Abstract
The cereal aleurone layer is a model system for studying the regulation of transcription by gibberellin (GA) and abscisic acid (ABA). GA stimulates and ABA prevents the transcription of genes for α-amylases and other secreted hydrolytic enzymes, but how GA and ABA affect the transcription of other genes is largely unknown. We characterized gene expression in rice (Oryza sativa) aleurone using a half-genome rice microarray. Of the 23,000 probe sets on the chip, approximately 11,000 hybridized with RNA from rice aleurone treated with ABA, GA, or no hormone. As expected, GA regulated the expression of many genes, and 3 times as many genes were up-regulated by GA at 8 h than were down-regulated. Changes in gene expression resulting from ABA treatment were not consistent with the hypothesis that the role of ABA in this tissue is primarily to repress gene expression, and 10 times more genes were up-regulated by ABA at 8 h than were down-regulated by ABA. We also measured transcript abundance in aleurone of dwarf1 (d1) mutant rice. The d1 protein is the sole α-subunit of heterotrimeric G-proteins in rice. Genes up-regulated by GA or ABA had higher expression in wild type than in d1 aleurone, and genes down-regulated by GA had lower expression in wild type relative to d1 aleurone. The d1 mutation did not result in a decrease in sensitivity to GA at the level of transcription. Rather, changes in transcript abundance were smaller in the d1 mutant than in wild type.
The aleurone layer of small cereal grains, such as barley (Hordeum vulgare), rice (Oryza sativa), wheat (Triticum aestivum), and wild oats (Avena fatua), has been used extensively to characterize the response of a plant tissue to GA and abscisic acid (ABA). The power of the aleurone layer for the study of gene expression lies in the simplicity of the tissue and the strength of the response. Aleurone layers contain a single cell type that is not photosynthetic and neither grows nor divides following grain maturation. Because of this uniformity, the cells in the aleurone layer respond nearly synchronously to exogenously applied GA and ABA. For some genes, these responses are dramatic. It has been estimated that transcripts for α-amylases may make up 20% of all newly transcribed mRNAs 24 h after GA addition, and α-amylase protein represents as much as 60% of all newly synthesized protein (Higgins et al., 1982). These rapid, large, hormone-dependent changes reflect the function of the aleurone layer, which is to produce a battery of hydrolytic enzymes that breaks down the stored carbohydrate and protein reserves in the dead starchy endosperm. The resulting sugars, small peptides, and amino acids are used to support postgerminative growth of the seedling. In the intact grain, GA produced by the embryo triggers the production of these hydrolases (Jacobsen et al., 2002). In the event that environmental conditions are unfavorable for germination or continued seedling growth, ABA produced by the embryo and aleurone layer prevents the synthesis of hydrolases and the break down of stored endosperm reserves (Jacobsen and Chandler, 1987).
In parallel to studies on hormone-regulated transcription in cereal aleurone layers, other work has focused on identifying signal transduction elements in this tissue. Some of these elements, such as changes in the concentration of cytosolic Ca2+, are not directly regulated at the level of transcription. A few, such as GAMyb, are transcriptionally regulated (Gubler et al., 1997). In rice, dwarf1 (d1) is a hormone-signaling mutant that lacks the sole Gα-subunit of heterotrimeric G-proteins in the rice genome. Mutant plants are semidwarf and deep green. Aleurone layers from d1 grain are less sensitive to GA than those from wild-type grain with regard to α-amylase mRNA accumulation, secreted α-amylase activity, and Ca2+-ATPase mRNA accumulation (Ueguchi-Tanaka et al., 2000). Previous experiments with d1 have led to the bypass model for GA signaling in which heterotrimeric G-proteins are important components of the GA signal transduction network at low concentrations of GA, but are much less important at high concentrations of GA when alternative signaling pathways are presumed to be paramount (Ueguchi-Tanaka et al., 2000; Iwasaki et al., 2003).
With the advent of microarray technology, it is now possible to examine changes in transcript abundance for thousands of genes within a single experiment. Data from microarray experiments therefore have the potential to add substantial depth to our understanding of how genes are regulated in response to a defined signal. In particular, it is now possible to compare the responses of selected, individual genes to the responses of large groups of genes that have not been identified in advance. Global or large-scale changes in transcription have been determined for cereals on a few occasions (Potokina et al., 2002; Lin et al., 2003; Rabbani et al., 2003; Close et al., 2004; Schnable et al., 2004; Yang et al., 2004; Yazaki et al., 2004). In most cases, however, RNA was isolated from complex tissues or after relatively long treatment times. For example, RNA for hybridization with cDNA or oligonucleotide-based microarrays has been extracted from developing rice seedlings (Lin et al., 2003; Rabbani et al., 2003); rice shoots, leaves, or roots (Kawasaki et al., 2001; Ozturk et al., 2002; Yang et al., 2004); barley leaves (Close et al., 2004); or maize (Zea mays) embryos (Lee et al., 2002). Interpretation of the data for these experiments is complicated because different tissues and cell types are expected to show distinct, yet overlapping, patterns of gene expression. A small increase in the measured amount of a transcript may reflect a small increase in all cells and tissues, or a large increase in a tissue that makes up a small percentage of the total tissue mass. Likewise, the observed change in transcription of a gene within a small gene family will be reduced if different tissues or cells utilize different family members. An alternative has been to isolate RNA from rice callus (Yazaki et al., 2004), which eliminates some of the difficulties associated with multiple or complex tissues, but complicates attempts to compare the microarray data with transcriptional changes that occur in planta.
We have used an oligonucleotide microarray containing 23,780 probe sets representing approximately 21,000 rice genes (Zhu et al., 2003) to examine transcript profiles in rice aleurone. Approximately one-half of the genes in the rice genome are represented on this array (Goff et al., 2002). Our data confirm the gene expression data that have been published previously for the rice aleurone layer and reveal novel aspects of transcriptional control by GA and ABA. We have also used d1 grain to determine how loss of Gα influences transcript abundance in tissue treated with GA or ABA and to test the bypass model of GA signaling. Finally, we have identified some of the genes in rice aleurone that are likely to be hormonally and/or temporally regulated.
RESULTS
Rice Half-Grains Contain Transcripts for Approximately One-Half of the Rice Genome
To learn more about hormonal control of gene expression in rice aleurone layers, we monitored the transcriptome of imbibed, de-embryonated rice grains (half-grains) treated with 20 mm CaCl2 alone (no hormone) for 0, 0.5, 1, 3, 6, or 8 h, or CaCl2 and either 5 μm GA or 5 μm ABA for 0.5, 1, 3, 6, or 8 h. The data in Table I make it clear that rice half-grains contained transcripts for a large number of genes. Regardless of hormone treatment, 45% to 50% of the probe sets on the microarray detected transcripts from the sample. Approximately 9,762 probe sets, or 41.1% of the total number of probe sets, had present calls for all treatments at all times, and this percentage may be an estimate for the percentage of the genome required for proper function of the rice aleurone.
Table I.
Number of probe sets called present when RNA samples from rice half-grains were hybridized to the rice oligonucleotide microarray
| Treatment | Probe Sets Present | Percentage of Total Probe Sets |
|---|---|---|
| No hormone | 11,506 | 48.4% |
| ABA (5 μm) | 10,899 | 45.8% |
| GA (5 μm) | 11,126 | 46.8% |
Expression Profiles of Rice Aleurone Genes Reveal Patterns of Gene Expression
Hormone-dependent changes in the abundance of specific mRNAs are likely to be associated with responses of rice aleurone layers to GA or ABA and, hence, to aleurone layer function. We identified subsets of the expressed genes that were enriched for probe sets with hormone-responsive changes in abundance. As an initial step, we removed probe sets from the data for which there was little or no evidence for a hormone-dependent change in transcript abundance. To do this, we computed an F statistic by comparing the expression values for the five samples not treated with hormone with the five samples treated with ABA or GA at 0.5, 1, 3, 6, and 8 h. Probe sets with a P value >0.05 do not have a hormone-dependent change in variance and are unlikely therefore to have a statistically meaningful, hormone-dependent change in expression within 8 h of hormone treatment. Using this procedure, we obtained datasets containing approximately 1,300 probe sets that were enriched for those with GA-dependent differences in gene expression and 1,350 probe sets that were enriched for those with ABA-dependent differences in gene expression relative to controls that were not treated with hormone.
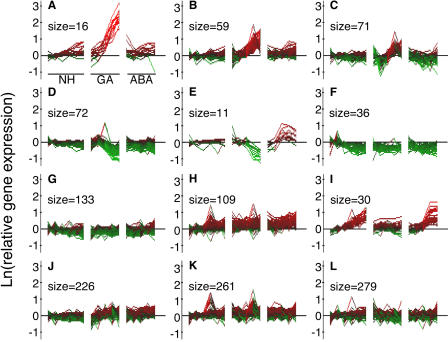
To visualize the nature of global changes in gene expression in rice aleurone, we used k-means clustering. The datasets enriched for GA- or ABA-responsive genes were sorted into 12 clusters based on their time- and hormone-dependent pattern of expression. The clusters for the probe sets enriched for GA-regulated genes are shown in Figure 1. For each cluster, the transformed probe set intensity is shown at each time of RNA extraction for tissue treated with no hormone (left six points), GA (middle five points), and ABA (right five points). Three clusters showed various degrees of up-regulation in GA-treated tissue. These are seen in Figure 1, A to C, and the probe sets in each cluster are presented in Supplemental Table I, A to C. The most strongly up-regulated cluster (Fig. 1A) contains 10 secreted hydrolases and includes α-amylases and endo-1,4-β-xylanases, as expected. This cluster also includes two probe sets for K+ transporters and one for a calcineurin B-like (CBL) protein. The clusters in Figure 1, B and C, show genes that are up-regulated by GA, but to a lesser extent than those in Figure 1A. Figure 1B has several probe sets for gene functions associated with the mobilization of stored reserves, including gluconeogenesis (lipase, acyl CoA oxidase), proteolysis (two carboxypeptidases, Cys proteinase), and phytate hydrolysis (acid phosphatases). Also in this cluster are two sugar transporters and a peptide transporter. Genes with functions in protein synthesis and posttranslational modification include BiP, calnexin, sec61B, a protein disulfide isomerase, and glucosyl-, acyl-, and xylosyl-transferases. Potential signaling elements include GAMyb, a CBL protein, and a protein phosphatase 2C. Cluster C in Figure 1 contains probe sets with similar putative functions, including Glc and peptide transporters, oryzain, vignain, acetyltransferase, and several cytochrome oxidase-related proteins (Supplemental Table IC). Two clusters show GA-dependent down-regulation of expression (Fig. 1, D and E; Supplemental Table I, D and E) with the data in Figure 1E also showing up-regulation by ABA. This cluster contains probe sets for dehydrin and late embryogenesis abundant (LEA) genes, among others. Genes down-regulated in all treatments relative to zero time (Fig. 1F), up-regulated in all treatments (Fig. 1H), or up-regulated with ABA or no hormone, but not strongly up-regulated by GA (Fig. 1I; Supplemental Table I, I), are also represented by clusters of at least 30 probe sets. Figure 1, J to L, shows little obvious pattern of regulation, except for a trend toward an increase in expression with time in ABA-treated samples (Fig. 1J), an increase in expression with GA and ABA at intermediate time points (Fig. 1K), and a decrease in expression with time in ABA-treated samples (Fig. 1L).
Figure 1.
Patterns of gene expression in a dataset containing 1,311 probe sets enriched for those that were GA responsive. The natural logarithm (ln) of probe set intensity at each time relative to the average intensity with no hormone (NH) at 0, 0.5, and 1 h is shown for each of the 16 microarrays used in this experiment. Samples were extracted from rice half-grains treated with NH for 0, 0.5, 1, 3, 6, and 8 h; GA for 0.5, 1, 3, 6, and 8 h; and ABA for 0.5, 1, 3, 6, and 8 h. Red indicates an increase in transcript abundance, and green indicates a decrease. The number of probe sets in each k-means cluster is indicated as its size.
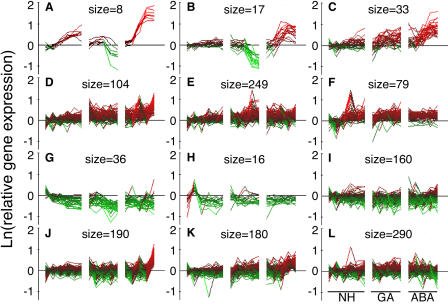
The same analysis was used to visualize changes in expression for the probe sets contained in the gene list enriched for ABA-responsive genes. Genes up-regulated by ABA and down-regulated by GA are seen in Figure 2, A and B, and Supplemental Table II, A and B, and genes up-regulated by ABA and less strongly up-regulated by no hormone and GA are seen in Figure 2C. The genes represented by Figure 2, A and B, include several dehydrins and LEA proteins. Genes that are up-regulated in all treatments relative to time zero are represented by the clusters shown in Figure 2, D to F. Genes down-regulated in all treatments are contained in the clusters shown in Figure 2, G to I. Two clusters show little or no obvious patterns of expression except for an up-regulation of expression at late times with ABA (Fig. 2, J and K). There are no clusters that show down-regulation by ABA in the absence of a change with GA or no hormone.
Figure 2.
Patterns of gene expression in a dataset of 1,369 probe sets enriched for those that are ABA-responsive. The natural logarithm (ln) of probe set intensity relative to the average intensity with no hormone (NH) at 0, 0.5, and 1 h is shown for each of the 16 microarrays used in this experiment. Samples were extracted from rice half-grains treated with NH for 0, 0.5, 1, 3, 6, and 8 h; GA for 0.5, 1, 3, 6, and 8 h; and ABA for 0.5, 1, 3, 6, and 8 h. Red indicates an increase in transcript abundance, and green indicates a decrease. The number of probe sets in each k-means cluster is indicated as its size.
Genes Specifically Regulated by GA or ABA Are Rare
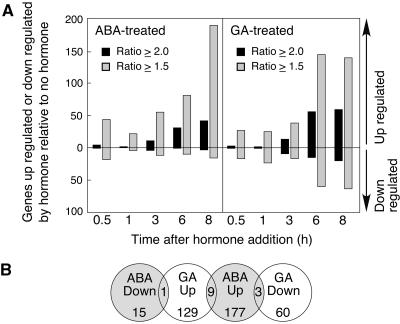
The data in Figures 1 and 2 suggested to us that few genes were regulated specifically by GA or ABA relative to no-hormone controls. To quantify hormonally regulated changes in gene expression, we identified the probe sets that were called present and whose signal intensity increased or decreased 2-fold or 1.5-fold relative to no hormone. Previous work by others and our own analyses (Supplemental Fig. 1) indicate that the false positive rate for the rice array is 0.5% (Zhu et al., 2003) or less (0.37%; Supplemental Fig. 1), with a 2-fold cutoff and approximately 1.9% with a 1.5-fold cutoff (Supplemental Fig. 1). Regardless of the expression ratio used, the number of hormonally regulated genes was relatively small. As seen in Figure 3A, fewer than 60 genes were specifically up-regulated or down-regulated by either ABA or GA at any of the five time points relative to no-hormone controls when a 2-fold cutoff was used. Even with the less stringent criterion of 1.5-fold, fewer than 200 genes showed an increase or decrease in expression in response to hormone treatment. A comparison of the genes up- or down-regulated by GA or ABA at 8 h shows that there is little evidence to support the hypothesis that many genes up-regulated by one hormone are down-regulated by the other when the hormones are added individually (Fig. 3B). Only one gene was down 2-fold with ABA and up 2-fold with GA, and this gene did not have any hits against other genes with a known function. Three genes were up with ABA and down with GA: a LEA protein and two dehydrins. Nine genes were up 2-fold with both hormones. No genes were specifically down-regulated by both GA and ABA.
Figure 3.
Number of genes specifically up- or down-regulated in rice half-grains by ABA or GA is small. A, RNA was extracted from rice half-grains treated with 5 μm ABA, 5 μm GA, or no hormone for 0.5, 1, 3, 6, or 8 h, and probe sets with a fold change in intensity >1.5 or 2 between hormone-treated and untreated were identified at each time point. B, Distribution of probe sets that were hormonally responsive by 1.5-fold or more at 8 h into categories of decreased in abundance with ABA (ABA down), increased in abundance with GA (GA up), increased in abundance with ABA (ABA up), and decreased in abundance with GA (GA down). The number of probe sets in two adjacent categories is indicated where the circles overlap.
The data in Figure 3 illustrate three important findings related to the global nature of gene regulation by ABA and GA in rice aleurone layers. First, the number of genes up-regulated by ABA is comparable to the number of genes up-regulated by GA. Second, GA down-regulates about one-third as many genes as it up-regulates. For example, 8 h after hormone treatment, 59 genes were specifically up-regulated and 20 genes were specifically down-regulated by GA. Third, GA down-regulates many more genes than ABA down-regulates. Indeed, the maximal number of genes down-regulated by ABA does not exceed the expected number of false positives.
The Gα-Protein in Rice Amplifies Transcriptional Changes
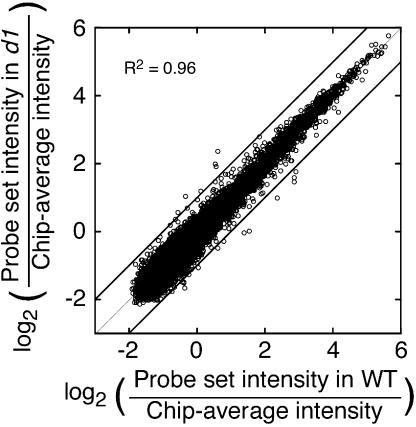
The mutated protein in d1 rice is the sole Gα-subunit in rice, and a role for Gα in GA signaling has been demonstrated previously (Ueguchi-Tanaka et al., 2000). We used transcriptional profiling and d1 grain to determine how loss of Gα function influences transcript abundance on a global scale. To show that the approach was suitable for this analysis, we compared transcript abundance for 13,818 probe sets called present in arrays hybridized with RNA from imbibed wild-type and d1 half-grains. As shown in the scatter plot in Figure 4, there was an extremely tight correspondence (r2 = 0.962) between probe set intensities in wild type and d1 at time zero. Intensity values for only 0.17% of the probe sets differed by more than 2-fold when the data from wild-type half-grains were compared with the data from d1 half-grains. This further confirms the reliability of the method as a tool for genome-scale comparisons and shows that there are small errors in the experimental system.
Figure 4.
Scatter plot of probe set intensities (13,818 probe sets) for wild-type rice and d1 mutant rice half-grains shows that few probe sets vary in intensity by more than 2-fold (diagonal lines) at time zero.
To assess the extent to which Gα influences transcript abundance, we compared expression in wild type with expression in d1 rice half-grains 8 h after treatment with either 5 μm GA or 100 nm GA. We chose 100 nm GA for one treatment because previous experiments demonstrated a large difference between wild type and d1 in the transcription of the α-amylase genes at this concentration (Ueguchi-Tanaka et al., 2000). We used 5 μm GA to facilitate comparisons with the time-course experiment and because there was a small difference between wild type and d1 in the amount of transcript for the α-amylase genes at this concentration (Ueguchi-Tanaka et al., 2000). Systematic differences in transcription between wild type and mutant are easily seen when the probe sets are ordered from those whose signal intensity increased most to those whose signal intensity decreased most 8 h after treatment of half-grain with 5 μm GA. Probe sets were grouped into bins of 25 probe sets to facilitate statistical analysis. Only those groups of 25 probe sets whose average expression at 8 h was 1.5-fold higher or lower than that at 0 h were included in this analysis.
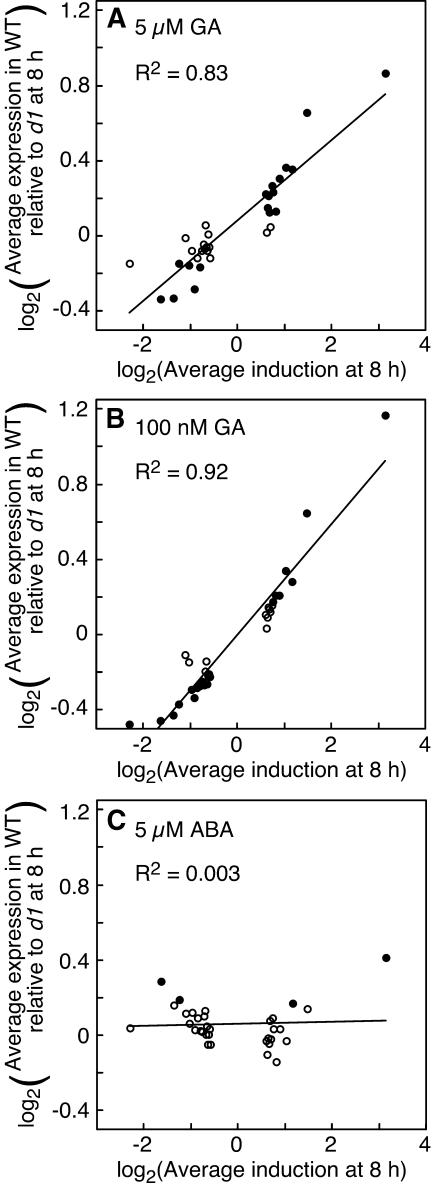
As seen in Figure 5, there is a clear, genome-wide effect of the Gα-subunit on transcript abundance. In this figure, ratios are plotted as log2 values so that relative changes in magnitude are easily compared and equal weight is given to comparable increases and decreases in intensity (e.g. a ratio of 2 and a ratio of 0.5). Genes that had the largest relative increase in expression in GA-treated samples (Fig. 5, data points farther to the right) had higher expression in wild-type half-grains relative to d1 half-grains (i.e. log2 ratio of expression > 0). In samples treated with 5 μm GA, the 25 probe sets most strongly induced by GA (Fig. 5A, data point farthest to the right) had an average expression in wild type that was almost twice that in d1 at 8 h. The ratio of wild-type to d1 expression for the same set of genes in samples treated with 100 nm GA was 2.2 (Fig. 5B). Conversely, probe sets that decreased in intensity relative to time zero had lower ratios of wild-type to d1 expression. Genes most strongly repressed with GA (Fig. 5, data points farther to the left) had an average expression in wild type that was as much as 30% less than that in d1 half-grains when samples were treated with 100 nm GA (Fig. 5B). These changes in wild-type expression relative to d1 expression were related to changes in mRNA abundance. For the data from half-grains treated with 5 μm GA, there was a good correlation (r2 = 0.83) between gene expression at 8 h relative to 0 h and the ratio of wild-type to d1 expression (Fig. 5A). For half-grains treated with 100 nm GA, this correlation was even better (r2 = 0.92). It is particularly noteworthy that these correlations hold for genes that show small changes in mRNA abundance during the 8 h of the experiment, as well as for genes showing large changes in abundance. Thus, the Gα-protein amplifies transcriptional changes, but is not required for most transcriptional changes. These data also suggest that the Gα-protein influences the abundance of many transcripts in GA-treated samples, regardless of whether they are up- or down-regulated. The differences in mRNA abundance between wild type and d1, however, were small. Only those probe sets with strong up- or down-regulation with GA had ratios of expression between wild type and d1 that were over 50%.
Figure 5.
Changes in RNA abundance are reduced in d1 half-grains compared to wild-type half-grains at both 5 μm GA (A) and 100 nm GA (B). Each point in A to C represents 25 probe sets, with the 25 probe sets having the highest induction with GA at 8 h represented by the rightmost point and the 25 probe sets with the highest repression with GA represented by the leftmost point. Black circles have an average expression ratio different from 1.0 at the level of P = 0.001. The d1 mutation does not affect the expression of the same sets of probes in half-grains treated with ABA (C). Note that the graphs in A to C are plotted on a log2-log2 scale.
To show that these relatively small differences were statistically significant, we asked whether each group of 25 probe sets had a mean ratio of wild-type to d1 expression that was different from 1.00. Groups of 25 genes in Figure 5 that have P values from one-tailed t tests <0.001 are indicated by black data points. Those groups of 25 genes where P > 0.001 are indicated by white circles. Overall, for the 33 groups of 25 probe sets used in this analysis, 19 had a P value <0.001 for samples treated with 5 μm GA and 22 had a P value <0.001 for samples treated with 100 nm GA. In both cases, the observed number is much greater than the expected number, which for this P value is less than 1.
As a control for this analysis, we looked at the intensity values for the same groups of GA-regulated genes in ABA-treated samples. In this case, there was very little difference in average expression between wild type and d1 (Fig. 5C) and little correlation between the ratio of wild-type to d1 expression and changes in gene expression with GA (Fig. 5C; r2 = 0.003).
Effects of Gα-Protein on Transcription Are Not Specific for GA
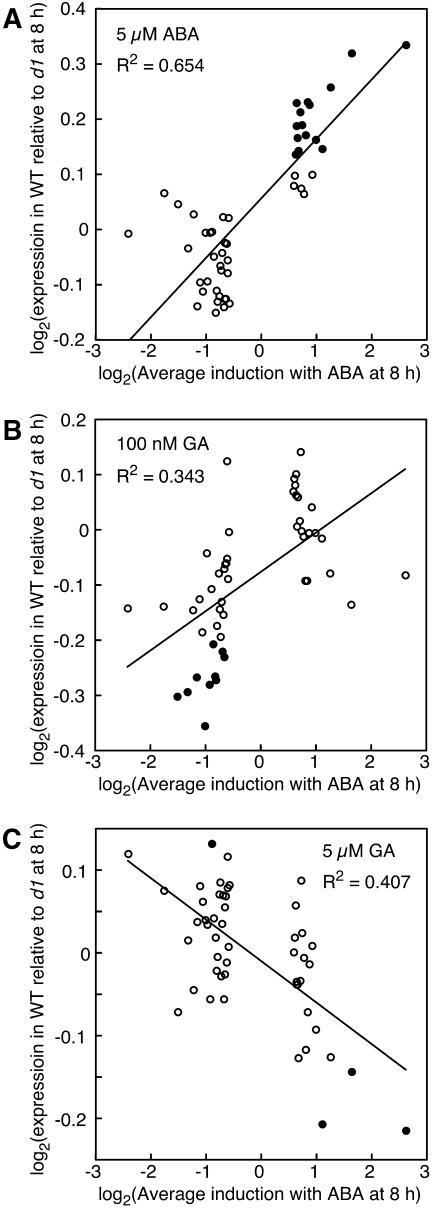
To see whether Gα-protein affected transcription of ABA-regulated genes, we compared the signal intensities of ABA-regulated genes in wild-type and d1 half-grains. Using the same approach as that described above, genes from ABA-treated samples were sorted and grouped by expression 8 h after treatment of wild-type samples with 5 μm ABA relative to expression at 0 h. As seen in Figure 6A, there was a good correlation between wild-type to d1 expression ratio and changes in intensity in samples treated with ABA (r2 = 0.65). As for the GA-treated samples, probe sets with larger changes in expression at 8 h relative to 0 h had larger differences in expression in wild type relative to d1. These differences in expression between wild-type and d1 grains were small, but are likely to be statistically meaningful. Of the 50 groups of 25 probe sets used in this analysis, 15 (black circles) had a P value <0.001 when one-tailed t tests were used to compare the ratio of wild-type to d1 expression for the 25 probe sets to probe sets having a ratio of 1.00 (Fig. 6A). When the analysis was extended to look at the same probe sets in GA-treated samples, there were still weak correlations between the ratio of expression in wild type to d1 and changes in gene expression in ABA-treated tissue (Fig. 6, B and C). This most likely reflects the observation that many changes in gene expression are not hormone specific (see below).
Figure 6.
Changes in RNA abundance for many genes that are up- or down-regulated with ABA are reduced in d1 mutant half-grains relative to wild type. Each point in A to C represents 25 probe sets, with the 25 probe sets having the highest induction with ABA at 8 h represented by the rightmost point and the 25 probe sets with the highest repression with ABA represented by the leftmost point. Black circles have an average expression ratio different from 1.0 at the level of P = 0.001. Note that the graphs in A to C are plotted on a log2-log2 scale.
Loss of Gα-Protein Does Not Decrease the Sensitivity to GA for Most Genes
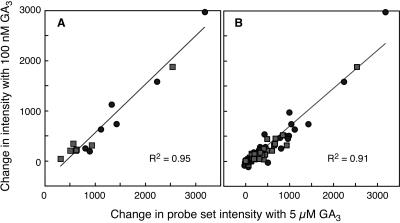
The d1 mutation results in decreased sensitivity to GA with respect to internode elongation and secreted α-amylase activity (Ueguchi-Tanaka et al., 2000). These data gave rise to the bypass model. We wished to know whether there was also a decrease in sensitivity to GA at the level of transcription. For this analysis, we examined two sets of genes. First, we looked at the seven α-amylases that were strongly up-regulated by GA (Fig. 1A; Supplemental Table IA). Second, we looked at the probe sets corresponding to the 59 genes that were specifically up-regulated 2-fold with GA in the time-course experiment 8 h after GA treatment (Fig. 3). These 59 probe sets are listed in Supplemental Table III and include five α-amylases, four proteases, an endo-β-xylanase, a GAMyb, two CBL proteins, and an acyl CoA oxidase. To look for changes in sensitivity to GA, we plotted the change in probe set intensity between 0 and 8 h with 5 μm GA against the change in intensity with 100 nm GA for tissue from wild-type and d1 mutant grains. If there is a difference in sensitivity between wild type and the d1 mutant, the data for changes in gene expression in wild type should fall on one line and the data for changes in gene expression in the d1 mutant should fall on a different line with a lower slope. The data for α-amylases are shown in Figure 7A. What is clear from this figure is that the line relating changes in gene expression at 5 μm GA to changes at 100 nm GA is no different for RNA samples from d1 mutants than for RNA samples from wild-type grains. Therefore, although the magnitude of the change observed with the d1 mutant is less than the magnitude of the change observed with wild type, there was no decrease in sensitivity to GA. For both wild type and d1 mutants, an α-amylase gene that had a large increase in transcript abundance with 5 μm GA had a large increase in transcript abundance with 100 nm GA. Conversely, for both wild type and d1 mutants, an α-amylase gene that had a moderate increase in transcript abundance with 5 μm GA had a moderate increase in transcript abundance with 100 nm GA. The data for the 59 probe sets that are up-regulated by GA show a similar pattern and again provide no evidence to support the hypothesis that the loss of the Gα-subunit results in a decrease in the sensitivity to GA at the level of transcription (Fig. 7B).
Figure 7.
Sensitivity of transcription to GA is not reduced in the d1 mutation. A, Changes in probe set intensity for the seven probe sets corresponding to α-amylase genes. B, Changes in probe set intensity for a group of GA-regulated genes. Data from wild-type half-grains in A and B are indicated by circles, and data from d1 half-grains are indicated by squares.
Time-Dependent Changes in Gene Expression Occur in the Absence of Hormone Treatment
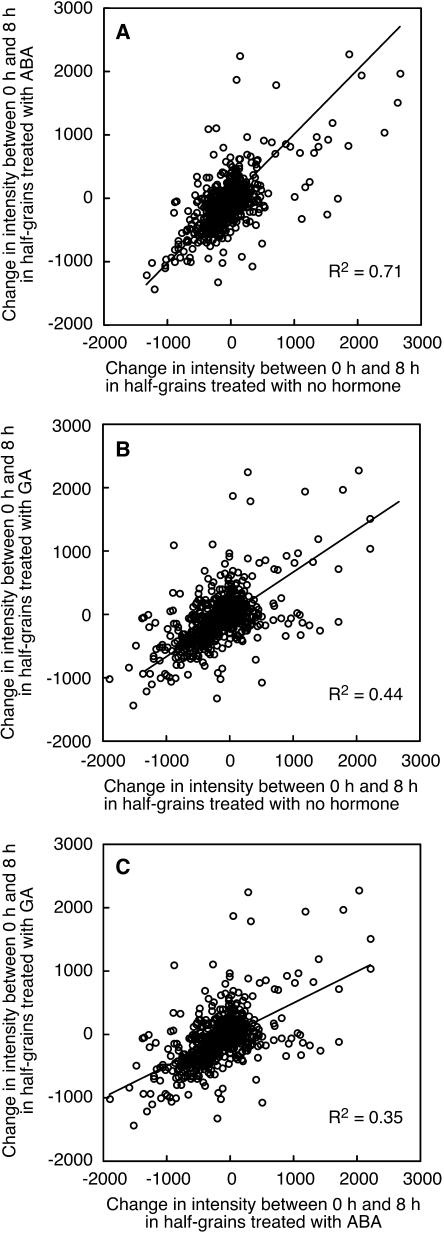
Time-dependent changes in gene expression may be as important as those that are hormone dependent. To identify genes in rice half-grains that have a temporal increase or decrease in mRNA, we compared signal intensity values for 9,762 probe sets called present 8 h after treating wild-type rice grains with GA or ABA with signal intensity values at 0 h. In the case of ABA-treated half-grains, the changes are very similar to those that occur in the absence of added hormone. As seen in Figure 8A, the differences in probe set signal intensity between 0 and 8 h are nearly equal for tissue treated with ABA and for tissue treated with no hormone, and the slope of the best-fit line is 1.0 (r2 = 0.71). In some cases, these changes are very large. Differences in hybridization signal intensity of over 2,000 units were observed in these data, where the mean signal intensity is 80. For 80% of the dataset, however, changes in transcription between 0 and 8 h are less than 25%. Likewise, changes in probe set signal intensity in GA-treated tissue mirror changes in tissue treated with no hormone (Fig. 8B). In this case, however, the correlation between changes in half-grains not treated with hormone and half-grains treated with GA was fairly low (r2 = 0.44) and this reflects the increases and decreases in mRNA abundance that occur with GA but do not occur in the absence of added hormone. The correlation between changes in probe set signal intensity for GA- and ABA-treated half-grains was lower (r2 = 0.35), but there was still a general trend such that probe sets that increased in intensity in one treatment increased in intensity in the other (Fig. 8C).
Figure 8.
Many changes in gene expression are not dependent on hormone treatment. A to C, Changes in intensity between 8 and 0 h for 9,762 probe sets in rice half-grains treated with 5 μm ABA and no hormone (A), 5 μm GA and no hormone (B), and 5 μm GA and 5 μm ABA (C). Note that the average intensity value for all probe sets on each microarray is 80.
Identification of Genes That Have Time-Dependent Changes in Transcription
To identify genes that were up-regulated or down-regulated with time, we found the probe sets whose signal intensities were up- or down-regulated in wild-type half-grains by 2-fold or more in both the rice time-course experiment and the d1 experiment. Over the 8-h time course of the experiments, 37 genes showed a 2-fold increase in expression in GA-treated tissue between 0 and 8 h (these are listed in Table II). As expected, hydrolytic enzymes are prominent and the probe sets identified include those for amylases (four times), endo-1,4-β-xylanase, endo-1,3-β-glucosidase, and proteases and carboxypeptidases (four times). Several up-regulated probe sets corresponded to genes that are involved in gluconeogenesis, including a lipase and acyl CoA oxidase. Two probe sets for CBL genes are also included in the list of genes up-regulated in GA-treated half-grains.
Table II.
Probe sets up-regulated with time in GA-treated rice half-grains
Induction is the ratio of expression at 8 h relative to 0 h. Similar proteins are identified by accession numbers and a brief description.
| Probe Set | Induction at 8 h in Time Course | Induction at 8 h in d1 Experiment | Accession Nos. | Description |
|---|---|---|---|---|
| Hydrolases | ||||
| OS000411_i_at | 30.4 | 6.5 | gi|113766|sp|P17654| | AMY1_ORYSA α-amylase 1 |
| OS000626_s_at | 10.72 | 23.6 | gi|113680|sp|P27937| | AM3B_ORYSA α-amylase isozyme 3B |
| OS022194_at | 9.96 | 20.95 | gi|113683|sp|P27934| | AM3E_ORYSA α-amylase isozyme 3E |
| OS000501_at | 7.59 | 22.01 | gi|113683|sp|P27934| | AM3E_ORYSA α-amylase isozyme 3E |
| OS023228_at | 9.69 | 11.59 | P14768 | XYNA_PSEFL endo-1,4-β-xylanase A |
| OS000920_s_at | 5.22 | 3.69 | P15737 | E13B_HORVU glucan endo-1,3-β-glucosidase GII |
| OS006213_at | 2.6 | 5.03 | gi|7262670|gb|AAF43928.1|AC012188_5| | Contains similarity to acid phosphatase and contains a sterol desaturase domain (Arabidopsis) |
| OS000590_f_at | 4.57 | 2.27 | Q06015 | CHI3_ARAHY endochitinase 3 |
| Proteolysis | ||||
| OS017015_s_at | 2.88 | 2.66 | gi|7671402|emb|CAB89316.1| | Carboxypeptidase (Arabidopsis) |
| OS020092.1_at | 2.1 | 3.32 | gi|3859606|gb|AAC72872.1| | Contains similarity to Cys proteases (Arabidopsis) |
| OS009235.1_at | 2.93 | 2.34 | P37891 | CBP3_ORYSA Ser carboxypeptidase III |
| OS012349_at | 2.21 | 2.36 | gi|3859607|gb|AAC72873.1| | Contains similarity to Cys proteases (Arabidopsis) |
| OS000667.1_at | 2.44 | 2.08 | gi|7635921|emb|CAB88392.1| | Bowman-Birk trypsin inhibitor (rice) |
| Lipid mobilization | ||||
| OS008739.1_at | 4.76 | 12.81 | gi|4587543|gb|AAD25774.1|AC006577_10| | Lipase/acylhydrolase with GDSL-motif family (Arabidopsis) |
| OS001843_f_at | 3.19 | 2.34 | P10974 | NLTB_RICCO nonspecific lipid-transfer protein B |
| OS012669.1_at | 2.68 | 2.46 | gi|3044212|gb|AAC13497.1| | Acyl-CoA oxidase (Arabidopsis) |
| Membrane transport | ||||
| OS006069.1_at | 2.43 | 5.75 | gi|7262678|gb|AAF43936.1|AC012188_13| | Contains similarity to UDP-galactose transporter (Arabidopsis) |
| OS005993.1_at | 3.85 | 3.09 | gi|5091611|gb|AAD39600.1|AC007858_14 gb|U43629| | Integral membrane protein from Beta vulgaris and member of the sugar transporter family (rice) |
| OS019839_s_at | 2.96 | 3.76 | gi|4249409|gb|AAD13706.1| | Sugar transporter (Arabidopsis) |
| Signaling | ||||
| OS015683_at | 5.66 | 3.2 | gi|4938494|emb|CAB43852.1| | CBL protein (Arabidopsis) |
| OS009519_at | 4.45 | 3.5 | gi|1277092|gb|AAB03774.1| | n-Acetyltransferase hookless1 |
| OS015049_i_at | 4.28 | 2.13 | gi|3309086|gb|AAC26010.1| | CBL protein 3 (Arabidopsis) |
| Other | ||||
| OS013253_at | 2.4 | 7.69 | gi|4584534|emb|CAB40764.1| | Cytochrome P450-like protein (Arabidopsis) |
| OS015512.1_at | 2.73 | 2.92 | gi|6863054|dbj|BAA90487.1| | Heat shock protein 90 (rice) |
| OS000907_s_at | 2.44 | 3.1 | gi|585960|sp|P38389| | S61B_ARATH protein transport protein sec61 β-subunit |
| OS011960.1_s_at | 3.06 | 2.41 | gi|2052381|gb|AAC49696.1| | Calreticulin |
| OS024781_at | 9.56 | 11.63 | gi|4972056|emb|CAB43924.1| | Putative protein (Arabidopsis) |
| OS006047_i_at | 9.02 | 6.91 | gi|3297809|emb|CAA19867.1| | Putative protein (Arabidopsis) |
| OS003244.1_s_at | 3.54 | 6.67 | gi|7378611|emb|CAB83287.1| | Putative protein (Arabidopsis) |
| OS_ORF013995_at | 3.98 | 4.11 | Open reading frame | |
| OS019454_f_at | 4.88 | 2.06 | F21B7.3 (Arabidopsis) | |
| OS010175.1_at | 2.32 | 3.85 | gi|7269851|emb|CAB79710.1| | Putative protein (Arabidopsis) |
| OS001396_at | 3.27 | 2.75 | gi|5123547|emb|CAB45313.1| | Putative protein (Arabidopsis) |
| OS_ORF015460_at | 2.37 | 3.4 | Open reading frame | |
| OS006045.1_at | 2.11 | 3.27 | gi|7485404|pir‖T02648| | Hypothetical protein F12C20.9 (Arabidopsis) |
| OS009650_at | 2.22 | 2.77 | F17L21.8 (Arabidopsis) | |
| OS001908_i_at | 2.01 | 2.65 | gi|7430871|pir‖T00843| | Basic blue protein (Arabidopsis) |
The genes that we observed to be up-regulated in GA-treated tissue echo previous reports on aleurone cell biology. Less is known about genes down-regulated in GA-treated tissue. We therefore determined the probe sets that showed a decrease in expression between 0 and 8 h after GA treatment. Forty-seven genes were down-regulated more than 2-fold in both experiments. As seen in Table III, these include many genes for metabolic enzymes and genes associated with stress responses. The down-regulation of two Cys proteinase inhibitors is consistent with the biology of the aleurone cell because GA treatment increases the activity of secreted and vacuolar Cys proteinases. It is notable that few signaling molecules are present in this list. We did not identify any strong candidates for protein kinases, protein phosphatases, or transcription factors, with the possible exception of a gene having similarity to nucleic acid-binding proteins.
Table III.
Probe sets down-regulated with time in GA-treated rice half-grains
Induction is the ratio of expression at 8 h relative to 0 h. Similar proteins are identified by accession numbers and a brief description.
| Probe Set | Induction at 8 h in Time Course | Induction at 8 h in d1 Experiment | Accession Nos. | Description |
|---|---|---|---|---|
| Stress related | ||||
| OS000677.1_f_at | 0.27 | 0.3 | gi|3980385|gb|AAC95188.1| | Small heat shock protein (Arabidopsis) |
| OS000130_f_at | 0.28 | 0.48 | P27777 | HS11_ORYSA 16.9-kD class I heat shock protein |
| OS000148.1_at | 0.48 | 0.28 | gi|971280|dbj|BAA09947.1| | RAB24 protein (rice) |
| OS000170_f_at | 0.33 | 0.44 | P31673 | HS12_ORYSA 17.4-kD class I heat shock protein |
| OS009060_at | 0.35 | 0.42 | gi|2944088|gb|AAC05216.1| | Glutathione S-transferase (rice) |
| OS011595_r_at | 0.48 | 0.3 | gi|445133|prf‖1908434C | Chilling tolerance-related protein (rice) |
| OS009441_f_at | 0.45 | 0.35 | gi|5080803|gb|AAD39312.1|AC007258_1| | Glutathione transferase (Arabidopsis) |
| Metabolism | ||||
| OS009718.1_at | 0.27 | 0.13 | gi|5103836|gb|AAD39666.1|AC007591_31| | Member of the glyoxalase family (Arabidopsis) |
| OS000253_at | 0.38 | 0.19 | gi|1778821|gb|AAC05590.1| | S-adenosyl-l-Met synthetase (rice) |
| OS001932.1_at | 0.37 | 0.26 | P43279 | MAOC_ORYSA malate oxidoreductase, chloroplast |
| OS005778.1_at | 0.41 | 0.34 | P34799 | URID_CANLI uricase II clone PCCLNUO-02 |
| OS014904_at | 0.37 | 0.4 | gi|294285|gb|AAA33840.1| | 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase |
| OS002747_at | 0.46 | 0.34 | gi|322787|pir‖JC1481| | Pyruvate kinase, cytosolic (Solanum tuberosum) |
| OS006190_at | 0.35 | 0.45 | gi|6729036|gb|AAF27032.1|AC009177_22| | Glucose and ribitol dehydrogenase homolog (Arabidopsis) |
| OS009600.1_at | 0.49 | 0.39 | gi|3738320|gb|AAC63661.1| | Cinnamoyl CoA reductase (Arabidopsis) |
| OS010021_at | 0.49 | 0.45 | gi|2739000|gb|AAB94588.1| | CYP71D10p (Glycine max) |
| Proteolysis | ||||
| OS000768_i_at | 0.46 | 0.3 | gi|2245006|emb|CAB10426.1| | Cys proteinase inhibitor-like protein (Arabidopsis) |
| OS009331.1_at | 0.41 | 0.41 | gi|2641619|gb|AAC12662.1| | Ubiquitin-conjugating enzyme protein E2 (maize) |
| OS004395.1_r_at | 0.43 | 0.43 | gi|4263720|gb|AAD15406.1| | Cys proteinase inhibitor C (Arabidopsis) |
| Oleosome | ||||
| OS006526_at | 0.27 | 0.29 | gi|944830|emb|CAA43183.1| | Soybean 24-kD oleosin isoform (G. max) |
| OS000217_s_at | 0.26 | 0.3 | gi|1171352|gb|AAC02239.1| | 16-kD oleosin (rice) |
| OS005956_f_at | 0.31 | 0.31 | gi|4455257|emb|CAB36756.1| | Oleosin, 18.5K (Arabidopsis) |
| Membrane transport | ||||
| OS004773_at | 0.33 | 0.33 | gi|2655098|gb|AAC32034.1| | Peptide transporter (barley) |
| OS012445_at | 0.48 | 0.46 | gi|3319374|gb|AAC28223.1| | Chloroplast triose phosphate translocators (Arabidopsis) |
| Other | ||||
| OS004203.1_f_at | 0.23 | 0.15 | gi|2058502|gb|AAC49884.1| | Hemoglobin 2 (rice) |
| OS005002.1_f_at | 0.24 | 0.21 | gi|544880|gb|AAB29661.1| | Multicystatin; PMC (S. tuberosum) |
| OS012817.1_at | 0.24 | 0.24 | gi|224647|prf‖1109273A| | Nodulin 35 (G. max) |
| OS003629.1_at | 0.3 | 0.21 | gi|2894603|emb|CAA17137.1| | Putative protein (Arabidopsis) |
| OS001028.1_at | 0.29 | 0.29 | Hypothetical protein (Z97343; rice) | |
| OS000786.1_at | 0.38 | 0.27 | gi|4105683|gb|AAD02495.1| | Unknown (rice) |
| OS_ORF002741_at | 0.44 | 0.26 | Open reading frame | |
| OS015813_f_at | 0.37 | 0.34 | gi|6175183|gb|AAF04909.1|AC011437_24| | RNA-binding protein (Arabidopsis) |
| OS_ORF014778_at | 0.36 | 0.36 | Open reading frame | |
| OS010428.1_f_at | 0.44 | 0.3 | P45592 | COF1_RAT cofilin, non-muscle isoform |
| OS003584_at | 0.38 | 0.39 | gi|4263648|gb|AAD15370.1| | Hypothetical protein (Arabidopsis) |
| OS000702_at | 0.32 | 0.47 | gi|4056503|gb|AAC98069.1| | Unknown protein (Arabidopsis) |
| OS009202_at | 0.45 | 0.37 | gi|6223644|gb|AAF05858.1|AC011698_9 | Unknown protein (Arabidopsis) |
| OS011577_s_at | 0.49 | 0.37 | gi|4091080|gb|AAC98962.1| | Nucleic acid-binding protein (rice) |
| OS011620_at | 0.49 | 0.38 | gi|6815055|dbj|BAA90342.1| | Unknown protein (rice) |
| OS001718.1_i_at | 0.49 | 0.38 | gi|439273|emb|CAA80984.1| | 54 amino acids (barley) |
| OS014974_s_at | 0.44 | 0.44 | gi|5281030|emb|CAB45966.1| | Putative protein (Arabidopsis) |
| OS011954_at | 0.42 | 0.47 | gi|2130118|pir‖S67993 | Amylogenin (maize) |
| OS009626.1_at | 0.44 | 0.47 | gi|4454458|gb|AAD20905.1| | Unknown protein (Arabidopsis) |
| OS025228.1_at | 0.45 | 0.46 | gi|2262116|gb|AAB63624.1| | Cellulose synthase isolog |
| OS012207_f_at | 0.43 | 0.49 | gi|8843793|dbj|BAA97341.1| | Unknown protein (Arabidopsis) |
| OS010104.1_s_at | 0.48 | 0.46 | gi|5262762|emb|CAB45910.1| | Putative protein (Arabidopsis) |
Genes specifically down-regulated by ABA are rare (Fig. 3), but many genes are down-regulated with time in half-grains treated with ABA (Fig. 8). We compared probe set signal intensities at 8 h with those at 0 h for wild-type half-grains treated with ABA in two separate experiments. Signal intensities for the 35 probe sets listed in Table IV were down-regulated by 2-fold or more in both experiments. These down-regulated genes have functions in basic cellular processes (e.g. pyruvate dehydrogenase, pyruvate kinase, and malate oxidoreductase) or are related to the mechanisms of transcription and translation, the proteosome, and cellular redox status. Three glutathione S-transferases were identified in the latter category. Genes for hemoglobin were identified twice. We did not identify transcription factors or signaling molecules in this analysis.
Table IV.
Probe sets down-regulated with time in ABA-treated rice half-grains
Induction is the ratio of expression at 8 h relative to 0 h. Similar proteins are identified by accession numbers and a brief description.
| Probe Set | Induction at 8 h in Time Course | Induction at 8 h in d1 Experiment | Accession Nos. | Description |
|---|---|---|---|---|
| Stress related | ||||
| OS000677.1_f_at | 0.25 | 0.4 | gi|3980385|gb|AAC95188.1| | Small heat shock protein (Arabidopsis) |
| OS011595_r_at | 0.43 | 0.27 | gi|445133|prf‖1908434C| | Chilling tolerance-related protein (rice) |
| OS002696_at | 0.27 | 0.5 | gi|6746590|gb|AAF27638.1|AF217458_1| | Heat shock protein 70 (Arabidopsis) |
| OS009441_f_at | 0.38 | 0.41 | gi|5080803|gb|AAD39312.1|AC007258_1| | Glutathione transferase (Arabidopsis) |
| OS009060_at | 0.45 | 0.39 | gi|2944088|gb|AAC05216.1| | Glutathione S-transferase (rice) |
| OS010527_f_at | 0.48 | 0.49 | Q28514 | GTP_MACMU glutathione S-transferase P |
| Redox related | ||||
| OS000156_at | 0.24 | 0.46 | gi|505136|dbj|BAA06456.1| | Ferredoxin (rice) |
| OS000729_s_at | 0.42 | 0.35 | gi|516839|dbj|BAA05494.1| | Catalase (rice) |
| Metabolism | ||||
| OS004308_at | 0.14 | 0.14 | gi|4894182|emb|CAB43506.1| | 12-Oxophytodienoate reductase (Lycopersicon esculentum) |
| OS002747_at | 0.36 | 0.23 | gi|322787|pir‖JC1481| | Pyruvate kinase cytosolic (Solanum tuberosum) |
| OS001932.1_at | 0.35 | 0.25 | P43279 | MAOC_ORYSA malate oxidoreductase, chloroplast |
| OS022578_at | 0.42 | 0.37 | gi|2924784|gb|AAC04913.1| | Myrosinase-binding protein (Arabidopsis) |
| OS013868_at | 0.48 | 0.45 | P32588 | Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 |
| OS004744_at | 0.49 | 0.48 | gi|3850999|gb|AAC72192.1| | Pyruvate dehydrogenase E1 β-subunit isoform 1 (maize) |
| Other | ||||
| OS004203.1_f_at | 0.14 | 0.05 | gi|2058502|gb|AAC49884.1| | Hemoglobin 2 (rice) |
| OS001666.1_at | 0.36 | 0.11 | gi|2058498|gb|AAC49882.1| | Hemoglobin 1 (rice) |
| OS000124_at | 0.41 | 0.11 | gi|7489465|pir‖T04144 | DNA-binding protein (rice) |
| OS005499.1_at | 0.33 | 0.24 | gi|7076779|emb|CAB75894.1| | Glycoprotein-like (Arabidopsis) |
| OS000701_at | 0.29 | 0.41 | gi|1155263|gb|AAA91170.1| | Eukaryotic release factor 1 homolog |
| OS005611.1_at | 0.44 | 0.29 | gi|949980|emb|CAA61258.1| | Open reading frame (maize) |
| OS009331.1_at | 0.46 | 0.28 | gi|2641619|gb|AAC12662.1| | Ubiquitin-conjugating enzyme protein E2 (maize) |
| OS017078_at | 0.32 | 0.45 | gi|7658344|gb|AAF66134.1| | Unknown protein (Arabidopsis) |
| OS003018.1_at | 0.37 | 0.46 | gi|2760362|gb|AAB95261.1| | 15.9-kD subunit of RNA polymerase II (Arabidopsis) |
| OS005587_at | 0.41 | 0.47 | gi|3169182|gb|AAC17825.1| | Unknown protein (Arabidopsis) |
| OS010228_at | 0.44 | 0.45 | gi|4803958|gb|AAD29830.1|AC006202_8 | Unknown protein (Arabidopsis) |
| OS012990.1_at | 0.48 | 0.43 | gi|7671481|emb|CAB89322.1| | Putative protein (Arabidopsis) |
| OS001505_at | 0.48 | 0.46 | gi|4836932|gb|AAD30634.1|AC006085_7 | Unknown protein (Arabidopsis) |
| OS003584_at | 0.49 | 0.45 | gi|4263648|gb|AAD15370.1| | Hypothetical protein (Arabidopsis) |
We used a similar approach to look at genes with increased expression in ABA-treated half-grains. The 26 probe sets listed in Table V had increased intensity in wild-type half-grains in both the time-course and d1 experiments. Many of the ABA up-regulated genes that we identified fell into three general categories: seed maturation, ABA and stress related, and proteolysis. Dehydrins and LEA proteins are known to be ABA up-regulated in cereals, and genes corresponding to several LEAs and dehydrins were up-regulated by ABA in these experiments. Fluorescence intensity from two probe sets for protease inhibitors and two tonoplast intrinsic proteins also increased more than 2-fold in ABA-treated half-grains.
Table V.
Probe sets up-regulated with time in ABA-treated rice half-grains
Induction is the ratio of expression at 8 h relative to 0 h. Similar proteins are identified by accession numbers and a brief description.
| Probe Set | Induction at 8 h in time Course | Induction at 8 h in d1 experiment | Accession Nos. | Description |
|---|---|---|---|---|
| Seed maturation | ||||
| OS001162_at | 2.58 | 14.5 | P46518 | LE14_GOSHI LEA protein LEA14-A |
| OS003541_at | 5.89 | 5.22 | Q00742 | DH15_WHEAT dehydrin RAB 15 |
| OS001970.1_r_at | 2.78 | 6.43 | gi|5802240|gb|AAD51623.1|AF169020_1| | Seed maturation protein PM35 (Glycine max) |
| OS003416.1_r_at | 3.62 | 5.1 | P28640 | DHN2_PEA dehydrin DHN2 |
| OS001415_at | 2.25 | 6.08 | gi|5802250|gb|AAD51628.1|AF169025_1| | Seed maturation protein PM41 (G. max) |
| OS003879.1_s_at | 2.81 | 3.47 | gi|1486503|gb|AAC03364.1| | LEA-like protein (rice) |
| OS001562.1_r_at | 2.23 | 3.3 | gi|1399913|gb|AAB03330.1| | Dehydrin (rice) |
| OS008919.1_f_at | 2.05 | 3.35 | gi|100706|pir‖JH0324| | RAB 16b protein (rice) |
| OS004708_at | 2.61 | 2.72 | gi|1526424|dbj|BAA11017.1| | LEA protein in group 3 (Arabidopsis) |
| ABA and stress related | ||||
| OS018179_f_at | 5.13 | 15.08 | gi|440847|gb|AAA21819.1| | Cold acclimation-induced WCS66 |
| OS016473_f_at | 5.87 | 13.1 | P46526 | CS66_WHEAT cold shock protein CS66 |
| OS000966_s_at | 3.3 | 8.24 | gi|1724112|gb|AAB38504.1| | ABA-induced plasma membrane protein PM 19 (wheat) |
| OS016003_f_at | 2.46 | 2.3 | gi|22460|emb|CAA41854.1| | ABA inducible (maize) |
| Proteolysis | ||||
| OS003094.1_at | 3.49 | 2.15 | gi|4678299|emb|CAB41090.1| | Cys proteinase (Arabidopsis) |
| OS008030.1_s_at | 2.73 | 2.12 | gi|7438264|pir‖S72492| | Probable proteinase inhibitor (L. esculentum) |
| OS000988_f_at | 2.25 | 2.04 | gi|475253|emb|CAA55588.1| | Proteinase inhibitor (maize) |
| Membrane transport | ||||
| OS009470_f_at | 4.17 | 4.55 | gi|2244974|emb|CAB10395.1| | Pore protein homolog (Arabidopsis) |
| OS003998.1_at | 3.94 | 3.2 | P26587 | TIPA_ARATH tonoplast intrinsic protein-α |
| OS007184_at | 2.68 | 3.54 | gi|2605714|gb|AAB84183.1| | β-Tonoplast intrinsic protein (Arabidopsis) |
| Other | ||||
| OS005119_at | 8.78 | 2.47 | gi|4105681|gb|AAD02494.1| | Unknown (rice) |
| OS000552_at | 6.17 | 2.28 | P33044 | THHR_HORVU antifungal protein R |
| OS006670_at | 2.6 | 5.06 | gi|4432835|gb|AAD20684.1| | Unknown protein (Arabidopsis) |
| OS003185.1_f_at | 2.22 | 5.2 | gi|2253578|gb|AAC69143.1| | Hypothetical protein (Arabidopsis) |
| OS002033.1_at | 2.08 | 4.63 | gi|7268686|emb|CAB78894.1| | Hypothetical protein (Arabidopsis) |
| OS003245_i_at | 2.6 | 2.95 | gi|5262190|emb|CAB45787.1| | Inositol 1,3,4-trisphosphate 5/6-kinase-like protein (Arabidopsis) |
| OS_ORF003709_f_at | 2.91 | 2.08 | Open reading frame |
The lists of temporally regulated genes contained in Tables II to V have little overlap. The only significant overlap was between the lists of probe sets down-regulated in ABA-treated tissue and down-regulated in GA-treated tissue. Nine probe sets were in common between these two lists and these include two glutathione S-transferases, hemoglobin, a small heat shock protein, a malic enzyme, a ubiquitin-conjugating enzyme, cofilin, a chilling tolerance-related protein, and an unknown protein. None of the probe sets that increased in GA- or ABA-treated tissues appeared in any of the other lists.
DISCUSSION
The cereal aleurone layer is a model system for studying the regulation of transcription by GA and ABA in part because of the dramatic stimulation of transcription that these hormones bring about for the α-amylases and other secreted hydrolases. Although much has been learned about the signaling elements and trans-acting factors required for high rates of α-amylase transcription, the scope of previous studies has been restricted to examining the responses of a few, usually highly expressed genes. Here we have used an oligonucleotide microarray to characterize global changes in transcription that occur in rice half-grains treated with GA, ABA, or no hormone. The data presented here include transcript profiles during an 8-h time course for approximately one-half of all rice genes. These data give us a much more complete picture of GA- and ABA-regulated gene expression in this tissue and have revealed heretofore unknown aspects of transcript regulation in rice aleurone layers. In particular, we have shown that the transcriptome in imbibed rice half-grains contains RNAs for approximately 11,000 of the 23,000 probe sets on the microarray (Table I). This extensive set of RNAs is dynamic, with large changes in transcript abundance occurring with time in the presence or absence of exogenous GA or ABA (Fig. 8). Despite this, genes specifically up- or down-regulated by GA or ABA alone are rare (Fig. 3). We have shown that the Gα-subunit of heterotrimeric G-proteins amplifies changes in transcript abundance in GA- or ABA-treated half-grains, but that this effect is small for all but a handful of genes (Figs. 5 and 6). Finally, we have shown that loss of the Gα-subunit does not result in reduced sensitivity to GA at the level of transcription (Fig. 7).
Our microarray data include transcript profiles for several well-characterized groups of proteins. In all cases, the microarray data are entirely consistent with previously published data. For example, we see large, GA-stimulated increases in transcription of genes for α-amylases, endo-1,4-β-xylanases, and proteases (Fig. 1, A and B; Supplemental Tables I, A and B, and III). Similarly, dehydrin genes and LEA genes have been shown on numerous occasions to be induced by ABA, and the data presented here show this as well (Fig. 2, A and B; Supplemental Table II, A and B). The down-regulation of these same genes by GA is also illustrated in Figures 1E and 2, A and B.
The analysis presented in Figures 1 and 2 allowed us to visualize common patterns of gene expression in rice aleurone layers treated with no hormone, GA, or ABA. These patterns were based on a subset of the data that had been enriched for probe sets that were hormone responsive, with 16 data points for each probe set. As a result, the patterns shown in Figures 1 and 2 are relatively insensitive to random variation in intensity at any one point. Specific expression values for the individual genes contained in each cluster were omitted from Supplemental Tables I and II, however, in recognition of the fact that only one hybridization was done at each time point for each hormone treatment.
We have found that rice half-grains contain a surprisingly large number of mRNA species. At all time points in all treatments (i.e. in the data from 16 separate microarray hybridizations), 9,762 probe sets were called present out of 23,000 possible probe sets on the chip. This percentage of the probe sets hybridizing with RNA from the samples (41%) is similar to the percentage of the genome transcribed in a barley leaf (45%; Close et al., 2004), although the identity of the individual mRNAs is likely to be different. For example, genes for endo-1,4-β-xylanases and LEA proteins are highly expressed in rice aleurone, but are not expected to be abundant in leaves. Because the aleurone layer is the only living tissue in the rice half-grain, it is highly likely that these mRNAs have their origin in the aleurone layer, although a few preexisting mRNAs may be present in the dead starchy endosperm (Potokina et al., 2002).
Despite the large number of mRNAs present in rice half-grains, very few were hormonally responsive. We looked for changes in transcript abundance that were associated specifically with GA and ABA treatment by comparing transcript profiles of tissue treated with these hormones to profiles from tissue incubated for the same length of time without hormone (Figs. 1–3). At most, 59 genes were specifically up-regulated 2-fold in GA-treated tissue and 42 genes in ABA-treated tissue (Fig. 3). Regulation of transcription by GA or ABA has been examined by others using microarrays, and, in general, larger numbers of hormone-responsive genes have been identified. In rice callus, 206 out of 20,500 genes were up-regulated with GA and 110 genes were up-regulated with ABA 3 d after hormone treatment (Yazaki et al., 2004). For 2-week-old rice seedlings, 43 of 1,700 cDNAs were induced by ABA (Rabbani et al., 2003). For rice shoots, 29 of 4,000 cDNAs were up with GA and 42 were down with GA 24 h after hormone treatment (Yang et al., 2004), and 30 of 6,144 cDNAs were up with ABA and seven down with ABA within 12 h of hormone treatment (Lin et al., 2003). Accounting for these differences is difficult, but with the exception of experiments with rice callus, the larger number of hormone-responsive genes may reflect the greater number of cell types involved.
Most previously published data on aleurone cells supported the view that GA up-regulated a large number of genes in this tissue. ABA antagonized this effect of GA for a few genes, and ABA had been shown to up-regulate several genes on its own. Our data add substantial breadth to this simple model for gene regulation in the aleurone layer. In particular, we showed that GA specifically down-regulated about one-third as many genes as it up-regulated (Fig. 3). Surprisingly, ABA up-regulated as many genes as did GA, and it is noteworthy that virtually no genes were specifically down-regulated by ABA (Fig. 3). With a few exceptions, transcripts that decreased in abundance in ABA-treated tissues also decreased in one of the other treatments to a similar extent (Figs. 1, 2, and 8). Because of this, we speculate that the promoters of genes that are strongly down-regulated in ABA-treated aleurone cells must contain regulatory elements for binding factors that are not dependent on ABA perception or concentration.
Many genes showed a temporal change in transcript abundance during the 8 h of the experiment (Fig. 8). In many cases, these changes were not dependent on hormone treatment (Fig. 8). We hypothesize that many of these changes result from imbibition of the half-grains and rehydration of the aleurone cells. We identified those genes that increased or decreased 2-fold or more 8 h after hormone treatment in two separate experiments (Tables II–V). Virtually none of the genes identified were obvious signal transduction components such as protein kinases or transcription factors. A notable exception was a CBL gene that we have characterized in detail (Hwang et al., 2005). This analysis includes northern blots for this CBL that confirm the expression pattern seen in the array data (Hwang et al., 2005). A GAMyb was among the 59 genes specifically up-regulated by GA (Supplemental Table III). Other previously identified signaling elements whose transcription is regulated by GA or ABA, such as some of the WRKY-domain transacting factors, are present on the microarray. The expression of these genes, however, does not change enough for them to be flagged by our analyses as hormonally or temporally regulated genes. Overall, our data suggest that aleurone cell function during this period utilizes preexisting signaling components or posttranslational modifications rather than large changes in transcription for the regulation of cellular activity.
One of the GA-signaling mutants that has been characterized in rice is the d1 mutant. We used d1 to see how widespread the effect of the Gα-protein was on transcript abundance and to determine the effect that GA has on transcript accumulation at high (5 μm) and low (100 nm) concentrations of GA. Our data make it clear that transcript abundance for many genes is affected by Gα (Figs. 5–7) and that the relative difference is larger as the gene is more strongly up- or down-regulated with GA (Fig. 5). This is true for tissue treated with both high and low GA concentrations (Fig. 5, A and B). Gα also influenced transcript accumulation in ABA-treated tissue (Fig. 6). As for GA-regulated genes, the effect was larger for those groups of genes that were most strongly up-regulated with ABA.
We also tested the hypothesis that the Gα-protein affects the sensitivity of transcription to GA concentration. Previously published data show clearly that the Gα mutation reduces the sensitivity to GA of α-amylase secretion and internode elongation. Our data, however, show that the sensitivity of transcription to GA is no different in d1 mutant half-grains than in wild type half-grains. This was demonstrated for both the α-amylases (Fig. 7A) and the GA-regulated genes that had been identified in an independent experiment (Fig. 7B). In both cases, there was a linear relationship between changes in transcript abundance with 5 μm GA and changes in transcript abundance with 100 nm GA, and this relationship was the same for wild-type and d1 mutant half-grains. It should be noted that our experiments looked at changes in expression that occurred within 8 h of treating rice half-grains with GA, whereas expression data after 4 d gave rise to the bypass model (Ueguchi-Tanaka et al., 2000). It is possible that differences in transcript accumulation that occur at later times are mediated by the Gα-protein and that these differences are more sensitive to GA concentration at later times than at earlier times.
Although the d1 mutation affects transcript abundance for many genes, most of the changes were small (Figs. 5 and 6). Because of this, we propose that the Gα-protein is used to fine tune changes in transcript accumulation. Additional roles for Gα are also possible. The microarray data, however, raise fundamental questions about how the d1 phenotype arises. In particular, does the phenotype result from the subtle changes in gene expression that we observed, or are most of these changes in gene expression a consequence of the dwarf phenotype?
MATERIALS AND METHODS
Plant Material
Wild-type and d1 mutant rice (Oryza sativa L. sp. Japonica cv Nipponbare) grains were a gift from Makoto Matsuoka (Nagoya University). Some grains were used immediately. Others were germinated and seedlings were grown to maturity in a glass house in Berkeley, California, under natural light conditions. Grains were harvested from the primary inflorescence and tillers of these plants as they ripened. The d1 mutant is in the Nipponbare background, and grains from d1 mutant plants grown in Berkeley were harvested from plants grown alongside wild-type plants.
Hormone Treatment of Grains
Grains were cut transversely to remove the embryo, surface sterilized with 1% sodium hypochlorite and 0.01% Tween X-20 for 10 min with shaking, and washed extensively with distilled water. Residual hypochlorite was removed by soaking the grains in 0.01 n HCl for 10 min followed by additional washes with distilled water. De-embryonated grains (half-grains) were placed in sterile, tall 10-cm-diameter petri dishes containing 15 mL of sterile distilled water at 28°C. After overnight (18 h) incubation designed to minimize changes in transcription caused by cutting, sterilizing, or shaking the grain, distilled water was replaced by 20 mL of 10 mm CaCl2. Some petri dishes also received ABA or GA3 to a final concentration of 5 μm and, in the case of the d1 mutant experiment, 100 nm GA3. Each petri dish was shaken gently at 100 rpm at 28°C for the indicated time prior to RNA extraction.
RNA Extraction and Hybridization
RNA extraction was performed by the method described by Hwang et al. (1999), followed by re-extraction using the Qiagen Plant RNeasy kit (Qiagen). For the rice time-course experiment, total RNA was extracted 0.5, 1, 3, 6, and 8 h after hormone addition for samples treated with ABA, GA, or no hormone. No-hormone samples were also isolated at the time of hormone addition (time zero). Typical extractions started with 70 half-grains. For the d1 experiment, RNA was isolated 8 h after treatment of wild-type and mutant grains with 5 μm GA, 100 nm GA, or 5 μm ABA. RNA was also isolated from wild-type and d1 grains that were not treated with either GA or ABA (no hormone) at 0 h. For each hybridization, RNA from two or more biological replicates was pooled in order to minimize variability in the data. For both the time-course experiment and the d1 experiment, single hybridizations were done for each of the 21 combinations of genotype, hormone treatment, and time. The data reported here, therefore, come from 24 separate microarrays.
All microarray experiments were done using the custom-built Genechip described by Zhu et al. (2003). Preparation of cDNA and cRNA, fluorescent labeling, hybridization to the chip, and quality control steps were done exactly as described previously (Zhu et al., 2003). Briefly, double-strand cDNAs were synthesized from 5 μg total RNA using an oligo(dT)24 primer containing a 5′ T7 RNA polymerase promoter sequence and SuperScript II reverse transcriptase (Invitrogen). Biotinylated complementary RNAs were transcribed in vitro from synthesized cDNA by T7 RNA polymerase (ENZO Biochem). Labeled cRNAs were applied to the rice GeneChip microarray, and subsequent hybridization, washing, and staining were conducted according to the Affymetrix recommended protocol. The image files (*.DAT) and raw files (*.CEL) were acquired using Microarray Analysis Suite 5.0 (Affymetrix). The signal intensity of a probe set was computed using the Robust Multi-Array Average method (Irizarry et al., 2003). If the difference is greater than 2, a present call will be assigned; if the difference is less than 0, an absent call will be assigned; a marginal call will be assigned if the difference is between 0 and 2.
Data Analysis
k-means clustering was done using EPCLUST software from the European Bioinformatics Institute (http://ep.ebi.ac.uk/EP/EPCLUST). To better visualize hormone-dependent responses, the data from the complete rice time-course experiment were enriched for GA-responsive genes or for ABA-responsive genes. Probe sets that had minimum intensity values <5 as well as maximum intensity values <25 were removed from the initial dataset of 23,780 probe sets because these genes are unlikely to be expressed at any time in rice half-grains. From the remaining 18,144 probe sets, we removed those probe sets that had P > 0.05 for an F test that compared samples treated with no hormone at 0.5, 1, 3, 6, and 8 h with samples treated with either GA (for GA-responsive genes) or ABA (for ABA-responsive genes) at 0.5, 1, 3, 6, and 8 h. To reduce the effect of expression intensity on the final clusters, the data were transformed by taking the natural log of the intensity value at each time point divided by the average intensity of the 0-, 0.5-, and 1-h samples treated with no hormone. These first three time points rarely showed differences and were used to give a more accurate estimate of mRNA abundance at the start of the experiment. Euclidean distance was used to cluster the data, and individual probe sets that repeatedly showed up in single member clusters were excluded from the final dataset. Other statistical analyses were done using Microsoft Excel. For one-tailed t tests, we compared intensity data for groups of 25 probe sets with the 1,000 probe sets near the middle of the dataset whose average ratio of wild-type to d1 expression equaled 1.00.
For scatter plots comparing wild-type to d1 expression ratios, average induction by GA or ABA was computed as follows. Induction for a probe set was the intensity value at 8 h in samples treated with 5 μm GA (Fig. 5) or 5 μm ABA (Fig. 6) divided by the intensity value at 0 h. For each probe set, two values of induction were computed, one for samples from wild-type half-grains and one for samples from d1 mutant half-grains. The average induction for each group of 25 genes was computed as the mean of 50 individual induction values, 25 from wild-type and 25 from d1 mutant half-grains. This allowed the probe sets to be sorted from most up-regulated with GA to most down-regulated with GA without a strong bias for either wild-type or d1 mutant grains.
Public Access to Microarray Data
All of the *.chp files for all of the experiments described in this article will be made available through the Internet. Proprietary probe set sequences and gene annotations for all of the genes (included in Figs. 1, A–E, I, and 2, A and B; Tables II–V; and Supplemental Tables I, A–E, I, II, A and B, and III) are included as supplemental data. This list of approximately 895 probe sets contains all of the genes described in the “Results” section of this article.
Supplementary Material
This work was supported in part by grants from the National Science Foundation, the Division of Agricultural and Natural Resources of the University of California, and the Torrey Mesa Research Institute.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Paul C. Bethke (pcbethke@nature.berkeley.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074435.
References
- Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise RP (2004) A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol 134: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang RL, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Gubler F, Watts RJ, Kalla R, Matthews P, Keys M, Jacobsen JV (1997) Cloning of a rice cDNA encoding a transcription factor homologous to barley GAMyb. Plant Cell Physiol 38: 362–365 [DOI] [PubMed] [Google Scholar]
- Higgins TJV, Jacobsen JV, Zwar JA (1982) Gibberellic-acid and abscisic-acid modulate protein synthesis and messenger RNA levels in barley aleurone layers. Plant Mol Biol 1: 191–216 [DOI] [PubMed] [Google Scholar]
- Hwang YS, Bethke PC, Cheong YH, Chang HS, Zhu T, Jones RL (2005) A gibberellin-regulated calcineurin B in rice localizes to the tonoplast and is implicated in vacuole function. Plant Physiol 138: 1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang YS, Thomas BR, Rodriguez RL (1999) Differential expression of rice alpha-amylase genes during seedling development under anoxia. Plant Mol Biol 40: 911–920 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Iwasaki Y, Fujisawa Y, Kato H (2003) Function of heterotrimeric G protein in gibberellin signaling. J Plant Growth Regul 22: 126–133 [Google Scholar]
- Jacobsen JV, Chandler PM (1987) Gibberellin and abscisic acid in germinating cereals. In PJ Davies, ed, Plant Hormones and Their Role in Plant Growth and Development. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 164–193
- Jacobsen JV, Pearce DW, Poole AT, Pharis RP, Mander LN (2002) Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol Plant 115: 428–441 [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13: 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-M, Williams ME, Tingey SV, Rafalski JA (2002) DNA array profiling of gene expression changes during maize embryo development. Funct Integr Genomics 2: 13–27 [DOI] [PubMed] [Google Scholar]
- Lin F, Xu SL, Ni WM, Chu ZQ, Xu ZH, Xue HW (2003) Identification of ABA-responsive genes in rice shoots via cDNA macroarray. Cell Res 13: 59–68 [DOI] [PubMed] [Google Scholar]
- Ozturk ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ (2002) Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 48: 551–573 [DOI] [PubMed] [Google Scholar]
- Potokina E, Sreenivasulu N, Altschmied L, Michalek W, Graner A (2002) Differential gene expression during seed germination in barley (Hordeum vulgare L.). Funct Integr Genomics 2: 28–39 [DOI] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Hochholdinger F, Nakazono M (2004) Global expression profiling applied to plant development. Curr Opin Plant Biol 7: 50–56 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M (2000) Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA 97: 11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GX, Jan A, Shen SH, Yazaki J, Ishikawa M, Shimatani Z, Kishimoto N, Kikuchi S, Matsumoto H, Komatsu S (2004) Microarray analysis of brassinosteroid- and gibberellin-regulated gene expression in rice seedlings. Mol Genet Genomics 271: 468–478 [DOI] [PubMed] [Google Scholar]
- Yazaki J, Shimatani Z, Hashimoto A, Nagata Y, Fujii F, Kojima K, Suzuki K, Taya T, Tonouchi M, Nelson C, et al (2004) Transcriptional profiling of genes responsive to abscisic acid and gibberellin in rice: phenotyping and comparative analysis between rice and Arabidopsis. Physiol Genomics 17: 87–100 [DOI] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Chen WQ, Provart N, Chang HS, Guimil S, Su WP, Estes B, Zou GZ, Wang X (2003) Transcriptional control of nutrient partitioning during rice grain filling. Plant Biotechnol J 1: 59–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.