Abstract
Vesicular stomatitis virus (VSV) infection depends on the fusion of viral and cellular membranes, which is mediated by virus spike glycoprotein G at the acidic environment of the endosomal compartment. VSV G protein does not contain a hydrophobic amino acid sequence similar to the fusion peptides found among other viral glycoproteins, suggesting that membrane recognition occurs through an alternative mechanism. Here we studied the interaction between VSV G protein and liposomes of different phospholipid composition by force spectroscopy, isothermal titration calorimetry (ITC), and fluorescence spectroscopy. Force spectroscopy experiments revealed the requirement for negatively charged phospholipids for VSV binding to membranes, suggesting that this interaction is electrostatic in nature. In addition, ITC experiments showed that VSV binding to liposomes is an enthalpically driven process. Fluorescence data also showed the lack of VSV interaction with the vesicles as well as inhibition of VSV-induced membrane fusion at high ionic strength. Intrinsic fluorescence measurements showed that the extent of G protein conformational changes depends on the presence of phosphatidylserine (PS) on the target membrane. Although the increase in PS content did not change the binding profile, the rate of the fusion reaction was remarkably increased when the PS content was increased from 25 to 75%. On the basis of these data, we suggest that G protein binding to the target membrane essentially depends on electrostatic interactions, probably between positive charges on the protein surface and negatively charged phospholipids in the cellular membrane. In addition, the fusion is exothermic, indicating no entropic constraints to this process.
Entry of enveloped animal viruses into their host cells always involves a step of membrane fusion, which is mediated by viral envelope glycoproteins (20, 23, 51). Two general mechanisms have been defined for the fusion reaction: (i) surface fusion between the viral envelope and host cell plasma membrane and (ii) fusion of the endosomal membrane with the viral envelope after virus particle internalization by receptor-mediated endocytosis. In the first case, a well-characterized fusion mechanism is that mediated by the human immunodeficiency virus (HIV) gp120 and gp41 glycoproteins (7). Interaction between gp120, the cellular CD4 molecule, and a coreceptor protein leads to the insertion of the gp41 hydrophobic fusion peptide into the plasma membrane. gp41 forms a trimeric coiled-coil containing two interacting α-helical peptides that acquire a six-helix bundle structure (8, 28).
Fusion at the endosome is triggered by conformational changes in viral glycoproteins induced by the low pH of this cellular compartment. The best-studied low-pH-activated viral fusion protein is the influenza virus glycoprotein hemagglutinin (HA). The X-ray structure of influenza virus HA was determined at both neutral and fusogenic pHs (5, 53). The conformational changes observed suggest that the hydrophobic fusion peptide moves to the tip of the molecule and is delivered toward the target membrane (5). The conformational transition occurs within a narrow pH range, corresponding to the optimal pH of fusion, in which the protein acquires the ability to interact with detergent micelles and lipid vesicles (46). This interaction leads to the insertion of the fusion peptide into the membrane, where a pore is formed (4, 48).
Vesicular stomatitis virus (VSV) enters the cell by endocytosis, followed by low-pH-induced membrane fusion mediated by its spike glycoprotein, named the G protein (12, 30). This protein is a trimeric type I glycoprotein of 67 kDa, which is anchored in the viral membrane via a single transmembrane anchor sequence close to the C terminus (39). Unlike most viral fusion proteins, VSV G protein does not contain an apolar amino acid sequence similar to the fusion peptides (23). Most of the studies on G protein-mediated fusion have focused on the description of the amino acids important for the low-pH-induced conformational change (9, 19, 45, 56). We have recently shown that, at the fusogenic pH, a dramatic conformational change on VSV G protein takes place, including loss and reorganization of its secondary and tertiary structures (6). Our results also indicated that the G protein interacts with target membranes through the formation and/or exposure of a hydrophobic domain at pHs close to 6.0, although the mechanism and the nature of protein-lipid interactions during fusion still remain unclear.
Here we describe a study of VSV-membrane interaction by force spectroscopy, isothermal titration calorimetry (ITC), and fluorescence spectroscopy. We show that VSV-membrane interactions as well as the fusion reaction mediated by the virus are highly dependent on the presence of negative charges on the vesicle surface. In addition, both VSV binding to phospholipid vesicles and the VSV-induced membrane fusion are enthalpy-driven reactions, suggesting the involvement of electrostatic interactions. This was confirmed by the lack of binding and fusion reaction at high ionic strength. Our results show that, although we cannot discard hydrophobic contributions in both processes, VSV interaction with the target membrane is probably driven by electrostatic interactions and H-bond formation.
MATERIALS AND METHODS
Virus propagation and purification.
VSV strain Indiana was propagated in monolayer cultures of BHK-21 cells. The cells were grown at 37°C in roller bottles containing 150 ml of Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Cultilab, Campinas, SP, Brazil), 100 μg of ampicillin, and 5 μg of gentamicin per ml. When the cells reached confluence, the medium was removed, and the cell monolayer was infected with VSV at a multiplicity of 5 PFU/ml. The cultures were kept at 37°C for 16 to 20 h, and the virus were harvested and purified by differential centrifugation, followed by equilibrium sedimentation in a sucrose gradient as described elsewhere (13). Purified virions were stored at −70°C.
Preparation of liposomes.
Phospholipids were dissolved in chloroform and evaporated under nitrogen. The lipid film formed was resuspended in 20 mM MES (morpholineethanesulfonic acid)-30 mM Tris buffer (pH 7.5 or 6.0) at a final concentration of 1 mM. The suspension was vortexed vigorously for 5 min. Small unilamellar vesicles were obtained by sonicating the turbid suspension with a Branson Sonifier (Sonic Power Company, Danbury, Conn.) equipped with a titanium microtip probe. Sonication was performed in an ice bath, alternating cycles of 30 s at 20% full power with 60-s resting intervals until a transparent solution was obtained (approximately 10 cycles). The vesicles used in this study were composed of phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) at a 3:1:1 ratio, with 10% cholesterol; PC and PS at different ratios, as indicated in the figure legends; PC and cardiolipin (CL), 3:1; and PC only. For fusion assays, 1% 1-hexadecanoyl-2-(1-pyrenedecanoyl)-sn-glycero-3-phosphocholine (10-PyPC; Molecular Probes Inc., Eugene, Oreg.) was incorporated in PC-PS vesicles by vortexing for 10 min. Phospholipids were purchased from Sigma Chemical Co.
Atomic force microscopy.
The atomic force microscope used in this work was built in collaboration with the Ludwig-Maximilians-Universität Lehrstuhl für Angewandte Physik in Munich, Germany. For all the experiments, the atomic force microscope was used in force spectroscopy mode (18, 21, 58). Mica coverslips were glued to magnetic stainless steel punches and mounted in a fluid cell without using the O-ring. The mica surfaces were incubated with vesicles before transfer to the fluid cell (25, 35). Since the presence of calcium ions appears to facilitate as well as to increase the rate of planar membrane formation from vesicles (34, 38), mica surfaces were incubated with 20 μl of the vesicle suspension, containing 1 mM phospholipids, plus 10 μl of 20 mM MES-30 mM Tris buffer, pH 7.4, containing 1 mM CaCl2, for approximately half an hour at room temperature (25 ± 0.5°C). After incubation, the slips were washed repeatedly with the same buffer used to prepare vesicles.
All experiments were performed at room temperature using standard V-shaped cantilevers, containing a silicon nitride tip with a 4-μm2 pyramidal base (Digital Instruments Inc.). The cantilevers have a spring constant of 0.06 N/m (manufacturer's data) and were incubated with VSV as follows. The cantilevers were immersed in a virus suspension (total protein concentration, 0.28 mg/ml) for 24 h at 4 to 6°C. The instrument allows the performance of “approach-retraction” cycles, in which the maximal contact force, interaction time and the approach-retracting rates can be controlled independently. The maximal force was limited to approximately 3 nN, the interaction time was set to zero, and the approach-retracting rate was set to 7,500 nm/s.
Calorimetric measurements.
The binding of VSV to lipid vesicles and the membrane fusion mediated by the G protein were studied at 35°C in an MCS-ITC microcalorimeter from MicroCal, Llc. (Northampton, Mass.). The implementation of ITC was previously described by Wiseman et al. (54). For the binding experiments, the samples were prepared at pH 7.5, and after equilibration at 35°C, several preparations (2 to 10 μl each) of a solution containing virus (28 μg of protein/ml) were injected into the cell (volume = 1.38 ml) containing the vesicles. The heat of dilution of the virus was measured by injecting the same solution of VSV into buffer only. The calorimetric thermograms (δ Q/δ t as a function of time) were analyzed by integrating the area under each peak to determine the heat (Q) of injection.
For the membrane fusion experiments, the protein concentration in the VSV sample was 10-fold higher than that used for binding, and after a single 10-μl injection the fusion process was followed for 30 min. The experiments were done at pH 6.0 and pH 7.5, and the data were analyzed by integrating the calorimetric thermogram in order to obtain the heat released (−Q) as a function of time, which allows the analysis of the kinetics of fusion. Due to the high concentration of the virus suspension, the heat of VSV dilution was very intense, making it difficult to subtract from the raw data for fusion. In this case, the data were analyzed after the heat effect for the VSV dilution. In all the ITC experiments, the syringe was rotated at 400 rpm. The samples were degassed under vacuum prior to the titration. The data were analyzed with the Origin 5.0 software provided by MicroCal. The changes in enthalpy and entropy for the association (ΔHass and the ΔSass, respectively) were calculated according to Hyre and Spicer (24).
Intrinsic fluorescence measurements.
G protein conformational changes during VSV interaction with membranes of different phospholipid composition were monitored by the changes in virus intrinsic fluorescence. VSV (final protein concentration, 70 μg/ml) was incubated with a liposome suspension containing 1 mM phospholipid in 20 mM MES-30 mM Tris buffer, pH 6.0. Intrinsic fluorescence data were recorded using a Hitachi F-4500 fluorescence spectrometer, exciting the samples at 280 nm and collecting emissions between 300 and 420 nm.
Liposome fusion assay.
A suspension of liposomes of different phospholipid compositions containing equal amounts of unlabeled vesicles and vesicles labeled with 10-PyPC were prepared in 20 mM MES-30 mM Tris buffer, pH 6.0 or 7.5, with a final phospholipid concentration of 0.1 mM. The emission spectrum of pyrene-labeled vesicles exhibited a broad excimer fluorescence peak with maximal intensity at 480 nm and two sharp peaks at 376 and 396 nm due to monomer fluorescence emission (not shown). The fusion reaction was initiated by addition of purified VSV. Fusion was followed by the decrease in the 10-PyPC excimer/monomer fluorescence intensity ratio, which was measured by exciting the sample at 340 nm and collecting the fluorescence intensities of excimer and monomer at 480 and 376 nm, respectively.
RESULTS
VSV binding to membranes.
Although the precise cellular receptor for VSV is still unknown, the finding that PS specifically inhibits VSV cell binding and infectivity suggested that PS is at least an important component of the VSV binding site (43). To study VSV binding to PS, we evaluated the interaction between the virus and membranes of different phospholipid composition using force spectroscopy (Fig. 1). This technique allows the direct determination of ligand-receptor interactions by measuring rupture forces between the cantilever and the surface, each of them covered with the molecules of interest.
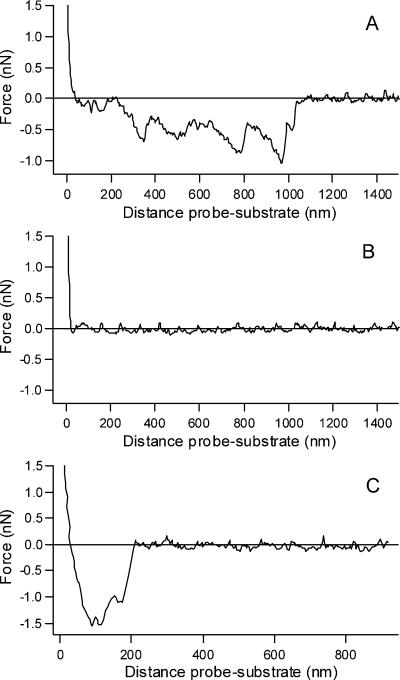
FIG. 1.
Force-distance curves for VSV interaction with membranes. Force-distance curves were recorded on lipid-covered mica substrates. Retracting curves were obtained with VSV adsorbed on the tip and mica substrates covered with PC-PS (3:1) (A), PC only (B), or PC-CL (3:1) (C). The negative values for the force peaks in panels A and C indicate adhesion and are absent in panel B. Data were collected in 20 mM MES-30 mM Tris, pH 7.5, at room temperature.
For this study, we chose three different membrane compositions: PC-PS (3:1), PC only, and PC-CL (3:1). VSV was adsorbed on the cantilever, and several approach-retraction cycles were performed at a fixed rate of 7,500 nm/s. The contact between the tip containing adsorbed VSV and the PC-PS surface gave rise to force-distance curves with negative peaks, indicative of adhesion (Fig. 1A). At least 200 curves could be collected with the same tip. In order to ensure the reproducibility of the data, six to eight curves were collected with several tips and substrate.
Table 1 summarizes the values of the force of the adhesion peaks obtained with six different cantilevers with VSV adsorbed and three mica surfaces covered with PC-PS (3:1). The mean adhesion force was 690 pN, with a variation range of 200 to 1,920 pN. Most of the curves present peaks occurring from 200 nm onwards, which corresponds to the virus length. This result suggests that the whole virus particle bridges the tip and the lipid film. No interaction between the virus and the membrane of PC only was observed (Fig. 1B). Of 120 curves obtained with mica substrate covered with 100% PC, only two showed a single small adhesion peak at 293 and 310 pN, both located very close to the surface, indicating nonspecific interactions. Again, to ensure reproducibility, the experiments were also performed using different tips and substrates, and the same results were obtained. No interaction was observed between bare tips and mica surfaces covered with PC or PC-PS membranes (not shown).
TABLE 1.
Force of adhesion peaks obtained with VSV and PC-PS (3:1) films on mica
| Expt | Force (pN)
|
|||||
|---|---|---|---|---|---|---|
| Substratea 1
|
Substrate 2
|
Substrate 3
|
||||
| Cantb 1 | Cant 2 | Cant 3 | Cant 4 | Cant 5 | Cant 6 | |
| 1 | 1,240 | 1,580 | 500 | 1,280 | 552 | 957 |
| 2 | 540 | 1,920 | 425 | 980 | 332 | 717 |
| 3 | 1,100 | 770 | 416 | 1,171 | 354 | 587 |
| 4 | 402 | 825 | 200 | 810 | 420 | 761 |
| 5 | 405 | 875 | 670 | 259 | 464 | 521 |
| 6 | 503 | 927 | 667 | 301 | 266 | 518 |
| 7 | 450 | 1,560 | 302 | 345 | ||
| 8 | 410 | |||||
Substrate, mica surface covered with membranes.
Cant, cantilever.
In order to verify whether the interaction observed in force spectroscopy experiments was due to a specific binding between PS and VSV or due to an electrostatic interaction between positive charges in G protein and negatively charged phospholipids, we substituted PS with another negatively charged phospholipid, CL (Fig. 1C). The force-distance curve presented in Fig. 1C is representative of several experiments and showed a strong interaction between the virus and PC-CL membranes, as found for PC-PS membranes. The mean adhesion force was 1,500 pN, ranging from 620 to 2,900 pN. Multiple peaks were also obtained. This result suggests that, rather than being specific to PS, the interaction between VSV and the membranes probably depends on the presence of negatively charged phospholipids.
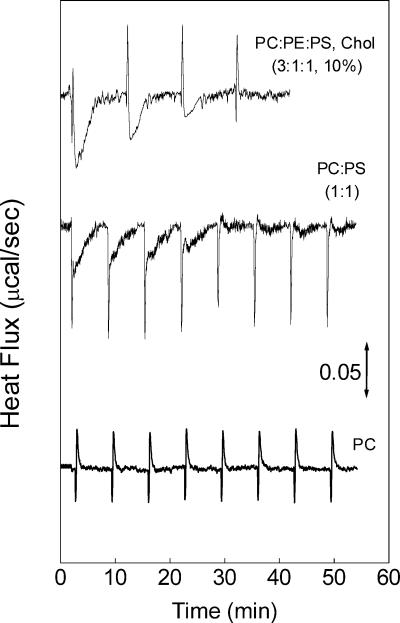
The interaction between VSV and vesicles of different phospholipid compositions was also studied by ITC at pH 7.5, at which G protein-induced membrane fusion is negligible (33, 52). As shown in the calorimetric traces in Fig. 2, each injection of VSV into vesicles of PC-PE-PS the cholesterol (3:1:1 and 10%) or PC-PS (1:1) results in a two-component reaction, one sharp exothermic peak followed by a broader exothermic component. A control experiment done by injecting VSV into buffer showed that the sharp peaks were due to the heat of dilution of the virus suspension. Injection of buffer only into a cell containing vesicles gave rise to negligible heat effects and was not considered for subtraction. On the other hand, VSV injection into PC vesicles gave rise to sharp peaks similar to those observed in the control experiment of virus dilution (Fig. 2). Since no binding was observed with PC vesicles, the broader exothermic component was related to the binding of VSV to the PS-containing membranes. Several injections of VSV were done until saturation.
FIG. 2.
Calorimetric measurement of VSV binding to liposomes at 35oC. Typical calorimetric traces (heat flow as a function of time) obtained for four to eight injections (5 μl each) of a VSV suspension (28 μg/ml) into the cell containing unilamellar vesicles of PC-PE-PS with cholesterol (3:1:1 and 10%), PC-PS (1:3), or PC only, in 20 mM MES-30 mM Tris, pH 7.5, at 35°C. The sharp peaks are due to the VSV dilution, as seen in control experiments of the injection of virus into buffer (not shown). The phospholipid concentration was 1 mM.
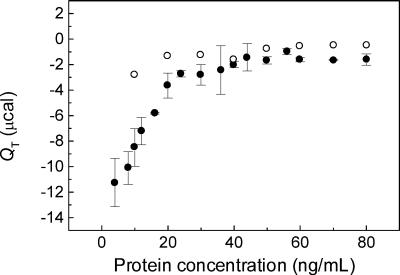
The plot of the total heat (QT) calculated for each peak shows that similar binding isotherms can be obtained with vesicles containing 25, 50, or 75% PS (Fig. 3). The exothermic nature of the binding indicates that this is an enthalpically driven reaction, probably derived from electrostatic interactions, as suggested by the need for negatively charged lipids for the binding to occur (Fig. 1). The QT calculated from the injections of VSV into PC-only vesicles was negligible in comparison with those obtained with vesicles containing PS (Fig. 3).
FIG. 3.
Binding isotherms. The total heat (QT) was calculated for each peak of the calorimetric thermograms resulting from the injection of VSV into vesicles (see Fig. 1). QT is plotted as a function of the protein concentration in each injection, with the mean ± standard error (SE) for five different experiments with PC-PS (•) and the mean of two experiments with PC-only vesicles (○) obtained with the same VSV preparation. The data were essentially the same for the PC-PS vesicles containing 25, 50, or 75% PS. The conditions were the same as in Fig. 1. Bar, 0.05 μcal s−1.
Interactions between G protein and PS drive the conformational changes involved in membrane fusion.
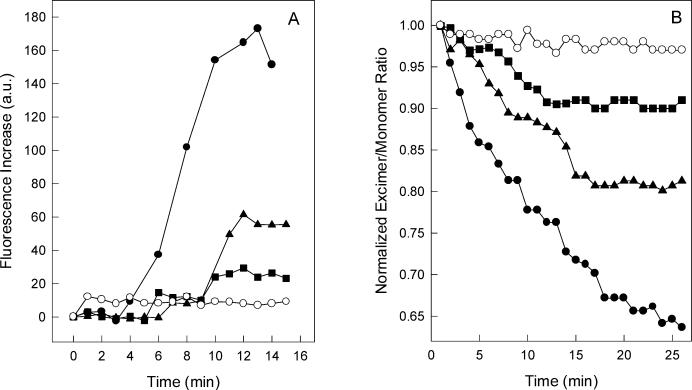
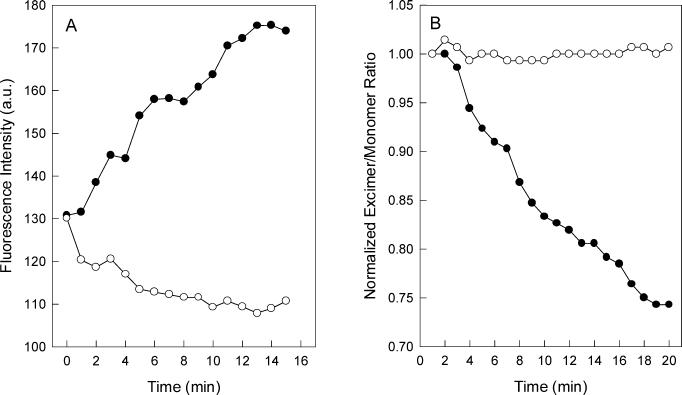
It is well established that VSV induces membrane fusion at acidic pH. The fusion reaction depends on VSV G protein and was characterized by using isolated virus to promote fusion of model cells or liposomes in vitro (33, 52). We have recently shown that G protein interaction with liposomes at pH 6.0 resulted in dramatic protein conformational changes, which can be followed by intrinsic fluorescence (6). In the presence of vesicles composed of PC and PS, a great increase in the tryptophan fluorescence of G protein occurred upon acidification of the medium, while a pH decrease led to intrinsic fluorescence quenching in the absence of liposomes (6). The time course of fluorescence increase after VSV incubation with liposomes of different PS content, at pH 6.0, is shown in Fig. 4A. The extent of fluorescence increase was strongly dependent on the amount of PS in the vesicle, and no increase in fluorescence was observed when the experiment was performed with vesicles of PC only. This result indicates that the G protein conformational changes that take place during protein-lipid interaction are mediated by VSV binding to PS at acidic pH, suggesting the requirement for negative charges in the membrane surface.
FIG. 4.
VSV G protein conformational change during virus incubation with vesicles of different phospholipid compositions. (A) Intrinsic fluorescence of VSV was recorded after virus incubation with small unilamellar vesicles of PC-PS (1:3) (•), PC-PS (1:1) (▴), PC-PS (3:1) (▪), and PC only (○). The vesicles were prepared in 20 mM MES-30 mM Tris buffer, pH 6.0, in a final phospholipid concentration of 0.1 mM. The excitation wavelength was 280 nm, and the emission was collected at 334 nm. The final protein concentration was 70 μg/ml. (B) Purified virus was added to a sample containing equal amounts of unlabeled vesicles and vesicles labeled with 10-PyPC. VSV-induced membrane fusion was measured by the decrease in the 10-PyPC excimer/monomer fluorescence intensity ratio. Vesicles used were PC-PS (1:3) (•), PC-PS (1:1) (▴), and PC-PS (3:1) (▪) at pH 6.0 and PC-PS (1:3) at pH 7.5 (○). 10-PyPC was excited at 340 nm, and the intensities were collected at 480 and 376 nm for the excimer and monomer, respectively. Experimental conditions were the same as described in the legend to panel A.
VSV-induced liposome fusion can be quantified by measuring the decrease in pyrene phospholipid excimer fluorescence (33). The ability of VSV to mediate fusion was highly dependent on PS content in the liposome (Fig. 4B), suggesting that interactions between G protein and negatively charged phospholipids are also involved in the fusion reaction.
In order to investigate the role of electrostatic interactions in G protein conformational changes during membrane fusion, we evaluated the effect of high ionic strength on G protein conformational changes and VSV fusion activity during incubation of the virus with PC-PS liposomes at pH 6.0. The kinetics of increase in VSV intrinsic fluorescence was followed in the presence of 250 mM KCl (Fig. 5A). At this salt concentration, the interaction between VSV and the vesicles was completely abolished. In addition, VSV-induced membrane fusion was also inhibited at this ionic strength (Fig. 5B). Taken together, these results corroborate the electrostatic nature of G protein-lipid interactions during VSV-induced membrane fusion.
FIG. 5.
Effect of high ionic strength on VSV G protein conformational changes during interaction with liposomes. (A) Intrinsic fluorescence of VSV was recorded after virus incubation with vesicles composed of PC-PS (1:3) in the absence (•) and in the presence (○) of 250 mM KCl. The vesicles were prepared in 20 mM MES-30 mM Tris buffer, pH 6.0, in a final phospholipid concentration of 0.1 mM. The excitation wavelength was 280 nm, and the emission was collected at 334 nm. The final protein concentration was 70 μg/ml. (B) VSV-induced membrane fusion measured as in Fig. 4 after virus incubation with vesicles composed of PC-PS (1:3) in the absence (•) and in the presence (○) of 250 mM KCl. Experimental conditions were the same as described in the legend to panel A.
Calorimetric studies of VSV-induced membrane fusion.
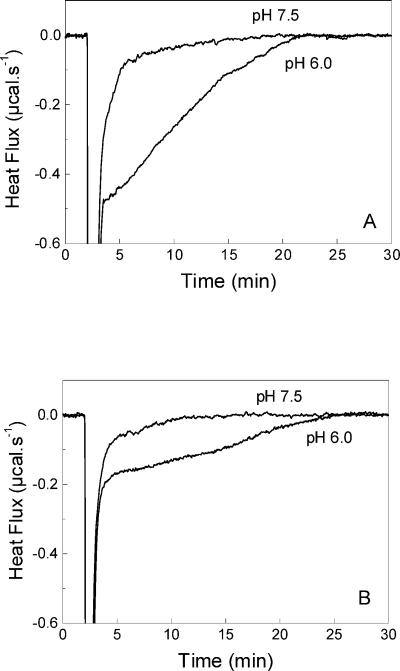
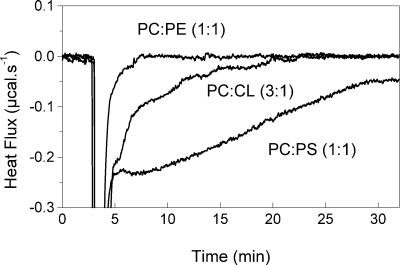
Membrane fusion was also studied by ITC at 35°C by following the heat effect after injection of VSV into liposomes. The fusion was studied with membranes of different compositions in order to show the importance of negatively charged phospholipids in this process. At pH 6.0, at which the fusion occurs, there is a displacement of the heat flow to negative values relative to the baseline after the heat effects for the dilution (Fig. 6). This effect was related to the VSV fusion to the liposomes, which is a slow process and can be followed for several minutes (6, 33). The heat flow always returns to the baseline level, suggesting that the fusion is complete. The rate of the fusion reaction was dependent on the virus concentration. The negative heat effect was not observed with liposomes lacking negatively charged lipids, such as PC-PE (Fig. 7) or PC only (not shown) vesicles. In agreement with the force spectroscopy experiments, fusion can be studied with CL-containing vesicles (Fig. 7).
FIG. 6.
Calorimetric traces of the fusion of VSV with vesicles of different PS content at 35oC . The calorimetric traces were obtained after the injection of 10 μl of VSV solution (0.28 mg/ml) into the cell containing 1 mM vesicles of PC-PS (1:3) (A) and PC-PS (3:1) (B) at pH 6.0 or pH 7.5, as indicated in each panel. After the heat due to the VSV dilution, there is a negative heat effect that can be associated with the fusion process. The return to the baseline level indicates that the fusion was complete. At pH 7.5, only the heat effect associated with the VSV dilution and binding to the vesicles is observed. The samples were prepared in 20 mM MES-30 mM Tris buffer, pH 7.5 or 6.0.
FIG. 7.
Calorimetric traces of the fusion of VSV with vesicles of different PS content at 35oC . The calorimetric traces were obtained after the injection of 10 μl of VSV solution (0.28 mg/ml) into the cell containing 1 mM vesicles of PC-PE (1:1), PC-CL (3:1), and PC-PS (1:3). The vesicles were prepared in 20 mM MES-30 mM Tris buffer, pH 6.0.
A control experiment was done with the same liposomes at pH 7.5 (Fig. 6). At this pH, essentially no fusion is observed (52), although several vesicles can bind to the virus surface (43). In this case, with the same vesicles studied before, the negative heat effect was not observed and a return of the heat flow to the baseline level was observed soon after the heat of VSV dilution. The calorimetric thermograms are similar to those obtained with PC or PC-PE liposomes, showing that the exothermic peak is a feature of systems in which fusion can be achieved.
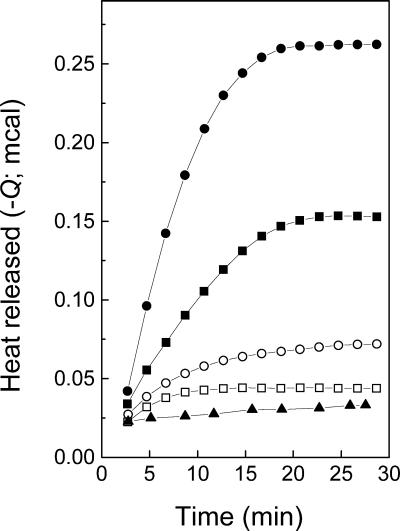
In Fig. 8, we show the integration of the calorimetric thermograms obtained with PC-PS vesicles at both pH 6.0 and pH 7.5 (from Fig. 6) and with PC only vesicles at pH 6.0 after injection of VSV. At pH 6.0, as the PS content in the membrane increased, there was an increase in the rate of the exothermic reaction. In this particular case, the rate calculated in the steady state was 8.06 mcal min−1 for 25% PS, increasing to 20.75 mcal min−1 for 75% PS. The process reached a plateau at 18 min with 75% PS and 21 min with 25% PS. At pH 7.5, however, the initial rate of the exothermic process was 1.04 and 2.34 mcal min−1 for 25 and 75% PS, respectively, reaching a plateau at around 10 min. For PC-only vesicles at pH 6.0, the heat effect was less intense than those observed with PS-containing vesicles at pH 7.5. This is in agreement with the results found by force spectroscopy, where no binding was observed with PC only (Fig. 1B). Furthermore, when the virus was inactivated by incubation at 50°C for 10 min, no heat effect related to the fusion could be observed (not shown).
FIG. 8.
Kinetics of VSV fusion with membranes at 35oC . The heat released after injection of VSV into the cell containing vesicles at pH 6.0 (•, ▪) or at pH 7.5 (○, □) was calculated by integrating the calorimetric traces shown in Fig. 6 for PC-PS at ratios of 1:3 (•, ○) and 3:1 (▪, □). Kinetics of the heat effects after VSV injection into vesicles of PC only at pH 6.0 (▴) was obtained from thermograms similar to those in Fig. 6 (not shown).
DISCUSSION
Enveloped-virus infection depends on a series of events, which comprise cell recognition, interaction between a viral surface protein and a cellular membrane, and membrane fusion induced by viral fusion proteins. Cell recognition by viruses is mediated by the interaction between a viral surface protein and a cellular receptor. Receptors used by viruses belong to different classes of macromolecules, including proteins, carbohydrates, and lipids (2), and virus-receptor interactions may determine the cell tropism and the viral host range. Membrane fusion is always mediated by viral glycoproteins and occurs either directly at the cell surface after virus binding to its receptor or at the acidic environment of the endosomal compartment.
In the case of VSV, cell recognition is mediated by its surface glycoprotein G, which also interacts with the target membrane at acidic pH and catalyzes the fusion reaction. VSV has a broad host range, extending from nearly all mammals to insects, suggesting that the VSV receptor is a widely distributed molecule. Several efforts to identify the VSV binding site on the cell surface pointed to a lipid as the VSV receptor. Schlegel and coworkers found that VSV binds with high affinity in a saturable site on Vero cells (42) and that the binding could be inhibited by a membrane extract, which was resistant to protease, neuraminidase, and heating, and also inactivated by treatment with phospholipase C (43). These findings, together with the observation that only PS among various purified lipids was able to inhibit VSV binding, led the authors to suggest that PS could participate in the cellular binding site for VSV. Similar results were obtained with erythrocytes at acidic pH: only the lipid moiety of the cell membrane, specifically the negatively charged PS, phosphatidylinositol, and GM3 ganglioside, inhibited VSV attachment to cells (29). Removal of the charged groups from these molecules greatly reduced their inhibitory activities, suggesting an important role of electrostatic interactions during cell recognition by VSV.
Here we show direct evidences that VSV interacts very strongly with membranes containing negatively charged phospholipids at neutral pH. Force-distance curves obtained by using atomic force microscopy showed that VSV did not interact with membranes composed of PC only, whereas forces as strong as 1,900 to 2,900 pN were observed when PS or CL, both negatively charged phospholipids, was present in the lipid film. This suggests that electrostatic interactions between positively charged G protein amino acid residues and the negative charges present in the membrane surface are important for membrane recognition.
Similar results were found in the ITC studies, where binding isotherms were only observed with vesicles containing PS or CL. The binding gives rise to negative peaks, indicating that this process is enthalpically driven. ITC is the only technique that allows the direct thermodynamic analysis of biomolecular interactions, providing the binding constant and stoichiometry in addition to the enthalpy and entropy of binding. Several ITC studies showed that electrostatic binding is usually driven by enthalpy (3, 26, 31, 41, 50). However, it is important to point out that the calorimetric enthalpy is actually a sum of all the heat effects, endothermic and exothermic, taking place during the interaction. Nevertheless, another evidence that electrostatic interactions make a major contribution in the VSV interaction with membranes is the lack of binding and fusion at high ionic strength, as discussed below.
The requirement for electrostatic interactions for VSV binding to the cell surface has also been raised by Bailey et al., who showed that DEAE-dextran, a polycation, increased both VSV binding to BHK cells and G protein-mediated membrane fusion (1). It is possible that DEAE-dextran interacts with the negative charges on virus surface, increasing the density of the positive charges involved in binding to the host cell.
Identification of the amino acid residues involved in the membrane binding site requires further investigation. Although it is possible that the binding domain is formed in the three-dimensional structure of the G protein, recent studies demonstrated that the p2 peptide, a sequence presenting heptad repeats found in all rhabdovirus G proteins, binds PS (11). These heptad repeats (abcdefg) contain two hydrophobic amino acid residues at positions a and d, followed by a sequence containing positively charged amino acid residues, and are located in the amino-terminal part of the glycoproteins (10). For VSV, this sequence comprises the region between amino acid residues 134 and 161.
The components of biological membranes are asymmetrically distributed between the membrane surfaces, and PS is highly segregated to the inner leaflet of plasma membranes (40), suggesting that the G protein-PS interaction is a very improbable event. However, recent findings showing that the binding of a fragment of a salmonid rhabdovirus G protein to model membranes induces PS translocation from the inner to the outer leaflet of the membrane (16) indicate that VSV binding to PS could present more physiological relevance than was expected. Another possibility is that other negatively charged molecules, such as glycosaminoglycans and gangliosides, could act as the physiological binding site for VSV. The role of these molecules in VSV binding to cells and in the membrane fusion process is now under investigation.
The force involved in G protein interaction with membranes containing PS or CL was much stronger than that expected for single-molecule interactions (≈100 pN) (35). This result could be explained if we consider that several G protein molecules interact with the membrane at a given point. Indeed, G protein is densely distributed in the viral envelope, suggesting that multiple binding occurs. It should be pointed out that several studies used force spectroscopy to determine the strength between molecular bonds (58) as well as to probe the adhesion forces between cells and surfaces (37). However, to our knowledge, the interaction between a virus and a target membrane has never been analyzed by this technique.
Viruses are much smaller than whole cells, and it is difficult to control the exact number of particles close to the tip apex. Considering the VSV dimensions (approximately 180 nm long and 65 nm wide), it should be expected that four to eight particles will adsorb on the sides of the tip apex. This also explains the multiple interaction peaks observed for the curves obtained with PC-PS and PC-CL membranes. Control experiments using VSV adsorbed on the tip and a clean mica surface (mica is negatively charged) showed many interacting peaks, indicating binding to the surface (not shown). The average force obtained in these experiments was 281 pN, which is enough to ensure that virus is adsorbed on the tip and sufficiently low to reinforce the specificity of the VSV interaction with membranes containing negative charges (much higher force values).
An attempt to calculate the calorimetric enthalpy (ΔHcal) by dividing QT (from the data in Fig. 2) by the amount of G protein present in each injection showed that ΔHcal values seems to be far from real, since they can be as low as −1,400 kcal/mol. As discussed before for the results with force spectroscopy, this is probably due to the fact that the G protein is densely distributed on the virus surface. Therefore, its local concentration in the binding reaction is actually much higher than that used for the calculations. If we consider the force spectroscopy data, at least seven proteins are involved in the binding, suggesting that the local concentration of G protein is at least seven times higher than that used for the calculations of ΔHcal. Therefore, the values found for ΔHcal will be decreased to around −200 kcal/mol. Although this value for enthalpy is large, it can be due to changes in the protein conformation, which may contribute to values as large as −150 kcal/mol (47).
Kozlov and Lohman (27) also showed that the SSB protein-DNA interaction gives rise to a large negative enthalpy that was related to stacking interactions of aromatic amino acid residues and lysine-phosphate or arginine-phosphate interactions. There is also a possibility that protonation of the protein upon binding also contributes to a large negative ΔHcal (14). From the analysis of the curve of ΔHcal as a function of G protein concentration, it was found a large contribution for the association enthalpy (ΔHass = −1,185 kcal/mol) and entropy (ΔSass = −3,839 cal/mol · K). The thermodynamic parameters for the association were also calculated considering the local concentration of G protein as seven times higher, with ΔHass = −166 kcal/mol and ΔSass = −525 cal/mol · K. The unfavorable entropy can be due to different factors, such as the exposure of hydrophobic surfaces to the solvent as well as the decrease in conformational motion in the protein and/or the membrane (49). Nevertheless, there are some examples in the literature where hydrophobic interactions are driven by enthalpy and not by entropy, in the so-called nonclassical hydrophobic effect (for a review, see reference 44). Seelig (44) explains this binding enthalpy of hydrophobic solutes into lipid bilayers as possibly derived from (i) the van der Waals interaction energy and (ii) the increased hydration of the lipid-water interface.
VSV binding to membranes at neutral pH did not induce changes in VSV intrinsic fluorescence, suggesting that the binding itself did not alter the G protein tryptophan environment. On the other hand, G protein conformational changes induced when the pH was decreased after VSV binding to membranes can be followed by the increase in intrinsic fluorescence (6). Here, we found that the extent of these conformational changes depends on the number of negative charges in the target membrane. G protein-mediated membrane fusion can also be correlated to the PS content in the vesicles, probably because it is driven by G protein conformational changes. Indeed, the electrostatic nature of VSV-membrane interactions was also demonstrated by the inhibition of G protein conformational change and membrane fusion at high ionic strength. An increase in ionic strength can abolish electrostatic interactions by reducing charge-charge intermolecular or intramolecular interactions or even by decreasing the fraction of free water available to solvate the protein and/or the ligand. Although all these results unequivocally demonstrate the importance of electrostatic interactions for VSV fusion, we cannot discard the possibility that hydrophobic interactions are also involved in the VSV interaction with membranes. In fact, exposure of hydrophobic domains has already been shown to occur in G protein at low pH (6, 15).
The electrostatic nature of VSV interaction with membranes during fusion is an interesting result considering that for most of the viruses studied so far it is suggested that the binding to membrane occurs through hydrophobic interactions (22, 36). Very similar structures occur among several viral envelope glycoproteins, such as those of influenza virus, HIV-1, Moloney murine leukemia virus, and respiratory syncytial virus, which form a coiled-coil trimer that is inserted into the target membrane (5, 8, 17, 55, 57). Actually, since the ΔHcal results from every single event, endothermic or exothermic, that is taking place during the binding, it is possible that hydrophobic interactions also occur in the case of VSV binding, but they are not the dominant energetic contribution for the overall process.
The ITC studies of VSV fusion to vesicles were done at pH 6.0. In these conditions, we clearly observed an exothermic effect following the heat effects due to the VSV dilution. This process was slow and could be followed for several minutes. The calorimetric pattern is related to fusion, and aggregation contributions can be excluded because the same pattern was not observed at pH 7.5. At this pH, the presence of multiple binding sites both on the virus surface, represented by G proteins, and in the vesicles containing negative charges probably causes aggregation. In fact, it was shown before by electron microscopy that several vesicles can bind to the surface of a single VSV (43). Nevertheless, the ITC experiments in such condition showed that, after the heat effects due to VSV dilution and binding to vesicles, there is a return to the baseline level, and the slow exothermic process is not observed.
It is interesting that our result is the opposite of that found in calorimetric studies of membrane fusion induced by influenza virus HA (32). This study showed that the fusion is an endothermic process, which could be explained by the increase in entropy upon both lipid mixing and the hydrophobic insertion of the fusion peptide into the lipid bilayer. It should be taken into account that the fusion process probably comprises several steps, such as the virus-membrane interaction, organization and/or destabilization of the outer monolayers of the membranes, and also the formation of a fusion pore, and other events (23). Thus, the heat released during the fusion process can be reflecting the result of all the changes that take place during this process as well as the mixing of virus and vesicle contents that results from the fusion. Nevertheless, our data suggest an alternative mechanism involved in VSV-induced membrane fusion compared to the calorimetric data for influenza virus HA-induced fusion, which is also supported by the fact that G protein does not contain an apolar fusion peptide. However, our results do not discard the participation of hydrophobic interactions during the VSV-induced fusion reaction.
Taken together, our results suggest that the interaction between VSV G protein and its target membrane seems to be more electrostatic than hydrophobic at both neutral and fusogenic pHs. We show that membrane recognition by VSV, G protein conformational changes induced by its interaction with the membranes, and the membrane fusion reaction itself are driven by electrostatic interactions between the viral G protein and negatively charged phospholipids present in the target membranes.
Acknowledgments
This work was supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Programa de Apoio ao Desenvolvimento Científico e Tecnológico (PADCT), Volkswagen Foundation, and Fundação Universitária José Bonifácio (FUJB).
We thank Leopoldo de Meis for the ITC facilities and Adalberto Vieyra for the use of the fluorometer. We also thank Paulo H. dos Santos for technical assistance.
REFERENCES
- 1.Bailey, C. A., D. K. Miller, and J. Lenard. 1984. Effects of DEAE-dextran on infection and hemolysis by VSV. Evidence that nonspecific electrostatic interactions mediate effective binding of VSV to cells. Virology 133:111-117. [DOI] [PubMed] [Google Scholar]
- 2.Baranowski, E., C. M. Ruiz-Jarabo, and E. Domingo. 2001. Evolution of cell recognition by viruses. Science 292:1102-1105. [DOI] [PubMed] [Google Scholar]
- 3.Bathaie, S. Z., A. A. Moosavi-Movahedi, and A. A. Saboury. 1999. Energetic and binding properties of DNA upon interaction with dodecyl trimethylammonium bromide. Nucleic Acids Res. 27:1001-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnafous, P., and T. Stegmann. 2000. Membrane perturbation and fusion pore formation in influenza hemagglutinin-mediated membrane fusion. A new model for fusion. J. Biol. Chem. 275:6160-6166. [DOI] [PubMed] [Google Scholar]
- 5.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 6.Carneiro, F. A., A. S. Ferradosa, and A. T. Da Poian. 2001. Low pH-induced conformational changes in vesicular stomatitis virus glycoprotein involve dramatic structure reorganization. J. Biol. Chem. 276:67-72. [DOI] [PubMed] [Google Scholar]
- 7.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 8.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 9.Cleverley, D. Z., and J. Lenard. 1998. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA 95:3425-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coll, J. M. 1995. Heptad-repeat sequences in the glycoprotein of rhabdovirus. Virus Genes 10:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coll, J. M. 1997. Synthetic peptides from the heptad repeats of the glycoproteins of rabies, vesicular stomatitis and fish rhabdoviruses bind phosphatidylserine. Arch. Virol. 142:2089-2097. [DOI] [PubMed] [Google Scholar]
- 12.Da Poian, A. T., A. M. O. Gomes, and T. Coelho-Sampaio. 1998. Kinetics of intracellular viral disassembly and processing probed by Bodipy fluorescence dequenching. J. Virol. Methods 70:45-58. [DOI] [PubMed] [Google Scholar]
- 13.Da Poian, A. T., A. M. O. Gomes, R. J. N. Oliveira, and J. L. Silva. 1996. Migration of vesicular stomatitis virus glycoprotein to the nucleus of infected cells. Proc. Natl. Acad. Sci. USA 93:8268-8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Haseth, P. L., T. M. Lohman, and M. T. Record. 1977. The nonspecific binding of lac-repressor to DNA, an association reaction driven by counterion release. Biochemistry 16:4783-4790. [DOI] [PubMed] [Google Scholar]
- 15.Durrer, P., Y. Gaudin, W. H. Ruigrok, R. Graf, and J. Brunner. 1995. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J. Biol. Chem. 270:17575-17581. [DOI] [PubMed] [Google Scholar]
- 16.Estepa, A. M., A. I. Rocha, V. Mas, L. Pérez, J. A. Encinar, E. Nuñez, A. Fernandez, J. M. G. Ros, F. Gavilanes, and J. M. Coll. 2001. A protein G fragment from the salmonid viral hemorrhagic septicemia rhabdovirus induces cell-to-cell fusion and membrane phosphatidylserine translocation at low pH. J. Biol. Chem. 276:46268-46275. [DOI] [PubMed] [Google Scholar]
- 17.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 3:465-469. [DOI] [PubMed] [Google Scholar]
- 18.Florin, E. L., V. T. Moy, and H. E. Gaub. 1994. Adhesion forces between individual ligand-receptor pairs. Science 264:415-417. [DOI] [PubMed] [Google Scholar]
- 19.Fredericksen, B. L., and M. Whitt. 1995. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J. Virol. 69:1435-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudin, Y., R. W. H. Ruigrok, and J. Brunner. 1995. Low-pH induced conformational changes in viral fusion proteins: implications for the fusion mechanism. J. Gen. Virol. 76:1541-1556. [DOI] [PubMed] [Google Scholar]
- 21.Gergely, C., J.-C. Voegel, P. Schaaf, B. Senger, M. Maaloum, J. K. H. Hörber, and J. Hemmerlé. 2000. Unbinding process of adsorbed proteins under external stress studied by atomic force microscopy spectroscopy. Proc. Natl. Acad. Sci. USA 97:10802-10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harter, C., P. James, T. Bachi, G. Semenza, and J. Brunner. 1989. Hydrophobic binding of the ectodomain of influenza hemagglutinin to membranes occurs through the “fusion peptide.” J. Biol. Chem. 264:6459-6464. [PubMed] [Google Scholar]
- 23.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 24.Hyre, D. E., and L. D. Spicer. 1995. Thermodynamic evaluation of binding interactions in the methionine repressor system of Escherichia coli using isothermal titration calorimetry. Biochemistry 34:3212-3221. [DOI] [PubMed] [Google Scholar]
- 25.Jass, J., T. Tjärnhage, and G. Puu. 2000. From liposomes to supported, planar bilayer structures on hydrophilic and hydrophobic surfaces: an atomic force microscopy study. Biophys. J. 79:3153-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, E., V. Katritch, W. K. Olson, M. Kharatisvili, R. Abagyan, and D. S. Pilch. 2000. Aminoglycoside binding in the major groove of duplex RNA: the thermodynamic and electrostatic forces that govern recognition. J. Mol. Biol. 298:95-110. [DOI] [PubMed] [Google Scholar]
- 27.Kozlov, A. G., and T. M. Lohman. 1998. Calorimetric studies of E. coli SSB protein-single-stranded DNA interactions. Effects of monovalent salts on binding enthalpy. J. Mol. Biol. 278:999-1014. [DOI] [PubMed] [Google Scholar]
- 28.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 29.Mastromarino, P., C. Conti, P. Goldoni, B. Hauttecoeur, and N. Orsi. 1987. Characterization of membrane components of the erythrocyte involved in vesicular stomatitis virus attachment and fusion at acidic pH. J. Gen. Virol. 68:2359-2369. [DOI] [PubMed] [Google Scholar]
- 30.Matlin, K. S., H. Reggio, A. Helenius, and K. Simons. 1982. Pathway of vesicular stomatitis virus entry leading to infection. J. Mol. Biol. 156:609-631. [DOI] [PubMed] [Google Scholar]
- 31.Messana, I., M. Angeletti, M. Castagnola, G. De Sanctis, E. Di Stasio, B. Giardina, S. Pucciarelli, and M. Coletta. 1998. Thermodynamics of inositol hexabisphosphate interaction with human oxyhemoglobin. J. Biol. Chem. 273:15329-15334. [DOI] [PubMed] [Google Scholar]
- 32.Nebel, S., I. Bartoldus, and T. Stegman. 1995. Calorimetric detection of influenza virus induced membrane fusion. Biochemistry 34:5705-5711. [DOI] [PubMed] [Google Scholar]
- 33.Pal, R., Y. Barenholz, and R. R. Wagner. 1988. Pyrene phospholipid as a biological fluorescent probe for studying fusion of virus membrane with liposomes. Biochemistry 27:30-36. [DOI] [PubMed] [Google Scholar]
- 34.Puu, G., and I. Gustafson. 1997. Planar lipid bilayers on solid supports from liposomes—factors of importance for kinetics and stability. Biochim. Biophys. Acta 1327:149-161. [DOI] [PubMed] [Google Scholar]
- 35.Puu, G., E. Artursson, I. Gustafson, M. Lundstro, and J. Jass. 2000. Distribution and stability of membrane proteins in lipid membranes on solid supports. Biosensors Bioelectronics 15:31-41. [DOI] [PubMed] [Google Scholar]
- 36.Rabenstein, M., and Y.-K. Shin. 1995. A peptide from the heptad repeat of human immunodeficiency virus gp41 shows both membrane binding and coiled-coil formation. Biochemistry 34:13390-13397. [DOI] [PubMed] [Google Scholar]
- 37.Ratazos, A., Y.-L. Ong, M. M. Sharma, and G. Georgiou. 1998. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Natl. Acad. Sci. USA 95:11059-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reviakine, I., and A. Brisson. 2000. Formation of supported phospholipid bilayers from unilamellar vesicles investigated by atomic force microscopy. Langmuir 16:1806-1815. [Google Scholar]
- 39.Rose, J. K., and C. J. Gallione. 1981. Nucleotide sequences of the mRNA's encoding the vesicular stomatitis virus G and M proteins determined from cDNA clones containing the complete coding regions. J. Virol. 39:519-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothman, J. E., and J. Lenard. 1977. Membrane asymmetry. Science 195:743-753. [DOI] [PubMed] [Google Scholar]
- 41.Sanderson, N. M., B. Guo, A. E. Jacob, P. S. Handley, J. G. Cunniffe, and M. N. Jones. 1996. The interaction of cationic liposomes with the skin-associated bacterium Staphylococcus epidermidis: effects of ionic strength and temperature. Biochim. Biophys. Acta 1283:207-214. [DOI] [PubMed] [Google Scholar]
- 42.Schlegel, R., M. C. Willingham, and I. H. Pastan. 1982. Saturable binding sites for vesicular stomatitis virus on the surface of Vero cells. J. Virol. 43:871-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlegel, R., T. S. Tralka, M. C. Willingham, and I. Pastan. 1983. Inhibition of VSV binding and infectivity by phosphatidylserine: is phosphatidylserine a VSV-binding site? Cell 32:639-646. [DOI] [PubMed] [Google Scholar]
- 44.Seelig, J. 1997. Titration calorimetry of lipid-peptide interactions. Biochim. Biophys. Acta 1331:103-116. [DOI] [PubMed] [Google Scholar]
- 45.Shokralla, S., Y. He, E. Wanas, and H. P. Ghosh. 1998. Mutations in a carboxy-terminal region of vesicular stomatitis virus glycoprotein G that affect membrane fusion activity. Virology 242:39-50. [DOI] [PubMed] [Google Scholar]
- 46.Skehel, J. J., P. M. Bayley, E. B. Brown, S. R. Martin, M. D. Waterfield, J. M. White, I. A. Wilson, and D. C. Wiley. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. USA 79:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spolar, R. S., and M. T. Record, Jr. 1994. Coupling of local folding to site-specific binding of proteins to DNA. Science 263:777-784. [DOI] [PubMed] [Google Scholar]
- 48.Stegmann, T., J. M. Delfino, F. M. Richards, and A. Helenius. 1991. The HA2 subunit of influenza hemagglutinin inserts into the target membrane prior to fusion. J. Biol. Chem. 266:18404-18410. [PubMed] [Google Scholar]
- 49.Tame, J. R. H., R. O'Brien, and J. E. Ladbury. 1998. Isothermal titration calorimetry of biomolecules, p. 27-38. In J. E. Ladbury and B. Z. Chowdhry (ed.), Biocalorimetry: applications of calorimetry in the biological sciences. John Wiley & Sons Ltd., London, England.
- 50.Thomas, P. G., and J. Seelig. 1993. Binding of the calcium antagonist flunarizine to phosphatidylcholine bilayers: charge effects and thermodynamics. Biochem. J. 291:397-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrinson, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 52.White, J., K. Matlin, and A. Helenius. 1981. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 54.Wiseman, T., S. Williston, J. F. Brandts, and L. N. Lin. 1989. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179:131-137. [DOI] [PubMed] [Google Scholar]
- 55.Yu, Y. G., D. S. King, and Y.-K. Shin. 1994. Insertion of a coiled-coil peptide from influenza virus hemagglutinin into membranes. Science 266:274-276. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, L., and H. P. Ghosh. 1994. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J. Virol. 68:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zlatanova, J., S. M. Lindsay, and S. H. Leuba. 2000. Single molecule force spectroscopy in biology using the atomic force microscope. Prog. Biophys. Mol. Biol. 74:37-61. [DOI] [PubMed] [Google Scholar]