Abstract
Histones are modified posttranslationally, e.g., by methylation of lysine and arginine residues, and by phosphorylation of serine residues. These modifications regulate processes such as gene expression, DNA repair, and mitosis and meiosis. Recently, evidence has been provided that histones are also modified by covalent binding of the vitamin biotin. Aims of this study were to identify biotinylation sites in histone H3, and to investigate the crosstalk among histone biotinylation, methylation, and phosphorylation. Synthetic peptides based on the sequence of human histone H3 were used as substrates for enzymatic biotinylation by biotinidase; biotin in peptides was probed using streptavidin peroxidase. These studies provided evidence that K4, K9, and K18 in histone H3 are good targets for biotinylation; K14 and K23 are relatively poor targets. Antibodies were generated to histone H3, biotinylated either at K4, K9, or K18. These antibodies localized to nuclei in human placental cells in immunocytochemistry and immunoblotting experiments, suggesting that lysines in histone H3 are biotinylated in vivo. Dimethylation of R2, R8, and R17 increased biotinylation of K4, K9, and K18, respectively, by biotinidase; phosphorylation of S10 abolished biotinylation of K9. These observations are consistent with crosstalk between biotinylation of histones and other known modifications of histones. We speculate that this crosstalk provides a link to known roles for biotin in gene expression and cell proliferation.
Keywords: Biotin, biotinidase, histone H3, lysine
Abbreviations: DAPI, 4’,6-diamidino-2-phenylindole; Fmoc, N-fluoren-9-ylmethoxycarbonyl
INTRODUCTION
Histones are small proteins (11 to 22 kDa) that mediate the folding of DNA into chromatin. The following five major classes of histones have been identified in eukaryotic cells: H1, H2A, H2B, H3, and H4 [1]. DNA is wrapped around octamers of core histones, each consisting of one H3-H3-H4-H4 tetramer and two H2A-H2B dimers, to form the nucleosomal core particle. Histone H1 associates with the DNA connecting nucleosomal core particles. Nucleosomes are stabilized by electrostatic interactions between negatively charged phosphate groups in DNA and positively charged ɛ-amino groups (lysine residues) and guanidino groups (arginine residues) in histones.
Histones consist of a globular C-terminal domain and a flexible N-terminal tail [1]. The amino terminus of histones protrudes from the nucleosomal surface; lysines, arginines, serines, and glutamates in the amino terminus are targets for acetylation, methylation, phosphorylation, ubiquitination, poly (ADP-ribosylation), and sumoylation [1–5]. These modifications play important roles in chromatin structure, regulating processes such as transcriptional activation or silencing of genes, DNA repair, and mitotic and meiotic condensation of chromatin.
Recently, a novel covalent modification of histones has been identified in human cells: biotinylation of lysine residues [6, 7]. Two enzymes can independently catalyze biotinylation of histones: biotinidase [8] and holocarboxylase synthetase [9]. Biotinidase belongs to the nitrilase superfamily of enzymes [10]; biotinylation of histones by biotinidase depends on the hydrolytic cleavage of biocytin (biotinyl-ɛ-lysine), coupled to the transfer of the biotinyl residue to free amino groups in histones [11]. In contrast, biotinylation of histones by holocarboxylase synthetase depends on ATP and biotin [9]. Preliminary studies suggest that biotinylation of histones might play a role in processes such as gene silencing [12], cell proliferation [6, 9], and DNA repair or apoptosis [12, 13]. These observations could have important implications for human health, based on the following lines of reasoning. First, preliminary evidence has been provided that biotinylation of K12 in histone H4 decreases rapidly in response to double-stranded DNA breaks caused by the cancer drug etoposide [13]. This observation is consistent with the hypothesis that alterations in the biotinylation pattern of histones might be an early signaling event in response to DNA damage. Second, mutations of the genes encoding biotinidase [14–16] and holocarboxylase synthetase [17] have been documented; some of these mutations are fairly common [18, 19]. Fibroblasts from individuals with mutated holocarboxylase synthetase are deficient in histone biotinylation [9]. Likewise, in-vitro studies provided evidence that mutated biotinidase is not capable of catalyzing biotinylation of histones [8]. Future studies might unravel abnormal patterns of gene silencing [12], cell proliferation [6, 9], and DNA repair or apoptosis [12, 13] in individuals carrying mutations of genes coding for biotinidase and holocarboxylase synthetase.
Although all five major classes of histones appear to be biotinylated in human cells [6], only two biotinylation sites have been identified so far: K8 and K12 in histone H4 [7]. This gap in our understanding of histone biotinylation has created a significant obstacle for investigating roles of biotinylated histones in cell biology, based on the following lines of reasoning. As long as biotinylation sites remain unknown, no site-specific antibodies to biotinylated histones can be generated. Such antibodies are invaluable tools (i) to study the cross-talk among modifications of histones, e.g., biotinylation and acetylation of lysine residues [7]; (ii) to investigate cellular distribution patterns of biotinylated histones by using immunocytochemistry; and (iii) to investigate roles for biotinylation of histones in the regulation of transcriptional activity of genes by using chromatin immunoprecipitation assays.
Recently we have developed a peptide-based procedure to identify biotinylation sites in histones [7]. In the present study we applied this procedure to identify biotinylation sites in human histone H3. As secondary goal we investigated interactions among histone biotinylation, methylation, and phosphorylation. Histone H3 was chosen as a model because of its pivotal role in regulating gene expression [20–22].
RESULTS
Biotinylation sites in histone H3
The N-terminal tail of histone H3 was efficiently biotinylated by biotinidase. The binding of biotin was substantially greater in peptide N1–25 compared to peptide N15–39, if equal amounts of both peptides were incubated with biotinidase and biocytin for 45 min (Fig. 1; compare lanes 1a and 1b with lanes 2a and 2b). The peptide (C116–136) based on the C-terminus of histone H3 was not biotinylated if incubated with biotinidase (lanes 3a and 3b). This is consistent with previous observations that biotinylation and other modifications of histones cluster in the N-terminal region [2, 7]. Also these findings suggest that the primary targets for biotinylation are located in the region spanning the 25 N-terminal amino acids. Thus, subsequent studies focused on this region in the histone H3 molecule.
FIGURE 1.
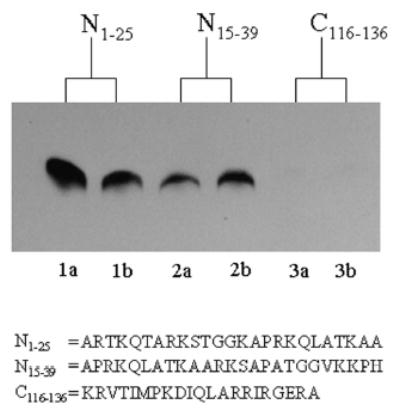
Biotinylation targets amino acids in the N-terminal tail of human histone H3. Synthetic peptides based on the N- and C-terminal region in histone H3 were biotinylated enzymatically, and biotin was probed using gel electrophoresis and streptavidin peroxidase. N1–25 = peptide spanning amino acids 1–25 in histone H3 (lanes 1a and b); N15–39 = peptide spanning amino acids 15–39 in histone H3 (lanes 2a and b); C116–136 = peptide spanning amino acids 116–136 in histone H3 (lanes 3a and b). Duplicate analyses are depicted.
Previous studies suggested that lysine residues in histones are targets for biotinylation [7]. Thus, we sub-divided the N-terminal 25 amino acids into four synthetic peptides to allow for easier identification of biotinylated lysines in histone H3: N1–9 (including K4 and K9), N9–16 (including K9 and K14), N16–23 (including K18 and K23), and N18–25 (including K18 and K23); subscripts denote the amino acid residues in the histone H3 sequence. These peptides were incubated with biotinidase and biocytin for up to 45 min; at timed intervals aliquots were collected and biotinylated peptides on transblots were probed using streptavidin peroxidase. Apparently, peptide N18–25 was a better substrate for biotinylation than peptides N1–9, N9–16, and N16–23 [Figs. 2A and B (lanes 2-5)]. The minor apparent differences in biotinylation signal among the peptides loaded in lanes 2 - 4 are caused by intra-assay variation, and are not observed if biotinylation of the same peptides is quantified by gel densitometry using multiple independent gels (compare Fig. 2A). Peptide N1–25 was used as a reference and was heavily biotinylated (Fig. 2B, lane 1): 100% relative biotinylation after 45 min of incubation. Peptide C116–136 was used as a negative control and was not biotinylated after 45 min (lane 6). These findings suggest that either K18, K23, or both are targets for biotinylation (see below). However, further below we provide evidence that modifications of arginines may substantially enhance the biotinylation of histone H3 by biotinidase, and that K4 and K9 may also be targets for biotinylation in vivo. All subsequent enzymatic biotinylations were conducted for 45 minutes.
FIGURE 2.
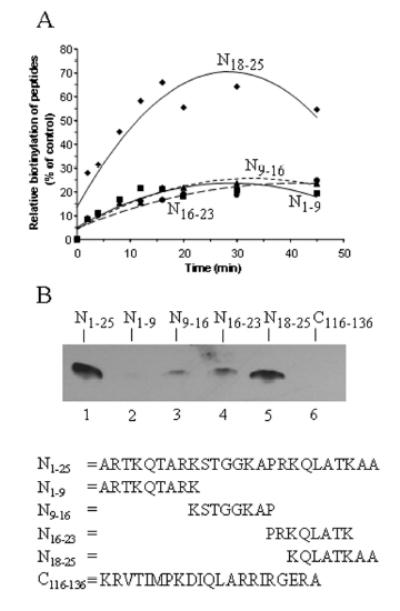
Biotinylation of peptides based on the N-terminal tail in histone H3. Panel A: Synthetic peptides were biotinylated enzymatically, and biotin was probed using gel electrophoresis and streptavidin peroxidase. N1–9 = peptide spanning amino acids 1–9 in histone H3; N9–16 = peptide spanning amino acids 9–16 in histone H3; N16–23 = peptide spanning amino acids 16–23 in histone H3; and N18–25 = peptide spanning amino acids 18–25 in histone H3. Each data point represents the mean of three independent measurements. Panel B: Representative gels, depicting peptides that were incubated with biotinidase and biocytin for 45 min. N1–25 (lane 1) = peptide spanning amino acids 1–25 in histone H3; N1–9, N9–16, N16–23, and N18–25 (lanes 2-5) = as described for panel A; C116–136 (lane 6) = peptide spanning amino acids 116–136 in histone H3.
The next series of experiments focused on K4, K9, and K14. Peptide N1–25 was used as a positive control and was heavily biotinylated (Fig. 3, lane 1). As expected, if both lysines (K4 and K9) in a peptide spanning amino acids 1 to 9 in histone H3 were substituted by alanine (K4,9A1–9), no binding of biotin was detectable (lane 2). This is consistent with previous studies, suggesting that lysines rather than other amino acids are targets for biotinylation [7]. If K4 was substituted with alanine (K4A1–9), biotinylation of K9 was barely detectable (lane 3). In contrast, if K9 was substituted with alanine (K9A1–9), K4 was biotinylated considerably (lane 4). These findings suggest that K4 is a target for biotinylation.
FIGURE 3.
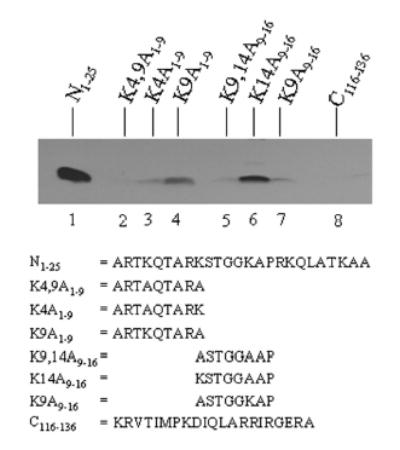
Biotinylation of K4, K9 and K14 in the N-terminal tail in histone H3. Synthetic peptides were biotinylated enzymatically, and biotin was probed using gel electrophoresis and streptavidin peroxidase. N1–25 = peptide spanning amino acids 1–25 in histone H3 (lane 1); K4,9A1–9, K4A1–9, K9A1–9, K9,14A9–16, K14A9–16, K9A9–16 = substitutions of K4, K9, and K14 in histone H3; and C116––136 = peptide spanning amino acids 116–136 in histone H3.
Next, variations of a peptide spanning amino acids 9 to 16 in histone H3 (i.e., including K9 and K14) were tested. If both K9 and K14 were substituted with alanine (K9,14A9–16), no binding of biotin was detectable (lane 5). If K14 was substituted with alanine (K14A9–16), K9 was heavily biotinylated (lane 6). This is in contrast to the findings described above, which suggested that K9 is a poor target for biotinylation (peptide K4A1–9 in lane 3). We offer the following explanation for these apparently contradictory observations: peptide K14A9–16 is lacking the positively charged and bulky arginine residue in position 8; in contrast peptide K4A1–9 includes R8. Biotinylation of K14A9–16 can not be explained by biotinylation of K14, given that K14 is a poor target for biotinylation (peptide K9A9–16, lane 7). These findings are consistent with the hypothesis that K9 might be a good target for biotinylation if R8 is modified covalently; this hypothesis was further tested in dimethylation experiments described below. Peptide C116–136 was used as a negative control; no biotinylation was detectable (lane 8).
The following series of experiments focused on K18 and K23. Peptide N1–25 was used as a positive control and was heavily biotinylated (Fig. 4, lane 1). As expected, if both lysines (K18 and K23) in a peptide based on amino acids 16 to 23 in histone H3 were substituted with alanine (peptide K18,23A16–23), no binding of biotin was detectable (lane 2). Likewise, biotinylation of K18 was weak if K23 was substituted with alanine (K23A16–23; lane 3), and biotinylation of K23 was weak if K18 was substituted with alanine (K18A16–23; lane 4). This is in apparent contrast to the findings presented in Figure 2, which suggested that K18 or K23 are good targets for biotinylation (peptide N18–25 in Figure 2). Based on the following lines of reasoning we hypothesize that R17 in peptide K23A16–23 interfered with biotinylation of K18 in the experiments depicted in Fig. 4: (i) Peptide N18–25 (Fig. 2) starts with K18, i.e., does not include R17; (ii) peptide K23A16–23 (Fig. 4) starts with A16, i.e., this peptide includes R17; (iii) experiments involving K9 suggested that arginine residues may interfere with biotinylation (see above). This hypothesis was tested as follows. Peptides were synthesized that started with K18 in histone H3; hence, these peptides did not include R17 but did include both K18 and K23 unless noted otherwise. No biotinylation was detected if both K18 and K23 were substituted with alanine (K18,23A18–25; lane 5). If K23 was substituted with alanine (K23A18–25), K18 was heavily biotinylated (lane 6). In contrast, if K18 was substituted with alanine (K18A18–25), biotinylation of K23 was barely detectable (lane 7). Peptide C116–136 was used as a negative control; no biotinylation was detectable (lane 8). These findings are consistent with the hypothesis that K18 is a target for biotinylation if R17 is modified; this hypothesis was further tested as described below. Also, these findings suggest that K23 is a poor target for biotinylation.
FIGURE 4.
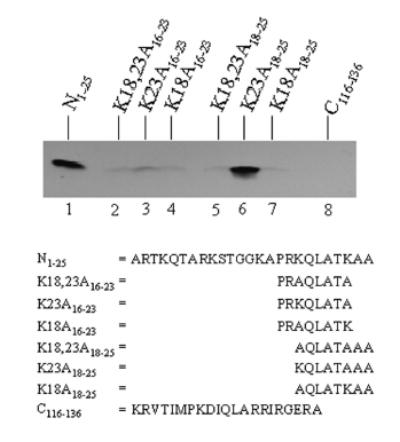
Biotinylation of K18 and K23 in the N-terminal tail in histone H3. Synthetic peptides were biotinylated enzymatically, and biotin was probed using gel electrophoresis and streptavidin peroxidase. N1–25 = peptide spanning amino acids 1–25 in histone H3 (lane 1); K18,23A16–23, K23A16–23, K18A16–23, K18,23A18–25, K23A18–25, K18A18–25 = substitutions of K18 and K23 in histone H3; and C116–136 = peptide spanning amino acids 116–136 in histone H3.
Arginine residues such as R2 and R17 in human histone H3 are modified by mono-and di-methylation; various lysine residues in histones are modified by mono-, di-, or tri-methylation [2, 23]. Here we determined whether naturally occurring modifications of arginines render lysines a better target for biotinylation in histone H3. Peptide N16–23 was used as a control; this peptide includes K18 and K23, and an arginine residue (R17) that is not di-methylated. Peptide N16–23 was a moderate target for biotinylation by biotinidase (Fig. 5, lane 1), confirming findings presented above. Likewise, peptides N1–9 (including K4 and K9) and N9–16 (including K9 and K14) were relatively poor targets for biotinylation (data not shown; see also Fig. 2). Dimethylation of R2 and R8 (combined or individually) moderately increased the enzymatic biotinylation of K4 and K9 by biotinidase (compare lanes 2 - 4 to lane 1). Dimethylation of R17 (peptide dmeR1716–23) substantially increased the enzymatic biotinylation of K18 (compare lanes 1 and 5). Note that peptide dmeR1716–23 also contains K23; however, studies presented above suggested that K23 is a poor target for biotinylation.
FIGURE 5.
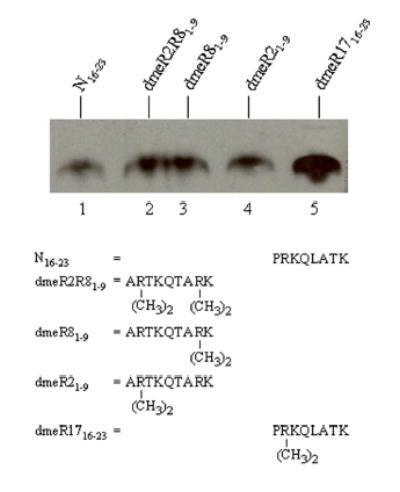
Effects of arginine dimethylation on the biotinylation of lysine residues in the N-terminal tail in histone H3. Synthetic peptides were biotinylated enzymatically, and biotin was probed using gel electrophoresis and streptavidin peroxidase. N16–23 = peptide spanning amino acids 1–23 in histone H3 (lane 1); dmeR2R81–9, dmeR81–9, dmeR21–9, dmeR1716–23 = dimethylation of R2, R8, or R17 in histone H3 (lanes 2 - 5).
Effects of arginine residues on biotinylation of lysines were further corroborated in the following series of experiments. The synthetic peptide N6–13 (including R8 and K9) was used as a control; this peptide was a moderate target for biotinylation (Table 1). If R8 was substituted with an alanine (peptide R8A6–13) biotinylation increased considerably, suggesting that unmodified arginines interfere with biotinylation of lysines by biotinidase. Substitution of arginine with ornithine leaves intact the positive charge in position 8. If R8 was substituted with an ornithine (peptide R8O6–13) biotinylation increased considerably, suggesting that the positive charge of arginine is not responsible for inhibiting biotinylation of lysines. If a negative charge was introduced by phosphorylation of S10 during peptide synthesis [S10S(p)6–13], K9 became a poor target for biotinylation. This suggests that the naturally occurring phosphorylation of S10 [2] may play a role in decreasing the availability of K9 for biotinylation. If K9 was substituted with an alanine (peptide K9A6–13), no biotinylation was observed (negative control). Finally, changing the sequence of amino acids 7 and 8 from AR to RA did not substantially affect biotinylation of K9.
Table 1.
Amino acid modifications affect biotinylation of K9 by biotinidasea
| Identifier | Amino acid sequence | Relative biotinylation |
|---|---|---|
| N6–13b | TARKSTGG | ++ |
| R8A6–13 | TAAKSTGG | +++ |
| R8O6–13 | TAOKSTGG | +++ |
| S10S(p)6–13 | TARKS(p)TGG | — |
| K9A6–13 | TARASTGG | — |
| AR7,8RA6–13 | TRAKSTGG | + |
Peptides are denoted by using one-letter amino acid code.
TARKSTGG represents the native unmodified peptide, based on the amino acid sequence in position 6 - 13 in histone H3.
Polyclonal antibody
Polyclonal antibodies were generated to determine whether histone H3 is biotinylated at K4, K9, and K18 in vivo. First, we determined whether the antibodies were specific for biotinylation sites. Transblots of the following biotinylated peptides were probed with the newly developed antibodies in all possible combinations: N1–13bioK4, N1–13bioK9, and N13–25bioK18 (see Materials and Methods for sequence information). The following observations were made with regard to antibody specificities. The antibody raised against histone H3 (biotinylated at K4) reacted with N1–13bioK4 and cross-reacted with N1–13bioK9, but bound only very weakly to N13–25bioK18 (Fig. 6A, lanes 1 - 3). No signal was detectable if non-biotinylated peptide (N1–25) was used as a target (lane 4), or if N1–13bioK4 was probed using pre-immune serum (lane 5). The antibody raised against histone H3 (biotinylated at K9) reacted with N1–13bioK9, but cross-reacted only very weakly with N1–13bioK4 and N13–25bioK18 (lanes 6 - 8). No signal was detectable if non-biotinylated peptide (N1–25) was used as a target (lane 9), or if N1–13bioK9 was probed using pre-immune serum (lane 10). The antibody raised against histone H3 (biotinylated at K18) reacted with N13–25bioK18, but did not bind to N1–13bioK4 and cross-reacted only very weakly with N1–13bioK9 (lanes 11 - 13). No signal was detectable if non-biotinylated peptide (N1–25) was used as a target (lane 14), or if N13–25bioK18 was probed using pre-immune serum (lane 15). Peptides N1–13bioK4, N1–13bioK9, and N13–25bioK18 produced equal signals if biotin was probed with streptavidin-peroxidase (data not shown). This is consistent with the notion that equal amounts of peptide were loaded per lane in specificity experiments.
FIGURE 6.
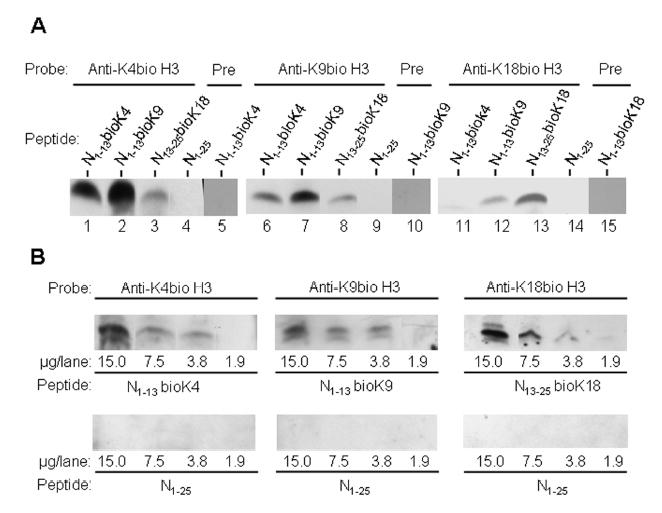
Biotinylation site specificity of antibodies against histone H3, biotinylated at K4, K9, or K18. Synthetic peptides based on histone H3 were chemically biotinylated either at K4 (denoted “N1–13bioK4”), K9 (“N1–13bioK9”), or K18 (“N13–25bioK18”); a non-biotinylated peptide spanning amino acids 1 – 25 was used as a negative control (“N1–25”). Panel A: Peptides were probed using antibodies to histone H3, biotinylated at K4 (lanes 1 - 4), biotinylated at K9 (lanes 6 - 9), biotinylated at K18 (lanes 11 - 14), or pre-immune sera (denoted “Pre”, lanes 5, 10, and 15) to test for cross-reactivity. Panel B: Peptides were titrated using antibodies against biotinylated K4, K9, and K18; equal amounts of the non-biotinylated peptide N1–25 were used as negative control.
Finally, we titrated biotinylated and non-biotinylated peptides by using antibodies to biotinylated histones. If biotinylated peptides were used as targets, the chemiluminescence signal paralleled the mass of peptide loaded per lane (Fig. 6B, top row). In contrast, no signal was detectable if non-biotinylated peptides were used as target (Fig. 6B, bottom row). These findings suggest that the affinities of antibodies for biotinylated peptides were at least seven times greater than for non-biotinylated peptides: detection limits were about 1.9 – 3.8 μg/lane for biotinylated peptides versus >15 μg/lane for non-biotinylated peptides if the autoradiography films were exposed to membranes for about five seconds.
Analysis of biotinylated histone H3 in human cells by immunoblotting and immunocytochemistry
Histone H3 in human JAr cell nuclei is biotinylated at K4, K9, and K18, as judged by Western blot analysis (Fig. 7A, top row). Recombinant (non-biotinylated) histone H3 was used as a negative control and did not produce a signal (bottom row). If human histone extracts or recombinant histone H3 were probed with pre-immune sera no signal was detectable (data not shown).
FIGURE 7.
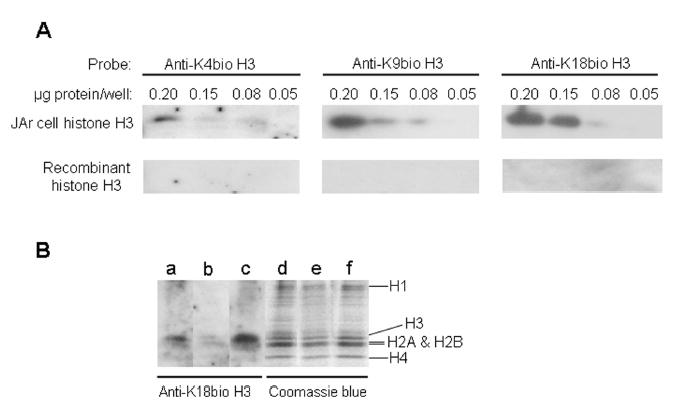
Biotinylated histones H3 are present in extracts from human cell nuclei, but biotinylation is reduced in biotinidase- and holocarboxylase synthetase-deficient cells. Panel A: Histones were extracted from JAr cell nuclei and biotinylated histones H3 were titrated using antibodies against biotinylated K4, K9, and K18; equal amounts of recombinant (non-biotinylated) histone H3 were used as negative control. Panel B: Histones were extracted from biotinidase- and holocarboxylase synthetase-deficient human skin fibroblasts (lanes a and b, respectively); IMR-90 fibroblasts from a healthy human were used as a control (lane c). Histones were probed using an antibody to K18-biotinylated histone H3. Equal loading was confirmed by staining with coomassie blue (lanes d – f).
Biotinylation of histone H3 depended on biotinidase and holocarboxylase synthetase. For example, biotinylation of K18 in histone H3 was less abundant in fibroblasts from biotinidase- and holocarboxylase synthetase-deficient individuals compared with normal fibroblasts (Fig. 7B, lanes a – c); equal loading of lanes was confirmed by staining with coomassie blue (lanes d – f). Of note, the antibodies against biotinylated histone H3 did not show significant cross-reactivity with histones H1, H2A, H2B, and H4.
Finally, biotinylated species of histone H3 were visualized in JAr cells by using immunocytochemistry. Antibody to K4-biotinylated histone H3 localized primarily to the cell nucleus (Fig. 8, image a - d); pre-immune serum did not generate a detectable signal (image e). Likewise, staining with antibodies to K9-biotinylated and K18-biotinylated histone H3 was consistent with nuclear localization of biotinylated histones (images f –o). No signal was detectable if cells were stained with secondary antibody alone (data not shown). Staining with an antibody to K12-biotinylated histone H4 [7] also produced a nuclear signal (positive control; data not shown). Collectively, these findings suggest that human cells contain histone H3, biotinylated at K4, K9, and K18.
FIGURE 8.
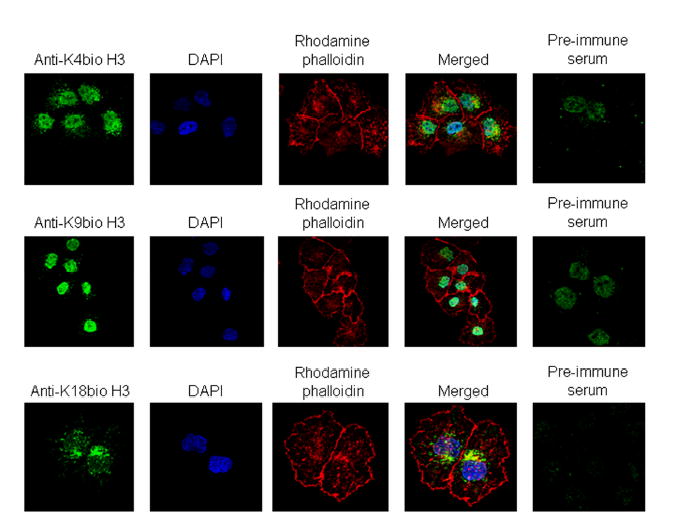
Biotinylated histones H3 localize to the nucleus in JAr cells. Cells were stained with antibodies to K4-biotinylated histone H3 (top panel), K9-biotinylated histone H3 (middle panel), and K18-biotinylated histone H3 (bottom panel). The nuclear compartment was stained using DAPI, and the cytoplasm was stained using rhodamine phalloidin. Images entitled “Merged” were created by overlaying images obtained by staining with antibody, DAPI, and rhodamine phalloidin. Pre-immune sera were used as negative controls.
DISCUSSION
This study provides evidence (i) that K4, K9, and K18 in histone H3 are good targets for biotinylation by human biotinidase; (ii) that K14 and K23 are relatively poor targets for biotinylation; (iii) that human cells contain histone H3, biotinylated in positions K4, K9, and K18; and (iv) that dimethylation of arginine residues in histone H3 enhances biotinylation of adjacent lysine residues, whereas phosphorylation of serine residues is likely to abolish biotinylation of adjacent lysine residues.
The following observations suggest that biotinylation of K4, K9, and K18 in histone H3 is physiologically important. First, evidence has been provided that biotinylation of histones might play a role in the cellular response to DNA damage [12, 13]. Second, biotinylation of histones might be associated with gene silencing [12]. Third, K4 and K9 are targets for both methylation [2] and biotinylation; methylation and biotinylation of the same lysine residue are mutually exclusive. Methylation of K4 is associated with transcriptionally active chromatin whereas methylation of K9 is associated with transcriptionally silent chromatin [3, 24]. Thus, biotinylation of K4 and K9 is likely to affect transcriptional activity of chromatin. Fourth, K18 is a target for both acetylation [2, 23] and biotinylation. Acetylation of K18 is associated with transcriptionally active chromatin [23]. It is unknown whether biotinylation of K18 affects acetylation-dependent activation of chromatin.
Modifications of arginine residues in histones affect biotinylation of adjacent lysine residues. The following lines of evidence support this notion. (i) Dimethylation of R2, R8, and R17 increased biotinylation of K4, K9, and K18, respectively, by biotinidase. Dimethylation of R2 and R17 in histone H3 has been shown to occur in vivo [2, 23], suggesting that the findings presented here are physiologically relevant. (ii) Substitution of R8 with ornithine was associated with increased biotinylation of K9. This is of potential physiological significance, given that monomethyl- and dimethyl-arginines in histones can be hydrolyzed to produce citrulline and, perhaps, ornithine [25]. Formally, we cannot exclude the possibility that free amino groups in ornithine and citrulline are substrates for biotinylation rather than enhancing biotinylation of adjacent lysines. However, our investigations of biotinylation motifs suggested that ornithine is not biotinylated by biotinidase, and that citrulline is only a relatively poor target for biotinylation (unpublished information).
Finally, the present study provides evidence that phosphorylation of serine residues may prevent biotinylation of adjacent lysine residues. This may be important for processes such as mitotic and meiotic chromosome condensation (phosphorylation of S10 and S28 in histone H3), transcriptional activation of chromatin (phosphorylation of S10 and S28 in histone H3), and DNA repair (phosphorylation of S14 in histone H2B) [23, 26].
What are the limitations of the studies presented here? First, it is unknown whether posttranslational modifications such as acetylation and methylation co-exist with biotinylation on the same histone molecule. For example, does methylation of K9 coexist with biotinylation of K4? These uncertainties are currently being addressed in our laboratory by mass spectrometrical analysis of histone extracts from human cells. Second, we have identified some modifications that affect biotinylation of histones, e.g., dimethylation of arginines and phosphorylation of serines. It is unknown as to whether this is a bi-directional interaction. For example, does biotinylation of K9 prevent phosphorylation of S10? Third, both biotinidase and holocarboxylase synthetase have biotinyl histone transferase activity [8, 9]. In the present study only biotinidase was used to identify biotinylation sites in histone H3. Theoretically, holocarboxylase synthetase might target distinct amino acid residues for biotinylation.
Taken together, the present study has revealed three new modifications of human histone H3: biotinylation of K4, K9, and K18. Previous studies suggested that K8 and K12 in histone H4 are also biotinylated [7], bringing to a total of five the known biotinylation sites in human histones. Undoubtedly, additional biotinylation sites will be identified in future studies, given that all five major classes of human histones contain streptavidin-reactive material [6]. The availability of site-specific antibodies to biotinylated histones H3 and H4 [7] is likely to generate novel insights into roles for histone biotinylation in eukaryotic cells.
MATERIALS AND METHODS
Peptide synthesis
Synthetic peptides were used as substrates for biotinidase to identify biotinylation sites in histone H3; the amino acid sequences in these peptides were based on human histone H3 (GenBank accession number NP_066403). Peptides were synthesized using N-fluoren-9-ylmethoxycarbonyl (Fmoc) chemistry by a standard solid-phase method [27] as described for our previous studies [7]; L-isomers of amino acids were used in all syntheses. One-letter annotation is used for denoting amino acids throughout this paper [28]. Chemically modified peptides were synthesized by using biotinylated, dimethylated, and phosphorylated Fmoc-ɛ-NH2-d-biotinyl-l-lysine, Fmoc-dimethyl-l-arginine, and Fmoc-phospho-l-serine. Identities of synthetic peptides were confirmed by using mass spectrometry [7].
Posttranslational modifications of histone H3 cluster in the N-terminal region of the molecule (amino acids 1 to 36), e.g., methylation of K4 and K9, acetylation of K9, K18, K23, and K36, phosphorylation of S10, and mono- or dimethylation of R17 [2]. In pilot studies we used the following synthetic peptides to determine whether biotinylation of histone H3 also takes place in the N-terminal region: (i) N-terminus of histone H3, spanning amino acids 1 to 25 (ARTKQTARKSTGGKAPRKQLATKAA; this peptide was denoted “N1–25”), and (ii) a peptide based on amino acids 15 to 39 in histone H3 (APRKQLATKAARKSAPATGGVKKPH; denoted “N15–39”). As negative control we used a peptide spanning the C-terminus of histone H3, i.e., amino acids 116 to 136 (KRVTIMPKDIQLARRIRGERA; denoted “C116–136”). Pilot studies using these peptides and previous studies of histone H4 [7] suggested that lysines located in the N-terminus of histone H3 are the primary targets for biotinylation (see below). Thus, the studies presented below focused on lysine residues in the N-terminal region; the amino acid sequences of the synthetic peptides used to identify biotinylation sites are provided in section “Results.”
Enzymatic biotinylation of peptides
Synthetic peptides were incubated with biotinidase for enzymatic biotinylation as described previously [7, 8]; biocytin (biotinyl-ɛ-lysine) was used as a biotin donor.
Gel electrophoresis
After enzymatic biotinylation, peptides were resolved using 16% tricine polyacrylamide gels according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Peptides were electroblotted onto polyvinylidene fluoride membranes (Millipore, Bedford, MA); peptide-bound biotin was probed with streptavidin-peroxidase [6, 7]. In previous studies both HPLC and mass spectrometry were used to confirm covalent biotinylation of peptides [7].
Polyclonal antibody
The following polyclonal antibodies to human histone H3 were generated using a commercial facility (Cocalico Biologicals, Reamstown, PA): anti-H3 (biotinylated at K4), anti-H3 (biotinylated at K9), and anti-H3 (biotinylated at K18). In order to raise these antibodies, the following peptides were custom-synthesized by the University of Virginia Biomolecular Research Facility: (i) N1–13bioK4 = ARTK(biotin)QTARKSTGGC (amino acids 1 – 13 in histone H3); (ii) N1–13bioK9 = ARTKQTARK(biotin)STGGC (amino acids 1 – 13); and (iii) N13–25bioK18 = GKAPRK(biotin)QLATKAAC (amino acids 13 – 25). Peptide identities were confirmed by mass spectrometry. Peptides were conjugated to keyhole limpet hemocyanin by utilizing the C-terminal cysteine [7]; these peptide conjugates were injected into white New Zealand rabbits. Booster injections were given after 14, 21, and 49 days. Serum was collected before immunization (pre-immune serum) and 2 days after each booster injection. Serum collected after the third booster injection was used for the assays described below; pre-immune serum was used as a control. For assessment of antibody specificities, electroblots of peptides N1–13bioK4, N1–13bioK9, and N13–25bioK18 were probed with the anti-histone H3 antibodies and a monoclonal mouse anti-rabbit IgG peroxidase conjugate as described [7]; non-biotinylated peptide (N1–25) was used as a control.
Analysis of biotinylated histone H3 in human cells by immunoblotting and immunocytochemistry
JAr human choriocarcinoma cells were cultured as described [29]. For Western blot analysis, nuclear histones were extracted by using hydrochloric acid as described [6]. Recombinant (non-biotinylated) histone H3 was purchased from Upstate, Inc. (Lake Placid, NY) and served as negative control. Equal amounts of JAr cell histone H3 and recombinant histone H3 (as judged by staining with coomassie blue and gel densitometry) were loaded onto 18% Tris glycine gels (Invitrogen) for electrophoresis. Proteins were electroblotted and probed with antibodies to biotinylated histone H3 as described [7]. Primary antibodies (rabbit serum) were diluted 250 fold, and the secondary antibody (mouse monoclonal anti-rabbit IgG peroxidase conjugate; Sigma, St. Louis, MO) was diluted 20,000 fold.
Biotinylation of histones is mediated by biotinidase and holocarboxylase synthetase [8, 9]. We obtained biotinidase-deficient human skin fibroblasts (strain code WG1371) and holocarboxylase synthetase-deficient human skin fibroblasts (strain code WG2215) from the Cell Repository at Montreal Children’s Hospital (Quebec, Canada) to determine whether deficiency is associated with decreased biotinylation of histone H3. Human IMR-90 fibroblasts were used as control (ATCC clone CCL-186; Manassas, VA). Nuclear histones from fibroblasts were analyzed by immunoblotting as described above.
Finally, biotinylated histones H3 in JAr cells were visualized by standard procedures of immunohistochemistry [26]. Primary antibodies (serum) were diluted 250 fold. Pre-immune sera were used as negative controls. As secondary antibody we used Cy2-conjugated AffiniPure Donkey anti-Rabbit IgG (Jackson ImmunoResearch, West Grove, PA) at an 80-fold dilution. The nuclear compartment was stained using 4’,6-diamidino-2-phenylindole (DAPI), and the cytoplasm was stained using rhodamine phalloidin (Molecular Probes, Eugene, OR). Images were obtained using Olympus FV500 confocal microscope equipped with an oil immersion lens.
Acknowledgments
This work was presented in part at Experimental Biology 2004, Washington DC [Sarath G, Kobza K, Rueckert B, Camporeale G, Zempleni J, and Haas E (2004) Biotinylation of human histone H3 and interactions with biotinidase. FASEB J 18:A103, 2004].
This work was supported by NIH grants DK 60447 and DK 063945, by a grant from the Nebraska Tobacco Settlement Biomedical Research Enhancement Funds and in part by NIH Grant Number 1 P20 RR16469 from the BRIN Program of the National Center for Research Resources. This paper is a contribution of the University of Nebraska Agricultural Research Division, Lincoln, NE 68583 (Journal Series No. 14924)
References
- 1.Wolffe, A. (1998) Chromatin, 3th edn, Academic Press, San Diego, CA.
- 2.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Boulikas T, Bastin B, Boulikas P, Dupuis G. Increase in histone poly (ADP-ribosylation) in mitogen-activated lymphoid cells. Exp Cell Res. 1990;187:77–84. doi: 10.1016/0014-4827(90)90119-u. [DOI] [PubMed] [Google Scholar]
- 5.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–5429. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 7.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur J Biochem. 2004;271:2257–2263. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 8.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem Mol Med. 1995;56:76–83. doi: 10.1006/bmme.1995.1059. [DOI] [PubMed] [Google Scholar]
- 9.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum Mol Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- 10.Brenner C. Catalysis in the nitrilase superfamily. Curr Opin Struct Biol. 2002;12:775–782. doi: 10.1016/s0959-440x(02)00387-1. [DOI] [PubMed] [Google Scholar]
- 11.Zempleni, J. (2005) Uptake, localization, and noncarboxylase roles of biotin, Annu. Rev. Nutr. (in press). [DOI] [PubMed]
- 12.Peters DM, Griffin JB, Stanley JS, Beck MM, Zempleni J. Exposure to UV light causes increased biotinylation of histones in Jurkat cells. Am J Physiol Cell Physiol. 2002;283:C878–C884. doi: 10.1152/ajpcell.00107.2002. [DOI] [PubMed] [Google Scholar]
- 13.Kothapalli N, Zempleni J. Double strand breaks of DNA decrease biotinylation of lysine-12 in histone H4 in JAr cells. FASEB J. 2004;18:A103–104. [abstract]. [Google Scholar]
- 14.Swango KL, Demirkol M, Huner G, Pronicka E, Sykut-Cegielska J, Schulze A, Mayatepek E, Wolf B. Partial biotinidase deficiency is usually due to the D444H mutation in the biotinidase gene. Hum Genet. 1998;102:571–575. doi: 10.1007/s004390050742. [DOI] [PubMed] [Google Scholar]
- 15.Wolf B, Jensen K, Huner G, Demirkol M, Baykal T, Divry P, Rolland MO, Perez-Cerda C, Ugarte M, Straussberg R, Basel-Vanagaite L, Baumgartner ER, Suormala T, Scholl S, Das AM, Schweitzer S, Pronicka E, Sykut-Cegielska J. Seventeen novel mutations that cause profound biotinidase deficiency. Mol Genet Metab. 2002;77:108–111. doi: 10.1016/s1096-7192(02)00149-x. [DOI] [PubMed] [Google Scholar]
- 16.Moslinger D, Muhl A, Suormala T, Baumgartner R, Stockler-Ipsiroglu S. Molecular characterisation and neuropsychological outcome of 21 patients with profound biotinidase deficiency detected by newborn screening and family studies. Eur J Pediatr. 2003;162:S46–49. doi: 10.1007/s00431-003-1351-3. [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Aoki Y, Li X, Sakamoto O, Hiratsuka M, Kure S, Taheri S, Christensen E, Inui K, Kubota M, Ohira M, Ohki M, Kudoh J, Kawasaki K, Shibuya K, Shintani A, Asakawa S, Minoshima S, Shimizu N, Narisawa K, Matsubara Y, Suzuki Y. Structure of human holocarboxylase synthetase gene and mutation spectrum of holocarboxylase synthetase deficiency. Hum Genet. 2001;109:526–534. doi: 10.1007/s004390100603. [DOI] [PubMed] [Google Scholar]
- 18.Wolf, B. & Heard, G. S. (1991) Biotinidase deficiency in Advances in Pediatrics (Barness, L. & Oski, F., eds) pp. 1–21, Medical Book Publishers, Chicago, IL. [PubMed]
- 19.Wolf B. Worldwide survey of neonatal screening for biotinidase deficiency. J Inher Metab Dis. 1991;14:923–927. doi: 10.1007/BF01800475. [DOI] [PubMed] [Google Scholar]
- 20.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, Cruz CC dl, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 21.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 22.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 23.Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 24.Bird A. Methylation talk between histones and DNA. Science. 2001;294:2113–2115. doi: 10.1126/science.1066726. [DOI] [PubMed] [Google Scholar]
- 25.Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 26.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 27.Fields, G. B. (1998) Solid-phase peptide synthesis in Molecular Biomethods Handbook (Rapley, R. & Walker, J. M., eds) pp. 527–545, Humana Press, Inc., Totowa, NJ.
- 28.Garrett, R. H. & Grisham, C. M. (1995) Biochemistry, Saunders College Publishing, Fort Worth, TX.
- 29.Crisp SERH, Camporeale G, White BR, Toombs CF, Griffin JB, Said HM, Zempleni J. Biotin supply affects rates of cell proliferation, biotinylation of carboxylases and histones, and expression of the gene encoding the sodium-dependent multivitamin transporter in JAr choriocarcinoma cells. Eur J Nutr. 2004;43:23–31. doi: 10.1007/s00394-004-0435-9. [DOI] [PubMed] [Google Scholar]


