Abstract
The Epstein-Barr virus nuclear antigen 3C (EBNA3C), encoded by Epstein-Barr virus (EBV), is essential for mediating transformation of human B lymphocytes. Previous studies demonstrated that EBNA3C interacts with a small, nonhistone, highly acidic, high-mobility group-like nuclear protein prothymosin alpha (ProTα) and the transcriptional coactivator p300 in complexes from EBV-infected cells. These complexes were shown to be associated with histone acetyltransferase (HAT) activity in that they were able to acetylate crude histones in vitro. In this report we show that ProTα interacts with p300 similarly to p53 and other known oncoproteins at the CH1 amino-terminal domain as well as at a second domain downstream of the bromodomain which includes the CH3 region and HAT domain. Similarly, EBNA3C also interacts with p300 at regions which include the CH1 and CH3/HAT domains, suggesting that ProTα and EBNAC3C may interact in a complex with p300. We also show that ProTα activates transcription when targeted to promoters by fusion to the GAL4 DNA binding domain and that this activation is enhanced by the addition of an exogenous source of p300 under the control of a heterologous promoter. This overall activity is down-modulated in the presence of EBNA3C. These results further establish the interaction of cellular coactivator p300 with ProTα and demonstrate that the associated activities resulting from this interaction, which plays a role in acetylation of histones and coactivation, can be regulated by EBNA3C. Furthermore, this study establishes for the first time a transcriptional role for ProTα in recruitment or stabilization of coactivator p300, as well as other basal transcription factors, at the nucleosomes for regulation of transcription.
Epstein-Barr virus (EBV) is a lymphotropic virus associated with numerous human cancers including Burkitt's lymphoma, nasopharyngeal carcinoma, gastric carcinoma, and, more controversially, breast carcinoma (4, 21, 24, 32, 45). EBV belongs to the Gammaherpesvirinae subfamily of viruses, all having similar colinear genomic organizations (24). Genetic analysis of the EBV genome has demonstrated that EBV nuclear antigen 3C (EBNA3C; also referred to as EBNA6) is critical for EBV-mediated immortalization of human primary B lymphocytes. Introduction of a stop codon inserted at position 365 of the open reading frame of EBNA3C rendered the recombinant EBV transformation incompetent (24, 34, 40). Biochemical studies have now linked the role of EBNA3C to regulation of transcription through down-modulation of EBNA2-mediated transactivation (35, 36). EBNA2 is tethered to the major EBV latent promoters through RBP-Jκ, a known cellular transcription repressor (18, 25, 47). EBNA3C disrupts RBP-Jκ binding to its cognate sequence by competing with EBNA2 for binding, thereby regulating the interaction of EBNA2 with RBP-Jκ (28, 33, 35, 36). EBNA3C is a large transcription factor encoded by EBV which belongs to a family of proteins transcribed about 110 kb from the major latent promoter (24, 37). EBNA3C is similar to the c-Jun and c-Fos transcription factors, with leucine zipper domains, acid-rich regions, and glutamine-rich domains. These molecules are known to be involved in transcription activation and contain nuclear localization signals important for mediating translocation to the nucleus (43) (Fig. 1). These domains are known to be involved in the repression and activation activities associated with EBNA3C (3, 28, 33, 46).
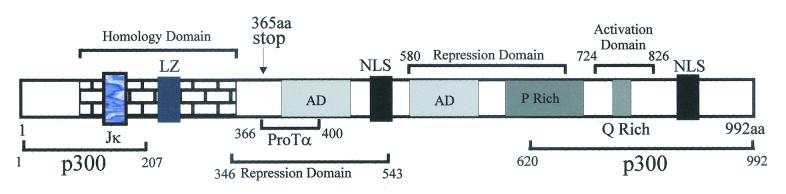
FIG. 1.
Schematic of basic structure of the EBNA3C protein with the identified motifs. Domains for interaction with p300 at both the amino and carboxy termini of the molecule are indicated. ProTα specifically interacts with the region between aa 366 and 400, a region shown previously to be essential for B-lymphocyte transformation (40). Arrow (at aa 365), insertion of an amber stop codon in the three reading frames, which demonstrated that EBNA3C is essential for B-lymphocyte transformation (40). The interaction domain for RBP-Jκ (Jκ) at the amino-terminal region and the leucine zipper (LZ) motif, acidic domains (AD), and proline (P)-rich and glutamine (Q)-rich domains are indicated (37, 40). The regions involved in activation and repression and the region of homology for the EBNA3 family members at the amino terminus are also shown (33, 34, 36, 37). NLS, nuclear localization signal.
Previous studies have demonstrated that, when tethered to promoters through fusions of specific domains in frame to the GAL4 DNA binding domain, EBNA3C contains two domains with potential for repression of transcription (3). Additionally, the glutamine-rich domain was shown to be capable of activating transcription through EBV latent promoters (28). Recent experiments have shown that the activation functions of EBNA3C are linked through its interactions with transcription factors SpiA and SpiB, which bind to the cognate sequences on the latent membrane protein 1 (LMP1) major latent promoter (46).
Studies with Raji cells, which contain an EBV genome with a deletion in the EBNA3C open reading frame, indicated that EBNA3C expression in these cells could reverse the block of these cells at the G0/G1 transition point of the cell cycle (2). LMP1 levels in these cells were also increased, suggesting that EBNA3C enhances transcription of LMP1 (1). Additionally, in vitro binding assays have demonstrated that EBNA3C can interact with the retinoblastoma protein (Rb) (2). Studies so far have not demonstrated this interaction in cells expressing these proteins under the control of heterologous promoters or in EBV-infected cells. However, the interaction data are intriguing, and further studies will be crucial for delineating the ability of EBNA3C to interact with Rb. Other studies have shown that the repression function of EBNA3C is associated with its ability to complex with histone deacetylase 1 in deacetylating histones (31).
Prothymosin alpha (ProTα) is a small, acid-rich, nonhistone nuclear protein, which was shown to be highly up-regulated in cells where the c-myc oncogene, induced under the control of a regulatable heterologous promoter (11, 16, 17). Moreover, ProTα is capable of transforming Rat-1 cells in culture, inducing the formation of foci in monolayers and foci in soft-agar assays (30). ProTα localizes to regions of the nucleus actively involved in transcription and associates with cellular histones, including nucleosome linker protein histone H1 (23, 41, 42). Moreover, ProTα removes histone H1 from the nucleosomes, which allows access to micrococcal nuclease (23). These activities suggest that ProTα may be similar to the human high-mobility group (HMG) proteins and that it may be HMG-like in its functions in the nucleus (5, 6, 8, 9, 23). Additionally, ProTα was shown to be associated with PML and cleavage bodies associated with gene regulation and transcriptional activities in the nucleus (41). These studies suggest that ProTα is involved in chromatin unwinding and remolding to allow access to transcription or replication factors. Recent studies in our laboratory have shown that ProTα also interacts with acetyltransferase p300 and that it is involved in regulating the acetylation of core histones through interactions with the p300 acetyltransferase (7). This interaction and the resulting acetylation activity are further regulated by the interaction of these complexes with EBNA3C, as EBNA3C interacts with both p300 and ProTα (7). It is possible that these interactions form part of a larger mega-activation complex involved in regulating chromatin remodeling.
Studies investigating the role of EBNA3C in regulating transcription have demonstrated that EBNA3C is involved with acetylases as well as deacetylases (31). These studies have also demonstrated the direct interaction of EBNA3C with these cellular targets, which results in acetylation or deacetylation of core histones (7, 31, 35). Therefore EBNA3C is capable of regulating cellular events requiring acetylation or deacetylation of the core histones localized to promoters of specific viral and cellular genes potentially involved in regulation of cell cycle events.
In this study we show that the interaction of ProTα and EBNA3C with p300 occurs at two distinct regions, the CH1 region and a region downstream of the bromodomain, which includes the CH3/HAT domain. These interactions are similar to the interactions of p53 with p300. The affinity for the amino-terminal CH1 domain of p300 is less than that seen with the CH3/HAT domain. Moreover, we show that the interactions of ProTα with EBNA3C are downstream of amino acid (aa) 365, a region potentially important for EBV-mediated transformation of human primary B lymphocytes. Additionally, p300 interacts with EBNA3C at the amino-terminal 207 aa, which includes the RBP-Jκ binding site and the leucine zipper motif, and at the carboxy terminus, which includes the proline-rich and glutamine-rich domain, which is known to be involved in transcriptional activation. We also show that ProTα can activate transcription when tethered to its promoter by fusing the entire ProTα cDNA (encoding 112 aa) to the GAL4 DNA binding domain and testing its activity on a multimerized GAL4-responsive reporter construct. Additionally, we have shown that p300 enhances the transcriptional activity mediated by ProTα on this GAL4-targeted promoter, indicating that ProTα complexed with p300 can functionally increase the activity of the basal transcriptional machinery. Moreover, EBNA3C modulates this effect on transcription and on HAT activity, demonstrating that in this context EBNA3C regulates the activity of ProTα and p300.
MATERIALS AND METHODS
Cell lines.
Burkitt's lymphoma-derived cell line BJAB was obtained from Elliott Kieff and was cultured in RPMI 1640 medium (Gibco-BRL, Gaithersburg, Md.) supplemented with 5 μg of penicillin/ml, 5 U of streptomycin/ml, and 10% fetal bovine serum purchased from Gibco-BRL. Human embryonic kidney (HEK) fibroblast cell line 293 was also obtained from Elliott Kieff and was grown in Dulbecco modified Eagle medium (Gibco-BRL) supplemented as described above.
Plasmids.
The p300 expression construct was a generous gift from Gary Nabel (12, 20). All glutathione S-transferase (GST)-p300 constructs were made by fusing the specific regions to the GST prokaryotic vector and are described below. The coding sequences for p300 fragments P1 to P5, corresponding to aa 1 to 672, 672 to 1193, 1069 to 1459, 1459 to 1892, and 1893 to 2414, respectively, were amplified by PCR and cloned into pGex6P2 (Clontech). The primers used were 5′-TAGCTAGCTCGAGGAATGGCCGAGAATGTGGTGGAACC-3′ and 5′-ATCGATCAAGCTTTTACCCTGGATTCATGGAAACTGGAACC-3′ for GST-p300-1, 5′-TAGCTAGCTCGAGGAGGGCCTAACATGGGACAGCCGC-3′ and 5′-ATCGATCAAGCTTTTAACTGTAATAAGTGGCATCACGAGG-3′ for GST-p300-2, 5′-TAGCTAGCTCGAGGAGATCCAGAATCCCTTCCCTTTCG-3′ and 5′-ATCGATCAAGCTTTTACTTGGGTATCTTCTGGTCAGGAG-3′ for GST-p300-3, 5′-TAGCTAGCTCGAGGAAAGCCCAAGCGACTGCAGGAATG-3′ and 5′-ATCGATCAAGCTTTTAAGCTTGAGTCCTGGGCAAGTAGG-3′ for GST-p300-4, and 5′-TAGCTAGCTCGAGGAGCTGCTGGCCCTGTGTCCCAG-3′ and 5′-ATCGATCAAGCTTTTACTAGTGTATGTCTAGTGTACTCTGTG-3′ for GST-p300-5. The coding sequence for GST-p300-HAT (aa 1197 to 1674) was amplified by PCR and cloned into pGex6P2. The primers used were 5′-CTCGATCGTCGACAGGTATCATTTCTGTGAGAAGTG-3′ and 5′-GATCGAGGTCGACTCACTTGCATTCATTGCAGGTGTAG-3′.
The full-length EBNA3C construct was described previously (35, 37). Truncated versions of EBNA3C, made with pSG5 under the control of the simian virus 40 promoter, including those in which the arginine-rich region was deleted, the leucine zipper region was deleted, the amino terminus 365 aa were deleted, and the carboxy terminus from aa 366 to 992 was deleted. The truncated EBNA3C with the leucine zipper deletion was constructed by deleting the region corresponding to bp 99061 to 99548 (aa 207 to 368), which includes the basic region. This deletion was created by digesting the EBNA3C pBluescript clone with an HpaI-SpeI fragment, blunting with mung bean nuclease, and religating the fragments. The deletion of the arginine-rich region corresponding to bp 98402 to 99062 (aa 12 to 207) involved deletion of the polyarginine motif and was performed by digesting the EBNA3C coding fragment with XbaI, end filling the fragment with Klenow fragments, digesting the fragment with HpaI, and religating the fragment and vector. EBNA3C Δ5′ construct is truncated at the 5′ end up to EBV bp 99537 with the first in frame codon. This clone was created by digestion with SpeI and religation of the largest fragment. The EBNA3C Δ3′ SpeI construct has a truncation at the 3′ end from bp 99537 (corresponding to amino acid 368) and was created by digestion of the EBNA3C pBS clone with SpeI and religation of the largest fragment. The EBNA3C Δ3′ HpaI construct has a truncated 3′ end from EBV bp 99061 (corresponding to amino acid 207) and was created by digesting the EBNA3C cDNA clone with HpaI and HindIII, end filling with Klenow fragments, and ligating the largest fragment.
The GAL4-ProTα construct was made by fusing the ProTα cDNA amplified by Vent polymerase into the pM-GAL4 expression plasmid. Sense and antisense primers were purchased from Gibco-BRL. Sense primer 5′-GAAGGATCCATGTCAGACGCAGCCGTAGAC-3′ and antisense primer 5′-AGCGATATCCTAGTCATCCTCGTCGGTCTT-3′ contained BamHI and EcoRV restriction sites, respectively. ProTα was amplified from the cDNA clone in pA3M vector with Vent polymerase from New England Biolabs (Beverley, Mass.), digested with BamHI and EcoRV, and ligated into the pM vector prepared by digestion with HindIII, blunting with T4 polymerase, and further digestion with BamHI. The cloned product was sequenced with the Thermocycle Sequenase sequencing system (Amersham Inc.).
Preparation of GST fusion proteins.
GST fusions were prepared as described previously (7). Briefly, TP10 cells were transformed with the plasmid constructs for each fusion, and single colonies were selected and grown in Luria-Bertani (LB) medium with 100 μg of ampicillin/ml. Five hundred milliliters of LB medium was inoculated at a dilution of 1:200 and allowed to shake at 30°C until mid-exponential growth phase. Cells were then induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 12 h with continuous shaking. Cells were harvested and sonicated, and the proteins were solubilized in the presence of protease inhibitors. Solubilized proteins were incubated with GST-Sepharose beads for 4 h at 4°C with rotation and then collected by centrifugation and washed three times in NETN buffer with protease inhibitors (7). GST fusion proteins bound to beads were used for binding assays and stored in NETN buffer with protease inhibitors and 1 mM phenylmethylsulfonyl fluoride at 4°C.
In vitro binding assays.
To determine binding of GST-p300 fusions to EBNA3C and ProTα, proteins were translated in vitro with [S35]Met-Cys translabel from NEN-Dupont by using the TNT system (Promega Inc.) in accordance with the manufacturer's instructions. Briefly, labeled EBNA3C and ProTα proteins were incubated with equivalent amounts of GST-p300 fusions bound to beads and rotated at 4°C for 4 h, followed by four washings in 1 ml of NETN with protease inhibitors. Bound proteins were eluted from the beads in sodium dodecyl sulfate (SDS) lysis buffer by heating at 95°C for 10 min and then fractionated by SDS-10% polyacrylamide gel electrophoresis. All binding experiments were done essentially as previously described (39).
For competition assays in vitro-translated ProTα or EBNA3C was incubated with a GST-p300 fusion protein for 1 h at 4°C and placed on ice and increasing amounts of EBNA3C or ProTα were added to the reaction mixtures. The reaction was then allowed to proceed for an additional 2 h, and then the bound products were collected by cenrifugation, washed four times in binding buffer, solubilized in SDS lysis buffer, dried, and electrophoresed on an SDS-15 or 8% polyacrylamide gel electrophoresis system. Dried gels were analyzed with a PhosphorImager (Molecular Dynamics Inc.), and signals were quantified with ImageQuant software.
Transfections and luciferase assays.
Ten million 293 cells were transfected with the pFr-GAL4-luciferase reporter, the pM-GAL4 DNA binding domain fusion protein, or the pM-GAL4-ProTα fusion protein expression plasmid. Mock transfection (no DNA) was used as a control, using a Bio-Rad electroporation apparatus at 210 V and 975 μF. Cells were resuspended in 10 ml of Dulbecco modified Eagle medium prepared as described above. Cells were washed once in 1× phosphate-buffered saline (PBS). All transfections were balanced for amounts of DNA and normalized for transfection efficiency. Increasing amounts of pM-GAL4-ProTα, from 5 to 20 μg in 5-μg increments, were added. Alternatively, increasing amounts of p300- and EBNA3C-expressing plasmids were added to the pM-GAL4-ProTα reaction to determine the effects on the GAL4-responsive luciferase reporter. All transfectants were incubated for 20 h, harvested, washed in 10 ml of PBS, and then resuspended in reporter lysis buffer and counted in a luminometer (MGM Instruments Inc.). All transfections were performed three times, and the means were plotted.
In vivo HAT assay.
Immunoprecipitates were collected with antibodies to ProTα or to the region of p300 encoded by the 5′ region of p300 from lysates of EBV-infected LCLs (LCL1 and LCL2) and B cells expressing EBNA3C or no EBNA as a control. For transient transfections BJAB cells were transfected at 210 V and 960 μF with a Bio-Rad electroporator. Expression constructs containing EBNA2 or EBNA3C were mixed with BJAB cells, electroporated, and then incubated for 24 h. Cells were collected by centrifugation and lysed in radioimmunoprecipitation assay (RIPA) buffer. Protein complexes containing acetylases were obtained by immunoprecipitation with polyclonal antibodies against the 5′ region of p300 or ProTα. Immunoprecipitates were washed twice in RIPA buffer (same as that used above except that it contained 0.5% Nonidet P-40) and then twice in histone acetyltransferase (HAT) assay buffer (50 mM Tris-HCl [pH 8.0], 10 mM sodium butyrate, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). Immunoprecipitates were incubated in HAT buffer with 25 μg of crude histones (Sigma) and 500 nCi of [3H]coenzyme A for 1 h at 30°C. After brief centrifugation to pellet protein A-Sepharose beads, the supernatant was spotted onto p-81 filters (Whatman) and washed in 0.2 M sodium carbonate three times by using vacuum filtration. Filters were allowed to dry under a heat lamp before being placed in ScintiVerse scintillation cocktail and counted in a Beckman LS3801 scintillation counter.
RESULTS
ProTα activates transcription when targeted to a GAL4-responsive promoter.
To determine if ProTα can affect the transcriptional activity of the basal transcription machinery, we cloned ProTα in frame with the GAL4 DNA binding domain as a fusion and tested its ability to influence transcription. HEK 293 cells were transfected with a GAL4-responsive luciferase reporter alone or in the presence of GAL4-ProTα. The transfections were incubated for 24 h then harvested. The harvested cells were lysed, and the cell lysate was clarified by centrifugation. Twenty-microliter aliquots were measured in a luminometer for 10 s and plotted.
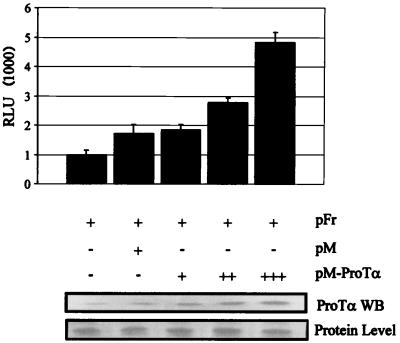
ProTα activated the GAL4-responsive promoter in a dose-responsive manner with increasing amounts of ProTα transfected in HEK 293 (Fig. 2). The total DNA was balanced for all transfections and normalized for transfection efficiency. Increments of 5 μg of DNA resulted in a gradual increase in activity up to approximately fivefold that for the reporter alone. This increase in activity continued as increasing amounts of pM-ProTα were introduced into the system (data not shown). The results suggest that ProTα is capable of associating with the basal transcription machinery to activate transcription when targeted to promoters by tethering to the GAL4 DNA binding domain. The activity observed was consistent with ProTα having activation functions, as increasing amounts of GAL4-ProTα resulted in a general increase in transcription activity (Fig. 2). Western blotting shows the increasing levels of ProTα, as indicated by the increasing intensity of the signal compared to the protein level.
FIG. 2.
Luciferase assay demonstrating that ProTα fused to GAL4 activates transcription when tethered to the promoter in 293 cells. 293 cells were transfected with increasing amounts of pM-ProTα in 5-μg increments. Transfections were balanced for amounts of DNA and normalized for transfection efficiencies. The pFr vector contains the luciferase gene downstream of a promoter element with GAL4 binding sites, which is targeted by the GAL4 DNA binding domain. The relative intensities are expressed as luciferase units (RLU). The pM vector contains only the GAL4 DNA binding domain not fused to a cDNA and was used as a control in this assay. Western blot (WB) shows increasing levels of ProTα with increasing amounts of ProTα DNA. The membrane was blotted with a rabbit polyclonal antibody against ProTα. A comparative stain of protein transferred to the membrane demonstrating equivalent levels of protein loading is shown at the bottom.
The p300 coactivator enhances transcriptional activation mediated by ProTα.
Previous studies by our group have shown that ProTα interacts with the acetyltransferase p300 and can recruit the activity mediated by p300 for acetylation of crude histones in vitro. Immunoprecipitation of complexes associated with ProTα using antibodies against ProTα contained molecules capable of acetylating histones (7). Additionally, we showed that ProTα interacts with linker histone H1 in uninfected and infected cells as well as in vitro, suggesting that there is a direct link to ProTα and its association with cellular histones and regulation of transcriptional activity in EBV-infected cells. Therefore we wanted to determine the effects of p300 on the ability of ProTα to activate transcription in a transient luciferase reporter assay.
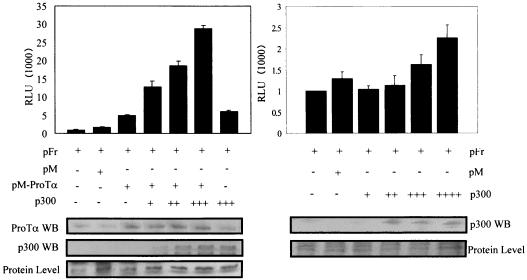
A p300 expression construct under the control of a heterologous promoter was transfected with pM-ProTα in HEK 293 cells. In this assay we show that the transcription activity mediated by ProTα was enhanced in the presence of p300 and that this activity gradually increased when increasing amounts of p300 were introduced in the system. Specifically, the activity was increased by threefold, fourfold, and sixfold with increments of 5 μg of p300 (Fig. 3, left). These results suggest that p300 cooperates with ProTα in enhancing basal transcription activity when targeted to the GAL4 promoter. Additionally, p300 by itself does not dramatically enhance the levels of activation, as it does when cooperating with ProTα, suggesting that the increased activity is dependent on the association of ProTα with p300 (Fig. 3, right). Western blots showing increasing levels of ProTα and p300 suggest that the activity is dependent on the levels of protein expressed compared to background level of a nonspecific protein.
FIG. 3.
The p300 coactivator enhances the transcription activation mediated by ProTα. The p300 expression vector was transfected in increasing amounts along with pM-ProTα in a luciferase assay. All transfections were normalized for transfection efficiencies and total DNA. Transfected cultures were harvested after 24 h of incubation at 37°C. The p300 expression construct was added in increments of 5 μg without pM-ProTα. Relative luciferase units (RLU) were measured for 10 s with an MGM luminometer (Optocomp1), and means of three experiments are shown. A Western blot (WB) assay for ProTα and p300 levels and for those of the protein loading control used for comparison is shown at the bottom.
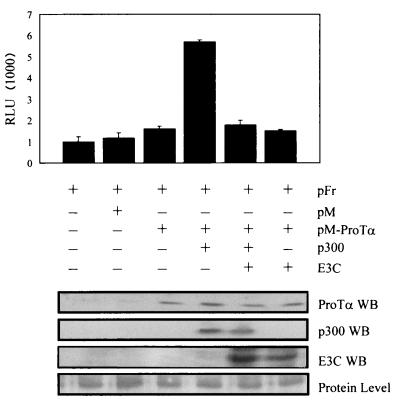
Previous work showed that EBNA3C is capable of activating and repressing transcription when targeted to promoters (3, 28). Additionally, it was shown that EBNA3C also modulated the transactivation activity of EBNA2 on the LMP1 promoter and thus that EBNA3C can be a modulator of transcription activation mediated by ProTα and p300 (35). Therefore we introduced EBNA3C in the transient reporter assay to determine its effects on transcription mediated by ProTα and p300. As expected, EBNA3C down-modulated the increased activity of ProTα and p300 when ProTα was tethered to promoters (Fig. 4). A slightly lower pM-ProTα activity than in the previous experiment was seen in this assay. However, the activity always increased with increasing amounts of ProTα expressed. EBNA3C did not produce any detectable changes in activity when introduced alone in the assay, suggesting that the modulatory activity observed is most likely due to the interaction with ProTα and p300. The levels of protein expression based on the transfected amounts are shown in Fig. 4.
FIG. 4.
Luciferase assay shows that EBNA3C down-modulates the transcription activity mediated by ProTα and p300 in 293 cells. Cells were transfected with pFr, pM-ProTα, and p300, followed by addition of EBNA3C, and harvested at 24 h after transfection. The results show that EBNA3C modulates the increased transcription activity mediated by pM-ProTα and p300. The results are the mean of the relative luciferase units (RLU), normalized for total DNA content and efficiency of transfection, from three independent transfections. Similar amounts of EBNA3C, when transfected alone with the reporter, did not have any detectable effects compared to vector alone in this assay. Western blotting (WB) shows levels of each protein transfected in each lane compared to the equivalent levels of protein in each lane.
ProTα and EBNA3C interact with regions of p300 similar to those of other known cellular and viral oncoproteins.
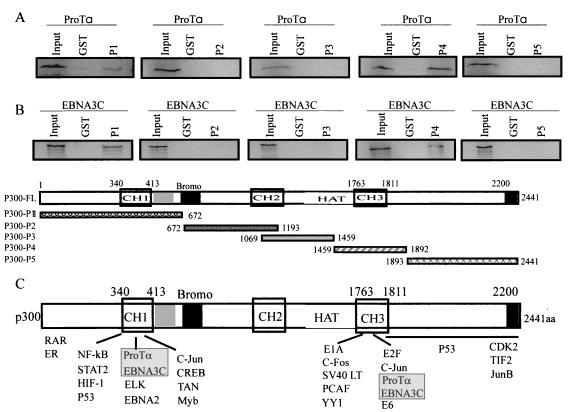
We previously demonstrated that ProTα bound to the regions of p300 encoded by 3′ and 5′ regions of the associated gene but that the affinity for the product of the 5′ region was much less than that for the product of the 3′ region (7). We therefore wanted to map more closely the binding sites for ProTα to determine if the sites were similar to those observed for the known regulators of transcription and cellular oncoproteins. ProTα was translated in vitro and incubated with five GST-p300 fusion proteins mapping to distinct domains, including the CH1, CH2, and CH3 domains; the bromodomain; and the HAT domain. Based on our results we show that ProTα interacts with two distinct regions of p300. The affinity of the observed interactions at the amino terminus of p300 at the CH1 domain was lower than that of those at the CH3/HAT domain downstream from the bromodomain and that of those at the CH2 domains (Fig. 5A). These sites map to similar regions known to bind transcription regulators, including NF-κB, c-myb, CREB, HIF-1, c-Jun, and STAT-2, which are associated with the amino terminus of p300 and the large T antigen of simian virus 40, E1A, and human papillomavirus E6, c-Fos, and E2F, which bind within the CH3 and HAT domains (Fig. 5A and C) (13, 14, 22, 26, 38). These data show that ProTα binds to domains of p300 similarly to those bound by known cellular and viral oncoproteins associated with human malignancies.
FIG. 5.
In vitro binding assay shows that EBNA3C and ProTα interact with p300 at two distinct domains, the CH1 and CH3/HAT domains. S35-labeled in vitro-translated ProTα and EBNA3C were incubated with GST fusions overlapping the entire p300. P1 through P5 were prepared and purified by binding to glutathione-Sepharose beads. Beads bound to the GST fusion proteins were washed and incubated with labeled in vitro-translated products. (A) Results of the binding assay with S35-labeled ProTα, where binding was seen at the amino terminus and CH3/HAT domain. (B) In vitro-translated S35-labeled EBNA3C similarly bound to the same p300 fusion. (C) Schematic indicating the specific domains of p300 and the known interacting molecules. Note that other cellular and viral transcription regulators interact with p300 at the CH3/HAT domain and at the CH1 domain (22, 38).
Since we also previously demonstrated that ProTα and EBNA3C bound strongly to the 3′ region-encoded end of p300 as well as to the 5′ region-encoded 1,100-aa region (7), we wanted to map more closely the regions involved in binding with EBNA3C to determine if similar regions of p300 were involved in the interactions with ProTα and with EBNA3C.
EBNA3C was translated in vitro with [S35]Cys-Met label from NEN-Dupont and then incubated with the five fusions of GST with p300 described above, which represent the different regions of the p300 molecule. We demonstrate that EBNA3C interacts with similar affinity to a region of p300 downstream of the bromodomain, which includes a portion of the HAT domain and the CH3 region. Additionally, we observed interaction again with the amino-terminal CH1 region of p300. These regions were similar to the regions that bound with ProTα shown above (Fig. 5A and B). Interestingly, EBNA3C and ProTα bind to p300 at the 5′ CH1 domain similarly to EBNA2, NF-κB, STAT, c-myb, and c-Jun (22, 38, 44) and to the CH3/HAT domain of p300, as seen in previous studies with E1A, E6, and c-Jun (Fig. 5) (22, 38).
ProTα and p300 interact with EBNA3C at distinctly separate domains.
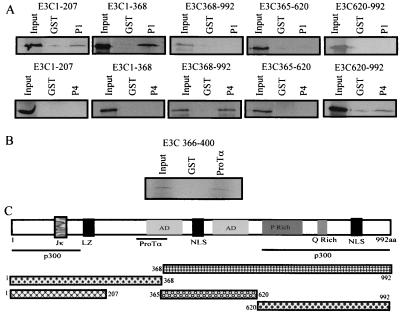
To demonstrate more precisely the domains of EBNA3C interacting with p300, we translated in vitro truncated versions of specific domains of EBNA3C and incubated these truncated polypeptides with GST-p300-1 and with GST-p300-4, which was shown above to interact with the full-length EBNA3C polypeptide. The binding assays were performed as described above. Here we demonstrated that the CH1 domain of p300 interacts with amino-terminal aa 1 to 207 of EBNA3C (Fig. 6A). In addition, the CH3/HAT domain of p300 interacts with the carboxy-terminal 370 aa, a region which includes the proline-rich and glutamine-rich domains of EBNA3C (Fig. 6A and C).
FIG. 6.
The p300 coactivator interacts with EBNA3C at two distinct domains, the amino-terminal 207 aa and the carboxy-terminal 372 aa. ProTα interacts specifically with residues located between aa 366 and 400 of EBNA3C. (A) The S35-labeled in vitro-translated regions of EBNA3C bind to the amino terminus of the p300-P1 fusion protein, whereas the carboxy-terminal region, aa 620 to 992, interacts with the p300-P4 fusion protein, which includes the CH3/HAT domain. (B) Specific interaction of ProTα with the region comprising aa 366 to 400 of EBNA3C. All input lanes are equivalent to 10% of the total used in the experimental lanes. (C) Schematic of the different regions of EBNA3C used in the binding experiments and the interaction domains for p300 and ProTα. NLS, nuclear localization signal.
Our previous work mapped the interacting domain of ProTα with EBNA3C downstream of aa 365 (7). Moreover, fusion proteins downstream of aa 393 did not associate with ProTα. Here we show that a short 34-aa stretch from aa 366 to 400 interacts with ProTα in vitro (Fig. 6B and C). This region lies just downstream of aa 365, which was shown to be critical for EBV-mediated transformation of primary B lymphocytes (40). These results indicate that ProTα and p300 interact with EBNA3C at distinctly different domains.
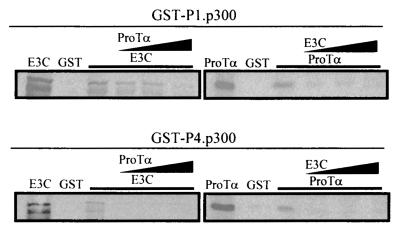
We were curious as to whether or not the interactions of EBNA3C and ProTα with p300 were independent or if EBNA3C and ProTα can compete for binding with the specific domains as shown in the above binding experiments, since previous experiments suggest that they may be in a complex with p300. We show that ProTα can possibly compete with EBNA3C for binding to the P4 region of p300 more efficiently than for binding to the P1 amino-terminal region of p300. EBNA3C, however, competes with ProTα for binding to p300 at the amino-terminal CH1 domain and P4 domain, which includes the CH3/HAT region, with similar efficiencies (Fig. 7). EBNA3C appears to be able to compete with greater efficiency than ProTα (Fig. 7, left), suggesting that EBNA3C may have a higher affinity for p300.
FIG. 7.
ProTα and EBNA3C compete for binding to p300 at the amino-terminal and CH3/HAT domains. Increasing amounts of ProTα and EBNA3C were added to the reaction mixture 1 h after allowing the GST-p300 fusion protein to bind in vitro. (Top) Effect of competition with the amino-terminal region; (bottom) competition with the CH3/HAT domain. EBNA3C and ProTα were translated in vitro and labeled with the S35 translabel for visualization of the protein by autoradiography.
EBNA3C modulates ProTα- and p300-enhanced HAT activity.
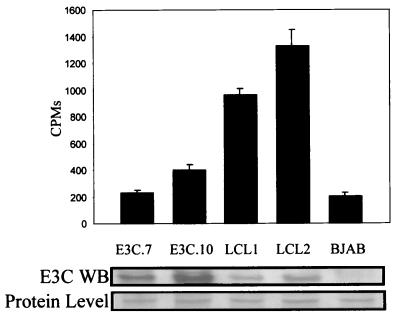
To determine if the transactivation effects seen above correlated with acetylation of nucleosomes, we wanted to determine the levels of acetylation mediated by complexes associated with ProTα. Burkitt's lymphoma cells lines overexpressing EBNA3C, human primary B lymphocytes transformed by EBV expressing the latent EBV nuclear antigens, and an EBNA3C- and EBV-negative cell line (as a control) were used. Cells were harvested and lysed to solubilize the nuclear proteins. Complexes associated with ProTα were immunoprecipitated with a polyclonal antibody against ProTα, washed in RIPA buffer and in HAT buffer, and then incubated with crude histones. The results demonstrate that there was a five- to sevenfold-greater level of HAT activity in EBV-infected cells than in cells overexpressing EBNA3C (Fig. 8), suggesting that other factors including other latent nuclear antigens expressed by EBV during establishment of latency may play a role in enhancing the HAT activity associated with ProTα. Little or no increase in HAT activity was seen in the B-cell lines overexpressing only EBNA3C compared to that for the control EBV-negative cell line, which does not express EBNA3C or any other EBV latent antigen (Fig. 8).
FIG. 8.
Histone acetylation assay showing that EBV infection increases the overall acetylation activity associated with cellular molecules which coimmunoprecipitate and form complexes with ProTα in the infected cell. EBNA3C cell lines stably overexpressing the E3C protein are E3C.7 and E3C.10. LCL1 and LCL2 are EBV-infected B-lymphoblastoid cell lines expressing the entire panel of latent EBV nuclear antigens. BJAB is an EBV-negative cell line. Cells were lysed, and complexes associated with ProTα were immunoprecipitated with polyclonal rabbit serum overnight, washed, and used in the HAT assay. The results, total 3H counts, are the averages of three independent assays. Western blotting (WB) shows the expression of EBNA3C in each assay.
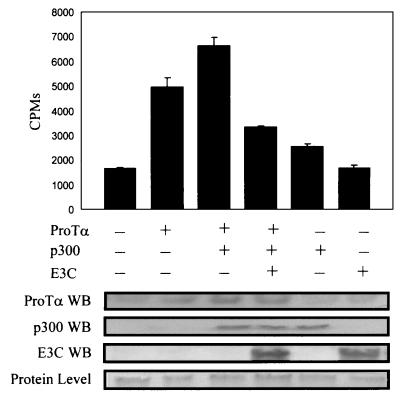
To demonstrate that EBNA3C modulates HAT activity when exogenously expressed in HEK epithelial cells, we transiently transfected HEK 293T cells with expression constructs containing ProTα, ProTα plus p300, and ProTα, p300, and EBNA3C into 15 million cells. The transfected cells were incubated for 24 h, harvested, and washed once in PBS. The cells were then lysed in RIPA buffer, and incubated with an anti-ProTα polyclonal antibody for 4 h. The complexes were collected and incubated with crude histones to determine the acetylation activity of the complex. We show that coimmunoprecipitation of protein complexes with ProTα resulted in activity 3.5-fold greater than that with the vector control alone (Fig. 9). When p300 was transfected under the control of a heterologous promoter, the activity was enhanced to a level 4.6-fold greater than that with the vector (Fig. 9). This enhancement of activity was abolished when EBNA3C was introduced into cells exogenously expressing ProTα and p300 (Fig. 9). These results suggest that EBNA3C can modulate the HAT activity when complexed with ProTα and p300. When p300 alone was transiently transfected, there was an increase in HAT activity, sometimes as much as the increase seen with ProTα, indicating that some level of endogenously expressed ProTα can associate with the exogenously transfected p300, resulting in increased activity. However, it is clear that no further HAT activity is obtained when EBNA3C alone is expressed from a heterologous promoter. These results suggest that modulation of the HAT activity mediated by p300 and ProTα is through association of these cellular complexes with EBNA3C.
FIG. 9.
Histone acetylation assay showing that EBNA3C modulates the activity mediated by ProTα and p300 in transfected HEK 293T cells. Transfected cells were harvested after 24 h posttransfection and lysed. Cleared lysate was immunoprecipitated with a ProTα antibody, washed, and used in the HAT assay. EBNA3C was transfected along with a p300 expression vector and pA3M-ProTα. All transfections were balanced with vector DNA for the total amount of DNA per transfection and normalized for transfection efficiencies. The results are the means of three separate assays. The p300 expression construct and the EBNA3C expression construct were transfected independently in the absence of pA3M-ProTα to determine their effects alone on the endogenous HAT activity associated with ProTα. The levels of protein expressed were evaluated by Western blotting (WB) using specific antibodies against ProTα, p300, and EBNA3C.
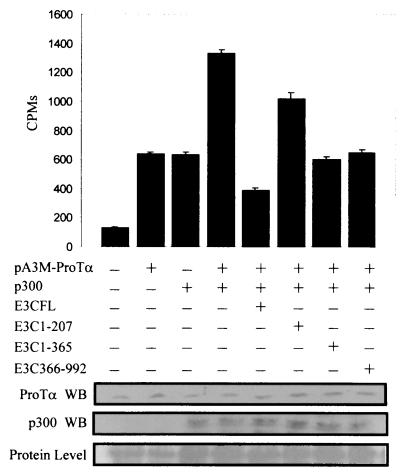
EBNA3C is a large transcription factor, which functions as an activator or a repressor of transcription (37). It was shown that two domains are involved in transcriptional repression and that the glutamine-rich domain can activate responsive promoters when targeted to that promoter (3, 28). In this regard we wanted to test the abilities of different domains of EBNA3C to modulate the HAT-associated activity of ProTα in a complex with EBNA3C. In this assay immunoprecipitation with an anti-ProTα polyclonal antibody demonstrated that as expected ProTα increased the HAT activity approximately sixfold over that with the vector alone and that p300 enhanced this activity when cotransfected with ProTα. Additionally, the full-length EBNA3C molecule again down-modulated this increased effect three- to fourfold. A truncated version of EBNA3C which contains the first 207 aa had little effect on the HAT activity, whereas truncated versions of EBNA3C containing the first 365 aa and the carboxy-terminal 727 aa reduced the effect by more than twofold (Fig. 10). These results indicate that both the amino terminus (which lies between aa 207 and 365) and the carboxy terminus (aa 365 to 992) of EBNA3C contain domains that are involved in modulating HAT activity and can associate with p300 complexed with ProTα in these transfected cells.
FIG. 10.
The amino and carboxy termini of EBNA3C reverse the HAT activity mediated by ProTα and p300. HEK 293T cells were transfected with the first 207 aa of EBNA3C, which contain the RBP-Jκ interacting domain, aa 1 to 365 and 365 to 992 of EBNA3C, along with similar levels of full-length EBNA3C for comparison. All transfections were balanced as described in Materials and Methods for equivalent amounts of DNA. Each truncated EBNA3C construct and full-length EBNA3C were introduced into the reaction mixture with pA3M-ProTα and p300. All transfectants were incubated for 24 h and harvested, and the acetylation complex was immunoprecipitated with a polyclonal antibody against ProTα. Complexes were washed and used in the HAT assay. The results are the means of three independent assays. (Bottom) Levels of p300 and ProTα expressed compared to equivalent protein loading control. WB, Western blot.
DISCUSSION
Previous studies have demonstrated that EBNA3C interacts with ProTα in regulating histone acetylation (7). ProTα was shown to interact and associate with histone H1 in EBV-infected cells (7, 8). Moreover, ProTα also interacted with acetyltransferase p300 in EBV-infected cells, suggesting that these interactions are important for transcriptional regulation and gene expression of the transformed cells. Here we further demonstrate these interactions and more precisely map the binding domains for each interaction. Our results show that ProTα interacts with p300 at two specific domains, a region downstream of the bromodomain, the CH3/HAT domain, and another region at the amino terminus of p300, referred to as the CH1 domain. Additionally, EBNA3C, an essential EBV molecule, interacts with p300 at the same amino-terminal domain interacting with ProTα. These interaction studies clearly indicate that EBNA3C exists in a complex with ProTα and p300 and that this complex may be critical for EBV-mediated regulation of viral and cellular gene expression.
To date the functional activity of ProTα at promoters has not been established. Here we show that ProTα can activate transcription of promoters when targeted to promoters using a GAL4-ProTα fusion on a GAL4-responsive promoter element. This result for the first time demonstrates that ProTα can affect the transcriptional machinery when tethered to promoters. Moreover, ProTα can interact with known transcriptional coactivator p300, which is well established as an effector of transcriptional activation (10, 29). ProTα interacts with histone H1 and removes H1 bound to the nucleosomes, suggesting that it is involved in chromatin remodeling to allow access to transcription and replication factors (8, 15, 23). If histone H1 promotes condensation of the chromatin into higher-order structures that are transcriptionally inactive, then association with ProTα can reverse this process and so allow for the decondensation of the chromatin as histone H1 is stripped from the nucleosomes, which allows for access by the transcriptional machinery. Similarly, this can be a mechanism by which the replication machinery can gain access to the chromatin structure. ProTα then is an antagonist of histone H1 by virtue of its ability to regulate the condensation of the higher-order structure of chromatin associated with histone H1 involved in gene regulation and possibly replication.
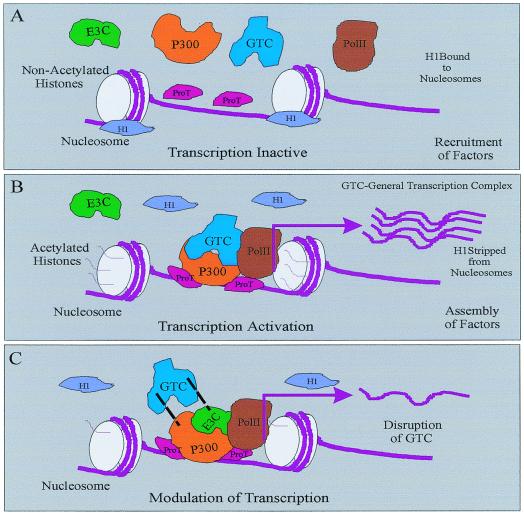
In our studies we also show that ProTα also interacts with transcription coactivator p300 and that this interaction results in the synergistic activation of transcription. The fact that ProTα interacts with p300 suggests that it may be in a complex with other transcription factors and that it may be an essential component of the larger basal transcription complex. We have now mapped the domains interacting with p300 to the amino terminus CH1 and the region downstream of the bromodomain, the CH3/HAT domain. These regions have been the subject of numerous investigations, which have shown that other known transcription factors associate with the CH1 domain and that viral and cellular factors including E6 and E1A associate with the CH3/HAT domain (14, 22, 38). This suggests that EBNA3C and ProTα are involved in regulating functions similar to those regulated by other cellular and viral factors, which include acetylation and transcription activation. Evidence provided in this study indicates that both ProTα and EBNA3C are involved in regulating the acetylation and transcriptional activity of p300. The modulation of these effects by EBNA3C is most likely important for regulation of cellular and viral gene expression required for the transformation process mediated by EBV. Based on this evidence we suggest a potential model whereby ProTα removes histone H1 from the nucleosomes and may be involved in the recruitment of other general transcription factors. ProTα may therefore be involved in the decondensation of the chromatin and the acetylation of core histones as it interacts with p300 at two specific domains complexed with other transcriptional activators including RNA polymerase II. This results in transcription activation. EBNA3C may not be a component of this complex during activation, or maybe it is sequestered by other transcription factors with access to the general transcription complex. However, when EBNA3C is recruited to this complex, the activity mediated by p300 and the general transcription activation complex is modulated by displacement of specific components and/or modification of the large transcription activation complex (Fig. 11).
FIG. 11.
Model showing EBNA3C regulation of transcription and HAT activity associated with p300 and ProTα. ProTα removes histone H1 from the nucleosomes, allowing access for transcription and coactivation complexes, which are functionally similar to some HMG molecules (8, 9, 19, 23). ProTα may then recruit and stabilize general transcription complexes and associates with p300 at two distinct contact points at the amino-terminal CH1 domain and at the CH3/HAT domain. The interaction with the HAT domain may be involved in regulation of HAT activity, as the core histones are acetylated and also cooperate with a number of transcription factors in a large general transcription complex. EBNA3C modulates this transcription activity by displacing and modifying the general transcription complex as the activity of p300 and ProTα is regulated. Overall, ProTα interacts with histone H1, decondensing the nucleosomes as the required transcription regulatory factors are recruited. The resulting activity is activation of transcription, which is modulated as EBNA3C is introduced into the complex.
Chromatin remodeling is a critical aspect of gene regulation (22, 27). ProTα is involved in the remodeling of chromosomes, is expressed ubiquitously in numerous cell types, and is up-regulated when the cell is undergoing oncogenesis induced by a known cellular oncogene (11, 30). Here we show that ProTα and EBNA3C interact specifically with p300 at regions identical to those at which E1A and E6, as well as c-Jun and E2F, all regulators of transcription, interact (22, 38). The fact that the interaction domains are the same suggests a role for ProTα in transcription regulation, possibly in recruitment and stabilization of complexes involved in activation or repression as they are brought to nucleosomes. EBNA3C, then, through its interaction with ProTα can regulate the resulting transcriptional activity generated by the activation complex. Importantly, more studies will be required to investigate the role of ProTα in chromatin remodeling and to identify other factors associated with this transcription complex.
Acknowledgments
We thank Fernando Dominguez for the ProTα construct and Gary Nabel for the full-length p300 expression construct. We thank Elliott Kieff for the EBNA3C expression construct.
This work was supported by grants from the Leukemia and Lymphoma Society of America and the National Cancer Institute (grant CA072150 to E.S.R.). E.S.R. is a Leukemia and Lymphoma Society of America Scholar. C.S. is a postdoctoral fellow of the Lady Tata Memorial Trust, London.
REFERENCES
- 1.Allday, M. J., D. H. Crawford, and J. A. Thomas. 1993. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J. Gen. Virol. 74:361-369. [DOI] [PubMed] [Google Scholar]
- 2.Allday, M. J., and P. J. Farrell. 1994. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J. Virol. 68:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain, M., R. J. Watson, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, M., J. M. Guinebretiere, E. Kremmer, V. Grunewald, E. Benhamou, G. Contesso, and I. Joab. 1999. Detection of Epstein-Barr virus in invasive breast cancers. J. Natl. Cancer Inst. 91:1376-1381. [DOI] [PubMed] [Google Scholar]
- 5.Bustin, M. 2001. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci. 26:431-437. [DOI] [PubMed] [Google Scholar]
- 6.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, M. A., II, and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz-Jullien, C., A. Perez-Estevez, G. Covelo, and M. Freire. 1996. Prothymosin alpha binds histones in vitro and shows activity in nucleosome assembly assay. Biochim. Biophys. Acta 1296:219-227. [DOI] [PubMed] [Google Scholar]
- 9.Ding, H. F., M. Bustin, and U. Hansen. 1997. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol. Cell. Biol. 17:5843-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckner, R. 1996. p300 and CBP as transcriptional regulators and targets of oncogenic events. Biol. Chem. 377:685-688. [PubMed] [Google Scholar]
- 11.Eilers, M., S. Schirm, and J. M. Bishop. 1991. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 10:133-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felzien, L. K., C. Woffendin, M. O. Hottiger, R. A. Subbramanian, E. A. Cohen, and G. J. Nabel. 1998. HIV transcriptional activation by the accessory protein, VPR, is mediated by the p300 co-activator. Proc. Natl. Acad. Sci. USA 95:5281-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Rodriguez, C., and A. Rao. 1998. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J. Exp. Med. 187:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giles, R. H., D. J. Peters, and M. H. Breuning. 1998. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 14:178-183. [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Marquez, J., and P. Rodriguez. 1998. Prothymosin alpha is a chromatin-remodelling protein in mammalian cells. Biochem. J. 333:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez-Marquez, J., and F. Segade. 1988. Prothymosin alpha is a nuclear protein. FEBS Lett. 226:217-219. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Marquez, J., F. Segade, M. Dosil, J. G. Pichel, X. R. Bustelo, and M. Freire. 1989. The expression of prothymosin alpha gene in T lymphocytes and leukemic lymphoid cells is tied to lymphocyte proliferation. J. Biol. Chem. 264:8451-8454. [PubMed] [Google Scholar]
- 18.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrera, J. E., K. L. West, R. L. Schiltz, Y. Nakatani, and M. Bustin. 2000. Histone H1 is a specific repressor of core histone acetylation in chromatin. Mol. Cell. Biol. 20:523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hottiger, M. O., and G. J. Nabel. 1998. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J. Virol. 72:8252-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai, S., J. Nishikawa, and K. Takada. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 72:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobson, S., and L. Pillus. 1999. Modifying chromatin and concepts of cancer. Curr. Opin. Genet. Dev. 9:175-184. [DOI] [PubMed] [Google Scholar]
- 23.Karetsou, Z., R. Sandaltzopoulos, M. Frangou-Lazaridis, C. Y. Lai, O. Tsolas, P. B. Becker, and T. Papamarcaki. 1998. Prothymosin alpha modulates the interaction of histone H1 with chromatin. Nucleic Acids Res. 26:3111-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2395. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 25.Ling, P. D., D. R. Rawlins, and S. D. Hayward. 1993. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 90:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, R. X., and D. C. Dean. 1999. Chromatin remodeling and transcriptional regulation. J. Natl. Cancer Inst. 91:1288-1294. [DOI] [PubMed] [Google Scholar]
- 28.Marshall, D., and C. Sample. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 30.Orre, R. S., M. A. Cotter II, C. Subramanian, and E. S. Robertson. 2001. Prothymosin alpha functions as a cellular oncoprotein by inducing transformation of rodent fibroblasts in vitro. J. Biol. Chem. 276:1794-1799. [DOI] [PubMed] [Google Scholar]
- 31.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rickinson, A., and E. Kieff 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 33.Robertson, E. S. 1997. The Epstein-Barr virus EBNA3 protein family as regulators of transcription. Epstein-Barr Virus Rep. 4:143-150. [Google Scholar]
- 34.Robertson, E. S., and E. Kieff. 1995. Genetic analysis of Epstein-Barr virus in B lymphocytes. Epstein-Barr Virus Rep. 2:73-80. [Google Scholar]
- 35.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson, E. S., J. Lin, and E. Kieff. 1996. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J. Virol. 70:3068-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sample, J., L. Young, B. Martin, T. Chatman, E. Kieff, and A. Rickinson. 1990. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 64:4084-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shikama, N., J. Lyon, and N. LaThangue. 1997. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 7:230-235. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian, C., M. A. Cotter II, and E. S. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat. Med. 7:350-355. [DOI] [PubMed] [Google Scholar]
- 40.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vareli, K., M. Frangou-Lazaridis, I. van der Kraan, O. Tsolas, and R. van Driel. 2000. Nuclear distribution of prothymosin alpha and parathymosin: evidence that prothymosin alpha is associated with RNA synthesis processing and parathymosin with early DNA replication. Exp. Cell Res. 257:152-161. [DOI] [PubMed] [Google Scholar]
- 42.Vareli, K., O. Tsolas, and M. Frangou-Lazaridis. 1996. Regulation of prothymosin alpha during the cell cycle. Eur. J. Biochem. 238:799-806. [DOI] [PubMed] [Google Scholar]
- 43.Vinson, C. R., P. B. Sigler, and S. L. McKnight. 1989. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 246:911-916. [DOI] [PubMed] [Google Scholar]
- 44.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanai, H., K. Takada, N. Shimizu, Y. Mizugaki, M. Tada, and K. Okita. 1997. Epstein-Barr virus infection in non-carcinomatous gastric epithelium. J. Pathol. 183:293-298. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimber-Strobl, U., L. J. Strobl, C. Meitinger, R. Hinrichs, T. Sakai, T. Furukawa, T. Honjo, and G. W. Bornkamm. 1994. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 13:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]