Abstract
Replication of flock house virus (FHV) RNA1 and production of subgenomic RNA3 in the yeast Saccharomyces cerevisiae provide a useful tool for the dissection of FHV molecular biology and host-encoded functions involved in RNA replication. The replication template activity of RNA1 can be separated from its coding potential by supplying the RNA1-encoded replication factor protein A in trans. We constructed a trans-replication system in yeast to examine cis-acting elements in RNA1 that control RNA3 production, as well as RNA1 and RNA2 replication. Two cis elements controlling RNA3 production were found. A proximal subgenomic control element was located just upstream of the RNA3 start site (nucleotides [nt] 2282 to 2777). A short distal element also controlling RNA3 production (distal subgenomic control element) was identified 1.5 kb upstream, at nt 1229 to 1239. Base pairing between these distal and proximal elements was shown to be essential for RNA3 production by covariation analysis and in vivo selection of RNA3-expressing replicons from plasmid libraries containing random sequences in the distal element. Two distinct RNA1 replication elements (RE) were mapped within the 3′ quarter of RNA1: the intRE (nt 2322 to 2501) and the 3′RE (nt 2735 to 3011). The 3′RE significantly overlaps the RNA3 region in RNA1, and this information was applied to produce improved RNA3-based vectors for foreign-gene expression. In addition, replication of an RNA2 derivative was dependent on RNA1 templates capable of forming the long-distance interaction that controls RNA3 production.
Viruses with positive-strand RNA genomes include numerous human, animal, and plant pathogens of medical and economic importance. Following binding and entry into a permissive cell, the viral genome is translated to produce nonstructural proteins that assemble with genomic RNA and presumably some host factors to form RNA replication complexes. Replication proceeds through a negative-strand RNA intermediate, which serves as a template for multiple rounds of positive-strand synthesis (reviewed in reference 11). Later in infection, nascent genomic positive strands interact with viral structural components to form new virus particles. The competing processes of genome translation, replication, and packaging are coordinated in part via differential viral gene expression. Mechanisms regulating viral gene expression include translational control such as readthrough at frameshifts or nonsense codons, postranslational control such as proteolytic cleavage of polyprotein precursors, and transcriptional control via production of subgenomic RNA (sgRNA). sgRNAs are colinear with the 3′ region of genomic RNA and are produced during replication via one of several possible mechanisms (reviewed in reference 38). Since ribosomal scanning takes place in a 5′-to-3′ direction, sgRNAs permit gene expression from downstream open reading frames (ORFs) that are not translatable from genomic RNA. Understanding the mechanisms that control sgRNA production should reveal novel aspects of viral gene regulation, permit its manipulation, and aid in the discovery of antiviral agents.
Flock house virus (FHV) is a Nodavirus of insects. Several characteristics of FHV make it a useful model system for understanding positive-strand RNA virus biology, including a simple genome structure with facile genetics, robust levels of RNA replication, and the ability to replicate in a wide variety of eukaryotic cell types (reviewed in reference 7). Nodavirus genomes consist of two copackaged, 5′-capped, nonpolyadenylated RNA segments, RNA1 and RNA2 (7). RNA1 (Fig. 1A) encodes protein A, a putative RNA-dependent RNA polymerase, which is the only viral protein needed for RNA replication. Hence, RNA1 is capable of autonomous replication in cells (20). RNA1 templates that do not express functional protein A can be complemented in trans (4, 42). RNA2 encodes viral capsid protein precursor α, which is required for virion formation and virus spread. Replication of RNA2 requires protein A expression and replication of RNA1 in trans (9). In addition to these two genome segments, a 387-nucleotide (nt) sgRNA is produced during RNA1 replication (19, 20, 22). This sgRNA, RNA3 (Fig. 1A), encodes proteins B1 and B2, small accessory proteins of unknown function, in separate reading frames. While protein B1 is expressed poorly and has no demonstrated function, mutant RNAs that lack protein B2 expression do not passage efficiently, and there is strong selective pressure for maintenance of this ORF (5). Presently, little information regarding the signals controlling RNA3 production is available. Mutation of the sgRNA initiation site, RNA1 nt 2721, decreases the accumulation of RNA3 positive strands (5, 42), with little effect on RNA3 negative strands (42). Production of RNA3 is downregulated by replication of RNA2 (20, 55). Likewise, little is known about the cis-acting RNA elements controlling RNA1 replication except that terminal extensions or deletions inhibit RNA replication (5).
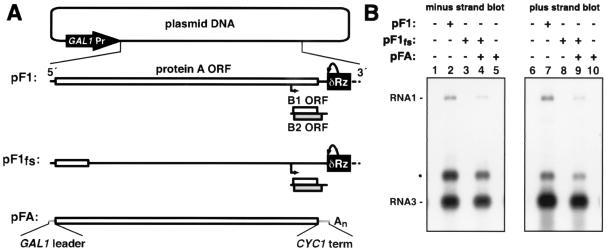
FIG. 1.
trans replication of FHV RNA1 in yeast. (A) Plasmids used to initiate cis and trans replication of FHV in yeast. Thick black (FHV sequences) and gray (yeast sequences) lines, RNA transcripts produced from the GAL1 promoter; open boxes, ORFs; bent arrow, subgenomic RNA3 start site. Transcripts are terminated via cis cleavage (curved arrow) of the HDV antigenomic ribozyme (Rz; black box) or a polyadenylation signal (An). (B) Northern blot analysis of FHV-specific RNA products. Total yeast RNA was electrophoresed and blotted as described in Materials and Methods and hybridized with a 32P-labeled probe complementary to RNA1 and RNA3 negative strands (lanes 1 to 5) or positive strands (lanes 6 to 10). •, slower-migrating form of RNA3.
A remarkable feature of nodavirus replication is that, once introduced into the cytoplasm, viral RNA replicates in insect (16, 20), mammalian (3, 6), and plant (47) cells, as well as in the budding yeast Saccharomyces cerevisiae (43). Thus, host factors involved in this process must be highly conserved. With appropriate FHV derivatives, yeast genetics should be useful for accessing host factors involved in FHV RNA replication. Our laboratory has described a system for initiating RNA1 replication in yeast via DNA plasmids containing RNA1 cDNA under control of the inducible GAL1 promoter (42). An authentic 3′ end is generated by a cis-cleaving hepatitis delta virus (HDV) antigenomic ribozyme immediately downstream of the RNA1 sequence. RNA1 templates containing a protein A frameshift can be replicated in trans by coexpressing protein A from a separate RNA1 derivative, pF1Δ3′, with a 5-nt 3′ truncation, which renders it a poor template for replication (42). In addition, these studies demonstrated expression of green fluorescent protein (GFP) from RNA3, although replication of such RNA1 derivatives was significantly reduced (42).
We sought to characterize the cis-acting signals that control RNA3 production to better understand the mechanisms of FHV RNA synthesis and gene regulation, as well as to improve RNA3-based foreign gene expression systems for genetic studies of yeast. Based on trans replication of RNA1, which allows the cis-encoded template functions of RNA1 to be separated from its coding potential, regions that control RNA3 production were mapped by deletion analysis. These studies revealed proximal and distal sgRNA control elements as well as two distinct regions in the 3′ one-fourth of RNA1 that are important for RNA replication. The ability of the proximal and distal sgRNA control elements to regulate RNA3 production through long-distance base pairing was identified and characterized via extensive covariation analyses. Second-generation RNA3-based vectors for foreign gene expression were developed and were shown to be useful in yeast genetic experiments designed to examine this long-distance interaction. In addition, we examined the ability of altered RNA1 templates to support the replication of RNA2 in trans.
MATERIALS AND METHODS
Yeast, media, and culturing conditions.
S. cerevisiae strain YPH500 (MATα ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1) (48) was transformed with plasmid DNAs by using the Frozen-EZ II yeast transformation kit (Zymo Research, Orange, Calif.) or according to an expedited one-step protocol (13). YBDL11, which expressed protein A from a chromosomally integrated form of pFA, was created by integration of pBDL11 into the leu2-Δ1 locus of YPH500 and selection for growth on Ura-deficient media. To examine pF1fsdsGFP libraries, YBDL11 was transformed as described previously (1). Following transformation, cells were grown and maintained on selective media containing 2% dextrose. For induction of FHV replication, cells were subcultured into 2% galactose-containing selective media.
Plasmid constructions.
Standard molecular cloning techniques were used throughout (2, 46). Oligodeoxynucleotides used are listed in Table 1. Unless otherwise noted, primed DNA synthesis was performed with Klen TaqLA DNA polymerase (Sigma, St. Louis, Mo.), and sequences of PCR-generated products were confirmed by sequence analysis. Numbering of FHV RNA1 sequences is based on GenBank accession no. X77156. In the following subparagraphs, laboratory designations for plasmids are in parentheses.
TABLE 1.
Oligodeoxynucleotides used in this study
| Name | Sequence |
|---|---|
| 1025 | CTTCTTATTCAAATGTAATAAAAGTGTTTTCGAAACAAATAAAACAG |
| 1026 | CTGTTTTATTTGTTTCGAAAACTCTTTTATTACATTTGAATAAGAAG |
| 1048 | CAGCCTCAGCCTTCCAACAACCGGAAGTGACTCGAGCATGGTACCAAGCTTA |
| 1049 | CCGGTAAGCTTGGTACCATGCTCGAGTCACTTCCGGTTGTTGGAAGGCTGAGGCTGAGCT |
| 1098 | CGCGTACTGCAGATCAACAAAAAATTGTTAATATACCTCTATACTTTAACGTCAAGG |
| 1118 | CGTCGACTGCAGTGTACATGGCTCTCCCTTAGCCATC |
| 1126 | CTGGTGTTCTCCTAGGATAACCTTAAGAGTCATTATAGTTTTTTCTCCTTGACGTTAAAGTATAGAG |
| 1146 | TCGAGAGCTCCCTAAAAAAACCTTCTCTTTGGAAC |
| 1147 | GCATGCGTCGACGAATTCTTTTATTACATTTGAATAAGAAGTAATAC |
| 1148 | GAATTCGTCGACGCATGCTCATGTAATTAGTTATGTCACGC |
| 1149 | GTCGACTCGAGGGTACCGGCCGCAAATTAAAGC |
| 1175 | TTTCGGGCTAGAACGGGTGT |
| 1299 | GCGGGATCCAATACCAGGACGAAACCAAACC |
| 1305 | GCGGGATCCTCAACGCTAGGCTTATCGGTATG |
| 1306 | CAGTGGGCCCGTAGAGTCGTAG |
| 1316 | CCATCTAGACGCGCTCGAGCAGCATCCGCAAGACGC |
| 1317 | CTGCTCGAGCGCGTCTAGATGGCCATGATCCGTCACAAG |
| 1330 | GCGGGATCCATACACATCCATGATTGTCCAG |
| 1333 | GCGGGATCCAACTGTGTATAAACCTACAATGCC |
| 1334 | GCGGGATCCTGCCACGCGTCCATTGGCCAG |
| 1335 | GCGGGATCCGTAACCAGTGACGCAGATGTACC |
| 1360 | GCGAGATCTACTCTGCGAAAACTACATCTTACAAC |
| 1361 | GCGAGATCTATCTTACAACAAGAGATCCAACGATAC |
| 1364 | GCCGTTCAATCGTTTCTTCA |
| 1376 | GCGCGAGATCTCACTGGTTACTGGCCAATG |
| 1377 | GCGCGAGATCTGTTACTGGCCAATGGACG |
| 1378 | GCATGAGATCTGGCCAATGGACGCGTG |
| 1379 | CGTACGACGCGTCCATACCGGTGTAACCAGTGACGCAGA |
| 1380 | GATCAACCGGTCAGCATCCGCAAGACGC |
| 1381 | CTGACCGGTTGATCCGTCACAAG |
| 1383 | GCGAGATCTACGATACCATTAGCTGATGCGGCTTGC |
| 1384 | GCGAGATCTGGCTTGCGACCGTGTCGAAGGCTATCT |
| 1385 | GCGAGATCTGCTATCTCTGTACCGATGCGCTTACTC |
| 1389 | GCGTCTAGATGGCCATGATCCGTCACAAGTCAAC |
| 1409 | CGTACGACGCGTCCATNNNNNNGTAACCAGTGACGCAGA |
| 1425 | CGTACGGCGCGCGGTTTGGCCAGTAACCAGTGACGCAGA |
| 1427 | TAACGATTCGCGGTCGGAGTTCGGC |
| 1428 | CGTACGACGCGTCCATTGGCCATGATCCGTCGTAACCAGTGACGCAGA |
| 1440 | GGAATTGTTTACACTTTTATTACATTTGAATAAGAAGTAATACAAACC |
| 1441 | TAAAAGTGTAAACAATTCCAAGTTCCAAAATG |
| 1442 | GCCGACCCACCTTAGTCTGTTGAC |
| 1446 | GATCATCTAGACAGCATCCGCAAGACGC |
| 1447 | CTGTCTAGATGATCCGTCACAAG |
| 1452 | CGTACGACGCGTCCATTCTAGAGTAACCAGTGACGCAGA |
| 1454 | GTACGTCCATTGGCCA |
| 1455 | GATCTGGCCAATGGAC |
| 1483 | CGTACGACGCGTCCATVHHDDBGTAACCAGTGACGCAGA |
| 1720 | CTTGTGACGGATCATGGCCACGCCCATTTGATGAAGC |
| 1721 | GCTTCATCAAATGGGCGTGGCCATGATCCGTCACAAG |
| 1722 | CTTGTGACGGATCATGGCCACGAATCGTTACCAATG |
| 1723 | CATTGGTAACGATTCGTGGCCATGATCCGTCACAAG |
| 1724 | GAAGCCCTACTGGTTGACAGCATCCGCAAGACGC |
| 1725 | GCGTCTTGCGGATGCTGTCAACCAGTAGGGCTTC |
| 1726 | CGTAGAAGCCGGAATAACAGCATCCGCAAGACGC |
| 1727 | GCGTCTTGCGGATGCTGTTATTCCGGCTTCTACG |
| 1728 | CTTGTGACGGATCATGGCCACGATGCCAAGCAAACTC |
| 1729 | GAGTTTGCTTGGCATCGTGGCCATGATCCGTCACACAAG |
| 1730 | CTTGTGACGGATCATGGCCAGCTAATCCAGGAACTTC |
| 1731 | GAAGTTCCTGGATTAGCTGGCCATGATCCGTCACAAG |
| 1732 | CTTGTGACGGATCATGGCCAAGCAGCCATGGGAATGAG |
| 1733 | GAGTTTGCTTGGCATCGTGGCCATGATCCGTCACAAG |
(i) pFA (pBDL7).
pFA is a yeast centromeric plasmid containing the LEU2 gene and the protein A ORF flanked by the GAL1 promoter/leader and the CYC1 polyadenylation signal. pGEM/link was created by ligating annealed oligonucleotides 1048 and 1049 into pGEM-3Zf(-) (Promega, Madison, Wis.). The yeast CYC1 polyadenylation signal was subcloned into pGEM/link from p423GALS (40) via common KpnI and XhoI sites to create pGEM/link/cyc. pBDL6 was generated via ligation of a 296-bp SacI/HindIII fragment of pGEM/link/cyc and 5,976-bp XbaI/HindIII and 3,652-bp XbaI/SacI fragments of CLp+G13′-5R (B. D. Price and P. Ahlquist, unpublished data), a yeast centromeric plasmid similar to pF1Δ3′ (42). pBDL7 was created by mutually primed extension of oligonucleotides 1098 and 1126 with an 8:1 (vol/vol) mixture of Taq (Promega) and Vent (New England Biolabs, Beverly, Mass.) DNA polymerases, cleavage with PstI and BsiWI, and insertion into pBDL6 cut with these same enzymes.
(ii) pBDL11.
pBDL11, which was used to integrate the protein A expression cassette into the leu2-Δ1 locus, was created via a multistep cloning procedure. First, 3,269-bp AatII/blunt SalI (filled in with Klenow fragment) and 2,208-bp AatII/blunt Acc65I (filled in with Klenow fragment) fragments of pRS305 (48) were ligated to produce pint1. URA3 was inserted via ligation of a HpaI/HindIII fragment from Yep352 (25) into Ecl136II/HindIII-cleaved pint1 to yield pint2. The protein A gene expression cassette from pFA was then inserted into pint2 via common EcoRI and Acc65I sites.
(iii) pF1fsΔ1 to pF1fsΔ5 (pBDL202 to pBDL206).
These deletion variants were created via PCR amplification of pF1 by using forward primer 1389 and reverse primers 1360, 1361, 1383, 1384, and 1385, respectively, and by ligating the products into pF1fs via common HindIII and MscI sites.
(iv) pF1fsΔ6 to pF1fsΔ15.
Primers 1330 and 1175 were used to amplify mutually priming first-round PCR products generated from template pF1fsΔ1. The forward first-round PCRs utilized forward primer 1330 and reverse primer 1725 (pF1fsΔ6 [pBDL209]), 1727 (pF1fsΔ7 [pBDL210]), 1721 (pF1fsΔ8 [pBDL207]), 1723 (pF1fsΔ9 [pBDL208]), 1729 (pF1fsΔ10 [pBDL211]), 1731 (pF1fsΔ11 [pBDL212]), or 1733 (pF1fsΔ12 [pBDL213]). Reverse first-round PCRs utilized forward primer 1724 (pF1fsΔ6), 1726 (pF1fsΔ7), 1720 (pF1fsΔ8), 1722 (pF1fsΔ9), 1728 (pF1fsΔ10), 1730 (pF1fsΔ11), or 1732 (pF1fsΔ12) and reverse primer 1175. Second-round PCR products were digested with BglII and BlpI and cloned into pF1fs cut with these same enzymes. Deletions in the RNA3 region were generated by recircularizing pF1fsΔ16 (described below) after cleavage with BsmI/NcoI (pF1fsΔ13 [pBDL188]), NcoI/PpuMI (pF1fsΔ14 [pBDL189]), or PpuMI/EcoNI (pF1fsΔ15 [pBDL190]) and blunt ending with T4 DNA polymerase (pF1fsΔ13) or Klenow fragments (pF1fsΔ14 and pF1fsΔ15). Variants of RNA3 with the deletions were subsequently subcloned into pF1fsΔ1 by using common Bsp120I/BsrGI sites.
(v) pF1ds1 (pBDL13), pF1ds2 (pBDL12), and pF1fsdsGFP (pBDL35).
Double-subgenomic RNA1s were created by ligation of 2,021-bp SphI/blunt SfcI (Klenow fragment filled) and 7,429-bp SphI/SmaI (pF1ds1) or 7,818-bp SphI/SmaI (pF1ds2) fragments of pF1. pF1fsdsGFP was constructed by first ligating a 2,752-bp SphI/blunt SfcI (Klenow fragment filled) fragment of pF1fsGFPN2 (42) and a 7,433-bp SphI/MscI fragment of pF1fs to create pBDL27. An improved GAL1 promoter-FHV RNA1 junction was then subcloned as a 1,009-bp EcoRI/BamHI fragment from pBDL4 into similarly digested pBDL27 to yield pF1fsdsGFP. pBDL4 is a pF1fs derivative containing a deletion of six extraneous base pairs between the GAL1 transcriptional start site (28) and FHV RNA1. It was created by extending primers 1025 and 1026 around a pF1 template with Pfu DNA polymerase, digesting the wild-type template (prepared in DH5α) with DpnI, and subcloning the mutant promoter junction into pF1fs by using common AgeI and PflMI sites.
(vi) pF1/DI-A and pF1fsΔ16 to pF1fsΔ36.
pF1/DI-A was constructed by ligation of a 6,627-bp PflMI/ApaI fragment of pF1fs and a 633-bp PflMI/ApaI fragment of pF1-DI[1,0] (kind gift from L. A. Ball, University of Alabama). pF1fsΔ16 (pΔBS) was created by recircularizing pF1fs after cleavage with BamHI and SmaI and Klenow fragment fill in. pF1fsΔ22 (pΔB′M) was created by ligation of 5,232-bp ClaI/blunt BglII (Klenow fragment filled) and 2,624-bp ClaI/MscI fragments of pF1fs. pF1fsΔ23 (pΔPS) was constructed by recircularization of pF1fs after cleavage with PshAI and SmaI. pF1fsΔ24 (pΔMS) was created by ligation of 5,011-bp ClaI/MscI and 3,013-bp ClaI/SmaI fragments of pF1fs. pF1fsΔ17 to pF1fsΔ21 were constructed by ligation of pF1fs digested with BamHI/MluI (pF1fsΔ17 and pF1fsΔ18) or BamHI/PshAI (pF1fsΔ19 to pF1fsΔ21) and similarly digested PCR fragments that had been generated with forward primer 1299 (pF1fsΔ17 [pBDL37]), 1305 (pF1fsΔ18 [pBDL43]), 1333 (pF1fsΔ19 [pBDL55]), 1334 (pF1fsΔ20 [pBDL56]), or 1335 (pF1fsΔ21 [pBDL57]) and reverse primer 1306, with pF1fs as a template. pF1fsΔ25 (pBDL96) and pF1fsΔ26 (pBDL97) were created by blunt end ligation of pF1fs cut with BamHI and MluI and filled in with Klenow fragments (pF1fsΔ25) or chewed back with mung bean nuclease (pF1fsΔ26). pF1fsΔ27 (pBDL116) is a variant of pF1fsΔ26, containing an additional 1-bp deletion, presumably due to overdigestion with mung bean nuclease. pF1fsΔ28 (pBDL90) was created by ligation of a 6,873-bp blunt BamHI (Klenow fragment filled)/Bsp120I fragment and a 1,194-bp Bsp120I/MscI fragment of pF1fs. pF1fsΔ29 (pBDL98), pF1fsΔ30 (pBDL99), and pF1fsΔ31 (pBDL100) were created by ligation of PCR fragments cut with SphI and BglII into similarly digested pF1fs. These PCR fragments were generated by amplifying pF1 with forward primer 1376 (pF1fsΔ29), 1377 (pF1fsΔ30), or 1378 (pF1fsΔ31) and reverse primer 1175. pF1fsΔ32 (pBDL145) was created by ligating annealed oligonucleotides 1454 and 1455 into pF1fs digested with BsiWI and BglII. pF1fsΔ33 (pBDL146) and pF1fsΔ34 (pBDL147) were created by ligating a 423-bp PstI/MluI fragment of pF1fsΔ19 and a similarly digested fragment of pF1fsΔ29 (to make pF1fsΔ33) or a similarly digested fragment of pF1fsΔ31 (to make pF1fsΔ34). pF1fsΔ35 (pBDL148) and pF1fsΔ36 (pBDL149) were created by ligating a 403-bp PstI/MluI fragment of pF1fsΔ20 and a similarly digested fragment of pF1fsΔ29 (to make pF1fsΔ35) or a similarly digested fragment of pF1fsΔ31 (to make pF1fsΔ36).
(vii) pF1fsmut1 to pF1fsmut8.
A PCR fragment amplified from pF1 with forward primer 1425 and reverse primer 1364 was cut with BssHII and BglII and cloned into the MluI and BglII sites of pF1fs to create pF1fsmut1 (pBDL123). pF1fsmut2 (pBDL124) was created by cloning a second-round PCR product into the Bsp120I and BlpI sites of pF1fs. The second-round PCR product was produced by mutually primed extension of first-round PCR products generated with forward primer 1305 and reverse primer 1427 or forward primer 1428 and reverse primer 1175 by using pF1 as a template. A 231-bp MluI/BglII fragment of pF1fsmut1 was ligated into similarly digested pF1fsmut2 to create pF1fsmut3 (pBDL125). A PCR fragment generated by amplifying pF1 with forward primer 1379 and reverse primer 1364 was cloned into the MluI and BglII sites of pF1fs to create pF1fsmut4 (pBDL93). pF1fsmut5 (pBDL94) was created by cloning a second-round PCR product into the Bsp120I and BlpI sites of pF1fs. The second-round PCR product was produced by mutually primed extension of first-round PCR products generated with forward primer 1305 and reverse primer 1381 or forward primer 1380 and reverse primer 1175 by using pF1 as a template. A 231-bp MluI/BglII fragment of pF1fsmut4 was ligated into similarly digested pF1fsmut5 to create pF1fsmut6 (pBDL95). pF1fsmut7 (pBDL138) was generated by cloning a second-round PCR product into the Bsp120I and BlpI sites of pF1fs. The second-round PCR product was produced by mutually primed extension of first-round PCR products generated with forward primer 1305 and reverse primer 1447 or forward primer 1446 and reverse primer 1175 by using pF1 as a template. pF1fsmut8 was generated by amplifying pF1 with forward primer 1452 and reverse primer 1364 and cloning a 231-bp MluI/BglII fragment into pF1fsmut7.
(viii) pF1fsdsGFPmut4, pF1fsdsGFPmut5, and pF1fsdsGFPran1 to pF1fsdsGFPran3.
A 231-bp MluI/BglII fragment of pF1fsmut4 was cloned into pF1fsdsGFP cut with these same enzymes, generating pF1fsdsGFPmut4 (pBDL154). A 2,271-bp BamHI/BseRI fragment of pF1fsmut5 was ligated to 7,604-bp BamHI/XbaI and 301-bp XbaI/BseRI fragments of pF1fsdsGFP to create pF1fsdsGFPmut5 (pBDL153). Random libraries were generated by amplifying pF1 with forward primer 1409 and reverse primer 1364 and cloning a MluI/BglII fragment of this PCR product into pF1fsdsGFPmut4 to generate pF1fsdsGFPran1 (pBDL155) or into pF1fsdsGFPmut5 to generate pF1fsdsGFPran2 (pBDL156). pF1fsdsGFPran3 (pBDL157) was generated in a similar fashion but was generated with forward primer 1483 and cloned into pF1fsdsGFPmut4. Products of library ligations were transformed into electrocompetent Escherichia coli strain DH5α and plated on ampicillin, and DNA was extracted from a minimum of 6.5 × 105 pooled colonies.
(ix) pF2/DI634.
pF2/DI634 is a His-selectable, 2μm plasmid that expresses RNA2-DI634 from the GAL1 promoter. The GAL1 promoter-RNA2-DI634 and RNA2-DI634-ribozyme junctions were created by PCR amplification of pF1fs with primers 1146 and 1440 or 1443 and 1118, respectively. The RNA2-DI634 region was amplified from pB*/DI (55) by using primers 1441 and 1442. The resulting products were extended by mutual priming and amplification with primers 1146 and 1118 and then cloned into pBDL10 via common AgeI and SphI sites. pBDL10 was created by amplifying the GAL1 promoter from pF1 with primers 1146 and 1147 and amplifying the CYC1 polyadenylation signal from pGEM/link/cyc with primers 1148 and 1149. The resulting products were mutually extended and amplified by using primers 1146 and 1149 and cloned into pRS423 (14) via common SstI and XhoI sites.
RNA analysis.
Total yeast RNA was prepared by extraction with hot phenol (33) and resuspended in H2O. RNA concentrations were determined spectrophotometrically, and 1 μg was dried in a centrifugal evaporator. RNA was resuspended in 10 μl of sample loading buffer consisting of 20 mM MOPS (3-[N-morpholino]propanesulfonic acid, pH 7.0, 5 mM sodium acetate, 1 mM EDTA, 46% (vol/vol) deionized formamide, 6.4% (vol/vol) formaldehyde, 0.5% glycerol, 0.005% bromphenol blue, heated for 5 min at 65°C, and cooled on ice. RNAs were electrophoresed through gels containing 1.1% (wt/vol) agarose and 1 μM ethidium bromide in 20 mM MOPS, pH 7.0-5 mM sodium acetate-1 mM EDTA with buffer recirculation. Following electrophoresis, gels were equilibrated in 10× SSC (1.5 M sodium chloride, 0.15 M sodium citrate, pH 7.0), and blotted to Nytran Supercharge (Schleicher & Schuell, Keene, N.H.) by upward capillary transfer. RNAs were UV cross-linked to the solid support with a Stratalinker 2400 (Stratagene, La Jolla, Calif.), and membranes were prehybridized in hybridization buffer (50 mM sodium phosphate [pH 7.0], 50% [vol/vol] deionized formamide, 0.8 M sodium chloride, 1 mM EDTA, 10× Denhardt's solution [2 mg of Ficoll 400, 2 mg of polyvinylpyrrolidone, and 2 mg of bovine serum albumin/ml], 0.25 mg of salmon sperm DNA/ml, 0.5% sodium dodecyl sulfate). RNAs were detected via 32P-labeled probes specific for positive- or negative-strand FHV RNA1 and RNA3 as previously described (42). Positive-strand RNA2-DI634 was detected by 32P-labeled T7 transcripts from pB*/DI.
Flow cytometry.
Four days postinduction, yeast cells were diluted to 1.4 × 106 cells/ml in a solution consisting of 8 mM dibasic sodium phosphate (pH 7.0), 3 mM monobasic potassium phosphate, 147 mM sodium chloride, 5.4 mM potassium chloride, and 1% heat-inactivated fetal calf serum (to prevent clumping). Cell sorting was preformed at the University of Wisconsin Comprehensive Cancer Center flow cytometry facility on a FACStar Plus (Becton Dickinson, San Jose, Calif.) equipped with an argon laser tuned to 488 nm. Approximately 1.2 × 105 cells were collected from the gated areas and regrown in selective media containing galactose. For some experiments, cells were subjected to a second round of sorting and regrowth, with similar results.
Computation.
RNA folding calculations were performed on an SGI Octane computer (250 MHz R10000 and 1.1 GB of RAM) with the program mfold, version 2.2 (56) compiled to handle large genome sequences and were verified with mfold, version 3.1.
RESULTS
trans complementation of RNA replication by helper plasmid pFA.
To study cis-acting functions of RNA1 replicons independently of their protein-coding potential, protein A must be provided in trans. Previously, protein A was provided in trans from pF1Δ3′, expressing an RNA1 derivative whose replication was inhibited by deletion of the last 5 nt (42). Because we detected residual template activity in this minimally 3′-truncated RNA1, we constructed a new protein A mRNA by replacing the 5′ and 3′ noncoding regions of RNA1 with the yeast GAL1 leader and CYC1 polyadenylation sequences, respectively. The structures of wild-type RNA1 expression plasmid pF1 (42), protein A ORF frameshift derivative pF1fs (42), and protein A expression plasmid pFA are illustrated in Fig. 1A. As previously described, the first two plasmids contained the yeast 2μm origin of replication and the yeast TRP1 gene for selection. pFA was maintained in yeast via an inserted centromere and the selectable yeast LEU2 gene.
FHV RNA1 and RNA3 accumulation was examined by Northern blot analysis 72 h after galactose induction of cells containing FHV expression plasmids or empty vector controls. As expected, negative- and positive-strand RNA1 and subgenomic RNA3 were observed in cells that contained autonomously replicating wild-type RNA1 (Fig. 1B, lanes 2 and 7). In addition, a third RNA form was observed to migrate above RNA3. Slower-migrating forms of Pariacoto virus RNA1 and RNA2 have been tentatively identified as covalently linked head-to-tail concatemers (27). Alternatively, the slower-migrating RNA3 band we observe could be a double-stranded form that remained undenatured since (i) it comigrates with double-stranded RNA3 created by annealing synthetic negative- and positive-strand RNA3 transcripts in high salt, (ii) strand-specific Northern blotting results indicate that the amounts of negative- and positive-strand RNAs in this band are equivalent, and (iii) this product can be gel purified and separated into products that coelectrophorese with single-strand RNA3 after further denaturation (data not shown). It is unclear why some RNA3 would remain undenatured during electrophoresis, although two G/C-rich subdomains in RNA3 (FHV RNA1 nt 2947 to 2967 and 3037 to 3048) could provide local stability to double-stranded forms of RNA3.
FHV RNA accumulation was not observed in cells containing pF1fs alone (Fig. 1B, lanes 3 and 8) or in cells containing pFA alone (lanes 5 and 10). A very low level of polyadenylated protein A mRNA, transcribed from pFA, could be detected on longer exposures of positive-strand blots. RNA replication products accumulated in cells containing both pF1fs and pFA (lanes 4 and 9), confirming that replication of RNA1 templates from pF1fs was dependent on expression of protein A from pFA. During trans replication with pFA, the RNA1 positive strand accumulated to 65% of the level for cis-replicating RNA1. Moreover, the ratios of positive- to negative-strand RNA1 and the ratios of RNA3 to RNA1 for cis and trans replication were similar. Accordingly, pFA was used as a helper plasmid in all testing described below.
Sequences in the 3′ one-fourth of RNA1 controlling RNA3 production and RNA1 replication.
Based on determinants controlling sgRNA production for other positive-strand RNA viruses (38), we investigated whether sequences near the FHV RNA3 initiation site at nt 2721 were important for RNA3 production. Therefore, deletion constructs were created to map regions in the 3′ portion of RNA1 that were required for efficient production of RNA3. The design of these deletion constructs, their average relative levels of RNA1 and RNA3 positive strand accumulation, and a representative Northern blot are presented in Fig. 2. All deletion derivatives were tested multiple times, and similar results were also obtained by using negative-strand probes (data not shown). Based on pF1fsΔ1 and additional constructs (not shown), deletion of residues 1464 to 2282 had minimal effects on RNA1 accumulation and RNA3 production (Fig. 2B, pF1fsΔ1), while RNA3 levels in deletion constructs pF1fsΔ2 and pF1fsΔ3 were specifically diminished (Fig. 2B). Thus, sequences immediately downstream of nt 2282 were important for RNA3 production. Additional deletion constructs pF1fsΔ4 and pF1fsΔ5 showed dramatic decreases in both RNA1 and RNA3 accumulation (Fig. 2B), indicating that the 5′ boundary of a region important for RNA1 replication lies between nt 2322 and 2342.
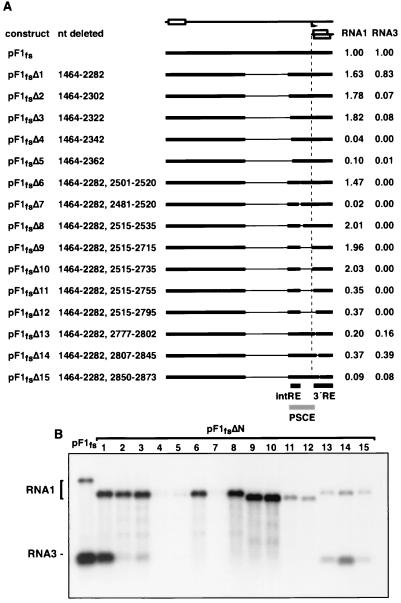
FIG. 2.
Mapping cis-acting elements in the 3′ one-fourth of RNA1. (A) Constructs used to map cis-acting replication elements in RNA1. The RNA1 frameshift derivative is illustrated at the top, as in Fig. 1A. Regions that are present (thick bars) or absent (thin lines) in several deletion constructs are illustrated below. Positions of the intRE and 3′RE are shown at the bottom. To the right of each construct are the average levels of RNA1 and RNA3 positive-strand accumulation, normalized to that of pF1fs, determined from three independent experiments. (B) A representative Northern blot was probed for RNA1 and RNA3 positive strands, as in Fig. 1B.
Additional deletions were used to further define regions important for RNA1 replication and RNA3 production. Construct pF1fsΔ6, which lacked residues 1464 to 2282 and 2501 to 2520, demonstrated normal RNA1 accumulation but no RNA3 production (Fig. 2B, lane 6). pF1fsΔ7 (residues 1464 to 2282 and 2481 to 2520 deleted) was a poor replication template and did not produce RNA3 (Fig. 2B). This replication defect was also observed in additional constructs having larger deletions with the same 3′ boundary as pF1fsΔ7 (data not shown). Thus, an internal cis-acting replication element (intRE) between nt 2322 to 2501 was important for efficient RNA1 replication (compare pF1fsΔ3 and pF1fsΔ7). Additional deletions in pF1fsΔ7 that extended from nt 2515 toward the 3′ end of RNA1 were created. pF1fsΔ8 (residues 1464 to 2282 and 2515 to 2535 deleted), pF1fsΔ9 (residues 1464 to 2282 and 2515 to 2715 deleted), and pF1fsΔ10 (residues 1464 to 2282 and 2515 to 2735 deleted) yielded RNA1 replication at levels higher than those for the parental construct, pF1fs, but failed to produce RNA3. In contrast, pF1fsΔ11 (residues 1464 to 2282 and 2515 to 2755 deleted) through pF1fsΔ15 (residues 1464 to 2282 and 2850 to 2873 deleted) showed diminished levels of RNA1 accumulation, although pF1fsΔ13 (residues 1464 to 2282 and 2777 to 2802 deleted) through pF1fsΔ15 produced RNA3 and showed a normal RNA3 to RNA1 ratio. Together these data suggest that sequences controlling efficient RNA3 production lie between nt 2282 and 2777, which we term the proximal subgenomic control element (PSCE). However, a subset of this region, nt 2322 to 2777, could support a low level of RNA3 production. Furthermore, a second, downstream region with a 5′ boundary between nt 2735 and 2755 was important for RNA1 replication. This element may be contiguous with the 3′ end of RNA1, as a 3′ boundary was not revealed by deletion constructs pF1fsΔ13 through pF1fsΔ15. We therefore refer to this as the 3′ replication element (3′RE).
Double-subgenomic RNA1 constructs and construction of improved RNA3-based expression vectors.
To further examine the organization of the PSCE, intRE, and 3′RE, the following pF1 derivatives with tandem duplications in the 3′ region were created: pF1ds1, which contained a duplication of nt 2518 to 3053, and pF1ds2, containing a duplication of nt 2129 to 3053 (Fig. 3A). Because both duplications encompassed the entire RNA3 region, the resultant constructs were referred to as double subgenomics. Based on the above mapping data, both contained one presumably interrupted 3′RE and a single complete 3′RE. pF1ds1 also retained one copy of the intRE, while pF1ds2 had two. Both double-subgenomic constructs replicated in cis to levels comparable to that for wild-type RNA1 (Fig. 3B, lanes 1 to 3), further indicating that the intRE and 3′RE were functionally separable elements. Interestingly, both constructs expressed reduced amounts of subgenome length RNA3 and high levels of longer RNA3 forms RNA3L1 and RNA3L2, which correspond to transcripts produced from the upstream subgenomic start sites. Consistent with the above deletion analysis, expression of subgenome length RNA3 by pF1ds1 suggested that the functional local sequence element directing basal RNA3 expression was contained within nt 2518 to 2777. Despite the fact that the RNA3 region of pF1ds2 was preceded by the full PSCE, production of RNA3L2 was favored over that of subgenome length RNA3. Similar results were observed for triple-subgenomic RNA1 derivatives (data not shown). Polarity effects on initiation site preference have also been described for other positive-strand RNA virus genomes expressing multiple subgenomic mRNAs (17, 29, 32, 44).
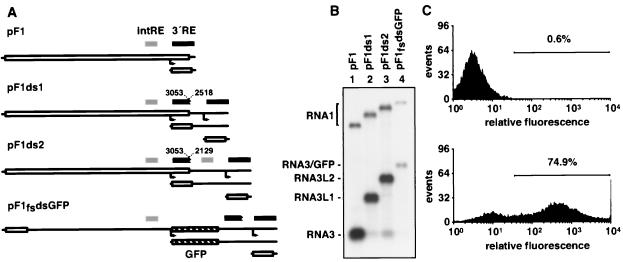
FIG. 3.
Double-subgenomic constructs and RNA3-based vector design. (A) The designs of double-subgenomic constructs are illustrated (as in Fig. 1A) below the map of wild-type RNA1, showing locations of the intRE (gray bars) and 3′RE (black bars). Also shown is the design of pF1fsdsGFP, which expresses a GFP gene (hatched rectangle). (B) Northern blot analysis for RNA1 and RNA3 positive strands, as in Fig. 1B. (C) Fluorescence profiles collected from 104 yeast cells that had been cotransformed with pF1fsdsGFP and an empty vector (top) or pFA (bottom).
It has been shown previously that a synthetic gene encoding GFP can be inserted in RNA3 and expressed in a replication-dependent manner (42). However, the levels of RNA1 and RNA3 accumulation from one such derivative, pF1fs-GFPN2, were diminished ∼20-fold (42). The above results indicated that the 3′RE significantly overlapped the subgenomic region of RNA1, suggesting that insertions in this region could perturb the structure of the 3′RE, which might explain the reduced levels of RNA replication seen with pF1fs-GFPN2. Based on their design, the double-subgenomic constructs of Fig. 3A should permit the expression of foreign genes from the upstream subgenomic mRNA without disrupting the 3′RE. To test this, pF1fsdsGFP was constructed by inserting GFP into the upstream subgenomic mRNA3 of pF1ds1 (Fig. 3A). As for pF1fs-GFPN2 (42), the GFP gene was translated from the B2 start codon, disrupting the protein A ORF. Therefore this construct was created in the context of trans-replicating replicon pF1fs. In cells expressing protein A in trans, pF1fsdsGFP expressed an RNA1 derivative that replicated to ∼20% of the level of wild-type RNA1 and that produced coordinately reduced levels of the GFP-encoding subgenomic mRNA, RNA3/GFP (Fig. 3B, lanes 1 and 4). This reduction can be explained in part by the reduced levels of RNA accumulation observed for replication in trans (Fig. 1). The identity of subgenomic RNA3/GFP was confirmed by hybridization with a GFP-specific probe (data not shown). GFP protein expression via pF1fsdsGFP was analyzed by fluorescence-activated flow cytometry. In the absence of protein A, only a basal level of fluorescence was detected, which was typical for yeast lacking any GFP sequences. Coexpression of protein A with pF1fsdsGFP led to a large increase in fluorescence (Fig 3C). In this case, two fluorescent cell populations were apparent: ∼75% of cells were brightly fluorescent, while the remainder were dimly fluorescent yet shifted relative to control cells lacking protein A. The dim population probably represents cells that recently lost one or both FHV expression plasmids, as plasmids segregate unequally in yeast (12, 14, 36, 51). Consistent with this explanation, differential plating experiments indicated that ∼20% of the total cell populations had lost the Leu+ or Trp+ phenotype.
Mapping a distal element controlling subgenome production.
During our studies with deletion mutants, it became apparent that a region 1.5 kb upstream of the subgenomic region of RNA1 was important for RNA3 accumulation. This is illustrated by comparing RNA3 accumulation in cells replicating pF1fs, pF1fs/DI-A, and pF1fsΔ16 (Fig. 4A and C, lanes 1 to 3). pF1/DI-A expresses the RNA1 of a defective-interfering genome originally obtained from persistently infected Drosophila melanogaster cells (R. Dasgupta, personal communication) containing deletions of nt 380 to 940 and 1295 to 2272. As previously described for trans replication in mammalian cells, a similar derivative replicated to high levels and expressed normal levels of RNA3 (9). In contrast, pF1fsΔ16, with a single deletion of nt 390 to 2128, replicated well but did not express RNA3. Thus, the region between nt 940 and 1294 contains a determinant for RNA3 production.
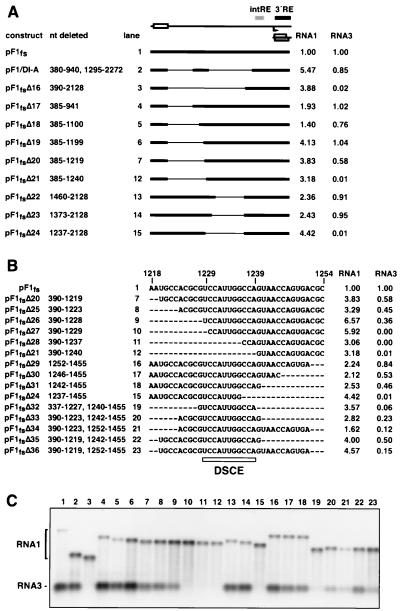
FIG. 4.
Mapping of a distal region involved in RNA3 accumulation. (A) Constructs used for low-resolution deletion mapping of the DSCE and the average relative levels of RNA1 and RNA3 accumulation are illustrated as in Fig. 2A. For simplicity, only the protein B2 ORF in RNA3 is shown. (B) Constructs used to map the DSCE at higher resolution. Dashes, deleted sequences. Right, average relative levels of RNA1 and RNA3 accumulation, calculated as for Fig. 2A. Note that some constructs from panel A are also shown for reference. Open rectangle, location of the core DSCE. (C) A representative Northern blot of RNA1 and RNA3 positive-strand accumulation.
Additional deletions were created to map the 5′ boundary of this RNA3 determinant. Constructs pF1fsΔ17, pF1fsΔ18, and pF1fsΔ19 demonstrated that nt 385 to 1199 were dispensable for RNA3 production (Fig. 4C, lanes 4 to 6). RNA3 production was modestly reduced in pF1fsΔ20 (Fig. 4C, lane 7) and was completely inhibited in pF1fsΔ21 (Fig. 4C, lane 12). Finer mapping (Fig. 4B) indicated that deletion constructs pF1fsΔ25 and pF1fsΔ26 were similar to pF1fsΔ20, with slightly reduced RNA3 accumulation (Fig. 4C, lanes 8 and 9). However, deletion of an additional nucleotide, as in pF1fsΔ27, abolished RNA3 accumulation (Fig. 4C, lane 10), as did deletion of seven additional nucleotides, as in construct pF1fsΔ28 (Fig. 4C, lane 11). Thus, the 5′ boundary of a distal element controlling RNA3 production was mapped to nt 1229.
The 3′ boundary of the distal element controlling RNA3 production was mapped in a similar fashion. At low resolution pF1fsΔ22 and pF1fsΔ23 showed RNA3 accumulation (Fig. 4C, lanes 13 and 14), while pF1fsΔ24 did not (Fig. 4C, lane 15). This suggested that residues between 1237 and 1373 were needed for RNA3 production. At a finer scale (Fig. 4B), constructs pF1fsΔ29, pF1fsΔ30, and pF1fsΔ31 all exhibited only a slight reduction in RNA3 accumulation (lanes 16 to 18), indicating that the 3′ boundary of a distal element controlling RNA3 production lies between nt 1237 and 1242. Additional constructs were created in order to delineate the minimal distal element controlling RNA3 production. pF1fsΔ32, pF1fsΔ33, pF1fsΔ34, pF1fsΔ35, and pF1fsΔ36 all accumulated RNA3, albeit at reduced levels. Thus, a small distal element, centered on an 11-nt core, was shown to be necessary for RNA3 production. We refer to this as the distal subgenomic control element (DSCE). The reduced levels of RNA3 seen in pF1fsΔ32, pF1fsΔ33, pF1fsΔ34, pF1fsΔ35, and pF1fsΔ36 could be due to the absence of the flanking regions, which may contribute to proper conformation of the DSCE.
Base pairing between the DSCE and sequences proximal to the RNA3 start site.
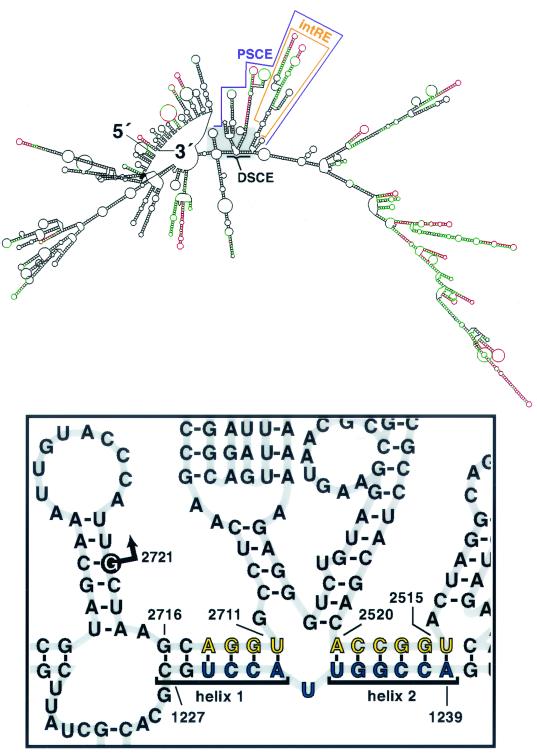
To explore how such a short, distal sequence element might exert its effect on subgenome production from 1.5 kb upstream of the subgenomic start site, we analyzed potential RNA1 secondary structures. Computer-assisted prediction of the lowest-free-energy secondary structure of positive-strand RNA1 (Fig. 5) indicated that the DSCE and sequences within the PSCE could potentially base pair to form two helices. The proximity of helix 1 to the RNA3 start site at position 2721 was interesting. Although nt 2515 to 2520 are located far from the subgenomic region in the primary sequence, the predicted RNA1 secondary structure places both helices near the subgenomic start site. This prediction also included values for pnum, the number of base pairing alternatives, to estimate the confidence level for a given base's predicted structure (26). Although the DSCE had high pnum values, indicating that these residues could base pair with many alternate partners, 7 of the top 10 RNA1 structure predictions (i.e., those with the lowest energy) retained the DSCE base pairing reflected in Fig. 5. Furthermore, 9 of the top 10 predictions had the same structure for residues 2520 to 2711.
FIG. 5.
Potential base pairing of the DSCE to regions proximal to the subgenomic region start site. (Top) Predicted secondary structure of FHV RNA1 with the lowest energy. Bases are colored according to their pnum values within a range of 9 kcal: red, low (<3% of a statistical pnum maximum value at infinite energy); green, moderate (<6%); black, high (>6%). The intRE is bracketed. Gray box, region around the subgenomic region start site. (Bottom) Magnification of the region contained within the gray box. Blue, DSCE residues; yellow, PSCE residues that potentially base pair to the DSCE. Putative helices 1 and 2 are bracketed. Arrow, RNA3 start site at nt 2721.
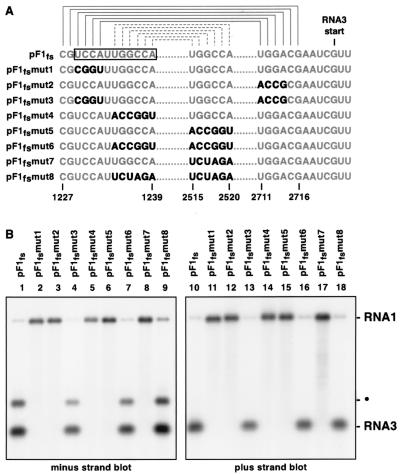
To determine if the predicted base pairing between the DSCE and PSCE is involved in RNA3 production in vivo, we varied and covaried the relevant positions by creating substitution mutations in pF1fs. In pF1fsmut1 the DSCE sequences involved in the first putative helix were varied, while in pF1mut2 the cognate PSCE positions were varied (Fig. 6A). Both mutations completely abolished both positive- and negative-strand RNA3 accumulation (Fig. 6B, lanes 2, 3, 11, and 12). Interestingly, these mutations also led to increased RNA1 accumulation (see Discussion). Combining the two mutations, which restored base pairing potential in pF1fsmut3 (Fig. 6A), also restored positive- and negative-strand RNA3 accumulation (Fig. 6B, lanes 4 and 13). pF1fsmut4 contained mutations in the DSCE that blocked the potential base pairing in helix 2 (Fig. 6A) and lacked the ability to produce RNA3 (Fig. 6B, lanes 5 and 14). Similarly, mutation of the PSCE sequences predicted to form helix 2, as in pF1fsmut5 or pF1fsmut7 (Fig. 6A), abolished RNA3 production (Fig. 6B, lanes 6, 8, 15, and 17). However, RNA production was restored (Fig. 6B, lanes 7, 9, 16, and 18) by compensatory mutations that restored the ability to form helix 2, as in pF1fsmut6 and pF1fsmut8 (Fig. 6A). Thus the potential to base pair was more important than the primary sequence of either region, indicating that RNA3 production is dependent on interaction between the DSCE and PSCE.
FIG. 6.
RNA3 accumulation is dependent on long-distance base pairing between the DSCE and PSCE. (A) Potential base pairing of the DSCE (open box) and PSCE residues to form helix 1 (solid brackets) and helix 2 (dashed brackets). Mutated nucleotides affecting these potential base pairing interactions are in boldface. (B) Northern blot analysis of FHV RNA1 and RNA3 negative (lanes 1 to 7) and positive strands (lanes 8 to 14). •, slower-migrating form of RNA3 (which was also noted in Fig. 1). This band is visible in longer exposures of the plus strand blot.
Randomization of DSCE sequences and in vivo selection of RNA3/GFP-expressing replicons.
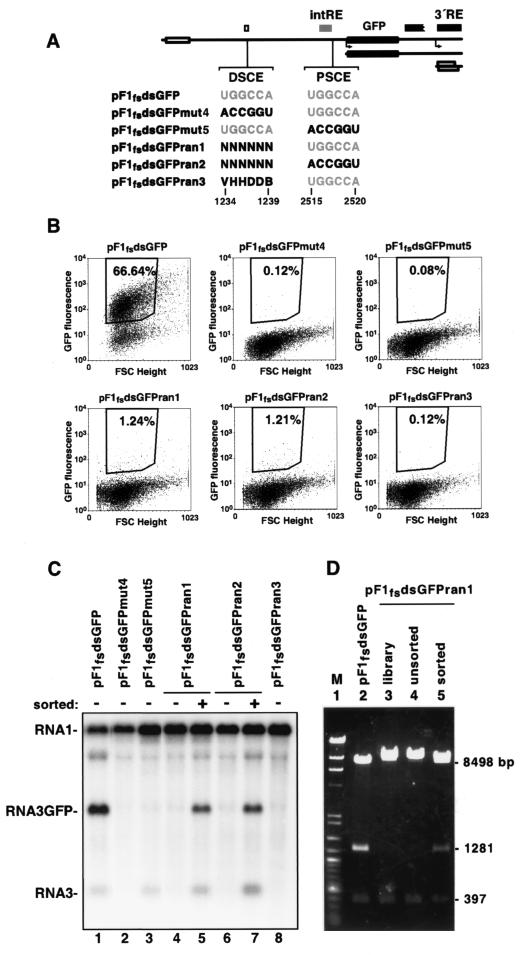
Covariation of DSCE and PSCE sequences provided a powerful genetic argument for the importance of base pairing between these two regions in controlling RNA3 expression. However, this approach was limited to only those mutations that were chosen for analysis. To encompass all possible variations in a relevant sequence, we used a randomization and in vivo selection approach. As controls, the second helix of the long-distance base pairing interaction was disrupted by mutating a 6-nt portion of the DSCE or the cognate PSCE region proximal to RNA3/GFP in pF1fsdsGFP, as described above for pF1fsmut4 and pF1fsmut5 (Fig. 7A). In addition, the same 6 nt of the DSCE were randomized to create small plasmid libraries that retained the wild-type PSCE region, as in pF1fsdsGFPran1, or that had mutations in the PSCE, as in pF1fsdsGFPran2 (Fig. 7A). An additional construct, pF1fsdsGFPran3, was similar to pF1fsdsGFPran1 but contained a smaller subset of sequences represented in the DSCE; it was randomized in such a way as to prevent the wild-type sequence from re-forming in the population (Fig. 7A). These plasmids were transformed into yeast expressing protein A from a chromosomally integrated form of pFA. For each construct, at least 5 × 104 cells were pooled to yield populations that should represent all possible sequences in the randomized positions. Following induction of FHV replication, cells were analyzed on a fluorescence-activated cell sorter. As described for Fig. 3, a majority of cells containing pF1fsdsGFP expressed a high level of fluorescence, while the pF1fsdsGFPmut4 and pF1fsdsGFPmut5 populations demonstrated >500-fold reductions in the number of brightly fluorescent cells (Fig. 7B). Populations containing the pF1fsdsGFPran1 and pF1fsdsGFPran2 libraries had ∼10-fold more GFP+ cells than the pF1fsdsGFPmut4 library-containing cells, while the number of GFP+ cells for pF1fsdsGFPran3 was similar to that for pF1fsdsGFPmut4 (Fig. 7B). These data indicated that the pF1fsdsGFPran1 and pF1fsdsGFPran2 populations contained a small number of cells that expressed GFP, and bright cells were collected from them by sorting and expanded in culture for analysis.
FIG. 7.
In vivo selection of GFP-expressing replicons. (A) Organization of pF1fsdsGFP and derivatives containing variations in part of the DSCE and/or PSCE, illustrated as in Fig. 3A. In accordance with standard International Union of Pure and Applied Chemistry nomenclature, N = any nucleoside; V = A, C, or G; H = A, C, or U; D = A, G, or U; and B = C, G, or U. (B) Cell fluorescence versus forward scattering (FSC), as plotted for 104 cells of the indicated cultures. The gates used for cell sorting and percentages of the total within these bounds are shown. (C) Northern blots of RNAs extracted from the indicated bulk cultures were probed for RNA1 and RNA3 positive strands as indicated in Fig. 1B. (D) MscI digestion of pF1fsdsGFP or the indicated pF1fsdsGFPran1 plasmid pools: library, original plasmid library prior to yeast transformation; unsorted, plasmids after recovery from an unsorted yeast population; sorted, plasmids after recovery from a yeast population sorted as for panel B. M, DNA size standards, HindIII-cut bacteriophage λ genome, and 100-bp ladder (New England Biolabs).
RNA was prepared from all unsorted populations as well as the sorted pF1fsdsGFPran1 and pF1fsdsGFPran2 populations and analyzed by Northern blotting. As expected, pF1fsdsGFP expressed both RNA1 and RNA3/GFP. In contrast, RNA3/GFP expression was strongly inhibited in the pF1fsdsGFPmut4, pF1fsdsGFPmut5, and unsorted pF1fsdsGFPran1, pF1fsdsGFPran2, and pF1fsdsGFPran3 populations (Fig. 7C). These results confirm the lack of GFP expression for these constructs (Fig. 7B) and are consistent with the results presented in Fig. 6B. In the sorted pF1fsdsGFPran1 and pF1fsdsGFPran2 cell populations, both RNA3 and RNA3/GFP expression was evident (Fig. 7C). Thus, fluorescence-based sorting had selectively enriched for those cells in the original population that expressed RNA3L.
Plasmid pools were recovered for analysis from both sorted and unsorted pF1fsdsGFPran1- and pF1fsdsGFPran2-containing populations. The wild-type DNA sequence of the DSCE positions that had been randomized was TGGCCA, which corresponds to the recognition site for restriction endonuclease MscI. Digestion of pF1fsdsGFP with this enzyme yielded the expected products of 8,498, 1,281, and 397 bp (Fig. 7D). Digestion of the original pF1fsdsGFPran1 library before or after recovery from unsorted yeast gave rise to products of 10,779 and 397 bp, which is consistent with the virtual elimination of the MscI site in the DSCE region from the plasmid pool. In contrast, band intensities indicate that about one-half of the plasmid pool rescued from the sorted pF1fsdsGFPran1 population regained the wild-type restriction pattern, implying that the previously randomized positions in these plasmids were now enriched for sequence TGGCCA.
Individual plasmids were isolated from the plasmid pools by bacterial transformation, and the sequence of the DSCE region was determined for ∼90 independent clones from each pool. The results of one experiment are represented in Table 2. Both unsorted populations contained relatively even distributions of sequences at the randomized positions, although a slight bias for T or G was present at all positions, perhaps due to inefficient mixing of deoxynucleoside phosphoramidites during oligonucleotide synthesis. After the pF1fsdsGFPran1 population was sorted, a strong bias for consensus sequence TGGCCA emerged at the randomized positions, thus restoring base pairing potential, and 46 of 91 plasmids contained the canonical MscI restriction site. The other clones contained sequences in these positions that clustered around this consensus sequence but that also contained some mismatch. Some plasmids also contained the consensus sequence, but the sequence was shifted slightly from its normal position (e.g., from nt 1234 to 1239 to nt 1233 to 1238). To determine whether clones containing sequences other than TGGCCA at nt 1234 to 1239 could confer RNA3-directed GFP expression, these plasmids were individually tested by retransforming them back into protein A-expressing yeast, inducing FHV replication with galactose, and analyzing them by flow cytometry. Of 45 clones tested, 40 yielded cells with bright fluorescence, indicating that helix 2 can tolerate some level of mismatch, while 5 clones yielded cells with little or no fluorescence. These nonexpressing clones could have been derived from cells that were spuriously scored positive during the sorting procedure, as for pF1fsdsGFPmut4 (Fig. 7B).
TABLE 2.
Sequences of DSCEs in rescued plasmidsa
| Construct | Type of cell population/(no. of sequences) | nt | No. of sequences with nt at position:
|
|||||
|---|---|---|---|---|---|---|---|---|
| 1234 | 1235 | 1236 | 1237 | 1238 | 1239 | |||
| pF1fsdsGFPran1 | Unsorted (90) | T | [29] | 34 | 27 | 28 | 35 | 30 |
| C | 21 | 14 | 23 | [21] | [18] | 16 | ||
| G | 28 | [28] | [26] | 32 | 26 | 32 | ||
| A | 12 | 14 | 14 | 9 | 11 | [12] | ||
| Sorted (91) | T | [77] | 14 | 5 | 11 | 12 | 21 | |
| C | 2 | 3 | 1 | [59] | [72] | 12 | ||
| G | 7 | [72] | [84] | 21 | 2 | 5 | ||
| A | 5 | 2 | 1 | 0 | 5 | [53] | ||
| pF1fsdsGFPran2 | Unsorted (91) | T | 40 | 34 | 31 | 37 | 36 | [34] |
| C | 17 | [16] | [13] | 24 | 17 | 9 | ||
| G | 20 | 24 | 27 | [14] | [26] | 31 | ||
| A | [14] | 17 | 20 | 16 | 12 | 17 | ||
| Sorted (87) | T | 19 | 12 | 17 | 11 | 28 | [44] | |
| C | 11 | [65] | [57] | 27 | 10 | 6 | ||
| G | 18 | 2 | 10 | [45] | [32] | 30 | ||
| A | [39] | 8 | 3 | 4 | 17 | 7 | ||
Plasmid DNA was recovered from yeast cell populations, prior to or after sorting by fluorescence-activated cell sorter, and isolated in bacterial host DH5α. The sequences corresponding to FHV RNA positions 1234 to 1239 were determined for 96 independent plasmid preparations from each starting population. Only those sequences that were free of ambiguities in this region were tabulated, and their numbers are shown in parentheses. The most abundant nucleotide at each position is in boldface, and the numbers of sequences complementary to positions 2515 to 2520 are in brackets.
By contrast, in the sorted pF1fsdsGFPran2 plasmid population, the consensus sequence that emerged in DSCE positions 1234 to 1239 was ACCGGT (Table 2). Thus, the sequences in the DSCE that were selected restored complementarity to the mutated PSCE (Fig. 7A), although some mismatch was again tolerated.
RNA2 replication requires the long-distance interaction in RNA1.
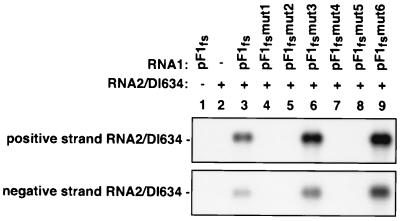
RNA2 replication requires RNA1 replication in trans (9), although the basis for this requirement is unknown. We tested several of the above RNA1 derivatives to see if they could also support replication of an RNA2 derivative. The GAL1 promoter was used to express RNA2-DI634, a defective-interfering RNA2 derivative containing two large internal deletions (55). This template was chosen because of its small size (634 nt), replication similar to that of wild-type RNA2, and lack of capsid protein α expression, which would affect the level of RNA2 accumulation. As expected, RNA2-DI634 was not replicated when coexpressed with protein A alone (Fig. 8, lane 2). Although a small amount of positive-stand RNA2-DI634 was observed, this probably represents transcripts made by RNA polymerase II because a similar amount of this RNA was seen in cells lacking protein A expression, and negative-strand RNA2-DI634 was undetectable (Fig. 8). Replication of RNA2-DI634 required coexpression with protein A and a functional RNA1 template, such as pF1fs (Fig. 8, lane 3). Consistent with this, several replication-defective RNA1 deletion derivatives failed to support RNA2-DI634 replication in trans (data not shown). Interestingly, pF1fsmut1, pF1fsmut2, pF1fsmut4, and pF1fsmut5, which lacked complementarity between the DSCE and PSCE, also did not support RNA2-DI634 replication in trans (Fig. 8, lanes 4, 5, 7, and 8). However, RNA2-DI634 was replicated when coexpressed with pF1fsmut3 and pF1fsmut6, the RNA1 derivatives containing restored DSCE-PSCE base pairing. In this experiment, RNA2-DI634 levels with these mutants were higher than those with wild-type RNA1. However, this effect was not seen in repeats of this experiment. Thus, replication of an RNA2 template can be controlled by the same long-distance interaction in RNA1 that controls RNA3 expression.
FIG. 8.
RNA2 replication depends on DSCE-PSCE interaction in RNA1. Shown is accumulation of positive- and negative-strand RNA2-DI634 in yeast cells harboring the indicated RNA1 and RNA2 derivatives. -, cells transformed with an appropriate control plasmid.
DISCUSSION
In this study, we have used trans replication of RNA1 to dissect cis-acting elements controlling RNA3 production and RNA1 replication, independent of the coding potential of RNA replication templates. Sequences proximal to the RNA3 start site and a short distal element, 1.5 kb upstream, were identified as important determinants of FHV sgRNA production. Furthermore, two regions important for efficient RNA1 accumulation were identified in the 3′ one-fourth of RNA1. Insights into cis-encoded functions were applied to the design of improved RNA3-based vectors for foreign gene expression, which have many potential applications. Finally, these tools were used to identify and characterize long-distance base pairing between the proximal and distal subgenomic control elements, which is essential for RNA3 production and for RNA2 replication in trans.
FHV RNA1 trans replication and cis-acting elements.
Previous results with FHV RNA1 (5) and RNA2 (8) transcripts containing 5′ or 3′ terminal extensions or deletions revealed the critical role of FHV RNA termini for directing RNA replication. It is unclear how far terminal cis elements extend into RNA1. For RNA2, it appears that terminal cis replication elements are contained within the first 14 nt and last 50 to 60 nt (4, 8). An internal region, nt 538 to 616, is also important for replication of RNA2 (8). Prior to the work presented here, no information about internal cis-acting replication signals within RNA1 was available and little was known about sequences controlling RNA3 production.
Deletion analysis was used to map cis-acting elements controlling RNA3 production and RNA1 replication. Because RNA3 is produced during RNA1 replication, we first consider the elements controlling RNA1 accumulation. These analyses indicated that the intRE (nt 2322 to 2501) was necessary for efficient RNA1 replication (Fig. 2). This region was separable from the 3′RE because intervening sequences could be deleted (Fig. 2) or extended, as in double-subgenomic constructs (Fig. 3). Furthermore, this element appears to function in cis, since the wild-type intRE that was present in the protein A mRNA was unable to support replication of templates with deletions in the intRE in trans (Fig. 2). Within the predicted RNA1 secondary structure, the intRE is predicted to fold into a long multibranched stem-loop (Fig. 5A, orange bracketed region). Similar structures were predicted in this region for other nodavirus genomes, such as those for black beetle virus, Boolarra virus, and Nodamura virus (J.-Y. Sgro, unpublished data).
Internal cis-acting replication elements have been described for a number of positive-strand RNA viruses. Phage Qβ replicase, in association with 30S ribosomal protein S1, initially binds to an internal region of the genome called the M site prior to negative-strand initiation (10, 37, 39). Picornavirus genomes contain internal cis replication elements (21, 34, 35) that are crucial for templating uridylylation of VPg prior to initiation of RNA synthesis (41, 45). The intergenic region of brome mosaic virus (BMV) RNA3 contains an important cis replication element (18) that functions in concert with the viral helicase and capping enzyme 1a to recruit this RNA from translation to viral replication (50). As in BMV, the FHV intRE is located upstream of the subgenomic region of RNA1. It is not yet clear how the FHV intRE functions, but, as for the examples just mentioned, it could be involved in template selection or other early steps of RNA replication.
Efficient RNA1 replication was also dependent on the 3′RE, a region whose 5′ boundary lies between nt 2735 and 2755 (Fig. 2). Several small deletions (constructs pF1fsΔ13, pF1fsΔ14, and pF1fsΔ15) did not reveal sequences that were dispensable within this 3′-terminal region (Fig. 2). Thus, the last 276 nt of RNA1 may function as a single cis replication element. This is much larger than the 50 to 60 nt that are required at the 3′ end of FHV RNA2 (4, 8), which could reflect a difference in recognition of these templates by the FHV replicase. Positive-strand RNA virus 3′ ends, which are often highly structured, direct initiation of negative-strand RNA synthesis (11). For FHV RNA1, only a series of stem-loops are predicted for this region of the genome.
Deletion analysis also indicated that sequences between nt 2302 and 2777, the PSCE, are important for efficient RNA3 production (Fig. 2). Given that heterologous sequences can be inserted at the AUG of the B1 ORF with little effect on RNA3 accumulation (42), the 3′ boundary of the PSCE should only extend to nt 2730 at most. Thus, ∼428 nt of RNA1 are needed for an efficient PSCE, although a subset of this region with a 5′ border of nt 2518 was capable of directing a basal level of RNA3 production in double-subgenomic constructs (Fig. 3).
Since RNA3 synthesis arises during the course of RNA1 replication, expression of foreign genes from RNA3 provides a useful way to monitor FHV replication. Because the 3′RE largely overlaps the RNA3 region of RNA1, insertions disrupt this replication element. Double-subgenomic replicons, which contain two copies of the RNA3 region, make it possible to manipulate the upstream subgenomic region while maintaining the downstream 3′RE, as well as the protein B2 ORF. Consistent with this, pF1fsdsGFP replicated to approximately one-third the level of pF1fs, which is a notable increase over the replication of pF1fs-GFPN2 (42). Based on the double-subgenomic design, general-purpose FHV RNA3-based expression vectors have been created (B. D. Lindenbach and P. Ahlquist, unpublished data) and should be useful in dissecting the nodavirus life cycle in yeast. Furthermore, the broad range of eukaryotic cell types that support nodavirus RNA replication suggests that these FHV derivatives could be potentially useful vectors for foreign-gene expression in numerous other cell types.
RNA3 production is controlled by long-distance base pairing in RNA1.
For several other positive-strand RNA viruses, the region surrounding the subgenomic start site contains cis elements controlling subgenome synthesis (reviewed in reference 38). During the course of our mapping studies, we encountered a region in RNA1 located 1.5 kb upstream of the subgenomic start site that was important for RNA3 production. A similar observation was previously noted in mammalian cells: RNA3 was produced during cis replication of an FHV RNA1 derivative with deletions of residues 313 to 941 and 1261 to 2283 but not by a derivative lacking residues 746 to 1830 (9). We further mapped this region to an 11-nt sequence element, the DSCE, that was necessary for RNA3 production (Fig. 4). Insight into how this element operates at a long distance came from the observation that it could potentially base pair with two regions within the PSCE (Fig. 5). Covariation analyses confirmed that both helices were important, as RNA3 production was completely abolished by mutations that disrupted either helix and was fully restored by compensatory mutations (Fig. 6). Moreover, complementarity between these regions was more important than the primary sequence of either region. These conclusions were further supported by in vivo selection of pF1fsdsGFP derivatives containing randomized DSCE sequences. Restoration of the wild-type sequence in the randomized positions of pF1fsdsGFPran1 restored the ability to base pair with the PSCE, although some tolerance for mismatches was seen in the selected sequences (Table 2). This confirmed the importance of this region in directing RNA3 synthesis. Selection of a different DSCE sequence in pF1fsdsGFPran2, which also restored the base pairing interaction, indicated that it was the base pairing potential that was important, rather than the sequences per se, in these positions (Table 2). These results support the model in which the DSCE controls RNA3 production by base pairing to sequences located proximal to the subgenomic start site. As for the intRE, this element appears to function in cis, since the wild-type DSCE that was present in the protein A mRNA did not support RNA3 production from pF1fsmut1 or pF1fsmut4 in trans (Fig. 6).
The experiments presented here show that base pairing between the DSCE and PSCE is necessary for RNA3 production. RNA3 synthesis must be regulated in additional ways, however, since the PSCE encompasses sequences in addition to those of the base-paired region. The fully active PSCE includes some or all of the intRE region located just upstream. Double-subgenomic constructs made a reduced level of RNA3 from their 3′-proximal subgenomic start sites, suggesting that proximity to the intRE is not an absolute requirement and that a minimal PSCE has a 5′ boundary at nt 2518. Although this minimal PSCE boundary excludes the last three base pairing partners of helix 2, additional mutagenesis of these base pairs showed reduced levels of RNA3 production (data not shown). This observation is consistent with the tolerance for some mismatch within DSCE sequences selected in vivo (Table 2). The PSCE also includes an intervening region (nt 2521 to 2710) between the base pairing residues (Fig. 5). Deletion constructs pF1fsΔ8 and pF1fsΔ9 did not reveal any sequences within this intervening region that were dispensable for RNA3 production (Fig. 2). As depicted in Fig. 5, the intervening region is predicted to form a long multibranched structure (downstream of the intRE), which might bind viral or host factors involved in RNA3 synthesis. Similar structures, flanked by long-distance base pairs, are also predicted in other nodavirus genomes, such as those of Nodamura virus, Boolarra virus, and black beetle virus (J.-Y. Sgro, unpublished data).
Long-distance RNA1 base pairing and RNA2 replication.
Our data demonstrate that, in addition to controlling RNA3 production, base pairing between the DSCE and PSCE is also a necessary feature of RNA1 templates that can support RNA2 replication in trans (Fig. 8). This long-distance interaction could induce a structure in RNA1 that interacts with a viral or host protein required for RNA2 replication. It has also been hypothesized that RNA2 can directly base pair to a short region within the PSCE of RNA1 (7, 15), although experimental confirmation of this interaction is needed. Long-distance cis base pairing within RNA1 could therefore affect this putative RNA1-RNA2 trans interaction. Alternatively, the long-distance interaction in RNA1 could also be required if RNA2 replication were dependent on the production of RNA3. Competition between RNA2 and RNA3 for a structure within RNA1, such as that due to long-distance base pairing, might also explain how RNA3 accumulation is downregulated by RNA2 replication. While the dependence of RNA2 replication on RNA1 replication requires further study, the results presented here indicate that the long-distance interaction in RNA1 controls multiple processes in the viral life cycle.
Long-distance interactions that regulate viral processes for several positive-strand RNA viruses have been described. Translation initiation and ribosomal frameshifting of the uncapped, nonpolyadenylated barley yellow dwarf virus genome are mediated via long-distance base pairing (23, 41). Flavivirus genome replication depends on complementarity of cyclization sequences located 10 kb apart (24, 30, 53). Potato virus X sgRNA synthesis is regulated via base pairing between the 5′ noncoding region and an octamer sequence located upstream of both viral sgRNAs (31). Synthesis of the tomato bushy stunt virus sgRNA2 is controlled via base pairing of 12-nt sequences located 1 kb apart (54). Perhaps the ultimate long-distance base pairing that is known to control sgRNA synthesis is the interaction of red clover necrotic mosaic virus genome segments 1 and 2 in trans, which is required for sgRNA3 synthesis (49). A trans base pairing interaction is also thought to direct the discontinuous transcription of nidovirus subgenomic mRNAs during negative-strand synthesis (52). Why do viruses use distal base pairing to control sgRNA synthesis? Clearly viral RNAs are structured in solution, and perhaps long-distance base pairings evolved simply as a means of forming a surface that interacts with the viral replicase or a host factor. In addition, long-distance base pair formation could be modulated by the structure of intervening sequences, which could include internal replication elements such as the FHV intRE. Thus, these structures could provide a conformationally dependent molecular switch to direct genome versus sgRNA synthesis. In this regard, RNA1 accumulated to higher levels in DSCE and PSCE mutants that had lost the ability to form distal interactions than in the wild type and RNA1 levels were reduced to normal when the base pairing potential was restored (Fig. 6).
Based on observations that RNA3 negative-strand accumulation is independent of RNA3 positive-strand accumulation, it has been suggested that the mechanism of FHV sgRNA production may involve an internal termination step during negative-strand synthesis (42, 55). Yet evidence that the RNA3 negative strand serves as a template for positive-strand synthesis is lacking. It is noteworthy that disruption of the DSCE-PSCE interaction abolished both negative- and positive-strand RNA3 accumulations (Fig. 6), suggesting that their synthesis is coordinated via a common mechanism, which could include a precursor-product relationship. Also of note is that long-distance base pairing in the positive strand of RNA1 was predicted, although these same Watson-Crick pairs could potentially form in the negative strand. The structure of the PSCE region in the negative strand of RNA1 could interact with the replicase, acting as a promoter for the synthesis of positive-strand RNA3. Alternatively, the structure of the PSCE in the positive strand of RNA1, involving a long-distance base pairing interaction as well as the spacer region, could act as a terminator to produce RNA3-sized negative strands, which would then serve as templates for synthesis of RNA3-positive strands. Experiments designed to distinguish between these possibilities are in progress.
Acknowledgments
We thank Janelle M. Bailey for technical assistance, as well as Billy Dye, Dan Loeb, and David Miller, for critically reviewing the manuscript.
This work was supported by National Institutes of Health grant GM35072. P.A. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Agatep, R., R. D. Kirkpartrick, D. L. Parchaliuk, R. A. Woods, and R. D. Gietz. October 1998, posting date. Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ssDNA/PEG) protocol. Tech. Tips Online 1:no. 51. [Online.] http://research.bmm.com/tto.
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Ball, L. A. 1992. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J. Virol. 66:2335-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, L. A. 1994. Replication of the genomic RNA of a positive-strand RNA animal virus from negative-sense transcripts. Proc. Natl. Acad. Sci. USA 91:12443-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball, L. A. 1995. Requirements for the self-directed replication of flock house virus RNA 1. J. Virol. 69:720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball, L. A., J. M. Amann, and B. K. Garrett. 1992. Replication of Nodamura virus after transfection of viral RNA into mammalian cells in culture. J. Virol. 66:2326-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ball, L. A., and K. L. Johnson. 1998. Nodaviruses of insects, p. 225-267. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Press, New York, N.Y.
- 8.Ball, L. A., and Y. Li. 1993. cis-acting requirements for the replication of flock house virus RNA 2. J. Virol. 67:3544-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ball, L. A., B. Wohlrab, and Y. Li. 1994. Nodavirus RNA replication: mechanism and harnessing to vaccinia virus recombinants. Arch. Virol. Suppl. 9:407-416. [DOI] [PubMed] [Google Scholar]
- 10.Barrera, I., D. Schuppli, J. M. Sogo, and H. Weber. 1993. Different mechanisms of recognition of bacteriophage Qβ plus and minus strand RNAs by Qβ replicase. J. Mol. Biol. 232:512-521. [DOI] [PubMed] [Google Scholar]
- 11.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bugeja, V. C., M. J. Kleinman, P. F. Stanbury, and E. B. Gingold. 1989. The segregation of the 2 mu-based yeast plasmid pJDB248 breaks down under conditions of slow, glucose-limited growth. J. Gen. Microbiol. 135:2891-2897. [DOI] [PubMed] [Google Scholar]
- 13.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 14.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 15.Dasmahapatra, B., R. Dasgupta, A. Ghosh, and P. Kaesberg. 1985. Structure of the black beetle virus genome and its functional implications. J. Mol. Biol. 182:183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dasmahapatra, B., R. Dasgupta, K. Saunders, B. Selling, T. Gallagher, and P. Kaesberg. 1986. Infectious RNA derived by transcription from cloned cDNA copies of the genomic RNA of an insect virus. Proc. Natl. Acad. Sci. USA 83:63-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.French, R., and P. Ahlquist. 1988. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J. Virol. 62:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friesen, P. D., and R. R. Rueckert. 1982. Black beetle virus: messenger for protein B is a subgenomic viral RNA. J. Virol. 42:986-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallagher, T. M., P. D. Friesen, and R. R. Rueckert. 1983. Autonomous replication and expression of RNA1 from black beetle virus. J. Virol. 46:481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guarino, L. A., A. Ghosh, B. Dasmahapatra, R. Dasgupta, and P. Kaesberg. 1984. Sequence of the black beetle virus subgenomic RNA and its location in the viral genome. Virology 139:199-203. [DOI] [PubMed] [Google Scholar]
- 23.Guo, L., E. Allen, and W. A. Miller. 2000. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA 6:1808-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 25.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 26.Jaeger, J. A., D. H. Turner, and M. Zuker. 1989. Improved predictions of secondary structures for RNA. Proc. Natl. Acad. Sci. USA 86:7706-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, K. N., J. L. Zeddam, and L. A. Ball. 2000. Characterization and construction of functional cDNA clones of Pariacoto virus, the first Alphanodavirus isolated outside Australasia. J. Virol. 74:5123-5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston, M., and R. W. Davis. 1984. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1440-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly, L., W. L. Gerlach, and P. M. Waterhouse. 1994. Characterisation of the subgenomic RNAs of an Australian isolate of barley yellow dwarf luteovirus. Virology 202:565-573. [DOI] [PubMed] [Google Scholar]
- 30.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, K. H., and C. L. Hemenway. 1999. Long-distance RNA-RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA 5:636-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koev, G., B. R. Mohan, and W. A. Miller. 1999. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J. Virol. 73:2876-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2303-2314. [DOI] [PubMed] [Google Scholar]
- 34.Lobert, P. E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mead, D. J., D. C. Gardner, and S. G. Oliver. 1986. The yeast 2 micron plasmid: strategies for the survival of a selfish DNA. Mol. Gen. Genet. 205:417-421. [DOI] [PubMed] [Google Scholar]
- 37.Meyer, F., H. Weber, and C. Weissmann. 1981. Interactions of Qβ replicase with Qβ RNA. J. Mol. Biol. 153:631-660. [DOI] [PubMed] [Google Scholar]
- 38.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 39.Miranda, G., D. Schuppli, I. Barrera, C. Hausherr, J. M. Sogo, and H. Weber. 1997. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J. Mol. Biol. 267:1089-1103. [DOI] [PubMed] [Google Scholar]
- 40.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price, B. D., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raju, R., and H. V. Huang. 1991. Analysis of Sindbis virus promoter recognition in vivo, using novel vectors with two subgenomic mRNA promoters. J. Virol. 65:2501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Selling, B. H., R. F. Allison, and P. Kaesberg. 1990. Genomic RNA of an insect virus directs synthesis of infectious virions in plants. Proc. Natl. Acad. Sci. USA 87:434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan, M., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Sand, S. T., W. Greenhalf, D. C. Gardner, and S. G. Oliver. 1995. The maintenance of self-replicating plasmids in Saccharomyces cerevisiae: mathematical modelling, computer simulations and experimental tests. Yeast 11:641-658. [DOI] [PubMed] [Google Scholar]
- 52.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, G., V. Slowinski, and K. A. White. 1999. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA 5:550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong, W., and R. R. Rueckert. 1993. Flock house virus: down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand. J. Virol. 67:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]