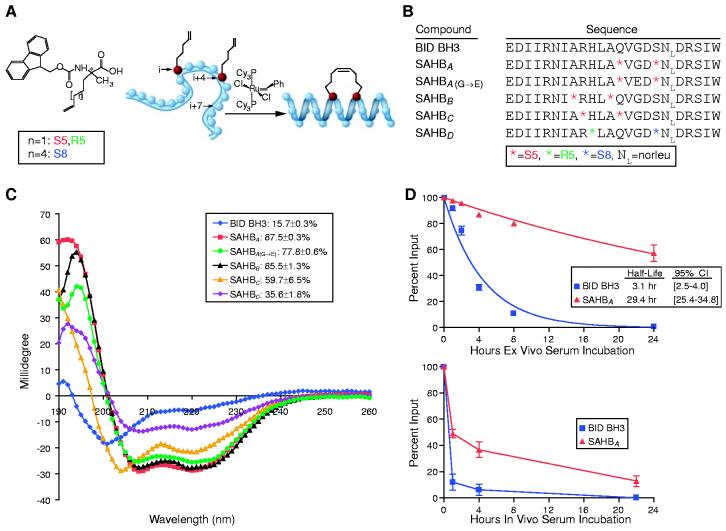
Most approaches to covalent helix stabilization involve polar or labile cross-links (12-15), which leave the peptides vulnerable to decomposition or unable to penetrate cells, if not both. We have developed an alternative strategy in which we use α,α-disubstituted non-natural amino acids containing olefin-bearing tethers to generate an all-hydrocarbon “staple” by ruthenium-catalyzed olefin metathesis (16, 17) (Fig. 1A). A panel of hydrocarbon-stapled peptides, referred to as stabilized alpha-helix of BCL-2 domains (SAHBs), was designed to mimic the BH3 domain of BID (Fig. 1B). BID is a pro-apoptotic BH3-only protein that, in response to death receptor signaling, interconnects the extrinsic and core intrinsic apoptotic pathways. Activated BID (tBID) can be bound and sequestered by anti-apoptotic proteins (such as BCL-2 and BCL-XL), but also triggers activation of the multidomain pro-apoptotic proteins BAX and BAK, resulting in cytochrome c release and a mitochondrial program of apoptosis (4-6, 8, 18, 19).
Fig. 1.
Enhanced helicity, protease resistance, and serum stability of hydrocarbon-stapled BID BH3 compounds. (A and B) α,α-disubstituted non-natural amino acids containing olefinic side chains of varying length were synthesized as previously reported (16, 31, 32). Non-natural amino acid substitutions were made to flank three (substitution positions i and i+4) or six (i and i+7) amino acids within the BID BH3 peptide, so that reactive olefinic residues would reside on the same face of the α helix. (C) Circular dichroism was used to measure the percentages of SAHB maintained in helical configuration when dissolved in aqueous potassium phosphate solution (pH7) (supporting online material). (D) Fluoresceinated SAHBA and BID BH3 peptide were incubated at 37°C in mouse serum or injected intravenously (10 mg/kg) into NOD SCID mice. Serum concentrations of SAHBA and BID BH3 peptide were measured at the indicated time points with a fluorescence-based high-performance liquid chromatography detection assay. Both assays demonstrated enhanced serum stability of SAHBA.
Circular dichroism revealed that a 23-amino acid BID BH3 peptide displays only 16% helicity in solution and thus predominantly exists as a random coil (Fig. 1C). SAHBs, however, demonstrated helical stabilization, with helical content ranging from 35 to 87%. In addition to reinforcing the biologically active secondary structure of the BID BH3 domain, peptide helix stabilization is expected to bury the amide backbone, shielding it from proteolysis. Insertion of the hydrocarbon staple did endow SAHBs with protease resistance and serum stability in vitro and in vivo, as exemplified by comparing SAHBA to the unmodified BID BH3 peptide in three distinct degradation assays (Fig. 1D and fig. S1).
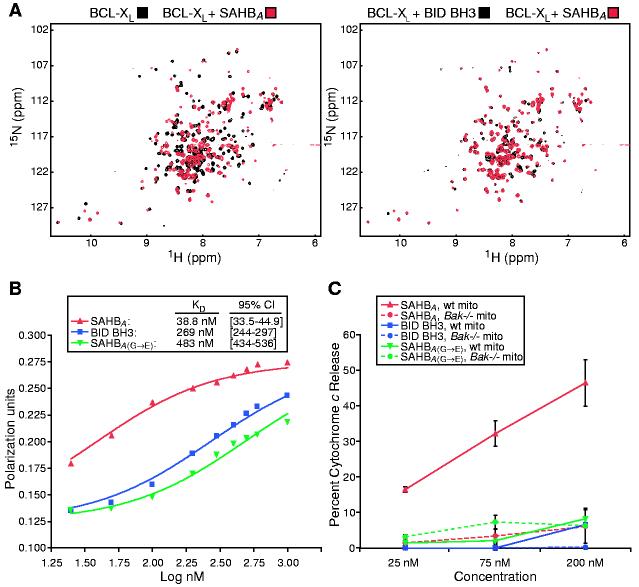
To determine whether SAHBA specifically interacts with the defined binding groove of an anti-apoptotic multidomain protein, we recorded a two-dimensional 15N-1H heteronuclear single-quantum correlation (HSQC) spectrum of 15N-labeled BCL-XL before and after the addition of SAHBA and compared the profile with the corresponding spectrum derived from addition of unmodified BID BH3 peptide (Fig. 2A). The overall similarity of the HSQC spectra indicates that the structural changes occurring in BCL-XL after the addition of SAHBA are nearly identical to those observed with BID BH3 peptide. A BCL-2 fluorescence polarization binding assay demonstrated more than sixfold enhancement in binding affinity of SAHBA (Kd, 38.8 nM) compared to that of unmodified BID BH3 peptide (Kd, 269 nM) (Fig. 2B). A Gly-to-Glu mutation (8) [SAHBA(G→E)] (Kd, 483 nM), reduced high-affinity binding and served as a useful control.
Fig. 2.
SAHBA targets the binding pocket of BCL-XL, displays enhanced BCL-2 binding affinity, and specifically activates cytochrome c release from mitochondria in vitro. (A) HSQC experiments show similar spectral changes in 15N-BCL-XL upon binding SAHBA or BID BH3 peptide. (B) Kd's for binding of individual peptides to glutathione S-transferase-BCL-2 were determined by fluorescence polarization. (C) Mouse liver mitochondria (wild-type or Bak-/-, 0.5 mg/ml) were incubated for 40 min with 25 to 200 nM concentrations of BID BH3 peptide, SAHBA, or SAHBA(G→E), and cytochrome c was measured in the supernatant and sedimented mitochondria by an enzyme-linked immunosorbent assay.
The in vitro biological activity of SAHBA was investigated by assaying peptide-induced cytochrome c release from purified mouse liver mitochondria. Measured at equilibrium, SAHBA caused a dose-dependent increase in cytochrome c release, in contrast to the negligible effects of BID BH3 peptide and the SAHBA(G→E) point mutant in the dose range from 25 to 200 nM (Fig. 2C). The identical experiment was performed on isolated Bak-/- mouse liver mitochondria, which lack both BAK and cytosolic BAX and do not release cytochrome c in response to tBID (5, 18). The inability of SAHBA to induce cytochrome c release from Bak-/- mitochondria confirms that it functions through the defined pathway of apoptosis.
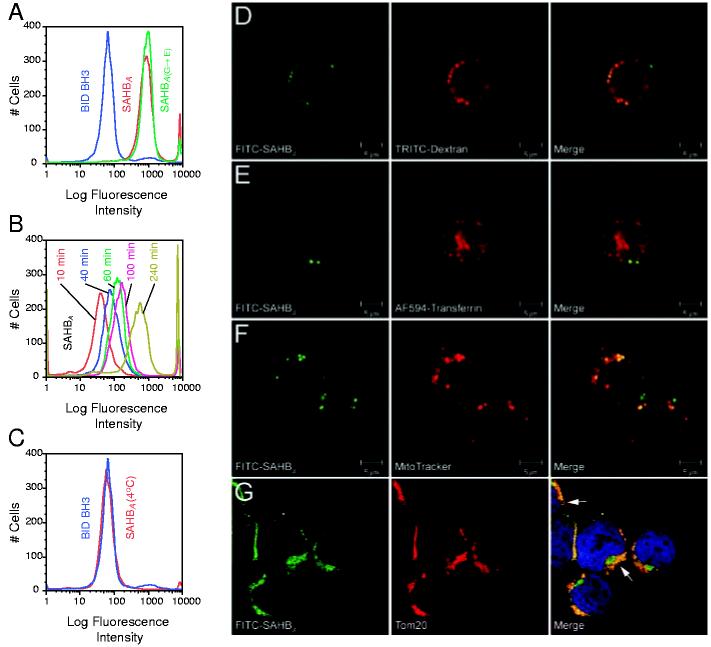
With the exception of select positively charged peptides (20-22), the electrostatic charge of amino acid side chains and the polarity of the peptide backbone generally impede the transduction of peptides across cellular membranes. We tested whether our approach to stabilizing helical structure by the insertion of a hydrocarbon staple would confer lipophilic and potentially membrane-penetrating properties on SAHB compounds. We incubated Jurkat T cell leukemia cells in culture with fluorescein isothiocyanate (FITC)-labeled BID BH3 peptide, SAHBA, or SAHBA(G→E), followed by confocal microscopy and fluorescence-activated cell sorting (FACS) analyses. Whereas FITC was not detected in BID BH3-treated cells, SAHBA-treated cells displayed fluorescent labeling, with a discrete cytoplasmic localization evident within the cells on confocal images (Fig. 3 and fig. S2). FITC-labeled Jurkat cells initially excluded propidium iodide (as determined by FACS analysis), indicating that SAHBA compounds do not function as nonspecific permeabilizing agents. The cellular uptake of FITC-SAHBA was time-dependent (Fig. 3B) and was inhibited at 4°C (Fig. 3C) and by a combination treatment with sodium azide and deoxyglucose (23), suggesting an energy-dependent endocytosis mechanism for cellular import. Whereas binding to cell surface glycosaminoglycans, such as heparin, has been implicated in the targeting and import mechanism of positively charged cell penetrating peptides such as human immunodeficiency virus transactivator of transcription (TAT) and Antennapedia (Antp) (24-26), cellular uptake of SAHBA was not inhibited in a dose-responsive manner by soluble heparin (fig. S3). Live cell confocal microscopy performed 4 hours after SAHB treatment demonstrated an initial colocalization of FITC-SAHBA with 4.4- or 70-kD dextran-labeled endosomes (Fig. 3D) but not transferrin-labeled endosomes (Fig. 3E), which is consistent with cellular uptake by fluid-phase pinocytosis (27), the endocytic pathway determined for TAT and Antp peptides (28). At a 24-hour time point, intracellular FITC-SAHBA showed increased colocalization with MitoTracker-labeled mitochondria in live cells (Fig. 3F), consistent with the mitochondrial colocalization observed in fixed cells when an antibody to Tom20 (29), a mitochondrial outer membrane protein, was used (Fig. 3G).
Fig. 3.
SAHBA penetrates Jurkat leukemia cells by fluid-phase endocytosis and localizes to the mitochondrial membrane. Jurkat leukemia cells were incubated with FITC-labeled peptides for 4 hours at 37°C, followed by FACS analysis (A). FITC-SAHBA uptake occurred in a time-dependent manner at 37°C (B), but no FITC-SAHBA labeling was evident by 4 hours, when the experiment was performed at 4°C (C). Live confocal images demonstrated a colocalization of FITC-SAHBA with 4.4-kD dextran-labeled endosomes (D) but not transferrin-labeled endosomes (E) at 4 hours. A mitochondrial colocalization was evident by 24 hours, as demonstrated by the merged images of FITC-SAHBA and MitoTracker in live cells (F) and those of FITC-SAHBA and Tom20 (a mitochondrial outer-membrane marker) in fixed cells (G). Arrows highlight sites of colocalization corresponding to the surface of mitochondria cut in cross section (G).
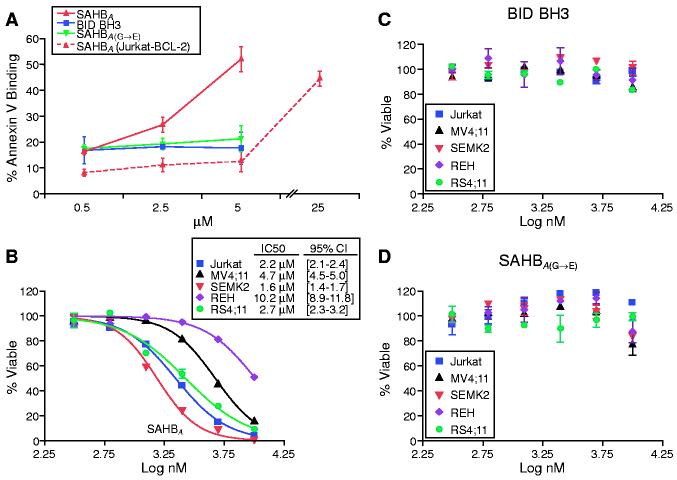
The ability of SAHBA to activate apoptosis was assessed in Jurkat T cell leukemia cells. Fifty percent of cells treated with 5 μM SAHBA demonstrated staining for annexin V after 20 hours, whereas cells treated with BID BH3 peptide or SAHBA(G→E) showed no increase in apoptosis in this dose range (Fig. 4A). Jurkat cells overexpressing BCL-2 were resistant to 5 μM SAHBA. However, the protective effect of BCL-2 could be overcome at higher concentrations of SAHBA; the rightward shift in dose response is consistent with SAHBA functioning at the BCL-2 control point in intact cells. In concert, these data indicate that SAHBA can penetrate leukemia cells and selectively trigger the apoptotic pathway.
Fig. 4.
SAHBA triggers apoptosis in Jurkat cells and inhibits a panel of human leukemia cells. FACS analysis of annexin V-treated cells was used to monitor apoptosis of Jurkat cells treated with 0.5 to 5 μM concentrations of BID BH3 peptide, SAHBA, or SAHBA(G→E) for 20 hours (A). Jurkat, REH, MV4;11, SEMK2, and RS4;11 leukemia cells were treated with serial dilutions of SAHBA (B), BID BH3 peptide (C), or SAHBA(G→E) (D), and MTT assays were performed at 48 hours to measure viability.
To investigate whether SAHBA would inhibit a wider panel of leukemia cells, 3-(4,5-dimethylthiazol-2-yl)2,5-diphenyl tetrazolium bromide (MTT) assays were performed on T cell (Jurkat), B cell (REH), and mixed lineage leukemia (MLL) cells (cell lines MV4;11, SEMK2, and RS4;11) in culture. SAHBA inhibited the proliferation of leukemia cells at median inhibitory concentrations of 2.2 (Jurkat), 10.2 (REH), 4.7 (MV4;11), 1.6 (SEMK2), and 2.7 (RS4;11) μM (Fig. 4B). Neither the BID BH3 peptide nor the SAHBA(G→E) point mutant had an effect in this dose range (Fig. 4, C and D).
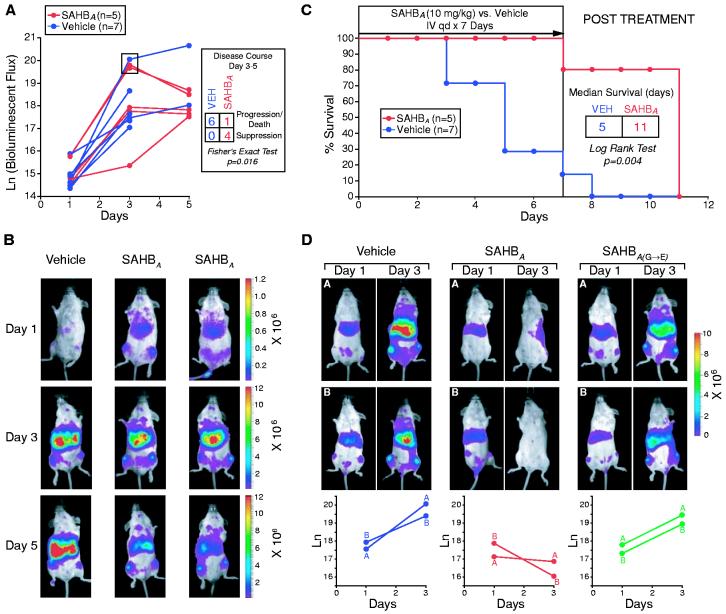
In four cohorts of immunodeficient mice bearing established human leukemia xenografts, SAHBA treatment consistently suppressed leukemia growth in vivo. On day-3 of experimentation, severe combined immunodeficient (SCID) beige mice were subjected to 300 cGy of total body irradiation, followed by intravenous injection of 5 × 106 RS4;11 leukemia cells that stably expressed luciferase. Leukemia burden was monitored using the In Vivo Imaging System (IVIS, Xenogen), which quantitates total body luminescence after intraperitoneal injection of D-luciferin (30). On day 1, leukemic mice were treated intravenously with SAHBA [10 mg per kg of body weight (mg/kg)] or with vehicle [5% dimethyl sulfoxide (DMSO) in 5% dextrose water] daily for 7 days. Mice were monitored daily for survival and were imaged on days 1, 3, and 5 to measure leukemia burden. Control mice demonstrated progressive leukemic growth as quantitated by increased luminescence from days 1 through 5 (Fig. 5A). In this cohort, SAHBA treatment usually suppressed the leukemic expansion after day 3, and tumor regression was frequently observed by day 5. Imaging showed progressive leukemic infiltration of the spleen and liver in control mice, but often showed regression of disease at these anatomical sites in SAHBA-treated mice by day 5 of treatment (Fig. 5B). The median time to death in this cohort was 5 days for control animals, but 11 days for SAHBA-treated animals (Fig. 5C). In a similar experiment comparing the effects of SAHBA and SAHBA(G→E), animals receiving the point mutant SAHB did not exhibit regression of leukemia (Fig. 5D). Histologic examination of SAHBA-treated mice showed no obvious toxicity of the compound to normal tissue.
Fig. 5.
SAHBA suppresses growth of human leukemia cells in vivo, prolonging the survival of leukemic mice. (A) Leukemic SCID beige mice [with a day-1 natural logarithm (ln) bioluminescence range of 14.4 to 15.9] were treated with intravenous injections of 10 mg/kg SAHBA or vehicle (5% DMSO in D5W) daily for 7 days and were monitored for survival; leukemia burden was quantified by total body luminescence (photons/s/mouse) on days 1, 3, and 5. The disease course from days 3 to 5 differed between SAHBA-treated animals and controls (P = 0.016, Fisher's exact test [box in (A)], as illustrated by representative Xenogen images of bioluminescent leukemic mice (B); red signal represents the highest level of leukemia on the colorimetric scale. (C) Median survival was prolonged in SAHBA-treated animals as compared to controls (P = 0.004, log rank test). (D) To compare SAHBA with SAHBA(G→E), leukemic mice (with a day 1 ln bioluminescence range of 17.1 to 17.9) were treated daily with SAHB (10 mg/kg) or vehicle, and animals were imaged on days 1 and 3 to measure total body luminescence.
Insertion of an all-hydrocarbon staple into the BID BH3 peptide yielded a marked enhancement of peptide α-helicity, stability, and in vitro and in vivo biological activity. SAHBs that engage the pocket of multidomain BCL-2 members could serve as prototypes for the development of therapeutics for cancer and perhaps other diseases. Intracellular protein-protein interactions constitute major control points in many signaling pathways, yet have frequently proven a difficult target for small-molecule chemistry, often reflecting a protein interface that is extensive, shallow, and hydrophobic. Such endogenous control points are typically regulated by other protein domains or their modifications. Synthetic approaches such as hydrocarbon stapling that reinforce native peptide sequences provide an alternative strategy to probe protein-protein interactions and manipulate biological pathways.
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/305/5689/1466/DC1 SOM Text Figs. S1 to S3 References and Notes
References and Notes
- 1.Bakhshi A, et al. Cell. 1985;41:899. doi: 10.1016/s0092-8674(85)80070-2. [DOI] [PubMed] [Google Scholar]
- 2.Cleary ML, Sklar J. Proc. Natl. Acad. Sci. U.S.A. 1985;82:7439. doi: 10.1073/pnas.82.21.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Science. 1985;228:1440. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- 4.Danial NN, Korsmeyer SJ. Cell. 2004;116:205. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 5.Wei MC, et al. Science. 2001;292:727. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scorrano L, et al. Science. 2003;300:135. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 7.Chittenden T, et al. EMBO J. 1995;14:5589. doi: 10.1002/j.1460-2075.1995.tb00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. Genes Dev. 1996;10:2859. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 9.Muchmore SW, et al. Nature. 1996;381:335. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 10.Sattler M, et al. Science. 1997;275:983. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 11.Kussie PH, et al. Science. 1996;274:948. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 12.Phelan JC, Skelton NJ, Braisted AC, McDowell RS. J. Am. Chem. Soc. 1997;119:455. [Google Scholar]
- 13.Leuc A, et al. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11273. [Google Scholar]
- 14.Bracken C, Gulyas J, Taylor JW, Baum J. J. Am. Chem. Soc. 1994;116:6431. [Google Scholar]
- 15.Yan B, Liu D, Huang Z. Bioorg. Med. Chem. Lett. 2004;14:1403. doi: 10.1016/j.bmcl.2003.09.101. [DOI] [PubMed] [Google Scholar]
- 16.Schafmeister C, Po J, Verdine G. J. Am. Chem. Soc. 2000;122:5891. [Google Scholar]
- 17.Blackwell HE, Grubbs RH. Angew. Chem. Int. Ed. Engl. 1994;37:3281. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 18.Wei MC, et al. Genes Dev. 2000;14:2060. [PMC free article] [PubMed] [Google Scholar]
- 19.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Cell. 1998;94:481. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 20.Fawell S, et al. Proc. Natl. Acad. Sci. U.S.A. 1994;91:664. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derossi D, Joliot AH, Chassaing G, Prochiantz A. J. Biol. Chem. 1994;269:10444. [PubMed] [Google Scholar]
- 22.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. Science. 1999;285:1569. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 23.Walensky LD, Korsmeyer SJ. unpublished data [Google Scholar]
- 24.Drin G, Cottin S, Blanc E, Rees AR, Temsamani J. J. Biol. Chem. 2003;278:31192. doi: 10.1074/jbc.M303938200. [DOI] [PubMed] [Google Scholar]
- 25.Console S, Marty C, Garcia-Echeverria C, Schwendener R, Ballmer-Hofer K. J. Biol. Chem. 2003;278:35109. doi: 10.1074/jbc.M301726200. [DOI] [PubMed] [Google Scholar]
- 26.Richard JP, et al. J. Biol. Chem. 2003;278:585. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 27.Araki N, Johnson MT, Swanson JA. J. Cell Biol. 1996;135:1249. doi: 10.1083/jcb.135.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wadia JS, Stan RV, Dowdy SF. Nature Med. 2004;10:310. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 29.Schleiff E, Shore GC, Goping IS. J. Biol. Chem. 1997;272:17784. doi: 10.1074/jbc.272.28.17784. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong SA, et al. Cancer Cell. 2003;3:173. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 31.Williams RM, Im MN. J. Am. Chem. Soc. 1991;113:9276. [Google Scholar]
- 32. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; D, Asp; E, Glu; G, Gly; H, His; I, Ile; L, Leu; Q, Gln; R, Arg; S, Ser; V, Val; and W, Trp
- 33. We thank D. Brown and M. Salanga for assistance with confocal microscopy; D. Neuberg and M. Goldwasser for biostatistics support; E. Gillespie and W. Beavers for assistance with automated peptide synthesis and amino acid analysis; S. Armstrong for providing leukemia cell lines; R. Bronson for rodent necropsy evaluation; Q. Liao for assistance with mass spectrometry; S. Lux, B. Malynn, and F. Bernal for helpful discussions; and E. Smith for editorial and computer graphics assistance. L.D.W. is a Lymphoma Research Foundation Fellow and is also supported by National Heart, Lung, and Blood Institute grant no. K08HL074049, an American Society of Hematology Scholar Award, and the Lauri Strauss Leukemia Foundation. This work is supported in part by NIH grant no. R37CA50239, a Leukemia and Lymphoma Society deVilliers International Achievement Award to S.J.K., a grant from the Virginia and D. K. Ludwig Fund for Cancer Research to G.W., and a gift from Enanta Pharmaceuticals to G.L.V. Molecular interaction data have been deposited in the Biomolecular Interaction Network Database with accession codes 151034 and 151035