Abstract
Although biological and biochemical data have been accumulated on most hepatitis C virus proteins, the structure and function of the 63-amino-acid p7 polypeptide of this virus have never been investigated. In this work, sequence analyses predicted that p7 contains two transmembrane passages connected by a short hydrophilic segment. The C-terminal transmembrane domain of p7 was predicted to function as a signal sequence, which was confirmed experimentally by analyzing the translocation of a reporter glycoprotein fused at its C terminus. The p7 polypeptide was tagged either with the ectodomain of CD4 or with a Myc epitope to study its membrane integration, its subcellular localization, and its topology. Alkaline extraction studies confirmed that p7 is an integral membrane polypeptide. The CD4-p7 chimera was detected by immunofluorescence on the surface of nonpermeabilized cells, indicating that it is exported to the plasma membrane. However, pulse-chase analyses showed that only approximately 20% of endoglycosidase H-resistant CD4-p7 was detected after long chase times, suggesting that a large proportion of p7 stays in an early compartment of the secretory pathway. Finally, by inserting a Myc epitope in several positions of p7 and analyzing the accessibility of this epitope on the plasma membrane of HepG2 cells, we showed that p7 has a double membrane-spanning topology, with both its N and C termini oriented toward the extracellular environment. Altogether, these data indicate that p7 is a polytopic membrane protein that could have a functional role in several compartments of the secretory pathway.
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma throughout the world (22). HCV is a positive-stranded RNA virus that belongs to the Hepacivirus genus. Together with the genera Flavivirus and Pestivirus, HCV belongs to the Flaviviridae family (43, 52). The HCV genome encodes a single polyprotein precursor of approximately 3,000 amino acid residues. This polyprotein precursor is co- and posttranslationally processed by cellular and viral proteases to yield the mature structural and nonstructural proteins C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (see reference 41 for a recent review) (Fig. 1A).
FIG. 1.
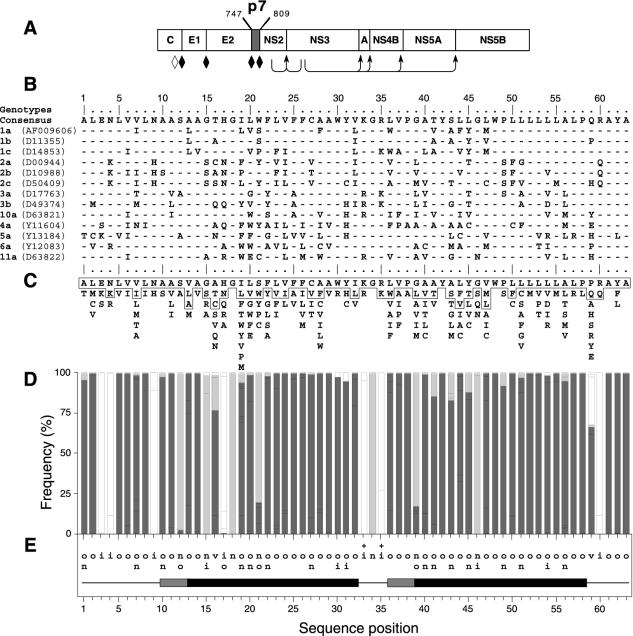
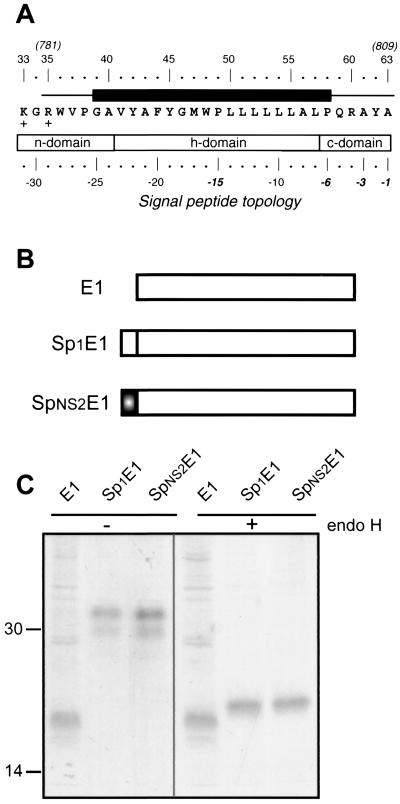
Sequence analyses of p7. (A) Position of p7 in the HCV polyprotein. p7 numbering is according to genotype 1a HCV H strain consensus cDNA (26) (accession number AF009606). Solid diamonds indicate cleavages of the HCV polyprotein precursor by an ER signal peptidase(s), and arrows indicate cleavages by the NS2-3 and NS3/4A proteases. The open diamond indicates further processing of the capsid protein by a cellular protease. (B) Alignment of p7 sequences representative of the principal HCV subtypes of clades 1 to 6. p7 numbering has been normalized from 1 to 63. The EMBL accession number of each sequence is indicated in parentheses. The consensus amino acid sequence for the 13 selected sequences is indicated on the top of the panel. Amino acids identical to those in the consensus sequence are represented by a dash. (C) Repertoire of amino acids per position in 289 HCV isolates of various genotypes. Amino acids are listed in decreasing order of observed frequency, from top to bottom. Amino acids within the box correspond to residues observed in more than 10% of the 289 sequences. Amino acids observed at a given position in fewer than two distinct sequences (<0.3%) were not taken into consideration. (D) Histogram showing the hydropathic character of residues at each p7 position. The height of each box in each bar indicates the number of sequences observed with a given residue at a given position. The boxes are presented in order of decreasing hydrophobicity, from bottom to top, according to the hydrophobicity scale of Black and Mould (F, I, W, Y, L, V, M, P, C, A, G, T, S, K, Q, N, H, E, D, R). Each box is colored according to the hydrophobic character of the residue: dark grey for hydrophobic (F, I, W, Y, L, V, M, P, C, and A), light grey for neutral (G, T, and S), and white for hydrophilic (K, Q, N, H, E, D, and R). (E) Consensus hydropathic pattern deduced from the data in panel D. o, hydrophobic residue; n, neutral residue; i, hydrophilic residue; v, variable residue; +, fully conserved positively charged residues. The black boxes indicate predicted minimal transmembrane segments deduced by various prediction methods (see Materials and Methods section). The grey boxes indicate that the corresponding amino acid positions were predicted to be membranous but not for all the HCV isolates.
Recently, it has been reported that an additional HCV protein can be produced by a ribosomal frameshift in the N-terminal region of the polyprotein (55). The structural proteins, i.e., capsid, E1, and E2, are released from the polyprotein by the endoplasmic reticulum (ER) signal peptidase(s) of the host cell. Further processing occurs at the C terminus of the capsid; however, the protease involved in this cleavage has not been identified (34). The nonstructural proteins (NS3, NS4A, NS4B, NS5A, and NS5B) are released from the polyprotein after cleavage by HCV proteases NS2-3 and NS3/4A. The cleavage between p7 and NS2 is supposed to be mediated by a cellular signal peptidase (30, 35). Although most cleavages in the polyprotein precursor proceed to completion during or immediately after translation, cleavages are delayed at the E2/p7 and p7/NS2 sites, leading to the production of an E2-p7-NS2 precursor (12). In addition, processing between E2 and p7 is incomplete and results in the production of fully processed E2 and uncleaved E2-p7 (30, 35).
The p7 polypeptide of HCV is a small hydrophobic protein (30) which has not been well characterized yet. Indeed, the structure of this 63-amino-acid-long polypeptide has not been determined, and its putative function(s) remains unknown. In the Pestivirus genus, a polypeptide called p7 is similar to HCV p7. It is a small 70-amino-acid-long polypeptide consisting mostly of hydrophobic residues which is also located between E2 and NS2 on the pestivirus polyprotein (14, 21). As observed for HCV, cleavage between E2 and p7 is also incomplete and results in the production of E2-p7, E2, and p7.
Functional data on pestivirus p7 have been obtained by introducing mutations into an infectious cDNA clone of bovine viral diarrhea virus (BVDV). A large in-frame deletion in p7 has shown that RNA replication is not affected, but no infectious virus is produced (21). In addition, p7 provided in trans is capable of functional complementation. Taken together, these data suggest that the pestivirus p7 is essential for the production of progeny virus.
To gain more information on the HCV p7 polypeptide, we coupled extensive amino acid sequence analyses and structure prediction to experimental investigations. We determined the topology of p7 and its subcellular localization and showed that a fraction of p7 is exported to the cell surface. We demonstrated that p7 is a polytopic membrane protein that crosses the plasma membrane twice and has its N and C termini oriented toward the extracellular environment. Finally, we demonstrated that the C-terminal transmembrane domain of p7 is a signal sequence.
MATERIALS AND METHODS
Sequence analyses and structure predictions.
All analyses were made using the Institut de Biologie et Chimie des Protéines HCV database website facility (HCVDB; http://hepatitis.ibcp.fr), which contains all reported HCV sequences from the EMBL database. The p7 sequence of the HCV H strain consensus cDNA (26) (accession number AF009606) was used to retrieve all reported isolates by using the FASTA homology search program (40). Incomplete sequences were removed from the list of matching sequences. A final set of 289 sequences of all genotypes were analyzed to construct Fig. 1.
Multiple sequence alignments and the consensus sequence determination were carried out with the Clustal W program (51). Selection of p7 sequences of clade 1 (genotypes 1a, 1b, and 1c) was done using the HCVSRS tool (HCV Sequence Retrieval System) of HCVDB. Visualization of sequence alignments and plotting of the most frequently represented amino acid residues at each position were done using the MPSA program (2). At each amino acid sequence position, the residue types and their respective frequencies were computed using a program developed at IBCP (C. Combet, unpublished data).
Various methods were combined for the prediction of transmembrane sequences: PHDhtm (45) (http://www.embl-heidelberg.de/predictprotein/), TMHMM (50) (http://www.cbs.dtu.dk/services/TMHMM-1.0/), DAS (8) (http://www.sbc.su.se/≈miklos/DAS/), and TopPred2 (4) (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html).
Plasmid constructs.
HCV sequences were amplified from clones derived from the H strain (16). Briefly, DNA sequences of proteins of interest were introduced into plasmid pTM1 (38) by PCR with appropriate oligonucleotides and templates. Plasmids expressing chimeric proteins were constructed in two steps by introducing successively the sequences of domains from two different proteins. A unique restriction site was introduced at the junction between the sequences of the protein domains.
Plasmid pTM1/CD4 has been described previously (6). Plasmid pTM1/CD4-p7 contains the signal sequence of CD4 followed by the sequence of its ectodomain (encoding amino acids 1 to 371) in fusion with p7 (encoding amino acids 747 to 809; positions on the HCV polyprotein). Between these two sequences, there is a junction sequence encoding two additional amino acids (Gly and Ser). Plasmid pTM1/p7NT contains p7 with its signal sequence and a Myc epitope (EQKLISEEDL) surrounded by three glycine residues inserted at the N terminus of p7, downstream of the signal peptide cleavage site. Plasmid pTM1/p7KR contains p7 with its signal sequence and a Myc epitope surrounded by three glycine residues inserted between lysine 779 and arginine 781 of p7. Plasmid pTM1/p7CT contains p7 with its signal sequence, a junction containing three glycine residues, and a Myc epitope inserted at the C terminus of p7. The last residue (alanine) at the carboxy terminus of p7 of pTM1/p7CT was replaced with an arginine to avoid signal sequence cleavage.
Plasmid pTM1/CE1E2p7CT contains the N terminus of the HCV polyprotein ending at p7 with a Myc epitope tag at the C terminus of p7 introduced as described for pTM1/p7CT. Plasmid pTM1/NS2-3-4A-4B-5A-5B contains the sequence of the C terminal two-thirds of the HCV polyprotein. Plasmid pTM1/E1 contains the entire sequence of E1 without its signal sequence. Plasmid pTM1/Sp1E1 contains the entire sequence of E1 with its signal sequence. Plasmid pTM1/SpNS2E1 contains the sequence of the second transmembrane domain of p7 (encoding amino acids 781 to 809) in fusion with the entire sequence of E1. Plasmids containing sequences amplified by PCR were verified by sequencing.
Cell culture.
The HepG2, CV-1, and 143B (thymidine kinase-deficient) cell lines were obtained from the American Type Culture Collection, Rockville, Md. Cell monolayers were grown in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% fetal bovine serum (Gibco-BRL).
Generation and growth of viruses.
Vaccinia virus recombinants were generated by homologous recombination as previously described (25) and plaque purified twice on thymidine kinase-deficient 143B cells under bromodeoxyuridine (50 μg/ml; Sigma) selection. Stocks of vTF7-3 (a vaccinia virus recombinant expressing the T7 DNA-dependent RNA polymerase) (17), the wild-type vaccinia virus strain (Copenhagen) and its thermosensitive derivative ts7 (10), and vaccinia virus recombinants expressing HCV proteins or chimeric proteins were grown and titrated on CV-1 cell monolayers. The genes of HCV proteins expressed in this work are under the control of a T7 promoter, and expression of the proteins of interest is achieved by coinfection with vTF7-3.
Antibodies.
Monoclonal antibodies (MAbs) A4 (anti-E1 [12]), OKT4 (anti-CD4; ATCC CRL-8002 [42]), and MYC 9E10 (anti-Myc; ATCC CRL-1729 [15]) were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer.
Sodium carbonate extraction of membranes.
At 5 h postinfection, infected cells were washed twice with 0.25 M sucrose-5 mM HEPES buffer (pH 6.8), resuspended in the same buffer, and disrupted using a Dounce homogenizer (30 strokes). The homogenates were centrifuged for 10 min at 2,000 rpm to remove intact cells and nuclei. Supernatants were spun for 15 min at 65,000 rpm in a Beckman TL-100 centrifuge. Membrane pellets were resuspended in 0.5 ml of 0.1 M sodium carbonate, pH 11.3, using a Dounce homogenizer (10 strokes) and incubated for 30 min on ice. The extracted proteins were separated from membranes by an additional centrifugation for 15 min at 65,000 rpm. Membranes were again resuspended in 0.5 ml of 0.1 M sodium carbonate, pH 11.3. After neutralization to pH 7 by the addition of 1 N HCl, samples were mixed with 2× Laemmli sample buffer and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and membrane-bound and soluble proteins were revealed by Western blotting.
Western blotting.
After separation by SDS-PAGE under reducing conditions, proteins were transferred to nitrocellulose membranes (Hybond C; Amersham) by using a Trans-Blot apparatus (Bio-Rad) and detected with the specific anti-Myc MAb (dilution 1:200) followed by rabbit anti-mouse immunoglobulin conjugated to peroxidase (dilution 1:2,500; Rockland). Proteins were revealed by enhanced chemiluminescence detection (ECL system) as recommended by the manufacturer (Amersham).
Indirect immunofluorescence.
Subconfluent HepG2 cells grown on coverslips were infected with the appropriate vaccinia virus recombinants at a multiplicity of infection of 3 PFU/cell. At 6 h postinfection, cells were fixed for 10 min with paraformaldehyde (4% in phosphate-buffered saline [PBS]) and then permeabilized or not for 30 min with PBS containing 0.1% Triton X-100. Cells were stained with anti-CD4 MAb OKT4 (dilution 1:250) or anti-Myc MAb (dilution 1:100) followed by rhodamine-conjugated donkey anti-mouse (Jackson) immunoglobulin (dilution 1:1,000).
Metabolic labeling, immunoprecipitation, and endo H digestion.
HepG2 cells grown to confluence were infected with the appropriate vaccinia virus recombinants and metabolically labeled with 35S-Protein Labeling Mix (3.7 × 106 Bq/ml) as previously described (12). Cells were lysed with 0.5% Triton X-100 (Sigma) in 50 mM Tris-HCl (pH 7.4)-150 mM NaCl-5 mM EDTA. Immunoprecipitations were carried out as previously described (13). Immunoprecipitated proteins were eluted from protein A-Sepharose (Pharmacia) in 30 μl of dissociation buffer (0.5% SDS and 1% 2-mercaptoethanol) by boiling for 10 min. The protein samples were then divided into two equal portions for digestions with endo-β-N-acetylglucosaminidase H (endo H; New England Biolabs) or no digestion (control). Digestions were carried out for 1 h at 37°C in the buffer provided by the manufacturer. Digested samples were mixed with an equal volume of 2× Laemmli sample buffer and analyzed by SDS-PAGE. For quantitative experiments, the gels were analyzed with a PhosphorImager (Molecular Dynamics).
RESULTS
Sequence analyses of p7.
Sequence comparisons and structure predictions were performed to assess the degree of conservation of p7 sequence among different HCV isolates, to locate putative membrane sequences, and to identify putative structural and/or functional motifs. Alignment of p7 sequences representative of the major HCV subtypes of clades 1 to 6 showed a limited conservation of amino acids at most sequence positions (Fig. 1B). The p7 amino acid repertoire deduced from the analysis of 289 HCV isolates of various genotypes revealed that most positions are variable (Fig. 1C). However, most of the positions showing apparent variability are occupied by residues with similar hydropathic character, as illustrated in Fig. 1D, where residues were classified into three classes according to their hydrophobic character. A letter-coded motif summarizing the corresponding p7 hydropathic pattern is shown in Fig. 1E.
Typically, most p7 positions bear exclusively hydrophobic (o), neutral (n), or hydrophilic (i) residues, while a few positions bear two classes of residues, and only two positions bear the three classes of residues (named v for variable position). In addition, some positions bear specific residues (Gly 18, Gly 34, Tyr 42, Trp 48, Ala 61, and Ala 63) or very similar residues, in particular basic residues (Arg or Lys) at position 33 and 35, indicating that these residues are essential for the structure and/or function of p7. Overall, despite the apparent amino acid variability, the conservation of the hydropathic character at most positions and the full conservation of specific residues at several positions reveal a strong conservation of p7 structure and organization independent of the HCV genotype.
The p7 polypeptide is composed of two long hydrophobic stretches (segments 19 to 32 and 36 to 58) connected by a short basic segment (amino acids 33 to 35). Whatever the genotype and the method used (PHDhtm, TMHMM, DAS, and TopPred2; see Materials and Methods), these two hydrophobic segments were predicted to be transmembrane elements. With only 14 residues, the first hydrophobic segment (19 to 32, named TM1) is too short to form a transmembrane α-helix by itself. However, it is reasonable to assume that such a helix extends up to position 13 or even 10, as indicated by several transmembrane prediction methods and despite the presence of the highly conserved hydrophilic position 17 and several neutral or hydrophobic positions often bearing Ser residues (12, 15, 18, and 21).
The C-terminal transmembrane sequence 36 to 58, which we called TM2, was predicted to form a transmembrane α-helix beginning at position 36 or 39. It is essentially composed of hydrophobic or neutral residues, and its length (20 to 23 amino acids) is typical of transmembrane α-helices. At the carboxy-terminal side, TM2 is flanked by a variable position (position 59), while the following four positions are highly conserved. This conservation is likely due to the signal peptide function of this region (see below).
Assuming such an organization, the short basic segment 33 to 35 connecting TM1 to TM2 is likely located at the surface of the membrane and ensures the positioning of both transmembrane domains within the membrane. Analyses of the amino-terminal part of p7 (segment 1 to 12) revealed highly conserved hydrophilic and hydrophobic positions. This region was not predicted to be membranous, but the presence of short stretches of hydrophobic residues separated by hydrophilic ones suggests the presence of a stable secondary structure. One possibility is the formation of a short helix parallel with the lipid bilayer, where hydrophobic residues would intercalate the leaflet side of the membrane.
p7 is an integral membrane protein.
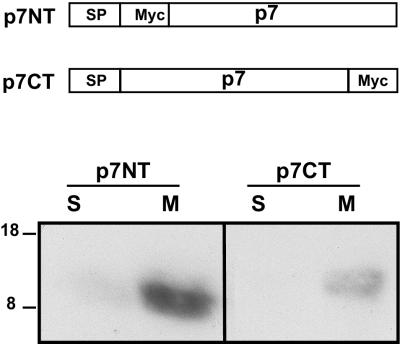
Previous attempts to generate p7-specific antisera have failed (30), suggesting that, because of its highly hydrophobic nature, p7 is poorly immunogenic. Similarly, our attempts to produce a specific antiserum were unsuccessful. Thus, p7 was tagged with a Myc epitope or the ectodomain of CD4 to allow its identification. As discussed above, p7 contains two hydrophobic sequences which might correspond to putative transmembrane domains, suggesting that it is a polytopic membrane protein. This possibility was examined by treating membranes with 0.1 M sodium carbonate, pH 11.3, as described by Howell and Palade (23) and Fujiki et al. (18). This procedure results in the extraction of lumenal and peripheral proteins, while integral membrane proteins remain bound to the membranes.
HepG2 cells were infected with vaccinia virus recombinants expressing p7 tagged at its N (p7NT) or C terminus (p7CT) with the Myc epitope and treated with sodium carbonate. After separation of membrane-bound (M) and soluble (S) proteins by SDS-PAGE, p7NT and p7CT were revealed by Western blotting with the anti-Myc antibody. As shown in Fig. 2, p7NT and p7CT were clearly detected in the membrane-associated fraction, in agreement with the prediction based on sequence analyses. Similar results were obtained by salt extraction (data not shown). The slight difference in mobility observed between p7NT and p7CT is likely due to an abnormal mobility in SDS-PAGE linked to the mutations introduced, as observed when the transmembrane domain of E1 was mutated (7, 16).
FIG. 2.
p7 is an integral membrane protein. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. At 5 h postinfection, membrane fractions were prepared as described in Materials and Methods and treated with 0.1 M sodium carbonate, pH 11.3. After separation of membrane-bound (M) and soluble (S) proteins, the presence of p7NT or p7CT in these fractions was revealed by Western blotting with the anti-Myc MAb. Sizes (in kilodaltons) of protein molecular mass markers are indicated on the left.
A fraction of p7 is transported to the plasma membrane.
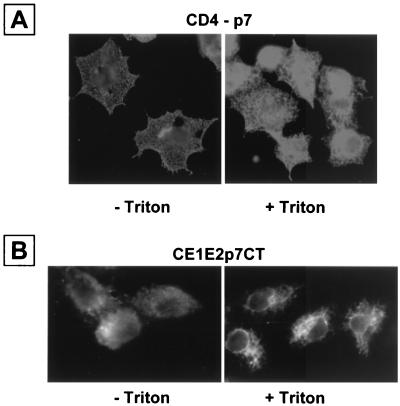
Most HCV proteins have been shown to associate with ER membranes (24, 37, 39). We therefore suspected that p7 would also be localized in the ER compartment. To determine the subcellular localization of p7, we expressed the fusion protein CD4-p7 in HepG2 cells and revealed its presence with the anti-CD4 antibody OKT4. Cells fixed with paraformaldehyde and permeabilized with Triton X-100 showed a staining pattern highly suggestive of localization of CD4-p7 in the ER (Fig. 3A), which was similar to what we have observed previously with HCV envelope glycoproteins expressed in HepG2 cells by using the same expression system (5, 6, 9, 12). Interestingly, in the absence of detergent treatment, a specific fluorescent signal was also detected, indicating that CD4-p7 is transported to the cell surface. Similar results were obtained when p7 tagged at its C terminus with a Myc epitope was expressed in the context of a CE1E2p7 polyprotein (Fig. 3B), indicating that the expression of HCV structural proteins does not modify the cell surface expression of p7.
FIG. 3.
CD4-p7 protein is exported to the cell surface. HepG2 cells were coinfected with vTF7-3 and a vaccinia virus recombinant expressing the ectodomain of CD4 in fusion with p7 (A) or a truncated form of HCV polyprotein (CE1E2p7CT) (B) at a multiplicity of infection of 3 PFU/cell. Cells were fixed with paraformaldehyde at 6 h postinfection, permeabilized or not with Triton X-100, immunostained with the anti-CD4 MAb OKT4 (A) or anti-Myc antibody (B), and analyzed by immunofluorescence.
In addition, we wondered whether the expression of all the HCV proteins would modify the subcellular localization of p7. For this purpose, HepG2 cells were coinfected with vaccinia viruses expressing CE1E2p7Myc and NS2-3-4A-4B-5A-5B at a multiplicity of infection of 5. In this context, p7 tagged at its C terminus with a Myc epitope was still detected at the cell surface (data not shown), indicating that the presence of all the HCV proteins does not alter the cell surface expression of p7.
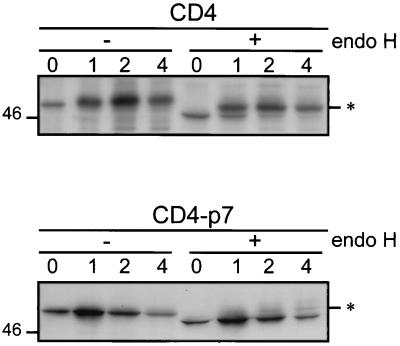
To follow the kinetics of transport of CD4-p7 out of the ER and determine the percentage of protein that leaves this compartment, the expression of CD4-p7 was analyzed in pulse-chase experiments followed by immunoprecipitation and deglycosylation by endo H. The CD4 protein contains two N-linked glycans, and only one of them becomes endo H resistant (49). Resistance to digestion with endo H indicates that the glycoprotein has been modified by enzymes present in the medial and/or trans-Golgi apparatus (44). Wild-type CD4 expressed in the same conditions was used as a control for transport through the secretory pathway.
During the pulse, CD4 was sensitive to endo H treatment, and its resistant form was detected after 1 h of chase or more (Fig. 4). In contrast, most of the CD4-p7 proteins remained sensitive to endo H treatment even after 4 h of chase. However, a faint band corresponding to endo H-resistant CD4-p7 was detected after 2 and 4 h of chase, suggesting that a fraction of CD4-p7, estimated to be ≈20%, is exported through the secretory pathway. Previous experiments expressing E1 and/or E2 under similar conditions by using the vaccinia virus/T7 expression system did not result in transportation of these proteins to the plasma membrane (5, 6, 9), indicating that cell surface detection of CD4-p7 (or P7CT) is not due to the high expression rate obtained in the chosen expression system. Altogether, these data indicate that a fraction of p7 is transported to the cell surface.
FIG. 4.
Analysis of the endo H sensitivity of CD4-p7. HepG2 cells were coinfected with vTF7-3 and a vaccinia virus recombinant expressing CD4 or CD4-p7 at a multiplicity of infection of 5 PFU/cell. At 4.5 h postinfection, infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with the anti-CD4 MAb and treated or not with endo H. Proteins were separated by SDS-PAGE (10% polyacrylamide). Endo H-resistant proteins are indicated by asterisks. The size (in kilodaltons) of a protein molecular mass marker is indicated on the left.
Second transmembrane domain of p7 is a signal sequence.
Like other viruses of the same family, HCV encodes a single polyprotein which is cleaved by cellular and viral proteases to generate its polypeptide products. In the HCV polyprotein, p7 is located downstream of the envelope glycoprotein E2 and upstream of the NS2 protein. The cleavages to generate these polypeptide products are supposed to be catalyzed by a host signal peptidase localized in the ER (reviewed in reference 41). We have recently confirmed that the C terminus of the envelope glycoprotein E2 contains a signal sequence (7; L. A. Cocquerel, O. de Beeck, M. Lambot, J. Roussel, D. Delgrange, A. Pillez, C. Wychowski, F. Penin, and J. Dubuisson, submitted for publication), supporting the idea that cleavage between E2 and p7 is catalyzed by a host signal peptidase. In addition, sequence analyses of the C-terminal half of p7 indicate that this domain exhibits the typical structural features of a signal peptide, suggesting that it might itself be a signal sequence for the NS2 protein (Fig. 5A).
FIG. 5.
Identification of a signal sequence in the C-terminal half of p7. (A) Sequence analysis of the C-terminal transmembrane domain of the p7 used in this work (HCV H strain), indicating that this domain has the characteristic structural features of a signal peptide (54). A typical signal peptide is composed of an N-terminal region (n-domain) encompassing between one and three positively charged residues (K33 and R35 here), a hydrophobic core region (h-domain) forming an α-helix (segment 41 to 57), and a more polar, flexible region (c-domain) containing the signal peptidase cleavage site (segment 58 to 63). Residues at positions −1 and −3 relative to the cleavage site are small neutral residues (A63 and A61) and form the recognition site for signal peptidase (53). Furthermore, an α-helix-destabilizing residue is frequently located at position −6 (P58) and/or in the middle of the h-domain (P49). The black box indicates the predicted minimal transmembrane segment (see Fig. 1E). (B) Schematic representation of the proteins used to identify the signal sequence function of the C-terminal half of p7. Sp1E1 corresponds to E1 with its signal sequence, and SpNS2E1 corresponds to the C-terminal half of p7 (residues 781 to 809 on the polyprotein) followed by the ectodomain and transmembrane domain of E1. (C) The C-terminal transmembrane half of p7 is a signal sequence. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. At 4.5 h postinfection, cells were labeled for 1 h with 35S-Protein Labeling Mix. Cell lysates were immunoprecipitated with MAb A4 (anti-E1) and treated or not with endo H. Samples were analyzed by SDS-PAGE (12% polyacrylamide) and autoradiography. Sizes (in kilodaltons) of protein molecular mass markers are indicated.
We thus tried to determine whether this sequence, called SpNS2, behaves as a signal sequence by analyzing the glycosylation of a reporter protein (HCV glycoprotein E1) fused to it as an indication that translocation has occurred. As shown in Fig. 5B and C, E1 expressed with SpNS2 migrated as two bands of 30 to 32 kDa when analyzed by SDS-PAGE. This pattern is similar to the migration profile observed for E1 expressed with its own signal sequence (Sp1), and it corresponds to different glycoforms of E1, as described previously (11). Indeed, these two bands disappeared after deglycosylation by endo H treatment, and a single fast-migrating band was detected by SDS-PAGE. In the absence of signal sequence, E1 migrated as an unglycosylated 20-kDa band of a slightly faster mobility than the endo H-treated proteins. This difference in mobility is likely due to the fact that endo H leaves the most proximal N-acetylglucosamine bound to the polypeptide backbone.
E1 expressed with the SpNS2 signal peptide had the same electrophoretic mobility as E1 expressed with its own signal sequence (Sp1), indicating that cleavage between SpNS2 and E1 has occurred. Interestingly, unlike the E2-p7-NS2 precursor observed in the context of the HCV polyprotein, no higher-molecular-weight precursor was observed when E1 was expressed with the SpNS2 signal peptide, suggesting that SpNS2/E1 cleavage is more efficient than the cleavage at the p7/NS2 site (30, 36).
Altogether, these data demonstrate that SpNS2 is a signal sequence, leading to the translocation of a reporter protein (E1) into the ER lumen.
Topology of p7.
To gain more information on the structure of p7, we determined its topology in cellular membranes. We took advantage of the expression of a fraction of p7 at the cell surface to determine the access of its N and C termini on unpermeabilized cells. Sequence analyses of p7 have predicted two putative transmembrane domains (Fig 1). In addition, the presence of a signal sequence immediately upstream of p7 (7) and the presence of a signal sequence in the second putative transmembrane domain, as shown in this work, suggest that the N and C termini are oriented towards the ER lumen and they should therefore also be accessible from the extracellular environment.
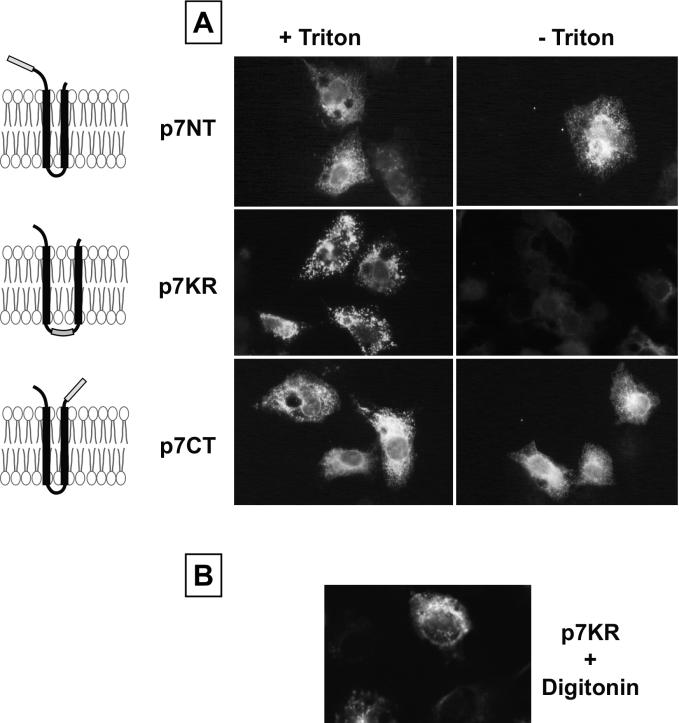
To confirm this prediction, we analyzed by immunofluorescence the accessibility of the Myc epitope fused at the N or C terminus of p7 (p7NT and p7CT, respectively) in unpermeabilized cells. An additional p7 polypeptide with a Myc epitope inserted between the two putative transmembrane domains (p7KR) was also used in this topological study. It is worth noting that these p7 polypeptides tagged with the Myc epitope contained at their N terminus the signal sequence present in the transmembrane domain of E2, which we suppose might influence the topology adopted by p7 in the context of the polyprotein.
HepG2 cells were infected by vaccinia virus recombinants expressing p7NT, p7KR, or p7CT proteins. After permeabilization with Triton X-100, a specific fluorescent signal was observed for each protein, indicating that they were expressed (Fig. 6A). However, as expected from our predictions, only p7NT- and p7CT-expressing cells were detected by the anti-Myc antibody in the absence of detergent treatment, indicating that the N and C termini of p7 are accessible from the extracellular environment. The absence of specific fluorescence with the anti-Myc antibody in unpermeabilized cells expressing p7KR suggests that the epitope is located in the cytosol. However, even if permeabilization with Triton X-100 confirmed the expression of this protein, we could not exclude that this particular chimeric protein was retained in an intracellular compartment, and if that were the case, the epitope would therefore not be accessible due the absence of expression of the protein at the cell surface.
FIG. 6.
Determination of the topology of p7 by epitope tagging. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 3 PFU/cell and analyzed by indirect immunofluorescence. Cells were fixed with paraformaldehyde at 6 h postinfection, permeabilized or not with Triton X-100 (A) or digitonin (B), and immunostained with the anti-Myc MAb. A schematic representation of the topology of the different p7 polypeptides tagged with a Myc epitope is presented on the left.
To circumvent this problem, the accessibility of the Myc epitope was determined by immunofluorescence in digitonin-permeabilized cells. Indeed, low concentrations of digitonin selectively permeabilize the plasma membrane because of its higher concentration of cholesterol compared with intracellular membranes (27), making the cytosol accessible to antibodies. A specific fluorescent signal was detected in cells expressing p7KR and treated with digitonin (Fig. 6B), confirming that the Myc epitope is accessible from the cytosol in the p7KR polypeptide. Although one cannot totally exclude that the insertion of an epitope alters the topology of a protein, the data obtained on the topology of p7 by tagging with a Myc epitope are in agreement with the presence of signal sequences immediately upstream of p7 (7) and in the second transmembrane domain (this work).
Altogether, these data indicate that p7 is a polytopic membrane protein which crosses the plasma membrane twice and has its N and C termini oriented towards the extracellular environment. In addition, these data fit very well with the prediction of two transmembrane helices connected by a short basic segment (33 to 35 or 38) accessible at the cytosolic side of the membrane. This consistency between the experimental and prediction data encouraged us to get further information on the structural properties of p7 by sequence analyses. As shown in Fig. 1C, the p7 amino acid repertoire deduced from the analyses of 289 HCV isolates of various genotypes has revealed that most positions are variable. However, this observation is partly due to an intergenotype variability.
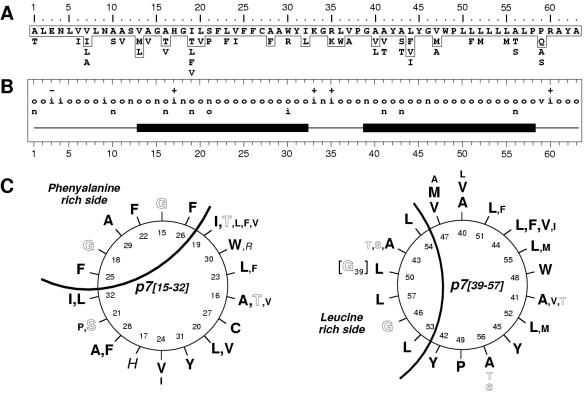
Indeed, by limiting our sequence analyses to HCV isolates of clade 1 (genotypes 1a, 1b, and 1c), for which a large number of p7 sequences have been reported (248 sequences), the variability and the repertoire of residues observed at each p7 positions were more limited (Fig. 7A and B). Specific residues were conserved in 50% of positions, and the hydropathic consensus showed that several classes of residues are only observed in 17% of positions. In addition, the latter positions mainly concern small residues that have similar conformational properties (A, S, and T, positions 1, 10, 43, and 46; A, T, and V, positions 16 and 21; S and P, position 21).
FIG. 7.
Structural prediction for transmembrane domains of p7. (A) Repertoire of amino acids per position in 248 HCV isolates of clade 1 (genotypes 1a, 1b, and 1c). Amino acids are listed in decreasing order of observed frequency, from top to bottom. Amino acids within the box correspond to residues observed in more than 10% of the 248 sequences. Residues observed in fewer than three sequences (<0.8%) are not presented. (B) Consensus hydropathic pattern deduced from the data in panel A. o, hydrophobic residue; n, neutral residue; i, hydrophilic residue; v, variable residue; + and −, positions where fully conserved positively or negatively charged residues, respectively, were observed. The black boxes indicate the predicted transmembrane segments deduced by various prediction methods (see Materials and Methods section). (C) Ideal α-helix projection of the putative p7 transmembrane segments TM1 (left) and TM2 (right). The variability of residues at each position is included, according to the data in panel A. The larger characters indicate the most frequently observed residues. Outlined, italic, and bold letters correspond to neutral, hydrophilic, and hydrophobic residues, respectively (for details, see legend to Fig. 1D).
Assuming that TM1 and TM2 form transmembrane α-helices, the corresponding ideal α-helix projections were constructed (Fig. 7C). This representation clearly highlights that both TM1 and TM2 have a side where amino acids are strictly conserved, whereas more variable amino acids were observed in the remaining positions, which strongly supports the α-helix prediction for both TM1 and TM2. The strictly conserved helix side of TM1 is rich in phenylalanine residues, while that of TM2 is rich in leucine residues. Interestingly, both the Phe- and Leu-rich sides include strictly conserved glycine residues (G15 and G17 in TM1 and G39 and G 46 in TM2). The presence of such residues, which are often observed in transmembrane domains involved in oligomerization, together with the strict conservation of Phe and Leu suggests that the corresponding helix faces are involved in specific helix-helix interactions (see Discussion).
Sequence analyses of p7 of other members of the Flaviviridae.
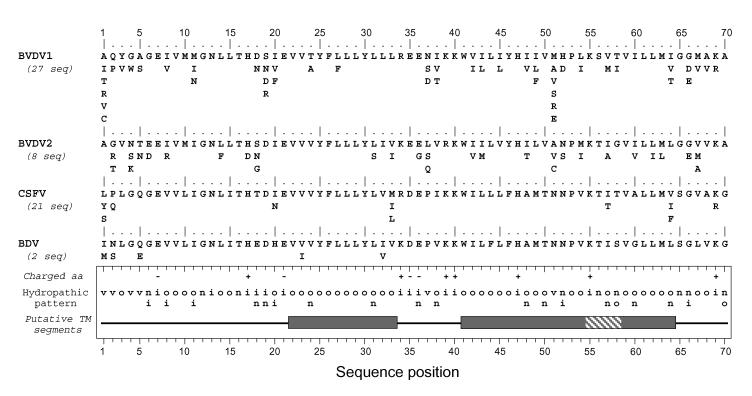
In the Pestivirus genus, a small 70-amino-acid-long polypeptide, called p7 and consisting mostly of hydrophobic residues, has also been described (14, 21). No polypeptide with similar characteristics has been reported for the Flavivirus genus. To determine to what extent p7 from the Hepacivirus and Pestivirus genera have similar structural features, structure predictions were performed on pestivirus p7 sequences reported for the four major pestivirus members (Fig. 8).
FIG. 8.
Conservation and comparison of amino acid sequences of p7 of the four main types of pestiviruses. The repertoires of residues per position for BVDV1, BVDV2, classical swine fever virus (CSFV), and border disease virus (BDV) (pestivirus types 1, 4, 2, and 3, respectively [1]) were deduced from the sequences of natural variants available in the EMBL database. Note that pestivirus variants exhibiting p7 sequences with a higher or lower number of residues were not taken into account in the reported repertoires. The number of sequences analyzed is indicated in parentheses. Amino acids are listed in decreasing order of observed frequency, from top to bottom. The bottom panel summarizes the consensus features for the four types of pestivirus. The hydropathic pattern is deduced from the repertoire analyses: o, hydrophobic residue; n, neutral residue; i, hydrophilic residue; v, variable residue (for details, see legend to Fig. 1D). Positions where fully conserved positively or negatively charged residues were observed are indicated by + and −, respectively. The boxes indicate possible transmembrane segments deduced from sequence analysis by various prediction methods (see Materials and Methods section). Dark grey sections show the consensus hydrophobic clusters of residues assumed to be involved in transmembrane segments. The shaded section indicates that the corresponding amino acid positions were predicted as membranous by some prediction methods and not for all the isolates.
Sequence comparisons and amino acid repertoires show homologies and similar structural organizations among the four pestivirus members, as summarized by the consensus hydropathic pattern. In contrast, predictions of transmembrane segments yielded different results depending on the pestivirus analyzed and the method used. To summarize, transmembrane segments were predicted in BVDV1 and BVDV2 by all the prediction methods used, while such segments were poorly predicted for the classical swine fever virus and not predicted for the border disease virus. However, three hydrophobic clusters common to the four pestiviruses can be defined (grey boxes in Fig. 8).
Using transmembrane prediction data and sequence examination, it is reasonable to assume that hydrophobic clusters 41 to 54 and 59 to 64 belong to a single transmembrane passage from 41 to 64 (called TM2). In contrast, the N-terminal hydrophobic cluster 22 to 33 is too short to form a transmembrane passage if folded into an α-helix. Indeed, a minimum of 17 residues are necessary to form a transmembrane helix, and the N-terminal segment flanking this hydrophobic cluster is too hydrophilic to assume that it can participate in the formation of a transmembrane helix. The prediction of the structure of this region remains enigmatic even though p7 is likely a polytopic membrane protein with two transmembrane passages, as observed for HCV p7. Compared to p7 of HCV, the transmembrane connecting segment supposed to be accessible on the cytosolic membrane side contains a larger number of charged residues. Another major difference is the presence of numerous polar and charged residues within and between the putative transmembrane passages in the pestivirus genus, some of them being strictly conserved.
DISCUSSION
The HCV genome encodes a single polyprotein which is co- and posttranslationally processed by cellular and viral proteases to produce at least 10 distinct polypeptides. Biological and biochemical data have been accumulated on all HCV proteins but p7, which has not attracted the interest of investigators yet. To gain more information on this polypeptide, its topology and subcellular localization were studied in this work. Prediction analyses and experimental data indicate that p7 is a polytopic membrane protein which crosses the plasma membrane twice and has its N and C termini oriented towards the extracellular environment. In addition, the C-terminal transmembrane domain of p7 has a signal sequence function. Finally, the export of a fraction of p7 at the plasma membrane suggests that this polypeptide might have a functional role in several compartments of the secretory pathway.
Studies on the subcellular localization of HCV proteins indicate that most of them associate with ER membranes (24, 37, 39). In the case of the envelope glycoproteins, it has been shown that they are strictly retained in the ER (reviewed in reference 39). The ER localization of HCV structural and nonstructural proteins is in agreement with the observation that both replication and assembly of the Flaviviridae seem to take place in association with modified membranes of the early secretory pathway (32, 43). As shown by immunofluorescence studies and the endo H sensitivity of CD4-p7, a large fraction of p7 is retained in an early compartment of the secretory pathway, which is likely the ER, suggesting that an ER retention signal is present in p7. The transmembrane domains of HCV envelope glycoproteins are signals for ER retention (for a review, see reference 39), and a similar localization signal might be contained in p7. However, the difference between the transmembrane domains of HCV envelope proteins and p7 is that the ER retention signal contained in p7 is leaky. Further experiments will be needed to identify the amino acid residues involved in the subcellular localization of p7.
If, as indicated for the pestivirus p7 (21), HCV p7 plays a role in production of progeny virus, localization of this polypeptide is expected in an early compartment of the secretory pathway, where viral assembly is supposed to occur. Why then is a fraction of p7 transported to the cell surface? Due to the low number of polypeptides encoded by the genomes of small positive-stranded RNA viruses like HCV and the huge task of redirecting a large number of cellular functions to its own profit, one can expect that the polypeptides encoded by such viruses have multiple functions. Targeting a protein to several subcellular localizations might therefore be important for this polypeptide to be involved in different tasks.
The C-terminal half of p7 (SpNS2) is a signal sequence. It has been suggested previously that cleavage between p7 and NS2 is catalyzed by a host signal peptidase (30, 36). This was suggested by the following lines of evidence: (i) the presence of a hydrophobic region immediately upstream of the p7/NS2 junction that resembles a signal sequence, (ii) the requirement for membranes in cleavage at this site, and (iii) the disruption of processing at this site by mutations known to inhibit signalase-dependent cleavages. However, the delayed cleavage at the p7/NS2 site raised some doubts on the identity of the enzyme involved in this cleavage.
Here, we showed that by itself, the C-terminal half of p7 is able to induce the translocation of a reporter protein (HCV glycoprotein E1) in the ER lumen, demonstrating that the C-terminal domain of p7 (TM2) has a signal sequence function. Together with the observations discussed earlier (30, 36), these data confirm that the cleavage between p7 and NS2 is catalyzed by a host signal peptidase. Interestingly, unlike what has been described for cleavage at the p7/NS2 site (30, 36), the cleavage between SpNS2 and E1 appeared not to be delayed. This suggests that the delayed cleavage at the p7/NS2 site in the HCV polyprotein might depend on the sequence present downstream of the cleavage site, e.g., NS2.
The p7 polypeptide is a polytopic membrane protein with its N and C termini oriented towards the extracellular environment. The presence of strictly conserved phenylalanine and leucine residues on one side of TM1 and TM2, respectively, strongly suggests that the corresponding helix faces are involved in specific helix-helix interactions. Interestingly, both the phenylalanine- and leucine-rich sides include strictly conserved glycine residues (G15 and G17 in TM1 and G39 and G 46 in TM2). It should be stressed that the presence of glycine residues is often observed in transmembrane α-helices involved in oligomerization (46).
By analyzing frequently occurring combinations of residues in a database of putative transmembrane domains, Engelman's group (48) identified a main pattern composed of small residues (Gly, Ala, and Ser) occupying i and i + 4 positions in association with large aliphatic residues (Ile, Val, and Leu) at neighboring positions (i.e., positions i ± 1 and i ± 2). From a structural point of view, the absence of side chain in the glycine residue creates a hole, allowing a large aliphatic residue to fit in and form a knob at the interacting face of the other helix (33). Such a “knob into hole” interaction is often reinforced by the presence of interhelix hydrogen bonds due to the presence of a polar residue (Asn, Ser, or Thr) within the hydrophobic core of the helix (48).
Although no canonical GxxxG motif was observed for TM1, the GxxG motif involving G15 and G18 yields the same topology, i.e., glycine residues on the same helix face. For TM2, the three small residues observed on the conserved leucine-rich face at positions 39 (Gly), 43 (Ala, Ser, or Thr), and 46 (Gly) form two successive i, i + 4 motifs, often encountered in interacting transmembrane helices. The presence of such motifs together with the strict conservation of Phe and Leu strongly suggests that the corresponding helix faces are involved in specific helix-helix interactions. Whether these interactions are intra- or intermolecular remains to be determined. Interestingly, preliminary data indicate that p7 can form homo-oligomers (our unpublished data), suggesting that intermolecular interactions exist in p7.
No function has yet been attributed to the p7 polypeptide. However, its double membrane-spanning topology with few residues accessible at one or the other side of the membrane suggests that it likely exerts its functions on membrane structures. Although the detailed analysis and comparison of p7 sequences from HCV and pestiviruses revealed substantial differences, a similar double membrane-spanning topology is conceivable for pestivirus p7, as proposed earlier (14). Functional data on pestivirus p7 indicate that this polypeptide is essential for the production of progeny virus (21). A large in-frame deletion in p7 introduced into an infectious cDNA clone of BVDV has shown that RNA replication is not affected but no infectious virus is produced (21). However, it is not known how pestivirus p7 helps in the production of infectious progeny virus.
A 60-amino-acid-long hydrophobic polypeptide (6K) with a topology potentially similar to that of p7 (28) is also produced after processing of the structural polyprotein of Alphaviruses, another genus of positive-stranded RNA viruses. Mutations in 6K result in greatly reduced virus yield, and many mutations tested result in multicored particles (19, 20, 29, 31), suggesting that this protein exerts some functions late in the assembly pathway, possibly during virus budding. In addition, expression of 6K in Escherichia coli has been shown to increase membrane permeability and cell lysis (47).
The recognition of virus proteins capable of enhancing membrane permeability has led to the description of a new family of virus proteins, called viroporins (3). Structurally, viroporins are generally short proteins containing about 50 to 120 amino acid residues. They are integral membrane proteins with at least one membrane-spanning domain and tend to form oligomers. Based on the structural features of HCV p7, it is tempting to include this protein in the viroporin family, as suggested for pestivirus p7 (21). However, experimental data will be needed to demonstrate whether, like the members of this family, HCV p7 is able to modify membrane permeability.
In conclusion, we have described some interesting biological and structural features of the p7 polypeptide of HCV, but in the absence of a tissue culture system that allows efficient production of HCV particles, it will be difficult to understand the role of p7 in the HCV life cycle. However, the resolution of the three-dimensional structure of p7 could provide a framework for a molecular understanding of its function. In addition, detailed functional studies on pestivirus p7 could also help in understanding the potential role of p7 in the HCV life cycle.
Acknowledgments
We thank Françoise Jacob-Dubuisson and Laurent Kremer for critical reading of the manuscript and Sophana Ung and André Pillez for excellent technical assistance. We are grateful to Bernard Moss for the gift of vaccinia virus recombinant vTF7-3.
This work was supported by the CNRS, the Institut Pasteur of Lille and EU grant QLK2-1999-00356. Laurence Cocquerel was successively supported by an MENRT and an FRM fellowship.
REFERENCES
- 1.Becher, P., M. Konig, D. J. Paton, and H. J. Thiel. 1995. Further characterization of border disease virus isolates: evidence for the presence of more than three species within the genus pestivirus. Virology 209:200-206. [DOI] [PubMed] [Google Scholar]
- 2.Blanchet, C., C. Combet, C. Geourjon, and G. Deléage. 2000. MPSA: integrated system for multiple protein sequence analysis with client/server capabilities. Bioinformatics 16:286-287. [DOI] [PubMed] [Google Scholar]
- 3.Carrasco, L. 1995. Modification of membrane permeability by animal viruses. Adv. Virus Res. 45:61-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 269:26898-26903. [DOI] [PubMed] [Google Scholar]
- 5.Cocquerel, L., S. Duvet, J.-C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocquerel, L., J.-C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a key role in the processing, subcellular localization and assembly of these envelope proteins. J. Virol. 74:3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in procaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 9.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drillien, R., D. Spehner, and A. Kirn. 1982. Complementation and genetic linkage between vaccinia virus temperature-sensitive mutants. Virology 119:372-381. [DOI] [PubMed] [Google Scholar]
- 11.Dubuisson, J., S. Duvet, J. C. Meunier, A. Op De Beeck, R. Cacan, C. Wychowski, and L. Cocquerel. 2000. Glycosylation of the hepatitis C virus envelope protein E1 is dependent on the presence of a downstream sequence on the viral polyprotein. J. Biol. Chem. 275:30605-30609. [DOI] [PubMed] [Google Scholar]
- 12.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubuisson, J., and C. M. Rice. 1996. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J. Virol. 70:778-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elbers, K., N. Tautz, P. Becher, D. Stoll, T. Rümenapf, and H.-J. Thiel. 1996. Processing in the pestivirus E2-NS2 region: identification of proteins p7 and E2p7. J. Virol. 70:4131-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fournillier-Jacob, A., A. Cahour, N. Escriou, M. Girard, and C. Wychowski. 1996. Processing of the E1 glycoprotein of hepatitis C virus expressed in mammalian cells. J. Gen. Virol. 77:1055-1064. [DOI] [PubMed] [Google Scholar]
- 17.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaedigk-Nitschko, K., M. X. Ding, M. A. Levy, and M. J. Schlesinger. 1990. Site-directed mutations in the Sindbis virus 6K protein reveal sites for fatty acylation and the underacylated protein affects virus release and virion structure. Virology 175:282-291. [DOI] [PubMed] [Google Scholar]
- 20.Gaedigk-Nitschko, K., and M. J. Schlesinger. 1991. Site-directed mutations in Sindbis virus E2 glycoprotein's cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology 183:206-214. [DOI] [PubMed] [Google Scholar]
- 21.Harada, T., N. Tautz, and H. J. Thiel. 2000. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J. Virol. 74:9498-9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houghton, M. 1996. Hepatitis C viruses, p. 1035-1058. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 23.Howell, K. E., and G. E. Palade. 1982. Hepatic Golgi fractions resolved into membrane and content subfractions. J. Cell Biol. 92:822-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 25.Kieny, M.-P., R. Lathe, R. Drillien, D. Spehner, S. Skory, D. Schmitt, T. Wiktor, H. Koprowski, and J.-P. Lecocq. 1984. Expression of rabies virus glycoprotein from a recombinant vaccinia virus. Nature 312:163-166. [DOI] [PubMed] [Google Scholar]
- 26.Kolykhalov, A. A., E. V. Agapov, K. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 27.Lange, Y. 1991. Disposition of intracellular cholesterol in human fibroblasts. J. Lipid Res. 32:329-339. [PubMed] [Google Scholar]
- 28.Liljestrom, P., and H. Garoff. 1991. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J. Virol. 65:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liljestrom, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, C., B. D. Lindenbach, B. M. Pragai, D. W. McCourt, and C. M. Rice. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loewy, A., J. Smyth, C. H. von Bonsdorff, P. Liljestrom, and M. J. Schlesinger. 1995. The 6-kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J. Virol. 69:469-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacKenzie, K. R., J. H. Prestegard, and D. M. Engelman. 1997. A transmembrane helix dimer: structure and implications. Science 276:131-133. [DOI] [PubMed] [Google Scholar]
- 34.McLauchlan, J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral Hepat. 7:2-14. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima, H., M. Hijikata, S.-I. Asabe, M. Hirota, K. Kimura, and K. Shimotohno. 1994. Two hepatitis C virus glycoprotein E2 products with different C termini. J. Virol. 68:6215-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizushima, H., M. Hijikata, Y. Tanji, K. Kimura, and K. Shimotohno. 1994. Analysis of N-terminal processing of hepatitis C virus nonstructural protein 2. J. Virol. 68:2731-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moradpour, D., P. Kary, C. M. Rice, and H. E. Blum. 1998. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology 28:192-201. [DOI] [PubMed] [Google Scholar]
- 38.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 39.Op De Beeck, A., L. Cocquerel, and J. Dubuisson. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 40.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 42.Reinherz, E. L., P. C. Kung, G. Goldstein, and S. F. Schlossman. 1979. Separation of functional subsets of human T cells by a monoclonal antibody. Proc. Natl. Acad. Sci. USA 76:4061-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice, C. M. 1996. Flaviviridae: viruses and their replication, p. 931-959. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 44.Robbins, P. W., R. B. Timble, D. F. Wirth, C. Hering, F. Maley, G. F. Maley, R. Das, B. W. Gibson, N. Royal, and K. Biemann. 1984. Primary structure of the Streptomyces enzyme endo-β-N-acetylglucosaminidase H. J. Biol. Chem. 259:7577-7583. [PubMed] [Google Scholar]
- 45.Rost, B., R. Casadio, P. Fariselli, and C. Sander. 1995. Transmembrane helices predicted at 95% accuracy. Protein Sci. 4:521-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russ, W. P., and D. M. Engelman. 2000. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296:911-919. [DOI] [PubMed] [Google Scholar]
- 47.Sanz, M. A., L. Perez, and L. Carrasco. 1994. Semliki Forest virus 6K protein modifies membrane permeability after inducible expression in Escherichia coli cells. J. Biol. Chem. 269:12106-12110. [PubMed] [Google Scholar]
- 48.Senes, A., M. Gerstein, and D. M. Engelman. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J. Mol. Biol. 296:921-936. [DOI] [PubMed] [Google Scholar]
- 49.Shin, J., R. L. Dunbrack, S. Lee, and J. L. Strominger. 1991. Signals for retention of transmembrane proteins in the endoplasmic reticulum studied with CD4 truncation mutants. Proc. Natl. Acad. Sci. USA 88:1918-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonnhammer, E. L. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (ed.), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the VIIth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 53.von Heijne, G. 1998. Life and death of a signal peptide. Nature 396:111-113. [DOI] [PubMed] [Google Scholar]
- 54.von Heijne, G. 1990. The signal peptide. J. Membr. Biol. 115:195-201. [DOI] [PubMed] [Google Scholar]
- 55.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. H. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]