Abstract
The replication complexes (RCs) of positive-stranded RNA viruses are intimately associated with cellular membranes. To investigate membrane alterations and to characterize the RC of mouse hepatitis virus (MHV), we performed biochemical and ultrastructural studies using MHV-infected cells. Biochemical fractionation showed that all 10 of the MHV gene 1 polyprotein products examined pelleted with the membrane fraction, consistent with membrane association of the RC. Furthermore, MHV gene 1 products p290, p210, and p150 and the p150 cleavage product membrane protein 1 (MP1, also called p44) were resistant to extraction with Triton X-114, indicating that they are integral membrane proteins. The ultrastructural analysis revealed double-membrane vesicles (DMVs) in the cytoplasm of MHV-infected cells. The DMVs were found either as separate entities or as small clusters of vesicles. To determine whether MHV proteins and viral RNA were associated with the DMVs, we performed immunocytochemistry electron microscopy (IEM). We found that the DMVs were labeled using an antiserum directed against proteins derived from open reading frame 1a of MHV. By electron microscopy in situ hybridization (ISH) using MHV-specific RNA probes, DMVs were highly labeled for both gene 1 and gene 7 sequences. By combined ISH and IEM, positive-stranded RNA and viral proteins localized to the same DMVs. Finally, viral RNA synthesis was detected by labeling with 5-bromouridine 5′-triphosphate. Newly synthesized viral RNA was found to be associated with the DMVs. We conclude from these data that the DMVs carry the MHV RNA replication complex and are the site of MHV RNA synthesis.
The replication complexes (RCs) of virtually all mammalian and plant positive-stranded RNA viruses have been shown to be intimately associated with cellular membranes. However, different RNA viruses seem to target or recruit distinct membranes for the assembly of their RCs. For example, poliovirus replicates its RNA on the surface of membranous vesicles (4) derived from vesicles of the anterograde membrane trafficking pathway (35). Brome mosaic virus and tobacco etch potyvirus have been shown to use endoplasmic reticulum (ER)-derived structures as the site of assembly for their RCs and for replication of their viral RNA (34, 36). Alphaviruses appear to use the cytosolic surface of endocytic organelles for the formation of their RCs (18), whereas rubella virus RCs have been identified as virus-modified lysosomes (30). For mouse hepatitis virus (MHV), a member of the order Nidovirales, the composition and site of assembly of the RC are not yet clear.
MHV is a prototype coronavirus with a positive-stranded RNA genome of 31.2 kb. Coronaviruses such as MHV and arteriviruses such as equine arteritis virus (EAV) are grouped in the order Nidovirales because of their discontinuous RNA synthesis, which results in the generation of a “nested set” of mRNAs (reviewed in references 25 and 43). The RNA-dependent RNA polymerase that mediates this unusual, discontinuous RNA synthesis is encoded by gene 1. For MHV, gene 1 encompasses 22 kb and contains two large open reading frames (ORF1a and ORF1b) that are joined via a ribosomal frameshifting mechanism to potentially produce an ORF1ab polyprotein of 800 kDa (6, 11, 26). This polyprotein is extensively processed by three viral proteinases encoded in ORF1a (reviewed in reference 50).
The MHV papain-like proteinase 1 (PLP1) autoproteolytically processes the amino-terminal protein, p28 (1), and can act in trans to release the adjacent protein, p65 (7, 8, 14). MHV papain-like proteinase 2 (PLP2) is encoded downstream of PLP1 and processes the ORF1a polyprotein to generate p210 and p150 (23). The p210 product also contains a hydrophobic domain (HD) that may be important for membrane association and assembly of the RC (21). The p150 product contains the MHV poliovirus 3C-like proteinase domain, 3CLpro, which can act in cis and in trans to generate seven proteolytic products from the p150 intermediate: membrane protein 1 (MP1; also called p44), 3CLpro (p27), membrane protein 2 (MP2), p10, p22, p12, and p15 (10, 23, 27-29, 37). The 3CLpro is also responsible for processing the ORF1b region to generate p100, the putative core polymerase region; p67, encoding helicase and zinc-binding domains; and three additional protein products of unknown function (15, 33, 41). The role of this extensive processing cascade in generating a functional RC and in regulating the mechanisms of MHV transcription, replication, and RNA recombination is not yet understood.
It has been shown that infection of cells with EAV results in the generation of ER-derived double-membrane vesicles (DMVs). These vesicles are the site of assembly of the EAV RC and the site where viral RNA synthesis occurs (32). In contrast, ultrastructural studies of the site of MHV RNA synthesis have been equivocal. One early study implicated membrane-bound inclusions visualized by electron microscopy as a possible site for MHV RNA synthesis (16). More recently, colocalization studies using cryoelectron microscopy techniques and endocytic tracers implicated late endosomes as the site of MHV RNA synthesis (48). In contrast to these findings, we and others have shown that translation products of the MHV gene 1 partially localized with Golgi and ER markers in human and murine cell lines (2, 41).
The present study characterizes the MHV RC by biochemical analysis and ultrastructural in situ labeling techniques. Biochemical fractionation experiments revealed that all MHV gene 1 products analyzed with available reagents sedimented with intracellular membranes, consistent with the formation of a membrane-associated complex. We removed peripheral proteins from the membrane-associated complex by treatment with detergent Triton X-114 and found that MHV ORF1a products p290, p210, p150, and p44 (MP1) are integral membrane proteins that may serve as scaffolding components for the complex. Ultrastructural studies revealed the presence of DMVs in MHV-infected cells. Immunoelectron microscopy studies showed that these vesicles contained MHV gene 1 products, positive-stranded RNA, and newly synthesized viral RNA. These results show that the DMVs are the site of MHV RNA synthesis.
MATERIALS AND METHODS
Viruses and cells.
MHV strains A59 and JHM-x were propagated as described previously (37). Cell lines used in this study included 17Cl-1 cells (45) for propagating virus stocks and electron microscopy studies, DBT cells (22) for determining the infectivity of virus stocks by plaque assay, and HeLa cells expressing the MHV receptor (HeLa-MHVR cells) (20) for all radiolabeling and the majority of the electron microscopy work.
Generation of anti-D14 serum.
The D14 region was generated by reverse transcription-PCR (RT-PCR) from RNA isolated from MHV JHM-x-infected HeLa-MHVR cells with primers FP38 (nucleotides [nt] 12274 to 12292; 5′-TATCTAGATGAATTTGTTAATATGGCT-3′) and FP39 (nt 12833 to 12851; 5′-TTAAGCTTGCAAAACGACCGTAGACAC-3′) using methods described previously (37). The underlined primer sequences were added for cloning purposes. The D14 region was cloned in-frame with glutathione S-transferase (GST) in the pGEX-KG vector, and fusion protein was induced by addition of 100 μM isopropyl-β-d-thiogalactopyranoside for 1 h. The GST-D14 fusion protein was isolated, purified, and injected into rabbits as previously described (23, 37).
Preparation of radiolabeled whole-cell lysates and immunoprecipitation.
MHV A59 infection, radiolabeling of proteins, treatment with 400 μg of protease inhibitor E64d (Matreya, Inc., Pleasant, Pa.), preparation of whole-cell lysates, and immunoprecipitation were performed as previously described for HeLa-MHVR cells (23). For most experiments, proteins in MHV A59-infected cells were metabolically radiolabeled with 100 μCi of Trans35S-label (ICN, Costa Mesa, Calif.) per ml for 2 h, and cells were harvested at 5.5 h postinfection (hpi).
The antibodies used in this study included rabbit anti-p28 (1), anti-D3, anti-D10, and anti-D12 (37), anti-D18 and anti-D23 (41), anti-D11 (23), anti-D14 (this paper), and mouse anti-V5 monoclonal antibody (Invitrogen, Carlsbad, Calif.).
Cell fractionation and sodium carbonate extraction.
Intracellular membranes were isolated from MHV A59-infected cells essentially as described by Fujiki et al. (19) and van der Meer et al. (49). Briefly, HeLa-MHVR cells were infected at a multiplicity of infection (MOI) of 10 with MHV A59 and metabolically radiolabeled with 100 μCi of Trans35S-label per ml from 3 to 5 hpi. The cell monolayers were washed twice with phosphate-buffered saline (PBS), scraped in 1 ml of PBS, and pelleted by centrifugation at 500 × g for 3 min. The cell pellet was resuspended at a final concentration of approximately 2 × 106 cells per ml in hypotonic buffer (1.0 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 15 mM NaCl, 2 μg of leupeptin, and 100 μg of phenylmethylsulfonyl fluoride per ml). The cells were broken open by 15 to 20 strokes with a Dounce homogenizer. Nuclei were pelleted by centrifugation at 1,500 × g for 5 min. The postnuclear supernatant was applied to a 6% sucrose cushion. Membranes were pelleted by centrifugation at 150,000 × g for 30 min. The supernatant fraction and membranous pellet were equalized for volume in hypotonic buffer, subjected to immunoprecipitation as described above, and analyzed by electrophoresis on sodium dodecyl sulfate-5.0 to 12.5% polyacrylamide gels and subjected to autoradiography.
Triton X-114 extraction.
Triton X-114 extractions were carried out as described by Bordier (9) and van der Meer et al. (49). Briefly, HeLa-MHVR cells were infected with MHV A59 at an MOI of 10, and newly synthesized proteins were labeled with Trans35S-label from 3.5 to 5.5 hpi. After labeling, cells were washed with PBS and lysed with an ice-cold solution of 1% Triton X-114 in PBS. The nuclear fraction was removed by centrifugation at 1,500 × g for 5 min. The cytosolic fraction was incubated on ice for 15 min and subsequently loaded onto a 6% sucrose cushion containing 1% Triton X-114. The phases were separated by incubation at 37°C for 5 min and centrifugation at room temperature at 2,700 × g for 3 min. The supernatant fraction was subjected to one additional round of Triton X-114 extraction. The detergent pellets were pooled and resuspended in lysis buffer A (4% SDS, 3% dithiothreitol, 40% glycerol, and 0.0625 M Tris-HCl, pH 6.8) (37). The immunoprecipitation conditions for both fractions were equalized by adding concentrated detergents to the supernatant fraction. Samples were diluted in radioimmunoprecipitation assay buffer (37) and used for immunoprecipitations as described above.
BrUTP labeling.
Newly synthesized viral RNA was labeled with 5-bromouridine 5′-triphosphate (BrUTP; Sigma, St. Louis, Mo.) as described (49) with some modifications. In short, HeLa-MHVR cells were infected with MHV A59 and incubated, after the absorption period, in complete medium. At 3.5 hpi, 5 μg of actinomycin D (Merck Sharp and Dohme, Whitehouse Station, N.J.) per ml was added to the culture medium to block cellular RNA synthesis. At 4.0 hpi, cells were incubated for 1 h with 10 mM BrUTP and Lipofectin (Invitrogen) according to the manufacturer's protocol for plasmid DNA transfection. Actinomycin D was present during the entire labeling period. Labeled RNA (BrU-RNA) was visualized by indirect immunocytochemical detection using a monoclonal antibody and gold-labeled secondary antibody as described below. Mock-infected cells processed in parallel were used as controls.
EM and IEM.
For conventional electron microscopy (EM), cells were fixed at the times indicated in 2.5% glutaraldehyde and 2% OsO4 and embedded in Poly/Bed 812 (Polysciences, Warrington, Pa.) according to standard protocols. For immunocytochemistry electron microscopy (IEM), cells were fixed and embedded in LRGold (Polysciences) at −20°C as described (4). For immunocytochemical labeling of viral proteins, polyclonal rabbit anti-D3 diluted 1:100 was used as the primary antibody. The grids were incubated at room temperature for 1 h, washed twice for 5 min, incubated with goat anti-rabbit immunoglobulin (Ig) antibody coupled to 15-nm gold particles (Amersham, Piscataway, N.J.), and washed as before.
BrU-RNA was detected by monoclonal antibromodeoxyuridine (BrdU) antibody (Bio Cell Consulting, Reinach, Switzerland) diluted 1:10 and containing 1 U of RNase inhibitor (Roche Molecular Biochemicals, Mannheim, Germany) per μl. After incubation for 45 min, grids were washed as above, incubated with goat anti-mouse Ig antibody coupled to 10-nm gold (Amersham) and washed again. For BrU-RNA detection, all incubation steps were carried out at 4°C. Further processing of the sections was performed as described previously (3).
EM-ISH, double EM-ISH, and EM-ISH combined with IEM.
For positive-stranded MHV RNA detection on LRGold sections, two nonoverlapping probes of negative polarity were prepared by in vitro transcription from linearized plasmid DNAs containing strain JHM-x sequences using T7 RNA polymerase. Probe 1, detecting subgenomic and genomic RNA, corresponded to nt 363 to 1418 of mRNA 7 (42). Probe 2 corresponded to nt 4790 to 7671 of ORF1a (6, 26) and is specific for genome-size RNA. During in vitro transcription, probe 1 was labeled with digoxigenin (DIG)-UTP (Roche), whereas probe 2 was labeled with biotin-UTP (Roche). Both probes were subjected to alkaline hydrolysis to generate fragments of approximately 500 nt (13). Purification of the probes and subsequent hybridization steps were done as described (3).
For EM in situ hybridization (EM-ISH), probe 1 was used, whereas for double EM-ISH, probes 1 and 2 were combined before the hybridization step. The hybridized probes were detected by 10-nm-gold-tagged sheep anti-DIG antibody (Aurion, Wageningen, The Netherlands) and 6-nm-gold-tagged goat antibiotin antibody (Aurion). When combining EM-ISH and IEM, incubation of the primary antibody for IEM was performed after hybridization with probe 1 and washing. The immunological detection steps of ISH and IEM were done simultaneously by incubation of the grids on a mixture of 10-nm-gold-labeled sheep anti-DIG and 15-nm-gold-tagged goat anti-rabbit Ig antibodies. The specificity of the ISH was tested by hybridizing the MHV-specific probes to mock-infected cells.
RESULTS
Identification of MHV ORF1ab precursors and products.
The processing of the MHV ORF1ab polyprotein is complex, with three distinct proteinase activities responsible for the release of at least 15 protein products. We hypothesize that ORF1a polyprotein precursors and/or intermediates are critical for embedding the MHV RC into cellular membranes. To test this hypothesis, we generated specific antisera to a number of distinct domains in the MHV ORF1ab polyprotein and then used the antisera in the biochemical characterization of the RC. Here, we describe the results of our analysis with a new antiserum, anti-D14, directed against a region of MHV ORF1a and the results of our analysis of precursors and products using the entire panel of antisera.
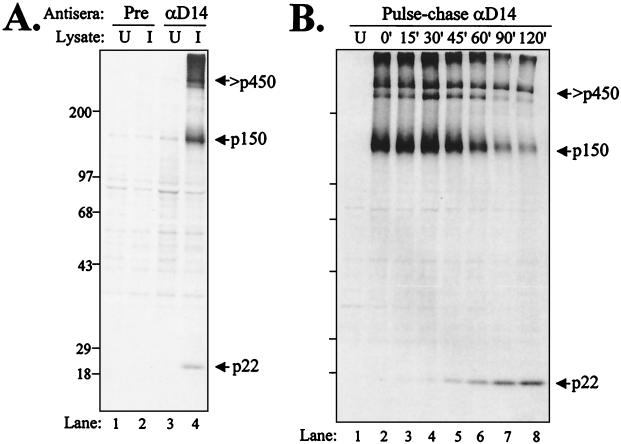
The anti-D14 serum was generated as described in Materials and Methods. The GST-D14 antigen encoded MHV amino acids glutamic acid-4021 to leucine-4212, a region bounded by 3CLpro cleavage sites (28). To determine if the anti-D14 serum recognized MHV-specific proteins, we tested the antiserum for the ability to immunoprecipitate radiolabeled MHV proteins. The anti-D14 antibodies specifically recognized MHV A59 proteins of 22 and 150 kDa and a high-molecular-weight protein (Fig. 1A, lane 4). Previously, Lu and coworkers had detected a 22-kDa protein using a polyclonal antiserum directed against a similar region, but no precursor had been identified (28).
FIG. 1.
Identification of MHV ORF1a products recognized by anti-D14 serum. (A) Detection of p150 and p22 ORF1a proteolytic products. HeLa-MHVR cells were infected with MHV A59, and newly synthesized proteins were labeled with Trans35S-label from 3.5 to 5.5 hpi. Total lysates of uninfected (U) or infected (I) cells were subjected to immunoprecipitation with preimmune (Pre) or anti-D14 serum. Products were analyzed by electrophoresis on an SDS-5.0 to 12.5% polyacrylamide gel and subjected to autoradiography. (B) Pulse-chase analysis of MHV JHM-x-infected cells. Proteins were pulse-labeled with Trans35S-label from 4.5 to 5.0 hpi. Labeling medium was then replaced with complete medium containing 10× methionine and cysteine. Cells were lysed at the chase times indicated, and total lysates were subjected to immunoprecipitation with anti-D14 serum. Products were analyzed on an SDS-7.5 to 12.5% polyacrylamide gel and subjected to autoradiography. Sizes are shown on the left in kilodaltons.
To determine if the p150 protein was a precursor to p22, pulse-chase analysis was performed. After the pulse for 30 min, MHV JHM-x-infected cells were washed with PBS and chased with medium containing excess methionine and cysteine. At the times indicated, cells were harvested and lysates were subjected to immunoprecipitation with anti-D14 serum. We found that the p150 protein was detected immediately upon labeling and that the p22 product was detected starting at 30 min into the chase (Fig. 1B). These data support the idea that p150 is a precursor to p22. Similar results were obtained from pulse-chase analysis of MHV A59-infected cells (data not shown). The processing of p22 from p150 is essentially identical to that in our previous pulse-chase results, showing the release of p27 and p15 (37) and p44 (23) from the p150 precursor in pulse-chase analysis. In each case, the processed product was detected approximately 30 min into the chase, and no stable processing intermediates were detected.
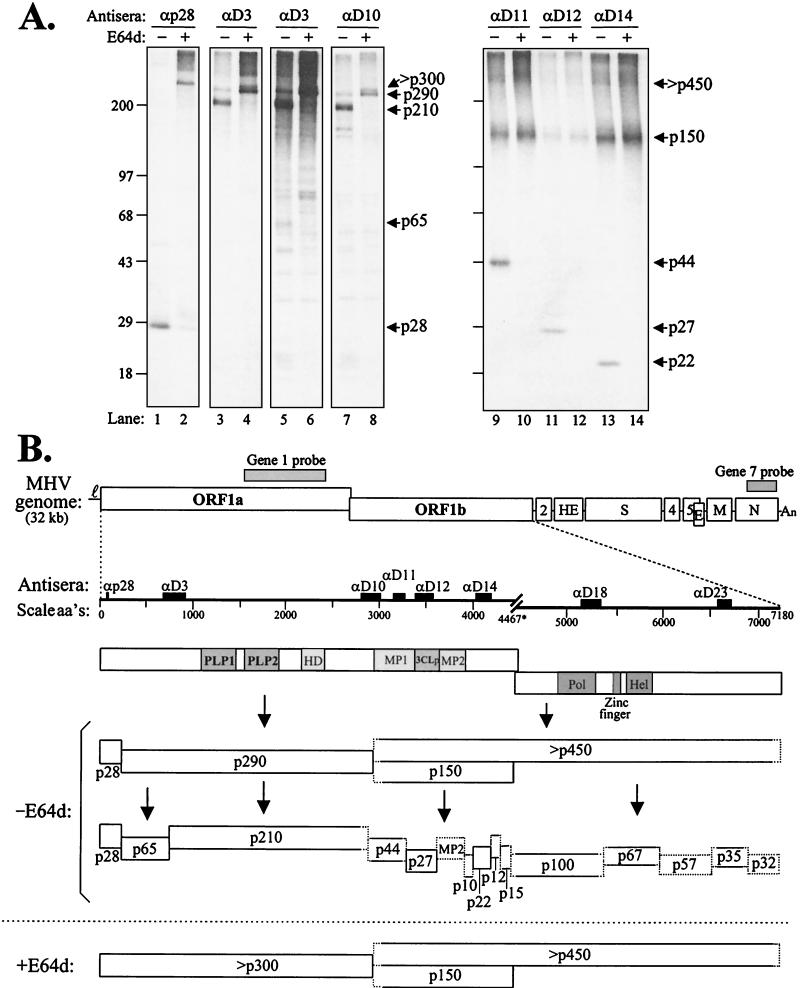
To further elucidate the precursor-product relationship of the MHV ORF1ab polyprotein and cleavage products, we radiolabeled proteins in the presence or absence of the cysteine protease inhibitor E64d (23, 24) and detected MHV-specific products by immunoprecipitation with our panel of previously characterized antisera. Products detected by immunoprecipitation are shown in Fig. 2A, and a schematic diagram of the complex processing of the MHV ORF1ab polyprotein is shown in Fig. 2B. In this experiment, E64d partially blocked PLP1 activity (cleavage of p28 and p65) and 3CLpro activity (processing at 11 sites as indicated in the diagram). As shown previously, E64d did not inhibit PLP2 activity (23). p150 was detected using anti-D11, anti-D12, and anti-D14 in these experiments, consistent with its role as a precursor to proteins p44 (MP1), p27 (3CLpro), p22 (Fig. 2A, lanes 10, 12, and 14), and additional products p10, MP2, p12, and p15 (not shown). We also noted the processing relationship between the p290 and p210 products. Antisera to D3 and D10 detected the p290 precursor in the presence of E64d (Fig. 2A, lanes 4 and 8), indicating that the protein is adjacent to p28 and extends to the PLP2 cleavage site.
FIG. 2.
Identification of polyprotein ORF1ab precursors and products of MHV A59-infected HeLa-MHVR cells labeled in the absence (−) or presence (+) of protease inhibitor E64d. (A) Infected cells were either untreated or treated with 400 μg of protease inhibitor E64d per ml at 3 hpi for 30 min. Then, proteins were labeled with Trans35S-label in the presence or absence of E64d for 2 h. Total lysates were prepared and subjected to immunoprecipitation with the indicated antisera. Products were analyzed by electrophoresis as in Fig. 1. Lanes 5 and 6 provide a longer exposure of the results depicted in lanes 3 and 4. Sizes are shown on the left in kilodaltons. (B) Schematic diagram depicting the MHV A59 genome and our current understanding of the proteolytic processing cascade of the RNA-dependent RNA polymerase (ORF1ab) polyprotein. The locations of antiserum determinants and riboprobes is indicated above the MHV genome. aa's, amino acids.
Next, we wanted to determine which ORF1ab proteins were associated with cellular membranes in a putative RC and whether any MHV product behaves as an integral membrane protein.
MHV ORF1ab products are associated with membranes in infected cells.
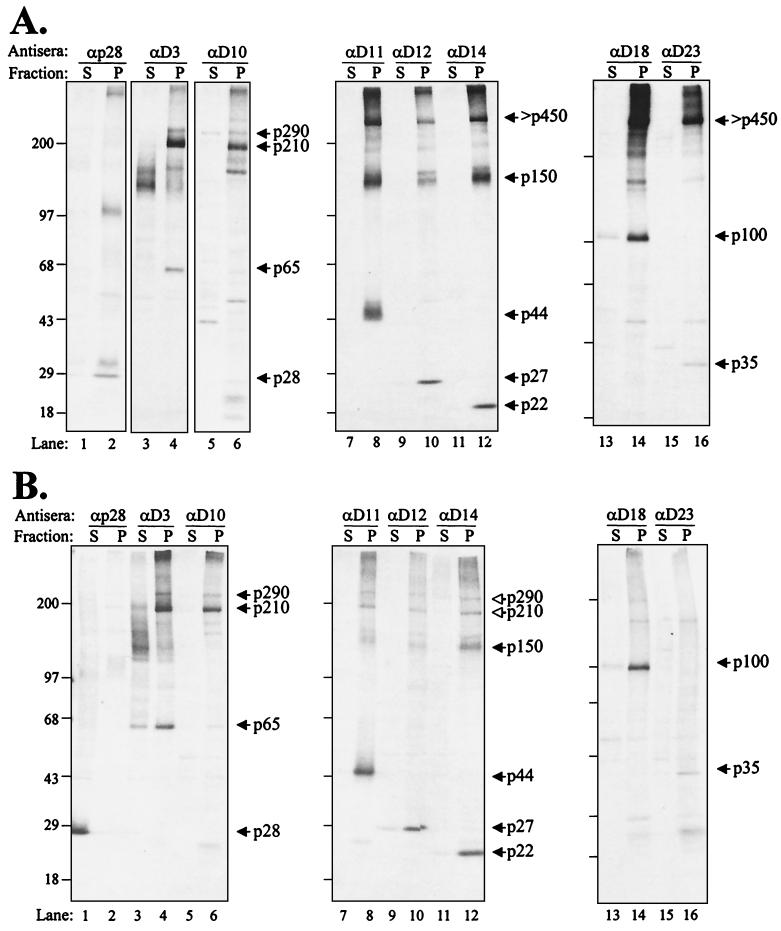
To characterize the MHV RC, we isolated intracellular membranes, which were treated with sodium carbonate at pH 7.0 or 11.0. The latter conditions strip microsomes of their peripheral and luminal proteins (19). Then, the remaining membrane sheets can be pelleted and analyzed for the presence of associated viral proteins. We found that essentially all the MHV ORF1ab products examined were associated with the pelleted membrane fraction at pH 7.0 (Fig. 3A, even-numbered lanes). Precursors like p290 and p150 were detected, and an additional high-molecular-weight protein was seen with antisera directed against the p150 and ORF1b regions. In addition, processed proteins such as p28, p65, p44, p27, p22, p100 (the putative core polymerase), and p35 were all clearly present in the membranous pellet.
FIG. 3.
MHV ORF1ab products detected from the postnuclear supernatant subjected to carbonate buffer at pH 7.0 (A) and pH 11.0 (B) and fractionated by centrifugation. HeLa-MHVR cells were infected with MHV A59, and proteins were metabolically labeled with Trans35S-label from 3.0 to 5.0 hpi. Cells were harvested and broken open with a Dounce cell homogenizer, and a postnuclear supernatant was prepared as described in Materials and Methods. One half of the sample was kept at pH 7.0, while the other half was treated with 100 mM sodium carbonate (pH 11.0) to remove peripheral proteins from the membranes (19). Samples were then subjected to ultracentrifugation to distinguish the soluble (S) and membranous pellet (P)-associated proteins. Soluble and pellet fractions were subjected to immunoprecipitation with anti-ORF1a and -ORF1b sera. Coprecipitating products are indicated by open arrowheads. Products were analyzed as described for Fig. 1. Sizes are shown on the left in kilodaltons.
We also detected some proteins that were smaller than the expected full-size products (for example, several proteins detected with anti-D3 in lane 3 of Fig. 3). These proteins may represent degradation products that partition to the soluble fraction. Interestingly, when the lysates were subjected to treatment with carbonate buffer at pH 11.0 and then pelleted, the majority of the MHV proteins remained associated with the membrane pellet fraction (Fig. 3B). Only p28 (lane 1) and to some extent p65 (lanes 3 and 4) were released from the pelleted complex. The remaining MHV ORF1ab proteins were detected in the pelleted fraction. Similar results were obtained from the analysis of EAV ORF1a- and ORF1b-encoded proteins, with the majority of proteins recovered in the membrane fraction and only minor differences between the pH 7.0 and pH 11.0 treatments (49).
The analysis of MHV proteins resistant to extraction with Triton X-114 was more revealing. The Triton X-114 extraction procedure is widely used to separate proteins based on their hydrophobicity (9). Integral membrane proteins partition into the detergent phase during extraction, whereas cytosolic and membrane-associated proteins remain in the aqueous phase. In the presence of Triton X-114, we found that p28, minor amounts of p65, and the majority of p27 (3CLpro), p22, p100 (the core polymerase), and p35 were extracted from the membrane fraction (Fig. 4). In contrast, MHV proteins containing significant hydrophobic domains, p290 and p210 (containing the HD), p150 (containing MP1 and MP2), and p44 (MP1) were detected in the pelleted detergent fraction. Interestingly, these same products appear to coprecipitate (Fig. 4, as indicated by the open arrowheads), suggesting that the integral membrane proteins remain associated under these conditions.
FIG. 4.
Identification of MHV A59 ORF1a integral membrane proteins from infected-cell lysates treated with Triton X-114. HeLa-MHVR cells were infected with MHV A59, and proteins were metabolically labeled with Trans35S-label from 3.5 to 5.5 hpi. Cells were lysed in PBS containing 1% Triton X-114, and nuclei were removed by low-speed centrifugation. The cytosolic fraction was separated by centrifugation through a 6% (wt/vol) sucrose cushion as described in Materials and Methods. The detergent pellet fraction (P) and the soluble fraction (S) were subjected to immunoprecipitation with the indicated antisera. Products were analyzed as described for Fig. 1. Coprecipitating products are indicated by open arrowheads. Sizes are shown on the left in kilodaltons.
This coprecipitation of p150 with p290/p210 was seen only when using antibodies to the membrane-associated products and thus was specific to the complex. Also, although p150, p210, and p290 appeared to be partially extracted into the soluble fraction, the p44 product (MP1) remained in the pellet (Fig. 4, lane 8). This is the first biochemical evidence of the membrane association of p44. The slightly “fuzzy” appearance of p44 and p150 after extraction may be due to conformation changes that occur during the treatment with Triton X-114. We predict that the putative MP2 protein would also act as an integral membrane protein, but reagents to detect that protein are not yet available. Overall, the extraction results support the hypothesis of an MHV membrane-associated RC with proteins p290, p210, p150, and p44 acting as membrane-associated anchors or scaffolds for the complex. These results are consistent with the finding reported by van der Meer et al. (49) for the identification of membrane-associated anchors for the EAV RC.
MHV induces DMVs in human and mouse cell lines.
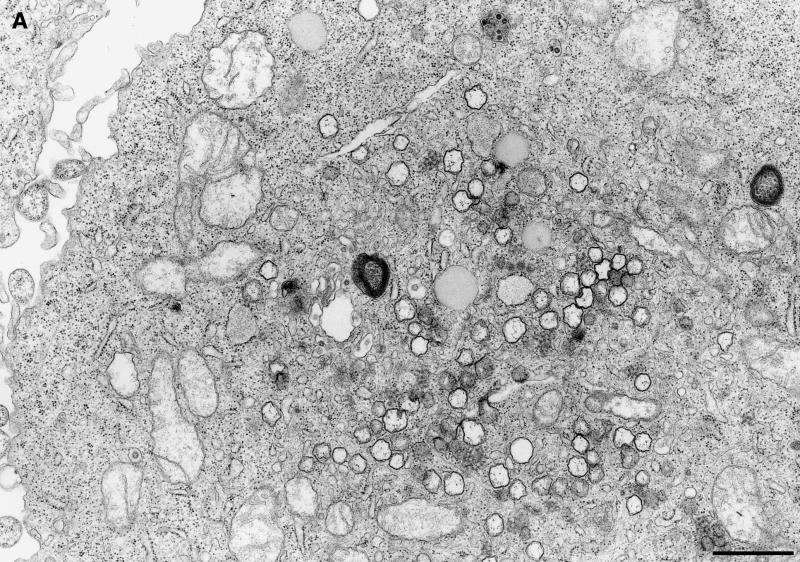
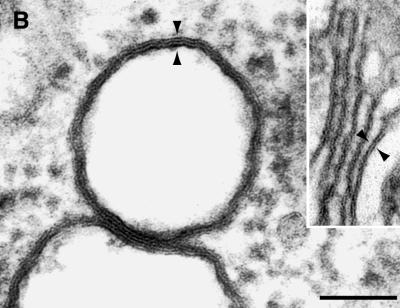
Electron micrographs revealed vesicular structures of 200 to 350 nm in diameter either as separate entities or in small clusters in the cytoplasm of MHV-infected HeLa-MHVR cells (Fig. 5A). Investigating a second MHV-infected cell line, murine 17Cl-1 cells, we found cytoplasmic vesicular structures that were indistinguishable from the vesicles found in MHV-infected HeLa-MHVR cells. The vesicles range in size from 200 to 350 nm and consist of a double membrane (Fig. 5B). Frequently, the two bilayers are fused into one trilayer. In both cell lines, the number of vesicles increased from 5 to 7 hpi (data not shown), the time of peak viral RNA synthesis. The vesicles do not appear to fuse with each other or to uniformly increase in size over time. Such vesicles were not found in mock-infected cells (data not shown). These data indicate that MHV infection induces the formation of vesicular structures in the cytoplasm of human and murine cells.
FIG. 5.
EM analysis of membrane alterations in MHV A59-infected cells. (A) Electron micrograph showing multiple DMVs in the cytoplasm of MHV-infected HeLa-MHVR cells at 5 hpi. Bar, 1 μm. (B) DMVs seen in MHV-infected 17Cl-1 cells at 7 hpi. The double membrane is fused into a trilayer. Inset: For comparison, Golgi apparatus composed of a bilayer membrane. Arrowheads indicate the thickness of the membranes. Bar, 100 nm.
MHV nonstructural proteins and positive-stranded RNA localize to the DMVs.
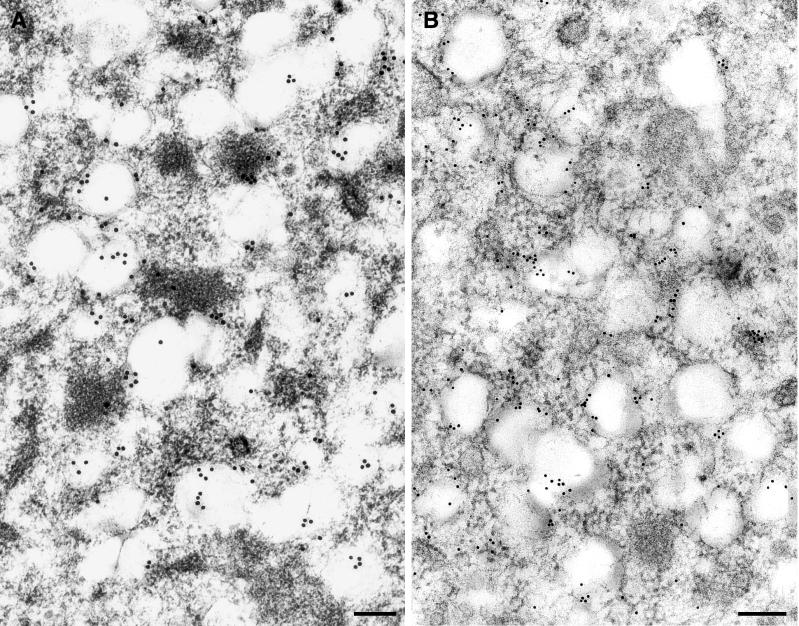
To determine the association of nonstructural proteins with the DMVs, IEM was performed with anti-D3, the only antiserum of a panel of antisera giving clear-cut results on ultrathin sections. Extensive labeling was always found associated with the DMVs on or near the vesicle membranes (Fig. 6A), indicating that ORF1a proteins localize to the DMVs. As shown in Fig. 2A, the anti-D3 polyclonal serum detected p290, p210, and p65. The labeling may represent detection of one or a combination of the proteins seen by immunoprecipitation. We also noted that some label associated with electron-dense regions near the DMVs. It is currently unclear if these areas represent ER in the process of translating the ORF1a polyprotein or, alternatively, misfolded protein that is targeted for degradation instead of incorporation into DMVs (see Discussion).
FIG. 6.
IEM and ISH analysis of MHV proteins and RNA in infected HeLa-MHVR cells at 5 hpi. (A) MHV ORF1a proteins are associated with DMVs. MHV ORF1a proteins were detected using IEM with rabbit anti-D3 serum and anti-rabbit Ig-15-nm gold conjugate. The majority of the protein detected is associated with DMVs, with some label associated with electron-dense regions. (B) MHV RNA is associated with DMVs. RNA was detected using a DIG-labeled riboprobe to MHV gene 7 and 10-nm-gold-tagged anti-DIG antibody. The majority of the RNA is associated with the DMVs, with some RNA labeled in a chain-like pattern. The riboprobe entirely hybridized to the target RNA covers a distance of approximately 160 nm. Bars, 200 nm.
To determine if viral RNA was associated with the DMVs, we performed in situ hybridization using a DIG-labeled MHV-specific RNA probe of negative polarity. This probe detects MHV gene 7 sequences present on the nested set of MHV mRNAs. The specificity of the ISH reaction was confirmed by using the MHV-specific probe on mock-infected HeLa-MHVR cells. This control showed only background labeling (data not shown). As shown in Fig. 6B, MHV-specific, positive-stranded RNA was found by ISH with a high labeling intensity on the DMVs, in some places in a chain-like pattern. The majority of the labeling was detected on or near the DMVs, although some labeling extends beyond the structures themselves.
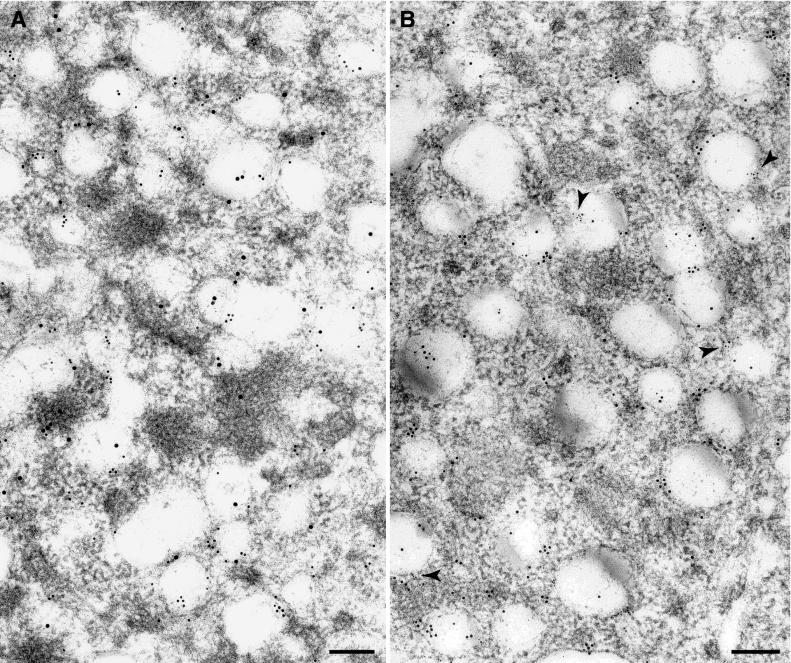
To determine if MHV ORF1a products and MHV mRNAs are associated with the same DMVs, we combined the immunocytochemistry and ISH-EM (Fig. 7A). We found multiple DMVs labeled with both gold-tagged antibodies, supporting the idea that the DMVs contain ORF1a proteins and viral RNA.
FIG. 7.
Combined IEM-ISH and double ISH performed on MHV-infected HeLa-MHVR cells at 5 hpi. (A) MHV ORF1a proteins detected by anti-D3 serum and secondary antibody coupled to 15-nm gold particles, and viral RNA detected by DIG-labeled riboprobe complementary to MHV gene 7 and anti-DIG antibody coupled to 10-nm gold particles localize to the same DMVs. (B) Subgenomic and genomic RNAs exist on the same DMVs. Probe 1, complementary to MHV gene 7 sequences, was detected using 10-nm-gold-tagged sheep anti-DIG antibody. Probe 2, complementary to a region of gene 1 sequences, was detected using 6-nm-gold-tagged antibiotin antibody. Some of the DMVs carrying both labels are indicated by arrowheads. Bars, 200 nm.
To determine whether subgenomic and genomic RNAs were found on the same DMV, we combined the DIG-labeled gene 7 riboprobe (shown by 10-nm gold particles) with a biotin-labeled gene 1 riboprobe (shown by 6-nm gold particles) and performed double in situ hybridization (Fig. 7B). The gene 7 probe detects all subgenomic RNAs and the genome-size RNA, whereas the gene 1 probe is specific for genome-size RNA. If RNA replication and subgenomic transcription take place at the same site, we would expect colocalization of both riboprobes. Double labeling was seen on the DMVs, suggesting that subgenome and genome-size RNAs exist on the same DMV.
This result is not a formal proof for replication and transcription occurring simultaneously on the same DMV because of the 3′ nested character of the MHV RNAs. We cannot exclude the possibility that both riboprobes detected solely genome-size RNA. However, with our double in situ hybridization performed at the peak of subgenomic RNA synthesis, MHV RNA was found exclusively on or in close proximity to the DMVs. Therefore, it is very likely that the DMVs are implicated in both transcription and replication of MHV RNA.
MHV RNA synthesis takes place at DMVs.
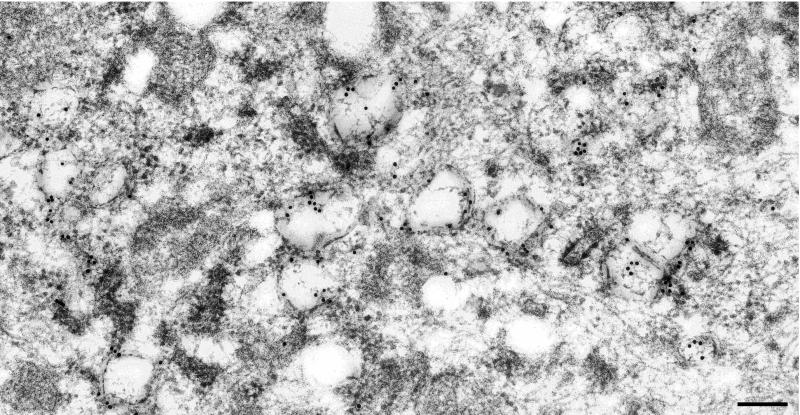
Our results supported the hypothesis that the DMVs contain the ORF1a proteins and viral RNAs, but do not directly address the site of MHV RNA synthesis. To determine if the DMVs were indeed the site of RNA synthesis, viral RNA was metabolically labeled with BrU under conditions in which cellular RNA synthesis was inhibited. Newly synthesized RNA was detected using an antibody that recognizes BrU-RNA in immuno-EM analysis (Fig. 8). BrU-RNA was associated with the DMVs, indicating that they are the sites of MHV RNA synthesis. The labeling was specific, as shown by the low background in the cytoplasm of the cells. The majority of the label was associated with membranes of the DMVs, indicating that the RC of MHV is associated with the membranes of DMVs.
FIG. 8.
Detection of newly synthesized viral RNA in MHV-infected HeLa-MHVR cells at 5 hpi. MHV-infected cells were incubated with BrUTP in the presence of actinomycin D for 1 h to allow incorporation of label into newly synthesized MHV RNA. BrU-RNA was detected using an anti-BrdU antibody and goat anti-mouse Ig antibody coupled to 10-nm gold particles. The BrU-RNA was detected in association with the DMVs. Bar, 200 nm.
DISCUSSION
In this study, we used a panel of polyclonal antisera to MHV gene 1 products to access the composition and site of assembly of the MHV RC. We found that all MHV gene 1 products examined pelleted with cellular membranes (Fig. 3), as expected for a membrane-bound RC. These results are consistent with our previous immunofluorescence studies that showed identical patterns of punctate, perinuclear expression for all the MHV gene 1 products examined (41). We also report that MHV ORF1a polyprotein processing intermediates p290 and p150 and products p210 and p44 (MP1) act as integral membrane proteins. We found that these proteins were not released from membranes by treatment with the detergent Triton X-114 (Fig. 4). These proteins must contain the signals required for insertion into intracellular membranes. Identification and analysis of these signal sequences may help us understand the mechanism used to drive the formation of MHV DMVs.
The ultrastructural studies reported here revealed that infection with MHV induced the formation of new cytoplasmic structures, DMVs. These DMVs were detected in both the human and murine cell lines examined (Fig. 5A and 5B), but not in mock-infected cells. Furthermore, MHV gene 1 products and MHV genome-size RNA detected with riboprobes corresponding to both ends of the genome and thus indicating subgenomic RNA were all associated with these DMVs (Fig. 6 and 7). Most importantly, we showed that newly synthesized viral RNA was detected in association with DMVs (Fig. 8). These data demonstrate that DMVs are the site of MHV RNA synthesis and thus harbor the RC of MHV.
MHV and EAV are united in the order Nidovirales because of their similar strategy of discontinuous viral RNA synthesis. It therefore seems reasonable to hypothesize that the site of assembly for the RC and the site of viral RNA synthesis would be similar if not identical for the two viruses. For EAV, Pedersen and coworkers used ultrastructural studies to show that arterivirus RNA synthesis takes place in association with intracellular, 80- to 100-nm DMVs (32). The EAV DMVs appear to arise by protrusion from the ER with separated inner and outer concentric membranes. In contrast, the MHV DMVs reported here were larger (200 to 350 nm), with tightly apposed membranes forming a lipid trilayer (Fig. 5B). These morphological differences in the EAV and MHV DMVs may reflect differences in replication of a 12.5- versus 31.2-kb genomic RNA or differences in recruiting membranes for the formation of the RC. Indeed, previous immunofluorescence studies using ER and Golgi markers indicated that MHV may generate DMVs from different compartments in different cell lines (41). Currently, it is not clear if membrane abundance or specific host factors contribute to preferential membrane utilization in these cell lines. The compartment(s) from which the MHV DMVs originate needs to be determined.
Our findings using riboprobes to detect MHV gene 1 and gene 7 sequences suggest that the DMVs harbor the RC that mediates both genome replication and subgenomic mRNA transcription. However, we cannot formally exclude the possibility that transcription of subgenomic RNA and replication of genome-size RNA occur separately on different DMVs. We saw several examples of “chaining” ISH signals, corresponding to the 500-nt riboprobe piece hybridized to the target viral RNA. Analysis at 5 hpi, the time of peak mRNA synthesis, revealed that the majority of RNA detected by both the gene 1 and 7 probes was associated with the DMVs. Thus, it is likely that both genomic and subgenomic RNAs are present on the same DMVs. These results are consistent with the observations of Sethna and Brian, who showed that both genomic and subgenomic RNAs are present in the same membrane fraction from MHV-infected cells (38).
Expression studies have shown that EAV ORF1a products nsp2 to nsp7 were sufficient for the generation of DMVs (32). This work was extended to show that coexpression of EAV nsp2 and nsp3 was sufficient for the induction and formation of DMVs (44). The MHV counterparts to nsp2 and nsp3 are p210 and p44, the proteins that we found to be integral membrane proteins. It will be interesting to determine if these proteins are sufficient for the formation of MHV DMVs, membrane recruitment, and the order of MHV RC assembly.
In contrast to our results, van der Meer and coworkers reported that late endosomes were the site of MHV RNA synthesis (48). A possibility for the differences between our results and those reported by van der Meer et al. is a difference in the labeling intensity of MHV replicase products and viral RNA. We found a higher intensity of both replicase protein and viral RNA labeling associated with the DMVs, facilitating the identification of DMVs as the site of RC assembly and viral RNA synthesis. Second, it is possible that some gene 1 products are not incorporated into DMVs; instead, some protein products may be shuttled to endosomes and degraded. The association of MHV replicase products with endocytic vesicles may reflect the consequences of aberrant assembly or turnover of the components of the RC. Further investigation will be required to determine if endosomes are indeed involved in some aspect of MHV replication.
Studies of the RC of poliovirus have shown that it is built up from individual membranous vesicles. The vesicles become tightly associated and assemble into higher-order structures (5). The formation of the poliovirus RC occurs in cis and allows essentially no rescue of lethal mutations by trans complementation (17, 46). In contrast, the DMVs harboring the RC of MHV present themselves as loosely arranged vesicles (Fig. 5). The formation of such structures might facilitate the replication of a 31.2-kb genome and raises several questions regarding the assembly and stability of the RC of MHV. Do gene 1 products act in trans as transcription factors for mRNA synthesis? Do some components of the RC remain associated with the nascent RNA and facilitate “jumping” to another RC-associated template? Alternatively, during discontinuous transcription, does the nascent RNA dissociate from the membrane-bound RC? What are the functions of the many processed products of the MHV gene 1 polyprotein?
Deletion analysis of full-length clones of EAV and human coronavirus has shown that expression of gene 1 is sufficient for genome replication and discontinuous mRNA transcription (31, 47). Thus, gene 1 products are capable of directing the localization and assembly of the viral RC and recruiting host factors required for viral RNA synthesis. For example, coprecipitation studies using the anti-D14 serum revealed that heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) is a host factor recruited into the MHV RC (40). This antiserum may be useful for detecting host factors that replace hnRNPA1 in cell lines in which it is not expressed (39). It may also be that some MHV gene 1 products are required to recruit or maintain other subunits in the RC. It was recently shown that the brome mosaic virus helicase-like 1a protein is required to direct the polymerase-like 2a protein to the endoplasmic reticulum to form the brome mosaic virus RC (12). However, it is currently unclear if the MHV RC assembles strictly in cis, or if trans association of subunits is permitted.
Experiments designed to determine the minimal regions required for MHV RC formation are in progress. Once such “scaffolding” components are identified, it may be possible to express MHV gene 1 products in trans and to determine if assembly and complementation assays can be used to identify the functions of the multitude of MHV gene 1 products. A better understanding of the mechanisms used to localize and assemble viral RCs may help us unravel the complex replication strategy of MHV, which is particularly challenging in view of the extreme size of the MHV genome.
Acknowledgments
We thank John Zaryczny for assistance with rabbit injection and serum collection.
This work was supported by Public Health Service Research Grant AI 45798 to S.C.B. and grant 31-055397.98/1 from the Swiss National Foundation to K.B. R.G. was supported in part by grants from the Novartis Stiftung and the Freiwillige Akademische Gesellschaft, Basel, Switzerland.
REFERENCES
- 1.Baker, S. C., C.-K. Shieh, L. H. Soe, M.-F. Chang, D. M. Vannier, and M. M. C. Lai. 1989. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 63:3693-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, W., J. D. Pinon, S. Hughes, P. J. Bonilla, K. V. Holmes, S. R. Weiss, and J. L. Leibowitz. 1998. Localization of mouse hepatitis virus open reading frame 1a derived proteins. J. Neurovirol. 4:594-605. [DOI] [PubMed] [Google Scholar]
- 3.Bienz, K., and D. Egger. 1995. Immunocytochemistry and in situ hybridization in the electron microscope: combined application in the study of virus-infected cells. Histochem. Cell Biol. 103:325-338. [DOI] [PubMed] [Google Scholar]
- 4.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 5.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonilla, P. J., A. E. Gorbalenya, and S. R. Weiss. 1994. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology 198:736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonilla, P. J., S. A. Hughes, J. D. Pinon, and S. R. Weiss. 1995. Characterization of the leader papain-like proteinase of MHV A59: identification of a new in vitro cleavage site. Virology 209:489-497. [DOI] [PubMed] [Google Scholar]
- 8.Bonilla, P. J., S. A. Hughes, and S. R. Weiss. 1997. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J. Virol. 71:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 10.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brierley, I., M. E. G. Boursnell, M. M. Binns, B. Bilimoria, V. C. Blok, T. D. K. Brown, and S. C. Inglis. 1987. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 6:3779-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox, K. H., D. V. DeLeon, L. M. Angerer, and R. C. Angerer. 1984. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev. Biol. 101:485-502. [DOI] [PubMed] [Google Scholar]
- 14.Denison, M. R., S. A. Hughes, and S. R. Weiss. 1995. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV A59. Virology 207:316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison, M. R., W. J. M. Spaan, Y. van der Meer, C. A. Gibson, A. C. Sims, E. Prentice, and X. T. Lu. 1999. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 73:6862-6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois-Dalcq, M., K. V. Holmes, and B. Rentier. 1984. Assembly of Coronaviradae, p. 99-216. In D. W. Kingsbury (ed.), Assembly of enveloped RNA viruses. Springer-Verlag, New York, N.Y.
- 17.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallagher, T. M. 1996. Murine coronavirus membrane fusion is blocked by modification of thiol buried within the spike protein. J. Virol. 70:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya, A. E., E. V. Koonin, and M. M. C. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses: identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirano, N., T. Murakami, K. Fujiwara, and M. Matsumoto. 1978. Utility of mouse cell line DBT for propagation and assay of mouse hepatitis virus. Jpn. J. Exp. Med. 48:71-75. [PubMed] [Google Scholar]
- 23.Kanjanahaluethai, A., and S. C. Baker. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 74:7911-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J. C., R. A. Spence, P. F. Currier, X. Lu, and M. R. Denison. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology 208:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, H.-J., C.-K Shieh, A. E. Gorbalenya, E. V. Koonin, N. La Monica, J. Tuler, A. Bagdzyadzhyan, and M. M. C. Lai. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180:567-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, X., Y. Lu, and M. R. Denison. 1996. Intracellular and in vitro-translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV A59. Virology 222:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, X. T., A. C. Sims, and M. R. Denison. 1998. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J. Virol. 72:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, Y., X. Lu, and M. R. Denison. 1995. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV A59. J. Virol. 69:3554-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magliano, D., J. A. Marshall, D. S. Bowden, N. Vardaxis, J. Meanger, and J.-Y. Lee. 1998. Rubella virus replication complexes are virus-modified lysosomes. Virology 240:57-63. [DOI] [PubMed] [Google Scholar]
- 31.Molenkamp, R., H. van Tol, B. C. D. Rozier, Y. van der Meer, W. J. M. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 32.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinon, J. D., H. Teng, and S. R. Weiss. 1999. Further requirements for cleavage by the murine coronavirus 3C-like proteinase: identification of a cleavage site within ORF1b. Virology 263:471-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H.-P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in the production of vesicles forming the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiller, J. J., A. Kanjanahaluethai, and S. C. Baker. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sethna, P. B., and D. A. Brian. 1997. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J. Virol. 71:7744-7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen, X., and P. S. Masters. 2001. Evaluation of the role of heterogeneous nuclear ribonucleoprotein A1 as a host factor in murine coronavirus discontinuous transcription and genome replication. Proc. Natl. Acad. Sci. USA 98:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi, S. T., P. Huang, H.-P. Li, and M. M. C. Lai. 2000. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 19:4701-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J.-W. Oh, and M. M. C. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 73:5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skinner, M. A., and S. G. Siddell. 1983. Coronavirus JHM: nucleotide sequence of the mRNA that encodes nucleocapsid protein. Nucleic Acids Res. 11:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snijder, E. J., and J. J. M. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 44.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Nonstructural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 45.Sturman, L. S., and K. K. Takemoto. 1972. Enhanced growth of a murine coronavirus in transformed mouse cells. Infect. Immun. 6:501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teterina, N. L., W. D. Zhou, M. W. Cho, and E. Ehrenfeld. 1995. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J. Virol. 69:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thiel, V., J. Herold, B. Schelle, and S. S. Siddell. 2000. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 75:6676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. M. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Meer, Y., H. van Tol, J. K. Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]