Abstract
Entry of mammalian reovirus virions into target cells requires proteolytic processing of surface protein σ3. In the virion, σ3 mostly covers the membrane-penetration protein μ1, appearing to keep it in an inactive form and to prevent it from interacting with the cellular membrane until the proper time in infection. The molecular mechanism by which σ3 maintains μ1 in this inactive state and the structural changes that accompany σ3 processing and μ1 activation, however, are not well understood. In this study we characterized the early steps in σ3 processing and determined their effects on μ1 function and particle infectivity. We identified two regions of high protease sensitivity, “hypersensitive” regions located at residues 208 to 214 and 238 to 244, within which all proteases tested selectively cleaved σ3 as an early step in processing. Further processing of σ3 was required for infection, consistent with the fact that the fragments resulting from these early cleavages remained bound to the particles. Reovirus type 1 Lang (T1L), type 3 Dearing (T3D), and T1L × T3D reassortant virions differed in the sites of early σ3 cleavage, with T1L σ3 being cleaved mainly at residues 238 to 244 and T3D σ3 being cleaved mainly at residues 208 to 214. These virions also differed in the rates at which the early cleavages occurred, with cleavage of T1L σ3 occurring faster than cleavage of T3D σ3. Analyses using chimeric and site-directed mutants of recombinant σ3 identified carboxy-proximal residues 344, 347, and 353 as the primary determinants of these strain differences. The spatial relationships between these more carboxy-proximal residues and the hypersensitive regions were discerned from the σ3 crystal structure. The results indicate that proteolytic processing of σ3 during reovirus disassembly is a multistep pathway with a number of molecular determinants.
Many viruses, both enveloped and nonenveloped, require proteolytic processing of surface proteins for initial steps in infection of target cells (12, 14, 20, 22, 28, 31, 36, 37, 39, 55). Mammalian orthoreovirus (reovirus), a nonenveloped virus from the Reoviridae family that has been widely used as a model to study nonenveloped virus entry into cells, has also been shown to require proteolytic processing of a surface protein prior to entry (2, 3, 9, 48, 50). The proteolytic activating steps described for many other viruses involve cleavage of a particular surface protein that switches it to an activated state and enables it to perform a function that was previously masked (20, 39, 55). The proteolytic step required for reovirus entry, in contrast, involves the processing of one surface protein, σ3, to achieve activation of a second protein, μ1 (24). In addition, the proteolytic processing of σ3 is suspected to involve not a single activating cleavage, but rather multiple cleavages that result in substantial degradation of the protein. Given these and other distinct features of the mechanism for proteolytic activation of reovirus particles, further studies to dissect this mechanism are needed.
Reovirus virions are 85-nm particles composed of eight different structural proteins organized in two concentric icosahedral capsids that surround the 10-segmented double-stranded RNA genome (16). The inner capsid particle, or core, contains five proteins, each found in several copies, and mediates the enzymatic activities necessary for viral mRNA synthesis following entry into target cells. The outer capsid contains three additional proteins—σ1, μ1, and σ3—and is important for particle stability and entry. The receptor-binding protein σ1 is a trimeric fiber found only at the icosahedral fivefold axes (19, 49). The majority of the outer capsid is formed by 600 copies each of μ1 and σ3 (16). Upon binding of σ1 to cell receptors, the virion is thought to be internalized by the cell via receptor-mediated endocytosis (8, 50). Acidification of the endocytic vacuole containing the virion particle activates lysosomal proteases that cleave σ3 and more fully expose the underlying μ1 protein (2, 3, 9, 48, 50). The resulting particles resemble the in vitro-generated intermediate subviral particles (ISVPs), whose σ3 surface has been proteolytically removed (7, 26, 42, 45). The exposed μ1 protein in ISVPs is thought to undergo conformational changes that result in interaction with the endosomal or lysosomal membrane and release of a core-like particle into the cytoplasm, where viral replication occurs (11, 23, 34, 41, 51).
Studies with cultured cell lines have shown that σ3 processing is necessary for productive infection (2, 9, 15, 48, 50). When added to the culture medium, NH4Cl (a weak base that raises the lysosomal pH, reducing the activity of acid-dependent proteases) or E-64 (a cysteine-protease-specific inhibitor) completely abrogates infection by virions, but not by ISVPs (2, 10, 15, 50). In addition, reconstitution of the σ3 coat on ISVPs by recoating them with recombinant σ3 completely blocks their capacity to initiate infection in the presence of NH4Cl or E-64 (24). The presence of σ3 on the virion surface is believed to prevent μ1 from undergoing conformational changes needed for membrane penetration and to prevent μ1 from engaging in those changes prematurely. The molecular mechanism by which σ3 maintains μ1 in an inactive state, however, is not well understood. Treatment of mice with inhibitors of alkaline proteases present in the gastrointestinal tract, a primary site of host infection, also blocks processing of the σ3 coat and prevents subsequent interaction with M cells and replication in the intestinal tissue (1, 5, 6). The mechanism by which the presence of σ3 blocks M-cell interaction by reovirus particles is not known but is thought to involve an effect on the receptor-binding activity of σ1.
The 41-kDa σ3 protein folds into a two-lobed, elongated structure, 76 Å in its long dimension (33, 43). The smaller lobe contains a CCHC-coordinated Zn2+ ion, which appears to be important for proper folding (35, 47). The smaller lobe and zinc-finger region also mediate interactions with μ1 and play the key role in anchoring σ3 to the virion surface (33, 43, 47). The larger lobe, which contains the binding site for monoclonal antibody 4F2 (27, 40, 53; J. Jané-Valbuena, unpublished data), protrudes from the virion surface above μ1 and is mostly surrounded by solvent on all four sides. In the virion, σ3 is organized into 60 P3 hexamers (each of these encircling a 42-Å-diameter channel) and 60 P2 tetramers (16, 38), with limited contacts between the σ3 subunits within each of these sets and no contacts between sets. Both cryoelectron microscopy reconstructions of virions and ISVPs (16) and the recently determined crystal structure of soluble μ1-σ3 heterohexamers (33) indicate that the 600 copies of μ1 in virions and ISVPs are organized as 200 trimers, much of the surfaces of which are directly covered by σ3 monomers. The crystal structure shows that the small lobe of each σ3 subunit sits between and interacts with the globular jelly-roll heads of two μ1 subunits within each trimer and is, thus, in position to help to hold the μ1 trimers together. This spanning effect of σ3 may be key to its maintaining μ1 in an inactive state. Given these new observations, questions about the specific sites at which σ3 is cleaved and the extent of σ3 cleavage that is necessary for μ1 activation come into focus.
In this study we analyzed the initial steps in σ3 processing by a variety of different proteases, including the lysosomal cysteine protease cathepsin L (Cat L), which has been suggested to be important for virion disassembly in some cells (3). We found that all of these proteases, as an early step in processing, cleaved σ3 within one or two well-defined and limited sequences of the protein, which we call hypersensitive regions (HSRs). Further processing, however, was found to be necessary for μ1 activation and infection. Comparisons of these early cleavage events in two reovirus strains and their derived reassortant viruses enabled us to identify three carboxy (C)-proximal residues of σ3 as the major determinants of the strain differences in cleavage. The recently determined crystal structures of σ3 (33, 43) allowed us to define the structural relationship between these C-proximal residues and the HSRs. The results indicate that the proteolytic processing of σ3 during reovirus disassembly is a multistep pathway with a number of molecular determinants.
MATERIALS AND METHODS
Cells.
Spinner-adapted L cells were grown in Joklik's modified minimal essential medium (Irvine Scientific Co., Irvine, Calif.) supplemented to contain 2% fetal bovine serum, 2% neonatal bovine serum (HyClone Laboratories, Logan, Utah), 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (Irvine Scientific). Spodoptera frugiperda clone 21 (Sf21) and Trichoplusia ni Tn High Five insect cells (Invitrogen, Carlsbad, Calif.) were grown in TC-100 medium (Gibco BRL, Gaithersburg, Md.) supplemented to contain 10% heat-inactivated fetal bovine serum.
Virions and ISVPs.
Virions of the reoviruses type 1 Lang (T1L) and type 3 Dearing (T3D) were purified by the standard protocol (24) and stored in virion buffer (150 mM NaCl, 10 mM MgCl2, 10 mM Tris; pH 7.5) at 4°C prior to use. ISVPs were obtained and purified as previously described (24). Particle concentrations were determined from the A260 (13).
Recombinant baculoviruses.
The recombinant baculovirus containing the wild-type S4 gene from reovirus T3D was described previously (24). To generate recombinant baculoviruses containing mutated and chimeric forms of that gene, mutagenesis and regional exchanges were performed as described below, and the modified genes were inserted into the pEV/35K/polybsmcr plasmid (32) at the EcoRI site under transcriptional control of the polyhedrin promoter. Recombinant baculoviruses containing the S4 gene were selected and amplified as described for the σ3 gene (21).
rcISVPs.
Purified ISVPs of reovirus T1L were incubated at room temperature for 30 to 45 min with lysate derived from Tn High Five cells or Sf21 cells infected with recombinant baculovirus. Infection and lysate preparation from Tn High Five cells were performed as described previously for Sf21 cells (24). Immediately after incubation, the sample was layered on a preformed CsCl density gradient (1.25 to 1.45 g/cm3 or 1.25 to 1.53 g/cm3) and subjected to centrifugation at 8°C in an SW 60 Ti or SW 41 Ti rotor (Beckman Coulter, Arlington Heights, Ill.) spun at 50,000 or 35,000 rpm, respectively, for at least 2 h. Particles were dialyzed extensively against virion buffer and concentrated using slide-A-lyzers (Pierce Chemical, Rockford, Ill.). Equivalent numbers of particles for cleavage reactions were determined by quantitation of σ3 band intensities after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). To generate recoated ISVPs (rcISVPs) containing [35S]methionine- and [35S]cysteine-labeled σ3, the infected Tn High Five cells were incubated with Grace's media (Gibco BRL) containing 50 mCi of Tran35S-Label (ICN Biomedicals, Irvine, Calif.) for 6 h prior to harvesting. Lysate preparation and generation of rcISVPs were done as described above. Prior to dialysis, particles extracted from the CsCl density gradients were placed on a second CsCl density gradient (1.25 to 1.45 g/cm3) and subjected to centrifugation in an SW 60 Ti or SW 50.1 rotor (Beckman) spun at 45,000 rpm for at least 12 h. Volumes of rcISVPs containing equivalent amounts of [35S]methionine- and [35S]cysteine-labeled protein were determined by quantitation of the σ3 band intensity on SDS-PAGE gels by phosphorimaging.
Proteolytic cleavage reactions.
Purified Na-p-tosyl-l-lysine chloromethyl ketone (TLCK)-treated α-chymotrypsin (CHT), trypsin (TRY), thermolysin (THL), and proteinase K (PRK) were obtained from Sigma Chemical Co. (St. Louis, Mo.); purified endoproteinase lys-C (EKC) and purified Cat L were obtained from Calbiochem (San Diego, Calif.). All reactions were performed in virion buffer with the exception of the Cat L reactions, which were performed in 400 mM sodium acetate, 4 mM EDTA, 8 mM dithiothreitol (DTT) (pH 5.5). Particle numbers, protease concentrations, temperatures, incubation times, and reaction volumes are specified in the figure legends. Reactions were stopped by removing samples to 4°C and adding inhibitors to the following final concentrations: CHT, TRY, and PRK, 1 mM phenylmethylsulfonyl fluoride; EKC, 2 mM TLCK; THL, 20 mM EDTA; and Cat L, 100 mM Tris (pH 7.5).
SDS-PAGE and immunoblotting.
SDS-PAGE sample preparation and protein visualization were performed as described previously (24). Acrylamide gels (13.5%) were used to visualize σ3-derived proteolytic fragments. For immunoblotting, protein samples were subjected to SDS-PAGE and transferred to nitrocellulose at 4°C for 30 to 40 min at 80 V in transfer buffer (25 mM Tris, 192 mM glycine; pH 8.3) using the Bio-Rad mini-transfer apparatus (Bio-Rad Laboratories, Hercules, Calif.). Rabbit- and rat-derived σ3-specific antibodies (see below) were used at 1/1,000 and 1/500 dilutions, respectively. Bound antibodies were detected by using a 1/3,000 dilution of alkaline phosphatase-coupled goat anti-rabbit immunoglobulin (Bio-Rad) or a 1/5,000 dilution of alkaline phosphatase-coupled goat anti-rat immunoglobulin (Pierce), with 300 μg of p-nitroblue tetrazolium chloride (Bio-Rad) and 150 μg of 5-bromo-4-chloro-3-indoyl phosphate p-toluidine (Bio-Rad) per ml of substrate buffer (100 mM Tris, 0.5 MgCl2; pH 9.5). Special electrophoresis and immunoblotting conditions were adopted for amino (N)-terminal sequencing (described below).
Antibodies.
Antibodies were raised against glutathione S-transferase (GST)-σ3 fusion proteins. To generate a fusion protein with full-length σ3, a DNA clone of the T1L S4 gene was inserted into the EcoRI site of the vector pGex-4T-2 (Pharmacia Biotech, Piscataway, N.J.), following the GST open reading frame. To generate a fusion protein with only an N-terminal portion of σ3, the pGex-4T-2 vector containing full-length T1L S4 was cleaved with FspI (New England Biolabs, Beverly, Mass.) to eliminate amino acids 105 to 365 but retain amino acids 1 to 104 of σ3 in the fusion protein. To generate a fusion protein with only a C-terminal portion of σ3, we used a pBluescript KS(+) (Stratagene, La Jolla, Calif.) construct containing the T1L S4 gene inserted at the EcoRI site. Oligonucleotide primers were used to amplify the sequence between nucleotide 864 of the T1L S4 gene and the EcoRI site located beyond the 3′ end of that gene by PCR. The upstream primer contained an EcoRI site at its 5′ end. The EcoRI-cleaved PCR product was then inserted into the EcoRI site of the pGex-4T-2 vector (Pharmacia), generating a GST-σ3 fusion protein containing amino acids 289 to 365 of σ3.
All three fusion proteins were expressed in Escherichia coli BL21 cells (Pharmacia) and were purified by first lysing the bacterial cells by sonication in phosphate-buffered saline (PBS; 137 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4; pH 7.5). Inclusion bodies were separated by centrifugation for 20 min at 16,000 × g. GST-σ3 fusion proteins were solubilized by resuspending inclusion body pellets in TE buffer (10 mM Tris, 1 mM EDTA; pH 8.0) with 6 M urea, followed by a 20-min centrifugation at 12,000 × g. The supernatant was subjected to SDS-PAGE using a 10%, 1.5 mM polyacrylamide gel. The gel was stained in cold 0.25 M KCl and 1 mM DTT (EM Science, Gibbstown, N.J.), and bands corresponding to the fusion proteins were excised. Gel slices were placed in dialysis tubing with a 25 mM Tris-192 mM glycine solution. The proteins were electroeluted by submerging the dialysis bags in a solution (25 mM Tris, 192 mM glycine, 0.1% SDS; pH 8.3) in a horizontal electrophoresis gel apparatus run at 10 mA for 6 h. Gel slices were removed from the buffer, and the resulting solution containing the GST-σ3 fusion protein was concentrated against polyethylene glycol (Sigma) and then dialyzed against PBS. One rabbit (σ3 full-length) and two rats (σ3 N terminus and σ3 C terminus) were inoculated with these proteins using standard protocols at the Animal Care Unit of the University of Wisconsin Medical School (Madison, Wis.).
Infectivity experiments.
Endpoint infectivity experiments with 20 and 30 mM NH4Cl and 100 μM E-64 were performed as described previously (24). Infectious titers were determined by using a TRY-overlay plaque assay as described previously (24).
Hemolysis.
The capacity of virions, ISVPs, and CHT-treated virions to lyse red blood cells was determined as described previously (11, 41).
T1L and T3D σ3 chimeras and site-directed mutants.
Chimeras and point mutants were generated using DNA clones of the T1L and T3D S4 genes inserted into pBluescript KS(+) at the EcoRI site. The 35-residue C-terminal chimeras were generated by cleaving the S4-containing pBluescript vectors with Eco0109I (New England Biolabs). This enzyme cleaves within both S4 genes in the position following amino acid 300 and within the vector sequence beyond the 3′ end of the σ3 coding sequence. The chimeras between amino acids 266 and 330 were generated using as template the StyI-EcoRV T1L and T3D S4 chimeras described previously (24) and performing identical Eco0109I cleavages as described above. Site-directed mutants were obtained using the QuikChange protocol (Stratagene), starting with the T3D S4 gene contained in pBluescript KS(+). All chimeras and point mutants were subcloned into the transfer vector pEV/35K/polybsmcr (32) to generate the corresponding recombinant baculoviruses.
N-terminal sequencing.
Discontinuous SDS-polyacrylamide gradient gels (5 to 20% acrylamide) or 13.5% polyacrylamide mini-gels were polymerized using a freshly deionized acrylamide-bisacrylamide solution (Fisher Scientific, Pittsburgh, Pa.). The solution was deionized by incubating with AG 501-8 Mixed Bed Resin (Bio-Rad) for 2 h. Gels were stored at 4°C overnight to ensure complete polymerization. Before utilization, gels were prerun for 10 min at 150 V with 2 mM thioglycolic acid (Fisher Scientific) in the upper buffer (125 mM Tris, 0.1% SDS; pH 6.8). Samples were disrupted by mixing with sample buffer (final concentrations: 125 mM Tris, 10% sucrose, 1% SDS, 2% β-mercaptoethanol, 0.01% bromophenol blue; pH 8.0) and incubating for 30 min at 65°C. After application of the samples, the gels were run at 100 V through the stacking gel and 300 to 350V through the resolving gel. After electrophoresis, the gel was soaked in transfer buffer for at least 15 min, and the proteins were transferred to a presoaked polyvinylidene difluoride membrane (Immobilon-PSQ; Millipore, Bedford, Mass.) at 4°C for 50 min at 80 V in transfer buffer, using the Bio-Rad mini-transfer apparatus. The presoak involved sequential treatments with methanol (3 s), double-distilled water (ddH2O) (3 min), and transfer buffer (at least 10 min). After transfer the membrane was washed in ddH2O for 10 min and stained with freshly prepared Coomassie brilliant blue R-250 (Sigma) for 5 min. Destaining of the membrane was achieved by incubating twice in 50% methanol-ddH2O for 5 min and twice in ddH2O for 5 min. The membrane was air dried and sent for analysis to Midwest Analytical (St. Louis, Mo.).
ESI-mass spectrometry (ESI-MS).
Protein bands were excised from gels and cut into 1-mm3 pieces. The pieces were washed and dehydrated with acetonitrile for 10 min, followed by removal of acetonitrile and drying in a Speed-Vac. Rehydration of the gel pieces was with 50 mM NH4HCO3 solution containing 12.5 ng of modified sequencing-grade TRY (Promega, Madison, Wis.)/μl at 4°C. After 45 min, the excess digest solution was removed and replaced with 50 mM NH4HCO3 solution to cover the gel pieces. Samples were incubated at 37°C overnight. Peptides were then extracted by removing the solution, followed by two 20-min washes with a solution containing 50% acetonitrile and 5% formic acid. The extracts were dried in a Speed-Vac and stored at 4°C until analysis. On the day of analysis the samples were reconstituted in 5 μl of solvent A containing 5% acetonitrile, 0.4% acetic acid, and 0.005% heptafluorobutyric acid. A nanoscale reverse-phase high-pressure liquid chromatography (HPLC) capillary column was created by packing 5-μm spherical C18 silica beads into a fused silica capillary (75 μm [inner diameter] by 12 cm [length]) with a flame-drawn tip (44). After the column was equilibrated, each sample was pressure-loaded off-line onto the column. The column was then reattached to the HPLC system. Peptides were eluted with increasing concentrations of solvent B containing 95% acetonitrile, 0.4% acetic acid, and 0.005% heptafluorobutyric acid. As each peptide eluted, it was subjected to electrospray ionization (ESI) and then entered into an LCQ DECA ion-trap mass spectrometer (ThermoFinnigan, San Jose, Calif.). Eluting peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences were determined by matching the acquired fragmentation pattern with protein databases using the software program Sequest (ThermoFinnigan) (18). Delta and cross-correlation values for the peptides noted in the text were 0.6 and 4.5, 0.7 and 4.3, and 0.6 and 3.4, respectively.
RESULTS
Limited CHT cleavage of T1L virion-bound σ3 yields two major fragments.
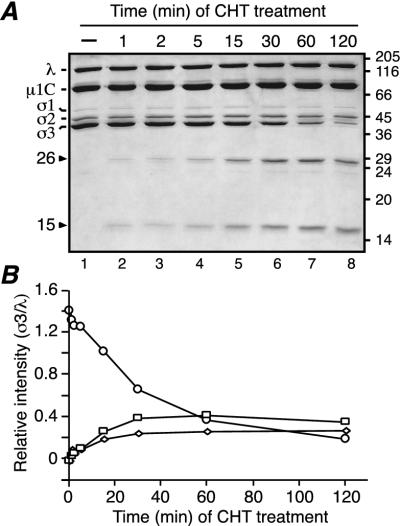
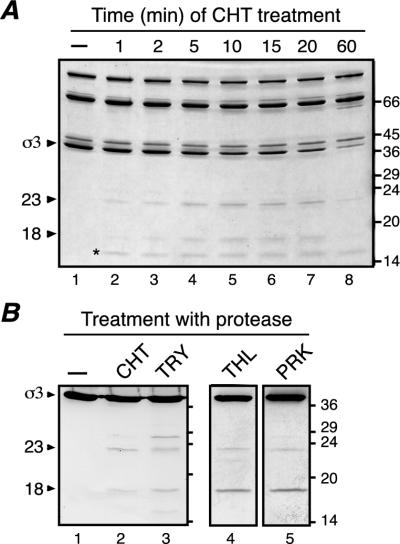
T1L virions were treated with CHT under conditions providing limited cleavage of σ3 (low enzyme concentration and decreased temperature) and were then subjected to SDS-PAGE. CHT was chosen due to its common use as a protease to generate ISVPs in vitro (26), as well as its probable involvement in processing of virions in the gastrointestinal tract of host animals (5, 6). Two major products were detected: a larger fragment of 26,000 Mr (26K) and a smaller fragment of 15,000 Mr (15K) (Fig. 1A, lane 6). Since the combined size of these fragments approaches that of full-length T1L σ3 (calculated mass, 41.2 kDa), they were hypothesized to represent complementary products of the same initial cleavage. In agreement, when T1L virions were analyzed over a time course of limited CHT treatment, both fragment bands appeared at early times and simultaneously (Fig. 1A, lane 2) and accumulated as the intensity of the full-length σ3 band decreased (Fig. 1B). Additional bands of lower intensity, corresponding to peptides of slightly smaller size than the 26K and 15K fragments, were observed at later times (Fig. 1A, lanes 6 to 8). These additional peptides may result from subsequent cleavages near the ends of the 26K and 15K products, or they may also result from early cleavages at alternative sites in full-length T1L σ3 that occur in a smaller proportion of the virion-bound molecules. In contrast to the initial accumulation of the two major σ3 fragments at early time points, at later times of protease treatment the intensity of both fragments decreased (Fig. 1A and B, time points 60 and 120 min), indicating that they were being further processed by CHT. Longer treatment of T1L virions under these conditions yielded ISVPs (data not shown).
FIG. 1.
Limited CHT digestion of T1L virions. (A) A total of 7.5 × 1010 purified T1L virions were treated with 10 μg of CHT/ml for different times at 8°C in a final volume of 20 μl. Reactions were stopped, and the samples were disrupted in sample buffer and analyzed by SDS-PAGE and Coomassie blue staining (lanes 2 to 8). Lane 1 corresponds to an equivalent number of mock-treated purified virions (incubated at 8°C in 20 μl for 1 h in the absence of CHT). Molecular mass markers are indicated at right (in kilodaltons). Reovirus proteins and the two major σ3 fragments are indicated at left. (B) Change over time of the relative band intensity (σ3/λ) of full-length σ3 (circles), the 26K fragment (squares), and the 15K fragment (diamonds).
The two major fragments correspond to N- and C-terminal regions of T1L σ3.
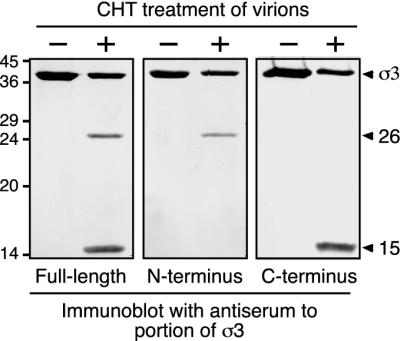
To establish the site of early CHT cleavage in σ3, we used antisera raised against full-length σ3, an N-terminal portion of σ3 (first 100 residues), or a C-terminal portion of σ3 (last 77 residues). The antiserum raised against full-length σ3 recognized full-length T1L σ3 as well as the 26K and 15K fragments, confirming that both fragments arise from σ3 (Fig. 2, left panel). The antiserum raised against the σ3 N terminus recognized full-length σ3 and the 26K fragment but failed to detect the 15K fragment (Fig. 2, middle panel). The antiserum raised against the σ3 C terminus recognized full-length σ3 and the 15K fragment but failed to detect the 26K fragment (Fig. 2, right panel). These results indicate that the 26K fragment represents an N-terminal portion of σ3, whereas the 15K fragment represents a distinct C-terminal portion, consistent with the hypothesis that the two fragments represent complementary products of an early cleavage of full-length T1L σ3.
FIG. 2.
Immunoblots of T1L σ3 fragments. A total of 7.5 × 1010 purified T1L virions were treated with 10 μg of CHT/ml for 1 h at 8°C in a final volume of 20 μl. Reactions were stopped, and the samples were disrupted in sample buffer and subjected to SDS-PAGE and immunoblot analysis. Antisera raised against full-length σ3 (left), the N-terminal 100 amino acids of σ3 (middle), or the C-terminal 77 amino acids of σ3 (right) were used in the immunoblotting. In all panels, the left lane corresponds to untreated virions (−) and the right lane corresponds to CHT-treated virions (+). Arrowheads indicate full-length σ3 and the two major σ3 fragments. Molecular mass markers are indicated at left (in kilodaltons).
To locate the exact site where CHT cleaves T1L σ3 under limited conditions, we identified the N-terminal residues of the 15K C-terminal fragment by Edman degradation. This analysis yielded the sequence Arg-Lys-Glu-Leu-Val (data not shown), which corresponds to residues 239 to 243 of the T1L σ3 sequence and identifies the peptide bond following Tyr238 as the CHT cleavage site. This site is consistent with the known cleavage specificity of CHT (30). Cleavage of T1L σ3 between Tyr238 and Arg239 would generate an N-terminal fragment of 27.2 kDa and a C-terminal fragment of 14.0 kDa in calculated mass, in reasonable agreement with the estimated Mr values of the two fragments. Localization of this site within the tertiary structure of σ3 (33, 43) revealed that it is surface exposed and, thus, potentially accessible to proteases (see Fig. 9 and Discussion).
FIG. 9.
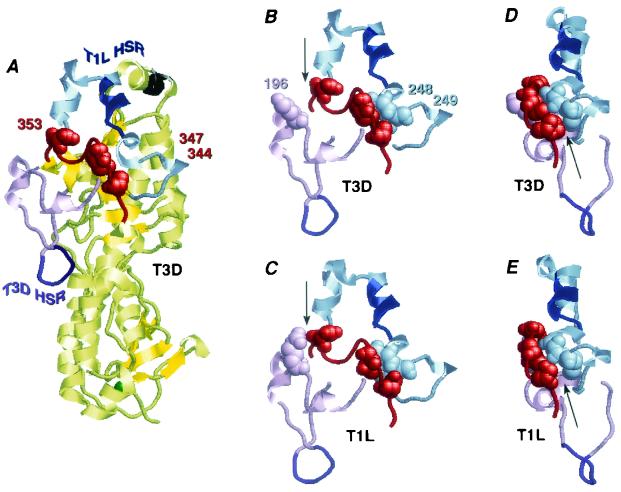
Identification of HSRs and C-proximal determinants of cleavage site and rate in the σ3 crystal structures. (A) The T1L HSR (blue) and flanking regions (light blue), the T3D HSR (purple) and flanking regions (light purple), and the C-proximal random-coil region (red) containing residues 344, 347, and 353 (space-filling format) are indicated in the crystal structure of soluble T3D σ3 (43). The structural zinc ion required for μ1 binding is shown in green at the base of the molecule, and the projecting epitope for monoclonal antibody 4F2 is shown in black at the top. (B to E) Structural context of the T1L HSR, the T3D HSR, and the C-proximal random-coil region, focusing on contacts between residues 344 and 249 (arrows in panels D and E), 347 and 248, and 353 and 196 (arrows in panels B and C) (space-filling format). Color coding is the same as for panel A. Portions of the crystal structures of soluble T3D σ3 (B and D) and T1L σ3 from the soluble T1L μ1-σ3 complex (33) (C) are shown from the same viewing perspectives. The perspective in panels B and C is the same as that in panel A. The view in panels D and E has been rotated to highlight the presence or absence of the interaction between residues 344 and 249.
To address whether, under the limited conditions, CHT had cleaved at other sites near the N or C terminus of the 26K fragment or near the C terminus of the 15K fragment, we excised the two gel bands and submitted them for in-gel TRY digestion and analysis by ESI-MS. Clearly identified were the N-terminal peptide acetyl-MEVCLPNGHQIVDLINNAFEGR from the 26K fragment and the C-terminal peptide FGDLDYPVMIGDPMILG from the 15K fragment. These findings provide evidence that, under the limited conditions, CHT had not cleaved near the N or C terminus of virion-bound σ3. The most C-terminal peptide clearly identified from the 26K fragment was SSLEWGVMVSELEHDPSK, ending at residue 234 and, thus, not accounting for residues 235 to 238 (GRAY). The missing region (235 to 238) contains an Arg residue and was thus likely cleaved by TRY in the excised band to generate the dipeptides GR and AY, which are too small to be identified by ESI-MS. However, since the missing region of residues 235 to 238 does not include a site for CHT cleavage, the findings provide evidence that under the limited conditions CHT had not cleaved near the C terminus of the 26K fragment. In sum, the data from ESI-MS suggest that a single endolytic cleavage after Tyr238 of virion-bound T1L σ3 was mediated by CHT to generate the 27-kDa N-terminal and 14-kDa C-terminal fragments.
The complementary N- and C-terminal fragments of T1L σ3 remain bound to virus particles.
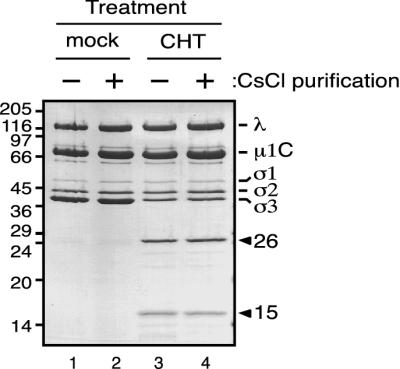
Since the 26K and 15K fragments are early products of a cleavage pathway that ultimately results in removal of σ3 from the particle surface, we investigated whether this early cleavage might alone release one or both fragments into solution. To determine if either the 26K or the 15K fragment was lost from the particle, purified T1L virions were treated with CHT under limited cleavage conditions (with approximately 70% of their full-length σ3 cleaved; Fig. 3, lane 3) and purified on a CsCl density gradient. After centrifugation, we observed a particle band equivalent in appearance to that of mock-treated virions (data not shown). SDS-PAGE analysis of the banded, CHT-treated particles revealed the presence of both the 26K N-terminal and the 15K C-terminal fragments of σ3 (Fig. 3, lane 4), indicating that both fragments remained particle associated. Moreover, quantitative measurements of the protein bands indicated that both major fragments were present in the same amounts before and after CsCl purification (data not shown; Fig. 3, lanes 3 and 4). Thus, CHT cleavage of T1L virion-bound σ3 between Tyr238 and Arg239 is not sufficient for disassociation of either resulting fragment from the particle. This continued particle association of both fragments is consistent with the tertiary structure of σ3 (43), which shows that the CHT cleavage site is not in a hinge region between domains that might allow one fragment to dissociate easily from the other upon cleavage (see Fig. 9 and Discussion).
FIG. 3.
CsCl purification of CHT-treated virions. A total of 7.5 × 1012 virions were digested with CHT at 10 μg/ml (CHT) or 0 μg/ml (mock) for 2 h at 8°C in a final volume of 700 μl. After the reactions were stopped, 50 μl of each mixture was removed and frozen immediately. The rest of each reaction mixture was placed on a 4.2-ml preformed CsCl gradient (1.25 to 1.53 g/cm3) and spun at 50,000 rpm for 22 h at 7°C in an SW 50.1 rotor. The purified particles were dialyzed extensively against virion buffer. Results shown are from 7.5 × 1010 mock-treated virions (lanes 1 and 2) and CHT-treated virions (lanes 3 and 4), before (−, lanes 1 and 3) and after (+, lanes 2 and 4) the CsCl gradient purification, after which samples were disrupted in sample buffer and subjected to SDS-PAGE and Coomassie blue staining. Molecular mass markers are indicated at left (in kilodaltons). Reovirus proteins and the two major σ3 fragments are indicated at right.
Limited cleavage of T1L virion-bound σ3 into its N- and C-terminal fragments is not sufficient for infection of L cells.
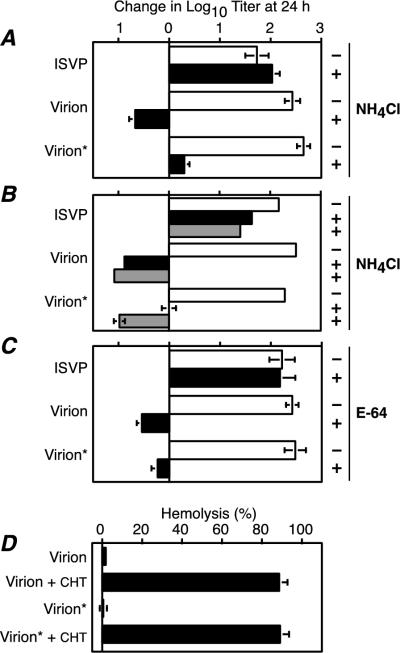
For infection of cells by reovirus virions, σ3 must be proteolytically processed (2, 3, 10, 50). This requirement for σ3 proteolysis is thought to reflect its blocking effect on the function of the underlying μ1 protein in viral penetration of the cellular membrane barrier (24). The extent to which σ3 must be processed, however, has not been determined. Although the 26K N-terminal and 15K C-terminal fragments generated by limited CHT cleavage of T1L virion-bound σ3 remain bound to the particle, it was possible that this early cleavage alone is sufficient to relieve the μ1 block. To test whether T1L virions subjected to limited CHT cleavage can infect L cells without further cleavage of σ3, L cells were infected with different types of viral particles in the presence or the absence of 20 mM NH4Cl or 100 μM E-64 (inhibitor concentrations which were previously shown to block infection by virions, but not ISVPs [2, 24, 50]). The types of particles used were purified T1L virions, T1L virions subjected to limited CHT cleavage to generate the 26K and 15K σ3 fragments (Fig. 3, lane 4; designated as “virion*s” in the figure and following text), and T1L ISVPs lacking σ3. Whereas infection by ISVPs was unaffected by the presence of inhibitor, infection by virion*s, like that by intact virions, was greatly diminished when either NH4Cl or E-64 was present (Fig. 4A and C). These results indicate that further processing of the particle-bound, CHT-generated, N-terminal and C-terminal fragments of σ3 is required for infection.
FIG. 4.
Viral growth in the presence of NH4Cl or E-64. (A to C) L cells were infected with ISVPs, purified T1L virions, or purified T1L virions containing partially cleaved σ3 (virion*) at 3 PFU/cell. After attachment for 1 h at 4°C, infected cells were incubated at 37°C for 0 to 24 h in the absence (−, open bars) or presence (+, solid bars) of 20 mM NH4Cl (A), 20 mM NH4Cl (black bars), or 30 mM NH4Cl (gray bars) (B), or 100 μM E-64 (C). Infectious titers were determined by plaque assay. The change in log10 titer after 24 h is expressed relative to the corresponding 0-h sample. Each bar represents the average of three independent infections, except for ISVPs and virions in panel B, where results are the average of two independent infections. (D) Hemolytic activity of viral particles. A 3% suspension of calf erythrocytes was incubated with purified virions, purified T1L virions containing partially cleaved σ3 (virion*), or ISVPs generated by CHT digestion of each (virion + CHT and virion* + CHT, respectively) at 37°C for 45 min in the presence of 200 mM CsCl. The extent of hemolysis is expressed as a percentage of that obtained by hypotonic lysis. Each bar represents the average of three independent incubations.
Interestingly, infection by virion*s was as completely inhibited as infection by intact virions in the presence of 100 μM E-64, but not in the presence of 20 mM NH4Cl. The finding with E-64 showed that ISVP-like particles were not present among the virion*s, which could have explained the limited bypass of NH4Cl inhibition (6). Instead, we hypothesized that the σ3 processing in virion*s was sufficient to have shifted the concentration dependence of these particles for sensitivity to NH4Cl. At 20 mM NH4Cl, enough lysosomal protease activity may be retained to allow enough further σ3 processing in a small subpopulation of virion*s for infection to proceed. We reasoned that if this hypothesis is correct, infections with virion*s in the presence of higher concentrations of NH4Cl should not show the limited bypass of inhibition observed with 20 mM NH4Cl. When infections were performed as above but with 30 mM NH4Cl, infection by virion*s was indeed inhibited to an equivalent level as intact virions (Fig. 4B). These results thus concur with the hypothesis that further processing of the 26K and 15K σ3 fragments is required for virion*s to initiate productive infection.
The failure of virion*s to mediate infection in the presence of NH4Cl or E-64 is probably due to the continued effect of bound σ3 fragments on the capacity of μ1 to promote membrane penetration (24). To investigate the effect of σ3 fragments on μ1 function in vitro, virion*s were examined for their capacity to lyse red blood cells. Purified T1L virions, virion*s, and ISVPs generated from each were incubated with red blood cells in the presence of CsCl, which has been shown to accelerate membrane interaction by ISVPs (11, 51). We found that virion*s, like intact virions, had no detectable capacity to lyse red blood cells (Fig. 4D), supporting the hypothesis that the function of μ1 is still blocked by the σ3 fragments that remain bound to the particle.
Cleavage of T1L virion-bound σ3 by other proteases generates fragments similar to those generated by CHT.
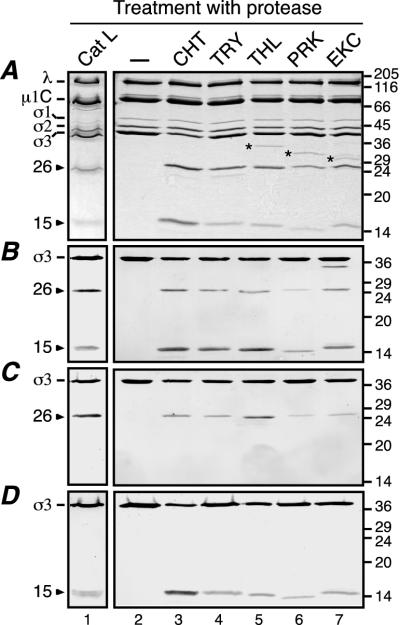
Given that CHT processing of T1L virion-bound σ3 includes an early cleavage that produces two major particle-bound fragments, we examined whether other proteases may cleave σ3 in a similar fashion. Several alkaline proteases were tested, as well as the acidic protease Cat L, a lysosomal protease suggested to be key for σ3 processing in some cells (2, 3, 29, 41). Purified T1L virions were treated under conditions that promote limited cleavage of σ3 and were then analyzed by SDS-PAGE (Fig. 5A) and immunoblot analysis using the antisera raised against full-length σ3 (Fig. 5B), the σ3 N terminus (Fig. 5C), or the σ3 C terminus (Fig. 5D). The results revealed that in all cases a larger fragment corresponding to an N-terminal portion of σ3 and a smaller fragment corresponding to a C-terminal portion of σ3 were produced. The Mr values of the N- and C-terminal fragments produced by the different proteases were very similar—25,000 to 27,000 and 13,000 to 15,000, respectively—suggesting that each of these proteases mediates an early cleavage within the same region of σ3 sequence.
FIG. 5.
Cleavage of T1L virions with different proteases. A total of 1.5 × 1012 purified T1L virions in a final volume of 750 μl were treated with Cat L (127 μU/μl for 1 h at 37°C). After the reaction was stopped, the particles were purified on a CsCl gradient as described for Fig. 3 and dialyzed extensively against virion buffer. A total of 1011 of the purified, Cat L-treated particles were disrupted in sample buffer for analysis by gel (lane 1). A total of 7.5 × 1010 purified T1L virions in a final volume of 20 μl were either left untreated (lane 2) or treated with CHT (10 μg/ml for 1 h at 8°C; lane 3), TRY (5 μg/ml for 10 min at 4°C; lane 4), THL (10 μg/ml for 1 h at 8°C; lane 5), PRK (10 μg/ml for 1 h at 15°C; lane 6), or EKC (0.3 U/ml for 1 h at 37°C; lane 7). After the reactions were stopped, the samples were disrupted in sample buffer. All samples were subjected to SDS-PAGE and Coomassie blue staining (A), or SDS-PAGE and immunoblot analysis using the antisera raised against full-length σ3 (B), the σ3 N terminus (C), or the σ3 C terminus (D). Molecular mass markers are indicated at right (in kilodaltons). Reovirus proteins and the two major σ3 fragments are indicated at left. Asterisks denote protease bands.
To determine the exact sites in σ3 at which several of the proteases cleave, we performed Edman degradation on the C-terminal fragments. The sequences obtained were Leu-Val-Thr-Pro-Ala (residues 242 to 246) and Glu-Leu-Val-Thr-Pro (residues 241 to 245) for the C-terminal fragments obtained with THL and EKC, respectively, identifying the peptide bond preceding Leu242 as the THL cleavage site and the peptide bond following Lys240 as the EKC cleavage site. Additional analysis of Cat L-treated particles on SDS-PAGE gels revealed that the C-terminal fragment band was composed of two fragments of similar sizes and amounts (data not shown). Edman degradation was performed on both fragments, and the sequences obtained were Lys-Glu-Leu-Val-Thr (residues 240 to 244) and Thr-Pro-Ala-Arg-Asp (residues 244 to 248), identifying the peptide bonds following Tyr238-Arg239 and Leu242-Val243 as the two Cat L cleavage sites. Each of these sites is consistent with the known cleavage specificities of THL, EKC, and Cat L (4, 25, 30). As suggested by the similar fragment sizes, these other protease cleavage sites are all within five peptide bonds of the CHT cleavage site, that is, within a defined region of the σ3 sequence. T1L virions subjected to limited cleavage with THL, EKC, or Cat L were also purified on CsCl density gradients and were found to contain the major N- and C-terminal σ3 fragments still bound to the particle (for THL and EKC, data are not shown; for Cat L results, see Fig. 5, lane 1). Furthermore, when these particles were used to infect L cells in the presence of 20 mM NH4Cl, their infectivities were greatly reduced (data not shown), suggesting that additional cleavages of σ3 are required for infectivity in each case.
Limited CHT processing of T3D virion-bound σ3 produces two major fragments distinct from those of T1L.
The discovery that several different proteases cleave T1L virion-bound σ3 to generate similar early products led us to examine if this pattern is specific to T1L virions or is shared by other reovirus strains. We therefore analyzed the early steps of CHT cleavage of σ3 in reovirus T3D virions. Purified T3D virions were treated with CHT under limited cleavage conditions for different times. Since cleavage of σ3 in the T3D virions occurred very slowly at 8°C (indicating a difference in the rate of cleavage of T1L and T3D σ3 [see below]), a higher temperature (25°C) was used to allow better visualization of the T3D σ3 fragments. The fragments obtained at 25°C were nevertheless equivalent to those obtained at 8°C (see Fig. 7 and 8). Two major fragments were observed: a larger fragment of 23,000 Mr (23K) and a smaller fragment of 18,000 Mr (18K) (Fig. 6A). The combined size of these fragments approaches that of full-length T3D σ3 (calculated mass, 41.1 kDa). Both fragments appeared simultaneously and at early times, and they accumulated with time as the σ3 band decreased in intensity (Fig. 6A). The levels to which the T3D fragments accumulated, however, were lower than those for the CHT-generated fragments of T1L σ3 (compare with Fig. 1A). Immunoblot analyses using the antisera raised against the σ3 N terminus and the σ3 C terminus demonstrated that the 23K fragment represents an N-terminal portion of T3D σ3, whereas the 18K fragment represents a distinct C-terminal portion (data not shown), consistent with the hypothesis that the two fragments represent complementary products of a single early cleavage of full-length T3D σ3. Also consistent with this hypothesis was the finding that the 23K fragment, but not the 18K fragment, was recognized in immunoblots with monoclonal antibody 4F2, which binds to a region of T3D σ3 containing residues 116 to 119 (27, 40; J. Jané-Valbuena, unpublished data). Edman degradation of the 18K C-terminal fragment yielded the sequence Asp-Ser-Arg-Ser-Ser, which corresponds to residues 211 to 215 of the T3D σ3 sequence and thus identifies the peptide bond following Phe210 as the CHT cleavage site within T3D σ3. Cleavage of T3D σ3 after Phe210 would generate an N-terminal fragment of 23.9 kDa and a C-terminal fragment of 17.2 kDa in calculated mass, in reasonable agreement with the estimated Mr values of the two major fragments. Following CsCl purification of the CHT-treated particles, both fragments remained particle bound (data not shown). In summary, the results indicate that although there is a difference between strains T1L and T3D with regard to the exact region of virion-bound σ3 within which the major early cleavage by CHT occurs, the pattern is similar in that the cleavage occurred within a defined region of sequence in each case. In each case as well, a larger N-terminal fragment and a complementary, smaller C-terminal fragment were generated, and both remained particle bound.
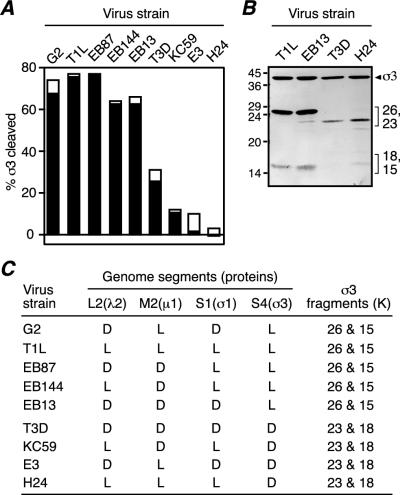
FIG. 7.
CHT treatment of T1L × T3D reassortants. (A) A total of 5 × 1010 purified virions of different T1L × T3D reassortants were either left untreated or treated with 10 μg of CHT/ml at 8°C for 1 h in a final volume of 20 μl. After the reactions were stopped, the samples were disrupted in sample buffer and subjected to SDS-PAGE and Coomassie blue staining. The intensities of the σ3 and λ protein bands of both treated and untreated samples were determined by gel densitometry. The extent of full-length σ3 cleavage after CHT treatment was expressed as {1−[(σ3/λ treated)/(σ3/λ untreated)]} × 100. Black and white superimposed bars represent the results from two cleavage reactions. (B) A total of 5 × 1010 purified virions were treated with 10 μg of CHT/ml in a final volume of 20 μl at 8°C for 30 min (T1L and EB13) or at 25°C for 30 min (T3D and H24). Reactions were stopped, and the samples were disrupted in sample buffer and subjected to SDS-PAGE followed by immunoblot analysis using the antiserum raised against full-length σ3. (C) Genetic origin of the outer-capsid proteins of the T1L × T3D reassortants shown in panel A and size of the σ3 fragments observed by SDS-PAGE after CHT treatment.
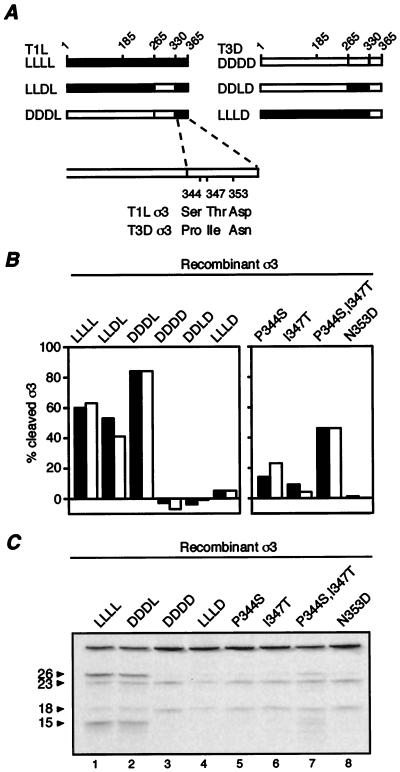
FIG. 8.
Identification of sequence determinants of the CHT cleavage rate difference between T1L and T3D σ3. (A) Schematic representation of the six recombinant σ3 proteins tested in panels B and C: T1L σ3 (LLLL), T3D σ3 (DDDD), and four chimeras generated by exchanging residues 265 to 330 or 331 to 365 between T1L and T3D σ3 (the origin of the regions of these chimeras are indicated by the letters L or D). Residue 185 indicates the limit of N-terminal (residues 1 to 185) and middle (residues 186 to 265) fragments exchanged in chimeras previously tested (24). Also indicated are the three sequence differences between the T1L and T3D σ3 proteins within their C-terminal 35 amino acids. T3D σ3 recombinant proteins containing single substitutions to the T1L residues at positions 344, 347, or 353, or a double substitution to the T1L residues at positions 344 and 347 were tested in panels B and C. (B) Purified rcISVPs were mock-treated or treated with 10 μg of CHT/ml at 8°C for 1 h. Reactions were stopped, and the samples were disrupted in sample buffer and subjected to SDS-PAGE and Coomassie blue staining. The percentage of full-length σ3 cleavage was determined as described for Fig. 7. The filled and open bars indicate measurements from two reactions. (C) Purified rcISVPs containing [35S]methionine- and [35S]cysteine-labeled recombinant σ3 proteins were treated with 10 μg of CHT/ml at 8°C for 15 min. Reactions were stopped, and the samples were disrupted in sample buffer and subjected to SDS-PAGE and phosphorimaging.
FIG. 6.
Protease treatments of T3D virions. (A) A total of 1011 purified T3D virions were treated with 10 μg of CHT/ml for different times at 25°C in a final volume of 35 μl. After the reactions were stopped, the samples were disrupted in sample buffer and subjected to SDS-PAGE and Coomassie blue staining. Molecular mass markers are indicated at right (in kilodaltons). Reovirus proteins and the two major σ3 fragments are indicated at left. The asterisk denotes the protease band. (B) A total of 1011 purified T3D virions in a final volume of 20 μl were left untreated (lane 1) or were treated with CHT (10 μg/ml for 30 min at 8°C; lane 2), TRY (5 μg/ml for 2 min at 8°C; lane 3), THL (10 μg/ml for 30 min at 25°C; lane 4), or PRK (10 μg/ml for 30 min at 25°C; lane 5). Reactions were stopped, and the samples were disrupted in sample buffer and subjected to SDS-PAGE and immunoblot analysis using the antiserum raised against full-length σ3. Molecular mass markers are indicated at right (in kilodaltons). Arrowheads indicate full-length σ3 and the two major σ3 fragments.
Cleavage of T3D virion-bound σ3 by other proteases generates fragments similar to those generated by CHT.
To address whether early cleavages of T3D virion-bound σ3 occur within the region near Phe210, we treated T3D virions with different proteases to observe the sizes of the early fragments. Purified T3D virions were treated with TRY, THL, or PRK and subjected to SDS-PAGE followed by immunoblot analysis using the antiserum raised against full-length σ3 (Fig. 6B). Cleavage with THL or PRK (Fig. 6B, lanes 4 and 5) generated two major fragments of complementary size which migrated similarly to the 23K and 18K fragments obtained with CHT (Fig. 6B, lane 2). Immunoblot analyses using the σ3 terminus-specific antisera identified the 23K fragment obtained with each protease as the N-terminal portion of σ3 and the 18K fragment as the C-terminal portion (data not shown). Edman degradation of the 18K C-terminal fragment obtained with THL yielded the sequence Phe-Asp-Ser-Arg-Ser, which corresponds to residues 210 to 214 of the T3D σ3 sequence and thus identifies the peptide bond preceding Phe210 as the early THL cleavage site in T3D σ3. Attempts to detect the early fragments of T3D σ3 resulting from Cat L cleavage at acidic pH were unsuccessful.
Limited TRY treatment of purified T3D virions (Fig. 6B, lane 3) generated two larger fragments, 26K and 23K, and two smaller fragments, 18K and 15K. The 26K and 23K fragments were detected by the antiserum raised against the σ3 N terminus and the monoclonal antibody 4F2, and the 18K and 15K fragments were detected by the antiserum raised against the σ3 C terminus (data not shown). The 15K fragment band was present at much lower intensity than the 18K fragment band and could not be further analyzed. Edman degradation of the 18K C-terminal band revealed that it contained two components present in equal amounts, one with the N-terminal sequence Tyr-Phe-Asp-Ser-Arg (residues 209 to 213) and one with the N-terminal sequence Ser-Ser-Leu-Glu-Trp (residues 214 to 218). These sequences identify the peptide bonds following Arg208 and Arg213 as early TRY cleavage sites that generate two C-terminal fragments of 17.3 kDa and 16.7 kDa in molecular mass, respectively. These sites are consistent with the known cleavage specificity of TRY (30). Notably, the additional presence of 26K and 15K fragments following limited TRY treatment of T3D virions (Fig. 6B, lane 3) suggests that, under these conditions, TRY also cleaves T3D σ3 within the 238-242 HSR identified in T1L σ3. The 26K and 15K fragments obtained from TRY cleavage of T3D σ3 were found to comigrate with those obtained from CHT cleavage of T1L σ3 (data not shown). The very low intensity of the 15K fragment band may indicate that this T3D σ3-derived fragment is unstable in the presence of TRY.
In summary, the results with T3D virions indicate that, as with T1L virions, the early stages of σ3 degradation involve cleavage within a defined HSR. In the case of T3D, this region comprises residues 208 to 214, which is separated by about 25 residues from that in T1L. The results with TRY cleavage of σ3 in T3D virions, however, suggest that the 238-244 HSR in T3D σ3 can also be subject to early cleavage.
The S4 genome segment determines the differences in site and rate of initial CHT cleavage of T1L and T3D virion-bound σ3.
The preceding results demonstrate a difference between T1L and T3D virions with regard to the region of σ3 in which early cleavages occur. In addition to this difference in cleavage sites, T1L and T3D virions differ in the rates at which σ3 is cleaved. When purified T1L and T3D virions were treated with CHT under limited cleavage conditions (8°C) for 1 h, a clear difference in the rate of conversion of full-length σ3 into its N- and C-terminal fragments was seen between the two strains. Densitometric analyses indicated that, while 76% of full-length σ3 was cleaved in T1L virions, only 28% was cleaved in T3D virions (Fig. 7A). Similar results were obtained with other alkaline proteases under conditions that allowed limited cleavage of virion-bound σ3; that is, T1L σ3 was cleaved at a much higher rate than T3D σ3 (data not shown).
Although the observed differences in site and rate of σ3 cleavage between the T1L and T3D proteins were most likely based within the two σ3 proteins, it was conceivable that they were determined by another protein such as μ1, with which σ3 interacts in virions. To determine which genome segment is responsible for the differences in σ3 cleavage between T1L and T3D virions, purified virions from a panel of T1L × T3D reassortants (Fig. 7C) were treated with CHT. The extent of σ3 cleavage was then determined following SDS-PAGE, and the sites of σ3 cleavage were inferred from the sizes of the gel-resolved fragments. The S4 genome segment, which encodes σ3, was found to segregate with both cleavage phenotypes. Virion-bound σ3 from all reassortants containing a T1L S4 was cleaved at a higher rate than virion-bound σ3 from all reassortants containing a T3D S4 (Fig. 7A). Equivalent results were obtained with EKC (data not shown). SDS-PAGE demonstrated that the virion-bound σ3 from all reassortants containing a T1L S4 was cleaved to yield 26K and 15K fragments, whereas virion-bound σ3 from all reassortants containing a T3D S4 was cleaved to yield 23K and 18K fragments (Fig. 7C). Thus, the major determinants of both differences in σ3 cleavage phenotype between T1L and T3D virions are likely contained within the σ3 protein itself.
Residues within the C-terminal 35 amino acids of σ3 determine rate and site of early cleavages of virion-bound σ3.
Comparison of the deduced amino acid sequences of the T1L and T3D σ3 proteins revealed only 11 positions at which the sequences differ: Ile/Val11, Ala/Glu108, Glu/Asp116, Val/Ile180, Arg/Lys196, Glu/Gly198, Asp/Glu300, Ser/Ala301, Ser/Pro344, Thr/Ile347, and Asp/Asn353. To identify which of these residues are responsible for the differences in cleavage phenotype between T1L and T3D σ3, we generated a series of T1L-T3D σ3 chimeras, which we then expressed in insect cells and used to generate rcISVPs (24). In a previous study, we used the same approach to show that the exchange of T1L and T3D σ3 residues 1 to 185 (chimeras DLL and LDD), 186 to 265 (chimeras LDL and DLD), and 266 to 365 (chimeras LLD and DDL) affected the rate of σ3 cleavage by EKC (24). These chimeras revealed that the last 100 amino acids of σ3 (residues 266 to 365) contain the primary determinants of the EKC cleavage rate difference, since the LLD chimera behaved similarly to T3D σ3 and the DDL chimera behaved similarly to T1L σ3 (24). As part of the present study, rcISVPs containing these chimeras were analyzed for limited cleavage by CHT at 8°C. Very similar results were obtained as in the previous study with EKC (data not shown). These results provided additional evidence that the difference in cleavage rate is based in the T1L and T3D σ3 proteins themselves and also showed that the primary determinants of this difference reside within the C-terminal 100 residues of σ3.
Since residues 266 to 365 contain 5 of the 11 sequence differences between T1L and T3D σ3, we generated and tested a new set of chimeras exchanging residues 266 to 330 (LLDL and DDLD) or 331 to 365 (LLLD and DDDL) to separate the effects of some of these differences (Fig. 8A). When rcISVPs containing these new chimeras were tested for limited cleavage by CHT at 8°C, the results revealed that the primary determinants of the CHT cleavage rate difference reside within the C-terminal 35 residues of the σ3 protein sequences (Fig. 8B, left panel). rcISVPs containing recombinant [35S]methionine- and [35S]cysteine-labeled T1L, T3D, DDDL, or LLLD σ3 protein were also treated with CHT under limited conditions (8°C for 15 min) and subjected to SDS-PAGE and phosphorimaging to identify the cleavage fragments. Both T1L (LLLL) and DDDL rcISVPs displayed two major cleavage products similar to the 26K and 15K fragments obtained from CHT treatment of T1L virions (Fig. 8C, lanes 1 and 2). In contrast, both T3D (DDDD) and LLLD rcISVPs displayed two major cleavage products similar to the 23K and 18K fragments obtained from CHT treatment of T3D virions (Fig. 8C, lanes 3 and 4). Interestingly, CHT treatment of T1L and DDDL rcISVPs also produced a low level of the two T3D-like cleavage products. Careful inspection of the cleavage pattern obtained with T3D and LLLD rcISVPs revealed a very low level of a higher-molecular-weight band similar to the 26K band obtained with T1L and DDDL rcISVPs as well. These results suggest that all four recombinant proteins are susceptible to cleavage in both the T1L and the T3D HSRs but that only T1L and DDDL σ3 are predominantly cleaved at the T1L HSR. The intensity of the 23K cleavage fragment obtained after CHT treatment of T1L and DDDL rcISVPs was similar to that of this fragment obtained after CHT treatment of T3D rcISVPs under identical conditions (Fig. 8C, lanes 1, 2, and 3), suggesting that the extent of cleavage within this region in T1L, DDDL, and T3D recombinant σ3 proteins was comparable. In conclusion, these results indicated that the determinants of the predominant CHT cleavage site in σ3 also reside within the C-terminal 35 amino acids of the σ3 protein sequence and suggested that their effect is to favor or disfavor cleavage within the T1L HSR.
The only residues that differ between T1L and T3D σ3 in the C-terminal 35 amino acids are those at positions 344, 347, and 353 (Fig. 8A). To analyze the role of each of these residues in determining the cleavage rate of σ3, we generated a series of mutations in the T3D σ3 protein background, using site-directed mutagenesis to replace the residues at these positions with those of T1L σ3: P344S, I347T, and N353D. A double-substitution mutant, P344S/I347T, was also created. Recombinant baculoviruses were generated for each of the mutations, and after σ3 expression the corresponding rcISVPs were obtained and tested for limited cleavage by CHT at 8°C (Fig. 8B, right panel). The N353D mutation caused no detectable increase in the CHT cleavage rate of the virion-bound protein. The P344S mutation, and to a lesser extent the I347T mutation, caused a small increase in the CHT cleavage rate of the virion-bound protein. Moreover, when combined in the double-substitution mutant P344S/I347T, these two mutations yielded a cleavage rate that was 75% of the cleavage rate of recombinant T1L σ3 and 50% of the cleavage rate of the DDDL chimera. The double mutant thus provided a substantial increase in the cleavage rate, although it did not provide the full extent of enhancement seen with the DDDL chimera. Since the only difference between the DDDL and double-mutant P344S/I347T proteins is the presence of Asn (as in T1L) instead of Asp (as in T3D) at position 353, we conclude that all three of the differences in the C-terminal 35 residues of σ3 contribute to determining the full extent of the cleavage rate difference between T1L and T3D σ3.
rcISVPs containing recombinant [35S]methionine- and [35S]cysteine-labeled P344S, I347T, N353D, or P344S/I347T σ3 proteins were also treated with CHT under limited conditions (8°C for 15 min) and subjected to SDS-PAGE and phosphorimaging to identify the cleavage fragments. All of these mutants displayed the same cleavage fragments as obtained with CHT treatment of T3D (DDDD) σ3 rcISVPs (Fig. 8C, lanes 5 to 8 versus 3, respectively). In addition, the double-substitution mutant P344S/I347T displayed a higher-molecular-weight fragment similar to, but at a lower intensity than, that obtained with CHT treatment of rcISVPs containing recombinant T1L (LLLL) σ3 protein (Fig. 8C, lanes 7 and 1, respectively). Careful inspection of the phosphorimaging results revealed that each of the single-substitution mutants also displayed this higher-molecular-weight fragment, but at a very low intensity similar to that seen with T3D rcISVPs (Fig. 8C, compare lanes 5, 6, and 8 with lane 3). These results suggest that all three sequence differences in the C-terminal 35 residues of σ3 contribute to the cleavage site difference between T1L and T3D σ3.
DISCUSSION
Early cleavage of particle-bound σ3 within HSRs.
Proteolytic processing of surface protein σ3 is required for infection by reovirus virions (2, 10, 50). The constituent steps in this processing have not been dissected, however. To begin to define these steps, we focused on the early stages of in vitro proteolysis of particle-bound σ3. The results demonstrate that all proteases we tested, both alkaline and acidic, cleave σ3 within one or two delimited HSRs as an early step in processing. Although in this study we were unable to detect the early fragments of T3D σ3 resulting from Cat L cleavage at acidic pH, we hypothesize that these fragments were not readily seen because they have very short half-lives and accumulate to only minor levels.
Previous studies have described long-lived fragments of σ3 that arise during processing of the particle-bound protein. Those fragments have not been extensively characterized, but they appear to be related to those described here. In particular, a σ3 fragment of approximately 28,000 Mr was seen after PRK treatment of T1L virions, ISVPs recoated with T1L σ3, and T1L μ1/σ3 complexes (46). Additionally, σ3 fragments were seen to accumulate during CHT treatment of T3D virions in the presence of the σ3-specific monoclonal antibody 10C1 (52). Recently, we have also observed similarly sized σ3 fragments accumulating at early times after reovirus infection of cells under certain conditions (L. A. Schiff et al., unpublished data). Thus, the stepwise processing of σ3 described in this article appears to be characteristic of that phenomenon in several different settings.
The proteases used in the present study have simple recognition determinants for cleavage and thus could have cleaved at numerous positions in σ3 based solely on primary sequence. CHT, for example, preferentially cleaves after aromatic residues not immediately followed by Pro (30). Both T1L and T3D σ3 sequences contain 30 such aromatic residues, yet CHT showed a strong preference for early cleavage after Tyr238 in T1L σ3 and after Phe210 in T3D σ3. Thus, there is something distinct about the σ3 structure in the regions near residues Phe210 and Tyr238 versus the regions containing the 29 other aromatic residues in each sequence. Recently determined crystal structures for T1L and T3D σ3 (33, 43) have produced new insights but also some new questions about this pattern of σ3 cleavage, as discussed in the following sections.
The HSR of particle-bound T3D σ3.
Residues 208 to 214, defined as the HSR of particle-bound T3D σ3 by cleavage with CHT, THL, and TRY in this study, are located on a loop that rests against the surface of the σ3 structure, near the midpoint of the molecule's long axis and near the constriction that separates the small and large lobes (Fig. 9A, purple) (33, 43). This loop is on the opposite face of σ3 from that which binds to μ1 (33), and it is exposed to solvent within the P2 and P3 channels that penetrate radially through the reovirus outer capsid (16, 38). The location of this loop on the solvent-exposed face of σ3 in virions is consistent with its accessibility to protease cleavage, but its location halfway down the radial length of σ3 raises some question as to how exposed to protease this loop may actually be. In fact, the narrowest width of the P3 channel at and above the particle radius at which the HSR of T3D σ3 is located is 40 to 45 Å. For comparison, the crystallographically defined widths of CHT, TRY, PRK, THL, and Cat L are 30 to 40 Å in one dimension (PDB files 1ACB, 1AUJ, 1PEK, 1HYT, and 1ICF, respectively). Thus, there appears to be sufficient room for each of these proteases to diffuse into the channel for cleavage in the T3D HSR.
Phe210, before and after which THL and CHT, respectively, cleave in T3D σ3, is almost fully buried in the T3D σ3 crystal structure (43). This observation suggests either (i) that a conformational change in this region occurs upon σ3 binding to μ1 or (ii) that this region, both in solution and when σ3 is bound to μ1, can adopt a different conformation from that observed in the crystal, making Phe210 more exposed and accessible to protease active sites. The recently solved crystal structure of the T1L σ3 protein in complex with T1L μ1 (33) shows few if any differences in this region of the σ3 structure from that in free T3D σ3 (43). Thus, it seems most likely that this region of σ3 can undergo conformational breathing in solution (see below).
The HSR of particle-bound T1L σ3.
Residues 238 to 244 (Fig. 9A, dark blue), defined as the HSR of particle-bound T1L σ3 by cleavage with CHT, THL, EKC, and Cat L in this study, overlap a short α-helix (residues 235 to 242) near the apex of the large lobe (43). This α-helix is part of a longer “structural arch” that spans residues 224 to 258 (Fig. 9B to E, blue) and includes two flanking regions of more extended structure (the “arch supports”). Much of this arch is surface exposed and thus potentially accessible to proteases. Moreover, according to the probable arrangement of σ3 in virions (33), the 238-244 HSR is projected above the top of μ1, outside the P2 and P3 channels, so that channel-based limitations to protease accessibility are not an issue for cleavage in this region. Notably, although much of the 224-258 region is surface exposed, Arg239 and Val243, after which respective cleavages by TRY and Cat L occur, are mostly buried. The structure of this region in μ1-bound T1L σ3 (33) shows subtle, though clear, differences relative to the free T3D σ3 structure (see below); however, Arg239 and Val243 remain buried. These observations suggest that unraveling of the α-helix, and thus conformational breathing, may be needed before cleavage in this HSR as well.
C-proximal sequences affect cleavage in the σ3 HSRs.
Since T1L and T3D σ3 contain identical sequences in both HSRs, there must be some variation between the T1L and T3D σ3 structures that determines their differences in preferred sites and rates of early CHT cleavage. T1L and T3D σ3 proteins have amino acid differences at 11 positions throughout their sequences, but the locations of these differences outside the HSRs indicate that their effects on cleavage must be structure based. C-proximal residues 344, 347, and 353, which we showed to be the primary determinants of the CHT cleavage site and rate differences between particle-bound T1L and T3D σ3, are quite distant from the HSRs in the primary sequence of σ3. In the σ3 structure, however, these three residues are found in an extended region of random-coil structure (residues 342 to 355) that rests on the surface of the large lobe (Fig. 9A, red) and is situated directly beneath the α-helix that contains the T1L HSR and across the arch supports that flank it (Fig. 9B to E). This extended coil structure containing residues 344, 347, and 353 is also directly above an extended surface loop (residues 186 to 220) that encompasses the T3D HSR (Fig. 9B to E). Prominent contacts are made between residues from the 342-355 region and residues from both HSRs and their flanking regions. Differences in these contacts provide likely explanations for the different cleavage patterns we observed for T1L and T3D σ3.
T1L σ3 exhibits many fewer interactions between the initial part of the extended coil region (residues 344 to 349) and the T1L HSR and arch support that follows it (residues 242 to 249) than does T3D σ3 (Fig. 9D and E). In particular, missing from T1L σ3 that are present in T3D σ3 are certain hydrophobic contacts by residue 344 (Ser in T1L, Pro in T3D) with Phe249 and by residue 347 (Thr in T1L, Ile in T3D) with Asp248, as well as by Lys348 with Arg247 and by Phe349 with Leu242. The likely result of these missing interactions would be less-firm tethering of the C-terminal side of the T1L HSR in T1L σ3, allowing greater conformational breathing and more efficient cleavage within that region. In T3D σ3, due to the presence of these additional interactions, cleavage in the 238-244 region would be less favored. On the other hand, in T1L σ3, a notable interaction is present between residue 353 and the part of the extended surface loop preceding the T3D HSR. In particular, H bonding and/or electrostatic interactions between Asp353 and Arg196 in T1L σ3 are not made by Asn353 and Lys196 in T3D σ3 (Fig. 9B and C). A possible effect of this additional interaction in T1L σ3 may be to draw the 342-355 random-coil region away from the T1L HSR and flanking arches, which may further promote conformational breathing within the T1L HSR.
Rate versus site of σ3 cleavage.
The finding that the C-proximal residues 344, 347, and 353 are responsible for the difference in cleavage rate as well as the difference in cleavage site between T1L and T3D σ3 suggests that these may be linked phenotypes. Analysis of the fragments obtained by CHT treatment of rcISVPs containing [35S]methionine- and [35S]cysteine-labeled T1L and T3D σ3 proteins provided evidence that both proteins are cleaved in the T3D HSR and accumulate the resulting fragments to a similar extent under the tested conditions (8°C for 15 min). In contrast, cleavage in the T1L HSR was greatly favored in T1L σ3 versus T3D σ3 (Fig. 8C, lanes 1 and 3). This observation suggests the hypothesis that the greater cleavage rate of full-length T1L σ3 is mainly due to the gain of cleavage in the T1L HSR of this protein. The results from CHT treatment of rcISVPs containing the σ3 chimeras as well as the single- and double-substitution mutants support this hypothesis. The increased cleavage rate of full-length DDDL σ3 versus T3D (DDDD) σ3 was accompanied by the gain of cleavage in the T1L HSR (Fig. 8C, lanes 2 and 3), whereas the decreased cleavage rate of full-length LLLD σ3 versus T1L (LLLL) σ3 was accompanied by the loss of cleavage in the T1L HSR (Fig. 8C, lanes 4 and 1, respectively). In addition, the single-substitution mutations in T3D σ3, which had minor effects on the cleavage rate, displayed a cleavage pattern indistinguishable from that of T3D σ3 (Fig. 8C, lanes 5, 6, and 8), whereas the double-substitution mutations at positions 344 and 347 in T3D σ3, which caused a substantial increase in the cleavage rate, displayed increased production of the 26K fragment representing cleavage in the T1L HSR (Fig. 8C, lane 7). The extent of accumulation of the fragments derived from σ3 cleavage within the T3D HSR was similar in the case of all of the rcISVPs we tested. These results thus suggest that the rate and site of σ3 cleavage are linked and that the differences in overall rate of full-length σ3 cleavage observed in this study were primarily determined by the extent of cleavage that occurred within the T1L HSR.
Other residues affecting the rate of T1L and T3D σ3 cleavage.
Although residues 344, 347, and 353 are the primary determinants of the cleavage rate difference between T1L and T3D σ3, other portions of the protein sequence appear to have more subtle effects. In previous work, we found that by exchanging residues 1 to 185 or 186 to 265 from T3D σ3 into the T1L background, we caused the particle-bound chimera to be more rapidly cleaved by EKC than wild-type T1L σ3 was. Reciprocally, we found that by exchanging either of these regions from T1L σ3 into the T3D σ3 background, we caused the particle-bound chimera to be more slowly cleaved by EKC than wild-type T3D σ3 was (24). Equivalent results were obtained with CHT as a part of this study (data not shown). Amino acid substitutions between T1L and T3D σ3 are found at four positions in the 1-185 region: positions 11, 108, 116, and 180. Residue 11 is located in the small lobe of σ3, on the surface suggested to interact with μ1 (33, 43). Residues 108, 116, and 180, in contrast, are in the larger lobe, close to the T1L HSR at residues 238 to 244. Interestingly, although residues 108 and 116 are on the surface of σ3 and on the opposite side of the structure from that containing the C-proximal extended region and both HSRs, residue 180 is an internal residue positioned directly beneath the T1L HSR and making contacts with Val243 and Thr244 within this region of both T1L and T3D σ3. Thus, Val/Ile180 is most likely the variable residue in the 1-185 region of T1L and T3D σ3 that affects the structure of the HSR(s) and thereby affects cleavage rate. The 186-265 region contains two amino acid substitutions between T1L and T3D σ3, at positions 196 and 198. These residues are located within the N-terminal surface loop that flanks the T3D HSR (see above) and are positioned in front of the extended region that contains residues 344, 347, and 353. Most notably, residue 196 makes contacts with residue 353 in T1L, but not in T3D. Thus, Arg/Lys196 is most likely the variable residue in the 186-265 region of T1L and T3D σ3 that affects the structure of the HSR(s) and thereby affects cleavage rate. The general conclusion is that substitutions at numerous positions in σ3 are capable of influencing its susceptibility to early cleavages within the HSRs and of thereby influencing the overall rate of particle activation for membrane penetration during reovirus entry. The capacity of a Tyr-to-His mutation at position 354 in T3D σ3 to enhance the rate of that protein's cleavage by CHT and within cells has been previously identified (17, 54), but the mechanism of action of this mutation remains to be defined.
Further proteolysis of the early σ3 cleavage fragments is required for fragment release and particle infection.
Analysis by SDS-PAGE of the progression of σ3 processing by CHT demonstrated that, whereas the N- and C-terminal fragments generated from early cleavage of T1L or T3D σ3 initially accumulated as the amount of full-length σ3 decreased (Fig. 1 and 6A), at later times the intensity of both fragment bands also decreased (Fig. 1 and 6A). Thus, in the case of both T1L and T3D σ3, the N- and C-terminal fragments resulting from the characterized early cleavage by CHT are subject to further processing as time proceeds. Longer digestion times under the same conditions result in the complete loss of fragments and the production of ISVPs (data not shown). Thus, it appears that once early cleavages have occurred within the HSR(s), proteolysis is extended to other exposed CHT cleavage sites within the two fragments. Cleavage at the HSR(s) could increase the accessibility of other cleavage sites in σ3 by increasing the conformational mobility of adjacent regions of the protein.
ISVPs, from which surface protein σ3 and its fragments have been removed, are strongly resistant to lysosomal protease inhibitors like NH4Cl and E-64 (2, 50). In this study, we found that particles in which the majority of full-length σ3 has been cleaved at least once in one of the HSRs remain sensitive to these inhibitors, most likely because the fragments remain particle bound and in position to inhibit μ1 functions in membrane penetration. Direct evidence for continued inhibition of μ1 functions in these particles was shown by their lack of hemolytic activity. Our hypothesis is therefore that additional cleavages of σ3 are required for μ1 activation for membrane penetration, and we are faced with the questions of how many and which additional cleavages of σ3 are sufficient in this regard and whether σ3 fragment elution from particles is a part of the requirement. Reasonable possibilities are that removal of the large, apical lobe of σ3 is required so that the top of μ1 can more closely approach the target membrane and that removal of the lower lobe of σ3 is also required for removing restraints on μ1 trimer dissociation (33).
Acknowledgments
We acknowledge our gratitude to A. M. Olland, S. Liemann, and S. C. Harrison for providing coordinates for the crystal structures of σ3 and μ1-σ3. We are also grateful to the University of Wisconsin Biotechnology Center DNA Sequencing Facility and the DF/HCC High-Throughput DNA Sequencing Facility at Harvard Medical School for DNA sequencing and to S. Gygi, L. Licklider, and R. Tomaino for performing the ESI-MS analyses at the Taplin Biological Mass Spectrometry Facility at Harvard Medical School. Additional thanks go to K. M. Coombs for advice concerning mass spectrometry, S. J. Harrison and E. Freimont for technical assistance and lab support, S. Liemann for suggestions for Fig. 9, and other members of our lab for helpful discussions. J.J.-V. would also like to thank G. A. Manji for insightful discussions and technical advice.
This work was supported by NIH grant R01 AI46440 and DARPA contract MDA 972-97-1-0005 to M.L.N. and by NIH grant R01 AI45990 to L.A.S. J.J.-V. was supported by a predoctoral fellowship from La Caixa d'Estalvis i Pensions de Barcelona and by a Steenbock/Wharton fellowship from the Department of Biochemistry, University of Wisconsin—Madison. M.L.N. was additionally supported as a Shaw Scientist from the Milwaukee Foundation and as a Vilas Associate from the University of Wisconsin Foundation.
Footnotes
Dedicated to the memory of Carles Martí Henneberg.
REFERENCES
- 1.Amerongen, H. M., G. A. R. Wilson, B. N. Fields, and M. R. Neutra. 1994. Proteolytic processing of reovirus is required for adherence to intestinal M cells. J. Virol. 68:8428-8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer, G. S., and T. S. Dermody. 1997. Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 71:4921-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer, G. S., D. H. Ebert, C. J. Chung, A. H. Erickson, and T. S. Dermody. 1999. Mutant cells selected during persistent reovirus infection do not express mature cathepsin L and do not support reovirus disassembly. J. Virol. 73:9532-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett, A. J., and H. Kirschke. 1981. Cathepsin B, cathepsin H and cathepsin L. Methods Enzymol. 800:535-561. [DOI] [PubMed] [Google Scholar]
- 5.Bass, D. M., D. Bodkin, R. Dambrauskas, J. S. Trier, B. N. Fields, and J. L. Wolf. 1990. Intraluminal proteolytic activation plays an important role in replication of type 1 reovirus in the intestines of neonatal mice. J. Virol. 64:1830-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodkin, D. K., M. L. Nibert, and B. N. Fields. 1989. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 63:4676-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borsa, J., T. P. Copps, M. D. Sargent, D. G. Long, and J. D. Chapman. 1973. New intermediate subviral particles in the in vitro uncoating of reovirus virions by chymotrypsin. J. Virol. 11:552-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borsa, J., B. D. Morash, M. D. Sargent, T. P. Copps, P. A. Lievaart, and J. G. Szekely. 1979. Two modes of entry of reovirus particles into L cells. J. Gen. Virol. 45:161-170. [DOI] [PubMed] [Google Scholar]
- 9.Borsa, J., M. D. Sargent, P. A. Lievaart, and T. P. Copps. 1981. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology 111:191-200. [DOI] [PubMed] [Google Scholar]
- 10.Canning, W. M., and B. N. Fields. 1983. Ammonium chloride prevents lytic growth of reovirus and helps to establish persistent infection in mouse L cells. Science 219:987-988. [DOI] [PubMed] [Google Scholar]
- 11.Chandran, K., and M. L. Nibert. 1998. Protease cleavage of reovirus capsid protein μ1/μ1C is blocked by alkyl sulfate detergents, yielding a new type of infectious subvirion particle. J. Virol. 72:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, S. M., J. R. Roth, M. L. Clark, B. B. Barnett, and R. S. Spendlove. 1981. Trypsin enhancement of rotavirus infectivity: mechanism of enhancement. J. Virol. 39:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coombs, K. M. 1998. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology 243:218-228. [DOI] [PubMed] [Google Scholar]
- 14.Cotten, M., and J. M. Weber. 1995. The adenovirus protease is required for virus entry into host cells. Virology 13:494-502. [DOI] [PubMed] [Google Scholar]
- 15.Dermody, T. S., M. L. Nibert, J. D. Wetzel, X. Tong, and B. N. Fields. 1993. Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J. Virol. 67:2055-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dryden, K. A., G. Wang, M. Yeager, M. L. Nibert, K. M. Coombs, D. B. Furlong, B. N. Fields, and T. S. Baker. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 122:1023-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebert, D. H., J. D. Wetzel, D. E. Brumbaugh, S. R. Chance, L. E. Stobie, G. S. Baer, and T. S. Dermody. 2001. Adaptation of reovirus to growth in the presence of protease inhibitor E-64 segregates with a mutation in the carboxy terminus of viral outer-capsid protein σ3. J. Virol. 75:3197-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eng, J., A. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 19.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert, J. M., and H. B. Greenberg. 1998. Cleavage of rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like particle-induced fusion from without. J. Virol. 72:4555-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillian, A. L., and M. L. Nibert. 1998. Amino terminus of reovirus nonstructural protein σNS is important for ssRNA binding and nucleoprotein complex formation. Virology 240:1-11. [DOI] [PubMed] [Google Scholar]
- 22.Greber, U., P. Webster, J. Weber, and A. Helenius. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766-1777. [PMC free article] [PubMed] [Google Scholar]
- 23.Hooper, J. W., and B. N. Fields. 1996. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J. Virol. 70:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jané-Valbuena, J., M. L. Nibert, S. M. Spencer, S. B. Walker, T. S. Baker, Y. Chen, V. E. Centonze, and L. A. Schiff. 1999. Reovirus virion-like particles obtained by recoating infectious subvirion particles with baculovirus-expressed σ3 protein: an approach for analyzing σ3 functions during virus entry. J. Virol. 73:2963-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jekel, P. A., W. J. Weijer, and J. J. Beintema. 1983. Use of endoproteinase Lys-C from Lysobacter enzymogenes in protein sequence analysis. Anal. Biochem. 134:347-354. [DOI] [PubMed] [Google Scholar]
- 26.Joklik, W. K. 1972. Studies on the effect of chymotrypsin on reovirions. Virology 49:700-715. [DOI] [PubMed] [Google Scholar]
- 27.Kedl, R., S. Schmechel, and L. Schiff. 1995. Comparative sequence analysis of the reovirus S4 genes from 13 serotype 1 and serotype 3 field isolates. J. Virol. 69:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klenk, H. D., R. Rott, M. Orlich, and J. Blodorn. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68:426-439. [DOI] [PubMed] [Google Scholar]
- 29.Kothandaraman, S., M. C. Hebert, R. T. Raines, and M. L. Nibert. 1998. No role for pepstatin-A-sensitive acidic proteinases in reovirus infections of L or MDCK cells. Virology 251:264-272. [DOI] [PubMed] [Google Scholar]
- 30.Kyte, J. 1995. Structure in protein chemistry, 1st ed. Garland Publishing, Inc., New York, N.Y.
- 31.Lee, W.-M., S. S. Monroe, and R. R. Rueckert. 1993. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J. Virol. 67:2110-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lerch, R. A., and P. D. Friesen. 1993. The 35-kilodalton protein gene (p35) of Autographa californica nuclear polyhedrosis virus and the neomycin resistance gene provide dominant selection of recombinant baculoviruses. Nucleic Acids Res. 21:1753-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liemann, S., K. Chandran, T. S. Baker, M. L. Nibert, and S. C. Harrison. 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell 108:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucia-Jandris, P., J. W. Hooper, and B. N. Fields. 1993. Reovirus M2 gene is associated with chromium release from mouse L cells. J. Virol. 67:5339-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mabrouk, T., and G. Lemay. 1994. Mutations in a CCHC zinc-binding motif of the reovirus sigma 3 protein decrease its intracellular stability. J. Virol. 68:5287-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maeda, T., K. Kawasaki, and S. Ohnishi. 1981. Interaction of influenza virus hemagglutinin with target membrane lipids is a key step in virus-induced hemolysis and fusion at pH 5.2. Proc. Natl. Acad. Sci. USA 78:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCune, J. M., L. B. Rabin, M. B. Feinberg, and M. Lieberman. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 53:55-67. [DOI] [PubMed] [Google Scholar]
- 38.Metcalf, P., M. Cyrklaff, and M. Adrian. 1991. The three-dimensional structure of reovirus obtained by cryo-electron microscopy. EMBO J. 10:3129-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moulard, M., and E. Decroly. 2000. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469:121-132. [DOI] [PubMed] [Google Scholar]
- 40.Nason, E. L., J. D. Wetzel, S. K. Mukherjee, E. S. Barton, B. V. V. Prasad, and T. S. Dermody. 2001. A monoclonal antibody specific for reovirus outer-capsid protein σ3 inhibits σ1-mediated hemagglutination by steric hindrance. J. Virol. 75:6625-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nibert, M. L. 1993. Ph.D. thesis, Harvard University, Cambridge, Mass.
- 42.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olland, A. M., J. Jané-Valbuena, L. A. Schiff, M. L. Nibert, and S. C. Harrison. 2001. Structure of the reovirus outer capsid and dsRNA-binding protein σ3 at 1.8 Å resolution. EMBO J. 20:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng, J., and S. P. Gygi. 2001. Proteomics: the move to mixtures. J. Mass Spectrom. 36:1083-1091. [DOI] [PubMed] [Google Scholar]
- 45.Shatkin, A. J., and A. J. LaFiandra. 1972. Transcription by infectious subviral particles of reovirus. J. Virol. 10:698-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepard, D. A., J. G. Ehnstrom, and L. A. Schiff. 1995. Association of reovirus outer capsid proteins σ3 and μ1 causes a conformational change that renders σ3 protease sensitive. J. Virol. 69:8180-8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shepard, D. A., J. G. Ehnstrom, P. J. Skinner, and L. A. Schiff. 1996. Mutations in the zinc-binding motif of the reovirus capsid protein σ3 eliminate its ability to associate with capsid protein μ1. J. Virol. 70:2065-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silverstein, S. C., C. Astell, D. H. Levin, M. Schonberg, and G. Acs. 1972. The mechanisms of reovirus uncoating and gene activation in vivo. Virology 47:797-806. [DOI] [PubMed] [Google Scholar]
- 49.Strong, J. E., G. Leone, R. Duncan, R. K. Sharma, and P. W. K. Lee. 1991. Biochemical and biophysical characterization of the reovirus cell attachment protein σ1: evidence that it is a homotrimer. Virology 184:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tosteson, M. T., M. L. Nibert, and B. N. Fields. 1993. Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc. Natl. Acad. Sci. USA 90:10549-10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Virgin, H. W., IV, M. A. Mann, and K. L. Tyler. 1994. Protective antibodies inhibit reovirus internalization and uncoating by intracellular proteases. J. Virol. 68:6719-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virgin, H. W., IV, R. Bassel-Duby, B. N. Fields, and K. L. Tyler. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J. Virol. 62:4594-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wetzel, J. D., G. J. Wilson, G. S. Baer, L. R. Dunnigan, J. P. Wright, D. S. Tang, and T. S. Dermody. 1997. Reovirus variants selected during persistent infections of L cells contain mutations in the viral S1 and S4 genes and are altered in viral disassembly. J. Virol. 71:1362-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiley, D. C., and J. J. Skehel. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365-394. [DOI] [PubMed] [Google Scholar]