Abstract
Expression of the entire complement of human immunodeficiency virus type 1 (HIV-1) viral proteins depends on the competing activities of viral RNA splicing and export into the cytoplasm by Rev. To investigate the possibility that modulation of viral RNA metabolism may alter Rev function, we analyzed the impact of multiple SR proteins on both processes. While overexpression of several of the SR factors altered splicing of HIV-1 env mRNA, they had disparate effects on Rev function that varied with the cell line used. Subsequent examination of exon splicing enhancer (ESE) and/or silencer (ESS) deletions suggests that the effects of the SR proteins on Rev function are not mediated through interaction with these elements. However, analysis of the deletions did indicate that the ESE and/or ESS does have significant effects on Rev function, with deletion of the ESS augmenting the magnitude of the response to Rev and deletion of the ESE significantly reducing it. In situ hybridization and reverse transcription-PCR indicated that the loss of Rev response upon deletion of the ESE was due to a failure of Rev to induce transport of the unspliced RNA into the cytoplasm. Together, the data indicate that cellular splicing factors and viral regulatory elements can have significant stimulatory and inhibitory effects on Rev function, raising the possibility that cells can be rendered permissive or nonpermissive for virus replication by modulation of splicing activities.
Human immunodeficiency virus type 1 (HIV-1) provides a useful model system for the study of RNA metabolism, as successful viral replication involves interplay between the processes of splicing and RNA transport. The primary viral RNA transcript is processed through alternative splicing into >25 different mRNA products (33) to generate the multiple proteins required for efficient virus assembly. The resulting viral mRNAs can be placed into three classes: the 9-kb unspliced RNA encoding gag and gag/pol proteins; the 4-kb singly spliced RNAs encoding env, vif, vpr, and vpu; and the 2-kb completely spliced RNAs which code for the regulatory proteins Tat, Rev, and Nef (45). In addition to undergoing splicing, viral RNAs of the 9- and 4-kb class are also exported from the nucleus in a process mediated by the interaction with the viral protein Rev (18, 29). The need for both splicing and Rev-mediated transport of the various viral RNAs suggests that these processes compete with one another. This hypothesis is supported by the observation that Rev function is dependent upon continued synthesis of viral RNA (20). If Rev-mediated export operates in competition with other RNA metabolic processes, then enhancing viral RNA splicing would be expected to inhibit HIV-1 replication by reducing the amount of incompletely spliced viral RNAs for Rev-mediated export and subsequent translation into structural proteins.
In addition to being regulated by the suboptimal signals present within the 3′ splice sites (3′ss) (28, 40), splicing of HIV-1 RNAs is also regulated by several cis-acting elements. Previous studies have mapped an exon splicing enhancer (ESE) in tat exon 3, while exon splicing silencers (ESS) are located in both tat exon 2 (ESS2) and 3 (ESS3) (1, 2, 8, 39). Although enhancer elements are poorly defined by sequence, studies have demonstrated that they function by enhancing recognition of adjacent splicing signals through interaction with members of the SR class of proteins (i.e., SF2/ASF, SC35, SRp40, and the SR-related proteins human Tra2α [hTra2α] and hTra2β) (16, 23, 37, 43, 44). By altering the balance of factors mediating ESE or ESS function, a shift in viral RNA splicing might be achieved, inhibiting new virion formation.
SR proteins are a family of arginine- and serine-rich proteins (14, 26) consisting of one or two amino-terminal RNA recognition motifs followed by a carboxy-terminal arginine- and serine-rich (RS) domain. The abundance of specific SR proteins varies among tissues and developmental stages (17), implicating them in establishing cell-type-dependent or developmentally regulated splicing patterns (17, 32, 36, 46). One example of how such variation can impact RNA splicing is the antagonism between SF2/ASF and hnRNP A1 for splice site selection. While SF2/ASF promotes use of a proximal 5′ss, hnRNP A1 promotes use of the distal site (7, 13).
Several pieces of evidence support the hypothesis that alterations in viral RNA processing can be achieved. Latent infection of a number of cell lines by HIV-1 has been demonstrated to be associated with predominant accumulation of multiply spliced viral RNAs (3, 22, 30, 35). Overexpression of SF2/ASF in vivo inhibited Rev function and viral replication (31). Furthermore, in vitro splicing studies using SF2/ASF have shown that SF2/ASF enhances the splicing of an HIV-1 tat substrate (4). In contrast to the effects of SF2/ASF, SC35 failed to alter HIV-1 env RNA splicing upon addition to SR-deficient S100 extracts (9, 26, 27), indicating that the alterations in splicing are specific to particular SR proteins. Although in vitro SELEX experiments had initially identified the optimal SF2/ASF binding sequence as a (GAA)n-repeat motif within the ESE (42) of the terminal exon of HIV-1 (39), subsequent experiments (43) identified hTra2α and hTra2β as the factors which directly bind the (GAA)n motif. Unlike classical SR proteins, hTra2α and hTra2β do not function in constitutive splicing in the absence of SR family proteins and cannot complement a splicing-deficient HeLa S100 extract.
The present study focused on identifying members of the SR protein family with the ability to alter HIV-1 env RNA splicing and/or inhibit Rev function. In preliminary experiments, we observed that both SF2/ASF and SC35 were able to significantly inhibit Rev function in a dose-dependent fashion, while hTra2α had a stimulatory effect. However, subsequent experiments to correlate effects on Rev function of the various SR proteins with changes in viral RNA splicing failed to yield a significant correlation. At doses of SF2/ASF or SC35 found to be strongly inhibitory of Rev function, either no change or an increase in relative levels of unspliced to spliced viral RNA were observed. Furthermore, the effect of these factors on Rev function was found to be dependent on the cell line used. Subsequent deletion studies to determine whether the observed effects are mediated through either the ESE or ESS demonstrated that removal of either one or both did not change the modulation of Rev function by these SR proteins. However, deletion of the ESS was observed to enhance the magnitude of the Rev response, while removal of the ESE resulted in significant loss of Rev-induced transport of viral RNA into the cytoplasm. Subsequent in situ hybridization experiments confirmed that deletion of the ESE resulted in an inhibition in Rev-mediated export of the unspliced RNA to the cytoplasm. Together, these observations demonstrate that multiple host factors are able to modulate Rev function and suggest that their relative activities could determine the permissiveness of a cell to support HIV-1 replication.
MATERIALS AND METHODS
Plasmids.
pET19bSF2 was a generous gift of Adrian Krainer. pDM128 was a gift from T. Parslow (19). pCMVhTra2α was obtained from William Mattox. The plasmids pgTat (25), pgTatΔESEΔESS, pgTatΔESE (34), and Vis CAT Env hygro (VCEhygro) (41) have been previously described. The Rev expression vector pSV5′H6Rev expresses a synthetic variant of Rev driven by the simian virus 40 early promoter in which the codons have been optimized for bacterial expression (11) and an amino-terminal six-histidine tag is included to permit purification. Cytomegalovirus (CMV) SF2/ASF was made using PCR primers 5′-CCCAAGCTTATGGGCCATCATCATCAT and 5′-GCTCTAGATTAGGTACGAGAACGGCT and pET19bSF2 as a template. The resulting amplicon was ligated into pCMVpA using the HindIII and XbaI sites to generate the new construct. CMVmycSC35 was generated by cloning the EcoRI fragment of pLexA-SC35 into the EcoRI site of CMVmyc. The resulting clones were confirmed by sequencing. CMVmyc was generated by PCR of pcDNA3 using 5′-CCGCTCGAGATATACGCGTTGACATTG and 5′-CCCAAGCTTCAGATCCTCTTCTGAGATGAGTTTTTGTTCCATAATTTCGATAAGCCAGTA and cloning the amplicon into the XhoI and HindIII sites of pBLSK. pLexA-SC35 was provided by Ben Blencowe.
To generate pgTatΔESS, the primers 5′-AAGGCGATTAAGTTGGGT and 5′-GCTCTAGACTTCTTCTTCTATTA were used on pSVHTSB (40) to generate an amplicon containing the 3′ portion of the env intron and adjacent exon lacking the ESS. The amplicon was digested with HindIII and XbaI, and the resultant fragment was ligated into the HindIII and XbaI site of pgTatx/x. pgTatx/x is derived from pgTat by the conversion of the XhoI site to a XbaI site. Construction of the VCEΔESS and VCEΔESEΔESS reporter plasmids was performed using a three-fragment ligation strategy. pBLSK was digested with XhoI and EcoRI and ligated with the XhoI and StuI fragment of the VCEhygro plasmid and the StuI and EcoRI fragment of either pgTatΔESS or pgTatΔESEΔESS. VCEΔESE was generated by site-directed mutation (Quick change site-directed mutagenesis kit; Stratagene) using VCEhygro as a template. Mutations were confirmed by sequencing.
Transfections and CAT assays.
Transfection into COS7 cells used the DEAE-dextran method as previously described (12). Transfections in HeLa and 293 cells used a calcium phosphate protocol (21). U937 cells were transfected with GenePorter (Gene Therapy Systems) according to the manufacturer's protocols. Forty-eight hours posttransfection, cells were rinsed with phosphate-buffered saline (PBS) and harvested, and chloramphenicol acetyltransferase (CAT) levels were assayed as previously described (15).
RNA analysis.
HeLa or 293 cells were transfected as described with 2 μg of pgTat and 8 μg of the test plasmid. Total RNA was subsequently isolated using the protocol of Chomczynski and Sacchi (10). RNase protection assays were performed using 10 μg of total RNA, according to the suggestion of the manufacturer (Ambion). RNA probes were generated by T7 transcription of a 400-bp probe that spans 200 nt on either side of the 5′ss intron-exon boundary of env.
Reverse transcription-PCR analysis of the effects of ESE or ESS deletions on env RNA splicing was performed as follows. cDNA was generated by incubation of 3 μg of total RNA with oligo(dT) and Moloney murine leukemia virus reverse transcriptase as outlined by the manufacturer (Gibco). PCR was performed using Taq polymerase, a 32P-labeled reverse primer (5′-GCAGAGGGGTGGACAGGGTAGT-3′), and the sense primers 5′-CGACCTGGATGGAGTGGGACA-3′ and 5′-AGCGGAGACAGCGACGAAGAG-3′ to detect unspliced and spliced RNA, respectively. Amplification was performed for 30 cycles as follows: 1 min at 94°C, 1 min at 56°C, and 2 min at 72°C. Amplicons were run on native polyacrylamide gels. Quantitation of unspliced and spliced RNA was performed following exposure of gels to phosphor screens and scanning with a Molecular Dynamics PhosphorImager.
Western blots.
COS cells were transfected with 5 μg of pDM128, 20 μg of CMV SF2/ASF, CMVhTra2α, CMVmycSC35, or CMVpA and 2 μg of pSV5′H6Rev. Cells were harvested and lysed in radioimmunoprecipitation assay buffer 48 h posttransfection. Fifty micrograms of total protein from each lysate was fractionated on sodium dodecyl sulfate-12.5% polyacrylamide gels and transferred to a polyvinylidene difluoride membrane. The blot was washed for 30 min at room temperature in 0.05% Tween-20-1× PBS and blocked for 1 h in 5% low-fat milk-0.2% Tween 20-1× PBS. The anti-Rev polyclonal antibody (6791) was used at a 1:100 dilution, and samples were incubated overnight at 4°C. After incubation, blots were washed in 0.05% Tween 20-1× PBS and then incubated with anti-rabbit horseradish peroxidase-conjugated antibody (Jackson Laboratories) at a 1:3,000 dilution for 1 h. The blot was washed as described above and developed with an Amersham ECL kit and exposure to Amersham Hyperfilm ECL.
Immunofluorescence.
HeLa cells were plated on glass coverslips and transfected as previously described. For localization of proteins, the cells were processed 48 h posttransfection, by washing with PBS and fixing in 4% paraformaldehyde for 30 min at room temperature. For shuttling assays, transfected cells were cocultured with mouse 3T3 cells, and cell fusion was initiated by treatment with polyethylene glycol as previously described (38). Fused cells were then incubated for 3 h prior to fixation as outlined above. The cells were washed twice with 10 mM glycine in PBS and then permeabilized with 1% Triton X-100 in PBS for 5 min. The cells were washed twice and then blocked with 3% bovine serum albumin for 1 h at room temperature or overnight at 4°C. Coverslips were inverted onto primary antibody and incubated for 1 h. After washing, coverslips were inverted over a fluorescently labeled secondary antibody and incubated for 1 h. Following washing, the coverslips were mounted onto slides using Prolong Antifade mounting buffer (Molecular Probes) containing 250 ng of 4,6-diamidino-2-phenylindole (DAPI) per μl. Images were obtained using OpenLab software on a Leica DMR microscope and captured with a Hamamatsu charge-coupled device camera. Anti-SF2/ASF antibody, AK-103, was provided by Adrian Krainer (17). Anti-SC35 was obtained from Sigma. Anti-myc antibody was obtained from Invitrogen. Polyclonal antibodies were raised against the Rev protein in rabbits.
In situ localization of unspliced env RNA.
HeLa cells were transfected with 5 μg of pgTat, pgTatΔESS, pgTatΔESE, or pgTatΔESEΔESS and 2 μg of either pSVH6Rev or pSVH6M10. Forty-eight hours posttransfection, cells were fixed for 10 min in 4% paraformaldehyde-1× PBS and in situ hybridization was carried out as previously described (34). To detect Rev/M10 within the cells, slides were incubated with rabbit polyclonal anti-Rev antibody followed by a Texas red-conjugated anti-rabbit antibody (Jackson Laboratories).
RESULTS
Effect of SR protein expression on Rev function and HIV-1 env RNA splicing.
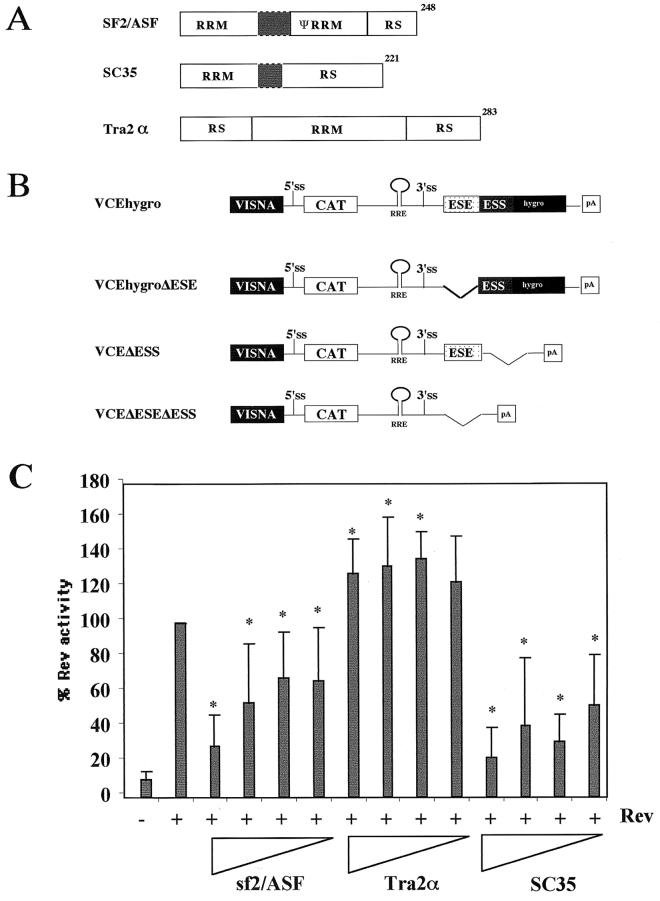
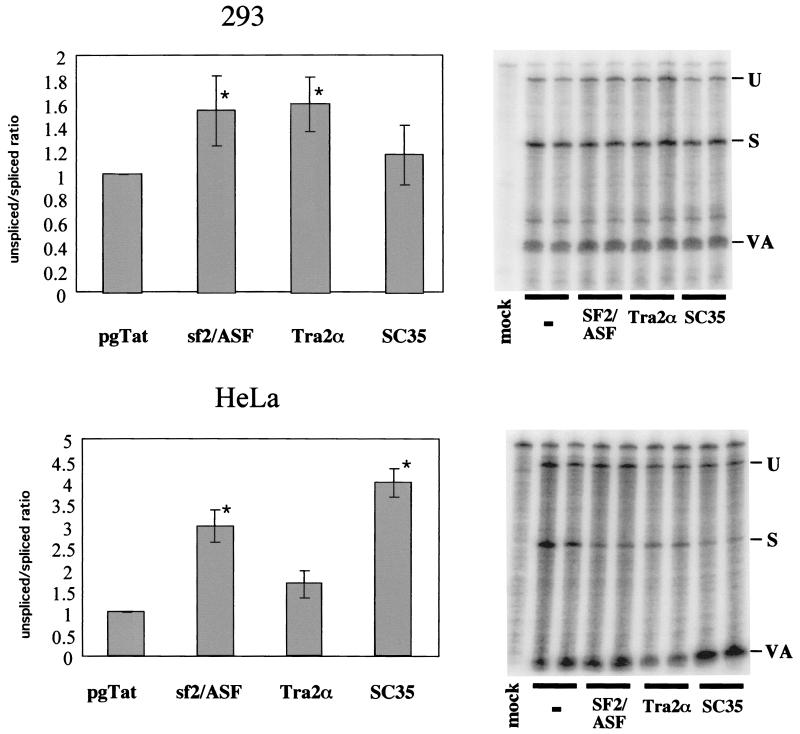
The presence of both an ESE and an ESS within the terminal exon of HIV-1 RNA suggested that the extent of viral RNA splicing and Rev function could be modulated by overexpression of factors mediating ESE or ESS function. To investigate the effect of SR proteins on Rev function, experiments were performed using increasing doses of SF2/ASF, hTra2α, or SC35, employing the Rev-dependent reporter VCEhygro. The vector VCEhygro has the CAT gene inserted into the env sequence such that expression of the CAT gene from this construct is dependent on Rev's ability to transport incompletely spliced RNA from the nucleus to the cytoplasm (41). As shown in Fig. 1, increasing quantities of both SF2/ASF and SC35 resulted in inhibition of Rev function. In contrast, over the dose range tested, hTra2α was found to be mildly stimulatory. In an attempt to correlate the effect of these factors on Rev activity to changes in HIV-1 RNA metabolism, RNase protection assays were performed. HeLa and 293 cells were transfected with pgTat and either SF2/ASF, SC35, or hTra2α, using a plasmid ratio at which maximum inhibition or stimulation of Rev function was observed, and the resulting RNase protection assays are shown in Fig. 2. To normalize for any differences in transfection efficiency or RNA recovery between samples, a plasmid expressing VA RNA was cotransfected. In previous in vitro studies performed using HeLa S100 extracts supplemented with purified SR proteins (4, 27), SF2/ASF was found to significantly augment HIV-1 env RNA splicing, while SC35 had no effect. hTra2α was expected to enhance viral RNA splicing due to its previously reported ability to stimulate RNA splicing through a GAA-repeat ESE similar to that found in the pgTat exon (43). SC35 was found to not affect HIV-1 env RNA splicing in vitro. In contrast, we found that overexpression of SF2/ASF and SC35 in HeLa cells resulted in an increased ratio of unspliced to spliced viral RNA, while in 293 cells, overexpression of the SR proteins tested only slightly altered viral RNA levels. Overexpression of hTra2α had little effect on viral RNA splicing in either HeLa or 293 cells. Consequently, a simple correlation between the ability of the SR proteins to alter Rev function and their effect on viral RNA metabolism does not appear to exist.
FIG. 1.
Effect of SR protein overexpression on Rev function. (A) Structural organization of SR proteins tested. Shaded areas indicate linker regions. (B) Schematic of Rev-dependent CAT reporter constructs. RRE, Rev-responsive element. (C) Effect of SR protein overexpression on Rev function. HeLa cells were transfected with VCEhygro in the presence and absence of Rev (0.2 μg) and various amounts (2.0, 1.0, 0.4, and 0.2 μg) of vector expressing SF2/ASF, SC35, or hTra2α. Forty-eight hours posttransfection, cells were harvested and the level of CAT expression was determined. All values were subsequently normalized to that observed upon cotransfection with Rev alone. Asterisks note values that were determined to be significant relative to Rev alone at a P of <0.005. Error bars, standard deviations.
FIG. 2.
Effects of SR protein expression on HIV-1 env RNA splicing. Cells (293 and HeLa) were transfected with 0.6 μg of VA expression plasmid, 2 μg of pgTat, and 8 μg of CMVpA, CMV SF2, CMV Tra2α, or CMV SC35. Forty-eight hours posttransfection, cells were harvested and total RNA isolated was used for RNase protection assays as described in Materials and Methods. Protected products were separated on denaturing polyacrylamide gels and levels of unspliced (U) and spliced (S) viral RNAs determined following exposure to phosphor screens. To normalize for transfection efficiency and sample recovery, samples were also probed for the presence of VA RNA (VA). Shown on the left is a summary of the data obtained from multiple data sets, while on the right is an example of the results from one such experiment. The ratio of unspliced to spliced RNA was set at 1.0 for pgTat alone to facilitate comparison between sample sets. Asterisks denote values that are statistically significant relative to pgTat alone at a P of <0.005. Error bars, standard deviations.
Effect of SR proteins on HIV-1 Rev function are cell type dependent.
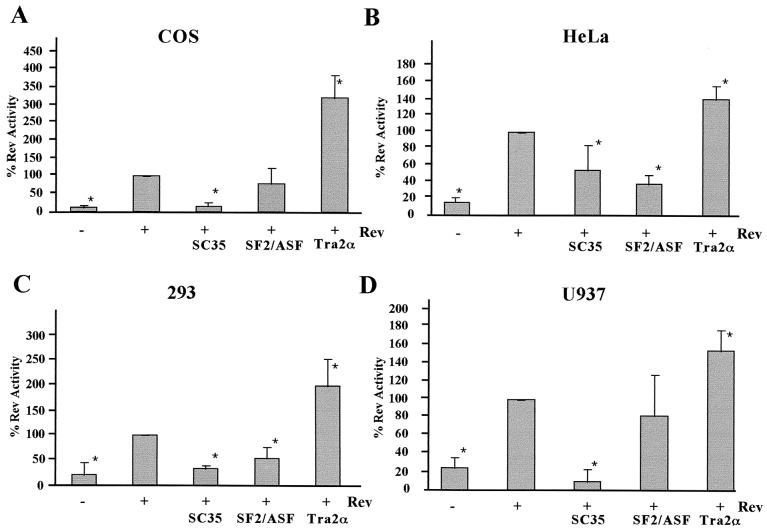
Previous work indicated that SF2/ASF inhibited Rev function and HIV-1 replication in vivo through a Rev-dependent interaction with the Rev-responsive element, possibly as a result of the increased splicing of the viral RNA (31). To test whether the response to SR protein overexpression was independent of the cellular context, assays were repeated in several cell lines (COS7, HeLa, 293, and U937). In agreement with previous studies (31), SF2/ASF was found to inhibit Rev function to a significant extent in both HeLa and 293 cell lines (Fig. 3). In contrast, overexpression of this factor in COS7 and U937 cells failed to have any significant effect on Rev function. A comparable study of SC35 showed that it was inhibitory in all cell lines tested. In contrast to the inhibitory effects of SF2/ASF and SC35, overexpression of hTra2α stimulated Rev function in all cell lines tested, although the magnitude of its effect varied (∼3.5-fold in COS7 to 1.5-fold in U937). Subsequent tests (data not shown) demonstrated that the stimulatory effect of hTra2α is not attributable to the ability of the factor alone to induce transport of unspliced RNA to the cytoplasm as it had no effect in the absence of Rev. In conclusion, the effects of these SR proteins on Rev function are diverse and cannot be correlated with their effect on viral RNA splicing. In addition, the magnitude and nature of the effects observed are often dependent upon the cell line used.
FIG. 3.
Effects of SR protein expression on Rev function are cell type dependent. Cells were transfected with VCEhygro in the presence (+) or absence (−) of pSV5′H6Rev and pCMVmycSC35, pCMVSF2/ASF, or pCMVhTra2α. Samples were collected 48 h posttransfection, and CAT assays were performed. Results shown are generated from transfections of COS7 (A), HeLa (B), 293 (C), or U937 (D) cell lines. The level of CAT expression observed in the presence of Rev alone was set at 100%, and all other values are expressed relative to that level of CAT activity. Asterisks denote values that are statistically significant relative to Rev alone at a P of <0.005. Error bars, standard deviations.
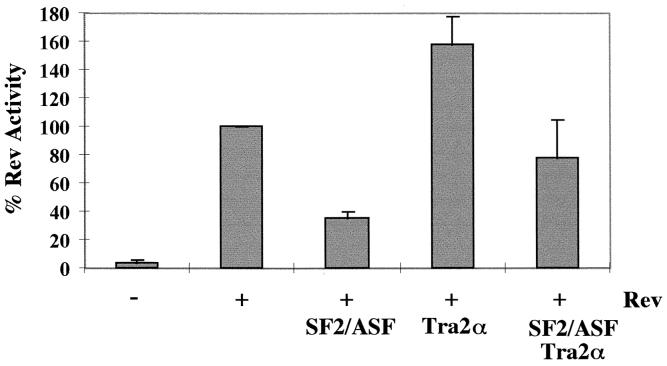
The differential effects of some of the factors in the various cell lines tested could be attributed to differences in endogenous levels of the various SR proteins. Such a hypothesis is credible given that the abundance of SR proteins can vary by cell type and developmental state (17, 32, 46). To investigate the possibility that the effect of these SR proteins may be attributed to the relative levels of individual factors, the effect of coexpression of an inhibitory SR protein with a stimulatory one was tested. Rev expression plasmid and reporter vectors were transfected into HeLa cells with SF2/ASF, hTra2α, or both. As seen previously, SF2/ASF inhibited the Rev response while hTra2α enhanced it (Fig. 4). Combination of the two factors resulted in a Rev response close to that found in the absence of the overexpressed proteins.
FIG. 4.
Effect of SR proteins on Rev function is dependent on the relative levels of SR protein expression. HeLa cells were transfected with reporter in the presence (+) or absence (−) of pSV5′H6Rev and 2.0 μg of pCMVSF2/ASF (+SF2/ASF), pCMVhTra2α (+Tra2α), or 1.0 μg of pCMVSF2/ASF and 1.0 μg of pCMVhTra2α (+SF2/ASF Tra2α). Samples were collected 48 h posttransfection, and CAT assays were performed. The level of CAT expression observed in the presence of Rev alone was set at 100%, and all other values are expressed relative to that level of CAT activity. Error bars, standard deviations.
Effect of SR proteins on Rev subcellular localization and shuttling.
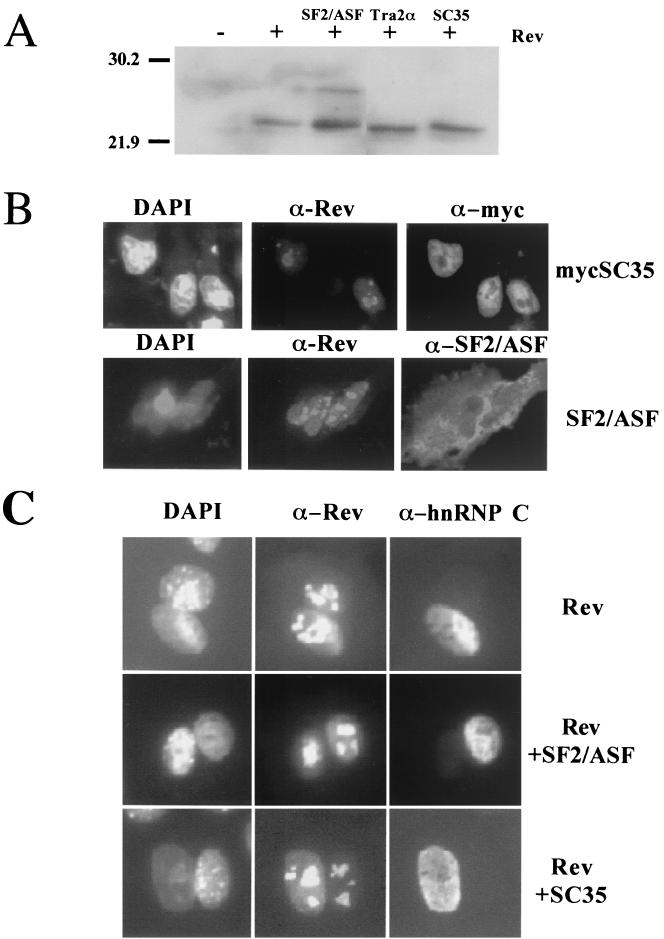
One means by which the SR proteins could alter Rev function is through changes in Rev localization or abundance. To test these possibilities, the various SR constructs were cotransfected at a 10:1 DNA ratio with the Rev expression vector pSV5′H6Rev in HeLa cells. Forty-eight hours later, the abundance and subcellular distribution of Rev and the SR proteins were analyzed by Western blotting and indirect immunofluorescence assay. None of the SR proteins were found to decrease the level of Rev expression (Fig. 5A). The slight increase in Rev protein levels detected upon addition of any of the proteins tested might explain the stimulatory effects of hTra2α but contrasts with the inhibitory effects of both SF2/ASF and SC35. Similarly, none of the factors appeared to affect Rev subcellular distribution. In the case of SF2/ASF, overexpression of the protein resulted in whole-cell staining, contrary to the association with the nuclear speckles normally seen for the endogenous protein (5) (Fig. 5B). Transfection of an SC35-expressing vector resulted in accumulation of the protein within the nucleus and within nuclear speckles as observed for the endogenous protein (14). However, no change in Rev subcellular distribution was apparent upon cotransfection with any of the SR proteins tested. Subsequent experiments to examine the effect of SF2/ASF and SC35 on Rev shuttling utilized heterokaryon assays. As shown in Fig. 5C, Rev readily redistributes from one nucleus to others upon cell fusion. In parallel experiments using HeLa cells cotransfected with Rev and SF2/ASF or SC35 expression vectors, a comparable level of accumulation of Rev in mouse nuclei was also observed. Consequently, overexpression of these proteins does not appear to have affected the capacity of Rev to shuttle.
FIG. 5.
Effect of SR protein overexpression on Rev abundance, subcellular distribution, and shuttling. Cells were transfected with pDM128 (19) in the presence of pSV5′H6Rev and the expression vectors for SF2/ASF, SC35, and hTra2α. Samples were then used in the assays outlined below. (A) Effect on Rev expression. Cells were harvested, and lysates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto a polyvinylidene difluoride membrane. Blots were subsequently probed for Rev as described in Materials and Methods. (B) Effect on Rev subcellular distribution in HeLa cells. Samples were processed 48 h posttransfection for subcellular localization of the SR proteins and Rev. Localization of SF2/ASF and SC35 was determined by indirect immunofluorescence assay using monoclonal AK-103 (1:5) and anti-myc antibody (α-myc), respectively. Localization of Rev was determined using a polyclonal anti-Rev (α-Rev) antibody. (C) Effect on Rev shuttling in HeLa cells. HeLa cells expressing Rev alone or also SF2/ASF or SC35 were fused to mouse 3T3 cells to monitor Rev shuttling between the human and mouse nuclei. Three hours postfusion, cells were fixed and Rev was localized using polyclonal α-Rev antibody. Staining with antibody specific to human hnRNP C (MAb 4F4) identified human nuclei. Also shown is a DAPI-stained image of the heterokaryon to detect the nuclei.
Effect of the ESE and ESS on Rev response to SF2/ASF, Tra2α, and SC35 expression.
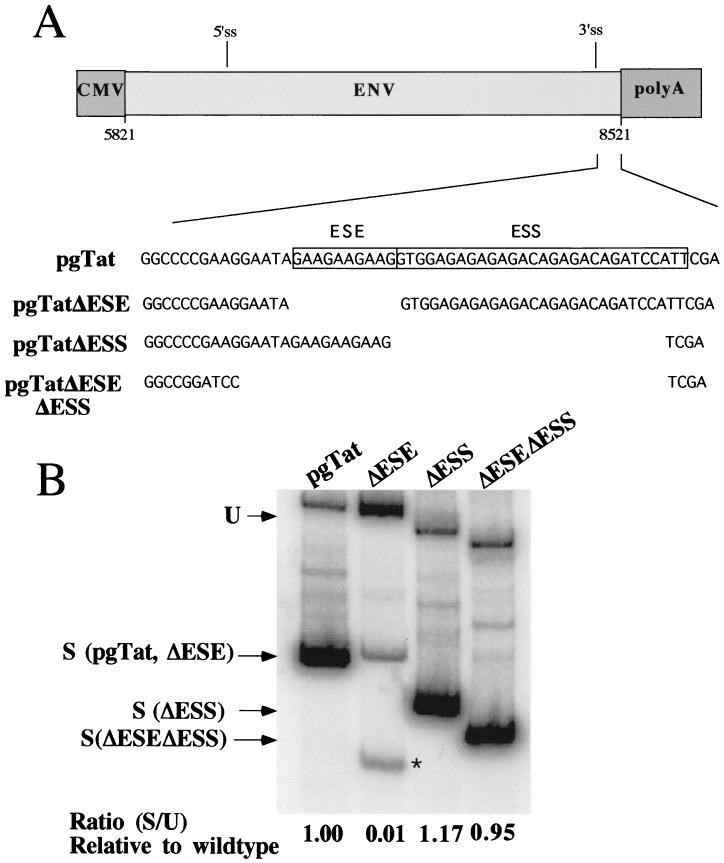
Having demonstrated that SR proteins tested can modulate Rev function, we were next interested in whether the effect was mediated by the splicing regulatory elements previously characterized. Prior to testing the effect of deleting the ESE and ESS within tat/rev exon 3 on SR protein modulation of Rev function, we first examined the effect of these mutations on viral RNA processing (Fig. 6). As reported previously (34, 39), deletion of the ESE results in a marked reduction in the level of correctly spliced RNA generated and induces the use of an alternative 3′ss downstream of the correct one (Fig. 6, pgTatΔESE). Deletion of both the ESE and ESS resulted in a splicing pattern comparable to that found for the wild-type RNA, although the quantity of spliced product was slightly reduced. In contrast to previous observations with truncated forms of env RNA lacking most of the intron sequence (39), removal of the ESS resulted in only a slight increase in the extent of splicing relative to the wild type.
FIG. 6.
Effect of ESE and/or ESS deletions on env RNA splicing. HeLa cells were transfected with 5 μg of pgTat, pgTatΔESE, pgTatΔESS, or pgTatΔESEΔESS. Forty-eight hours posttransfection, cells were harvested and used in reverse transcription-PCR assays for analysis of the relative levels of unspliced and spliced env RNA generated. Products were subsequently analyzed on polyacrylamide gels. Indicated are the positions of the products generated from the unspliced (U) and spliced (S) forms of the RNA generated from the various vectors. The asterisk marks an aberrant spliced product generated from pgTatΔESE.
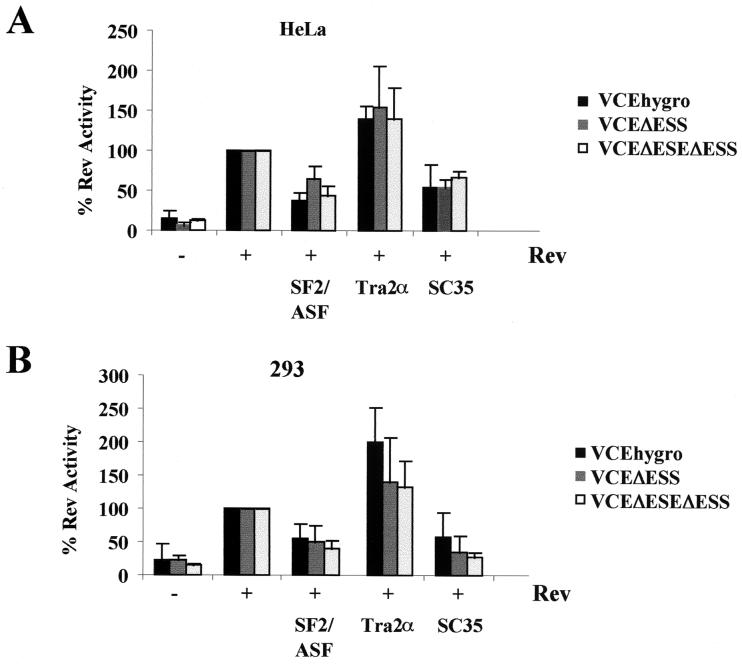
To examine what effect the ESE and ESS had on Rev function, reporter plasmids were constructed which delete either the ESS (VCEΔESS), the ESE (VCEΔESE), or the ESE and ESS in combination (VCEΔESEΔESS) (Fig. 1). These reporters were subsequently tested for changes in Rev response upon cotransfection of the various SR proteins. In order to facilitate the comparison of the responses of the various reporters used, results for each reporter were normalized to the level of CAT activity obtained upon cotransfection with Rev alone. As shown in Fig. 7, there was no significant difference in the response to SF2/ASF, hTra2α, or SC35 overexpression in the absence of either the ESS or both the ESE and ESS in the cell types tested. While deletion of the ESE and/or the ESS failed to alter the relative response to SR protein overexpression, the mutant RNAs did display altered Rev response characteristics within HeLa cells. Induction over baseline is about ninefold upon addition of Rev to the reporter containing both exonic elements in HeLa cells. However, addition of Rev to VCE ΔESS results in a 24.62 ± 6.42-fold induction over baseline. Subsequent deletion of both the ESE and ESS resulted in induction ratios close to that seen when both the ESE and ESS are present (5.71 ± 1.35 versus 8.91 ± 4.74, respectively). In contrast, deletion of the ESE was found to significantly impair the response to Rev, with the VCEΔESE yielding only a 2.56 ± 0.93-fold induction.
FIG. 7.
Contribution of ESE and ESS to SR protein modulation of Rev function. HeLa or 293 cells were transfected with 0.5 μg of VCEhygro, VCEΔESS, or VCEΔESEΔESS in the presence (+) or absence (−) of 0.2 μg of pSV5′H6Rev and 2.0 μg of pCMVSF2/ASF, pCMVhTra2α, or pCMVmycSC35. Samples were collected 48 h posttransfection, and CAT assays were performed. Results shown are generated from transfections of HeLa (A) and 293 (B) cell lines. To facilitate comparison among the various reporter vectors (VCE, VCEΔESS, and VCEΔESEΔESS), the value of CAT expression for each reporter in the presence of Rev alone was set at 100%, and all other values are expressed relative to that level of CAT activity. Error bars, standard deviations.
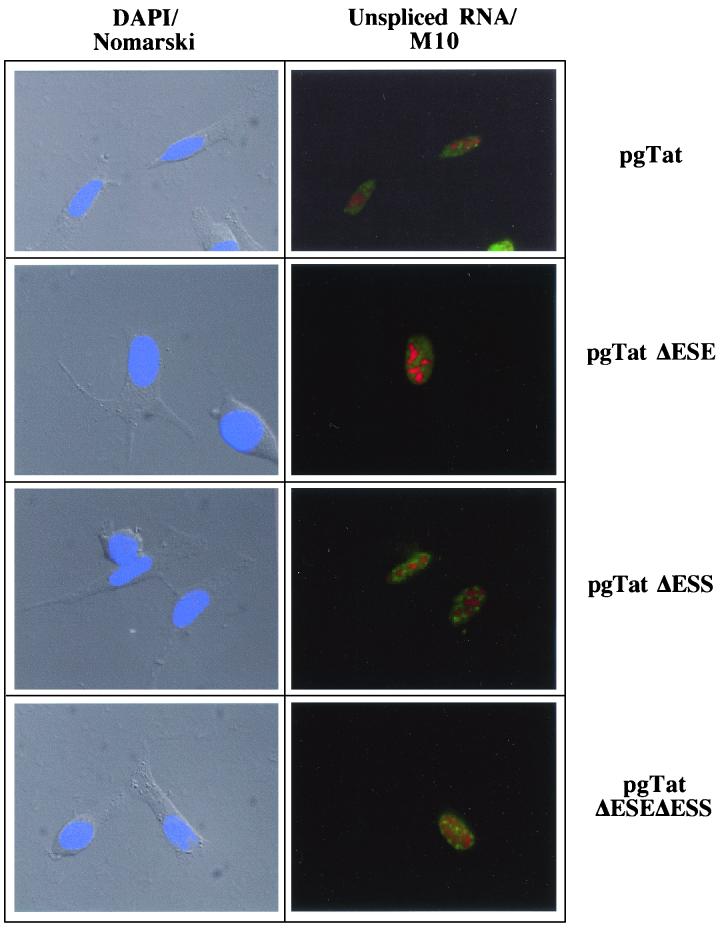
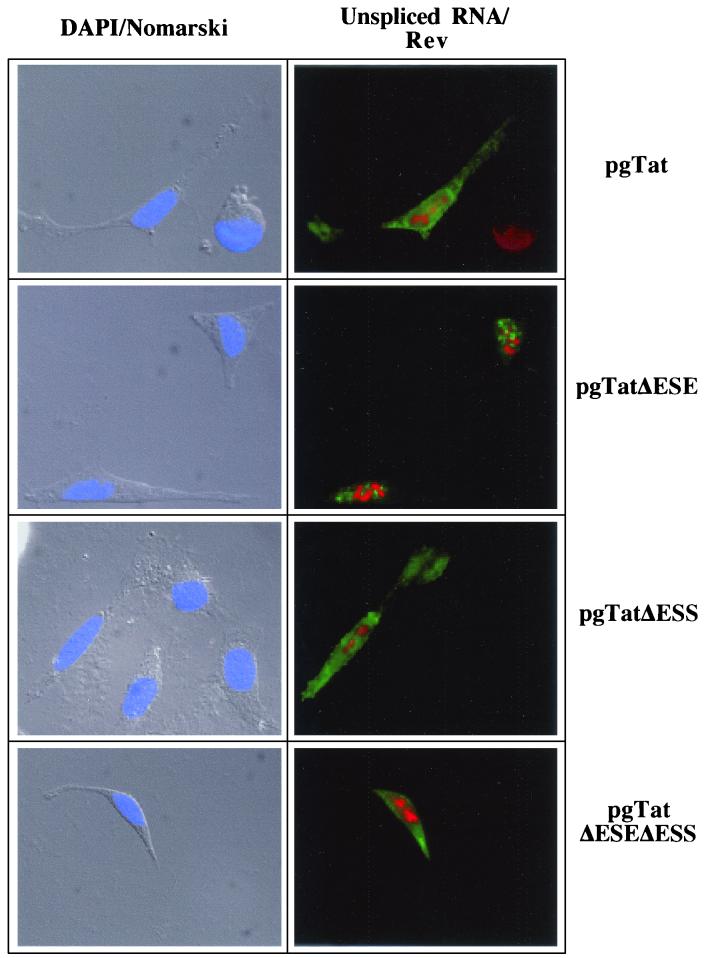
The changes in extent of Rev response among the various reporters cannot be easily accounted for by differences in the quantity of unspliced env RNA present (Fig. 6). Therefore, in situ hybridization was used to examine the subcellular distribution of the RNA in the absence and presence of Rev. As shown in Fig. 8, cotransfection with an inactive mutant of Rev (M10) (24) results in the accumulation of the unspliced viral RNA in discrete foci within the nucleus for all the RNA mutants tested (location of unspliced viral RNA is indicated by green staining; location of Rev or M10 is indicated by red staining). Consistent with the responses to Rev expression seen in the reporter assays, expression of Rev results in extensive cytoplasmic accumulation of unspliced env RNA generated from pgTat, pgTatΔESS, and pgTatΔESEΔESS (Fig. 9). In contrast, unspliced RNA from pgTatΔESE remained confined to discrete foci within the nucleus. Similar differences in distribution pattern between VCEhygro and VCEΔESE were observed in the presence of Rev (data not shown). Costaining for Rev confirmed expression of the protein in these cells.
FIG. 8.
Subcellular distribution of viral RNAs in the absence of Rev. HeLa cells were transfected with 2 μg of pSVH6M10 and 5 μg of pgTat, pgTatΔESE, pgTatΔESS, or pgTatΔESEΔESS. Forty-eight hours posttransfection, cells were fixed and processed for in situ hybridization as outlined in the text. Location of unspliced env was determined by staining with fluorescein-isothiocyanate-conjugated anti-digoxigenin antibody (green), while M10 (red) was localized using rabbit polyclonal anti-Rev antibody and Texas red-conjugated goat anti-rabbit antibody. Images were collected using a Leica DMR microscope at a magnification of ×630. Shown at left is an overlay of the DAPI staining (blue) of nuclei with the Nomarski image of the cell.
FIG. 9.
Effect of ESE and/or ESS deletions on viral RNA subcellular distribution in the presence of Rev. HeLa cells were transfected with 2 μg of pSVH6Rev and 5 μg of pgTat, pgTatΔESE, pgTatΔESS, or pgTatΔESEΔESS. Forty-eight hours posttransfection, cells were fixed and processed for in situ hybridization as outlined in the text. Location of unspliced env RNA was determined by staining with fluorescein isothiocyanate-conjugated anti-digoxigenin antibody (green), while Rev (red) was localized using rabbit polyclonal anti-Rev antibody and Texas red-conjugated goat anti-rabbit antibody. Images were collected using a Leica DMR microscope at a magnification of ×630. Shown at left is an overlay of the DAPI staining (blue) of the nuclei with the Nomarski image of the cell.
DISCUSSION
HIV-1 RNA splicing and transport are important points of control for the successful replication of the virus. However, the elements and factors that control these processes are only beginning to be evaluated. In previous work (20), we observed that viral RNA within the nucleus was not uniformly accessible for Rev-mediated export to the cytoplasm. Inhibition of viral RNA synthesis was found to result in an almost complete inhibition of the Rev response despite the presence of significant quantities of unspliced HIV-1 env RNA in the nucleus. It was hypothesized that these RNAs were rendered inaccessible for Rev-mediated export due to commitment to either splicing or sequestration in the nucleus. Consequently, we believed that by accelerating the commitment of HIV-1 RNAs to either splicing or nuclear sequestration, an inhibition of Rev function could be achieved. The mapping of an ESE and ESS within the terminal exon of HIV-1 suggested that one means by which alteration of HIV-1 RNA metabolism could be achieved would be through changes in abundance of the SR protein(s) which mediates the effect of the ESE (16, 37). In support of this model, it was previously determined that SF2/ASF overexpression alters both viral RNA splicing and Rev function (31). To test the model further, other members of the SR and SR-like family were tested for their effects, with particular focus on those (ASF/SF2, SC35, and hTra2α) which have been previously shown to bind to sequences within the ESE and ESS (27, 42, 47). In light of these previous findings, it was surprising to observe that overexpression of SF2/ASF and hTra2α did not universally result in increased viral RNA splicing (Fig. 2). In addition, SC35 was found to increase the ratio of unspliced to spliced viral RNA within both HeLa and 293 cell lines.
Furthermore, the effects of the factors on viral RNA splicing were not predictive of their modulation of Rev function. Overexpression of both SF2/ASF and SC35 resulted in significant inhibition of Rev function in HeLa and 293 cells despite failing to strongly stimulate viral RNA splicing in either. In addition, hTra2α, which was expected to augment viral RNA splicing, resulted in a stimulation of Rev function in the cell lines tested. The inhibition of Rev-mediated RNA export by SC35 is particularly intriguing in light of the previous finding that SC35 overexpression stimulated cytoplasmic accumulation of unspliced simian virus 40 RNA (47). These findings suggest that the effect of SC35 on RNA transport may be either pathway or template dependent. The subsequent finding that the nature and magnitude of the effect of the SR proteins (SF2/ASF in particular) on Rev function varied significantly from one cell line to the next supports the hypothesis that their function is context dependent. The demonstration that coexpression of the factors resulted in a balancing of their effects (Fig. 4) suggests that the proteins act at equivalent points in the RNA metabolic pathway but with opposing effects. The different effects of the various factors raise the possibility that a cell's ability to support Rev function could be modulated by changes in relative expression of SR proteins, rendering a cell permissive or nonpermissive for virus replication.
Attempts to define the mechanism by which SF2/ASF, SC35, and hTra2α alter Rev function have been inconclusive. All the factors failed to significantly affect Rev abundance, subcellular localization, or capacity to shuttle. Furthermore, the failure of ESE and ESS deletions to alter their activity suggests that these SR proteins function either in an indirect fashion or through binding to another region of the RNA. However, data clearly indicate that the ESE and ESS do play a role in Rev function, as the removal of either element changed the magnitude of Rev-induced gene expression (see above). Removal of the ESS resulted in a three- to fourfold increase in the level of induction by Rev relative to wild type, while deletion of the ESE significantly reduced Rev responsiveness. Deletion of both the ESE and ESS resulted in induction ratios comparable to that observed for the wild-type reporter. Analysis of the effect of these deletions on viral RNA splicing (Fig. 6) determined that deletion of the ESE inhibited viral RNA splicing, while RNA lacking both the ESE and ESS had a splicing pattern comparable to that of the wild type, as has been previously reported (34). The failure of the ESS deletion to have any significant effect on viral RNA splicing was surprising in light of previous data based on subregions of env which determined that deletion of the ESS stimulated RNA splicing (1, 39). One possible explanation for the difference could be the presence of functionally analogous ESS sequences within the intron which were deleted in the constructs used previously. Alternatively, the introns used in previous in vivo assays (39) were close to the minimum able to undergo splicing. Therefore, the ESE may stimulate the processing of this extremely suboptimal intron. Subsequent in situ analysis (Fig. 8 and 9) confirms the functional data and demonstrates that deletion of the ESE results in failure of Rev to export unspliced viral RNA into the cytoplasm. Comparison of the level of unspliced RNA among the various expression constructs (Fig. 6) failed to demonstrate significant differences, indicating that the observed effects cannot be ascribed to changes in abundance of this RNA. The inhibition of Rev-mediated export seen for pgTatΔESE cannot be ascribed to the inhibition of splicing alone. Addition of hnRNP A1 binding sites was found to elicit the same decrease in spliced RNA without a comparable inhibition of RNA transport (K. Asai and A. Cochrane, unpublished data). Therefore, differences in responsiveness to Rev must be attributed to the number or nature of the factors that are binding the ESS. Evidence supports a model in which factors binding the ESS direct the RNA into a complex or metabolic pathway that renders the RNA inaccessible to Rev-mediated transport into the cytoplasm. The ability of RNA lacking both the ESE and ESS to be spliced and transported to the cytoplasm by Rev to the same extent as wild type indicates that the ESE is not essential for either activity but rather serves to counteract the effect of the ESS. However, the finding that RNA containing only the ESE confers a greater Rev response without dramatically altering the extent of viral RNA splicing suggests that the ESE may enhance the commitment of the RNA to a pathway that is more conducive to Rev-mediated export.
Acknowledgments
J.P. is supported by a studentship from OHTN, K.A. is supported by an NSERC studentship, and A.C. is the recipient of an OHTN scientist award. Research was supported by grants from the CIHR and OHTN.
We thank B. Blencowe and C. M. Stoltzfus for critical review of the manuscript.
REFERENCES
- 1.Amendt, B., Z. Si, and C. M. Stoltzfus. 1995. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 15:4606-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendt, B. A., D. Hesslein, L.-J. Chang, and C. M. Stoltzfus. 1994. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 14:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butera, S., B. Roberts, L. Lam, T. Hodge, and T. Folks. 1994. Human immunodeficiency virus type 1 RNA expression by four chronically infected cell lines indicates multiple mechanisms of latency. J. Virol. 68:2726-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caceres, J., and A. Krainer. 1993. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 12:4715-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caceres, J., T. Misteli, G. Screaton, D. Spector, and A. Krainer. 1997. Role of modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caceres, J., T. Misteli, G. Screaton, D. Spector, and A. Krainer. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706-1709. [DOI] [PubMed] [Google Scholar]
- 8.Caputi, M., A. Mayeda, A. Krainer, and A. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler, S., A. Mayeda, J. Yeakley, A. Krainer, and X.-D. Fu. 1997. RNA splicing specificity determined by the coordinated action of the RNA recognition motifs in SR proteins. Proc. Natl. Acad. Sci. USA 94:3596-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Cochrane, A., C.-H. Chen, R. Kramer, L. Tomchak, and C. Rosen. 1989. Purification of biologically active human immunodeficiency virus rev protein from Escherichia coli. Virology 173:335-337. [DOI] [PubMed] [Google Scholar]
- 12.Cullen, B. R. 1987. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 152:684-704. [DOI] [PubMed] [Google Scholar]
- 13.Eperon, I., O. Makarova, A. Mayeda, S. Munroe, J. Caceres, D. Hayward, and A. Krainer. 2000. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 20:8303-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, X.-D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663-680. [PMC free article] [PubMed] [Google Scholar]
- 15.Gorman, C. M., L. F. Moffatt, and B. H. Howard. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanamura, A., J. Caceres, A. Mayeda, B. R. Franza, Jr., and A. Krainer. 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4:430-444. [PMC free article] [PubMed] [Google Scholar]
- 18.Hope, T. J. 1999. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365:186-191. [DOI] [PubMed] [Google Scholar]
- 19.Hope, T. J., X. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping of cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iacampo, S., and A. Cochrane. 1996. Human immunodeficiency virus type 1 Rev function requires continued synthesis of its target mRNA. J. Virol. 70:8332-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kriegler, M. 1990. Gene transfer and expression: a laboratory manual, 1st ed. Stockton Press, New York, N.Y.
- 22.Laughlin, M., and R. Pomerantz. 1994. Retroviral latency. R. G. Landes Company, Austin, Tex.
- 23.Liu, H., M. Zhang, and A. Krainer. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12:1998-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malim, M. H., S. Bohnlein, J. Hauber, and B. R. Cullen. 1989. Functional dissection of the HIV-1 rev trans-activator: derivation of a trans-dominant repressor of rev function. Cell 58:205-214. [DOI] [PubMed] [Google Scholar]
- 25.Malim, M. H., J. Hauber, S.-Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 26.Manley, J. L., and R. Tacke. 1996. SR proteins and splicing control. Genes Dev. 10:1569-1579. [DOI] [PubMed] [Google Scholar]
- 27.Mayeda, A., G. Screaton, S. Chandler, X.-D. Fu, and A. Krainer. 1999. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol. 19:1853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Reilly, M., M. McNally, and K. Beemon. 1995. Two strong 5′ splice sites and competing 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology 213:373-385. [DOI] [PubMed] [Google Scholar]
- 29.Pollard, V., and M. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 30.Pomerantz, R. J., D. Trono, M. B. Feinberg, and D. Baltimore. 1990. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral mRNA expression: a molecular model for latency. Cell 61:1271-1276. [DOI] [PubMed] [Google Scholar]
- 31.Powell, D., M. Amaral, J. Wu, T. Maniatis, and W. Greene. 1997. HIV Rev-dependent binding of SF2/ASF to the Rev response element: possible role in Rev-mediated inhibition of HIV RNA splicing. Proc. Natl. Acad. Sci. USA 94:973-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samford, J. R., and J. P. Bruzik. 2001. Regulation of SR protein localization during development. Proc. Natl. Acad. Sci. USA 98:10184-10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz, S., B. K. Felber, D. M. Benko, E.-M. Fenyo, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seguin, B., A. Staffa, and A. Cochrane. 1998. Control of HIV-1 RNA metabolism: the role of splice sites and intron sequences in unspliced viral RNA subcellular distribution. J. Virol. 72:9503-9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seshamma, T., D. Bagrasa, D. Trono, D. Baltimore, and R. Pomerantz. 1992. Blocked early-stage latency in the peripheral blood cells of certain individuals infected with human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 89:10663-10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinozaki, A., K. Arahata, and T. Tsukahara. 1999. Changes in pre-mRNA splicing factors during neural differentiation in P19 embryonal carcinoma cells. Int. J. Biochem. Cell Biol. 31:1279-1287. [DOI] [PubMed] [Google Scholar]
- 37.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 38.Soros, V., and A. Cochrane. 2001. Alterations in HIV-1 Rev transport in response to cell stress. Virology 280:199-210. [DOI] [PubMed] [Google Scholar]
- 39.Staffa, A., and A. Cochrane. 1995. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 15:4597-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staffa, A., and A. Cochrane. 1994. The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3′ splice site. J. Virol. 68:3071-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swenarchuk, L., P. Harakidas, and A. Cochrane. 1999. Regulated expression of HIV-1 Rev function in mammalian cell lines. Can. J. Microbiol. 45:480-490. [PubMed] [Google Scholar]
- 42.Tacke, R., and J. L. Manley. 1995. The human splicing factors ASF/SF2 and SC35 possess different, functionally significant RNA binding specificities. EMBO J. 14:3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tacke, R., M. Tohyama, S. Ogawa, and J. Manley. 1998. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell 93:139-148. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka, K., A. Watakabe, and Y. Shimura. 1994. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol. Cell. Biol. 14:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, H., K. L. Kuhen, and F. Wong-Staal. 1999. Lentivirus replication and regulation. Annu. Rev. Genet. 33:133-170. [DOI] [PubMed] [Google Scholar]
- 46.ten Dam, G. B., B. Wieringa, and L. G. Poels. 1999. Alternative splicing of CD45 pre-mRNA is uniquely obedient to conditions in lymphoid cells. Biochim. Biophys. Acta 1446:317-333. [DOI] [PubMed] [Google Scholar]
- 47.Wang, J., and J. Manley. 1995. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA 1:335-346. [PMC free article] [PubMed] [Google Scholar]