Abstract
The NS5B RNA-dependent RNA polymerase encoded by hepatitis C virus (HCV) plays a key role in viral replication. Reported here is evidence that HCV NS5B polymerase acts as a functional oligomer. Oligomerization of HCV NS5B protein was demonstrated by gel filtration, chemical cross-linking, temperature sensitivity, and yeast cell two-hybrid analysis. Mutagenesis studies showed that the C-terminal hydrophobic region of the protein was not essential for its oligomerization. Importantly, HCV NS5B polymerase exhibited cooperative RNA synthesis activity with a dissociation constant, Kd, of ≈22 nM, suggesting a role for the polymerase-polymerase interaction in the regulation of HCV replicase activity. Further functional evidence includes the inhibition of the wild-type NS5B polymerase activity by a catalytically inactive form of NS5B. Finally, the X-ray crystal structure of HCV NS5B polymerase was solved at 2.9 Å. Two extensive interfaces have been identified from the packing of the NS5B molecules in the crystal lattice, suggesting a higher-order structure that is consistent with the biochemical data.
Hepatitis C virus (HCV) belongs to the Flaviviridae family and is responsible for a significant proportion of acute and chronic hepatitis in humans worldwide (7, 8). Similar to other flaviviruses, HCV is a small positive-stranded RNA virus with a genome size of ≈9.6 kb encoding a single polyprotein (30). This viral polyprotein is processed by both host and virally encoded proteases to generate mature structural and nonstructural proteins essential for virus replication (see references 9, 34, and 39 for review). One of the nonstructural proteins, designated NS5B, is the virally encoded RNA-dependent RNA polymerase (RdRp), which contains the GDD signature motif of RNA polymerases (3).
It has been shown that NS5B is a membrane-associated protein, which contains a C-terminal domain comprising ≈21 hydrophobic amino acids that is responsible for membrane anchorage (41). NS5B may form a complex with cellular proteins (37) or other HCV nonstructural proteins, including NS3, the viral protease and helicase; NS4A, a cofactor of NS3 protease activity; and NS5A, a phosphoprotein containing a putative interferon sensitivity region (15). Although the HCV replication mechanism is not clearly understood, the essential role of NS5B polymerase in the HCV replication and infection process has been demonstrated in chimpanzees (22). Accordingly, it has been viewed as an attractive target for antiviral intervention.
Recombinant HCV NS5B polymerase has been produced and purified from both bacterial and insect cells by several groups (3, 11, 16, 19, 25, 27, 31, 41). The availability of highly purified protein has facilitated the biochemical characterization of HCV NS5B polymerase. Similar to other viral RdRps, purified HCV NS5B is able to synthesize RNA using various RNAs as templates in vitro (3, 11, 16, 19, 25, 27, 31, 41). In this regard, two RNA synthesis reaction modes have been described for this enzyme: RNA elongation using a preannealed primer and RNA initiation through a de novo mechanism (31, 20, 27, 35, 42). In addition to its polymerase activity, HCV NS5B protein can bind to ribosomes and to RNAs of various sizes (19, 26, 36).
Here, we describe the further biochemical characterization of HCV NS5B polymerase. Using various biochemical methods and the yeast cell two-hybrid system, we have demonstrated that HCV NS5B forms an oligomeric complex and, as such, catalyzes RNA synthesis in a cooperative manner. Evidence obtained from the analysis of the NS5B crystal structure that highlights the potential interfaces involved in NS5B protein-protein interaction is also presented.
MATERIALS AND METHODS
Materials.
Reagents and columns used for protein purification and homopolymeric RNAs used for the polymerase assay were purchased from Pharmacia. Radiolabeled and nonlabeled nucleotides were purchased from NEN and Gibco, respectively. Polyclonal antibodies against NS5B were generated in a rabbit using denatured purified recombinant HCV-1b NS5B. Chemical cross-linking reagents were purchased from Pierce.
Preparation of recombinant HCV NS5B proteins.
Molecular cloning and expression of a full-length nontagged HCV-1b NS5B protein in bacterial cells were performed as described previously (19). A truncated version of this protein (NS5B-ΔC51), in which the last 51 amino acids were deleted from the C terminus, was also overproduced as a nontagged protein. In addition, an active-site mutant form of this truncated NS5B protein, containing the double substitution of both Asp-319 and Asp-320 in the GDD motif to alanine (NS5B-Δ51GAA), was constructed by rapid PCR-mediated mutagenesis (40) and overexpressed under the conditions described previously (19). Mutations were confirmed by nucleotide sequencing.
The overexpressed full-length or truncated NS5B proteins were purified to homogeneity by sequential chromatography, including DEAE-cellulose, heparin-agarose, poly(U)-Sepharose, and Q-Sepharose as described previously (19). The identity of NS5B was confirmed by N-terminal amino acid sequencing and Western blot analysis using polyclonal antibodies against HCV NS5B peptide.
RNA synthesis assay.
The in vitro RNA polymerase assays for HCV NS5B were performed as described previously using RNA poly(A)/oligo(U)12 as the template/primer pair (3, 19). A typical assay was performed using 10 μg of poly(A) and 1 μg of oligo(U)12 per ml and 10 μM [α-32P]UTP as the substrate (specific activity, 2,000 to 10,000 cpm/pmol) in a total volume of 20 μl of reaction mix containing 20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 25 mM KCl, 1 mM dithiothreitol, and purified NS5B at the concentrations specified. Reactions were conducted at room temperature for the time specified.
A 25-μl aliquot of 100 mM EDTA in 2× SSC solution (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) was then added to the mix to terminate the reaction. Unincorporated radioactive substrate was removed by filtering the reaction mix through a nitrocellulose membrane. The membrane containing the captured RNA products was washed five times with 2× SSC buffer. The incorporated radioactivity was then quantified by liquid scintillation counting as described previously (19).
The de novo RNA synthesis reactions were carried out under the conditions described above, except that poly(C) was used as the template and GTP as the substrate and no primer was added (35). The initial RNA synthesis rates of the NS5B polymerase were calculated and fitted into an equation describing hyperbolic binding equilibria to obtain the apparent dissociation constant (Kd) for monomer-oligomer equilibrium of NS5B protein (24). A cooperativity parameter (Hill coefficient) was generated using the equation log(v/[Vmax − v]) = h log[E] − log K, described by Copeland (10), where v is the velocity at a given enzyme concentration [E] and h is the Hill coefficient.
Size exclusion chromatography.
NS5B-ΔC51 was first purified to homogeneity by sequential chromatography as described above. Purified NS5B-ΔC51 (1.4 mg/ml) was then applied to a Pharmacia Superdex-200 column (PC 3.2/30) preequilibrated with buffer A (25 mM HEPES, pH 7.5, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol), in the absence or presence of 0.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). Proteins were eluted using a Shimadzu SCL-10APV high-pressure liquid chromatograph with buffer A at a flow rate of 0.1 ml/min and detected by absorbance at 280 nm. Protein standards used for gel filtration were bovine serum albumin (66 kDa), catalase (232 kDa), and ferritin (440 kDa), which eluted from the column under the conditions described for NS5B.
Chemical cross-linking of the purified HCV NS5B proteins.
Disuccinimidyl suberate (DSS) and bis-maleimidohexane (BMH) were used as cross-linking reagents. DSS is reactive toward primary amines, and BMH is reactive toward thiol groups under mild conditions. Cross-linking reactions containing either 1 mM DSS or BMH were performed on ice in a total volume of 100 μl with purified NS5B at 10 μg/ml for 60 min in a buffer containing 20 mM Tris-HCl, pH 7.5, and 10% (vol/vol) glycerol. Reactions containing equivalent volumes of dimethyl sulfoxide (DMSO) were used as controls. After 5 min of boiling in the presence of sample buffer, reaction mixtures were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under denaturing conditions followed by silver staining.
Yeast two-hybrid analysis.
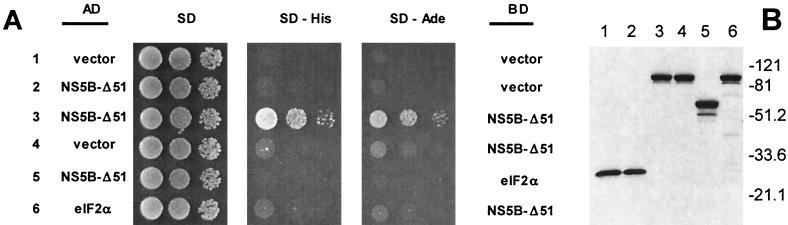
For expression of the C-terminally truncated version of NS5B (NS5B-ΔC51) as a Gal4 activation domain (AD) or Gal4 DNA-binding domain (BD) fusion protein in Saccharomyces cerevisiae, NS5B-ΔC51 was amplified by PCR and subcloned into the EcoRI and XhoI sites of pGADT7 (Clontech) or EcoRI and SalI sites of pGBKT7 (Clontech), respectively. For yeast cell two-hybrid analysis, plasmids were introduced into strain AH109 (Clontech), a derivative of pJ69-4A (18), as described previously (17). In order to screen for two-hybrid interactions, the resulting transformants were grown in synthetic complete medium lacking tryptophan and leucine (SC/Trp−Leu−) medium at 30°C for approximately 24 h; diluted to optical densities of 0.25, 0.025, and 0.0025 at 600 nm; and spotted onto SC plates as indicated in the legend to Fig. 7.
FIG. 7.
Identification of HCV NS5B self-interaction by the yeast two-hybrid system. (A) Interaction of NS5B-ΔC51 molecules in the yeast two-hybrid assay. The indicated combinations of plasmids were introduced into yeast strain AH109. The resulting transformants were grown in SC/Trp−Leu− medium and spotted onto plates lacking tryptophan, leucine, and histidine or onto plates lacking tryptophan and leucine and containing limiting amounts of adenine as described in Materials and Methods. Growth on plates lacking histidine or on plates containing limiting amounts of adenine is indicative of an interaction between the indicated activation domain (AD) fusion and DNA-binding domain (BD) fusion proteins. (B) Western blot analysis of extracts from the yeast strains shown in panel A. A total of 50 μg of proteins of each extract was first resolved by SDS-4 to 20% PAGE, transferred to nitrocellulose membranes, and detected using anti-DNA-binding domain antibodies as described in Materials and Methods.
For Western blot analysis, yeast cell extracts were prepared by growing each strain in SC/Trp−Leu− medium to mid-log phase and harvesting by centrifugation. Cell pellets were washed with ice-cold breaking buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM β-mercaptoethanol, 0.1% NP-40) containing protease inhibitors (Roche). The cell pellets were then resuspended in 250 μl of cold breaking buffer and vortexed with glass beads. Extracts were clarified by centrifugation for 20 min at 14,000 rpm. The protein concentration of each extract was determined as described previously (4) using bovine serum albumin as the standard. Protein samples (50 μg) were resolved by SDS-4 to 20% PAGE and transferred to nitrocellulose membranes. Membranes were blocked with TBS-T (20 mM Tris, pH 8.0, 150 mM NaCl, 0.1% Tween 20) containing 5% nonfat dry milk, probed with activation domain- or DNA-binding domain-specific antibodies (Santa Cruz), and immune complexes were visualized by enhanced chemiluminescence according to the manufacturer's protocol (Amersham).
Crystallization and structural determination.
Highly purified HCV-1b NS5B-ΔC21 was prepared as described previously (1) and concentrated to 10 mg/ml in 20 mM Tris (pH 7.5), 1 mM EDTA, 10 mM dithiothreitol, 10% glycerol, 500 mM NaCl, and 500 mM imidazole. Fresh dithiothreitol (final concentration, 5 mM) was added just prior to crystallization. Crystals were grown in a hanging drop plate using 2.0 M ammonium sulfate and 5% 2-propanol as the precipitant. Hexagonal crystals of dimensions 100 by 100 by 150 mm grew within 7 to 10 days. X-ray diffraction data were collected with a Mar charge-coupled device (CCD) detector at the IMCA beam line ID-17 at Advanced Photon Source in Argonne National Laboratories. The crystals belong to space group P6522 with unit cell dimensions of 110.815 Å and 238.573 Å. The structure was solved by molecular replacement using a similar molecule described by Ago and colleagues (PDB code 1QUV) as the search model. The structure was further refined in CNX to 2.9 Å resolution with the R factor of 25% and Rfree of 29%.
RESULTS
Oligomerization of purified HCV NS5B polymerase.
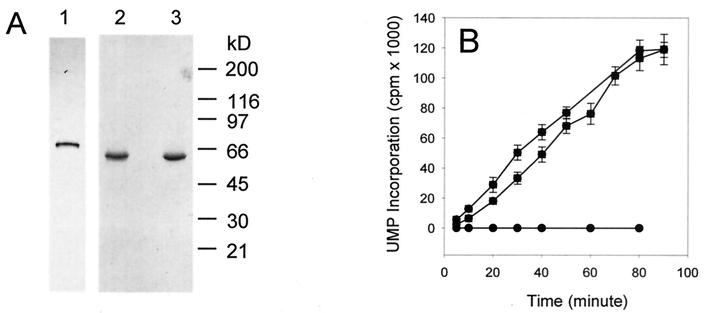
We recently reported the expression and purification of an active nonfusion form of HCV genotype-1b full-length NS5B (NS5B-FL) polymerase (19). When analyzed by denaturing SDS-PAGE, the highly purified HCV NS5B migrated as a single band with the expected molecular mass of ≈65 kDa (Fig. 1A). However, when the same sample was analyzed under nondenaturing gel electrophoresis conditions, NS5B protein migrated as a smeared band from the monomer position to the top of the gel, even in the presence of 0.5% Triton X-100 (data not shown), suggesting that HCV NS5B was highly oligomerized or aggregated.
FIG. 1.
Analysis of purified HCV NS5B proteins . (A) SDS-PAGE analysis of purified HCV NS5B proteins. Purified full-length and truncated forms of HCV NS5B proteins (≈100 ng) were separated on a 4 to 20% gradient gel followed by silver staining. Lane 1, NS5B-FL; lane 2, NS5B-ΔC51; lane 3, NS5B-ΔC51GAA. (B) RNA polymerase activity of the purified HCV NS5B proteins. Reactions were performed using poly(A)/oligo(U)12 as the template/primer and UTP as the substrate under the conditions described in the text using 1 μg of purified full-length NS5B (▪), NS5B-ΔC51 (•), and NS5B-ΔC51GAA (▴) per ml.
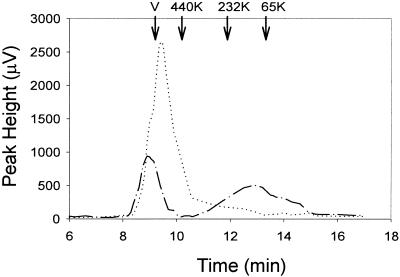
Since HCV NS5B protein contains a hydrophobic C terminus that is responsible for membrane association (41), we examined whether this observed oligomerization was due to the hydrophobic region in NS5B. Therefore, a truncated NS5B protein (NS5B-ΔC51), lacking the C-terminal 51 amino acids, was prepared and purified (Fig. 1A). Consistent with data reported previously by others (25), the purified NS5B-ΔC51 was catalytically active and demonstrated RNA polymerase activity in vitro (Fig. 1B). Surprisingly, the active NS5B-ΔC51 eluted from the size-exclusion column as an oligomer (Fig. 2). Monomeric NS5B could be detected in the presence of 0.5% nonionic CHAPS (Fig. 2), indicating that the larger apparent size in the absence of detergent did not result from an irregular shape, but rather from oligomerization that could occur even in the absence of the C-terminal hydrophobic domain.
FIG. 2.
Gel filtration analysis of purified HCV NS5B. Purified HCV NS5B-ΔC51 (70 μg) was loaded onto a Superdex-200 column and eluted in the absence (dotted line) and presence (dashed line) of 0.5% CHAPS as described in the text. The void volume (V) and the elution positions for bovine serum albumin (66 kDa), catalase (232 kDa), and ferritin (440 kDa) are labeled. NS5B-ΔC51 had a calculated mass of ≈60 kDa.
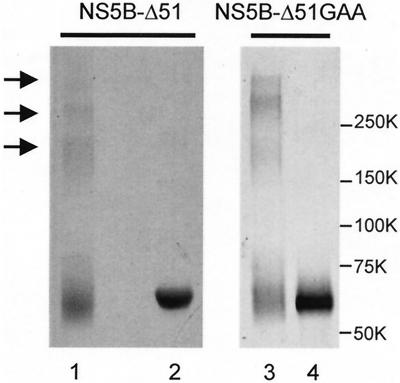
We then investigated HCV NS5B protein oligomerization using protein-protein cross-linking agents. Since HCV NS5B contains a number of lysine residues, we used the homobifunctional cross-linking agent DSS, which reacts with primary amines. As seen in Fig. 3, a number of DSS cross-linked species of NS5B-ΔC51 were observed. A major component appeared to have a molecular weight consistent with that of an NS5B trimer, although dimer and higher oligomers were also present (Fig. 3). Similar results were also obtained using the catalytically inactive NS5B-ΔC51 protein (NS5B-ΔC51GAA), as shown in Fig. 3 (lanes 3 and 4).
FIG. 3.
Chemical cross-linking of purified HCV NS5B. Cross-linking reactions were performed under the conditions described in the text. Cross-linked samples were separated on a 4 to 20% gradient gel under denaturing conditions, followed by silver staining. Lane 1, NS5B-ΔC51 plus DSS; lane 2, NS5B-ΔC51 plus DMSO control; lane 3, NS5B-ΔC51GAA plus BMH; lane 4, NS5B-ΔC51GAA plus DMSO control.
Western blot analysis revealed that the cross-linked proteins were recognized by the antibodies raised against the HCV NS5B (results not shown). Oligomerized NS5B-ΔC51 could also be detected when BMH, reactive with the thiol group of cysteine residues, was used as the cross-linker (data not shown). These results confirmed that NS5B-ΔC51 protein lacking the C-terminal hydrophobic region was capable of forming various-sized oligomers.
It has been shown that oligomerization can protect proteins from thermal denaturation (28). The activity of a monomeric enzyme as a function of temperature should be independent of enzyme concentration and dependent only on the intrinsic activity of the enzyme at that temperature. Since oligomerization is protective against thermal denaturation, oligomerized enzymes are likely to display increased activity at elevated temperatures when present at high concentrations that favor oligomerization.
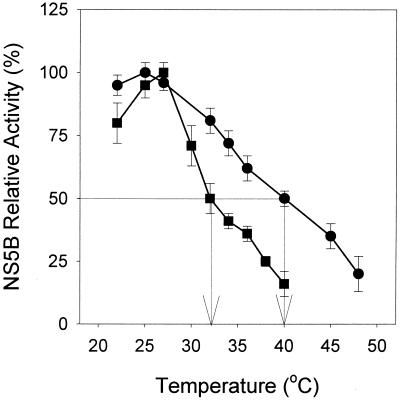
To determine whether this was the case for HCV NS5B, we examined the effects of temperature, ranging from 22 to 48°C, on its polymerase activity at low (8 nM) and high (80 nM) concentrations. The 8 nM samples were supplemented with bovine serum albumin so that the total protein concentration in both sets was 80 nM. As shown in Fig. 4, NS5B-ΔC51, when present at lower concentrations, was less active at the higher temperature, demonstrating increased thermal stability as a result of the formation of NS5B oligomers. T1/2, the temperature at which the enzyme displayed 50% of the maximal activity, was 32°C at 8 nM and 40°C at 80 nM NS5B-ΔC51 (Fig. 4).
FIG. 4.
Effect of HCV NS5B protein concentration on its enzymatic activity at increased temperature. RNA polymerase activity of NS5B-ΔC51 was examined as a function of temperature under the typical assay conditions described in the text. Activity was measured for 20 min at the temperature indicated using two different NS5B-ΔC51 concentrations, 80 nM (•) and 8 nM (▪), with the latter case containing 72 nM bovine serum albumin to make up the final protein concentration to 80 nM. The rate data from the two concentrations were normalized to percent maximal activity because the specific activities at high and low NS5B concentrations were different. The arrows indicate the values of T1/2 as 32°C and 40°C for 8 and 80 nM NS5B-ΔC51, respectively.
Cooperative RNA synthesis activity of HCV NS5B polymerase.
If HCV NS5B oligomers are more active than monomers, the RNA polymerase activity of the NS5B protein would increase exponentially with increased protein concentration due to the monomer-oligomer equilibrium shift. To examine if HCV NS5B displays such cooperativity, the polymerase activity of NS5B-ΔC51 was measured over a broad range of enzyme concentrations using poly(A)/oligo(U)12 as the template/primer and UTP as the substrate.
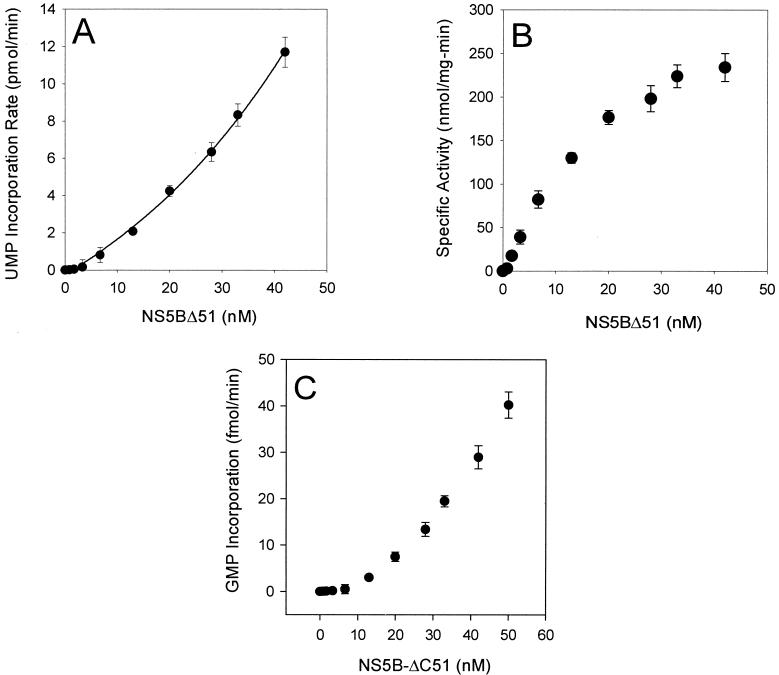
As shown in Fig. 5A, a nonlinear dependency of the RNA synthesis activity on HCV NS5B protein concentration was observed. When displayed as a plot of apparent specific activity versus protein concentration, as seen in Fig. 5B, the data exhibit a hyperbolic dependence, yielding a ≈85-fold increase in the apparent specific activity as the concentration of NS5B-ΔC51 was increased from 1 nM to 42 nM. From these data, the Hill coefficient, a measure of enzyme cooperativity, was determined to be ≈2.2, suggesting a minimum of two to three interaction sites on the oligomerized NS5B protein. A marked increase in the specific activity of NS5B-FL polymerase was also observed (data not shown). Taken together, these data suggest that NS5B catalyzed RNA elongation in a cooperative manner.
FIG. 5.
Dependence of RNA polymerase activity on HCV NS5B concentration. Polymerase activity was measured at different concentrations of NS5B-ΔC51 under the conditions described in the text. The exponential dependence of the NS5B-ΔC51 activity for RNA elongation (A) and RNA initiation (C) on enzyme concentration is shown. The specific activity of RNA elongation of the purified NS5B-ΔC51 protein is presented in panel B. The equilibrium value Kd was determined as ≈22 nM by using the data presented in panel B, and the equation was described previously (24).
Cooperative RNA initiation reaction catalyzed by HCV NS5B.
Using the conditions we established previously (35), the de novo RNA synthesis activity of the purified NS5B-ΔC51 was assayed in the absence of a primer over a broad range of enzyme concentrations using poly(C) as the template and GTP as the substrate. Similar to the RNA elongation catalyzed by this enzyme, the extent of de novo RNA synthesis was not proportional to the NS5B protein concentration; instead, an exponential dependence was observed (Fig. 5C). As a result, the apparent specific activity of the NS5B showed a greater than 51-fold increase when the concentration was increased from 4.2 nM to 50 nM, consistent with our hypothesis that more active NS5B oligomers were formed at higher protein concentrations.
Inhibition of HCV NS5B polymerase activity by inactive mutant NS5B protein.
To further investigate the cooperative nature of the polymerase activity associated with HCV NS5B protein, the activities of mixtures of wild-type and catalytically inactive mutant NS5B were measured. The catalytic activity of the wild-type NS5B would be affected by the inactive mutant if there was a cooperative interaction between these proteins and several active sites per oligomer were needed.
It has been shown previously by several groups that substitutions at the highly conserved GDD motif of NS5B result in catalytically inactive proteins (3, 25, 41). Therefore, we constructed a mutant NS5B-Δ51 protein (NS5B-Δ51GAA) in which the conserved metal nucleotide-binding motif GDD was changed to GAA and the protein was purified as shown in Fig. 1. As expected, the double mutation resulted in the abrogation of NS5B-Δ51GAA catalytic activity compared to the wild-type enzyme (Fig. 1B) but did not affect its self-oligomerization (Fig. 3, lanes 3 and 4) or interaction with the wild-type NS5B-Δ51 protein (results not shown).
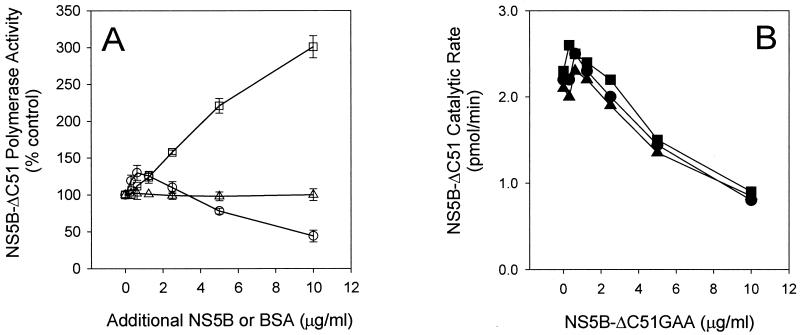
Next, the polymerase activity of the wild-type NS5B protein was measured in the presence of increasing amounts of catalytically inactive NS5B-Δ51GAA. To provide equal RNA template/primer binding activity for both mutant and wild-type NS5B proteins, we mixed the two protein samples on ice for 30 min and then initiated the reactions by addition of substrate. As shown in Fig. 6, a decrease in the rate of RNA synthesis was observed when mutant NS5B protein at high concentration was added to the reaction. In contrast, more RNA products were generated when increased amounts of active NS5B-Δ51 were added to the reaction (Fig. 6A), indicating that the decreased rate of the reactions containing NS5B-GAA was not a result of template titration.
FIG. 6.
Inhibition of NS5B polymerase activity by inactive NS5B-Δ51GAA protein. (A) Effect of added mutant and wild-type NS5B-ΔC51 on NS5B-ΔC51 activity. RNA polymerase activity of NS5B-ΔC51 (1.25 μg/ml) was measured for 5 min in the presence of excess NS5B-ΔC51 (□) or catalytically inactive NS5B-ΔC51GAA (○) at the concentrations indicated. Data were expressed as the percentage of the control reaction containing 1.25 μg of NS5B-ΔC51 per ml. Reactions performed using the same amounts of bovine serum albumin are also shown (▵). (B) Time-dependent inhibition of NS5B by NS5B-Δ51GAA. Reactions were performed as described above for different time points: 2.5 min (▪), 5 min (•), and 10 min (▴). Shown is the RNA synthesis rate of NS5B-ΔC51.
Inhibition of the wild-type NS5B polymerase activity by the mutant form was also observed from reactions performed for different time periods (Fig. 6B). No inhibition was seen when bovine serum albumin at the same concentration was mixed with the active polymerase (Fig. 6A). In addition, HCV helicase and human eIF4A, both RNA-binding proteins, did not show inhibition when tested under the same conditions (data not shown). Interestingly, at lower concentrations, NS5B-Δ51GAA could stimulate the wild-type NS5B-Δ51 activity (Fig. 6), suggesting to us that the catalytically inactive NS5B-Δ51GAA, through interacting with the active NS5B-Δ51, helped recruit NS5B-Δ51 to bind to the template/primer. Taken together, these results strongly suggest the formation of a complex between the wild-type and catalytic mutant NS5B proteins.
Interaction of HCV NS5B protein molecules in vivo.
Physical interactions among NS5B molecules were further demonstrated using the yeast cell two-hybrid system. NS5B-ΔC51 was efficiently expressed in yeast cells cells when fused to the Gal4 DNA-binding domain (Fig. 7B) or Gal4 activation domain (data not shown). Auxotrophic growth of yeast cells containing the indicated combinations of plasmids was scored by spotting on the plates lacking histidine or containing limiting amounts of adenine as described in Materials and Methods. Synthesis of His3p or Ade2p was dependent on reconstitution of a functional transcription factor via protein-protein interactions among NS5B molecules.
As can be seen in Fig. 7A, the strain expressing AD-NS5B-ΔC51 and BD-NS5B-ΔC51 displayed efficient growth under these conditions, whereas little or no growth was observed in the vector control strains, arguing for physical interactions between NS5B-ΔC51-containing fusion proteins. No interaction between NS5B-ΔC51 and human translation initiation factor eIF2α (Fig. 7), human lamin, or murine p53 (data not shown) was observed, suggesting that the observed NS5B self-interaction was specific under the conditions employed.
Molecular packing of HCV NS5B in crystal lattice.
Using a highly purified version of NS5B protein similar to that described previously (1), we obtained NS5B crystals under the conditions described in Materials and Methods and solved the X-ray structure at a resolution of 2.9 Å. Although this protein, containing the N-terminal 570 amino acids of HCV NS5B, had a typical polymerase folding, with three characteristic subdomains of fingers, palm, and thumb as reported by other groups (1, 5, 23), our structural analysis also revealed interesting features which will be reported in detail elsewhere.
For the purpose of this study, we examined the packing status of the NS5B molecules in the crystal lattice, as it could provide important insight on protein-protein interaction; for example, the interactions observed in the crystal structure of poliovirus polymerase contribute to functional oligomerization (13, 21). In our case, the NS5B crystal was packed in a space group with one NS5B molecule per asymmetric unit and six symmetry-related molecules in a unit cell, differing from other reported HCV NS5B structures in which one or two molecules were found in one unit cell (1, 5, 23).
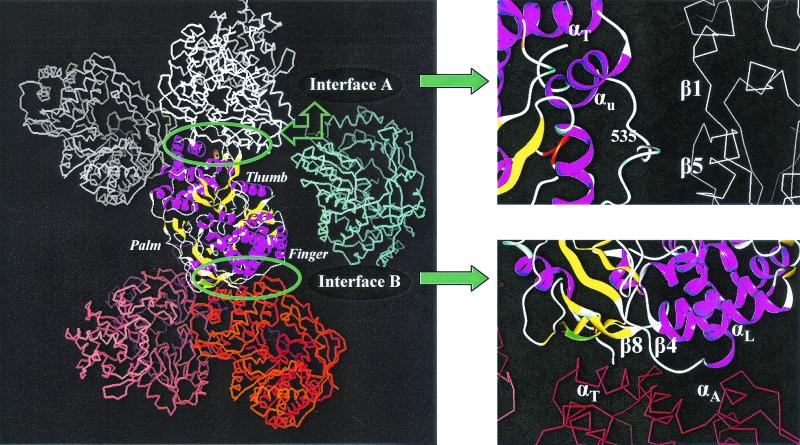
Analysis of the protein packing diagram in one unit cell revealed two extensive interfaces, designated A and B, between NS5B polymerase molecules (Fig. 8). Interface A is a head-to-tail interaction between the thumb subdomain of one molecule and the front of the finger subdomain of the adjacent NS5B molecule. In this interface, the β strands β1, β4, and β8 from one molecule are in close contact with αT and αU from the α finger subdomain of the other molecule (nomenclature as described in reference 5). Interface B is a head-to-side interaction between the back of the thumb subdomain in one molecule and the antiparallel β-strand of the finger subdomain from another molecule. The residues involved in the close contact in interface B are from β2, β5, and αB of one molecule and the loops between αT and αU plus the C-terminal region (amino acids 532 to 540) of another molecule.
FIG. 8.
Interactions between NS5B molecules in crystal lattice. The crystal structure for HCV NS5B-ΔC21 was solved at a resolution of 2.9 Å as described in the text. The two interfaces identified from the NS5B crystal lattice are highlighted and labeled interface A and interface B. Nomenclature for the key motifs involved in these interfaces is as described in reference 5.
DISCUSSION
Due to the essential role of NS5B protein in the HCV replication and infection process (22), this enzyme has been studied extensively. In addition, the crystal structure of purified HCV NS5B has been solved at high resolution by several groups (1, 5, 23), providing an in-depth understanding of this important enzyme. Recently, we and others have shown that NS5B is able to catalyze template-dependent RNA synthesis in the absence or presence of primer (20, 27, 31, 35, 42). In this report, we present both biochemical and structural data that demonstrate the oligomerization and cooperativity of HCV NS5B.
Our results suggest that HCV NS5B is able to oligomerize and catalyze both RNA initiation and elongation in a cooperative manner. The oligomerization of NS5B was demonstrated by gel filtration chromatography, chemical cross-linking, stability to thermal denaturation, and yeast two-hybrid methods. The data for HCV NS5B oligomerization were obtained in the absence of RNA molecules using both nontagged full-length and C-terminally truncated NS5B proteins. Therefore, this protein-protein interaction was not induced by RNAs and was not a result of nonspecific aggregation involved in the C-terminal hydrophobic region of the HCV NS5B.
Evidence for NS5B cooperative RNA polymerase activity was provided by the observed increase in its specific activity (per unit enzyme) at higher concentrations. This conclusion was further supported by the observed inhibition of the wild-type NS5B activity by catalytically inactive mutant NS5B-Δ51GAA, especially at high concentration, indicating that a mixed oligomer was formed. These results might help explain the biochemical data reported previously. For example, Carroll and colleagues reported that a large fraction of the purified HCV NS5B protein was catalytically incompetent (6). If inactive NS5B proteins can form a complex with the active enzyme, as seen in Fig. 6, it might impede the formation of more active complexes among active NS5B molecules. This could then explain the observed low catalytic efficiency, in general, associated with HCV polymerase compared to the poliovirus polymerase (38) and human immunodeficiency virus (HIV) reverse transcriptase (33).
Little is known regarding the stability of the NS5B protein complexes and the frequency with which the protein subunits in the complex exchange during the course of reaction. The data presented here suggest that the interaction between NS5B protein in the absence of RNA is not strong, since nonionic detergents could disrupt its oligomerization (Fig. 2). It is possible, therefore, that frequent exchange of the protein subunits in the complex might occur.
Interestingly, a transiently unstable model has been proposed recently for the HCV helicase functional oligomer (24). According to this model, the transient interaction between the wild-type and mutant helicase proteins resulted in a decrease in the rate of helicase unwinding activity (24), which is similar to the effect of inactive mutant NS5B on the wild-type NS5B-ΔC51 polymerase activity (Fig. 6). Therefore, the unstable-transient model proposed for HCV helicase might also apply to the HCV NS5B polymerase.
It remains to be determined whether only the HCV NS5B oligomers possess enzymatic activity or whether the NS5B monomer is also active but not catalytically efficient. Similar questions have been asked but have not yet been resolved for several helicases encoded by HCV and bacteria (21, 24, 29). It was found that the poliovirus 3D monomer could catalyze RNA synthesis but was much less active than were the oligomers (2, 32). For HCV NS5B, a definitive answer to this question should also be obtained from the site-directed mutagenesis approaches discussed above.
For poliovirus 3D polymerase, two interfaces, termed I and II, have been described (13) and proposed to be responsible for the cooperative RNA binding and polymerization observed for this enzyme (2, 32). Mutagenesis has revealed that interface I in poliovirus 3D polymerase is important for nucleic acid binding, while interface II is crucial for catalytic activity, and more importantly, the formation of these interfaces correlates with viral viability (14).
For HCV NS5B, analysis of the packing diagram of NS5B crystals has revealed the existence of two extensive interfaces, interfaces A and B (Fig. 8). The structural and functional relationships between interfaces A and B in HCV NS5B and interfaces I and II in poliovirus polymerase are now being evaluated. Experiments, including generation of mutant NS5B polymerases with specific amino acid substitutions at each of the two interfaces observed in the crystals and evaluation of their nucleic acid binding activity and catalytic efficiency, are currently ongoing. It is hoped that these approaches will help gain a better understanding of HCV NS5B polymerase oligomerization. If NS5B-NS5B interactions play an important role in the regulation of HCV RNA replication, as seen with poliovirus, this interaction can serve as an appropriate target for development of antiviral agents.
Acknowledgments
We thank Dayue Chen for suggestions and critical reading of the manuscript. Special thanks to Ouhong Wang for data processing and analysis.
REFERENCES
- 1.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold Des. 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 2.Beckman, M. T., and K. Kirkegaard. 1998. Site size of cooperative single-stranded RNA binding by poliovirus RNA-dependent RNA polymerase. J. Biol. Chem. 273:6724-6730. [DOI] [PubMed] [Google Scholar]
- 3.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Bressanelli, S., L. Tomei, A. Roussel, I, Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, S. S., V. Sardana, Z. Yang, A. R. Jacobs, C. Mizenko, D. Hall, L. Hill, J. Zugay-Murphy, and L. C. Kuo. 2000. Only a small fraction of purified hepatitis C RNA-dependent RNA polymerase is catalytically competent: implications for viral replication and in vitro assays. Biochemistry 39:8243-8249. [DOI] [PubMed] [Google Scholar]
- 7.Choo, Q.-L, G. Kuo, A. J. Weiner, R. L. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 8.Choo, Q. L., A. J. Weiner, L. R. Overby, G. Kuo, M. Houghton, and D. W. Bradley. 1990. Hepatitis C virus: the major causative agent of viral non-A, non-B hepatitis. Br. Med. Bull. 46:423-441. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, B. 1997. Molecular virology of hepatitis C virus. J. Gen. Virol. 78:2397-2410. [DOI] [PubMed] [Google Scholar]
- 10.Copeland, R. A. 1996. Enzymes, p. 279-296. Wiley-VCH Press, New York, N.Y.
- 11.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagedorn, C. H., E. H. van Beers, and C. de Staercke. 2000. Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase). Curr. Top. Microbiol. Immunol. 242:225-260. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Strucuture 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 14.Hobson, S. D., E. S. Rosenblum, O. C. Richards, K. Richmond, K. Kirkegaard, and S. C. Schultz. 2001. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 20:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishido, S., T. Fujita, and H. Hotta. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35-40. [DOI] [PubMed] [Google Scholar]
- 16.Ishii, K., Y. Tanaka, C. C. Yap, H. Aizaki, Y. Matsuura, and T. Miyamura. 1999. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology 29:1227-1235. [DOI] [PubMed] [Google Scholar]
- 17.Ito, H., Y. Fukada, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast cells. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, R. B., X. L. Sun, M. A. Hockman, E. C. Villarreal, M. Wakulchik, and Q. M. Wang. 2000. Specificity and mechanism analysis of hepatitis C virus RNA-dependent RNA polymerase. Arch. Biochem. Biophys. 377:129-134. [DOI] [PubMed] [Google Scholar]
- 20.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khu, Y.-L., E. Koh, S. P. Lim, Y. H. Tan, S. Brenner, S. G. Lim, W. J. Hong, and P.-Y. Goh. 2001. Mutations that affect dimer formation and helicase activity of the hepatitis C virus helicase. J. Virol. 75:205-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 24.Levin, M. K., and S. S. Patel. 1999. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 274:31839-31846. [DOI] [PubMed] [Google Scholar]
- 25.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann, V., A. Roos, F. Korner, J. O. Koch, and R. Bartenschlager. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108-118. [DOI] [PubMed] [Google Scholar]
- 27.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat. J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margosiak, A. P., D. L. Vanderpool, W. Sisson, C. Pinko, and C. C. Kans. 1996. Dimerization of the human cytomegalovirus protease: kinetic and biochemical characterization of the catalytic homodimer. Biochemistry 35:5300-5307. [DOI] [PubMed] [Google Scholar]
- 29.Mechanic, L. E., M. C. Hall, and S. W. Matson. 1999. Escherichia coli DNA helicase II is active as a monomer. J. Biol. Chem. 274:12488-12498. [DOI] [PubMed] [Google Scholar]
- 30.Miller, R. H., and R. H. Purcell. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. USA 87:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pata, J. D., S. C. Schultz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 33.Reardon, J. E. 1992. Human immunodeficiency virus reverse transcriptase: steady-state and pre-steady-state kinetics of nucleotide incorporation. Biochemistry 31:4473-4479. [DOI] [PubMed] [Google Scholar]
- 34.Reed, K. E., and C. M. Rice. 1998. Molecular characterization of hepatitis C virus, p. 1-37. In H. W. Reesink (ed.), Hepatitis C virus. Karger, Basel, Switzerland. [DOI] [PubMed]
- 35.Sun, X. L., R. B. Johnson, M. A. Hockman, and Q. Wang. 2000. De novo RNA synthesis catalyzed by HCV RNA-dependent RNA polymerase. Biochem. Biophys. Res. Commun. 268:798-803. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka, T., K. Sugiyama, M. Ikeda, A. Naganuma, A. Nozaki, M. Saito, K. Shimotohno, and N. Kato. 2000. Hepatitis C virus NS5B RNA replicase specifically binds ribosomes. Microbiol. Immunol. 44:543-550. [DOI] [PubMed] [Google Scholar]
- 37.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. C. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 38.Van Dyke, T. A., R. J. Rickles, and J. B. Flanegan. 1982. Genome-length copies of poliovirion RNA are synthesized in vitro by the poliovirus RNA-dependent RNA polymerase. J. Biol. Chem. 257:4610-4617. [PubMed] [Google Scholar]
- 39.Wang, Q. M., and B. A. Heinz. 2000. Recent advances in prevention and treatment of hepatitis C virus infections. Prog. Drug Res. 55:1-32. [DOI] [PubMed] [Google Scholar]
- 40.Weiner, M. P., and G. L. Costa. 1995. Rapid PCR site-directed mutagenesis, p. 613-621. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 42.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]