Abstract
Little is known about the function of flavivirus nonstructural protein NS2A. Two forms of NS2A are found in yellow fever virus-infected cells. Full-length NS2A (224 amino acids) is the product of cleavage at the NS1/2A and NS2A/2B sites. NS2Aα, a C-terminally truncated form of 190 amino acids, results from partial cleavage by the viral NS2B-3 serine protease at the sequence QK↓T within NS2A. Exchange of serine for lysine at this site (QKT→QST) blocks the production of both NS2Aα and infectious virus. The present study reveals that this defect is not at the level of RNA replication. Despite normal structural region processing, infectious particles containing genome RNA and capsid protein were not released from cells transfected with the mutant RNA. Nevertheless, production of subviral prM/M- and E-containing particles was unimpaired. The NS2A defect could be complemented in trans by providing NS1-2A or NS1-2Aα. However, trans complementation was not observed when the C-terminal lysine of NS1-2Aα was replaced with serine. In addition to true reversions, NS2Aα cleavage site mutations could be suppressed by two classes of second-site changes. The first class consisted of insertions at the NS2Aα cleavage site that restored its basic character and cleavability. A second class of suppressors occurred in the NS3 helicase domain, in which NS3 aspartate 343 was replaced with an uncharged residue (either valine, alanine, or glycine). These mutations in NS3 restored infectious-virus production in the absence of cleavage at the mutant NS2Aα site. Taken together, our results reveal an unexpected role for NS2A and NS3 in the assembly and/or release of infectious flavivirus particles.
The Yellow fever virus (YF) belongs to the genus Flavivirus within the family Flaviviridae. Members of the Flavivirus genus are typically transmitted to vertebrates by mosquitoes or ticks and frequently cause significant human morbidity and mortality (reviewed in reference 25). Human pathogens include dengue virus, Japanese encephalitis virus, tick-borne encephalitis virus, West Nile virus, and YF. The estimated 100 million cases of dengue virus infection per year worldwide (5) and the emergence and spread of West Nile virus in the Eastern United States underscore the need for continued efforts to develop effective and inexpensive flavivirus vaccines. For YF, a live attenuated vaccine strain (17D) has been used effectively for almost 65 years. However, YF remains an enduring global public health problem due to the endemic persistence of mosquito-borne disease in sub-Saharan Africa and South America and recent reports of six fatalities temporally associated with live-virus vaccination (22, 33).
The YF genome is a positive-sense RNA approximately 11 kb in length that is capped at the 5′ end but lacks a 3′ poly(A) tract. The RNA contains a single large open reading frame that is cleaved co- and posttranslationally by host cell and viral proteases (reviewed in reference 16). The polyprotein is arranged with the structural proteins at the amino terminus (C-prM-E), followed by the nonstructural (NS) proteins (NS1 through NS5). The arrangement of the proteins is NH2-C-(pr)M-E-NS1-NS2A-NS2B-NS3-NS4A-2K-NS4B-NS5-COOH. Most of the cleavages releasing the structural proteins are mediated by host cell signal peptidase. Exceptions include prM cleavage into pr and M by the host cell enzyme furin shortly before virus release and the cleavage generating the C terminus of the virion C protein by the virus-encoded serine protease (NS2B-3 protease). This serine protease, consisting of NS2B and the N-terminal part of NS3, produces the N termini of NS2B, NS3, NS4A, 2K, and NS5. The enzyme responsible for cleavage at the NS1/2A site is unknown.
Although all flavivirus proteins stem from a single polyprotein, the structural and NS proteins have been viewed as functionally distinct modules responsible for virion formation and RNA replication, respectively. For example, coexpression of prM and E is sufficient for secretion of subviral particles that mimic the subunit structure and fusogenic capabilities of the mature virion envelope (2, 29). Subgenomic replicons lacking the structural proteins replicate efficiently and can be packaged by trans expression of the structural proteins (11). Although initially thought to be involved in virion morphogenesis or release, secreted glycoprotein NS1 plays an essential role in RNA replication (18). These observations have reinforced a modular view of the flavivirus polyprotein with the dividing line between structural proteins and replicase drawn at the E/NS1 junction. The only known exception is the NS2B-3 serine protease-mediated cleavage at the C terminus of mature C, a cleavage that is a necessary prerequisite for signalase generation of the prM N terminus and virus production (4).
Specific functions have been attributed to many flavivirus proteins (reviewed in reference 16), but little is known about the function of NS2A. NS2A is a small hydrophobic protein of about 22 kDa (8). Consistent with a role in RNA replication, studies with Kunjin virus (KUN) have shown that NS2A colocalizes with double-stranded RNA in discrete cytoplasmic foci and interacts with the 3′ untranslated region of KUN RNA, as well as NS3 and NS5 (20). NS2A has also been implicated in the Japanese encephalitis virus-induced cytopathic effect (CPE) (10).
In YF-infected cells, 22- and 20-kDa forms of NS2A have been identified and both of these forms possess the same N-terminal sequence (8). The 20-kDa form (called NS2Aα) is believed to result from an additional internal cleavage by the NS2B-3 serine protease. Cleavage by the YF serine protease occurs at a consensus sequence consisting of two basic amino acids followed by an amino acid with a short side chain (RR↓S/G) (6). However, in the case of the NS4A/2K junction, the cleavage site is QR↓S (14). Based on the known cleavage sites, as well as substitutions at these sites that are tolerated (9, 15, 26), NS2A residues 189 to 191 (QK↓T) were identified as a possible target for the viral serine protease. Consistent with this hypothesis, replacement of Lys-190 with Ser resulted in loss of the 20-kDa protein (26). Inhibition of cleavage at the NS2A/2B site did not abrogate production of NS2Aα, indicating that processing at the NS2A/2B site was not a prerequisite for cleavage at the NS2Aα site (26). It was further shown that the NS2A Lys-190-Ser mutation blocked production of infectious virus (26).
Although the roles of NS2A and this additional cleavage in YF replication were unknown, a block at the level of YF RNA replication seemed most likely. In this report, we show that NS2Aα cleavage site mutants have unimpaired RNA replication but are unable to produce infectious virus. This defect in virus production can be complemented by NS2A or NS2Aα supplied in trans and compensated for by mutations at the NS2Aα cleavage site or by second-site changes in the helicase domain of NS3. These results reveal a more complex interplay between the NS proteins and virus production than previously suspected.
MATERIALS AND METHODS
Cell cultures and virus stocks.
BHK-21/J cells (kindly provided by Paul Olivo, Washington University, St. Louis, Mo.) have been described previously (18). Cells were grown in minimal essential medium (MEM) containing 7.5% fetal calf serum (FCS) and nonessential amino acids. Virus was harvested at the indicated times, clarified by centrifugation at 3,000 × g for 10 min, aliquoted, and frozen at −80°C.
Plasmid constructions.
Restriction enzyme digestions, subcloning, and other standard recombinant DNA procedures were done essentially as previously described (27). All NS2Aα cleavage site mutants were constructed and analyzed by using full-length YF 17D clone pACNR/FLYF (17; P. J. Bredenbeek, E. A. Kooi, B. D. Lindenbach, N. Huijkman, C. M. Rice, and W. J. M. Spaan, unpublished data). To facilitate mutagenesis of the NS2A-3 region, the 2,190-bp KpnI-BamHI fragment of pACNR/FLYF was subcloned into KpnI-BamHI-digested BluescriptII SK− (Stratagene). This subclone was used for QuikChange mutagenesis as described below. To generate the NS2Aα cleavage site mutants, the AvrII-SapI fragment from the mutagenized pBluescriptII plasmid was ligated with the appropriate MluI-AvrII and MluI-SapI fragments of pACNR/FLYF.
Plasmid pSINrep19/NS1-2A was kindly provided by Brett D. Lindenbach (18). To obtain pSINrep19/NS1-2A(K-S), the 388-bp AvrII-SphI fragment of pSINrep19/NS1-2A was subcloned and QuikChange mutagenesis was performed with primers PNACL1066 (CACCTCCATGCAGTCGACTATACCTCTG/+) and PNACL1067 (CAGAGGTATAGTCGACTGCATGGAGGTG/−). The AvrII-SphI fragment containing the mutation was ligated with the AflII-AvrII (4,522-bp) and AflII-SphI fragments of pSINrep19/NS1-2A. To generate pSINrep19/NS1-2Aα, a subregion of pSINrep19/NS1-2A was amplified by PCR with primers BRL685 (ATCGGCTTTGGGCTCAGG/+) and PNACL1046 (TACGACGCGTCACTTCTGCATGGAGGTGTCCTTG/−) and digested with AvaI and MluI. The resulting fragment was ligated with the AflII-AvaI (4,500-bp) and AflII-MluI fragments of pSINrep19/NS1-2A. pSINrep19/NS1-2Aα(K-S) was created by using the same strategy with primers BRL685 and PNACL1080 (TACGACGCGTCACGACTGCATGGAGGTGTCCTTG/−). To obtain pSINrep19/NS1ΔSK-2Aα, pSINrep19/NS1-2Aα was digested with SacI, treated with the Klenow fragment of DNA polymerase I, and then digested with BamHI. The 423-bp fragment was ligated with an 838-bp KpnI (blunt-end)-SphI fragment from pSINrep19/NS1-2A and BamHI-SphI-digested pSINrep19/NS1-2Aα.
Site-directed mutagenesis.
Mutagenesis was performed in accordance with the QuikChange site-directed mutagenesis method (Stratagene). The nucleotide exchanges introduced for the single NS2Aα cleavage site mutants are shown in Table 1. Mutagenized fragments were sequenced to confirm the presence of the desired mutation and the absence of other mutations.
TABLE 1.
Recovery of infectious virus after electroporation of NS2Aα cleavage site mutants
| Mutationa | Specific infectivityb | Titerc at:
|
Compensating mutationd | |
|---|---|---|---|---|
| 24 h | 48 h | |||
| None (WT; QKT) | 1 × 106 | NDf | 3.5 × 107 | None |
| 5.4 × 105 | 1.8 × 107 | 8.2 × 107 | None | |
| QST (AAG→TCG) | <50 | ND | <50 | No virus recovered |
| <50 | <50 | <50 | QKLEEGSTe | |
| <50 | <50 | <50 | No virus recovered | |
| <50 | <50 | 3.3 × 103 | QRRSTGST | |
| QKV (ACT→GTT) | 4.5 × 105 | ND | 1 × 106 | None |
| 2.5 × 105 | 1.1 × 105 | 1.7 × 106 | None | |
| QQT (AAG→CAG) | <50 | ND | 7.5 × 105 | QKT |
| <50 | <50 | 7 × 104 | QKT | |
| QET (AAG→GAG) | <50 | ND | 1.6 × 105 | QKT |
| <50 | <50 | 2.3 × 105 | QKT | |
| QRT (AAG→AGG) | 8.3 × 105 | ND | 1.6 × 107 | None |
| 4 × 105 | 1.3 × 107 | 4.5 × 107 | None | |
| QIT (AAG→ATC) | <50 | ND | 5.5 × 103 | QKRETIT |
| <50 | <50 | <50 | No virus recovered | |
| <50 | <50 | <50 | No virus recovered | |
| <50 | <50 | 6.3 × 103 | QRT | |
| SKT (CAG→TCG) | <50 | ND | 9.2 × 104 | NS3:D343V, A or G |
| <50 | 4.8 × 102 | 1.6 × 105 | NS3:D343V | |
| RRS (CAG-AAG-ACT→AGG-AGG-AGT) | 1.5 × 106 | ND | 2.1 × 107 | None |
| 5.4 × 105 | 2.8 × 107 | 2.4 × 107 | None | |
The mutated amino acid(s) at the NS2Aα cleavage site is in boldface. The codon (as DNA) of the original amino acid at that position and its change with the mutagenized nucleotide are underlined and in parentheses.
Determined by an infectious-center assay with BHK cells.
Determined by a plaque assay with BHK cells.
RNA isolated from cells reinfected with virus, obtained 3 days after electroporation, was used as the starting material for RT-PCR analysis of the viral genomes. Mutated residues are in boldface.
RNA isolated from cells reinfected with virus, obtained after one passage, was used as the starting material for RT-PCR analysis of the viral genomes.
ND, not determined.
In vitro transcription.
Plasmid DNAs were linearized with XhoI (for both pACNR/FLYF and pSINrep19 constructs) and transcribed with SP6 polymerase (Gibco-BRL) in the presence of cap analog m7G(5′)ppp(5′)G (New England Biolabs). To determine the RNA yield, incorporation of [5,6-3H]UTP (NEN) was determined by adsorption to DE-81 (Whatman) filter paper. Transcript integrity was confirmed by electrophoresis in agarose gels containing ethidium bromide at 100 ng/ml.
Transfections and selection of cell populations.
BHK21/J cells split the day before transfection and grown to about 80% confluency were trypsinized and washed twice in ice-cold phosphate-buffered saline (PBS; Bio-Whittaker). Cells were resuspended in PBS at a concentration of 2 × 107/ml. A 0.4-ml volume of this cell suspension was mixed with 3 μg of in vitro-transcribed RNA and placed in an electroporation cuvette (BTX Inc.) with a 2-mm gap. Electroporation was performed with a T820 electroporator (BTX Inc.) with five pulses of 960 V and a pulse length of 99 μs. After 10 min of recovery at room temperature, cells were mixed with 20 ml of complete growth medium and plated. To select for replicon-containing cells, the medium was changed at 12 h posttransfection to complete medium containing puromycin (Sigma Chemical Co.) at 5 μg/ml. After selection (2 to 3 days), cell populations were passaged and maintained in medium containing puromycin (5 μg/ml).
Purification of virions and subviral particles.
BHK-21/J cells were labeled from 14 to 24 h postelectroporation in methionine- and cysteine-deficient MEM containing 2% FCS, 1.25% MEM, and EXPRE35S35S protein labeling mix (NEN) at 50 μCi/ml. Clarification of the cell supernatants was performed by centrifugation at 16,000 × g for 30 min at 4°C. The particles were pelleted by centrifugation at 104,000 × g (28,000 rpm; Beckman SW28 rotor) for 1.5 h at 4°C, resuspended in TNE buffer (50 mM NaCl, 100 mM Tris-HCl [pH 8.5], 1 mM EDTA), and layered over a 5 to 25% glycerol gradient in TNE with bovine serum albumin (BSA) at 200 μg/ml. Centrifugation was done at 160,000 × g (36,000 rpm; Beckman SW41 rotor) for 1.45 h at 4°C. Portions of pooled fractions containing virions or subviral particles were layered over potassium tartrate gradients (heavy solution, 35% [wt/vol] potassium tartrate-25% [vol/vol] glycerol-100 mM Tris-HCl [pH 8.5]-1 mM EDTA-200 μg of BSA per ml; light solution, 25% glycerol-50 mM NaCl-100 mM Tris-HCl [pH 8.5]-1 mM EDTA-200 μg of BSA per ml). Equilibrium banding was performed by centrifugation overnight at 111,000 × g (30,000 rpm; Beckman SW41 rotor) at 4°C. Fractions were collected from the bottom to the top. A 200-μl volume of each fraction was mixed with 1 ml of Microscint 20 scintillation fluid (Packard), and radioactivity was measured in a TopCount NXT microplate scintillation counter (Packard). In parallel, fractions were analyzed for infectious virus by a plaque assay as described above.
Infectious-center assays and plaque assays.
For infectious-center assays, 10-fold serial dilutions of transfected cells were mixed with 5 × 105 untransfected cells and seeded in 35-mm-diameter dishes. Following attachment for 4 to 6 h, the medium was replaced with an agarose overlay (MEM with 0.6% agarose and 2% FCS). After a 3-day incubation at 37°C, cells were fixed with 7% formaldehyde and stained with 1% crystal violet in 5% ethanol. For plaque assays, 10-fold serial dilutions of culture supernatant were made in PBS containing 1% FCS. Cells at a density of 70 to 80%, seeded the day before, were inoculated with the serial dilutions. After infection at 37°C for 1 h, monolayers were overlaid with agarose, incubated for 3 days, fixed, and stained as described above.
RT-PCR and sequencing.
Total infected-cell RNA was isolated with Trizol (Gibco-BRL). Reverse transcription (RT)-PCR was performed as previously described (23). Subregions of YF17D were amplified with the following primers: nucleotides (nt) 1 to 1065, primers 8108 (AGTAAATCCTGTGTGCTA /+) and CMR45 (ACACACTTGTCTTGCT/−); nt 942 to 2639, primers CMR92 (TACTGGTCTTGGCTGTTGG/+) and CMR67 (GGGAGTCAACTGAATTTAGGC/−); nt 3381 to 4285, containing the NS2A region, primers BRL488 (AATGGTGTTGCCGCTCCT/+) and CMR74 (TCCAACTGCAATCGGAC/−); the region encoding the NS2Aα cleavage site, primers PNACL1057 (CTCCTGGGTTACATTTGGAGAAATACAT/+) and CMR75 (AGCGAGGTCATCACAA/−); nt 4316 to 5523, primers CMR231 (GGGAGGGTGGATGGG/+) and CMR7 (AAGATTGTTGCACTTTCA/−); nt 5427 to 6145, primers PNACL1285 (TCATTATGGATGAAGCC/+) and CMR82 (ACCAGGGGAAACTGGT/−); nt 5860 to 6817, primers PNACL1286 (GAGCGAGTGCTGGATTGC/+) and BRL804 (CCTTTGTTGCCCTGGCTC/−). Amplified subregions were purified from agarose gels by using the QIAEX II gel purification kit (Qiagen). Purified PCR fragments were sequenced either directly or after subcloning by cycle sequencing with ABI PRISM dye-labeled terminators (Perkin-Elmer).
Protein analyses.
BHK-21/J cells were labeled for 4 to 5 h in methionine- and cysteine-deficient MEM containing 2% FCS and 50 μCi of EXPRE35S35S protein labeling mix (NEN) per ml. For precipitation with YF hyperimmune ascitic fluid (HIAF) (8), cells were lysed in 0.5% Triton X-100-50 mM Tris-HCl (pH 7.5)-200 mM NaCl-1 mM EDTA and clarified by centrifugation at 16,000 × g for 5 min. Before the addition of 1 μl of HIAF, samples were diluted as described previously (8). For immune precipitation with rabbit antisera, cells were lysed in 0.5% sodium dodecyl sulfate (SDS)-50 mM Tris-HCl (pH 7.5)-1 mM EDTA and heated for 10 min at 75°C. Before the addition of 2 μl of undiluted rabbit antiserum, samples were diluted with 3 volumes of 50 mM Tris-HCl (pH 7.5)-1 mM EDTA-200 mM NaCl-0.67% Triton X-100-1.33 mg of BSA per ml. Cell culture supernatants were clarified by centrifugation at 3,000 × g for 10 min. SDS was added to a final concentration of 0.5%, and the mixture was heated at 75°C for 10 min and diluted as described above. In some experiments, to facilitate detection of C and (pr)M, the clarified supernatant was centrifuged at 95,000 × g (42,000 rpm; Beckman TLA45 rotor) for 2 h. The pellet was resuspended in SDS-lysis buffer, denatured, and diluted as described above. Washed Pansorbin (Calbiochem) was used for collection of the immunoprecipitates. SDS-polyacrylamide gel electrophoresis was carried out as described by Laemmli (12) or Schägger and Jagow (28). Gels were processed for fluorography by treatment with En3Hance (NEN). The following rabbit antisera were used for detection of YF proteins: αC (raised against a fusion protein containing residues 1 to 94 of the YF C protein and kindly provided by M. Bouloy, Institut Pasteur, Paris, France) and prM- and E-specific YF antisera, which have been described previously (7).
Viral RNA analysis.
Viral RNA synthesis was examined by metabolic labeling with [3H]uridine (50 μCi/ml; ICN) in the presence of actinomycin D (2 μg/ml). After labeling, total RNA was isolated from the cells with Trizol (Gibco-BRL). To analyze viral RNA in the cell culture supernatant, it was first clarified by centrifugation at 3,000 × g for 10 min. Clarified supernatant was mixed with 4 volumes of 40% (wt/vol) polyethylene glycol 8000. After 2 h of incubation at 4°C, the polyethylene glycol precipitate was pelleted at 14,000 × g and 4°C for 30 min. The pellet was resuspended in Trizol, and the viral RNA was extracted as recommended by the manufacturer. The RNA was denatured with glyoxal at 50°C for 30 min and analyzed on a 1% agarose gel in 10 mM sodium phosphate buffer (SPB) (27). After electrophoresis, the gel was soaked in 50 mM NaOH for 20 min, stained in SPB containing 0.5 μg of ethidium bromide per ml, and destained for 1 h in SPB. The gel was dehydrated by being soaked twice (for 1 h each time) in methanol and then incubated overnight in 2.5% 2,5-diphenyloxazole in methanol. After being washed with water, the scintillant-impregnated gel was dried and exposed to X-Omat film (Kodak).
RESULTS
A YF NS2Aα QST mutant does not form plaques despite efficient RNA replication.
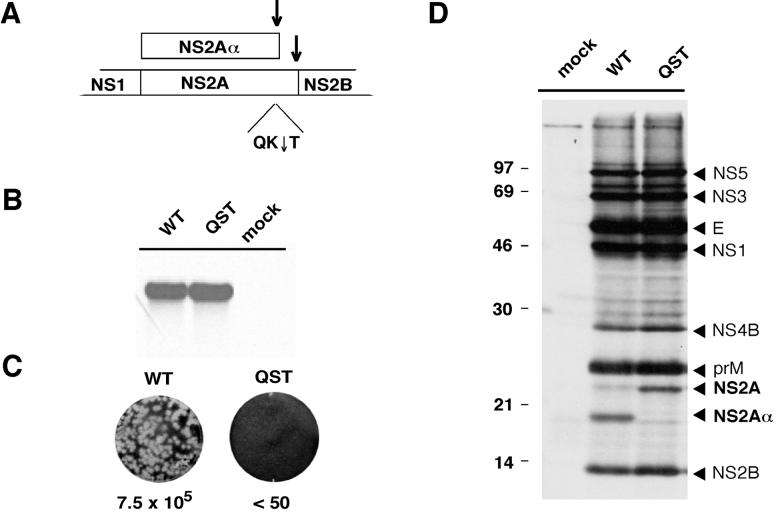
In the course of studying the effects of mutations that blocked processing at each of the NS2B-3 serine protease-dependent sites, we found that RNA replication was unimpaired for a mutation (QK↓T to QST) that inhibited cleavage at the NS2Aα cleavage site (data not shown; Fig. 1A). This observation was made with a subgenomic YF replicon and was surprising given that full-length RNAs harboring this QST mutation failed to yield infectious virus (26). To examine the RNA phenotype of the full-length mutant construct, we recreated the QST NS2Aα cleavage site mutant with a full-length YF 17D clone, pACNR/FLYF (17; Bredenbeek et al., unpublished). After electroporation of BHK cells and labeling with [3H]uridine in the presence of actinomycin D, RNA accumulation to a level comparable to that of parental YF 17D (wild-type [WT]) RNA was observed (Fig. 1B). Immunoprecipitation of [35S]methionine-cysteine-labeled proteins with HIAF recognizing several YF proteins confirmed that the NS2Aα QST mutant expressed only NS2A whereas both NS2Aα and NS2A were present in WT-transfected cells (Fig. 1D). Despite normal levels of RNA replication and protein production (other than NS2A-related proteins), the NS2Aα QST mutant failed to produce plaques. Figure 1C shows an infectious-center assay in which electroporated cells were serially diluted and seeded with naive indicator cells. The WT RNA produced 7.5 × 105 infectious centers/μg of RNA; no infectious centers were produced by the NS2Aα QST mutant. These data suggest that NS2A Lys-190 or cleavage at the NS2Aα site is important for plaque formation. These experiments did not, however, distinguish between effects on infectious-particle production and a YF-induced CPE, both of which are required for plaque formation.
FIG. 1.
Phenotype of the YF NS2Aα QST mutant. (A) NS2A processing and the NS2Aα cleavage site. Shown is a diagram of the YF polyprotein encompassing NS2A. In addition to full-length NS2A, a C-terminally truncated form (NS2Aα) is produced. Cleavages to generate the C termini of NS2A and NS2Aα are mediated by the NS2B-NS3 serine protease (↓). The NS2Aα cleavage site is defined by the sequence QK↓T. (B) RNA analysis. BHK cells were electroporated with WT or NS2Aα QST mutant RNA transcripts. From 20 to 28 h postelectroporation, the viral RNA was labeled with [3H]uridine in the presence of actinomycin D. Total RNA was isolated, denatured, and resolved by electrophoresis through a 1% agarose gel. The gel was treated for fluorography, dried, and exposed to X-ray film. (C) Infectious-center assay. BHK cells were electroporated with the WT or mutant (QST) transcript, and serial 10-fold dilutions were seeded on 35-mm-diameter dishes in the presence of untransfected cells and overlaid with agarose. After 3 days, cells were fixed and crystal violet staining was performed. The values below the dishes indicate the amounts (PFU per microgram) of in vitro-transcribed RNA. (D) NS2A processing. BHK cells were metabolically labeled 20 to 25 h after electroporation with [35S]methionine-cysteine. YF-specific proteins were immunoprecipitated from cellular extracts with YF-specific HIAF. Proteins were solubilized, denatured, and resolved by electrophoresis through an SDS-12% polyacrylamide gel (12). The values on the left are molecular masses of standard proteins in kilodaltons.
The NS2Aα QST mutation results in a defect in infectious-virus production.
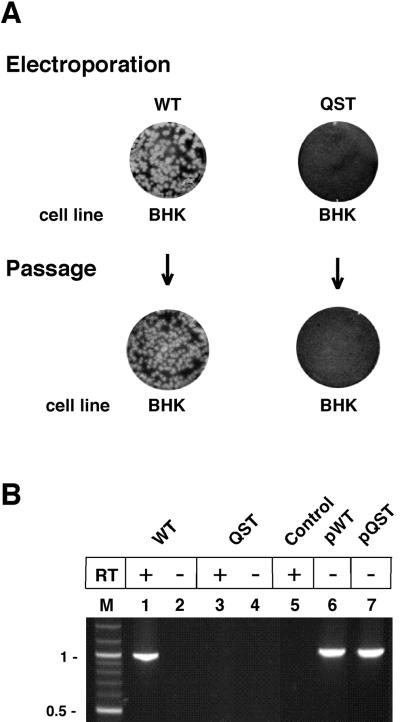
To determine if the NS2Aα QST mutant produced infectious but noncytopathic (ncp) virus, supernatants were harvested after electroporation and used to infect fresh BHK cells (Fig. 2A). To assess productive reinfection, total BHK RNA was isolated and the region encoding the NS2Aα cleavage site was amplified by RT-PCR. With the NS2Aα QST mutant, neither plaques (Fig. 2A) nor an RT-PCR product (Fig. 2B, lanes 3 and 4) was obtained after reinfection, suggesting that infectious particles were not released from mutant-transfected cells. In contrast, parallel analysis of the WT RNA resulted in efficient plaque formation after electroporation and subsequent reinfection (Fig. 2A). RT-PCR analysis of the total RNA from WT-reinfected cells yielded a PCR product of the expected size (Fig. 2B, lane 1).
FIG. 2.
Failure of the NS2Aα QST mutant to produce infectious virus. (A) Infectious-center and infectivity assays. WT or NS2Aα QST RNA transcripts were electroporated into BHK cells. Serial 10-fold dilutions were seeded on 35-mm-diameter dishes in the presence of the respective untransfected cells and overlaid with agarose. After 3 days, cells were fixed and crystal violet staining was performed. Supernatants from parallel liquid cultures were used to perform a plaque assay on new BHK cells. In addition, total cellular RNA was extracted from parallel cultures of reinfected cells and used for RT-PCR analysis. (B) RNA analysis. The region containing the NS2Aα cleavage site sequence was amplified from total cellular RNAs from the experiment described in panel A. As a control for DNA carryover, parallel reactions without reverse transcriptase (RT) were performed (lanes 2 and 4). Amplifications from the WT full-length plasmid (pWT) or the mutant full-length plasmid (pQST) served as positive controls (lanes 6 and 7). Lane M, DNA size markers (molecular sizes are in kilobases on the left). Control, no-template control.
The NS2Aα QST lesion blocks release of C protein and viral RNA but not E, prM, and M.
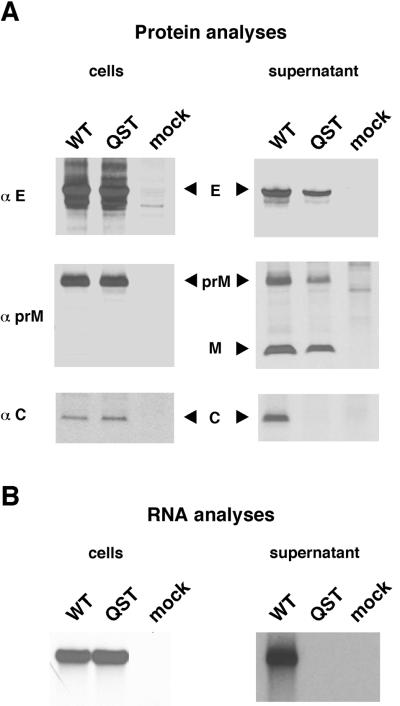
Our results indicated that the NS2Aα QST mutant did not produce infectious particles despite efficient RNA replication. This phenotype could be due to defects in particle assembly, release, or infectivity. To further define the step(s) that was blocked, we analyzed structural protein processing and the release of these proteins from cells transfected with NS2Aα QST mutant or WT RNA transcripts. YF-specific proteins were identified by metabolic labeling and precipitation with specific antibodies. With both the WT and the NS2Aα QST mutant, envelope proteins E and prM were detectable in the cells and the supernatant (Fig. 3A). During virion release, most of the prM is cleaved to release the mature M protein (16) and, as expected, M was detectable in the supernatants of both the WT and the mutant (Fig. 3A). A much different pattern was obtained with the C protein and viral RNA. Both the C protein and [3H]uridine-labeled RNA were readily detected in cells transfected with WT or mutant RNA (Fig. 3). However, whereas both of these species were released from WT-transfected cells, they were undetectable in mutant-transfected cells (Fig. 3). Thus, despite apparently normal structural region processing, the mutant failed to secrete immature or mature virus particles. The unimpaired release of prM/M and E into the culture supernatant suggested that production of subviral particles might be unimpaired.
FIG. 3.
Processing and release of structural proteins and viral RNA from WT- and NS2Aα QST-transfected cells. (A) Structural protein analysis. BHK cells were metabolically labeled 20 to 25 h postelectroporation with [35S]methionine-cysteine. YF-specific proteins were precipitated from either cellular extracts or supernatants with polyclonal rabbit sera against E, prM, or C. Precipitated proteins were resolved on a Tricine-SDS-10% (αE) or -14% (αprM and αC) polyacrylamide gel (28). (B) RNA analysis. BHK cells were labeled 15 to 22 h postelectroporation with [3H]uridine in the presence of actinomycin D. RNA was extracted either from the cells or from virus pelleted from the supernatant. The extracted RNA was resolved on an agarose gel under denaturing conditions.
The NS2Aα QST mutation prevents production of virus but not subviral particles.
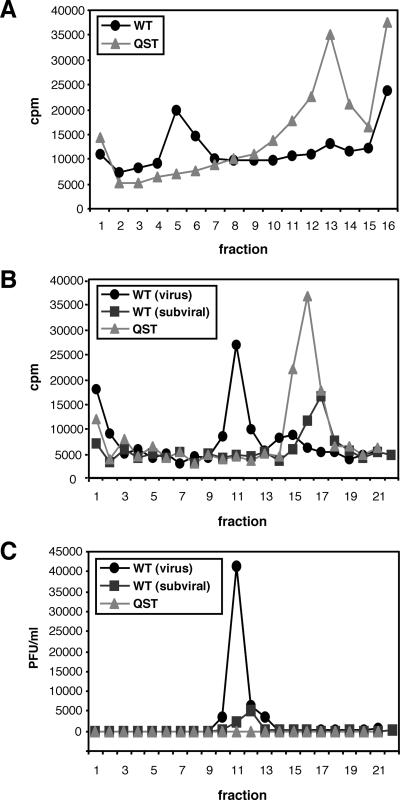
Two classes of particles are released from flavivirus-infected mammalian cells. Infectious virus includes the RNA genome complexed with multiple copies of the C protein surrounded by an envelope containing prM/M and E. Noninfectious subviral particles containing prM/M and E but lacking nucleocapsids are also secreted from infected cells. These subviral particles sediment more slowly than virus and are lower in density since they lack nucleocapsids (29, 30). Particles released from WT- and NS2Aα QST mutant-transfected cells were compared by rate-zonal and equilibrium density centrifugation. Metabolically labeled (with [35S]methionine-cysteine) particles released into the supernatant were concentrated by centrifugation, resuspended, and sedimented through a glycerol gradient. The profiles of the WT and the NS2Aα QST mutant are shown in Fig. 4A. A peak of 35S radioactivity, corresponding to the position expected for infectious virus, was present in fractions 5 and 6 for the WT but not for the mutant. With the NS2Aα QST mutant, a single peak of 35S radioactivity was present in fractions 12 to 14, consistent with the idea that slowly sedimenting subviral particles were released. A small peak at the same position was also seen with the WT, suggesting the production of both virus and subviral particles. YF E protein was found in the peak gradient fractions of both the WT and the mutant (data not shown). Pooled peak fractions from the glycerol gradients were then subjected to equilibrium centrifugation on tartrate gradients (Fig. 4B). Putative WT virus banded in tartrate fractions 10 to 12 at a higher density than the slowly sedimenting subviral material from either the WT (Fig. 4B, fractions 16 to 17) or the mutant (Fig. 4B, fractions 15 to 17). The presence of infectious virus in tartrate fractions 10 to 12 was confirmed by plaque assay; no infectivity was detected in any fraction of the mutant or in the low-density subviral peak of the WT (Fig. 4C). Thus, the NS2Aα QST mutant produces subviral particles but the assembly and/or release of infectious virus is blocked.
FIG. 4.
Characterization of particles released from WT- and NS2Aα QST-transfected cells. BHK cells were transfected with WT or NS2Aα QST mutant (QST) RNA transcripts, metabolically labeled with [35S]methionine-cysteine from 14 to 26 h posttransfection, concentrated by pelleting, and resuspended in buffer as described in Materials and Methods. (A) Rate-zonal sedimentation. WT or NS2Aα QST mutant particles were sedimented in parallel glycerol gradients as described in Materials and Methods. (B) Equilibrium banding. A portion of pooled fractions 5 and 6 [WT (virus)] and a portion of pooled fractions 12 to 14 [WT (subviral)] from the glycerol gradient shown in panel A were loaded on parallel tartrate gradients. For the NS2Aα QST mutant (QST), fractions 12 to 14 were pooled and analyzed. (C) Infectivity of the gradient fractions. Fractions of the gradients shown in panel B were analyzed for infectivity by plaque assay on BHK cells. In all panels, fractions are numbered from the bottom to the top.
Complementation of the NS2Aα QST defect in trans.
We next examined the ability of the NS2Aα QST defect to be complemented in trans. For this experiment, BHK cell populations expressing YF NS1-2A or other NS2A-related proteins were created with the ncp Sindbis virus (SIN) expression system (1). SIN, an Alphavirus, is an enveloped virus containing a single-stranded RNA with positive polarity. The ncp SIN vector system is a recombinant self-replicating SIN RNA that produces two subgenomic mRNAs. Foreign genes of interest are driven by one subgenomic promoter with the second promoter driving expression of the pac gene encoding puromycin-selectable puromycin N-acetyltransferase. Earlier studies showed that the ncp SIN expression system does not interfere with YF 17D replication and could be used for trans complementation of a YF NS1 deletion mutant (18).
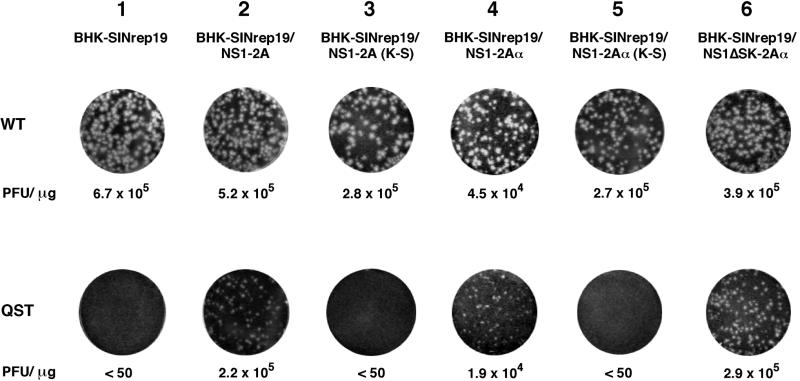
Either NS2Aα QST or WT RNA was electroporated into each BHK cell population, and then serial dilutions of the transfected cells were plated with the same nontransfected cell populations to quantify infectious centers (Fig. 5). As shown above for naive BHK cells, the NS2Aα QST mutant RNA was defective for plaque formation in BHK cells expressing the vector alone (SINrep19; Fig. 5, column 1) but small plaques were observed in NS1-2A-expressing BHK cells (Fig. 5, column 2). The supernatant from these cells did not form plaques on naive BHK cells, although RT-PCR analysis of reinfected BHK cells yielded a PCR fragment of the expected size that was cleaved with SalI, a marker for the NS2Aα QST mutation (data not shown). These results indicated that the YF NS2Aα QST defect in infectious-virus production could be complemented by supplying WT NS1-2A in trans.
FIG. 5.
trans-acting factors required for complementation. WT or NS2Aα QST RNA transcripts were electroporated into BHK-SINrep19 cells or into BHK-SINrep19 cells expressing the indicated YF proteins. Serial 10-fold dilutions were seeded on 35-mm-diameter dishes in the presence of the respective untransfected cells and overlaid with agarose. After 3 days, cells were fixed and stained with crystal violet. The values below the dishes indicate PFU per microgram of electroporated RNA.
The NS1-2A cassette consists of a signal sequence derived from the C terminus of E followed by the complete NS1-2A coding region. Processing of this cassette results in functional glycosylated NS1 (18) and, presumably, both NS2Aα and NS2A (see below). To further map the minimal requirements for trans complementation of the NS2Aα QST defect, additional constructs were tested with the ncp SIN expression system. trans complementation was not observed with NS1-2A containing the Lys-to-Ser substitution at the NS2Aα cleavage site [NS1-2A (K-S); Fig. 5, column 3]. Expression of NS1-2Aα was sufficient for trans complementation (Fig. 5, column 4), but replacement of the C-terminal Lys of NS2Aα with Ser abrogated its activity (Fig. 5, column 5). NS1 was not required in cis for trans complementation since a large in-frame deletion of NS1 sequences was also active (NS1ΔSK-2Aα; Fig. 5, column 6). These results indicate that NS2Aα is sufficient for trans complementation. Surprisingly, the NS2Aα C-terminal Lys residue also appears to be important for plaque formation, independently of its role in serine protease-dependent cleavage. It should be mentioned that NS1 expression levels were monitored in each cell population and were equivalent (except for the SINrep19 negative control and the NS1ΔSK-2Aα cassette). We do not have an antiserum specific for NS2A, and precipitation of NS2A and NS2Aα from YF-infected cells by HIAF (Fig. 1D) is dependent upon coexpression of the other YF proteins. Hence, the levels of NS2A and NS2Aα expression in the BHK populations could not be measured and compared. When trans complementation was observed, the specific infectivity of the mutant transcripts was within two- to threefold of the WT level. This is suggestive of true complementation rather than rare events like reversion, acquisition of compensating mutations, or recombination. The smaller plaques observed by trans complementation suggest that NS2A or NS2Aα may function more efficiently in cis. However, this small-plaque phenotype could also reflect suboptimal expression levels of NS2A-related proteins or heterogeneity in the complementing cell populations. Consistent with the small-plaque phenotype, trans complementation appeared to be inefficient since the titers of the NS2Aα QST mutant produced by NS1-2A-expressing cells were 1 to 2 logs lower than those of the WT virus.
Additional NS2Aα cleavage site mutations, revertants, and second-site changes.
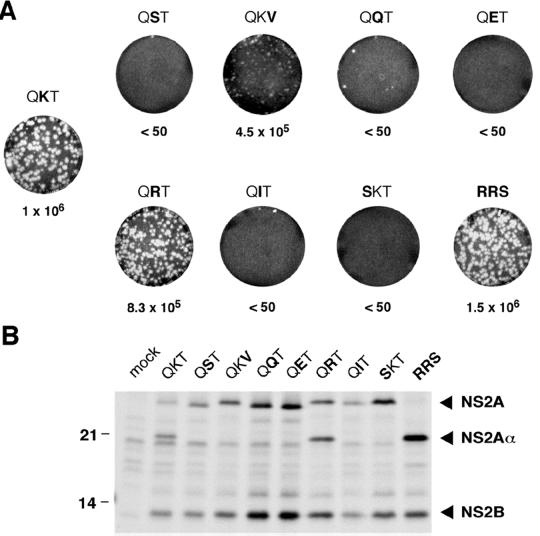
Given the importance of NS2A Lys-190 for virus production, we made additional substitutions for amino acids surrounding the NS2Aα cleavage site. These mutations are summarized in Table 1. Replacement of Lys-190 with Arg or introduction of the consensus YF serine protease cleavage site Arg-Arg-Ser yielded infectious plaque-forming transcripts that were indistinguishable from the WT in specific infectivity and plaque size (Fig. 6A). In contrast, electroporation of transcripts encoding Gln, Glu, or Ile at the NS2Aα P1 position (QQT, QET, or QIT mutants) or Ser at the P2 position (SKT mutant) failed to induce plaque formation (Fig. 6A). Electroporation of a mutant in which Thr at the P1′ position was changed to Val (QKV mutant) resulted in the formation of small plaques with a specific infectivity similar to that of the WT (Fig. 6A). Analysis of viral RNA and protein synthesis revealed levels similar to those of the WT for all of the mutants (Fig. 6B and data not shown). Analysis of NS2Aα production by immunoprecipitation revealed that NS2Aα was detectable only for the QRT and RRS mutants (Fig. 6B). For the RRS mutant, cleavage at the NS2Aα site was even more efficient than for the WT and appeared to be complete (Fig. 6B). Interestingly, despite production of infectious particles, NS2A cleavage was not observed in the QKV mutant (Fig. 6B). This was also the case for all of the other, non-plaque-forming mutants (Fig. 6B).
FIG. 6.
Analysis of additional NS2Aα cleavage site mutants. (A) Infectious-center assay. After electroporation of the indicated NS2Aα cleavage site mutants, an infectious-center assay was performed as described in the legend to Fig. 1C. (B) NS2A processing. BHK cells were metabolically labeled from 20 to 25 h postelectroporation with [35S]methionine-cysteine. Immunoprecipitation of YF-specific proteins from cellular extracts was performed with YF-specific HIAF as described in Materials and Methods. Precipitated proteins were resolved on an SDS-14% polyacrylamide gel (12). The values at the left are molecular masses of standard proteins in kilodaltons.
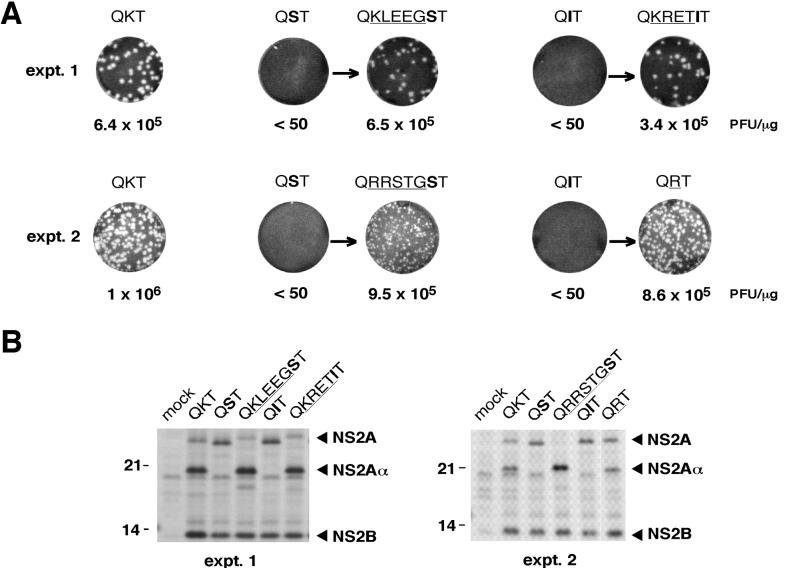
To identify mutations that could compensate for deleterious changes at the NS2Aα cleavage site, transfected cells were incubated and observed for evidence of a CPE, an indicator of release and spread of cytopathic virus. In the first experiment, a CPE developed after 48 to 72 h even with mutants that did not form plaques in the infectious-center assay (except for the QST mutant). Titration of the supernatant confirmed the presence of cytopathic plaque-forming virus, suggesting the presence of reversions or second-site changes (Table 1). Seventy-two-hour supernatants were used to infect naive cells, and total RNA was harvested and used for RT-PCR amplification and sequence analysis of the NS2A coding region. Direct sequencing of the RT-PCR products revealed that the QQT and QET mutants had reverted to the WT sequence (Table 1). Plaque-forming virus derived from the QIT mutant contained an insertion of four amino acids at the NS2Aα cleavage site (KRET; Table 1). The Gln-to-Ser substitution at the P2 position (SKT mutant) was still present. Since no other change was found in NS2A, this suggested the presence of a second-site mutation elsewhere in the genome.
In a second set of electroporations, a CPE developed again with the QQT, QET, and SKT mutants (Table 1). Sequencing of the NS2A region of these plaque-forming variants confirmed the results of the first experiment; QQT and QET had reverted to the WT, and the SKT mutation was stable (Table 1). No CPE was observed with the QST or QIT mutant, and the transfected cells were maintained and passaged. A CPE arose with the QST mutant after one passage, whereas no CPE was observed with the QIT mutant even after three passages. Sequencing of the variant derived from the QST transfection revealed an insertion of five amino acids at the NS2Aα cleavage site (KLEEG; Table 1). In a third experiment, no variants were recovered for the QST or QIT mutants, even after three passages, whereas a fourth repetition yielded infectious plaque-forming virus of both mutants. For the QST mutant, a virus with a five-amino-acid insertion at the NS2Aα cleavage site was recovered (RRSTG; Table 1); with the QIT mutant, mutation of the ATC codon to AGG resulted in Arg at the P1 position of the NS2Aα cleavage site (Table 1). As shown earlier, this substitution (QRT) is indistinguishable from the WT (Fig. 6).
Since limited sequencing of the plaque-forming virus derived from the SKT mutant had failed to uncover additional changes in NS2A, other parts of the YF genome were analyzed. Sequence analysis of the structural region (nt 1 to 2639) did not reveal any mutation. Direct sequencing of the NS2B-3-4A region (nt 4316 to 6817), however, uncovered an A-to-T substitution at nt 5598 in NS3. This was the predominant nucleotide substitution found in plaque-forming virus recovered from two independent electroporations of SKT mutant RNA. Subcloning of this region for one of the plaque-forming virus populations and analysis of 16 individual clones revealed 9 with T, 4 with C, and 3 with G at nt 5598. At the amino acid level, these changes lead to the replacement of NS3 Asp-343 with either Val, Ala, or Gly; all amino acids with relatively short, uncharged side chains.
Reconstruction of potential compensating mutations: insertions at the NS2Aα site or second-site changes in NS3 restore production of infectious particles.
We first analyzed the ability of the insertions identified at the NS2Aα cleavage site to restore production of infectious virus. Appropriate full-length plasmids and the corresponding RNA transcripts were tested in a standard infectious-center assay. As shown in Fig. 7A, the reconstructed insertion mutants, in contrast to the original substitution mutants, formed plaques with a specific infectivity similar to that of the WT RNA. Plaque size was comparable to that of the WT for the KLEEG and KRET insertions, whereas smaller plaques were obtained for the RRSTG insertion mutant. All of the insertions restored production of NS2Aα (Fig. 7B). Inspection of the amino acids in the inserted sequences revealed a typical serine protease-dependent cleavage site (3) only for the RRSTG insertion. In this case, cleavage probably occurs after the two Arg residues. As for the NS2Aα RRS mutant, NS2A cleavage for the RRSTG insertion is even more efficient than in the WT (Fig. 7B). For the other two insertion mutants, the actual NS2Aα processing site remains to be determined.
FIG. 7.
Phenotypes of reconstructed insertion mutants. (A) Infectious-center assays. Infectious-center assays of the parental NS2Aα cleavage site mutants (QST and QIT) and the corresponding reconstructed insertion mutants were performed as described in the legend to Fig. 1C. At the left, WT RNA was electroporated in parallel as a positive control for the electroporations shown in the same line. Two sets of experiments (expt. 1 and expt. 2) were performed. (B) NS2A processing. Samples were labeled and analyzed as described in the legend to Fig. 6B.
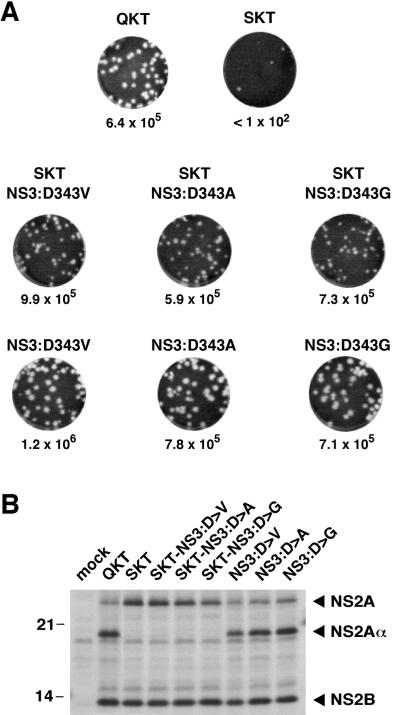
The most unexpected and interesting second-site changes were found for the SKT mutant in NS3 at position 343. Three different nucleotide substitutions led to the replacement of Asp-343 in the helicase domain of NS3 with Val, Ala, or Gly. To test the abilities of these substitutions to compensate for the SKT mutation, we constructed double mutants that contained the Gln-to-Ser exchange at the P2 position of the NS2Aα site and either Val, Ala, or Gly at position 343 of NS3. Electroporation of the reconstructed mutant RNAs resulted in the formation of plaques with a specific infectivity similar to that of the WT (Fig. 8A). Thus, the second-site mutations in NS3 do, indeed, rescue the SKT mutant. Analysis of NS2A cleavage for those mutants revealed that NS2A remained uncleaved (Fig. 8B). As seen for the QKV mutant (Fig. 6), these results demonstrate that cleavage at the NS2Aα site is not required for infectious-virus production. We also examined the phenotypes of mutants with each NS3 substitution in the absence of the SKT mutation. All three mutants formed plaques that were slightly larger than WT plaques with specific infectivities that were comparable to that of the WT (Fig. 8A). Compared to the WT, no alterations in cleavage efficiency at the NS2Aα site were noted (Fig. 8B).
FIG. 8.
Phenotypes produced by NS3 second-site mutations in the context of NS2Aα SKT or WT. (A) Infectious-center assays. Infectious-center assays of the NS2Aα SKT mutant or the WT with the reconstructed NS3 mutants were performed as described in the legend to Fig. 1C. For comparison, infectious-center assays for the WT and the NS2Aα SKT mutant are shown again at the top. (B) NS2A processing. Samples were labeled and analyzed as described in the legend to Fig. 6B. The values on the left are molecular sizes in kilodaltons.
DISCUSSION
This analysis of the NS2A Lys-190-to-Ser substitution that blocks cleavage at the NS2Aα site has revealed a defect in infectious-virus production despite apparently normal levels of RNA amplification and viral protein synthesis. No alterations of structural region processing or structural protein levels were noted. This block was selective for nucleocapsid-containing virions but did not affect secretion of prM/M- and E-containing subviral particles. Analyses of additional cleavage site mutants, second-site compensating changes, and trans-complementation data indicate that the sequence of NS2A in the region of the NS2Aα site is important for its role in infectious-virus production rather than cleavage by the NS2B-3 serine protease. Furthermore, a deleterious Gln-to-Ser substitution at the P2 position of the NS2Aα site could be compensated for by amino acid substitutions in the NS3 helicase domain. This suggests additional interactions involving NS3 and viral (or cellular) components required for virion assembly or release.
Substitutions at the NS2Aα site that preserved the properties of an NS2B-3 serine protease recognition site were tolerated. For instance, mutagenesis of QKT to either QRT or RRS produced transcripts with WT specific infectivity and plaque size. In the case of the RRS mutant, processing at the NS2Aα site was complete and uncleaved NS2A was undetectable, suggesting that unprocessed NS2A is not required for YF replication. Substitutions that blocked cleavage at the NS2Aα site but differed from the WT sequence by only a single base (QQT and QET) were unstable and reverted to the WT sequence. Mutations at the P1 position (QST and QIT) that differed by 2 nt from the WT codon yielded an interesting set of compensating changes. For the QST mutant, insertions of five amino acids (KLEEG and RRSTG) were recovered. In the case of the QIT mutant, an insertion of four residues (KRET) or replacement of Ile with Arg (QRT) was identified in infectious virus from two independent experiments. In all cases, the altered sequences mimicked NS2B-3 protease cleavage sites (Gln followed by one or two basic residues) and restored processing at or near the WT NS2Aα site. The origin of the inserted nucleotide sequences is unclear. One striking similarity of the inserted sequences was that all were purine rich. Perfect matches were not present in the YF genome or its complement, and the inserts were too short to be definitively assigned to specific cellular genes. Presumably, the insertions occurred during RNA replication, either as a consequence of two or more template switches, as seen for pestivirus recombinants (24), or by addition of nontemplated bases by the YF replicase.
Although the results just described are consistent with a requirement for serine protease-mediated processing at the NS2Aα site, three lines of evidence suggest otherwise. The Val substitution at the P1′ position (QKV) blocked detectable proteolytic cleavage but still allowed formation of small plaques. Released-virus titers of this mutant were, however, reduced by 1 to 2 logs compared to that of the WT. In the trans-complementation analyses, the QST defect could be complemented by expression of NS2Aα terminating with Lys-190 but not Ser-190. Both of these observations point toward a requirement for a basic residue at NS2A position 190 rather than cleavage per se. Basic residues (either Lys or Arg) are found in this region of NS2A from all sequenced flaviviruses. The second-site changes in NS3 that compensate for the P2 Gln-to-Ser substitution (SKT) are also consistent with this view: each of three substitutions (Val, Ala, or Gly) for NS3 Asp-343 restored production of infectious virus in the absence of NS2A cleavage. Given these findings, the significance of processing at the NS2Aα site remains a mystery. It is possible that interaction of this region of NS2A with the NS2B-3 serine protease via a suboptimal cleavage site is important for its function in virus assembly or release. Alternatively, this function of NS2A may be unrelated to its ability to interact with or be cleaved by the NS2B-3 protease.
A role for NS proteins in flavivirus structural protein maturation is not unprecedented. As mentioned earlier, the NS2B-3 serine protease is responsible for mediating the cleavage that produces the C terminus of the mature capsid protein. It was originally thought that this cleavage occurred after cotranslational signalase-mediated processing at the C-prM junction. It was later shown that this serine protease-dependent cleavage on the cytosolic side of the endoplasmic reticulum (ER) membrane was actually a prerequisite for efficient signalase cleavage in the ER lumen (3, 19, 31, 34, 35). Mutations that either blocked cleavage by the viral serine protease (4) or uncoupled signalase cleavage from this event (13) blocked production of infectious virus. Other than these coordinated processing events, we know very little about possible interactions between NS components in virion assembly. The results presented here unveil a role for NS2A and the helicase domain of NS3 in one or more steps in virion morphogenesis and release.
Several models can be envisioned. The most obvious is that in which NS2A and the NS3 helicase domain, as components of the NS2B-3 protease complex, modulate production of the mature C protein. However, we have not detected any difference in the electrophoretic mobility of the C protein or any other alteration in structural region processing. Another possibility is that the NS2A-3 region interacts with structural proteins or genome RNA to promote nucleocapsid assembly, glycoprotein maturation, or budding. Given that subviral particles are efficiently assembled and secreted from cells transfected with the QST mutant RNA, a role for NS2A in glycoprotein oligomerization or higher-order assembly seems unlikely. Rather, we favor a model in which the NS2A-3 region modulates nucleocapsid assembly or perhaps budding. Unfortunately, very little is known about the early events in flavivirus particle assembly, including nucleation and formation of nucleocapsids, budding into intracellular vesicles, resolution of discrete enveloped particles, and the movement of these particles through the secretory pathway. At the molecular level, how NS2A and NS3 might participate in these processes remains obscure. The topology of NS2A has not been investigated, but we presume that the NS2A N terminus is produced by cleavage in the ER lumen (21) whereas the C terminus is created by the viral serine protease on the cytosolic side of the membrane. A similar membrane-associated cytoplasmic localization can also be inferred for the NS2Aα cleavage site. This places this region of NS2A in a subcellular compartment where it could interact with NS3 (and the capsid protein). Attempts to compare the subcellular localization of YF capsid protein in WT- and QST mutant-transfected cells have been unsuccessful. NS3 Asp-343, the position of second-site mutations that compensate for the NS2Aα P2 Ser substitution, is localized in the helicase domain downstream of the DEAH motif. Amino acid sequence alignments based on ClustalX (version 1.8) (32) suggest that YF NS3 Asp-343 corresponds to hepatitis C virus (HCV) Thr-344. Assuming that HCV and YF NS3 have similar folds, the HCV NS3 structure (36) predicts that YF Asp-343 is solvent accessible. Thus, this domain of NS3 may well be accessible for interaction with NS2A or virus structural components. Binding of NS2A fused to glutathione S-transferase to NS3 has already been described for KUN (20). However, a direct interaction between YF NS2A and NS3 has yet to be demonstrated. Interestingly, we found that replacement of NS3 Asp-343 with Val, Ala, or Gly in the absence of the NS2Aα mutation did not inhibit plaque formation. Rather, these substitutions yielded plaques slightly larger that WT plaques. Determination of whether these substitutions act at the level of virus assembly or release, as opposed to enhanced RNA replication, requires additional study.
In conclusion, our genetic analysis shows that NS2A and determinants in the NS3 helicase domain can dramatically influence the production of infectious YF particles. These results suggest a more intimate relationship between NS proteins and virion maturation than previously thought. While the mechanisms involved remain to be elucidated, the ability of the NS2Aα QST defect to be complemented in trans should greatly facilitate such studies. In addition, the search for second-site suppressors of additional NS2A mutations blocking virus production may help to unravel the intermolecular interactions important for this process. It will also be of interest to see if the results for YF can be generalized to other members of the Flavivirus genus and perhaps to the other Flaviviridae genera, the pestiviruses and the hepaciviruses.
Acknowledgments
We thank Michèle Bouloy (Institut Pasteur) for generously providing YF anti-capsid protein serum. We are also grateful to many colleagues for helpful discussions during the course of this work and to Holly Hanson, Carine Logvinoff, Joe Marcotrigiano, and Tim Tellinghuisen for critical reading and editing of the manuscript.
This work was supported in part by grants from the Public Health Service (CA57973 and AI24134). B.M.K. was supported by a fellowship (KU 1201/2-1) from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Agapov, E. V., I. Frolov, B. D. Lindenbach, B. M. Prägai, S. Schlesinger, and C. M. Rice. 1998. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA 95:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., K. Stadler, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 69:5816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amberg, S. M., A. Nestorowicz, D. W. McCourt, and C. M. Rice. 1994. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J. Virol. 68:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amberg, S. M., and C. M. Rice. 1999. Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. J. Virol. 73:8083-8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe and P. M. Howley (ed.), Fields virology, fourth ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 6.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 7.Chambers, T. J., D. W. McCourt, and C. M. Rice. 1990. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology 177:159-174. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, T. J., D. W. McCourt, and C. M. Rice. 1989. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology 169:100-109. [DOI] [PubMed] [Google Scholar]
- 9.Chambers, T. J., A. Nestorowicz, and C. M. Rice. 1995. Mutagenesis of the yellow fever virus NS2B/3 cleavage site: determinants of cleavage site specificity and effects on polyprotein processing and viral replication. J. Virol. 69:1600-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. S., C. L. Liao, C. H. Tsao, M. C. Chen, C. I. Liu, L. K. Chen, and Y. L. Lin. 1999. Membrane permeabilization by small hydrophobic nonstructural proteins of Japanese encephalitis virus. J. Virol. 73:6257-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khromykh, A. A., M. T. Kenney, and E. G. Westaway. 1998. trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72:7270-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Lee, E., C. E. Stocks, S. M. Amberg, C. M. Rice, and M. Lobigs. 2000. Mutagenesis of the signal sequence of yellow fever virus prM protein: enhancement of signalase cleavage in vitro is lethal for virus production. J. Virol. 74:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, C., S. M. Amberg, T. J. Chambers, and C. M. Rice. 1993. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 67:2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, C., T. J. Chambers, and C. M. Rice. 1993. Mutagenesis of conserved residues at the yellow fever virus 3/4A and 4B/5 dibasic cleavage sites: effects on cleavage efficiency and polyprotein processing. Virology 192:596-604. [DOI] [PubMed] [Google Scholar]
- 16.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, fourth ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 17.Lindenbach, B. D., and C. M. Rice. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 73:4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindenbach, B. D., and C. M. Rice. 1997. trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobigs, M. 1993. Flavivirus premembrane protein cleavage and spike heterodimer secretion requires the function of the viral proteinase NS3. Proc. Natl. Acad. Sci. USA 90:6218-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203-215. [DOI] [PubMed] [Google Scholar]
- 21.Markoff, L. 1989. In vitro processing of dengue virus structural proteins: cleavage of the pre-membrane protein. J. Virol. 63:3345-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin, M., T. F. Tsai, B. Cropp, G. J. Chang, D. A. Holmes, J. Tseng, W. Shieh, S. R. Zaki, I. Al-Sanouri, A. F. Cutrona, G. Ray, L. H. Weld, and M. S. Cetron. 2001. Fever and multisystem organ failure associated with 17D-204 yellow fever vaccination: a report of four cases. Lancet 358:98-104. [DOI] [PubMed] [Google Scholar]
- 23.Meyers, G., N. Tautz, P. Becher, H.-J. Thiel, and B. M. Kümmerer. 1996. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J. Virol. 70:8606-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyers, G., and H.-J. Thiel. 1996. Molecular characterization of pestiviruses. Adv. Virus Res. 47:53-118. [DOI] [PubMed] [Google Scholar]
- 25.Monath, T. P., and F. X. Heinz. 1996. Flaviviruses, p. 961-1034. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, third ed., vol. 1. Lippincott-Raven Publishers, Philadelphia. Pa. [Google Scholar]
- 26.Nestorowicz, A., T. J. Chambers, and C. M. Rice. 1994. Mutagenesis of the yellow fever virus NS2A/2B cleavage site: effects on proteolytic processing, viral replication and evidence for alternative processing of the NS2A protein. Virology 199:114-123. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range of 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 29.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, T. M., and R. W. Schlesinger. 1965. Studies on the nature of dengue viruses. I. Correlation of particle density, infectivity, and RNA content of type 2 virus. Virology 27:103-112. [DOI] [PubMed] [Google Scholar]
- 31.Stocks, C. E., and M. Lobigs. 1995. Posttranslational signal peptidase cleavage at the flavivirus C-prM junction in vitro. J. Virol. 69:8123-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasconcelos, P. F., E. J. Luna, R. Galler, L. J. Silva, T. L. Coimbra, V. L. Barros, T. P. Monath, S. G. Rodigues, C. Laval, Z. G. Costa, M. F. Vilela, C. L. Santos, C. M. Papaiordanou, V. A. Alves, L. D. Andrade, H. K. Sato, E. S. Rosa, G. B. Froguas, E. Lacava, L. M. Almeida, A. C. Cruz, I. M. Rocco, R. T. Santos, and O. F. Oliva. 2001. Serious adverse events associated with yellow fever 17DD vaccine in Brazil: a report of two cases. Lancet 358:91-97. [DOI] [PubMed] [Google Scholar]
- 34.Yamshchikov, V. F., and R. W. Compans. 1994. Processing of the intracellular form of the West Nile virus capsid protein by the viral NS2B-NS3 protease: an in vitro study. J. Virol. 68:5765-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamshchikov, V. F., and R. W. Compans. 1993. Regulation of the late events in flavivirus protein processing and maturation. Virology 192:38-51. [DOI] [PubMed] [Google Scholar]
- 36.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure Fold. Des. 7:1353-1363. [DOI] [PubMed] [Google Scholar]