Abstract
In four distinct alstroemeria-infecting cucumber mosaic virus (CMV) isolates, additional sequences of various lengths were present in the 3′ nontranslated regions of their RNAs 2 and 3, apparently the result of intra- and intermolecular recombination events. Competition experiments revealed that these recombined RNA 2 and 3 segments increased the biological fitness of CMV in alstroemeria.
The genome of Cucumber mosaic virus (CMV), the type species of the genus Cucumovirus in the family Bromoviridae (38), consists of three single-stranded RNA molecules: RNAs 1 and 2 encode components of the viral RNA-dependent RNA polymerase (RdRp) (31, 32), while the bicistronic RNA 3 encodes the movement protein (MP) (43) and coat protein (CP) (39). The CP is translated from a subgenomic RNA, RNA 4, which is encoded by the 3′ half of the RNA 3 (19). A small overlapping gene (2b), residing in RNA 2 and implicated in suppression of gene silencing (4), was discovered more recently and is most likely expressed through a second subgenomic RNA (13). Based on their genomic sequences, CMV isolates have been classified into two major subgroups, I and II (2, 32, 35). A further division of subgroup I, into IA and IB, has been proposed based on the nucleotide sequences of 5′ nontranslated regions (NTRs) of RNA 3 (34, 37). With a host range of over 1,000 plant species, CMV is one of the most widespread plant viruses in the world (32). Also, the ornamental plant alstroemeria has been reported as a host of CMV (21), although few data on the interaction between CMV and this host are available.
RNA recombination is generally considered one of the major forces that favor the evolution and adaptation of RNA viruses (14, 24, 36, 41, 42). Most of the data collected from different experimental systems suggest that RNA recombination prefers replicase (RdRp)-driven template switching models or copy choice mechanisms (6, 30, 41). Alternatively, the breakage-and-ligation mechanism has been suggested and proven as a possible means of in vitro RNA recombination as well (12). RNA recombination has been classified according to the extent of involvement of homologous sequences (23, 24, 30).
Evidence from nucleotide sequence data suggests that recombinational events have occurred in satellite RNA associated with CMV strain Y (27) and defective RNAs of the Fny strain of CMV (20). Recombinational events have also been observed in artificial pseudorecombinant viruses composed of CMV and tomato aspermy cucumovirus (TAV) RNAs (16, 28). However, a systematic search of available CMV nucleotide sequences did not indicate that potential recombinational events among numerous analyzed CMV strains occur (7). No natural mixed infection and no genetic exchange between CMV subgroup I and II strains were found (17), indicating that most heterologous genetic recombinations seem to be at a competitive disadvantage (18), although a case of natural interspecies recombination between CMV and TAV has been reported (1). More recently, a homologous recombinational event between RNA 1 and RNA 2 or RNA 3 was demonstrated in vivo in subgroup I CMV when transgene sequences were used as donors (8).
In our previous work we described an alstroemeria-infecting subgroup II CMV isolate (ALS-CMV) that contained an additional sequence of 218 nucleotides (nt) in the 3′ NTR of RNAs 3 and 4 (10). This additional sequence appeared to be identical to parts of the 3′ NTRs of RNAs 1 and 2 of the homologous virus, suggesting that an RNA recombination event occurred between RNA 3 and RNA 1 or RNA 2. To investigate the commonness of additional sequences in the 3′ termini of alstroemeria-infecting CMV isolates, the genomic RNAs of five distinct ALS-CMV isolates were analyzed. Furthermore the biological relevance of the recombinational events encountered are tested by competition experiments.
The CMV strains used in this study, designated ALS-0 through -4 (Table 1), were all independently isolated from alstroemeria plants (ALS-CMV) and were stock material of the Bulb Research Center, Lisse, The Netherlands, the Inspection Service for Floriculture and Arboriculture (NAKtuinbouw), Roelofarendsveen, The Netherlands, and Plant Research International, Wageningen, The Netherlands. ALS-0, although originally isolated from alstroemeria, had been maintained on tobacco for an extended period (at least 5 years); the others were exclusively passaged on alstroemeria. For short-term propagation, all five ALS-CMV isolates were mechanically inoculated onto Nicotiana benthamiana, while alstroemeria plants (clone VV024-6) (25) were used for pathogenicity assays. Purifications of virions, viral RNA, and total RNA were carried out by using published protocols (26, 33, 40). Reverse transcriptase (RT)-PCR amplification of CMV genomic RNAs was performed as previously described (9) with degenerate primer sets for RNA 1 (corresponding to nt 1 to 23 and 1098 to 1108, nt 1074 to 1087 and 2238 to 2255, and nt 2238 to 2248 and 3307 to 3391), RNA 2 (corresponding to nt 1 to 22 and 1105 to 1118, nt 1105 to 1118 and 2062 to 2075, and nt 1937 to 1951 and 3025 to 3039), or RNA 3 (corresponding to nt 1 to 34 and 1120 to 1131 and nt 1087 to 1111 and 2195 to 2204). RT-PCR products were cloned into pGEM-T Easy (Promega), and inserts were sequenced in both directions. Programs of the University of Wisconsin Genetics Computer Group package were used for sequence analysis (Table 1).
TABLE 1.
Lengths and accession numbers of genomic RNAs of ALS-CMV isolates
| Isolate | RNA 1
|
RNA 2
|
RNA 3
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Length (nt)
|
Accession no. | Length (nt)
|
Accession no. | Length (nt)
|
Accession no. | ||||
| Full RNA | 3′ NTR | Full RNA | 3′ NTR | Full RNA | 3′ NTR | ||||
| ALS-0 | 3,391 | 317 | AJ304405 | 3,039 | 424 | AJ304396 | 2,204 | 321 | AJ304399 |
| ALS-1 | 3,442 | 317 | AJ304393 | 3,334 | 719 | AJ304394 | 2,419 | 539 | AJ304397 |
| ALS-2 | 3,391 | 317 | AJ304404 | 3,289 | 674 | AJ304395 | 2,419 | 539 | AJ304398 |
| ALS-3 | NDa | ND | ND | 1,398b | 719 | AJ304400 | 1,413b | 616 | AJ304402 |
| ALS-4 | ND | ND | ND | 1,261b | 582 | AJ304401 | 1,415b | 616 | AJ304403 |
ND, not determined.
Partial sequence.
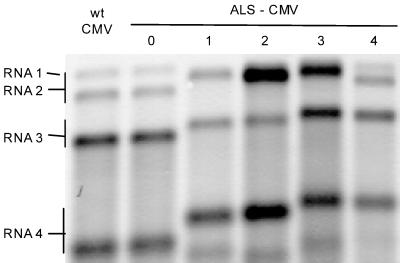
The sequences obtained from all ALS-CMV genomic RNAs appeared to be highly homologous (>97%) to those of reported subgroup II CMV strains. Whereas no large differences were found in the RNAs 1 of the three ALS-CMV isolates tested (ALS-0, ALS-1, and ALS-2), the 3′ NTRs of both RNA 2 and RNA 3 were found to vary significantly in length (Table 1). The RNA 2 molecules did not differ in their 5′ NTR (92 nt), 2a open reading frame (ORF) (2,523 nt), or 2b ORF (303 nt) but varied considerably in their 3′-NTR length (Table 1). Also, the RNA 3 molecules had similar lengths in their 5′ NTRs (96 nt), 3a ORF (840 nt), intercistronic regions (287 to 290 nt), and CP ORF (657 nt) but were notably different in 3′-NTR length (Table 1). The additional sequences were not PCR artifacts, as upon electrophoresis RNAs 3 and 4 of ALS-CMV strains 1 to 4 were visibly larger (Fig. 1) than those of a wild-type subgroup II CMV isolate. Furthermore, the size addition to RNA 2 made this RNA indistinguishable in size from RNA 1 in Northern blots (Fig. 1). ALS-0 had RNAs of wild-type size and sequence, and strikingly, this was the only alstroemeria strain maintained (for several years) on N. benthamiana, instead of its original host. Hence, it cannot be excluded that it originally also contained additional sequences.
FIG. 1.
RNA patterns of five alstroemeria-infecting CMV strains (ALS-0 to -4). Northern blot analysis used a mixture of two digoxigenin-labeled cDNA probes specific for the RNA 2-derived 218 nt in RNA 3 of ALS-1 and ALS-2 (10) and for the CP ORF of subgroup II CMV. The CP-specific probe was PCR amplified with a pair of degenerate primers corresponding to positions 1227 to 1240 and 1866 to 1883 on RNA 3 of ALS-0. A subgroup II isolate from Crocus (9) was used as a wild-type (wt) reference isolate. Size ranges of the genomic RNAs (and subgenomic RNA 4) are indicated on the left.
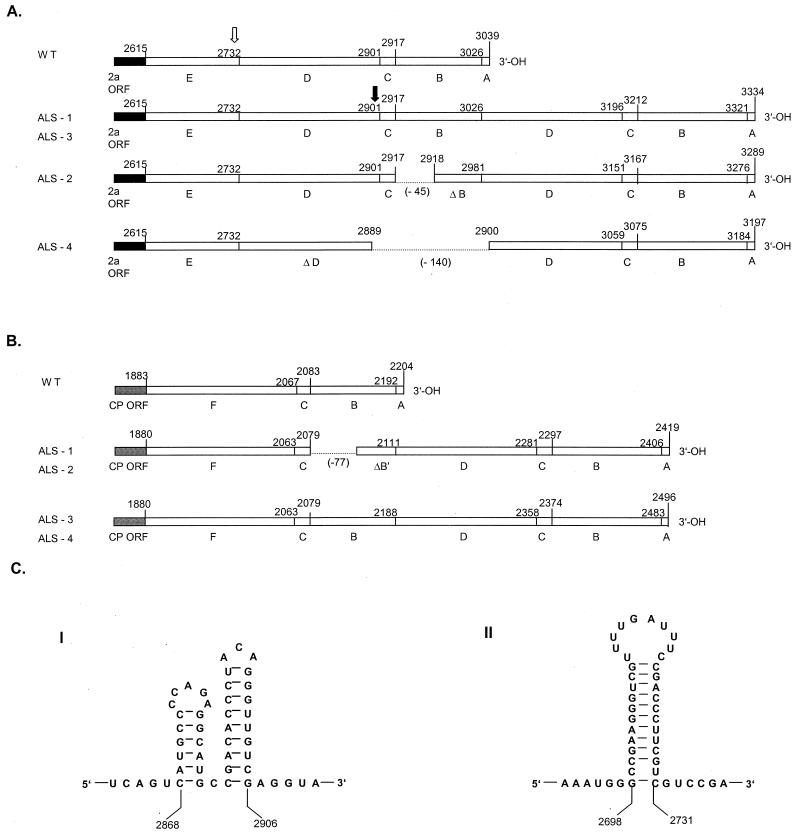
The wild-type (and ALS-0) 3′ NTR of RNA 2 can be arbitrarily divided into five regions (A through E) (Fig. 2A), and that of RNA 3 can be divided into four regions (A, B, C, and F) (Fig. 2B). Regions A through F are 13, 109, 16, 170, 116, and 183 nt long, respectively. Regions A, B, and C are common to all reported CMV segments and can be folded into a tRNA-like structure that is involved in viral minus-strand synthesis (3). In ALS-0, secondary structures consisting of stem-loop configurations are predicted between regions C and D (positions 2868 to 2906) and between regions D and E (positions 2698 to 2731) (Fig. 2C). Interestingly, in all four other ALS-CMV isolates the additional sequences were located in the central region of the 3′ NTRs of both RNA 2 and RNA 3. In the RNA 2 of both ALS-1 and ALS-3, the extra sequences were 295 nt long and represented an exact duplicate of regions D, C, and B (Fig. 2A). Shorter additional sequences of 250 and 158 nt were found in the RNAs 2 of ALS-2 and ALS-4, respectively, and seemed to have arisen from deletions following the original duplication that resulted in the RNAs 2 of ALS-1 and ALS-3.
FIG. 2.
Schematic representations of the 3′ NTR of RNA 2 (A) and RNA 3 (B) of alstroemeria-infecting CMV strains and predicted secondary structures between nt 2868 and 2906 (I) and nt 2698 and 2731 (II) of wild-type (WT) RNA 2 (C). (A and B) Black and gray boxes indicate parts of the ORFs of 2a and CP genes, respectively. The numbers above the empty frames are the nucleotide numbers of the various regions relative to the 5′ terminus. The letters under the diagrams indicate the arbitrary regions of various lengths: A, 13 nt; B, 109 nt; C, 16 nt; D, 170 nt; E, 116 nt; and F, 183 nt. Regions with the same letter are identical in nucleotide sequence and length. The dotted lines indicate deletions (lengths are in parentheses). Δ indicates partially deleted regions. The open arrowhead indicates stem loop structure II (shown in panel C), where replication of RNA 2 is proposed to halt prior to restarting downstream of that position to form a recombinant RNA 2. The solid arrowhead indicates the position of stem loop structure I (C), where replication termination is thought to occur prior to reinitiation on RNA 3.
In the 3′ NTR of RNA 3, the ALS-CMV isolates harbored an additional sequence of 218 nt (ALS-1 and -2) or 295 nt (ALS-3 and -4) (Fig. 2B). Strikingly, the additional 295 nt inserted in the RNA 3 of ALS-3 were completely identical to those of the homologous RNA 2, both in length and in sequence (Fig. 2).
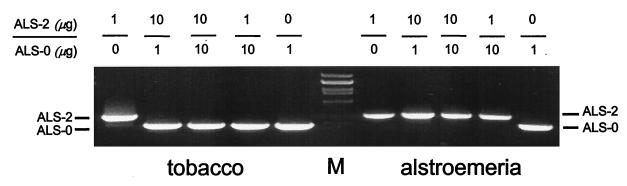
Their presence in the genome of four ALS strains maintained in their original host, and their absence in the only ALS strain not maintained in this host, suggests that the additional sequences may play a role in host adaptation. Indeed, based on visual observations, all isolates containing recombinant RNAs 2 and 3 (ALS-1 through -4) were more virulent than isolates lacking these repetitions, when inoculated into alstroemeria (data not shown), suggesting a greater fitness on this host. To verify this possibility, the ALS-CMV strains were tested in competitive inoculation assays. Tobacco and alstroemeria plants were inoculated with mixtures of various ratios of purified virions of wild-type ALS-0 and one of the recombinant isolates (ALS-2). In systemically infected leaves of tobacco plants, only nonrecombinant sequences could be amplified in subsequent RT-PCR analysis, except when ALS-2 was inoculated alone. In alstroemeria plants, however, the outcome was reversed: only RNAs of recombinant ALS-2 were detected in mixed-inoculation plants (Fig. 3). These results demonstrated a higher replicative ability of the nonrecombinant ALS-0 than of ALS-2 in tobacco plants, while the recombinant ALS-2 isolate was more competitive in alstroemeria.
FIG. 3.
Agarose gel electrophoresis of RT-PCR fragments amplified from total RNA of tobacco or alstroemeria leaves infected with CMV isolates ALS-0 and ALS-2 in various ratios. The degenerate primers used amplified the intercistronic region, CP ORF, and 3′ NTR of RNA 3, resulting in cDNA fragments of 1.1 kb (ALS-0; wild type) or 1.3 kb (ALS-2) (9). Lambda DNA digested with PstI was used as a size marker (M).
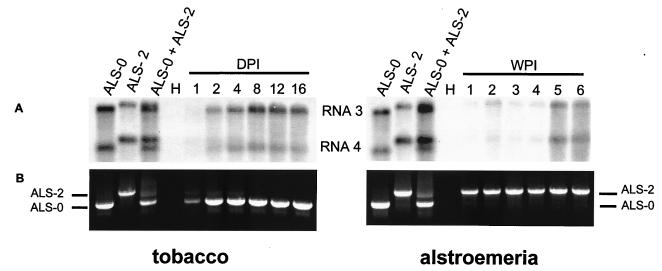
To further delineate the difference in replication speed, tobacco and alstroemeria plants were inoculated with mixtures of virus isolates in which the least fit virus was added in 10-fold excess. Viral RNA replication was monitored in the inoculated leaves by Northern blot analyses and RT-PCR assays (Fig. 4). In tobacco, the enlarged RNAs 3 and 4 of ALS-2 could be detected only until the fourth day postinoculation, while those of ALS-0 could be detected from the first day postinoculation, with the signals further increasing over time. However, on alstroemeria, ALS-0, even though inoculated in 10-fold excess, remained undetectable on inoculated leaves. This experiment indicated that ALS-0 and ALS-2 are outcompeted during the early stages of infection on alstroemeria and tobacco, respectively, in spite of the initial molecular excess over the competitor. Therefore, the additional sequences in the 3′ NTRs of RNAs 2 and 3, which were the only differences observed between these isolates, were responsible for the enhanced fitness of the virus in alstroemeria but produced an adverse effect in tobacco plants. The significance of this finding is further underscored by the fact that four different alstroemeria isolates have these extra sequences, indicating that the occurrence of such recombination is not a single incidence.
FIG. 4.
Competition between ALS-CMV recombinant (ALS-2) and nonrecombinant (ALS-0) isolates upon mixed infection of tobacco and alstroemeria. Total RNA was extracted at various times in days (DPI) or weeks (WPI) postinoculation. (A) RNA gel blot assay using the probes used to produce Fig. 1; (B) products of RT-PCR of the isolated RNA with the primers described in the legend to Fig. 3.
Though many cases of recombination events between RNA segments of viruses or between viral RNAs and their satellite RNAs have been described (e.g., see references 5, 28, and 29), naturally occurring stable recombinations are rare (17). An increase in the biological fitness of a virus by recombination within or between its segments has not been reported previously without applying selection pressure in favor of the recombinant virus. However, our conclusion that the 3′ NTRs of the CMV genome are important for viral fitness is confirmed by previous observations that an artificial pseudorecombinant of CMV and TAV, having RNAs 1 and 2 from CMV and RNA 3 from TAV, gained in fitness upon a recombinational event in which CMV RNA 2-derived 3′-NTR sequences were inserted in its TAV-derived RNA 3 (16).
How the recombinant RNAs in the alstroemeria CMV isolates may have arisen remains subject to further study. Some clues, however, can be obtained from the positions of two stem-loop structures (Fig. 2), in combination with the previously implicated involvement of the CMV RdRp in template switching (22), a mechanism commonly accepted as a major means of generating recombinant RNAs (6, 11, 30, 41, 44). The RNAs 2 of the recombinant isolates show the duplication of a contiguous internal 3′-NTR segment. The same RNA 2-derived sequence was found to be incorporated in the 3′ NTR of the RNAs 3 of the recombinant alstroemeria CMV isolates, some of which seem to have undergone further deletions. A complex secondary structure consisting of a stable hairpin (Fig. 2C, panel II) may have caused the RdRp to halt and disengage during minus-strand synthesis of the recombinant RNA 2. Subsequent restarting of replication at the 3′ end of a second RNA 2 template resulted in the initial duplication (Fig. 2A). While replicating the 3′ end of the recombinant RNA 2, the viral replication complex may have halted and separated at hairpin loop I (Fig. 2C). Subsequently, the RdRp continued replication on the highly homologous 3′ NTR of RNA 3, resulting in an RNA 3 molecule with a duplicated 3′ NTR of RNA 2.
Recombinational events leading to mutant viruses as observed here may be commonplace for CMV, as suggested by Canto and coworkers (8). These authors were readily able to generate recombinant CMV RNAs in a transgene setting, selecting for such events. Nonetheless, recombinant viruses do not occur frequently in nature, most likely due to reduced fitness (17). Our observations with the mixed infections of tobacco plants are in agreement with that notion. Alstroemeria plants, however, seem to be an exception to that rule, sustaining the presence of specific recombinant viruses. Additional sequences observed in the ALS-CMV isolates are all duplicated 3′ NTRs. These 3′ ends of positive-strand viral RNAs contain the origin of minus-strand synthesis and are the presumed site of the promoter elements that control this synthesis in cis (15). It can be hypothesized that a duplication within the NTRs enhances the recruiting of host proteins involved in the viral replication complex. If binding of these components is less efficient in alstroemeria than in other host plant species, it may be beneficial to duplicate the regions involved in recruiting these factors. Although the inserted sequences might interrupt the sequence and secondary structure of the 3′ termini, the alteration apparently offers biological advantages to alstroemeria-infecting CMV isolates. In addition, the slow infection process of CMV in alstroemeria and low accumulation of viral RNA (results not shown) could indicate that duplications in viral sequences are tolerated in this host, while in other hosts these viruses are increased in their sensitivity to host defense responses, such as gene silencing. The competitive inoculation tests demonstrated the merit of these recombination events, which enhance the fitness of ALS-CMV infecting alstroemeria.
Nucleotide sequence accession numbers.
The sequences of the genomic RNAs of ALS isolates used here have been made available in GenBank (accession numbers AJ304393 through AJ304405).
Acknowledgments
We thank R. van der Vlugt and I. Bouwen of Plant Research International and A. van Zaayen of NAKtuinbouw for providing the Alstroemeria isolates of CMV and M. de Jeu and J. B. Kim of the Laboratory of Plant Breeding of Wageningen University for providing Alstroemeria plants.
Y.-K.C. was financially supported by the Ministry of Education of Taiwan.
REFERENCES
- 1.Aaziz, R., and M. Tepfer. 1999. Recombination between genomic RNAs of two cucumoviruses under conditions of minimal selection pressure. Virology 263:282-289. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, B. J., P. M. Boyce, and C. L. Blanchard. 1995. RNA 4 sequences from cucumber mosaic virus subgroup I and II. Gene 161:193-194. [DOI] [PubMed] [Google Scholar]
- 3.Boccard, F., and D. Baulcombe. 1993. Mutational analysis of cis-acting sequences and gene function in RNA 3 of cucumber mosaic virus. Virology 193:563-578. [DOI] [PubMed] [Google Scholar]
- 4.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Bruyere, A., M. Wantroba, S. Flasinski, A. Dzianott, and J. J. Bujarski. 2000. Frequent homologous recombination events between molecules of one RNA component in a multipartite RNA virus. J. Virol. 74:4214-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bujarski, J. J., and P. D. Nagy. 1996. Different mechanisms of homologous and nonhomologous recombination in brome mosaic virus: role of RNA sequences and replicase proteins. Semin. Virol. 7:363-372. [Google Scholar]
- 7.Candresse, T., F. Revers, O. Le Gall, S. A. Kofalvi, J. Marcos, and V. Pallas. 1997. Systematic search for recombination events in plant viruses and viroids, p. 20-25. In M. Tepfer and E. Balazs (ed.), Virus-resistant transgenic plants: potential ecological impact. INRA/Springer-Verlag, Paris, France.
- 8.Canto, T., S. K. Choi, and P. Palukaitis. 2001. A subpopulation of cucumber mosaic virus RNA 1 contains 3′ termini originating from RNAs 2 or 3. J. Gen. Virol. 82:941-945. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Y. K., A. F. L. M. Derks, S. Langeveld, R. Goldbach, and M. Prins. 2001. Host-specific clustering of sequence variation in cucumber mosaic virus isolates infecting ornamental crops. Arch. Virol. 146:3301-3306. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y. K., M. Prins, A. F. L. M. Derks, S. A. Langeveld, and R. Goldbach. Alstroemeria-infecting cucumber mosaic virus isolates contain additional sequences in the RNA 3 segment. Acta Horticultura, in press.
- 11.Chetverin, A. B. 1999. The puzzle of RNA recombination. FEBS Lett. 460:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chetverin, A. B., H. V. Chetverina, A. A. Demidenko, and V. I. Ugarov. 1997. Nonhomologous RNA recombination in a cell-free system: evidence for a transesterification mechanism guided by secondary structure. Cell 88:503-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding, S. W., B. J. Anderson, H. R. Haase, and R. H. Symons. 1994. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198:593-601. [DOI] [PubMed] [Google Scholar]
- 14.Dolja, V. V., and J. C. Carrington. 1992. Evolution of positive-strand RNA viruses. Semin. Virol. 3:315-326. [Google Scholar]
- 15.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Cuartero, B., J. Burgyan, M. A. Aranda, K. Salanki, E. Moriones, and F. Garcia-Arenal. 1994. Increase in the relative fitness of a plant virus RNA associated with its recombinant nature. Virology 203:373-377. [DOI] [PubMed] [Google Scholar]
- 17.Fraile, A., J. L. Alonso-Prados, M. A. Aranda, J. J. Bernal, J. M. Malpica, and F. Garcia-Arenal. 1997. Genetic exchange by recombination or reassortment is infrequent in natural population of a tripartite RNA plant virus. J. Virol. 71:934-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Arenal, F., F. Escriu, M. A. Aranda, J. L. Alonso-Prados, J. M. Malpica, and A. Fraile. 2000. Molecular epidemiology of Cucumber mosaic virus and its satellite RNA. Virus Res. 71:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Gould, A. R., and R. H. Symons. 1982. Cucumber mosaic virus RNA 3: determination of the nucleotide sequence provides the amino acid sequences of protein 3A and viral coat protein. Eur. J. Biochem. 126:217-226. [PubMed] [Google Scholar]
- 20.Graves, M. V., and M. J. Roossinck. 1995. Characterization of defective RNAs derived from RNA 3 of the Fny strain of cucumber mosaic cucumovirus. J. Virol. 69:4746-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakkaart, F. A. 1986. Virusziekten van Alstroemeria. Jaarversl. Instituut Plantenziektenkundig Onderz. 1985:96. [Google Scholar]
- 22.Kim, M.-J., and C. Kao. 2001. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc. Natl. Acad. Sci. USA 98:4972-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King, A. M. Q. 1988. Genetic recombination in positive strand RNA viruses, p. 149-185. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA genetics, vol. II. CRC Press, Boca Raton, Fla. [Google Scholar]
- 24.Lai, M. M. C. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 56:61-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, H. S., M. J. de Jeu, and E. Jacobsen. 1998. Formation of shoots from leaf axils of Alstroemeria: the effect of the position on the stem. Plant Cell Tissue Organ Culture 52:165-169. [Google Scholar]
- 26.Lot, H., J. Marrou, J. B. Quiot, and C. Esvan. 1972. A contribution to the study on cucumber mosaic virus (CMV). II. Quick method of purification. Ann. Phytopathol. 4:25-38. [Google Scholar]
- 27.Masuta, C., S. Kuwata, T. Matzuzaki, Y. Takanami, and A. Koiwai. 1992. A plant virus satellite RNA exhibits a significant sequence complementarity to a chloroplast tRNA. Nucleic Acids Res. 20:2885.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuta, C., S. Uyeda, M. Suzuku, and I. Uyeda. 1998. Evolution of a quadripartite hybrid virus by interspecific exchange and recombination between replicase components of two related tripartite RNA viruses. Proc. Natl. Acad. Sci. USA 95:10487-10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy, P. D., J. Pogany, and A. E. Simon. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 18:5653-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagy, P. D., and A. E. Simon. 1997. New insight into the mechanisms of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 31.Nitta, N., C. Masuta, S. Kuwata, and Y. Takanami. 1988. Inoculation with RNAs 1 and 2 of cucumber mosaic virus induced viral RNA replicase activity in tobacco mesophyll protoplast. J. Gen. Virol. 69:2695-2700. [Google Scholar]
- 32.Palukaitis, P., M. J. Roossinck, R. G. Dietzgen, and R. I. B. Francki. 1992. Cucumber mosaic virus. Adv. Virus Res. 41:281-348. [DOI] [PubMed] [Google Scholar]
- 33.Palukaitis, P., and R. H. Symons. 1980. Purification and characterization of the circular and linear forms of chrysanthemum stunt viroid. J. Gen. Virol. 46:477-489. [Google Scholar]
- 34.Palukaitis, P., and Zaitlin, M. 1997. Replicase-mediated resistance to plant virus diseases. Adv. Virus Res. 48:349-377. [DOI] [PubMed] [Google Scholar]
- 35.Quemada, H., C. Kearney, D. Gonsalves, and J. L. Slightom. 1989. Nucleotide sequences of the coat protein gene and flanking regions of cucumber mosaic virus. J. Gen. Virol. 70:1065-1073. [DOI] [PubMed] [Google Scholar]
- 36.Roossinck, M. J. 1997. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 35:191-209. [DOI] [PubMed] [Google Scholar]
- 37.Roossinck, M. J., L. Zhang, and K. Hellwald. 1999. Rearrangements in the 5′ nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J. Virol. 73:6752-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rybicki, E. P. 1995. The Bromoviridae, p. 450-457. In F. A. Murphy, C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers (ed.), Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. Springer-Verlag, New York, N.Y.
- 39.Schwinghamer, M. W., and R. H. Symons. 1977. Translation of the four major RNA species of cucumber mosaic virus in plant and animal cell-free system and toad oocytes. Virology 79:88-108. [DOI] [PubMed] [Google Scholar]
- 40.Seal, S., and D. Coates. 1998. Detection and quantification of plant viruses by PCR, p. 469-485. In G. D. Foster and S. C. Taylor (ed.), Plant virology protocols, from virus isolation to transgenic resistance. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 41.Simon, A. E., and J. J. Bujarski. 1994. RNA-RNA recombination and evolution in virus-infected plants. Annu. Rev. Phytopathol. 32:337-361. [DOI] [PubMed] [Google Scholar]
- 42.Strauss, J. H., and E. G. Strauss. 1988. Evolution of RNA viruses. Annu. Rev. Microbiol. 42:657-683. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, M., S. Kuwata, J. Kataoka, C. Masuta, N. Nitta, and Y. Takanami. 1991. Functional analysis of deletion mutants of cucumber mosaic virus RNA3 using an in vitro transcription system. Virology 183:106-113. [DOI] [PubMed] [Google Scholar]
- 44.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80:2535-2543. [DOI] [PubMed] [Google Scholar]