Abstract
The vaccinia virus E3L gene encodes two double-stranded RNA binding proteins that promote viral growth and pathogenesis through suppression of innate immunity. To explore how E3L enables vaccinia virus to evade the interferon system, cells and mice deficient in the principal interferon-regulated antiviral enzymes, PKR and RNase L, were infected with wild-type vaccinia virus and strains of vaccinia virus from which E3L had been deleted (E3L-deleted strains). While wild-type virus was unaffected by RNase L and PKR, virus lacking E3L replicated only in the deficient cells. Nevertheless, E3L-deleted virus failed to replicate to high titers or to cause significant morbidity or mortality in triply deficient mice lacking RNase L, PKR, and Mx1. To investigate the underlying cause, we determined the effect of E3L on interferon regulatory factor 3 (IRF3), a transcription factor required for viral induction of subtypes of type I interferons. Results showed that IRF3 activation and interferon-β induction occurred after infections with E3L-deleted virus but not with wild-type virus. These findings demonstrate that E3L plays an essential role in the pathogenesis of vaccinia virus by blocking the interferon system at multiple levels. Furthermore, our results indicate the existence of an interferon-mediated antipoxvirus pathway that operates independently of PKR, Mx1, or the 2-5A/RNase L system.
Poxviruses, such as vaccinia virus (VV), are remarkable for the wide spectrum of factors they encode to evade host defenses. Included in the poxvirus repertoire are receptor mimics for tumor necrosis factor, interleukin 18 (IL-18), IL-1β, alpha interferon (IFN-α), IFN-β, IFN-γ, and chemokines, as well as chemokine homologs, complement control proteins, and inhibitors of IFN-induced antiviral enzymes (1, 2, 24, 38). Not surprisingly, VV is relatively resistant to IFN in a variety of cell lines and VV can even rescue IFN-sensitive viruses, such as vesicular stomatitis virus and encephalomyocarditis virus, from the antiviral effects of IFN (27, 43, 44, 48). However, it is unknown how VV mediates its anti-IFN effects in vivo.
The antiviral activities of IFNs are mediated by proteins encoded among the IFN-stimulated genes (38). The IFN-induced proteins include certain interferon regulatory factors (IRFs) that amplify or inhibit the IFN response, including IRF1, IRF2, IRF9/ISGR3γ (13), and IRF7 (31). Another member, IRF3, is constitutively expressed in a variety of tissues in an inactive form. Phosphorylation of IRF3 after virus infection on serine and threonine residues in the C-terminal region leads to nuclear translocation and binding to the coactivator CREB-binding protein (CBP)/p300 (20, 33, 42, 47). The activated IRF3 in turn induces the expression of a subset of type I IFN genes, and the production of these IFNs then triggers strong induction of IRF7 expression. Finally, IRF3 and IRF7 cooperate and result in induction of a wider range of IFN genes (22, 32). IFN-inducible pathways, such as the 2-5A/RNase L system, protein kinase PKR, and Mx proteins mediate the antiviral actions of IFNs (reviewed in reference 38). The 2-5A/RNase L system and PKR both require IFN for gene induction and double-stranded RNA (dsRNA) for enzyme activation. In the former pathway, IFN treatment of cells induces a group of 2-5A synthetases that produce 2-5A [px(A2′p)nA, where x = 1 to 3 and n ≥ 2] from ATP in response to dsRNA (17). 2-5A activates the latent endoribonuclease, RNase L, causing cleavage of viral and cellular RNA (9, 36, 49). PKR is activated by dsRNA, causing autophosphorylation and eIF-2α phosphorylation leading to inhibition of translation initiation (25). Furthermore, activation of either PKR or the 2-5A system leads to apoptosis, a process with the potential to eliminate virus-infected cells (6, 12, 14, 19, 50).
The ability of VV to cause disease in mice and to infect a broad range of cell types in vitro is dependent on the VV E3L gene that encodes 25- and 20-kDa proteins with C-terminal dsRNA binding domains (5, 7, 41). In contrast to wild-type VV, VV from which E3L is deleted (VVΔE3L) is sensitive to the antiviral action of IFNs and does not rescue vesicular stomatitis virus from IFN treatment (3, 4). Because PKR and 2-5A synthetase require dsRNA as allosteric effectors, sequestration of dsRNA by E3L can prevent activation of both enzymes (3, 7, 41). For instance, studies in which rRNA degradation was monitored during VV infections suggested that E3L was able to suppress activation of RNase L, presumably by preventing 2-5A synthesis (3, 4). In cell culture studies, the dsRNA binding domain of E3L is required for IFN resistance as shown with VV encoding different E3L mutants (8). However, the N-terminal domain of E3L binds to and antagonizes PKR, suggesting that inhibition of PKR by E3L occurs through an E3L:PKR:dsRNA complex (29, 35). Studies with mice support roles for both the C- and N-terminal domains of E3L for viral pathogenesis, highlighting the importance of in vivo experiments for dissecting specific functions of viral immunity factors (5). Recently, expression of E3L from a plasmid vector was shown to inhibit IRF3 and IRF7 phosphorylation, suggesting that E3L could also interfere with viral induction of IFNs (37). To explore how E3L functions as a viral immunity factor in vitro and in vivo, VV and VVΔE3L were used to infect cells and mice lacking Mx1 and either PKR or RNase L or both PKR and RNase L. While E3L prevented the antiviral action of RNase L and PKR in vitro, VVΔE3L failed to cause significant disease in triply deficient mice lacking Mx1, RNase L, and PKR. Moreover, VVΔE3L, but not wild-type VV, caused phosphorylation and nuclear translocation of IRF3 and induction of IFN-β.
MATERIALS AND METHODS
Viruses and cells.
Wild-type VV and VVΔE3L in both the Western Reserve (WR) strain and the Copenhagen strain were used (see legends to Fig. 1 through 5). The E3L gene was inserted into the parental VVΔE3L virus of strain WR by recombination of pMP E3L plasmid into the E3L locus of VVΔE3L, following a procedure previously described (18). The presence of the E3L gene in the recombinant virus was confirmed by sequencing and by monitoring protein expression in Western blots using primary antibodies specific for E3L. Immortalized mouse embryonic fibroblasts (MEFs) (50, 51), baby hamster kidney (BHK) cells, and human fibrosarcoma (HT1080) cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. African green monkey kidney cells (BSC40) were grown in the same media with 10% HyClone (Gibco/BRL) and antibiotics. Conditions for virus growth, infection, and plaque titration were as described previously (11). Briefly, BHK cells were inoculated with virus at a multiplicity of infection (MOI) of 0.1 and incubated at 37°C for 2 h. The inoculum was removed and replaced with fresh medium, and the infected cells were incubated at 37°C until the cytopathic effects were complete. Cells were harvested by centrifugation and resuspended in phosphate-buffered saline (PBS). The infected cells were frozen and thawed twice before the titers of the viruses were determined.
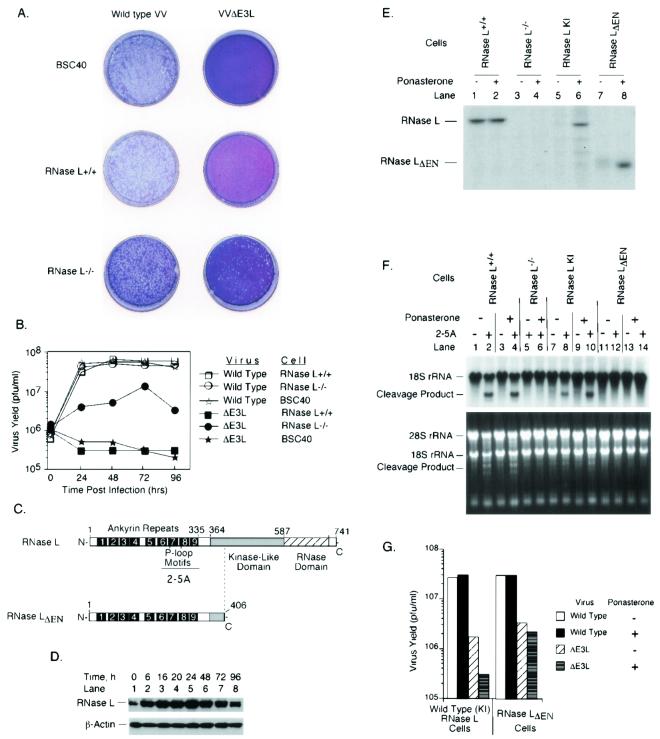
FIG. 1.
E3L allows VV to evade the antiviral activity of RNase L. Plaque assays (A) and one-step viral-growth curves (B) of wild-type VV and VVΔE3L (Copenhagen strain) are shown. (C) Diagram of RNase L and mutant RNase LΔEN. (D) Ponasterone induction of RNase L in comparison to β-actin as determined by probing a Western blot with antibodies. (E) Assay for RNase L and mutant RNase LΔEN levels by covalent cross-linking to a 32P-labeled 2-5A analog before and after ponasterone induction (5 μM for 24 h). (F) Assay for RNase L activity in intact cells as measured by specific cleavages in rRNAs. Cells were incubated in the absence or presence of ponasterone (5 μM) for 24 h and transfected with 2-5A [p3A(2′p5′A)3] (1 μM) for 3 h. Total RNA was isolated, electrophoresed in a formaldehyde-1.2% agarose gel, stained with ethidium bromide (lower panel), and probed with 32P-labeled 18S rRNA cDNA (upper panel). (G) RNase L−/− cells containing inducible cDNAs for RNase L and RNase LΔEN were incubated in the absence or presence of ponasterone (5 μM) for 24 h and infected at an MOI of 6 with wild-type VV or VVΔE3L (Copenhagen strain). At 24 h postinfection, viruses were harvested and virus yields were determined by plaque titration on BHK21 cells.
Plaque assays.
Confluent cells in 60-mm-diameter dishes were infected with serial dilutions of virus. After 30 min, the inoculum was removed and 4 ml of low-melting-point agarose and Dulbecco's modified Eagle medium was added per dish. In some instances, plaque assays were done under liquid medium instead of an agar overlay. After 4 days of incubation, dishes with agar overlay were stained with neutral red overnight, while dishes with liquid medium were stained with crystal violet for 1 h.
One-step viral-growth curves.
Viral-growth curves (one step) were generated as previously described (46). Briefly, cells were infected with virus at an MOI of 6 to ensure that only one cycle of growth was measured. At various times postinfection, viruses were harvested and the yield from each time point was quantified by plaque titration assay on BHK21 cells. The experiments were repeated three times with essentially similar results.
Construction of inducible RNase L clones.
A ponasterone-inducible mammalian cell expression system (Invitrogen) was used to control expression of either RNase L or a nuclease-deleted RNase L polypeptide, RNase LΔEN. The full-length coding sequence cDNA for human RNase L in plasmid ZC5/4T3 or a cDNA with a deletion of the endoribonuclease domain of RNase LΔEN (plasmid CΔ406) (16) was subcloned into plasmid vector pIND/Hygro between BamHI and NotI cloning sites to generate inducible RNase L clones pINDWT or pINDΔEN. Either pINDWT or pINDΔEN was cotransfected with plasmid pVgRXR (containing the heterodimeric ecdysone receptor and zeocin resistance genes) into the RNase L−/− MEF cell line using Lipofectamine Plus transfection reagent (Gibco/BRL). At 24 h posttransfection, cells were washed and fresh medium was added. At 48 h posttransfection, the cells were split into fresh selective medium containing zeocin and hygromycin at 150 μg/ml each. Fresh selective medium was added every 3 days. The cell colonies, which are resistant to zeocin and hygromycin, were expanded. Expression of full-length RNase L was verified by Western blotting using monoclonal antibody to RNase L (15), and expression of the C-deleted RNase LΔEN was identified by 2-5A binding assay (26).
Western blot analysis.
Proteins (200 μg per lane) in cell extracts were separated on 10% polyacrylamide/sodium dodecyl sulfate (SDS) gels, transferred to Immobilon-P membrane (Millipore), and incubated with monoclonal antibody against RNase L (15) or monoclonal antibody against β-actin (Boehringer Mannheim) for 1 h. Membranes were washed with PBS containing 0.1% Tween 20 and incubated with goat anti-mouse antibody tagged with horseradish peroxidase (Gibco BRL) for 1 h. Proteins were visualized by enhanced chemiluminescence (ECL) reagent (Amersham).
2-5A binding assay for RNase L.
The 32P-labeled and bromine-substituted 2-5A probe, p(A2′p)2(br8A2′p)2A[32P]Cp, was covalently cross-linked to RNase L in crude cell extracts under UV light as described previously (26). Briefly, 105 cpm (specific activity, ≥3,000 Ci/mmol) of 2-5A probe was incubated on ice for 1 h with cell extract containing 200 μg of protein. Cross-linking under a UV lamp (308 nm) was done on ice for 1 h. The proteins were separated on 10% polyacrylamide/SDS gels, and the gels were dried and then autoradiographed.
RNase assays.
To activate RNase L in intact cells, the tetramer form of 2-5A, p3A(2′p5′A)3 prepared with isolated human 2-5A synthetase (30), was transfected at a concentration of 1 μM for 3 h using Lipofectamine Plus. Total RNA, isolated from the transfected cells with Trizol reagent (Gibco/BRL), was denatured in formamide and electrophoresed through 1.2% formaldehyde-agarose gels. RNAs were transferred to Nylon membranes (Schleicher & Schuell), prehybridized, and hybridized to a 32P-labeled human 18S rRNA cDNA (American Type Culture Collection).
Virus infections of mice.
Seven-week-old mice were used, with four males plus four females per group. Wild-type C57BL/6 mice were from Jackson Laboratories. RNase L−/− and PKR−/− mice were from a C57BL/6 background, and double-knockout mice were of mixed background (51). Mice were anesthetized with isoflurane and inoculated by the intranasal (i.n.) route with 106 PFU of wild-type VV or 5 × 106 PFU of VVΔE3L in 10 μl. Mice were monitored daily for morbidity and mortality.
IRF3 phosphorylation assay.
Cells were infected with the WR strains of wild-type VV, VVΔE3L, and reconstituted wild-type VV at an MOI of 15. At various times postinfection, cells were collected and 40 μg (protein) of cell extract was used per lane in 10% polyacrylamide-SDS gels for Western blot analysis with polyclonal antibody against human IRF3 (kindly provided by Michael David, University of California at San Diego). The IRF3:antibody complex was detected with secondary goat anti-rabbit antibody (Gibco/BRL) by using ECL reagent (Amersham).
Detection of IRF3 in intact cells by indirect immunofluorescence.
HT1080 cells, fixed with paraformaldehyde for 30 min and permeabilized with 0.2% Triton X-100 for 15 min at room temperature, were washed and blocked with PBS containing 0.02% Tween 20, 3% bovine serum albumin (Gibco/BRL), and 3% goat serum for 2 h. Cells were incubated with polyclonal antibody to human IRF3 at a 1:5,000 dilution for 1 h. Washed cells were incubated with secondary goat anti-rabbit antibody Alexa Fluor 488 (Molecular Probes) at a 1:1,500 dilution for 1 h. All antibodies were diluted in PBS containing 3% bovine serum albumin. Washed cells were mounted to slides using Vectashield with 4′,6′-diamidino-2-phenylindole (DAPI) (Vecta Laboratories, Inc). Stained cells were monitored with a Leica fluorescence microscope (magnification, ×400).
IFN-β mRNA assay.
Plates (60 mm) of confluent HT1080 cells were mock infected or infected with either wild-type VV, VVΔE3L, or reconstituted wild-type VV at an MOI of 15. At different times postinfection, total RNA was purified from each dish using Trizol reagent (Gibco/BRL). RNA preparations were incubated with DNase I and then monitored on 1.2% agarose gels for integrity. IFN-β mRNA was assayed by reverse transcriptase (RT)-PCR as previously described (39). Briefly, RNA was reverse transcribed with Superscript RT enzyme by using random hexamer primers (Gibco/BRL). A negative control reaction (−RT) was carried out in the absence of reverse transcriptase. One-twentieth of the resulting cDNA was used as a template for PCRs containing [α-32P]dCTP and specific primers for IFN-β or glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) mRNAs. The products were analyzed after electrophoresis in 5% polyacrylamide gels by autoradiography.
RESULTS
Avoidance of the 2-5A/RNase L system by E3L.
To determine whether E3L exerts its interferon-suppressive function by inhibition of the 2-5A/RNase L pathway, RNase L−/− and RNase L+/+ MEFs were infected with wild-type VV or the E3L deletion mutant, VVΔE3L (Fig. 1). Wild-type VV formed plaques of similar sizes and numbers on African green monkey kidney cells (BSC40), RNase L+/+ MEFs, and RNase L−/− MEFs, suggesting that RNase L had no effect (Fig. 1A). In contrast, VVΔE3L did not form plaques on either RNase L+/+ MEFs or BSC40 cells but did form reduced numbers (10-fold fewer than did wild-type VV) of small plaques on RNase L−/− cells (Fig. 1A). To determine the kinetics of virus replication, one-step viral growth experiments were performed (Fig. 1B). Wild-type VV grew equally well in the three cell lines with titers peaking at 6 × 107 PFU/ml by 48 h and remaining constant thereafter, results consistent with the plaque assays. VVΔE3L, on the other hand, did not grow in RNase L+/+ or BSC40 cells (<200-fold the level of virus obtained with wild-type VV). However, VVΔE3L growth in RNase L−/− cells was measurable by 24 h and peaked by 72 h postinfection at levels that were severalfold lower than with wild-type VV. The yields of VVΔE3L from the RNase L−/− cells were about 20-fold higher than those from RNase L+/+ MEFs or BSC40 cells. These findings support the idea that E3L suppresses the 2-5A/RNase L system, while also indicating additional functions for E3L.
To confirm that E3L acts in part by preventing RNase L activity, we constructed stable cell lines expressing human RNase L or a mutant RNase LΔEN (this designation indicates the deletion of the endonuclease domain) (Fig. 1C). The wild-type and mutant enzymes were cloned in the RNase L−/− MEFs under control of a ponasterone-inducible promoter. Immunoblot assays showed that RNase L was induced by 6 h of treatment with ponasterone in the knock-in (KI) cells, with levels peaking at 24 h and declining by 96 h, while the β-actin levels monitored for comparison were unchanged (Fig. 1D). A relatively low level of expression of RNase L was observed without ponasterone treatment, indicating a leaky promoter. Because RNase LΔEN could not be detected with this antibody, which is against a deleted epitope, it was detected instead by using a radioactive 2-5A cross-linking assay (Fig. 1E, lane 8). The same assay was used to show that levels of RNase L in the wild-type cells were similar to the level in the induced, reconstituted (KI) cells (Fig. 1E, compare lanes 2 and 6). Specific rRNA cleavage products were visualized after transfections with 2-5A by Northern blotting for 18S rRNA and by staining with ethidium bromide (Fig. 1F). RNase L activity was observed in the RNase L+/+ parental cells following 2-5A transfection and was unaffected by ponasterone treatment (Fig. 1F, lanes 1 to 4). In contrast, rRNA was not degraded in 2-5A-treated RNase L−/− cells or in cells expressing RNase LΔEN (Fig. 1F, lanes 5, 6, and 11 to 14). A low level of RNase L activity was observed in the reconstituted (KI) cells in the absence of ponasterone (again indicating a slightly leaky promoter) (Fig. 1F, lane 8). However, levels of RNase L activity increased to wild-type levels at 24 h after induction (lane 10).
Induction of full-length RNase L did not affect yields of wild-type VV (4 × 107 PFU/ml at 24 h postinfection) (Fig. 1G). However, in the absence of ponasterone, VVΔE3L yields were 20-fold less than those of wild-type VV. Ponasterone induction of RNase L further reduced yields of VVΔE3L by an additional factor of 6, while induction of RNase LΔEN had no effect (Fig. 1G). Similar results were obtained by using Copenhagen and WR strains of VV and VVΔE3L (Fig. 1G and data not shown). These findings show that RNase L has no impact on the growth of wild-type VV but is able to partially suppress replication of VVΔE3L, an effect that is dependent on the endonuclease domain.
Combined effects of RNase L and PKR against VV in vitro and in vivo.
To determine the combined effect of the absence of RNase L and PKR on VV and VVΔE3L, experiments were performed with cells and mice lacking both RNase L and PKR (51). In addition, Mx1, responsible for IFN-induced suppression of some negative-RNA-stranded viruses, was also absent from these cells and mice (reviewed in reference 38). Absence of PKR and RNase L had no effect on yields of wild-type VV from the cultured cells (Fig. 2A). However, VVΔE3L again failed to replicate in wild-type cells (the titer shown in Fig. 2A, about 2 × 105 PFU/ml, is the input level of virus). In contrast, in the RNase L−/− PKR−/− cells, VVΔE3L yields were enhanced by a factor of about 20 compared to that obtained in the wild-type cells. The RNase L−/− PKR−/− cells produced VVΔE3L titers about fivefold lower than those obtained with wild-type VV in the same cells. Curiously, the combined effect on VVΔE3L growth of knockouts of both RNase L and PKR was not greater than that obtained by deleting only RNase L or only PKR (compare Fig. 1G and 2A; also data not shown). Findings indicating only a partial restoration of VVΔE3L growth in the absence of both RNase L and PKR suggest an alternative or additional host evasion function of E3L.
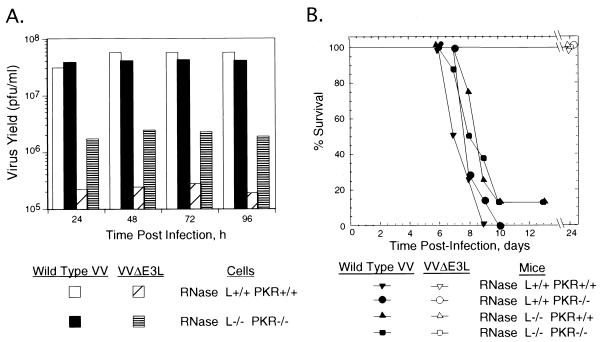
FIG. 2.
Impact of RNase L and PKR deficiencies on viral growth and pathogenesis. (A) One-step growth of VV and VVΔE3L (Copenhagen strain) in wild-type and RNase L−/− PKR−/− cells. Cells were infected at an MOI of 6. At different times postinfection, viruses were harvested and virus yields were determined by plaque titration on BHK21 cells. (B) VV but not VVΔE3L is lethal for mice regardless of the presence or absence of RNase L and PKR. Seven-week-old wild-type, RNase L−/−, PKR−/−, and RNase L−/− PKR−/− mice were injected i.n. with 106 PFU of wild-type VV or 5 × 106 PFU of VVΔE3L (WR strain). Survival was monitored daily, and the mouse survival was plotted.
To extend these findings to an animal model, wild-type VV and VVΔE3L were used to infect, by the i.n. route, wild-type mice and mice lacking either RNase L or PKR or lacking both (Fig. 2B). Wild-type VV (106 PFU) caused similar death curves for all of the different types of mice, regardless of the presence or absence of RNase L and PKR (nearly all mice died within 10 days). Generally, mice displayed signs of sickness within 4 days as characterized by ruffled fur and lack of activity, and death occurred within 7 to 10 days. The 50% lethal dose (LD50) for wild-type VV in wild-type mice was about 103 PFU (data not shown). Surprisingly, we found that VVΔE3L virus was not lethal to wild-type, PKR−/−, RNase L−/−, or RNase L−/− PKR−/− mice even at 5 × 106 PFU, a 5,000-fold-greater dose than the LD50 for wild-type VV (Fig. 2B). More than 50% of the mice infected with VVΔE3L appeared sick, regardless of genotype, at 4 days postinfection, but they all recovered after 7 to 8 days postinfection. In addition, viral growth was monitored by plaque assays of lung extracts. Two mice of each genotype were infected with either wild-type VV or VVΔE3L. Lungs were removed at 4 days after virus infection in all four kinds of mice. The organs were homogenized, and virus titers were determined by plaque assay. Viruses were detected in the lungs of mice infected with wild-type VV (at titers as high as 107 PFU) but not in mice infected with VVΔE3L, with the exception of one PKR−/− mouse that had 105 PFU in the lungs (data not shown). To rule out the possibility of second site mutations in VVΔE3L being responsible for the debilitation of this virus in the animal model, we inserted the E3L gene back into VVΔE3L. The revertant virus caused 100% mortality in wild-type mice at 104 PFU and fully restored pathogenicity (data not shown). Thus, we were able to demonstrate that the E3L gene is required for virulence of VV and that deletion of this gene produces a nonpathogenic virus.
E3L blocks IRF3 activation and induction of the IFN-β gene during VV infections.
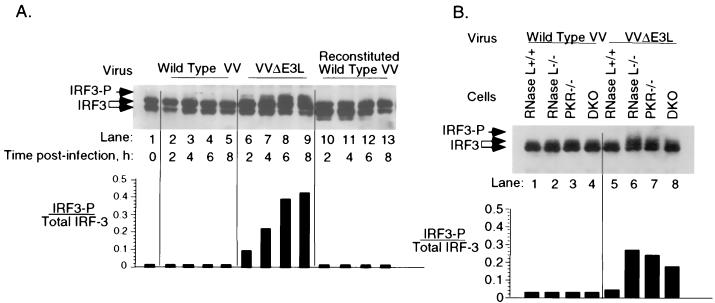
Our results indicate that there is an essential virulence role for E3L in addition to inhibition of PKR and RNase L. One possibility was that IRF3, a proximal transcription factor for a subset of type I IFN genes, was blocked by E3L. To determine if E3L was able to block IRF3 phosphorylation during VV infections, human HT1080 cells were infected with wild-type VV or VVΔE3L (Fig. 3A). It was previously established that the lower-mobility fraction of IRF3 represents the phosphorylated form of the protein (34). Immunoblot assays show that IRF3, present as a doublet of two isoforms, was not phosphorylated in response to infections with wild-type VV (Fig. 3A, lanes 2 to 5). In contrast, a large fraction (as high as 45%) of IRF3 was phosphorylated in cells infected with VVΔE3L (Fig. 3A, lanes 6 to 9). The increased phosphorylation of IRF3 was detected at 4 h postinfection and peaked by 6 to 8 h. IRF3 was not phosphorylated after infection with the reconstituted wild-type VV generated by cloning E3L into VVΔE3L (Fig. 3A, lanes 10 to 13). These results clearly indicate that E3L inhibits IRF3 phosphorylation during the course of VV infections.
FIG. 3.
E3L blocks phosphorylation of IRF3 during VV infections. HT1080 cells (A) or wild-type or mutant MEFs (B) were infected with VV, VVΔE3L, or reconstituted wild-type VV (WR strain; MOI of 15) as indicated. IRF3 phosphorylation was monitored in Western blots probed with polyclonal antibody against IRF3 (upper panels). The ratios of phosphorylated to total IRF3 was plotted as a function of time postinfection (lower panels).
To determine if RNase L or PKR could affect IRF3 phosphorylation, the various MEF cell lines were infected (Fig. 3B). Wild-type VV did not cause IRF3 phosphorylation in any of the cell types (Fig. 3B, lanes 1 to 4). VVΔE3L also did not cause IRF3 phosphorylation in the wild-type cells, probably because this virus does not replicate in these cells (Fig. 3B, lane 5). However, VVΔE3L induced IRF3 phosphorylation in the single- and double-gene knockout (DKO) MEFs (Fig. 3B, lanes 6 to 8). Therefore, the ability of VVΔE3L to induce IRF3 phosphorylation correlated with the ability to replicate in these cells. These findings show that neither PKR nor RNase L was required for IRF3 phosphorylation.
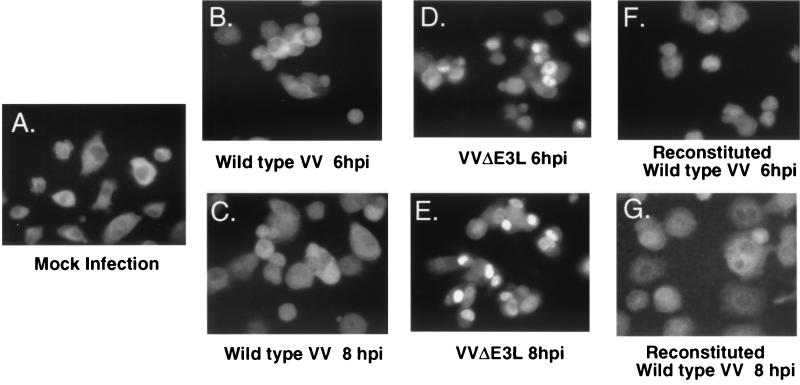
To verify that IRF3 was activated, we monitored its migration from the cytoplasm to the nucleus by indirect immunofluorescence with antibody against IRF3 (Fig. 4). In uninfected cells, IRF3 was observed principally in the cytoplasm (Fig. 4A). The cytoplasmic distribution of IRF3 was unchanged by infections with wild-type VV or reconstituted VV (Fig. 4B, C, F, and G). In sharp contrast, there was an obvious migration of IRF3 to nuclei in cells infected with VVΔE3L by 6 to 8 h postinfection (Fig. 4D and E). The migration of IRF3 to nuclei was first detected at 4 h postinfection with VVΔE3L (data not shown).
FIG. 4.
Cytoplasmic to nuclear translocalization of IRF-3 in HT1080 cells after virus infection occurs only in the absence of E3L. The subcellular localization of IRF3 is shown in mock-infected cells (A) and in cells infected with wild-type VV (B and E), VVΔE3L (C and E), or reconstituted wild-type VV (WR strain) (F and G). Infections were for 6 h (B, D, and F) or 8 h (C, E, and G). Fixed cells were stained with polyclonal anti-IRF3 antibody and then secondary Alexa 488 goat anti-rabbit antibody. Stained cells were monitored with a Leica fluorescence microscope (magnification, ×400).
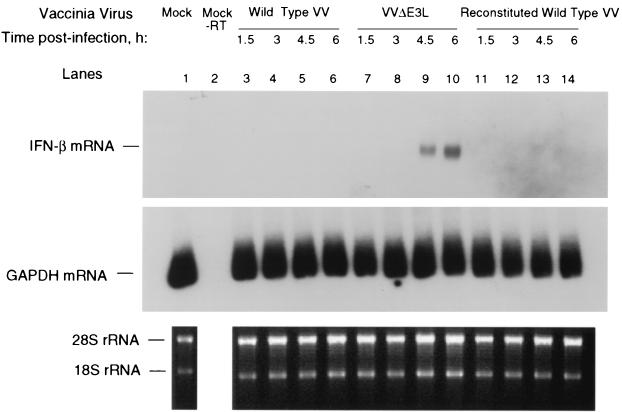
To determine the effect of IRF3 activation and nuclear migration on IFN synthesis, we measured induction of the IFN-β gene by RT-PCR of its mRNA. There was no detectable induction of IFN-β mRNA in cells infected with wild-type VV or the reconstituted wild-type VV (Fig. 5, lanes 3 to 6 and 11 to 14). However, IFN-β mRNA was observed in the VVΔE3L-infected cells at 4.5 and 6 h postinfection (Fig. 5, lanes 9 and 10). GAPDH mRNA was detected by RT-PCR in all of the RNA preparations. There was a close correlation between times when IFN-β mRNA was detectable in the VVΔE3L-infected cells and migration of IRF3 to the nuclei (Fig. 4 and 5 and data not shown). These findings show that E3L is a potent suppressor of the IFN-β gene in the context of an active VV infection.
FIG. 5.
Induction of IFN-β mRNA after VVΔE3L infection. HT-1080 cells were infected with either wild-type VV, VVΔE3L, or reconstituted wild-type VV (WR strain; MOI of 15) for the lengths of time indicated. The expression of IFN-β (upper panel) or GAPDH mRNA (middle panel) was detected by RT-PCR analysis. The products were analyzed on polyacrylamide gels and by autoradiography. Ethidium bromide staining was performed on the total RNA (lower panel).
DISCUSSION
E3L enables VV to evade the antiviral action of the 2-5A system.
The ability of RNase L to curtail viral growth is dependent on activation of 2-5A synthetases by virus-generated dsRNA (36, 45). VV infections are thought to generate dsRNA as the result of transcription from both strands of the viral DNA genome with subsequent annealing of cRNA strands (10, 23, 46). E3L probably prevents RNase L activation indirectly, by sequestering dsRNA from 2-5A synthetases. In this regard, previous studies have shown that rRNA is degraded in IFN-pretreated VVΔK3L- or VVΔE3L-infected L929 cells, suggesting involvement of the K3L and E3L genes in the inhibition of the 2-5A pathway (3, 4). In contrast to E3L, for which the mechanism of action likely involves dsRNA sequestration, it is unclear why K3L, which encodes a homolog of the PKR substrate eIF2α, should inhibit the 2-5A system. However, supporting evidence that E3L blocks the 2-5A system was provided by a study in which E3L prevented apoptosis by 2-5A synthetase and RNase L expressed from VV vectors (28). Our results show for the first time that constitutive levels of RNase L suppress VVΔE3L but not wild-type VV (Fig. 1). Accordingly, VVΔE3L was able to grow in RNase L−/− cells but not in RNase L+/+ cells (Fig. 1A and B). In addition, VVΔE3L yields declined about sixfold when full-length RNase L was expressed in RNase L−/− cells (Fig. 1G). Yields of VVΔE3L did not change after RNase LΔEN was expressed in the RNase L−/− cells, showing that the nuclease domain was required (Fig. 1G).
Combined deficiencies of RNase L, PKR, and Mx1 in cells and mice did not compensate for the absence of E3L.
Previous studies show that E3L suppresses PKR activation during the course of VV infections (7). Surprisingly, combined deficiencies in PKR and RNase L did not result in higher yields of VVΔE3L than those obtained by single deficiencies in either PKR or RNase L (Fig. 2A and data not shown). Yields of VVΔE3L were reduced about 20-fold by these antiviral enzymes. However, our studies with mice show that both PKR and RNase L are dispensable for the pathogenic effect of E3L, suggesting that viral dsRNA produced by VVΔE3L was signaling to additional host immunity factors (Fig. 2B). Indeed, i.n. administration of 5 × 106 PFU of VVΔE3L, 5,000 times the LD50 for wild-type VV, failed to result in death. This result confirmed a prior study showing the importance of the E3L gene in pathogenesis in a mouse model, while showing for the first time that E3L does more than inhibit the PKR and 2-5A systems (5).
E3L blocks the IFN pathway at an early step in the development of the antiviral state.
Because the E3L gene encodes proteins that bind to and sequester dsRNA, we hypothesized that the candidate targets for inhibition by E3L require dsRNA. IRF3 is a component of dsRNA-activated factor 1 (DRAF1) involved in dsRNA-mediated signal transduction (20, 33, 40, 42, 47). After viral infections, IRF3 is phosphorylated, migrates to the nucleus, and complexes with transcriptional coactivator CBP/p300. We observed IRF3 phosphorylation only in cells infected with VVΔE3L and not with wild-type VV or with a reconstituted wild-type VV (Fig. 3). Migration of IRF3 from the cytoplasm to the nucleus also occurred only when cells were infected with virus lacking E3L (Fig. 4). Further, IFN-β gene induction in response to viral infection was observed only in the absence of E3L (Fig. 5). Therefore, by blocking activation of IRF3, E3L effectively blocks the first wave of type I IFN synthesis that would otherwise serve as an IFN diversification and amplification event through IRF7 (22, 32). Similarly, E3L was reported to inhibit Newcastle disease virus-induced IRF3 phosphorylation following cotransfection of E3L and IRF3 plasmids (37). That report showed that virus induction of IRF3 was independent of PKR. We show that neither RNase L nor PKR is required for IRF3 activation (Fig. 3B). There are similarities between the functions of VV E3L and the NS1 dsRNA-binding protein of influenza virus. NS1 blocks IRF3 phosphorylation when expressed in trans, and IFN-β mRNA induction was prevented in wild-type influenza virus-infected cells but not in cells infected with a virus lacking the NS1 gene (39). In addition, NS1 is an inhibitor of PKR, suggesting that dsRNA sequestration is a strategy utilized by both RNA and DNA viruses to evade the IFN induction and action (21).
The inability of VVΔE3L to cause significant disease is presumably due, at least in part, to induction of type I IFN subtypes, including IFN-β, that lead to antiviral proteins, such as PKR and 2-5A synthetases. However, because VVΔE3L was not lethal to mice lacking PKR, RNase L, and Mx1, there must be an additional IFN-induced antiviral pathway(s) effective against this virus. These findings reinforce the idea of multiple pathways in the antiviral activities of IFNs. For instance, it was shown previously that the triply deficient mice retain the ability to mount antiviral defenses to encephalomyocarditis virus in response to IFN-α (51).
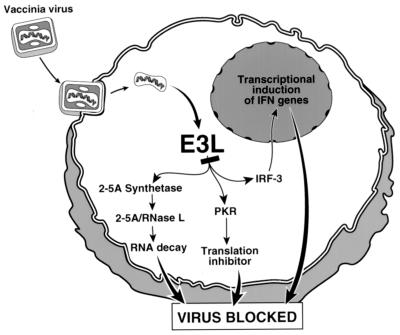
Our results demonstrate that E3L is a versatile viral immunity factor that blocks at least three host defense factors collectively involved in IFN induction and action. We propose that the primary role of E3L is to prevent activation of IRF3, RNase L, and PKR simultaneously (Fig. 6). Inhibition likely occurs through a combination of dsRNA sequestration and direct inhibition as a result of protein-protein interactions. In this regard, alternative dsRNA binding proteins, Staufen and DRBP76, were unable to inhibit IRF3/IRF7 phosphorylation while direct interactions and inhibition of PKR by E3L was reported (29, 35, 37). Our findings provide the first in vivo evidence that E3L is a versatile pathogenesis factor that allows VV to avoid the IFN system at multiple levels.
FIG. 6.
E3L of VV evades the IFN system by blocking IFN induction through IRF3 and IFN action through the 2-5A/RNase L pathway and protein kinase PKR.
Acknowledgments
We thank Michael David (University of California at San Diego) for the gift of IRF3 antibody; Aimin Zhou, Beihua Dong, Shoudong Li, Cristy Peters, Ganes Sen (Cleveland Clinic), and Curt Horvath (Mt. Sinai Medical School, New York, N.Y.) for valuable discussions; and Aimin Zhou for purifying the labeled 2-5A.
This investigation was supported by United States Public Health Service grants CA44059 from the Department of Health and Human Services, National Cancer Institute, to R.H.S and AI34039 from National Institute of Allergy and Infectious Disease to B.R.G.W.
REFERENCES
- 1.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcami, A., J. A. Symons, and G. L. Smith. 2000. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J. Virol. 74:11230-11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beattie, E., K. L. Denzler, J. Tartaglia, M. E. Perkus, E. Paoletti, and B. L. Jacobs. 1995. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol. 69:499-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beattie, E., E. Paoletti, and J. Tartaglia. 1995. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L- and E3L-mutant viruses. Virology 210:254-263. [DOI] [PubMed] [Google Scholar]
- 5.Brandt, T. A., and B. L. Jacobs. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 75:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castelli, J. C., B. A. Hassel, A. Maran, J. Paranjape, J. A. Hewitt, X. L. Li, Y. T. Hsu, R. H. Silverman, and R. J. Youle. 1998. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 5:313-320. [DOI] [PubMed] [Google Scholar]
- 7.Chang, H. W., J. C. Watson, and B. L. Jacobs. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 89:4825-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, H. W., L. H. Uribe, and B. L. Jacobs. 1995. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J. Virol. 69:6605-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens, M. J., and B. R. Williams. 1978. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell 13:565-572. [DOI] [PubMed] [Google Scholar]
- 10.Colby, C., and P. H. Duesberg. 1969. Double-stranded RNA in vaccinia virus infected cells. Nature 222:940-944. [DOI] [PubMed] [Google Scholar]
- 11.Condit, R. C., A. Motyczka, and G. Spizz. 1983. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology 128:429-443. [DOI] [PubMed] [Google Scholar]
- 12.Der, S. D., Y. L. Yang, C. Weissmann, and B. R. Williams. 1997. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc. Natl. Acad. Sci. USA 94:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz-Guerra, M., C. Rivas, and M. Esteban. 1997. Activation of the IFN-inducible enzyme RNase L causes apoptosis of animal cells. Virology 236:354-363. [DOI] [PubMed] [Google Scholar]
- 15.Dong, B., and R. H. Silverman. 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J. Biol. Chem. 270:4133-4137. [DOI] [PubMed] [Google Scholar]
- 16.Dong, B., and R. H. Silverman. 1997. A bipartite model of 2-5A-dependent RNase L. J. Biol. Chem. 272:22236-22242. [DOI] [PubMed] [Google Scholar]
- 17.Kerr, I. M., and R. E. Brown. 1978. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl. Acad. Sci. USA 75:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kibler, K. V., T. Shors, K. B. Perkins, C. C. Zeman, M. P. Banaszak, J. Biesterfeldt, J. O. Langland, and B. L. Jacobs. 1997. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. J. Virol. 71:1992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, S. B., and M. Esteban. 1994. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology 199:491-496. [DOI] [PubMed] [Google Scholar]
- 20.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 22.Mamane, Y., C. Heylbroeck, P. Genin, M. Algarte, M. J. Servant, C. LePage, C. DeLuca, H. Kwon, R. Lin, and J. Hiscott. 1999. Interferon regulatory factors: the next generation. Gene 237:1-14. [DOI] [PubMed] [Google Scholar]
- 23.Maran, A., and M. B. Mathews. 1988. Characterization of the double-stranded RNA implicated in the inhibition of protein synthesis in cells infected with a mutant adenovirus defective for VA RNA. Virology 164:106-113. [DOI] [PubMed] [Google Scholar]
- 24.McFadden, G., and P. M. Murphy. 2000. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr. Opin. Microbiol. 3:371-378. [DOI] [PubMed] [Google Scholar]
- 25.Meurs, E., K. Chong, J. Galabru, N. S. Thomas, I. M. Kerr, B. R. Williams, and A. G. Hovanessian. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62:379-390. [DOI] [PubMed] [Google Scholar]
- 26.Nolan-Sorden, N. L., K. Lesiak, B. Bayard, P. F. Torrence, and R. H. Silverman. 1990. Photochemical crosslinking in oligonucleotide-protein complexes between a bromine-substituted 2-5A analog and 2-5A-dependent RNase by ultraviolet lamp or laser. Anal. Biochem. 184:298-304. [DOI] [PubMed] [Google Scholar]
- 27.Paez, E., and M. Esteban. 1984. Resistance of vaccinia virus to interferon is related to an interference phenomenon between the virus and the interferon system. Virology 134:12-28. [DOI] [PubMed] [Google Scholar]
- 28.Rivas, C., J. Gil, Z. Melkova, M. Esteban, and M. Diaz-Guerra. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon-induced 2-5A synthetase enzyme. Virology 243:406-414. [DOI] [PubMed] [Google Scholar]
- 29.Romano, P. R., F. Zhang, S. L. Tan, M. T. Garcia-Barrio, M. G. Katze, T. E. Dever, and A. G. Hinnebusch. 1998. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol. 18:7304-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rusch, L., B. Dong, and R. H. Silverman. 2001. Monitoring activation of ribonuclease L by 2′,5′-oligoadenylates using purified recombinant enzyme and intact malignant glioma cells. Methods Enzymol. 342:10-20. [DOI] [PubMed] [Google Scholar]
- 31.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106-110. [DOI] [PubMed] [Google Scholar]
- 32.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF3 and IRF7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 33.Sato, M., N. Tanaka, N. Hata, E. Oda, and T. Taniguchi. 1998. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 425:112-116. [DOI] [PubMed] [Google Scholar]
- 34.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2000. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 35.Sharp, T. V., F. Moonan, A. Romashko, B. Joshi, G. N. Barber, and R. Jagus. 1998. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology 250:302-315. [DOI] [PubMed] [Google Scholar]
- 36.Silverman, R. H., J. J. Skehel, T. C. James, D. H. Wreschner, and I. M. Kerr. 1983. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J. Virol. 46:1051-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Ikappa B kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 38.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 39.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 41.Watson, J. C., H. W. Chang, and B. L. Jacobs. 1991. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology 185:206-216. [DOI] [PubMed] [Google Scholar]
- 42.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitaker-Dowling, P., and J. S. Youngner. 1983. Vaccinia rescue of VSV from interferon-induced resistance: reversal of translation block and inhibition of protein kinase activity. Virology 131:128-136. [DOI] [PubMed] [Google Scholar]
- 44.Whitaker-Dowling, P., and J. S. Youngner. 1986. Vaccinia-mediated rescue of encephalomyocarditis virus from the inhibitory effects of interferon. Virology 152:50-57. [DOI] [PubMed] [Google Scholar]
- 45.Williams, B. R., R. R. Golgher, R. E. Brown, C. S. Gilbert, and I. M. Kerr. 1979. Natural occurrence of 2-5A in interferon-treated EMC virus-infected L cells. Nature 282:582-586. [DOI] [PubMed] [Google Scholar]
- 46.Xiang, Y., D. A. Simpson, J. Spiegel, A. Zhou, R. H. Silverman, and R. C. Condit. 1998. The vaccinia virus A18R DNA helicase is a postreplicative negative transcription elongation factor. J. Virol. 72:7012-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youngner, J. S., H. R. Thacore, and M. E. Kelly. 1972. Sensitivity of ribonucleic acid and deoxyribonucleic acid viruses to different species of interferon in cell cultures. J. Virol. 10:171-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, A., B. A. Hassel, and R. H. Silverman. 1993. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72:753-765. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, A., J. Paranjape, T. L. Brown, H. Nie, S. Naik, B. Dong, A. Chang, B. Trapp, R. Fairchild, C. Colmenares, and R. H. Silverman. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, A., J. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]