Abstract
Enveloped viruses are released from infected cells after coalescence of viral components at cellular membranes and budding of membranes to release particles. For some negative-strand RNA viruses (e.g., vesicular stomatitis virus and Ebola virus), the viral matrix (M) protein contains all of the information needed for budding, since virus-like particles (VLPs) are efficiently released from cells when the M protein is expressed from cDNA. To investigate the requirements for budding of the paramyxovirus simian virus 5 (SV5), its M protein was expressed in mammalian cells, and it was found that SV5 M protein alone could not induce vesicle budding and was not secreted from cells. Coexpression of M protein with the viral hemagglutinin-neuraminidase (HN) or fusion (F) glycoproteins also failed to result in significant VLP release. It was found that M protein in the form of VLPs was only secreted from cells, with an efficiency comparable to authentic virus budding, when M protein was coexpressed with one of the two glycoproteins, HN or F, together with the nucleocapsid (NP) protein. The VLPs appeared similar morphologically to authentic virions by electron microscopy. CsCl density gradient centrifugation indicated that almost all of the NP protein in the cells had assembled into nucleocapsid-like structures. Deletion of the F and HN cytoplasmic tails indicated an important role of these cytoplasmic tails in VLP budding. Furthermore, truncation of the HN cytoplasmic tail was found to be inhibitory toward budding, since it prevented coexpressed wild-type (wt) F protein from directing VLP budding. Conversely, truncation of the F protein cytoplasmic tail was not inhibitory and did not affect the ability of coexpressed wt HN protein to direct the budding of particles. Taken together, these data suggest that multiple viral components, including assembled nucleocapsids, have important roles in the paramyxovirus budding process.
Enveloped viruses form at cellular membranes of infected cells by a budding process. Soluble viral components, along with integral membrane glycoprotein spikes, assemble at highly concentrated budding sites, followed by envelopment of the viral core and fission of the membrane to create progeny virions. Much progress has been made in identifying both viral and cellular components that drive this assembly process. For example, alphavirus budding has been found to require both soluble nucleocapsid cores and envelope glycoproteins (9, 25, 44, 45), and a specific interaction between the viral capsid protein and the cytoplasmic tail of the viral E2 glycoprotein is critical for assembly (54). In contrast, the assembly and budding of retroviruses does not require participation of envelope components, and expression of soluble Gag polyprotein in the absence of any other viral component results in efficient budding of virus-like particles (VLPs) that resemble immature virions (46). Several elements within the Gag protein sequence that are important for budding have been identified, including M domains which mediate membrane targeting (1, 31, 32), I domains which direct Gag-Gag interactions (3, 11), and L domains which recruit cellular machinery necessary for membrane fission and virus release (1, 12, 14, 34-36, 41, 43, 48, 49).
The parameters that influence the budding of negative-strand RNA viruses are not as well defined. A key role is played by soluble matrix (M) proteins that bind to the inner surfaces of plasma membranes and form an electron-dense layer underlying the virion envelope. Recombinant viruses that lack M proteins have been constructed in the cases of both rabies (27) and measles (4), and in both cases budding was drastically impaired. Furthermore, VLP release has been observed from cells transfected with plasmids encoding M proteins derived from vesicular stomatitis virus (VSV) (16, 21, 23), Ebola virus (15, 47), influenza A virus (13, 22), and human parainfluenza virus type 1 (hPIV-1) (5). VLP budding has been examined quantitatively for both VSV (21) and Ebola virus (47), and in both cases budding was remarkably efficient, with >20% of the M protein released from transfected cells into the culture medium. VLP budding directed by the M proteins of influenza A virus and hPIV-1 has at this point been examined only qualitatively (5, 13, 22). In all of these cases, budding of particles was observed upon expression of M protein in the absence of any other viral components. However, studies with recombinant viruses, including recombinant rabies virus (26), VSV (40), influenza A virus (20), and the paramyxoviruses Sendai virus (10) and simian virus 5 (SV5) (39) suggest that M proteins may not be sufficient to direct normal budding of a virus, since truncations or sequence alterations to glycoprotein cytoplasmic tails resulted in poor budding, despite the presence of unmodified matrix protein.
SV5, like other paramyxoviruses, consists of a core of genomic RNA encapsidated by nucleocapsid (NP) protein that is bound by a polymerase complex composed of large (L) protein and phosphoprotein (P). This core is surrounded by a lipid envelope that is coated with M protein on its inner surface and penetrated by spike glycoproteins that function for attachment (hemagglutinin-neuraminidase [HN]) and fusion (F). Minor structural components of SV5 include the SH protein, which blocks apoptosis in infected cells (17), and the V protein, which blocks interferon signaling and modulates the cell cycle (24, 50, 51). It is unclear at present which of these viral proteins are necessary to direct normal assembly and budding of virions.
Here we describe experiments in which we have transiently transfected mammalian cells with cDNAs encoding SV5 proteins in an attempt to reconstitute budding in a virus-free system. By using this approach, we found that efficient budding of SV5 VLPs required expression not only of the M protein but also of NP protein and a glycoprotein (either HN or F protein). Furthermore, we suggest that the cytoplasmic tails of the two SV5 glycoproteins have critical, but redundant, roles in budding.
MATERIALS AND METHODS
Plasmids.
SV5 DNA sequences corresponding to M, NP, HN, and F proteins from the SV5 infectious clone pBH276 (GenBank accession no. AF052755) (18) were subcloned into the eukaryotic expression vector pCAGGS (30) to generate pCAGGS-SV5 M, pCAGGS-SV5 NP, pCAGGS-SV5 HN, and pCAGGS-SV5 F, respectively. Cloning details will be provided upon request. Mutant SV5 HN proteins HNΔ2-9 and HNΔ2-13 containing truncations in the DNA region encoding the cytoplasmic tail have been described previously (39). Mutant SV5 F protein FΔ16 containing a truncation in the DNA region encoding the cytoplasmic tail was made by PCR mutagenesis of the F DNA sequence. The nucleotide sequence of the entire F gene was confirmed with an ABI Prism 310 genetic analyzer (Applied Biosystems, Inc., Foster City, Calif.). A cDNA encoding VSV glycoprotein (G) was provided by John K. Rose, Yale University Medical School, and pCAGGS-influenza HA has been previously described (53).
Generation of VLPs and immunoprecipitation.
293T cells in 6-cm-diameter dishes (70 to 80% confluent) grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum were transfected in duplicate with plasmid DNAs by using Lipofectamine-Plus reagents (Invitrogen, Carlsbad, Calif.). Plasmid quantities per dish were as follows unless otherwise indicated: pCAGGS-SV5 M, 0.4 μg; pCAGGS-SV5 NP, 50 ng; pCAGGS-SV5 HN, 0.75 μg; pCAGGS-SV5 HNΔ2-9, 1.5 μg; pCAGGS-SV5 HNΔ2-13, 1.5 μg; pCAGGS-SV5 F, 0.75 μg; pCAGGS-SV5 FΔ16, 0.75 μg; pCAGGS-influenza HA, 3.0 μg; and pCAGGS-VSV G, 0.75 μg. Alternatively, 293T cells in 6-cm-diameter dishes were infected in duplicate with rSV5 (SV5 recovered from infectious cDNA clone) or rSV5 HNΔ2-9 as previously described (39) at a multiplicity of infection of 1.0 PFU/cell. At 24 h posttransfection (p.t.) or postinfection (p.i.), the medium was replaced with 1.5 ml of DMEM containing one-tenth the normal amount of methionine and cysteine and 50 μCi of [35S]Promix (Amersham Pharmacia Biotech, Piscataway, N.J.)/ml. At 40 h p.t. or p.i., cells and culture media were collected. To analyze the VLPs or virions released from cells, culture media from duplicate dishes were combined and centrifuged at 5,000 × g for 5 min to remove cell debris and then layered onto 35% sucrose cushions (4 ml in NTE [0.1 M NaCl; 0.01 M Tris-HCl, pH 7.4; 0.001 M EDTA]) and centrifuged at 186,000 × g for 2 h. Pellets were resuspended in 0.5 ml of NTE and combined with 1.3 ml of 80% sucrose in NTE for flotation. Additional layers containing 50% sucrose (1.8 ml) and 10% sucrose (0.6 ml) were applied to the top of the sample, followed by centrifugation at 186,000 × g for 4 h. In some cases, six equal fractions (0.7 ml each) were collected from the top of the gradient for analysis of proteins by immunoprecipitation. In other cases, only the top 2.1 ml was collected as a single fraction from the gradient, and the remainder of the gradient was discarded.
To analyze intracellular proteins, cells were scraped from dishes, pelleted by centrifugation (5,000 × g for 5 min), resuspended in a hypotonic solution (25 mM NaCl; 25 mM HEPES, pH 7.3; 1 mM phenylmethylsulfonyl fluoride), and placed on ice for 10 min. Cells were then disrupted with 30 strokes of a Dounce homogenizer on ice. Nuclei and cell debris were removed by centrifugation (2,000 × g for 5 min). Proteins in half of the resulting cell lysate (equivalent of a single 6-cm-diameter dish) were analyzed by immunoprecipitation.
For immunoprecipitation analysis, cell lysates or sucrose gradient fractions were combined with an equal volume of 2× radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris, pH 7.4; 2% deoxycholate; 2% Triton X-100; 0.2% sodium dodecyl sulfate [SDS]) containing 0.3 M NaCl, 100 mM iodoacetamide, and 2 mM phenylmethylsulfonyl fluoride (33). Samples were incubated with antibodies for 3 h at 4°C, and immune complexes adsorbed to protein A-Sepharose beads for 30 min at 4°C. Antisera used were as follows: for SV5 M, HN, and NP, mouse monoclonal antibodies (MAbs) M-h, HN1b, and NP-a (37) (kindly provided by Richard Randall, St. Andrews University, St. Andrews, Scotland, United Kingdom) were used; for SV5 F protein, rabbit polyclonal anti-F2 peptide antiserum was used; for influenza A HA protein, goat serum raised to purified influenza A/Udorn/301/72 virus was used; and for VSV G protein, goat anti-VSV serum (kindly provided by John K. Rose, Yale University Medical School) was used. After incubation with antibodies, samples were washed three times with RIPA buffer containing 0.3 M NaCl; two times with RIPA buffer containing 0.15 M NaCl; and once with 50 mM Tris (pH 7.4), 0.25 mM EDTA, and 0.15 M NaCl. Samples were boiled in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer containing 2.5% (wt/vol) dithiothreitol and fractionated on 10% polyacrylamide-SDS gels (33). Quantitation was performed by using a Fuji BioImager 1000 (Fuji Medical System, Stanford, Conn.). The fraction of M protein in the floated media fraction was calculated as the counts in media/[2(the counts in lysate + the counts in medium)]. This calculation takes into account the fact that only half of each lysate was used for analysis.
Isolation of nucleocapsid-like structures.
Gradient purification of assembled nucleocapsid structures was performed by a method similar to that reported previously (2). 293T cells were transfected and metabolically labeled by the same method and plasmid amounts as for VLP generation, except that the labeling period was shortened to 8 h. Dounce-homogenized, postnuclear cell lysates (0.5 ml) were applied to the tops of CsCl density gradients containing 40% CsCl (0.8 ml), 30% CsCl (1.2 ml), 20% CsCl (1.2 ml), and 30% glycerol (0.4 ml). Gradients were ultracentrifuged for 16 h at 165,000 × g. Six 0.7-ml fractions were collected from the top of each gradient, and 0.5 ml of each fraction was analyzed by immunoprecipitation as described above.
Electron microscopy.
To generate virions for electron microscopy, MDBK cells were infected with rSV5 and virions were purified on sucrose gradients as described previously (33). To generate VLPs for electron microscopy, 293T cells in 6-cm-diameter dishes were transfected with pCAGGS-SV5 M plus pCAGGS-SV5 NP plus pCAGGS-SV5 HN or with pCAGGS-SV5 M plus pCAGGS-SV5 NP plus pCAGGS-SV5 F. At 16 h p.t., the medium was replaced with DMEM supplemented with 10% fetal bovine serum. At 40 h p.t., the culture medium was harvested. Media from seven identical transfections were pooled and centrifuged at 5,000 × g for 5 min to remove cell debris and then layered onto 35% sucrose cushions (10 ml in NTE) and centrifuged at 186,000 × g for 2 h. Pellets were resuspended in 0.25 ml of NTE by passaging 10 times through a 25-gauge needle. Insoluble material was removed by centrifugation at 14,000 × g for 5 min, and samples were concentrated to a ca. 75-μl volume by using a Microcon-100 centrifugal filter device (Millipore Corp., Bedford, Mass.). Drops of undiluted VLP preparations were allowed to absorb for ca. 30 s onto Parlodion-covered copper grids that previously had a thin layer of carbon evaporated upon them. Excess sample was drawn off, and the samples were stained for 30 s with 1% sodium silicotungstate (pH 7.1) and then air dried. Examples of SV5 were similarly prepared except the purified virus was diluted in NTE 1:20 and the grids were stained with 2% phosphotungstic acid (pH 6.6). Specimens were examined in a JEOL JEM-100CX II electron microscope operated at 80 kV.
To generate nucleocapsid-like particles for electron microscopy, 293T cells in 6-cm-diameter dishes were transfected with pCAGGS-SV5 NP. At 40 h p.t., cells were Dounce homogenized and cell lysates were fractionated on a CsCl density gradient as described above. Undiluted fractions were absorbed for 2 min onto Parlodion-covered copper grids that previously had a thin layer of carbon evaporated upon them. The grids were washed to eliminate CsCl by floating sample side down sequentially on three drops of phosphate-buffered saline (PBS) over a total time of 10 min. The nucleocapsid samples were then stained with 2% phosphotungstic acid (pH 6.6) and examined in a JEOL JEM-100CX II electron microscope operated at 80 kV.
Confocal microscopy.
CV-1 cells grown on glass coverslips were infected with rSV5 or rSV5 HNΔ2-9 at a multiplicity of infection of 0.2 PFU/cell. At 16 h p.i. monolayers were fixed with 1% methanol-free formaldehyde for 15 min and blocked with 1% bovine serum albumin in PBS. Cells were incubated for 30 min with the HN-specific MAb HN1b (immunoglobulin G2a [IgG2a] isotype), washed five times with PBS, incubated for 30 min with a IgG2a-specific R-phycoerythrin-conjugated goat antimouse secondary antibody (Southern Biotechnology Associates, Inc., Birmingham, Ala.), washed five times with PBS, and then fixed a second time and permeabilized with 0.1% saponin (Sigma-Aldrich Co., St. Louis, Mo.). Cells were then incubated for 30 min with the M protein-specific MAb M-h (IgG3 isotype), washed five times with PBS, incubated for 30 min with a IgG3-specific fluorescein isothiocyanate-conjugated goat antimouse secondary antibody, washed five times with PBS, and visualized with a model LSM 410 confocal microscope (Zeiss, Inc., Thornwood, N.Y.).
RESULTS
Coexpression of multiple SV5 proteins in transfected cells results in budding of particles.
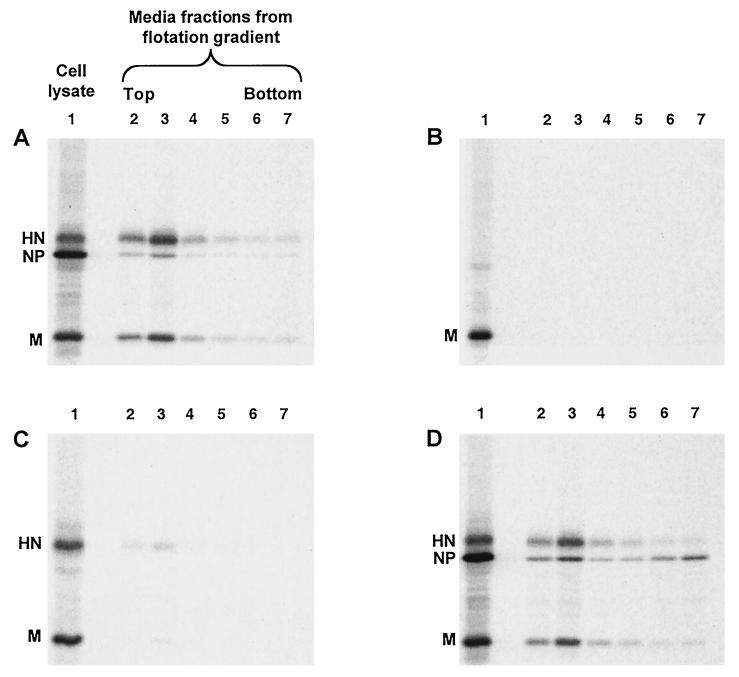
To determine whether the SV5 M protein can function to direct the budding of particles from transfected cells, M protein was expressed transiently from cDNA either by itself or in combination with cDNAs encoding other SV5 proteins. Cells were metabolically labeled for 24 h, followed by harvesting of both the cells and the culture media. Particles from the culture media were pelleted through 35% sucrose cushions and then applied to the bottoms of sucrose flotation gradients. Enveloped particles floated to the tops of the gradients upon ultracentrifugation, and SV5 proteins were detected by immunoprecipitation and SDS-PAGE. As a positive control for budding, cells were infected with SV5 and as expected, particles containing SV5 proteins were detected in the media and floated to the top of the gradient (Fig. 1A, lanes 2 and 3).
FIG. 1.
Coexpression of multiple SV5 proteins in transfected cells leads to budding of particles. 293T cells were infected with rSV5 (A), transfected with plasmid encoding SV5 M protein (B), transfected with plasmids encoding SV5 M and SV5 HN proteins (C), or transfected with plasmids encoding SV5 M, SV5 HN, and SV5 NP proteins (D). At 24 h p.i. or p.t., cells were radiolabeled for 16 h with [35S]Promix, followed by collection of both cells and culture media. Culture media was clarified by low-speed centrifugation, centrifuged through 35% sucrose cushions, and separated on sucrose flotation gradients into six equal fractions as described in Materials and Methods. Cells were disrupted by Dounce homogenization and centrifuged at low speed to remove nuclei and debris. SV5 proteins M, HN, and NP were immunoprecipitated from samples as described in Materials and Methods, and polypeptides were analyzed by SDS-PAGE on 10% gels. Lane 1, cell lysate; lanes 2 to 7, fractions from flotation gradient, with lane 2 representing proteins found at the top of the gradient and lane 7 representing proteins found at the bottom of the gradient.
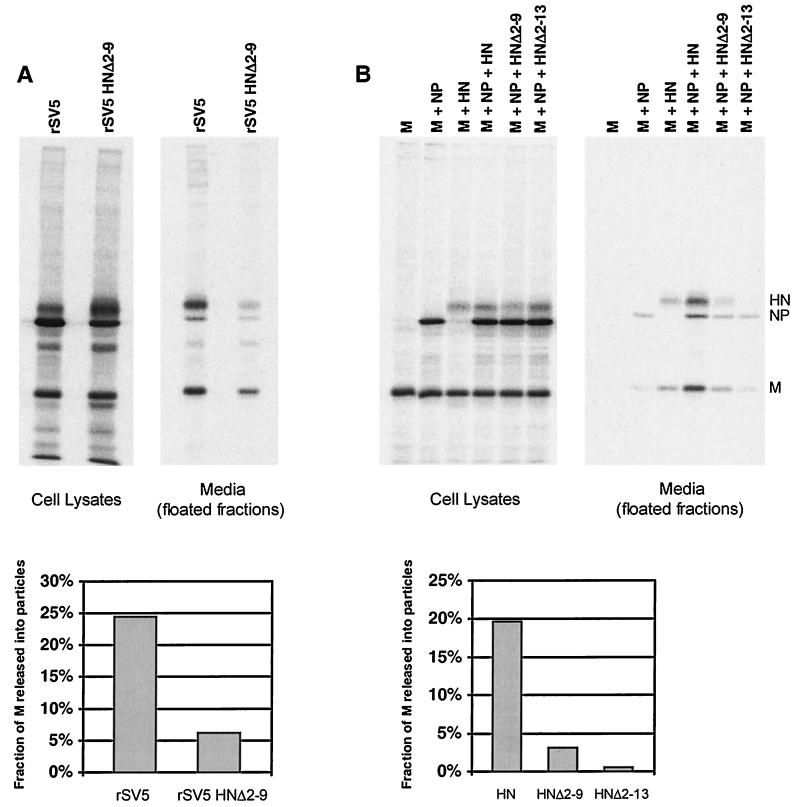
SV5 M protein was then expressed by itself in transfected cells, and the same budding assay was performed. No particles were detected in the culture media, even though the level of M protein detected in the transfected cell lysate was as great as the level of M protein in the SV5-infected cell lysate (Fig. 1B). Thus, SV5 M protein by itself did not direct substantial budding of particles, in contrast to the M proteins of other negative-strand RNA viruses. Similar results were obtained when particles were centrifuged through 20% sucrose cushions instead of 35% sucrose cushions or when the sucrose cushions were eliminated entirely (not shown), further indicating a lack of budding, as opposed to the budding of particles having insufficient density to pellet through sucrose.
We have shown previously that the HN glycoprotein of SV5 has an important role in the budding of virus, since deletions to the cytoplasmic tail of HN protein resulted in recombinant viruses that assembled poorly (39). The HN cytoplasmic tail most likely makes contact with soluble viral components such as M protein inside the cell, and these interactions appear to be important for the assembly process.
To investigate whether HN and M proteins may both be required to induce budding of particles, HN and M proteins were coexpressed in cells such that the two proteins accumulated to levels similar to those found in SV5-infected cells (Fig. 1C). This resulted in detectable budding of particles into the media, and although this level of budding varied from experiment to experiment, it was always quite low compared with budding of virions from infected cells (at least fourfold less). This result suggested that, although HN protein may be important for budding, additional viral components might be necessary for efficient budding.
Interactions between NP and M proteins could be important during the assembly process to facilitate budding and/or to increase the likelihood that budded particles contain a copy of the virus genome. Thus, NP protein was coexpressed together with HN and M proteins such that all three proteins accumulated to levels similar to those found in SV5-infected cells (Fig. 1D). Examination of the culture media after M+HN+NP coexpression showed an abundance of particles that floated to the top of the sucrose gradient, and these particles contained all three viral proteins. Thus, the efficiency of budding of SV5 particles increased upon inclusion of NP protein with coexpression of HN and M proteins (compare Fig. 1A to D).
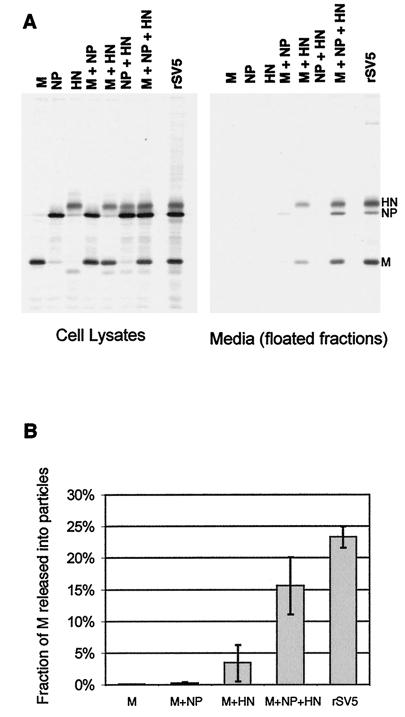
To examine further the requirements for budding, all possible combinations of M, HN, and NP proteins were coexpressed in cells, and budding was assayed as described above, except that only the top half of each flotation gradient was pooled and analyzed on a single lane of a gel (Fig. 2A). Budding efficiency was quantified as the portion of M protein detected in the floated medium fraction compared to the total M protein in the floated medium fraction plus the cell lysate (Fig. 2B). As described above, when M, HN, and NP proteins were coexpressed, the budding of particles was efficient: 16% of the total M protein was detected in the floated medium fraction compared to 23% for SV5-infected cells. When any of these three SV5 proteins were expressed individually, no budding of particles was detected. When pairwise combinations of these proteins were expressed that omitted either M or HN protein, virtually no budding was detected, and when NP protein was omitted, budding was detected but was relatively poor (3% of M protein detected in the floated media fraction). Thus, budding was efficient only when all three components were expressed, with the lack of NP protein resulting in ∼4.5-fold less budding and the lack of M or HN protein resulting in almost a complete lack of budding (Fig. 2B).
FIG. 2.
Requirements for the release of particles containing SV5 proteins from transfected cells. (A) 293T cells were either infected with rSV5 or transfected with the indicated plasmids. At 24 h p.i. or p.t., cells were radiolabeled for 16 h with [35S]Promix, followed by collection of both cells and culture media. Culture media was clarified by low-speed centrifugation, centrifuged through 35% sucrose cushions, and separated on sucrose flotation gradients into two equal fractions. The bottom fraction was discarded and proteins in the top (floated) fraction were analyzed by immunoprecipitation. Cells were disrupted by Dounce homogenization and centrifuged at low speed to remove nuclei and debris. SV5 proteins were immunoprecipitated from samples as described in Materials and Methods, and polypeptides were analyzed by SDS-PAGE on 10% gels. (B) Budding efficiency was quantified as the fraction of SV5 M protein detected in the media compared with the total SV5 M protein in the media plus the cell lysate. Values represent averages from three experiments, and the standard deviations are indicated.
The relative M and HN protein quantities in particles budded from transfected cells compared to SV5 virions were found to be very similar, with the M:HN ratio in particles released from transfected cells averaging 1.6:1 compared to 1.7:1 for virions in three different experiments. However, the relative abundances of M and NP proteins were found to be substantially different. The M:NP ratio in particles released upon transfection averaged 2.2:1 compared to 6.3:1 for virions, a difference of ∼3-fold. These numbers do not represent true polypeptide compositions, since they are based on immunoprecipitation of viral proteins, and while they do take into consideration different methionine and cysteine contents for the different proteins, they do not take into account differences in antibody affinities. However, from this analysis it does appear that particles budded from transfected cells differ from virions in that they contain substantially more NP protein relative to other viral proteins.
HN and F glycoproteins are interchangeable for budding of particles.
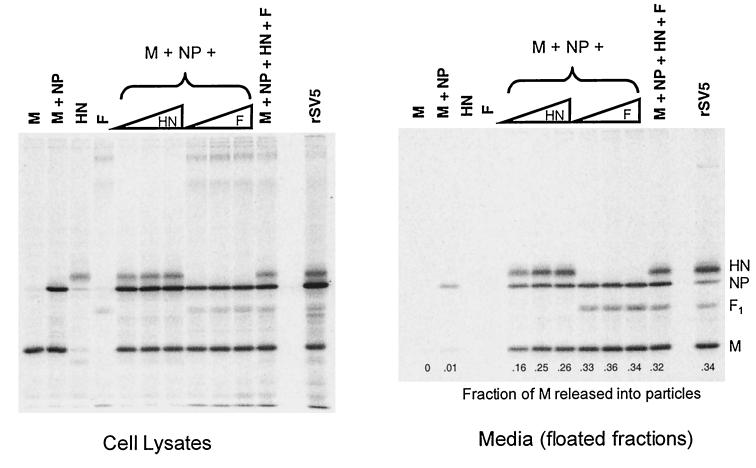
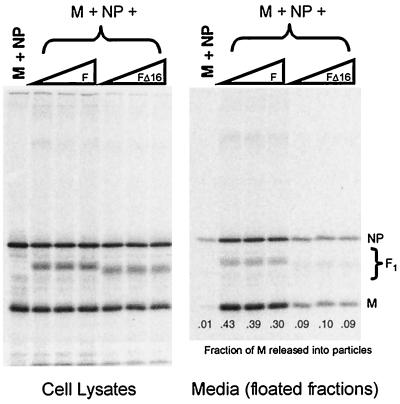
The HN glycoprotein is a type II membrane protein with a cytoplasmic tail that is 17 amino acids long, whereas the F glycoprotein is a type I membrane protein with a cytoplasmic tail that is 20 amino acids long. To test whether F protein, like HN protein, could function to direct budding, F protein was coexpressed in cells together with other viral components. In agreement with above data (Fig. 2), essentially no budding was detected when M and NP proteins were coexpressed in the absence of a glycoprotein, but with the inclusion of HN protein budding became efficient (Fig. 3). When F protein was coexpressed with the M and NP proteins (but without HN protein) budding was even more efficient: the fraction of M protein detected in the medium was ca. 1.4-fold higher than when HN protein was expressed. When both F and HN proteins were coexpressed with M and NP proteins, no further increase in budding was observed. These data indicate that the two SV5 glycoproteins likely contain separate and redundant elements necessary for efficient budding.
FIG. 3.
SV5 F protein can replace HN for budding of particles. 293T cells were either infected with rSV5 or transfected with plasmids as indicated. Plasmid quantities are indicated in Materials and Methods, except that increasing HN and F plasmid amounts represent 1.0, 1.5, and 2.0 μg of plasmid DNA. Budding assays were carried out as described in the legend to Fig. 2. Budding efficiency was calculated as the fraction of total SV5 M protein detected in the culture media.
Only specific glycoproteins can function for budding of SV5 particles.
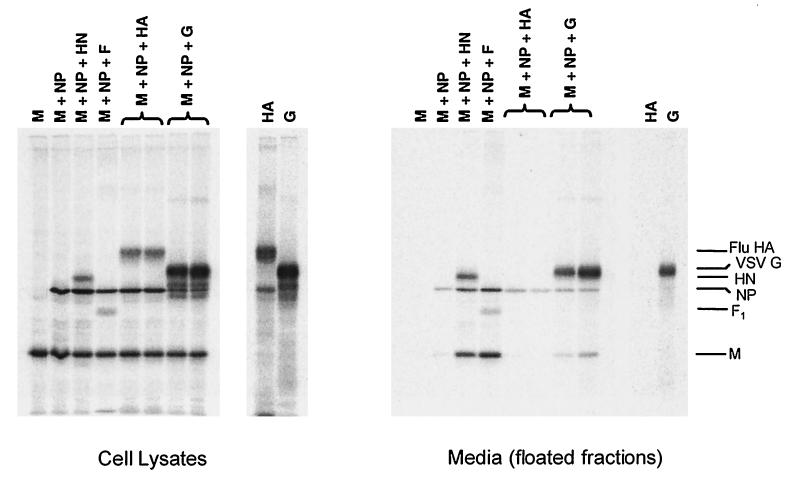
Since either of the two SV5 glycoproteins can function for budding, we investigated whether there was any specificity in the requirement for a glycoprotein by attempting to generate particles with glycoproteins from two other negative-strand RNA viruses distantly related to SV5: influenza A virus and VSV. Previous studies on the formation of influenza A VLPs have shown that, while glycoproteins are not required for budding driven by the influenza virus M protein, the glycoproteins hemagglutinin (HA) and neuraminidase (NA) are incorporated into these particles when the M protein and glycoproteins were coexpressed (13, 22). To determine whether an influenza glycoprotein could function for budding of SV5 particles, the influenza virus HA glycoprotein was coexpressed with the SV5 M and NP proteins. Budding of particles in this case was found to be very poor, similar to the level of budding observed when M and NP proteins were expressed without glycoprotein (Fig. 4). Thus, whereas SV5 glycoproteins can function together with SV5 M and NP proteins to bud particles, and influenza virus HA protein can function together with influenza virus M protein to form particles, influenza virus HA protein cannot function in conjunction with SV5 M and NP proteins to bud particles, indicating that some specificity exists in the requirement for a glycoprotein in the budding of SV5 particles.
FIG. 4.
Influenza A virus HA and VSV G proteins cannot replace SV5 glycoproteins for budding of particles. 293T cells were transfected with the indicated plasmids as described in Materials and Methods. Lanes designated M+NP+HA differ in the amount of influenza A virus HA plasmid: 1.5 μg (left) or 3.0 μg (right). Lanes designated M+NP+G differ in the amount of VSV G plasmid: 0.375 μg (left) or 0.75 μg (right). Budding assays were carried out as described in the legend to Fig. 2.
It has been found previously that expression of the VSV G glycoprotein at high levels can lead to blebbing (vesiculation) of the plasma membrane and secretion of G from cells (38) and, as shown in Fig. 4, efficient budding of G-containing particles occurred on expression of G in cells. To investigate whether coexpression of G in conjunction with SV5 proteins would result in efficient incorporation of SV5 proteins into these particles, G was coexpressed with the SV5 M and NP proteins. As shown in Fig. 4, particles containing G were released from cells, but incorporation of SV5 proteins into these particles was inefficient compared to incorporation of VSV G, suggesting that SV5 proteins could not function together with VSV G for budding of particles containing high levels of both VSV and SV5 proteins.
Particles released from transfected cells are morphologically similar to authentic SV5 virions.
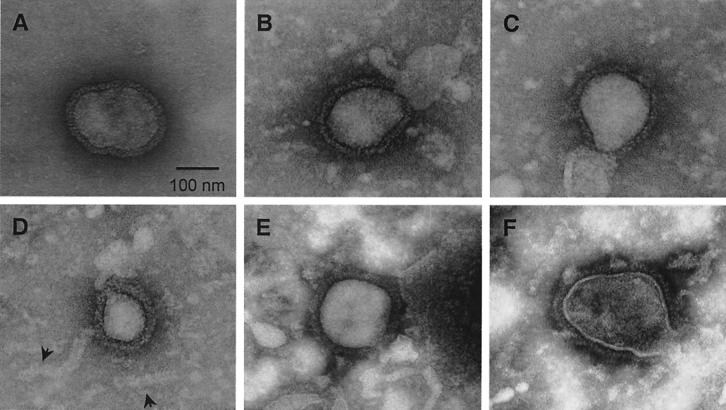
SV5 grown in tissue culture cells forms particles that have roughly spherical morphology and an average diameter of ca. 150 nm. The lipid envelope contains viral glycoproteins that form projections visible by electron microscopy as spikes (see Fig. 5A). To determine whether particles released from cells expressing M, NP, and HN (or F) proteins were similar in size and morphology to SV5 virions, concentrated medium fractions from transfected cells were examined by negative staining and electron microscopy. VLPs resembling typical paramyxovirus particles were found released from cells transfected with either M+NP+HN (Fig. 5B to D) or M+NP+F (Fig. 5E and F). The VLPs varied in size: many had diameters of between 100 and 200 nm, which is typical of SV5 virions, while others, such as the one shown in Fig. 5D, had diameters of between 50 and 100 nm and thus were smaller than typical virions. No overall differences in size were noted between VLPs made from HN- and F-protein-expressing cells. Spikes were clearly visible on the surfaces of the VLPs, and spike morphology was similar between VLPs and SV5 virions.
FIG. 5.
Particles released into the media upon coexpression of SV5 proteins resemble authentic SV5 virions. (A) MDBK cells were infected with rSV5, and virions were purified on sucrose gradients as described in Materials and Methods. (B to F) 293T cells were cotransfected with plasmids encoding the SV5 M, NP, and HN proteins (B to D) or M, NP, and F proteins (E and F). VLPs were pelleted from the culture media through 35% sucrose cushions. Virions and VLPs were adsorbed onto carbon-coated grids, negatively stained, and visualized by electron microscopy. Arrowheads indicate nucleocapsid-like structures that were occasionally observed in VLP preparations.
NP protein expressed in cells forms assembled nucleocapsid structures.
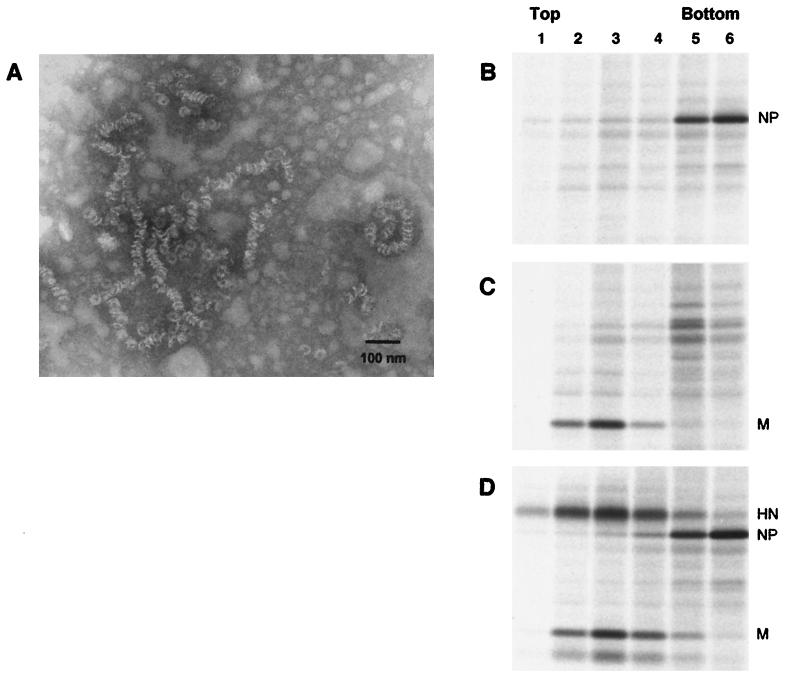
Examination of the VLPs by electron microscopy also indicated the presence of tubular structures which had a herringbone morphology characteristic of paramyxovirus nucleocapsid structures (Fig. 5D, arrowheads). Similar structures are frequently observed in purified virion preparations, presumably as a result of broken particles and the release of nucleocapsid structures. The structures observed from VLP preparations measured 15 to 20 nm in diameter, similar to the SV5 RNPs, but were much shorter and more variable in length than authentic RNPs which encapsidate the 15,246-nucleotide RNA genome of SV5. It has been found for several other paramyxoviruses that the NP protein expressed by itself in cells encapsidates nonspecifically cellular RNA and forms structures similar in morphology to viral RNPs (2, 5, 7, 8, 28, 42).
To investigate whether the nucleocapsid structures found on transient expression of SV5 NP protein were important for VLP budding, NP protein was expressed in cells and lysates were fractionated on a CsCl density gradient, which separates free NP protein from assembled nucleocapsid structures (2). Immunoprecipitation of NP protein from gradient fractions showed that nearly all of the NP protein had assembled into dense structures that sedimented toward the bottom of the gradient (Fig. 6B), and electron microscopy of these fractions revealed the presence of structures having the same herringbone morphology as had been observed in preparations of purified VLPs (Fig. 6A). These nucleocapsid-like structures varied in length from those of the single “washer” form to strands longer than 100 nm, observations consistent with previous studies on nonspecific assembly of paramyxovirus NP proteins with cellular RNA (7, 42). SV5 M protein, in contrast to NP protein, would not be expected to form this type of dense structure, and M protein did not sediment through a similar CsCl gradient when expressed alone (Fig. 6C). Furthermore, when M, NP, and HN proteins were coexpressed by using the same conditions known to generate VLPs, virtually all of the intracellular NP protein formed dense structures that sedimented through the CsCl gradient, whereas HN and M protein did not form such structures and remained at the top of the gradient (Fig. 6D). VLPs would not be expected to contribute significantly to the protein observed in Fig. 6D because VLPs bud from the cell, whereas the proteins examined are intracellular. Therefore, it is likely that assembled nucleocapsids contribute to the budding of VLPs, as under the conditions used to generate VLPs, very little of the NP protein in the cell remained unassembled.
FIG. 6.
SV5 NP protein expressed in cells assembles into nucleocapsid-like structures. (A) 293T cells were transfected with a plasmid encoding SV5 NP protein. At 40 h p.t., cells were disrupted by Dounce homogenization and centrifuged at low speed to remove nuclei and debris. The cell extract was layered onto a CsCl step gradient and fractionated as described in Materials and Methods. Structures from the bottom fraction were adsorbed onto carbon-coated grids, negatively stained, and visualized by electron microscopy. (B to D) 293T cells were transfected with plasmids encoding SV5 NP (B) or SV5 M (C) protein or cotransfected with plasmids encoding SV5 M, NP, and HN proteins (D). At 24 h p.t., cells were radiolabeled for 8 h with [35S]Promix, followed by Dounce homogenization of cells. Cell extracts were layered onto CsCl step gradients and centrifuged at 165,000 × g for 16 h, and gradients were divided into six equal fractions. SV5 proteins were immunoprecipitated from gradient fractions as described in Materials and Methods, and polypeptides were analyzed by SDS-PAGE on 10% gels.
Truncation of the HN cytoplasmic tail prevents efficient budding of both recombinant viruses and VLPs.
Previous studies have shown that SV5 budding requires the cytoplasmic tail of the HN glycoprotein: recombinant viruses with truncated HN cytoplasmic tails were found to be defective for the release of progeny virus particles (39). These findings made it possible to test whether the same factors that influenced the budding of virions from infected cells would also influence the budding of VLPs. Cells were transfected with M protein, NP protein, and either wild-type (wt) HN protein, cytoplasmic tail truncated HNΔ2-9 protein, or cytoplasmic tail truncated HNΔ2-13 protein. In parallel, cells were infected with either wt SV5 or recombinant SV5 harboring the HNΔ2-9 mutation. Previous work has shown that these alterations to the HN cytoplasmic tail do not substantially affect cell surface transport, endoglycosidase H cleavage kinetics, or the neuraminidase activities of the proteins (39). As found previously, rSV5 HNΔ2-9 was defective for budding: the fraction of M released into virus particles was fourfold less than for wt SV5 (Fig. 7A). In the VLP assay, replacement of wt HN with HNΔ2-9 led to a sixfold reduction in the fraction of M released into VLPs, and further truncation of the HN cytoplasmic tail (HNΔ2-13) led to even poorer VLP budding (Fig. 7B). Thus, the data from experiments with both recombinant viruses and VLPs are in agreement and indicate that truncation of the cytoplasmic tail of HN is detrimental to budding.
FIG. 7.
Truncation of the HN cytoplasmic tail results in defective budding of both recombinant viruses and VLPs. 293T cells were either infected with the indicated viruses (A) or transfected with the indicated plasmids (B). Budding assays were carried out as described in the legend to Fig. 2. Budding efficiency was calculated as the fraction of total SV5 M protein detected in the culture media.
Truncation of the F cytoplasmic tail prevents efficient budding of VLPs.
We next tested whether the cytoplasmic tail of F protein was important for the budding of VLPs. Cells were transfected with M protein, NP protein, and either wt F protein or cytoplasmic-tail-truncated FΔ16 protein, in which the 16 C-terminal amino acids of the F protein have been removed. FΔ16 protein is expressed at the cell surface, and its transport rate to the trans Golgi network is similar to that of wt F protein (David. L. Waning, Anthony P. Schmitt, and Robert A. Lamb, unpublished data). For VLP formation, expression of FΔ16 protein in place of wt F protein led to a fourfold reduction in the fraction of M protein released into VLPs (Fig. 8). Thus, truncation of the F cytoplasmic tail, similar to the effect of truncation of the HN cytoplasmic tail, prevented the protein from directing efficient budding of VLPs.
FIG.8.
Truncation of the F cytoplasmic tail results in defective budding of VLPs. 293T cells were transfected with the indicated plasmids. Plasmid quantities are indicated in Materials and Methods, except that increasing F and FΔ16 plasmid amounts represent 1.0, 1.5, and 2.0 μg of plasmid DNA. Budding assays were carried out as described in the legend to Fig. 2. Budding efficiency was calculated as the fraction of total SV5 M protein detected in the culture media.
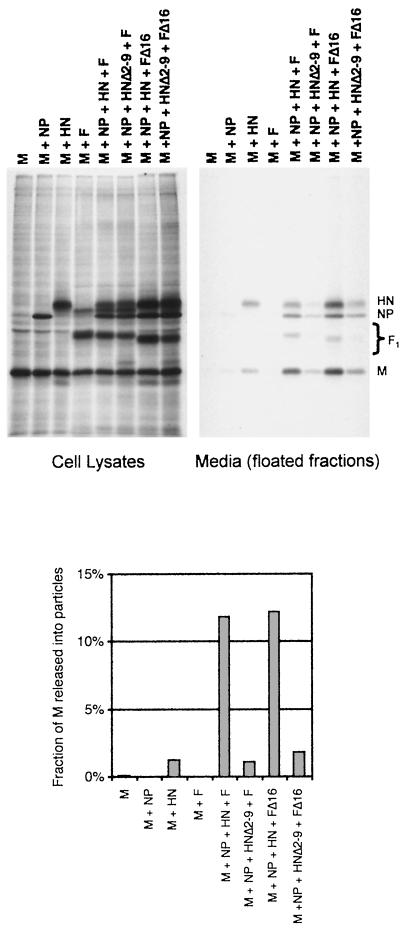
Presence of cytoplasmic tail-truncated HN prevents F from directing efficient VLP budding.
Either HN or F protein could function for VLP budding (Fig. 3), suggesting redundant roles for HN and F glycoprotein cytoplasmic tails in the budding of SV5. However, recombinant virus rSV5 HNΔ2-9, which has a truncated HN cytoplasmic tail but a normal F protein, budded poorly (Fig. 7A). Thus, we sought to investigate the conundrum of why the wt F protein expressed by rSV5 HNΔ2-9 did not direct normal budding, as it does for VLP budding when HN is not expressed. Therefore, VLP experiments were performed in which mutant HNΔ2-9 was coexpressed along with wt F protein, mimicking glycoprotein expression found in cells infected by mutant virus rSV5 HNΔ2-9. As shown in Fig. 9, it was found that VLP budding was greatly inhibited when these two glycoproteins were coexpressed together with M and NP, to a level similar to the level of VLP budding found on expression of M and NP protein together with the two mutant glycoproteins HNΔ2-9 and FΔ16. Thus, while budding occurred efficiently in the presence of F protein without HN, budding was poor in the presence of F protein together with mutant HNΔ2-9, indicating that this mutant protein had a negative effect on budding. When the converse experiment was performed and wt HN protein was coexpressed with mutant FΔ16, budding was found to be very efficient and reached nearly the same level as when wt HN and wt F proteins were coexpressed (Fig. 9). In this case, budding was presumably directed by the wt HN protein, since FΔ16 itself failed to direct efficient budding of VLPs (Fig. 8). However, while FΔ16 was not functional for budding, neither was it inhibitory, since its presence did not affect the ability of HN to induce VLP budding. Thus, while both HN and F depended on the presence of cytoplasmic tails to function for budding of VLPs, only HN was inhibitory toward budding when its cytoplasmic tail was removed.
FIG. 9.
The presence of a truncated HN cytoplasmic tail prevents F protein from directing efficient VLP budding. 293T cells were transfected with the indicated plasmids. Budding assays were carried out as described in the legend to Fig. 2. Budding efficiency was calculated as the fraction of total SV5 M protein detected in the culture media.
Truncation of the HN cytoplasmic tail affects an early step in the budding process.
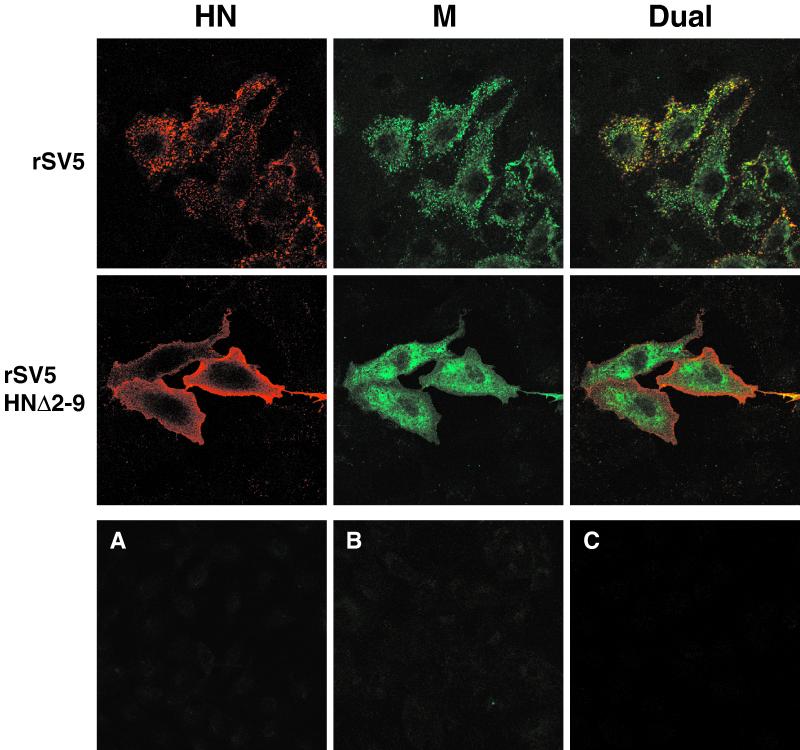
To examine further the budding defect caused by expression of mutant HN protein, cells infected with wt SV5 or budding-defective rSV5 HNΔ2-9 were examined by confocal microscopy. If budding of rSV5 HNΔ2-9 is inhibited at an early stage, such as targeting of viral components to the plasma membrane and assembly into concentrated budding patches, then it could be anticipated that viral proteins would appear to be mislocalized in infected cells. In contrast, if the block in virus budding of rSV5 HNΔ2-9 is at a late stage, such as bending of the plasma membrane and membrane fission to release particles, then it could be anticipated that targeting of viral proteins to budding sites on the plasma membranes of infected cells would still occur. Although we have reported previously a related experiment (39), it was not definitive since dual labeling was not used. Analysis of infected cells by dual labeling and confocal microscopy (Fig. 10) showed clear mislocalization of both HN and M proteins in cells infected with the budding-defective virus rSV5 HNΔ2-9 compared to wt SV5. In wt SV5-infected cells, surface staining of HN protein as well as intracellular staining of M protein revealed the organization of these two proteins into brightly stained patches, and dual labeling showed that both proteins were contained within the same patches. This staining pattern, which was not observed upon transfection of cells with either HN or M protein (not shown), is consistent with the idea that viral proteins concentrate at specific sites on the plasma membrane from which virus subsequently buds. In cells infected with rSV5 HNΔ2-9, HN and M proteins were clearly mislocalized and the organized staining pattern seen for wt SV5 was lost (Fig. 10). HNΔ2-9 protein was distributed uniformly across the cell surface, and M protein was found randomly dispersed throughout the cytoplasm of the cell. These results demonstrate an early defect in SV5 assembly upon truncation of the HN cytoplasmic tail and suggest that in the absence of presumed interactions between the glycoprotein cytoplasmic tails and M protein, organization and concentration of viral components into budding sites on the plasma membranes of infected cells cannot occur.
FIG. 10.
Recombinant SV5 with truncated HN cytoplasmic tail is defective at an early step in budding. CV-1 cells grown on glass coverslips were infected for 16 h, fixed with formaldehyde, and bound with the HN-specific MAb HN1b (IgG2a isotype), followed by an IgG2a-specific R-phycoerythrin-conjugated secondary antibody. Cells were then fixed a second time, permeabilized with 0.1% saponin, and bound with the M-specific MAb M-h (IgG3 isotype), followed by an IgG3-specific fluorescein isothiocyanate-conjugated secondary antibody. Fluorescence was examined with a Zeiss LSM 410 confocal microscope. (A) Mock-infected control cells treated as described above; (B) rSV5-infected cells bound with HN1b, followed by the IgG3-specific fluorescein isothiocyanate conjugate; (C) rSV5-infected cells permeabilized and bound with M-h, followed by the IgG2a-specific R-phycoerythrin conjugate.
DISCUSSION
VLP systems have proven to be useful tools for the study of virus budding. Retrovirus budding assays, for example, have played a key role in defining budding domains within Gag, and further study of these domains, e.g., the retroviral L domain, have provided important insights into how viruses may exploit cellular pathways to mediate their own exit from cells (1, 12, 35, 36, 48).
We have established here a system for generating SV5 VLPs from cells transfected with cDNAs. Unlike previous studies of VLP budding with other negative-strand RNA viruses such as VSV and Ebola virus in which efficient budding was observed upon expression of viral M proteins alone (15, 16, 21, 23, 47), we found a requirement for coexpression of multiple SV5 proteins in order to achieve efficient budding of particles. It is unclear at this point whether this functional difference between viral M proteins is related to the observation that some M proteins (such as VSV M and Ebola VP-40) contain L domain sequences identical to those found in many retroviral Gag polyproteins, whereas other M proteins (such as SV5 M) do not contain any of the known retroviral L domain sequences. It is also unclear whether VLP budding directed solely by influenza A virus M protein or hPIV-1 M protein occurs with an efficiency that is comparable to a virus infection. It is possible that coexpression of multiple viral proteins in these systems could improve budding efficiency, since the combination of proteins that proved effective for SV5 VLP budding (M protein, NP protein, and a viral glycoprotein) was not reported as having been tried in these studies of VLP budding. In our case, budding was found to be remarkably efficient when all of these components were included, with budding efficiency from transfected cells differing by <2-fold from that found for SV5-infected cells.
Efficient budding of SV5 VLPs required expression of NP protein. Paramyxovirus NP proteins have been found previously to assemble into nucleocapsid-like structures when synthesized in either mammalian cells or in insect cells as a result of uncontrolled encapsidation of cellular RNA (2, 5, 7, 8, 28, 42). We observed similar assembly of SV5 NP protein into nucleocapsid-like structures both when NP protein was expressed alone and when NP protein was coexpressed with other SV5 proteins. Furthermore, the efficiency of NP protein assembly into nucleocapsid-like structures was very high, such that very little NP protein in the transfected cells remained unassembled. This suggests that the requirement for NP protein for VLP budding might be due to a dependence of budding on assembled nucleocapsid structures. It is conceivable that such a requirement could favor the production of genome-containing particles over empty particles in a virus infection. Of course, to obtain infectious virions from virus-infected cells, packaging of encapsidated viral genomes occurs and packaging of cellular RNAs is largely avoided. It is thought that in virus-infected cells, uncontrolled encapsidation of cellular RNA must be inhibited, and that the paramyxovirus P protein may play a role in this inhibition (2, 6, 7). This could result in a situation where there is no requirement to discriminate between encapsidated cellular and viral RNAs at budding sites, because virtually all encapsidated products in the infected cell would contain viral RNA. On the other hand, it is possible that inhibition of nonspecific encapsidation in virus-infected cells is incomplete, and therefore active selection of encapsidated viral RNAs at budding sites would be important to maximize the yield of infectious particles. It may in the future be possible to extend the VLP system to include both P protein and paramyxovirus minigenome RNA expression to study further this possibility.
The SV5 VLPs were found to differ from virions in terms of NP protein content. The ratio of NP protein to M protein was ∼3-fold higher in VLPs formed on M+NP+HN coexpression than the ratio found in SV5 virions. Although it cannot be ruled out that the extra NP protein was released into the culture media independently of VLPs and not contained in the VLPs themselves, this seems unlikely given that the VLP isolation procedure included fractionation on a sucrose flotation gradient. Thus, while assembled nucleocapsid-like structures could have been released into the culture media from dead or damaged cells and would have copelleted with VLPs through a 35% sucrose cushion, they should not have been able to float with VLPs on a sucrose gradient unless they were associated with membranes. For this reason, we favor the possibility that the VLPs are packed with nonspecifically encapsidated cellular RNA even more densely than virions are packed with encapsidated viral RNA.
A role for glycoprotein cytoplasmic tails in the budding of negative-strand RNA viruses has been previously established, since a variety of recombinant viruses containing altered glycoprotein cytoplasmic tails have been found to bud poorly (10, 20, 26, 39, 40). Here, we show a role for glycoprotein cytoplasmic tails in the budding of paramyxovirus VLPs. Furthermore, we show that the same alterations to the SV5 HN cytoplasmic tail that lead to inefficient budding of a recombinant virus also lead to poor budding of VLPs. This suggests that the VLP system could be used to discriminate between viral proteins that are budding competent and budding defective and should allow future dissection of viral proteins to identify critical budding domains, similar to the approaches taken with retroviral Gag proteins to define M, I, and L budding domains.
The two SV5 glycoproteins, HN and F, were found to be interchangeable for VLP budding. This suggests that the assembly function performed by SV5 glycoproteins is highly important and that there is redundancy in the information needed for assembly. Such redundancy has a precedent with another negative-strand RNA virus, influenza A virus. Here, the cytoplasmic tails of the two glycoproteins, HA and NA, function to control not only budding efficiency but also particle size and shape. Removal of either cytoplasmic tail by itself in a recombinant virus resulted in only mild or moderate defects in virus budding and particle morphology. However, a virus lacking both glycoprotein cytoplasmic tails was found to bud very poorly, and particles were grossly deformed (19, 20, 29, 52). Thus, redundant assembly functions among multiple glycoprotein cytoplasmic tails may be beneficial to a wide variety of enveloped viruses.
Deletion of the HN cytoplasmic tail was found to be inhibitory toward budding. VLP budding was poor in the presence of both HNΔ2-9 and F proteins but was efficient in the presence of F protein alone. This observation is in agreement with the phenotype of recombinant virus rSV5 HNΔ2-9, which buds poorly despite coding for a wt F protein. Inhibition of budding appeared to be at an early step, based on examination of virus-infected cells by confocal microscopy, which showed mislocalization of HN and M proteins in mutant virus-infected cells. The fact that M protein was mislocalized in these cells may explain partly the negative effect on budding, since the presence of wt F protein in this case would not necessarily be expected to overcome mislocalization of M protein away from budding sites. One hypothesis is that M protein, once bound to the plasma membrane, assembles into budding sites partly as a result of interactions with glycoprotein cytoplasmic tails. Removal of the HN cytoplasmic tail may result in mislocalization of the glycoproteins and prevent M protein from organizing into appropriate sites at the plasma membrane.
Acknowledgments
This work was supported in part by Research Grant AI-23173 from the National Institute of Allergy and Infectious Disease. A.P.S. is an Associate and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchholz, C. J., D. Spehner, R. Drillien, W. J. Neubert, and H. E. Homann. 1993. The conserved N-terminal region of Sendai virus nucleocapsid protein NP is required for nucleocapsid assembly. J. Virol. 67:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cathomen, T., B. Mrkic, D. Spehner, R. Drillien, R. Naef, J. Pavlovic, A. Aguzzi, M. A. Billeter, and R. Cattaneo. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 17:3899-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronel, E. C., K. G. Murti, T. Takimoto, and A. Portner. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J. Virol. 73:7035-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curran, J., J. B. Marq, and D. Kolakofsky. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 69:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Errington, W., and P. T. Emmerson. 1997. Assembly of recombinant Newcastle disease virus nucleocapsid protein into nucleocapsid-like structures is inhibited by the phosphoprotein. J. Gen. Virol. 78:2335-2339. [DOI] [PubMed] [Google Scholar]
- 8.Fooks, A. R., J. R. Stephenson, A. Warnes, A. B. Dowsett, B. K. Rima, and G. W. Wilkinson. 1993. Measles virus nucleocapsid protein expressed in insect cells assembles into nucleocapsid-like structures. J. Gen. Virol. 74:1439-1444. [DOI] [PubMed] [Google Scholar]
- 9.Forsell, K., L. Xing, T. Kozlovska, R. H. Cheng, and H. Garoff. 2000. Membrane proteins organize a symmetrical virus. EMBO J. 19:5081-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouillot-Coriou, N., and L. Roux. 2000. Structure-function analysis of the Sendai virus F and HN cytoplasmic domain: different role for the two proteins in the production of virus particle. Virology 270:464-475. [DOI] [PubMed] [Google Scholar]
- 11.Franke, E. K., H. E. Yuan, K. L. Bossolt, S. P. Goff, and J. Luban. 1994. Specificity and sequence requirements for interactions between various retroviral Gag proteins. J. Virol. 68:5300-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Puertas, P., C. Albo, E. Perez-Pastrana, A. Vivo, and A. Portela. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538-11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, B., G. Y. Lin, J. E. Durbin, R. K. Durbin, and R. A. Lamb. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J. Virol. 75:4068-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, B., R. G. Paterson, C. D. Ward, and R. A. Lamb. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249-260. [DOI] [PubMed] [Google Scholar]
- 19.Jin, H., G. Leser, and R. A. Lamb. 1994. The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO J. 13:5504-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Justice, P. A., W. Sun, Y. Li, Z. Ye, P. R. Grigera, and R. R. Wagner. 1995. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J. Virol. 69:3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latham, T., and J. M. Galarza. 2001. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 75:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Y., L. Luo, M. Schubert, R. R. Wagner, and C. Y. Kang. 1993. Viral liposomes released from insect cells infected with recombinant baculovirus expressing the matrix protein of vesicular stomatitis virus. J. Virol. 67:4415-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez, S., J.-S. Yao, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1994. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J. Virol. 68:1316-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mebatsion, T., M. Konig, and K.-K. Conzelmann. 1996. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84:941-951. [DOI] [PubMed] [Google Scholar]
- 27.Mebatsion, T., F. Weiland, and K. K. Conzelmann. 1999. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J. Virol. 73:242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meric, C., D. Spehner, and V. Mazarin. 1994. Respiratory syncytial virus nucleocapsid protein (N) expressed in insect cells forms nucleocapsid-like structures. Virus Res. 31:187-201. [DOI] [PubMed] [Google Scholar]
- 29.Mitnaul, L. J., M. R. Castrucci, K. G. Murti, and Y. Kawaoka. 1996. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J. Virol. 70:873-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 31.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73. In A. Davidson and R. M. Elliott (ed.), Molecular virology: a practical approach. IRL Oxford University Press, Oxford, England.
- 34.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randall, R. E., D. F. Young, K. K. A. Goswami, and W. C. Russell. 1987. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 68:2769-2780. [DOI] [PubMed] [Google Scholar]
- 38.Rolls, M. M., P. Webster, N. H. Balba, and J. K. Rose. 1994. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell 79:497-506. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt, A. P., B. He, and R. A. Lamb. 1999. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J. Virol. 73:8703-8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnell, M. J., L. Buonocore, E. Boritz, H. P. Ghosh, R. Chernish, and J. K. Rose. 1998. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 17:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with Gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spehner, D., A. Kirn, and R. Drillien. 1991. Assembly of nucleocapsid-like structures in animal cells infected with a vaccinia virus recombinant encoding the measles virus nucleoprotein. J. Virol. 65:6296-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strauss, J. H., E. G. Strauss, and R. J. Kuhn. 1995. Budding of alphaviruses. Trends Microbiol. 3:346-350. [DOI] [PubMed] [Google Scholar]
- 45.Suomalainen, M., P. Liljestrom, and H. Garoff. 1992. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J. Virol. 66:4737-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Press, Cold Spring Harbor, N.Y. [PubMed]
- 47.Timmins, J., S. Scianimanico, G. Schoehn, and W. Weissenhorn. 2001. Vesicular release of ebola virus matrix protein VP40. Virology 283:1-6. [DOI] [PubMed] [Google Scholar]
- 48.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young, D. F., N. Chatziandreou, B. He, S. Goodbourn, R. A. Lamb, and R. E. Randall. 2001. Single amino acid substitution in the V protein of simian virus 5 differentiates its ability to block interferon signaling in human and murine cells. J. Virol. 75:3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, J., G. P. Leser, A. Pekosz, and R. A. Lamb. 2000. The cytoplasmic tails of the influenza virus spike glycoproteins are required for normal genome packaging. Virology 269:325-334. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao, H., B. Lindqvist, H. Garoff, C. H. von Bonsdorff, and P. Liljestrom. 1994. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 13:4204-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]