Abstract
It has been demonstrated that calcium plays a central role in mediating abscisic acid (ABA) signaling, but many of the Ca2+-binding sensory proteins as the components of the ABA-signaling pathway remain to be elucidated. Here we identified, characterized, and purified a 58-kD ABA-stimulated calcium-dependent protein kinase from the mesocarp of grape berries (Vitis vinifera × Vitis labrusca), designated ACPK1 (for ABA-stimulated calcium-dependent protein kinase1). ABA stimulates ACPK1 in a dose-dependent manner, and the ACPK1 expression and enzyme activities alter accordantly with the endogenous ABA concentrations during fruit development. The ABA-induced ACPK1 stimulation appears to be transient with a rapid effect in 15 min but also with a slow and steady state of induction after 60 min. ABA acts on ACPK1 indirectly and dependently on in vivo state of the tissues. Two inactive ABA isomers, (−)-2-cis, 4-trans-ABA and 2-trans, 4-trans-(±)-ABA, are ineffective for inducing ACPK1 stimulation, revealing that the ABA-induced effect is stereo specific to physiological active (+)-2-cis, 4-trans-ABA. The other phytohormones such as auxin indoleacetic acid, gibberellic acid, synthetic cytokinin N-benzyl-6-aminopurine, and brassinolide are also ineffective in this ACPK1 stimulation. Based on sequencing of the two-dimensional electrophoresis-purified ACPK1, we cloned the ACPK1 gene. The ACPK1 is expressed specifically in grape berry covering a fleshy portion and seeds, and in a developmental stage-dependent manner. We further showed that ACPK1 is localized in both plasma membranes and chloroplasts/plastids and positively regulates plasma membrane H+-ATPase in vitro, suggesting that ACPK1 may be involved in the ABA-signaling pathway.
The phytohormone abscisic acid (ABA) regulates many important events during both vegetative and reproductive growth of plants as well as in plants' adaptation to their environment. During vegetative growth, ABA is a central signal of plant response to various environmental challenges including drought, salt, and cold stresses (for review, see Koornneef et al., 1998; Leung and Giraudat, 1998; Finkelstein and Rock, 2002). In reproductive organ seeds, ABA is responsible for the seed storage reserve synthesis, acquisition of desiccation tolerance and dormancy, and induction of stress tolerance (for review, see Finkelstein et al., 2002). Fleshy fruits are also the essential portions of reproductive organs and economically important harvest organs, as are crop seeds. In fleshy fruits as in seeds, ABA regulates various processes concerning assimilate uptake and metabolism to enhance reserve accumulation in these economic sinks (Yamaki and Asakura, 1991; Rock and Quatrano, 1995; Wayne and John, 1996; Opaskornkul et al., 1999; Peng et al., 2003; Pan et al., 2005). Grape berry (Vitis vinifera and also Vitis labrusca) is one of the most widely cultivated fruit trees in the world and also one of the typical nonclimacteric fruits. The ripening of grape berry is considered to be independent of the hormone ethylene but to be triggered essentially by ABA (for review, see Coombe, 1992). However, the ABA-signaling pathway in regulating development of the fleshy fruits remains essentially elusive.
ABA signal transduction has been extensively studied in the past years. Numerous cellular components that modulate ABA responses have been identified, leading to considerable progress in understanding the ABA-signaling pathway (for review, see Finkelstein et al., 2002; Himmelbach et al., 2003; Fan et al., 2004). Reversible protein phosphorylation, catalyzed by protein kinases and phosphatases, has been believed to play central roles in ABA signal transduction. Calcium-modulated protein phosphatases (PPs) 2C ABI1 and ABI2 are the two most characterized, homologous negative regulators of ABA signaling (Leung et al., 1994, 1997; Meyer et al., 1994; Sheen, 1998; Gosti et al., 1999; Merlot et al., 2001). They interact with multiple cellular targets such as a calcineurin B-like (CBL) protein kinase CIPK15 and a CBL Ca2+-binding protein ScaBP5. CIPK15 and one of its homologs CIPK3 and ScaBP5 are all involved in ABA signaling as negative regulators (Guo et al., 2002; Kim et al., 2003), possibly providing PP2Cs ABI1 and ABI2 with the Ca2+ sensor (Pandey et al., 2004) when forming a protein complex for perceiving the upstream signal Ca2+ (Allen et al., 1999). A link has been established between CIPK15 and an AP2 transcription factor AtERT7 that negatively regulates ABA response as a kinase substrate of CIPK5 (Song et al., 2005). A nuclear-localized transcription factor homeodomain protein ATHB6 has also been identified as a more downstream component of the ABI1 and ABI2 (Himmelbach et al., 2002). In contrast to PP2Cs ABI1 and ABI2, a PP 2A, RCN1, has been identified as a positive regulator of ABA response involved in early events of ABA signaling (Kwak et al., 2002).
Two members of the family of SNF1-related protein kinase, PKABA1 from wheat (Triticum aestivum) and AAPK from broad bean (Vicia faba), were characterized as the ABA-stimulated protein kinases and positive regulators of ABA response (Anderberg and Walker-Simmons, 1992; Li and Assmann, 1996; Gomez-Cadenas et al., 1999, 2001; Li et al., 2000, 2002; Johnson et al., 2002). PKABA1 mediates ABA-suppressed gene expression (Gomez-Cadenas et al., 1999), and phosphorylates and activates the ABA-responsive basic-domain Leu-zipper (bZIP) transcription factor TaABF, a homolog of ABI5 of Arabidopsis (Arabidopsis thaliana; Johnson et al., 2002). AAPK mediates ABA-induced stomatal closure and anion channels (Li et al., 2000) and modulates an RNA-binding protein AKIP1 by phosphorylating it and inducing its translocation into subnuclear speckles in guard cells (Li et al., 2002). An AAPK ortholog, OST1, has also been identified as a regulator of stomatal response to ABA (Mustilli et al., 2002). Recently, three homologs of AAPK and OST1 in rice (Oryza sativa), SAPK8, SAPK9, and SAPK10, were shown to be activated by ABA, suggesting their potential role in ABA signaling (Kobayashi et al., 2004).
Particular members of another class of important protein kinases, mitogen-activated protein kinase (MAPK), have been reported to be also activated by ABA (Knetsch et al., 1996; Burnett et al., 2000; D'Souza and Johri, 2002; Lu et al., 2002; Xiong and Yang, 2003). An Arabidopsis MAPK, AtMPK3, and a rice MAPK, OsMAPK5, have been identified as ABA-activated MAPKs. The former mediates postgermination arrest of development by ABA (Lu et al., 2002), and the latter is involved in disease resistance and abiotic stress tolerance (Xiong and Yang, 2003).
It has been well accepted that calcium is a central regulator of plant growth and development (Hepler, 2005). Specific calcium signatures are recognized by different calcium sensors to transduce specific calcium-mediating signal into downstream events (Sanders et al., 1999; Harmon et al., 2000; Rudd and Franklin-Tong, 2001). In plants, there are several known classes of Ca2+-binding sensory proteins, including calmodulin (CaM) and CaM-related proteins (Zielinski, 1998; Luan et al., 2002), CBL proteins (Luan et al., 2002), and calcium-dependent protein kinases (CDPKs; Harmon et al., 2001; Cheng et al., 2002). Among them, CDPKs, a novel class of Ca2+ sensors having both kinase and CaM-like domain, are the best characterized and involved in most of calcium-stimulated protein kinase activities iden tified in plants (Harmon et al., 2001; Cheng et al., 2002). Encoded by a large multigene family with possible redundancy and/or diversity in their functions (Harmon et al., 2001; Cheng et al., 2002), CDPKs are believed to be important components in plant hormone signaling (Cheng et al., 2002; Ludwig et al., 2004). Using a protoplast transient expression system, two Arabidopsis CDPKs, AtCPK10 and AtCPK30, have been demonstrated to activate a stress- and ABA-inducible promoter, showing the connection of CDPKs to ABA-signaling pathway (Sheen, 1996). However, a previous report showed that ABA had no effect on the induction of AtCPK10 mRNA (Urao et al., 1994), and it has been unclear whether AtCPK30 is stimulated by ABA. This suggests complexity of CDPK signaling in mediating ABA signal. In addition, calcium-dependent histone-phosphorylating activity was reported to be activated by ABA in rice seedling, but the CDPK gene responsible for this activity was not identified (Li and Komatsu, 2000). In tobacco (Nicotiana tabacum), the transcripts of a CDPK gene, NtCDPK1, were enhanced unspecifically by various phytohormones ABA, indole-3-acetic acid (IAA), GA3, and synthetic cytokinin benzyladenine and other growth substances such as jasmonic acid, but it is unknown whether the NtCDPK1 is activated by ABA in its kinase activities (Yoon et al., 1999). Thus, information is currently lacking about ABA-activated CDPKs identified by combined biochemical and molecular approaches as potential ABA signal transducers. In this report, we identified, characterized, and purified a 58-kD ABA specifically stimulated CDPK from grape berry, designated ACPK1. We isolated the cDNA coding for the ACPK1 based on sequencing of the purified ACPK1 protein. Also, we showed that ACPK1 positively regulates plasma membrane (PM) H+-ATPase in vitro, suggesting that ACPK1 may be involved in ABA-signaling pathway in grape berry.
RESULTS
Characterization of a 58-kD Membrane-Associated CDPK in Grape Berry
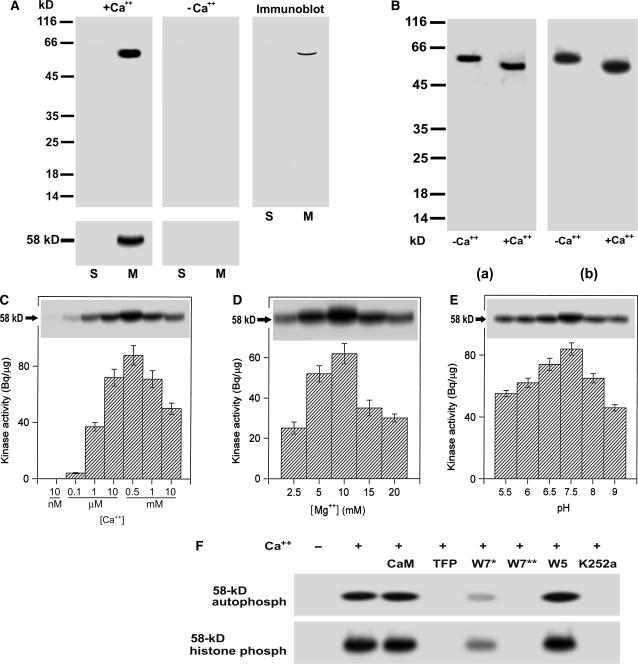
Grape is a berry fruit, comprising the fleshy portion and seeds. The fleshy portion covers three parts, i.e. pericarp and endocarp that both are several cell layers thick, and mesocarp that makes the large fleshy portion. Using in-gel assays, we detected in the microsomes of the grape mesocarp a calcium-dependent autophosphorylated protein with a molecular mass of 58 kD (Fig. 1A). This 58-kD protein phosphorylates histone III-S, one of the best exogenous substrates for assaying CDPKs (Roberts and Harmon, 1992), and this kinase activity depends also on the presence of Ca2+ (Fig. 1A). The 58-kD kinase was immunorecognized by an antiserum raised against the CaM-like domain of soybean (Glycine max) CDPKα (Bachmann et al., 1996; Fig. 1A). In the soluble fractions or in the absence of free Ca2+, neither the autophosphorylation nor histone-phosphorylating kinase activity was detected (Fig. 1A). These results suggest that the 58-kD membrane-associated kinase is a member of CDPK family.
Figure 1.
Biochemical characterization of a 58-kD membrane-associated Ca+-dependent protein kinase in grape berry. A, A 58-kD membrane-associated Ca2+-dependent protein kinase is present in grape berry. Soluble fraction (lanes labeled S, 40 μg of protein) and microsomes (lanes labeled M, 20 μg of protein) were separated on a 12% SDS-polyacrylamide gel. The separating gel for assaying kinase catalytic activity was polymerized in the presence of histone III-S. Autophosphorylation and kinase activities were assayed in the presence (+Ca2+) or absence (−Ca2+) of free Ca2+ as described in “Materials and Methods.” The molecular masses of protein standards are shown at the left of the sections in kD. Autophosphorylating (gels above) and histone-phosphorylating activity (gels below) of 58-kD kinase are shown in the left and middle sections, and immunoblotting in the right section. Immunoblotting was done with the antiserum directed against the CaM-like domain of soybean CDPKα as described in “Materials and Methods.” B, Ca2+-dependent electrophoretic mobility shift of the 58-kD kinase in both the in-gel autophosphorylation (a) and histone-phosphorylating activity (b) assays. Ca2+ or EGTA to a final concentration of 2 mm was added to the microsomal proteins dissolved in SDS-PAGE sample buffer. After SDS-PAGE, the in-gel phosphorylation assays were done in the presence of Ca2+. −Ca2+ and +Ca2+ indicate the absence and presence of Ca2+ in the SDS-PAGE sample buffer, respectively. C, Ca2+ dependence of the in-gel autophosphorylation (indicated by 58-kD with arrow) and in vitro activity (columnar figures) of the kinase in a medium pH at 7.5 and in the presence of Mg2+ at 10 mm and EGTA at 0.45 mm. The in vitro and in-gel histone III-S-phosphorylating activities and the in-gel autophosphorylation of the kinase were assayed as described in “Materials and Methods.” Values in the columnar figures are means ± se (n = 5). D, The same assay as in C but in different concentrations of Mg2+ and in the presence of 0.55 mm Ca2+. E, The same assay as in C but in different medium pHs and in the presence of 0.55 mm Ca2+ and 10 mm Mg2+. F, Inhibition of both the in-gel autophosphorylation (indicated by 58-kD autophosph) and histone-phosphorylating activity (indicated by 58-kD histone phosph) of the 58-kD kinase by CaM antagonists or kinase inhibitors. CaM was used at 5 μm; TFP or W5 at 250 μm; W7 at 100 (W7*) or 250 μm (W7**), and K252a at 10 μm. These reagents were added, respectively, to the phosphorylation reaction medium (buffer B as described in “Materials and Methods”) for a preincubation and a subsequent reaction incubation for 32P-labeling to the kinase or its substrate histone as described in “Materials and Methods;” − and + indicate the absence and presence of Ca2+ in the reaction buffer, respectively.
Ca2+-binding proteins such as CDPKs migrate in gels at different rates in the Ca2+-bound versus Ca2+-free state (Roberts and Harmon, 1992). To investigate this phenomenon, we added Ca2+ or EGTA to the protein sample just before electrophoresis and then the in-gel phosphorylation was analyzed in the presence of Ca2+. The assays of both in-gel autophosphorylation and histone-phosphorylating activity of the 58-kD kinase showed a clear mobility shift when the kinase migrates in the presence of Ca2+ (Fig. 1B). The in-gel autophosphorylation and in vitro histone-phosphorylating activities of the 58-kD kinase depended strongly on Ca2+ (Fig. 1C) and Mg2+ (Fig. 1D) concentrations and in a weaker extent on medium pHs (Fig. 1E). The assays of in-gel kinase activity gave the same results as the in vitro assays (data not shown).
We also analyzed the effects of the CaM antagonists trifluoperazine (TFP) and N-(6-aminohexyl)-5-chloro-1-naphthalene sulfonamide (W7) and inhibitor of Ser/Thr protein kinases K252a on the 58-kD kinase. The calcium-dependent in-gel autophosphorylation and histone-phosphorylating activity of the 58-kD kinase could be inhibited by TFP, W7, and K252a, and the dose dependence of W7 for the inhibition was observed (Fig. 1F). By contrast, CaM and N-(6-aminohexyl)-1-naphthalene sulfonamide (W5, an inactive analog of W7), had no apparent effect on autophosphorylation or phosphorylating activity of the 58-kD kinase (Fig. 1F). Taken together, all of the results consistently indicate that the 58-kD membrane-associated kinase is a CDPK.
ABA Stimulates the 58-kD CDPK
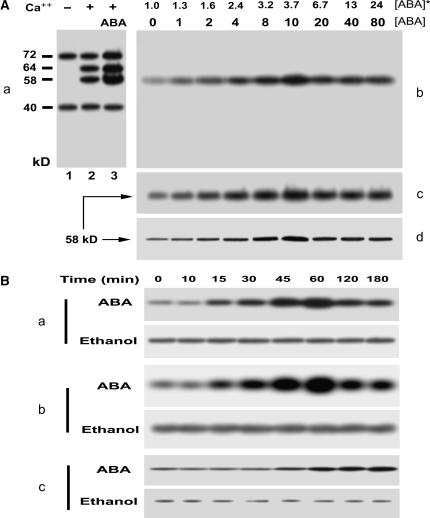
To investigate the effects of ABA on grape CDPK, we used throughout this study a system to incubate in vivo the berry tissues in the ABA-containing medium essentially according to the technique of Peng et al. (2003), as detailed in “Materials and Methods.” The assays of in vitro phosphorylation of the total microsomal proteins extracted from ABA-free incubated tissues allowed the detection of two bands of calcium-independent phosphoproteins (CIPs; Fig. 2A, a, lane 1) and two other bands of calcium-dependent phosphoproteins (CDPs; Fig. 2A, a, lane 2). The molecular masses of the CIPs are 72 (CIP72) and 40 kD (CIP40), and those of CDPs 64 (CDP64) and 58 kD (CDP58), respectively (Fig. 2A, a). ABA appeared to stimulate three of them, CIP72, CDP64, and CDP58, but not CIP40 (Fig. 2Aa). However, only one band of phosphorylation at the point of 58 kD was detected by in-gel phosphorylation (Fig. 2A, a and b), probably because the other in vitro-detected phosphoproteins may be only substrates of protein kinase, or that phosphorylation of diverse proteins may require the complex in vitro system where total membrane proteins are present and protein-protein interactions related to the phosphorylation may be realizable. The CDP58 phosphoprotein detected in the in vitro system should be the identified 58-kD kinase (Fig. 1). ABA strongly stimulated the 58-kD kinase in both its autophosphorylation and histone-phosphorylating kinase activities (Fig. 2A, b and c). In accordance with this, the amount of the 58-kD kinase, estimated by immunodetection with the antiserum to CaM domain of soybean CDPKα, was also enhanced by ABA treatment (Fig. 2A, d). The ABA-induced stimulation of the 58-kD kinase was dependent on the dose of ABA application, and the ABA concentration of 10 μm maximized the effects (Fig. 2A, b–d). When ABA at concentrations above 10 μm was applied, the ABA-induced effects declined but still remained at steady, higher levels (Fig. 2A, b–d). It is noteworthy that the concentrations of ABA within the treated tissues corresponding to the concentrations of applied ABA below 10 μm did not overpass the highest level of the physiological concentrations in grape berries (Fig. 2A; also see Fig. 8).
Figure 2.
ABA stimulates the 58-kD Ca2+-dependent protein kinase. A, ABA stimulates the 58-kD Ca2+-dependent protein kinase in a dose-dependent manner. a, ABA-stimulated in vitro phosphorylation in total microsomal proteins. The berry tissues were incubated in a medium (pH 5.5) containing 0 (control) or 10 μm (±)-ABA for 1 h. The microsomes prepared from the tissues were subjected to phosphorylation in vitro and separated by SDS-PAGE, and then phosphoproteins were detected by autoradiography. The detailed procedures of these assays were described in “Materials and Methods.” The phosphoproteins from the control tissues are shown after the in vitro phosphorylation in the absence (lane 1) or presence (lane 2) of Ca2+. The phosphorylation level of some phosphoproteins increases in the microsomes from the ABA-treated tissues after the in vitro phosphorylation in the presence of Ca2+ (lane 3). The calculated molecular masses according to the mobility rate of protein standards are shown at the left of the section in kD; − and + indicate the absence and presence of Ca2+, respectively. b to d, ABA-induced in-gel-stimulation of the 58-kD Ca2+-dependent protein kinase is dose dependent. The microsomal proteins prepared from the ABA-treated tissues were separated by SDS-PAGE and then subjected to in-gel autophosphorylation (b) and histone-phosphorylating activity (c) assays and immunoblotting analysis with rabbit polyclonal antibodies to the CaM-like domain of soybean CDPKα (d). The procedures of the assays were described in “Materials and Methods.” The numbers displayed in line [ABA] indicate the exogenous ABA concentrations applied to the berry tissues, and those in line [ABA]* show the ABA concentrations within the treated tissues determined by radioimmunoassay as described in “Materials and Methods.” The ABA-stimulated in-gel autophosphorylated protein (b) corresponds to the 58-kD phosphoprotein shown in section a. The molecular mass of the kinase phosphorylating histone III-S (c) or that of the signal immunodetected by anti-soybean CDPKα serum (d) is shown to be 58 kD. B, Time course of the ABA-induced stimulation of the 58-kD Ca2+-dependent protein kinase. Berry tissues were incubated for different durations of time from 0 to 180 min in the medium containing 10 μm ABA (ABA) or the same amount of ethanol for solubilizing ABA (Ethanol) as described in “Materials and Methods.” The microsomes prepared from the treated tissues were used for analysis of the 58-kD Ca2+-dependent protein kinase as described in A. a, In-gel autophosphorylation of the 58-kD kinase. b, In-gel histone-phosphorylating activity of the 58-kD kinase. c, 58-kD immunosignal detected by the rabbit antiserum directed against the calmudulin-like domain of soybean CDPKα.
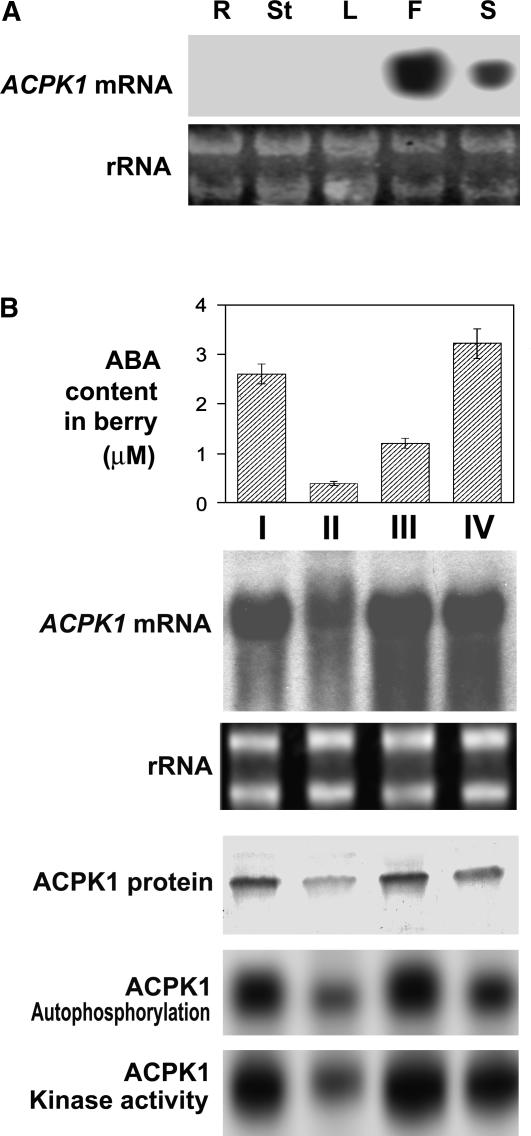
Figure 8.
Expression of ACPK1 in different tissues and during fruit development. A, Expression of ACPK1 gene in different tissues. Total RNA (20 μg) from root (R), young stem (St), leaf (L), flesh (F), or seed (S) of berry, transferred on a nylon membrane after electrophoresed in an agarose gel, was probed with the 32P-labeled cDNA fragment corresponding to the N-terminal variable domain of ACPK1 to detect specifically mRNA level of ACPK1 (ACPK1 mRNA). rRNA indicates the RNA samples stained with ethidium bromide. B, Expression of ACPK1 during berry development. Total RNA from berry tissues was probed as in A with the 32P-labeled cDNA fragment corresponding to the N-terminal variable domain of ACPK1 to detect specifically mRNA level of ACPK1 (ACPK1 mRNA). rRNA, RNA samples stained with ethidium bromide. Total microsomal proteins (100 μg) extracted from berry tissues were immunoprecipitated with 5 μg anti-ACPK1-N40 serum protein. The immunoprecipitated proteins were subjected to SDS-PAGE and then to the assays for detecting the levels of ACPK1 protein (ACPK1 protein, blotted with anti-ACPK1-N40 serum), autophosphorylation (ACPK1 Autophosphorylation), or histone-phosphorylating kinase activity (ACPK1 Kinase activity) as described in “Materials and Methods.” The concentrations of ABA in the berry tissues (columnar figures, ABA content in berry) were determined by radioimmunoassay (values in the columnar figures are means of five replications ± se). I, II, III, and IV indicate the sampling dates that are the early stage of berry growth (10 d after anthesis, stage I), first rapid growth phase (stage II, 10th–30th d after anthesis), lag growth phase (stage III, 30th–50th d after anthesis), and second rapid growth phase or ripening stage (stage IV, 50th–80th d after anthesis).
We further analyzed time course of the ABA-induced effects. ABA rapidly stimulated the 58-kD kinase in its autophosphorylation and kinase activity, and the ABA-induced effects appeared to be transient (Fig. 2B, a and b). An incubation of 15 min could induce the effects, and the effects were maximized with a 60-min incubation (Fig. 2B). A longer incubation over 60 min allowed still a steady, higher level of the 58-kD kinase stimulation, though the effects declined (Fig. 2B, a and b). It is noteworthy, however, that the increase of the apparent amount of the 58-kD kinase began at the point of 45-min incubation, significantly later than did its autophosphorylation and kinase activity, and this enzyme amount did not decrease after 60 min of incubation, which, contrasting with the enzyme activities, showed a typical saturation curve (Fig. 2B, c). These data suggest that the rapid phase (15–30 min) of the 58-kD kinase stimulation by ABA may be due only to a posttranslational modification, but the slow phase (45 min and later) may involve the events of both posttranslational regulation and de novo protein synthesis of the enzyme (Fig. 2B). Whatever mechanism is involved, all above results allow the identification of the 58-kD kinase as an ABA-stimulated CDPK, and so we designate it ACPK1.
ABA-Induced Stimulation of ACPK1 Is Specific to the Physiologically Active Form of ABA
To determine the stereo specificity of ABA in the ACPK1 stimulation, two ABA isomers (−)-2-cis, 4-trans-ABA [(−)-ABA], and 2-trans, 4-trans-(±)-ABA (trans-ABA), were used in the experiment. The used (±)-ABA in the experiments is a mixture of (+)-2-cis, 4-trans-ABA [(+)-ABA] and (−)-ABA with (+)-ABA being physiologically active.
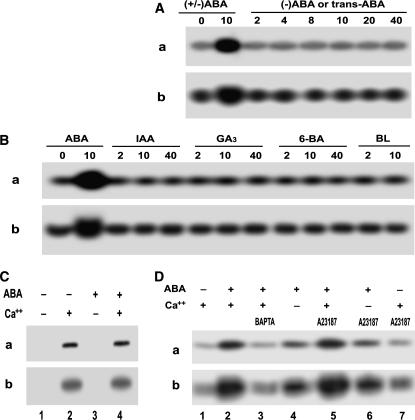
The two ABA isomers (−)-ABA and trans-ABA are structurally similar to physiologically active (+)-ABA, but functionally inactive (Balsevich et al., 1994; Hill et al., 1995; Walker-Simmons et al., 1997). We previously showed also that the two ABA isomers were unable to be bound to ABA-specific binding proteins that possess potential receptor nature (Zhang et al., 1999, 2001, 2002). The assays of both the in-gel autophosphorylation and histone-phosphorylating activities showed that the two ABA analogs were unable to stimulate the ACPK1 kinase (Fig. 3A), revealing that the ACPK1 kinase stimulation induced by ABA is stereo specific, only physiologically active (+)-ABA being effective.
Figure 3.
Some features of ABA-induced ACPK1 kinase stimulation. A, Specificity of the stimulation of the ACPK1 kinase to the physiologically active form of ABA. Berry tissues were incubated in the medium containing various concentrations (indicated by the numbers under lines as μm) of (±)-ABA or ABA isomers for 1 h as described in “Materials and Methods.” In-gel phosphorylation (a) and histone-phosphorylating activity (b) of the 58-kD kinase in the microsomes prepared from the tissues treated by (±)-ABA (as a control) or its isomers (−)-ABA or trans-ABA were analyzed. B, ACPK1 appears to be stimulated uniquely by ABA. In-gel phosphorylation (a) and histone-phosphorylating activity (b) of the 58-kD kinase were assayed in the microsomes prepared from the tissues treated as described in A by (±)-ABA (as a control) or other plant hormones auxin IAA, GA3, synthetic cytokinin N-benzyl-6-aminopurine (6-BA), and brassinolide (BL). The numbers under lines indicate the applied concentrations as μm. C, ABA stimulates ACPK1 kinase indirectly. Microsomes were prepared from nontreated tissues. In-gel phosphorylation (a) and histone-phosphorylating activity (b) of the 58-kD kinase were assayed in the microsomes in the presence or absence of 10 μm ABA or Ca2+ or both; − and + indicate the absence and presence, respectively. D, Effects of apoplasmic Ca2+ and influx of Ca2+ on ABA-induced stimulation of ACPK1 kinase. Berry tissues were preincubated in a medium (the equilibration buffer as detailed in “Materials and Methods”) containing 5 mm BAPTA (a Ca2+ chelator, lane 3) or 5 μm A23187 (a Ca2+ ionophore, lanes 5–7) in the presence of Ca2+ (lanes 3, 5, and 7), or 1 mm EGTA in the absence of Ca2+ (lanes 4 and 6), and then ABA (10 μm except for lane 7 where ABA was absent) was added to the medium for a further incubation of 1 h. The incubation in the presence of Ca2+ but absence of ABA (lane 1) was taken as a control, and that in the presence of both Ca2+ and ABA (lane 2) as another control. The microsomes were prepared from the treated tissues. In-gel autophosphorylation (a) and histone-phosphorylating activity (b) of the 58-kD kinase in the microsomes were analyzed in the presence of Ca2+; − and + indicate the absence and presence, respectively.
We further analyzed the effects of some other phytohormones on activities of the ACPK1 kinase. In the assayed substances auxin IAA, GA3, synthetic cytokinin N-benzyl-6-aminopurine, and brassinolide, none of them had any effect on ACPK1 in either its autophosphorylation or histone-phosphorylating kinase activity (Fig. 3B). This result indicates that the ACPK1 kinase appears to be stimulated specifically and exclusively by ABA.
ABA Indirectly Stimulates ACPK1, and Influx of Ca++ May Be Involved in This Stimulation
To investigate whether ABA directly acts on the ACPK1 kinase, ABA was added directly to the reaction medium of the in-gel phosphorylation instead of incubating the tissues in vivo in the ABA-containing medium. The results showed that ABA had no direct effect on the ACPK1 kinase in either its autophosphorylation or kinase activity (Fig. 3C), indicating that ABA indirectly stimulates the ACPK1 kinase.
We further investigate possible roles of Ca2+ influx in mediating the ABA-induced ACPK1 stimulation. Two Ca2+ chelators, 1,2-bis(2-aminophenoxy)-ethane-N,N,N,M-tetraacetic acid (BAPTA) and EGTA, were used to incubate the tissues in vivo together with ABA. This treatment of eliminating apoplasmic Ca2+ significantly reduced the ACPK1 stimulation by ABA, though BAPTA was more effective compared to EGTA (Fig. 3D). A further experiment was done with A23187, a Ca2+ ionophore that allows diffusion of Ca2+ through plasma membranes in an electrically neutral manner, thus creating an artificial influx of Ca2+ from apoplasm into cytoplasm. However, the treatment with A23187 had no significant effect on the ABA-induced ACPK1 stimulation (Fig. 3D, lanes 5 and 6). The treatment with A23187 alone did not stimulate ACPK1 (Fig. 3D, lane 7). These data suggest that influx of Ca2+ from apoplasm into cytoplasm may be involved in the ABA-induced events, which requires a mechanism such as calcium channels to mediate influx of Ca2+, but ABA signaling in these events may be more complex than dependent on influx of Ca2+.
Purification and Molecular Cloning of ACPK1
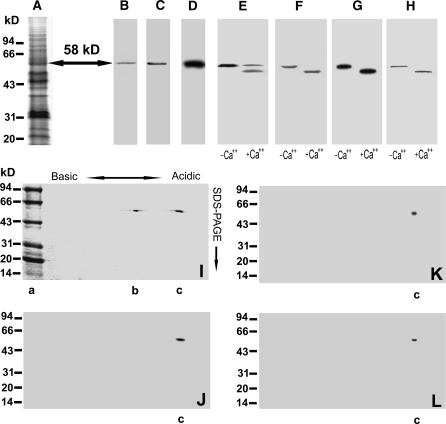
We adopted a technique to purify ACPK1 kinase from SDS-polyacrylamide gels after the total membrane proteins were separated by SDS-PAGE, because the approaches based on columnar chromatography that we used for this purification were unsuccessful. The exact position of the 58-kD kinase on gels (Fig. 4A) was determined by both autophosphorylation and immunodetection with antiserum against the CaM-like domain of soybean CDPKα, and thus the portions of the gels containing the 58-kD kinase were excised and eluted. The lyophilized eluate was analyzed for assessing the purification. A total amount of 400 μg purified proteins was obtained from 30 mg crude microsomal proteins, i.e. 50 gels × 10 lanes with 60 μg crude microsomal proteins on each lane.
Figure 4.
Purification of the ACPK1 kinase. The portions at the 58-kD point (arrow) of SDS-polyacrylamide gels (A, Coomassie Brilliant Blue R-252-stained gels) were excised, crushed, and eluted for purification of 58-kD proteins as described in “Materials and Methods.” The collected 58-kD proteins were subjected to SDS-PAGE (B, Coomassie Brilliant Blue R-252-stained gels), immunorecognized by anti-soybean CDPKα serum (C), and detected by in-gel autophosphorylation assay (D). The purified 58-kD proteins were further analyzed for their Ca2+-dependent electrophoretic mobility shift with the assays of SDS-PAGE (E, Coomassie Brilliant Blue R-252-stained gels), in-gel phosphorylation (F), histone-phosphorylating activity (G), and immunoblotting by anti-soybean CDPKα serum (H). IEF/SDS-PAGE two-dimensional electrophoresis of the purified 58-kD proteins displays two polypeptides at the 58-kD point but with different pI (I, Coomassie Brilliant Blue R-252-stained gels). One of the two polypeptides (lane c in sections I–L) is shown to be the 58-kD ABA-stimulated Ca2+-dependent protein kinase with the assays of in-gel autophosphorylation (J), histone-phosphorylating activity (K), and immunoblotting by anti-ACPK1-N40 serum (see “Materials and Methods”), an antibody specific to the 58-kD ACPK1 kinase (L). The molecular masses of protein standards are shown at the left of the sections in kD. Lane a in section I shows the protein standards on gel; −Ca2+ and +Ca2+ in sections E to H indicate the absence and presence of Ca2+, respectively.
The purified proteins migrated as a single protein band at the point of 58-kD on SDS-polyacrylamide gel (Fig. 4B). The purified 58-kD proteins could autophosphorylate (Fig. 4C) and phosphorylate histone III-s (Fig. 4D) in the presence of Ca2+, but both of their autophosphorylation and kinase activities were lost in the absence of Ca2+ (data not shown). It is particularly noteworthy that SDS-PAGE of the apparently purified protein in the presence of calcium displayed two bands (Fig. 4E), of which one possessing Ca2+-dependent electrophoretic mobility shift should be the 58-kD kinase, and another may be a protein other than this kinase. This nature of Ca2+-dependent electrophoretic mobility shift of the 58-kD kinase was further shown in the assays of autophosphorylation (Fig. 4F), histone-phosphorylating activity (Fig. 4G), and immunoblotting with antiserum against the CaM-like domain of soybean CDPKα (Fig. 4H).
The 58-kD proteins were further identified and purified by isoelectric focusing electrophoresis (IEF)/SDS-PAGE two-dimensional electrophoresis. Two protein bands were observed on SDS-polyacrylamide gel with the same molecular mass of 58 kD but different pIs (Fig. 4I, lanes b and c), which is consistent with the above results of SDS-PAGE. One of them (Fig. 4I, lane c) was identified as the 58-kD ACPK1 kinase by its features of Ca2+-dependent autophosphorylation (Fig. 4J), histone-phosphorylating activity (Fig. 4K), and immunorecognition by anti-ACPL1-N40 serum (Fig. 4L), a specific antibody to ACPK1 (see below).
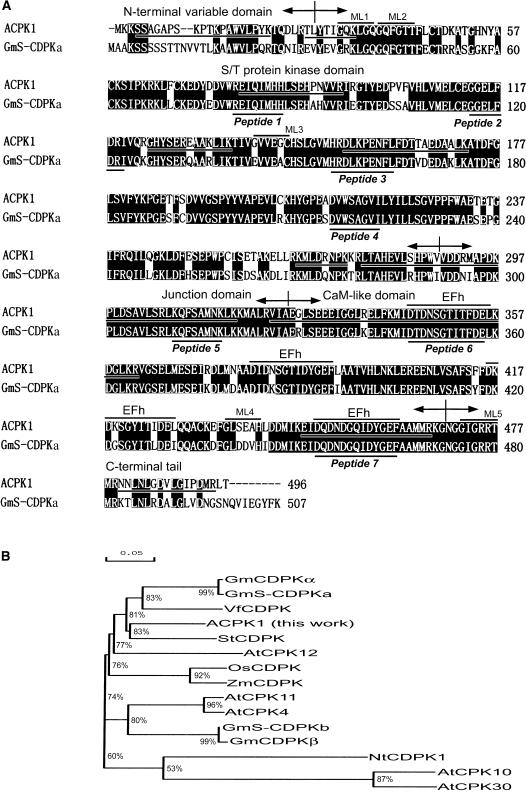
The IEF/SDS-PAGE-purified 58-kD kinase was sequenced by tandem mass spectrometry (TSM). The data from the first sequencing indicated that the sequence of the purified 58-kD kinase matches that of some plant CDPKs registered in GenBank in their conserved kinase, junction, and CaM-like domains. The sequences of matched peptides were: REIQIMHH, GGELFDRI, HRDLKPENFL, DVWSAGVI, KQFSAMNK, DTDNSGTITFDE, and DQDNDGQIDYGEF (Fig. 5A, indicated by Peptide 1–Peptide 7), which formed the basis for the cloning of ACPK1 by PCR. Using the designed degenerate primers that correspond to two of the matched sequences of the 58-kD kinase and also to the conversed domain of most CDPKs (-EIQIMHHL- and -KQFSAMNK-), a cDNA clone putatively coding for ACPK1 (Fig. 5A) was obtained and registered in GenBank. A second sequencing of the purified 58-kD kinase showed that the sequence of 58-kD kinase matched the predicted amino acid sequence from the cDNA clone in 10 pieces particularly covering all the N-terminal variable, conserved kinase, junction and CaM-like domains, and variable C-terminal tail domain (Fig. 5A; see also Supplemental Fig. 1; Supplemental Table I), demonstrating that the isolated cDNA clone encodes the 58-kD ACPK1 kinase. Based on the cDNA cloning, an ACPK1-specific antiserum against the N-terminal 40 amino acids covering the full variable domain and an adjacent portion of kinase domain (11 amino acids) of ACPK1 was prepared and recognized the 58-kD IEF/SDS-PAGE-purified kinase as mentioned above, confirming that the ACPK1 is encoded by the isolated cDNA clone.
Figure 5.
Alignment and phylogenetic relationships of ACPK1 and its related members of the CDPK family. We added the following prefixes to the names of some cited CDPKs in this paper for distinguishing them easily if their original names do not contain the prefixes: At for Arabidopsis, Gm and GmS (S indicates seed) for soybean, Os for rice, St for potato, Vf for broad bean, and Zm for maize. A, Alignment of deduced amino acid sequences of ACPK1 and one of its closest homologs, soybean seed CDPKa (GmS-CDPKa), shows the presence of conserved features in Ser/Thr (S/T) protein kinase domain, junction domain, and CaM-like domain and also variable features in N-terminal domain and C-terminal tail. Numbers on the right column indicate numbers of amino acid residues in the predicted sequences. Gaps, indicated by dashes (–), were introduced to maximize alignment. Identical amino acid residues are indicated by white letters on a black background. The boundaries of the N-terminal variable domain, S/T kinase domain, junction domain, CaM-like domain, and C-terminal tail are indicated by arrows. Four EF-hand motifs (EFh) and five potential myristoylation sites (ML1-5) are also shown. The matched sequences of seven peptides obtained in the first sequencing of the purified 58-kD ABA-stimulated CDPK (natural ACPK1 protein, see Fig. 7) by TMS are marked by Peptide 1 to Peptide 7 underneath the GmS-CDPKa sequence, respectively. The degenerate oligonucleotides corresponding to the conserved sequences of the peptide 1 in kinase domain (forward primer) and peptide 5 in junction domain (reverse primer) were used for cloning the putative ACPK1. The underlined sequences of ACPK1 are the matched sequences obtained in the second sequencing of the natural ACPK1 protein by TMS after the cDNA coding for the putative ACPK1 was isolated. B, Tree depicting phylogenetic relationships based on alignment of deduced amino acid sequences among top 11 of CDPKs having higher identity with ACPK1 and the other three ABA-responsive CDPKs, NtCDPK1 (Yoon et al., 1999), and AtCPK10 and AtCPK 30 (Sheen, 1996). Percentages of amino acid sequence identity are also indicated. The corresponding accession numbers or locus tag numbers for the genes encoding these CDPKs are given in the section “Accession Numbers” in “Materials and Methods.”
Sequence Features of ACPK1
ACPK1 is a full-length clone with an open reading frame (ORF) having 1,491 nucleotides and encoding a protein of 496 amino acids with a predicted molecular mass of 56 kD and a pI at 5.3. The predicted amino acid sequence exhibits the modular structure typical for CDPKs (Harmon et al., 2001) comprising an N-terminal variable region, a kinase domain, an autoinhibitory junction domain, and a CaM-like domain with four conserved EF-hand motifs implicated in Ca2+ binding. Five potential myristoylation sites are present in the full-length amino acid sequence of ACPK1, and two of them are localized in the N-terminal region, suggesting possible membrane-associated nature of ACPK1 via N-terminal myristoylation (Martin and Busconi, 2000; Lu and Hrabak, 2002; Rutschmann et al., 2002; Dammann et al., 2003). This modification by acylation may be partly responsible for the higher molecular mass of the biochemically identified 58-kD kinase than that of the predicted 56 kD. Comparison of the deduced ACPK1 amino acid sequence with a soybean CDPK from seeds (GmS-CDPKa) shows a strong homology that extends all over the conserved domains (Fig. 5A). The highest identity is shared with potato (Solanum tuberosum) StCDPK (83%), broad bean VfCDPK (81%), and two soybean CDPKs (81%), GmCDPKα (soybean CDPKα) and GmS-CDPKa (Fig. 5B). The two soybean CDPKs should be orthologs with amino acid sequence identity of 99% between them (Fig. 5B). The other closer homologs of ACPK1 are Arabidopsis AtCPK4, AtCPK11, and AtCPK12; soybean CDPKβ (GmCDPKβ); soybean seed CDPKb (GmS-CDPKb); rice OsCDPK; and maize (Zea mays) ZmCDPK (Fig. 5B).
ACPK1 has a lower amino acid sequence identity (53%) with the three previously identified ABA-responsive CDPKs (Fig. 5B; Sheen, 1996; Yoon et al., 1999). The analysis of phylogenetic relationships among the closet homologs of ACPK1 and the three ABA-responsive CDPKs based on amino acid sequence alignment shows that ACPK1 may be classified as a different group from the three previously identified ABA-responsive CDPKs (Fig. 5B).
Genomic Southern Analysis of ACPK1
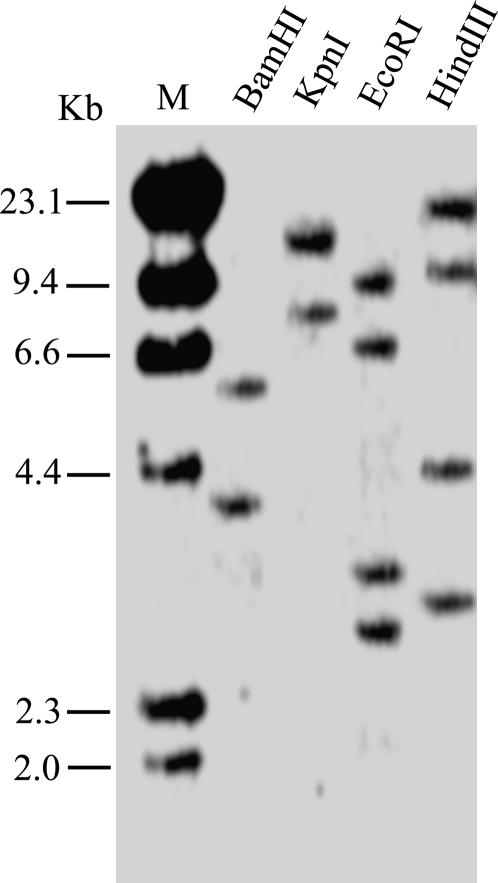
Genomic Southern-blot analysis was done with a probe corresponding to full-length ACPK1 cDNA. Genomic DNA was digested with BamHI, KpnI, EcoRI, and HindIII restriction enzymes. The probe generated two hybridizing bands in BamHI or KpnI digestion, and four bands in EcoRI or HindIII digestion (Fig. 6). Since there is a catalytic site within the cDNA sequence of ACPK1 for both EcoRI and HindIII, these results suggest that the grape genome contains two copies or more than one copy of ACPK1-related genes.
Figure 6.
Southern-blot analysis of the ACPK1 gene. A 10-μg portion of grape genomic DNA was digested with BamHI, KpnI, EcoRI and HindIII, electrophoresed in a 0.8% agarose gel, and transferred onto a nylon membrane. The membrane was hybridized with the 32P-labeled full-length cDNA of ACPK1. The DNA markers (M) are indicated at the left of the section in kb.
Characterization of the Products of the ACPK1 Gene and Up-Regulation of Both Its Products and Transcripts by ABA
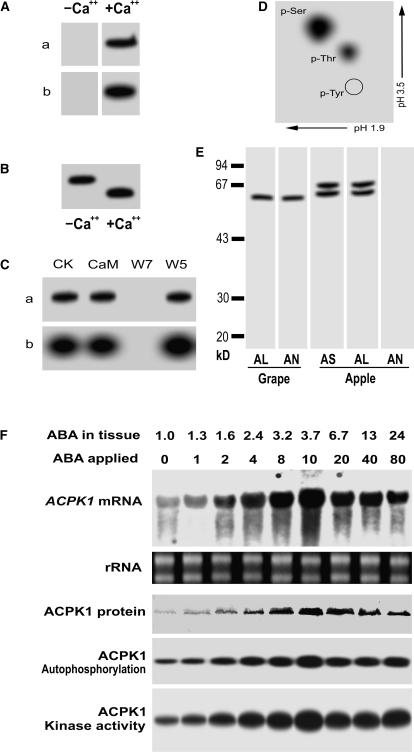
To confirm that ACPK1 encodes an active CDPK whose expression and activity are stimulated by ABA, the full-length ACPK1 (ACPK1L) and the truncated form specific to ACPK1 (residues 1–40; ACPK1-N40) containing only the N-terminal variable region and its adjacent portion in the kinase domain (as mentioned above) were expressed in Escherichia coli as fusion proteins. The ACPK1L protein showed Ca2+-dependent autophosphorylation and histone-phosphorylating activities (Fig. 7A), and also Ca2+-dependent electrophoretic mobility shift in SDS-PAGE, shown by its autophosphorylation (Fig. 7B). Both the autophosphorylation and histone-phosphorylating activities of the ACPK1L were inhibited by W7 but not by W5, and these activities were apparently not stimulated by CaM (Fig. 7C). Two-dimensional, thin-layer chromatography of the hydrolyzed autophosphorylated ACPK1L showed that 32P-labeled spots corresponded to the positions of phospho-Ser and phospho-Thr (Fig. 7D), suggesting that ACPK1 possesses Ser/Thr kinase activity. Taken together, these data confirmed the nature of ACPK1 as an active CDPK.
Figure 7.
Characterization of the products of ACPK1 gene and up-regulation of both its products and transcripts by ABA. A, The purified recombinant protein (10 μg) of the full-length ACPK1 (ACPK1L) was subjected to SDS-PAGE and then assayed for its in-gel autophosphorylation (a) and histone-phosphorylating activity (b) in the absence (−Ca2+) or presence (+Ca2+) of Ca2+. B, Ca2+-dependent electrophoretic mobility shift of the ACPK1L, shown by its in-gel autophosphorylation; −Ca2+ and +Ca2+ indicate the absence and presence of Ca2+, respectively. C, Neither the in-gel autophosphorylation (a) nor histone-phosphorylating activity (b) is activated by CaM, whereas both activities are inhibited by CaM antagonist W7 but not by W5. CK, Control. D, Phosphoamino acid analysis of the autophosphorylated ACPK1L. γ-32P-labeled ACPK1L was hydrolyzed with HCl and subjected to two-dimensional thin-layer electrophoresis. The positions of phospho-Ser (p-Ser), phospho-Thr (p-Thr), and phospho-Tyr (p-Tyr) are indicated. E, Specificity of the anti-ACPK1-N40 serum to ACPK1. The 58-kD ACPK1 immunosignal was detected in the microsomes of grape mesocarp (Grape) by both the anti-ACPK1L (indicated by AL) and -ACPK1-N40 (indicated by AN) sera. In the microsomes of apple flesh prepared with the same procedures as described in “Materials and Methods” for grape mesocarp, two immunosignals having molecular mass of about 60 and 67 kD, respectively, were detected by the antiserum against CaM-like domain of soybean CDPKα (indicated by AS) or anti-ACPK1L serum (AL), but not by anti-ACPK1-N40 serum (AN). F, ABA up-regulates gene expression, autophosphorylation, and kinase activity of ACPK1. Berry tissues were incubated in the medium containing different concentrations of (±)-ABA from 0 to 80 μm (indicated by ABA applied; unit: μm), and after the incubation, the concentrations of ABA present in the treated tissues (ABA in tissue; unit: μm) were determined by radioimmunoassay as described in “Materials and Methods.” Total RNA and microsomes were extracted from the treated tissues, respectively. Total RNA (20 μg), transferred on a nylon membrane after electrophoresed in an agarose gel, was hybridized with the 32P-labeled cDNA fragment corresponding to the N-terminal variable domain of ACPK1 to detect specifically mRNA level of ACPK1 (ACPK1 mRNA). rRNA indicates loading control of the RNA samples stained with ethidium bromide. Total microsomal proteins (50 μg) were immunoprecipitated with 3 μg anti-ACPK1-N40 serum protein. The immunoprecipitated proteins were subjected to SDS-PAGE and then to the assays for detecting the levels of ACPK1 protein (ACPK1 protein, blotted with anti-ACPK1-N40 serum), autophosphorylation (ACPK1 Autophosphorylation), or histone-phosphorylating kinase activity (ACPK1 Kinase activity) as described in “Materials and Methods.”
The antisera against either ACPK1L or ACPK1-N40 were prepared in rabbit. Both the anti-ACPK1L and -ACPK1-N40 sera recognized the ACPK1 signal from grape berry (Fig. 7E). The anti-ACPK1L serum can recognize other putative members of CDPK family as both this antiserum and the antiserum against the CaM-like domain of soybean CDPKα detected from apple fruit (Malus domestica) two immunosignals having molecular mass of about 60 and 67 kD, respectively, but the anti-ACPK1-N40 serum could not (Fig. 7E). This showed that the anti-ACPK1-N40 serum is specific to ACPK1. The same in vivo incubation assays as used above when testing the ABA-induced 58-kD kinase stimulation were performed to confirm the action of ABA on CDPK1, but the specifically immunoprecipitated ACPK1 protein by anti-ACPK1-N40 serum was used for the phosphorylation assays, and also ACPK1 mRNA was probed with 32P-labeled cDNA fragment corresponding to the variable domain of ACPK1 to assess the ACPK1 expression. The results showed that ABA stimulated the ACPK1 kinase in all levels of the gene transcription, mRNA translation and kinase autophosphorylation, and catalytic activities in a dose-dependent manner (Fig. 7F). It is noteworthy that in the immuno- and enzyme assays of ACPK1, the immunoprecipitated proteins with the anti-ACPK1L serum gave substantially the same results as those with the ACPK1-specific anti-ACPK1-N40 serum (data not shown), suggesting that ACPK1 may be a major CDPK having a predominant abundance in grape mesocarp as was observed above (Fig. 2A). All these assays based on molecular cloning of ACPK1 consistently confirmed the above-mentioned results from the biochemical approaches (Fig. 2A).
ACPK1 Expression Is Fruit Specific and Altered Accordantly with Endogenous ABA Concentrations during Fruit Development
Northern-blot analysis showed that ACPK1 expressed only in the grape berry covering two essential portions of the fleshy mesocarp and seeds with much higher abundance of ACPK1 mRNA in the mesocarp than in seeds, but ACPK1 mRNA was undetectable in roots, young stems, or leaves (Fig. 8A). The amounts of mRNA and protein of ACPK1 as well as its autophosphorylation and histone-phosphorylating activities were all altered with changes in the endogenous ABA concentrations in the grape mesocarp during fruit development at different stages: I (early growth stage), II (first rapid growth phase), III (lag growth phase), and IV (second rapid growth phase or ripening stage; Fig. 8B).
ACPK1 Is Localized in Both Plasma Membranes and Chloroplasts
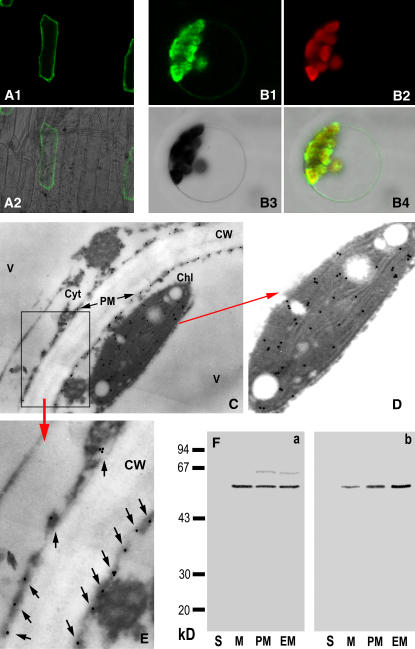
Transient expression of the green fluorescent protein (GFP)-ACPK1 fusion protein in chloroplast-free epidermis cells of onion (Allium cepa) showed that the fusion protein was targeted to the cell periphery, likely the plasma membrane (Fig. 9A, 1 and 2). A further assay of the transient expression of the fusion protein in Arabidopsis protoplasts showed that the fusion protein was localized to both plasma membranes and chloroplasts (Fig. 9B, 14). The localization of the enzyme in grape mesocarp by immunogold technique with the specific antiserum against ACPK1-N40 showed clearly the double-localized ACPK1 in both plasma membranes and chloroplasts (Fig. 9, C–E). No substantial immunosignal was detected in the controls without antiserum or using the preimmune serum instead of antiserum (data not shown), indicating the specificity and reliability of the immunolocalization.
Figure 9.
Subcellular localization of ACPK1. A1 and A2, Transient expression of ACPK1-GFP fusion protein in the epidermis cells of onion. The fusion protein is present in the cell outlines, shown by the ACPK1-GFP fluorescence image (A1) and merged image (A2) of laser-scanning confocal microscopy. B1 to B4, Transient expression of ACPK1-GFP fusion protein in the protoplast of Arabidopsis shows that the fusion protein is localized in both the plasma membranes and protoplasts. The laser-scanning confocal microscopy images are the ACPK1-GFP fluorescence (B1), chlorophyll autofluorescence (B2), bright field (B3), and merged (B4) images. C, Immunogold labeling of ACPK1 in the grape berry tissue with anti-ACPK1-N40 serum (antiserum against the N-terminal 40 amino acids of ACPK1) specific to ACPK1. Immunogold particles are distributed in plasma membranes (PM) and chloroplast (Chl) but not in other cellular compartments such as cell wall (CW), cytosol (Cyt), and vacuole (V). The arrows indicate the presence of the immunogold particles along the plasma membranes. D, An amplified portion of the image C, displaying clearly the chloroplast localization of the immunogold particles representing ACPK1. E, Blow up of the boxed-in area in C. Arrows indicate the immunogold particles localized to plasma membranes. F, Immunoblotting in different subcellular fractions with either anti-ACPK1L serum (antiserum against the full-length ACPK1; a) or anti-ACPK1-N40 serum (b). The fractions of soluble portion (lanes S, 40 μg protein), microsomes (lanes M, 20 μg protein), plasma membranes (lanes PM, 20 μg protein), and endomembranes (lanes M, 20 μg protein) were separated on a 12% SDS-polyacrylamide gel and then subjected to immunoblotting. The molecular masses of protein standards are shown at the left of the sections in kD.
The ACPK1-specific anti-ACPK1-N40 serum recognized the ACPK1 in the total membrane proteins (microsomes), purified plasma membrane, and endomembrane fractions, but not in the soluble fraction, whereas the antiserum against the full-length ACPK1 (ACPK1L) recognized in these membrane fractions not only the 58-kD kinase ACPK1 but also a signal having higher molecular mass than ACPK1 (Fig. 9F). This immunosignal may be the 64-kD phosphoprotein detected in the in vitro phosphorylation of the total membrane proteins (Fig. 2A, a), which may be a heavily modified form of 58-kD ACPK1 or a different protein kinase whose content is probably too low to be detectable in the crude extracts of membrane fractions (microsomes) but was sufficiently enriched in the purified membranes to be immunodetected by the anti-ACPK1L serum. These data from immunoblotting further demonstrated the specificity of the anti-ACPK1-N40 serum to ACPK1 and confirmed the plasma membrane-associated nature of ACPK1. The signal of ACPK1 immunodetected in the endomembrane fraction (Fig. 9F) should be the chloroplast-associated ACPK1. It is finally noteworthy that neither the transmembrane domains nor signal to target ACPK1 to plasma membranes or plastids were predicated for this kinase with several programs (data not shown). So, ACPK1 may be myristoylated in its N-terminal region for targeting to the membranes (Dammann et al., 2003 and refs. therein).
ACPK1 Positively Regulates PM H+-ATPase in Vitro
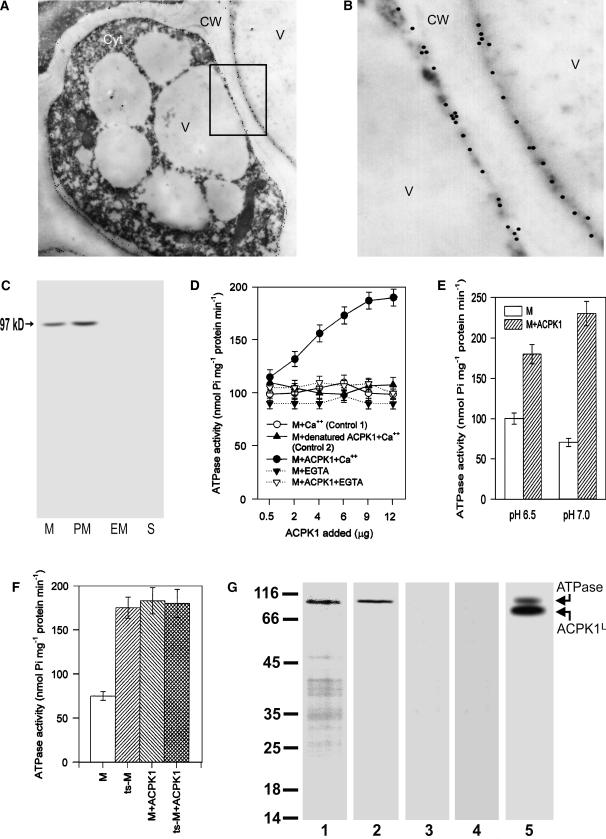
We previously reported that ABA activates plasma membrane (PM H+-ATPase) in apple fruit (Peng et al., 2003). The similar results were obtained in grape berry (data not shown). To assess whether ACPK1 is possibly involved in this ABA-induced event, we first showed the presence of a 97-kD plasma membrane-localized H+-ATPase in grape mesocarp by both immunogold subcellular localization (Fig. 10, A and B) and immunoblotting of the subcellular fractions (Fig. 10C). This plasma membrane colocalization of the ACPK1 kinase with PM H+-ATPase may facilitate possible interactions between the two proteins. A preincubation of the microsomal proteins with the recombinant full-length ACPK1 kinase ACPK1L significantly enhanced the activity of PM H+-ATPase in an ACPK1L dose-dependent manner with the activity being doubled in the peaks of the activation (Fig. 10D). The controls of the precipitation in the ACPK1L free- or denatured ACPK1L-containing medium showed no significant effect on PM H+-ATPase (Fig. 10D), indicating that the ACPK1-related effects were specific. Removal of free Ca2+ from the preincubation medium abolished the PM H+-ATPase activation by ACPK1 (Fig. 10D), showing that the PM H+-ATPase activation was Ca2+ dependent. This is consistent with the nature of the action of a CDPK.
Figure 10.
Presence of plasma membrane-localized H+-ATPase in grape berry and positive regulation of its activity by ACPK1. A, Immunogold labeling of PM H+-ATPase in the grape mesocarp with antiserum against Arabidopsis PM H+-ATPase AHA3, showing numbers of immunogold particles along the plasma membranes. Cyt, Cytosol; CW, cell wall; V, vacuole. B, Blow up of the boxed-in area in A, displaying clearly the presence of numbers of H+-ATPase molecules visualized by gold particles along the plasma membranes. C, A 97-kD (indicated by arrow) immunosignal was detected on SDS-polyacrylamide gel by antiserum against Arabidopsis PM H+-ATPase AHA3 in both the microsomes (M) and plasma membrane fraction (PM), but not in either the endomembrane (EM) or soluble fraction (S). D, ACPK1 activates PM H+-ATPase in a dose-dependent manner. The recombinant ACPK1L protein was added to the microsomes in the presence (M+ACPK1+Ca2+) or absence (M+ACPK1+EGTA) of free Ca2+ for a preincubation before measuring PM H+-ATPase activities as described in “Materials and Methods.” The assay with the ACPK1L-free microsomes preincubated in the presence (M+Ca2+, Control 1) or absence (M + EGTA) of free Ca2+ were taken as controls. The same amount of the microsomal proteins denatured by boiling was used instead of the ACPK1L protein as another control (M + denatured ACPK1 + Ca2+, Control 2). E, The same assay as described above in M + ACPK1 + Ca++ in D, but with the PM H+-ATPase activities measured in different medium pH. The microsomes only (M) or the microsomes together with ACPK1L protein (M + ACPK1) was used for the assay. F, The same assay was done as described above in M + ACPK1 + Ca++ in D, but the PM H+-ATPase activities were measured at pH 7.0, and the trypsin-treated microsomes only (ts-M) or the trypsin-treated microsomes together with ACPK1L protein (ts-M + ACPK1) was used for the assay where the nontreated microsomes (M) or the nontreated microsomes in the presence of ACPK1L protein was used as controls. G, ACPK1 phosphorylates PM H+-ATPase. The molecular masses of protein standards are shown at the left of the sections in kD. The partially purified PM H+-ATPase fraction was subjected to SDS-PAGE (lane 1), and an immunosignal of about 97 kD (lane 2) was recognized from the separated protein bands by the antiserum against Arabidopsis PM H+-ATPase AHA3, which is the grape PM H+-ATPase. The in vitro phosphorylation in the medium containing the partially purified PM H+-ATPase fraction and ACPK1L protein showed that the PM H+-ATPase was phosphorylated (lane 5, indicated by ATPase with arrow). ACPK1L with arrow in lane 5 indicates the autophosphorylated GST-tagged ACPK1L protein having a molecular mass of about 85 kD. The in vitro phosphorylation assay of the partially purified PM H+-ATPase fraction in the ACPK1L-free medium (lane 3) was taken as a control, and the same assay as described for lane 5 but in the absence of free Ca2+ was taken as another control (lane 4).
It has been reported that the posttranslational modification of PM H+-ATPase involves the C-terminal auto-inhibitory domain of the enzyme, and cleavage of the C-terminal autoinhibitory domain by mild trypsin treatment or genetic deletion activates the enzyme with its pH optimum shifted to more alkaline (Palmgren et al., 1990, 1991; Rasi-Caldognov et al., 1993; Sekler et al., 1994; Regenberg et al., 1995). In this experiment, the activation of PM H+-ATPase by ACPK1 was enhanced when the medium pH was shifted from 6.5 to 7.0 (Fig. 10E), and a mild treatment of the microsomal proteins by trypsin also induced a significant PM H+-ATPase activation, which was comparable to the ACPK1 treatment (Fig. 10F). The ACPK1 treatment did not induce any additional activating effect on PM H+-ATPase in the trypsin-treated microsomes (Fig. 10F). These data suggest that ACPK1 regulates PM H+-ATPase by acting on its C-terminal autoinhibitory domain.
To further assess the regulation of PM H+-ATPase by ACPK1, we partially purified PM H+-ATPase, which was tested by SDS-PAGE and immunoblotting (Fig. 10G, lanes 1 and 2). The assay of in vitro phosphorylation of the partially purified fraction in an ACPK1-free medium showed no phosphorylated protein band in the SDS-PAGE gel (Fig. 10G, lane 3), indicating that there was no phosphorylating activity in this partially purified fraction. Using this fraction, we showed that the PM H+-ATPase was phosphorylated by ACPK1 (Fig. 10G, lane 5). Removal of free Ca2+ from the in vitro phosphorylation medium completely abolished the PM H+-ATPase phosphorylation (Fig. 10G, lane 4), indicating the Ca2+ dependence of this phosphorylation event. These data suggest that ACPK1 activates PM H+-ATPase by phosphorylating the enzyme.
DISCUSSION
Identification of an ABA-Stimulated CDPK, ACPK1, from Grape Berry
Numerous studies have demonstrated that calcium is involved in ABA signal transduction (for review, see Finkelstein et al., 2002; Himmelbach et al., 2003; Fan et al., 2004). However, many of the Ca2+-binding sensory proteins as the components of the ABA-signaling pathway remain to be elucidated. In this study, we identified and purified a 58-kD ABA-stimulated CDPK from the mesocarp of grape berries (Figs. 1–4), designated ACPK1. Based on sequencing of the two-dimensional electrophoresis-purified ACPK1, we isolated a cDNA clone encoding a putative CDPK (Fig. 5). This cDNA clone was demonstrated to encode ACPK1 essentially by two mutually linked approaches. One is the second time of sequencing of the purified natural ACPK1 protein, showing that the predicted amino acid sequence from the cDNA clone matches the sequence of the natural ACPK1 protein (Fig. 5, see also Supplemental Fig. 1; Supplemental Table I), and another is the assay of immunoblotting where the purified 58-kD ACPK1 kinase was specifically immunorecognized with the anti-ACPK1-N40 serum specific to the full-length putative ACPK1 cDNA-encoded protein (Fig. 4). The analysis of the recombinant full-length protein expressed in E. coli further demonstrated that the isolated ACPK1 cDNA clone indeed encodes an active number of CDPK family (Fig. 7, A–D), and the further assays based on the molecular cloning of ACPK1 revealed that ABA stimulates the ACPK1 kinase in both protein (amounts and activities) and mRNA levels (Fig. 7F), confirming the ABA-induced effects of stimulating the ACPK1 kinase.
Biological Significance of ABA-Induced ACPK1 Kinase Stimulation
The features of the ACPK1 kinase stimulation by ABA are of importance to possible biological functions of ACPK1. The dose dependence of the ACPK1 stimulation induced by a physiological range of ABA concentrations (<3.7 μm) within the tissues (Figs. 3 and 7) is suggestive of action of endogenous ABA on ACPK1. For this action of endogenous ABA, further supporting evidence was provided by correlating the developmental changes of grape berries in their ABA concentrations with their ACPK1 expression and enzyme activities (Fig. 8B). The dose dependence of the ABA-induced ACPK1 stimulation essentially exhibited a saturation-like phenomenon with a slight decline of the ABA-induced effects with the over-physiological higher levels of ABA (>3.7 μm) resulting from excessively applied ABA (20–80 μm; Figs. 2 and 7). This phenomenon of saturation, difficult to explain, may be due to a possible CDPK turnover mechanism involving a degradation pathway induced by excessive expression of the enzyme as observed in tobacco NtCDPK2 by Romeis et al. (2001) or probably to a decline of ABA-signaling pathway resulting from a typical saturation-binding nature of ABA to its putative receptor in this fruit (Zhang et al., 1999).
ABA functions in mediating both rapid responses such as stomatal apertures and slow events involving gene expression to regulate growth and development (Finkelstein et al., 2002; Himmelbach et al., 2003; Fan et al., 2004). The rapid stimulation of the ACPK1 kinase in 15 min by ABA without increase of the ACPK1 protein (Fig. 2B) suggests involvement of ACPK1 in mediating some rapid ABA-responsive events, while a slow and steady state of the ABA-stimulated ACPK1 activities paralleling de novo ACPK1 synthesis (Fig. 2B) implicates roles of ACPK1 in regulating development-related ABA-signaling pathway. Although, in the rapid phase of the ACPK1 stimulation by ABA, ABA could probably act on posttranslational modification of the ACPK1 protein, the ABA signal must require a living cell-dependent mechanism to be transduced to its target ACPK1, indirectly acting on the enzyme (Fig. 3C). The pharmacological assays showed that influx of apoplasmic Ca2+ into cytoplasm may be involved in this event of ABA signal transduction (Fig. 3B) by promoting direct binding of Ca2+ to ACPK1 or activating a more complex mechanism to stimulate ACPK1. However, an artificial influx of Ca2+ created by Ca2+ ionophore A23187 could not enhance the ABA-induced effects, and it could not otherwise replace ABA to stimulate ACPK1 (Fig. 3D), implying that more complex signal cascades than a single Ca2+ signal may be involved in the ACPK1 stimulation.
Whether the effect of ABA on ACPK1 is specific to the physiological active form of ABA is fundamental in establishing the function of ABA. We showed that the ABA-induced ACPK1 stimulation is stereo specific to physiological active (+)-ABA, while other inactive structurally similar isomers (−)-ABA or trans-ABA are ineffective (Fig. 3A). Also, other phytohormones were shown to be ineffective in the ACPK1 stimulation (Fig. 3B). These findings of stereo specificity and exclusivity of ABA in stimulating ACPK1 support further the specific biological relevance of the ABA-induced ACPK1 stimulation.
It has been documented that PM H+-ATPase could be phosphorylated in its C terminus in a calcium-dependent manner (Schaller and Sussman, 1988; Camoni et al., 1998; Lino et al., 1998; De Nisi et al., 1999; Rutschmann et al., 2002), but complex positive or negative regulatory mechanisms to control its activity may occur (Lino et al., 1998; De Nisi et al., 1999; Kinoshita and Shimazaki, 1999; Sze et al., 1999; Morsomme and Boutry, 2000; Palmgren, 2001). We showed that ACPK1 is a plasmalemma and chloroplast/plastid double-localized protein (Fig. 9), suggesting that it may function both in the plasma membranes and intracellularly. It is particularly interesting that the activity of plasma membrane-colocalized H+-ATPase is up-regulated by ACPK1 in vitro most likely through phosphorylation of the PM H+-ATPase by ACPK1 (Fig. 10), showing that ACPK1 may be positively involved in ABA-signaling pathway where ABA stimulates PM H+-ATPase in grape berry.
What possible functions may ACPK1 have in regulating fruit development? ABA has been believed to regulate development of fleshy fruits, especially grape berry, essentially by promoting assimilate accumulation and triggering fruit ripening (Coombe, 1992; Rock and Quatrano, 1995; Wayne and John, 1996; Opaskornkul et al., 1999; Peng et al., 2003; Pan et al., 2005). The activation of plasma membrane ATPase by ACPK1 (Fig. 10) suggests that ACPK1 may be involved in ABA-stimulated assimilate accumulation by improving energization of the fruit cells to promote plasmalemma H+-ATPase-powered active uptake (Palmgren, 2001). The expression of ACPK1 exclusively in grape berry including the mesocarp and seeds (Fig. 8) appears to be associated with its possible specificity of biological function to this fruit. The expression pattern of ACPK1 during fruit development (Fig. 8) seems to be of particular significance. The lag growth phase stage III is a critical period when intracellular structural changes and active metabolism occur in spite of relatively lag appearance of berry growth, and this has been believed to prepare the onset of ripening (stage IV, see Fig. 8) during which nearly the totality of storage assimilates accumulates and fruit quality and yield are formed (Coombe, 1992; Zhang et al., 1997). The higher levels of ACPK1 in both its protein and mRNA amounts and enzyme activities in accordance with higher ABA concentrations (Fig. 8B) may be connected with its possible involvement in regulating postripening, triggering onset of ripening, and promoting rapid accumulation of carbon reserves during these two phases. It is difficult, however, to explain the high level of ACPK1 at stage I, an early stage after anthesis when berries begin growing, as the biological significance of the peak level of ABA at this stage remains currently unexplainable. Getting an insight into how ACPK1 works in fruit development will be of interest to understand ABA signal transduction in plants.
MATERIALS AND METHODS
Plant Materials and Chemicals
Grape (Vitis vinifera × Vitis Lubrusca L. cv Kyoho) berries and other organs were sampled from a commercial vineyard in the western suburb of Beijing. We divided the sampling period of grape berries into four stages, i.e. early stage of berry growth (10 d after anthesis, stage I), first rapid growth phase (stage II, 10th–30th d after anthesis), lag growth phase (stage III, 30th–50th d after anthesis), and second rapid growth phase or ripening stage (stage IV, 50th–80th d after anthesis; see Fig. 9). Sampling for most of the assays was done during the stage III unless otherwise mentioned. Grape berries were picked for immediate use or frozen in liquid nitrogen and kept at −80°C until use. Rabbit polyclonal antibodies to the CaM-like domain of soybean (Glycine max) CDPKα (Bachmann et al., 1996) were immunopurified on a column of immobilized soybean CDPKα and generously provided by Dr. Alice Harmon (University of Florida, Gainesville, FL). Rabbit polyclonal antibodies against Arabidopsis (Arabidopsis thaliana) PM H+-ATPase AHA3 (Pardo and Serrano, 1989) was also a generous gift from Dr. R. Serrano (Universidat Politecnica de Valencia, Valencia, Spain). [γ-32P]ATP was purchased from Amersham Pharmacia. All other chemicals were purchased from Sigma unless otherwise noted.
Preparation of Subcellular Fractions
The subcellular fractions were prepared as described by Shen et al. (2004). Grape pericarp was removed from berries to obtain mesocarp that was used to prepare microsomal and cytosolic fractions. Plasma membranes and endomembranes were isolated from the microsomes by an aqueous polymer two-phase system consisting of Dextran T500 and polyethylene glycol (PEG) 3350. Purity and biochemical activity of each subcellular fraction were evaluated by measuring the activity of the marker enzymes to ensure good quality of the subcellular fractions (Shen et al., 2004). Protein concentrations were determined by the method of Bradford (1976) with bovine serum albumin (BSA) as a standard.
Gel Electrophoresis and Immunoblotting
SDS-PAGE was carried out according to the method of Laemmli (1970). To detect Ca2+-induced electrophoretic mobility shifts of CDPK, CaCl2 or EGTA was added into protein samples in SDS-PAGE sample buffer to a final concentration of 2 mm. The protein samples were boiled for 2 min before being analyzed on a 12% SDS-polyacrylamide gel. IEF/SDS-PAGE two-dimensional electrophoresis of the purified CDPK (see Fig. 7) was done essentially according to the method of Zhang et al. (2002). The purified CDPK protein (50 μg) was subjected to IEF and then was applied to a 12% SDS-polyacrylamide gel for the electrophoresis of second dimension. Proteins in the gel were detected by Coomassie Brilliant Blue R-250 (Amersham Pharmacia).
Immunoblotting was done essentially as described by Harper et al. (1994). After SDS-PAGE or IEF/SDS-PAGE, the proteins on gels were electrophoretically transferred to nitrocellulose membranes (0.45 μm, Amersham Pharmacia). The membranes were blocked for 2 h at room temperature with 3% (w/v) BSA and 0.05% (v/v) Tween 20 in a Tris-buffered saline containing 10 mm Tris-HCl (pH 7.5) and 150 mm NaCl, and then were incubated with gentle shaking for 2 h at room temperature in the rabbit polyclonal antibodies (against the CaM-like domain of soybean CDPKα, or the full length of ACPK1 or N-terminal fragment of ACPK1, or Arabidopsis PM H+-ATPase AHA3; all diluted 1:1,000 in the blocking buffer). After being washed three times for 10 min each in the Tris-buffered saline containing 0.05% (v/v) Tween 20, the membranes were incubated with the alkaline phosphatase-conjugated antibody raised in goat against rabbit IgG (diluted 1:1,000 in the blocking buffer) at room temperature for 1 h, and then washed three times for 10 min each with 50 mm Tris-HCl (pH 7.5) buffer containing 150 mm NaCl and 0.1% (v/v) Tween 20. Protein bands were visualized by incubation in the color development solution using a 5-bromo-4-chloro-3-indolyl-P/nitroblue tetrazolium substrate system according to the manufacturer's protocol.
In-Gel Autophosphorylation and Kinase Assays
Autophosphorylation of proteins in polyacrylamide SDS gels was done essentially as described by Li and Assmann (1996) based on the method of Kameshita and Fujisawa (1989). After SDS-PAGE or IEF/SDS-PAGE as described above, the gels were washed twice with 50 mm Tris-HCl, pH 8.0, containing 20% (v/v) 2-propanol for 1 h per wash, and then with buffer A composed of 50 mm Tris-HCl, pH 8.0, 5 mm 2-mercaptoethanol, and 0.1 mm EDTA for 1 h at room temperature. Proteins in the gels were denatured by incubating the gels in buffer A containing 6 m guanidine hydrochloride for two incubations of 1 h each at room temperature. Proteins were then renatured using buffer A containing 0.05% (v/v) Tween 20 for six incubations of 3 h each at 4°C. After preincubation at room temperature for 30 min with buffer B composed of 40 mm HEPES-NaOH, pH 7.5, 10 mm MgCl2, 0.45 mm EGTA (1 mm in the Ca2+-free medium), and 2 mm dithiothreitol (DTT) in the absence or presence of 0.55 mm CaCl2, the gels were incubated with buffer B containing 50 μm ATP and 10 μCi/mL [γ32-P]-ATP (3,000 Ci/mmol; Amersham Pharmacia) for 1 h at room temperature. The gels were then washed extensively with 5% trichloroacetic acid (TCA) and 1% sodium pyrophosphate until radioactivity in the used wash solution was barely detectable. The gels were then stained with Coomassie Brilliant Blue R-250. After destaining, the gels were air dried between two sheets of cellophane, and autophosphorylated CDPK in gels was detected by autoradiography after exposition of the dried gels to Kodak X-Omat AR film for 2 to 3 d at −20°C. The in-gel kinase activity assay was performed as described above, except that the separating gel was polymerized in the presence of 0.5 mg mL−1 histone III-S as a substrate for kinases.
In Vitro Kinase Activity Assay
In the assays to determine Ca2+, Mg2+, and pH dependence of the 58-kD kinase activity (see Fig. 2), kinase activity was analyzed in vitro (in solution) essentially as described previously (Shen et al., 2004) based on the method of Yao et al. (1995). Briefly, microsomes (5 μg) were mixed with histone III-S (0.5 mg mL−1) essentially in the buffer B in the absence or presence of 0.55 mm CaCl2 with corresponding modifications in medium pHs and Ca2+ and Mg2+ concentrations when assaying their effects on kinase activity (see Fig. 2). The medium pH was modulated with 50 mm MES-NaOH at 5.5 to 6.5 and with 50 mm Tris-HCl at 7.5 to 9. The phosphorylation reaction (50 μL) was initiated by addition of 25 μm ATP containing 50 μCi mL−1 [γ-32P]ATP. After 15 min at room temperature, the reaction was stopped by transferring 20 μL reaction mixture to P81 filter treated by TCA. After an extensive wash in 5% TCA solution at 4°C, P81 filter was dehydrated in ethanol and air dried. The bound radioactivity in the P81 filter was measured with a liquid scintillation counter.
In Vitro Phosphorylation of Total Microsomal Proteins
The microsomes (100 μg proteins) were added to the buffer B (50 μL) described above in the absence or presence of 0.55 mm CaCl2. After turbination, the mixtures were incubated in a water bath at 30°C for 10 min. The reactions were initiated by addition of 25 μm ATP containing 50 μCi mL−1 [γ-32P]ATP. After further incubation at 30°C for 20 min, the reaction was stopped by addition of 10% (w/v) TCA. After the sample was centrifuged, the pellets were rinsed twice with ice-cold acetone. Precipitated proteins were dissolved in SDS-PAGE sample and boiled for 2 min. The samples were then electrophoresed on a 12% SDS-polyacrylamide gel. The gels were air dried between two sheets of cellophane, and phosphorylated proteins in gels were detected by autoradiography after exposition of the dried gels to Kodak X-Omat AR film for 3 d at room temperature.
In Vivo Incubation of Berry Tissues in the ABA-Containing Medium
The treatment of the tissues of grape berry with ABA by an in vivo incubation was done essentially as described by Peng et al. (2003). After washing with tap and distilled water, the freshly harvested berries were precooled to 4°C. Discs of berry mesocarp, 10 mm in diameter and 1 mm in thickness, were prepared with a cork borer. The discs were immediately immersed in the equilibration buffer for 30 min. The equilibration buffer consists of 5 mm MES-NaCl, pH 5.5, 250 mm mannitol, 10 mm MgCl2, 5 mm ascorbic acid, 1 mm EGTA, and 1 mm CaCl2. (±) ABA (or ABA analogs or other plant hormones in the assay to test the specificity of ABA-induced effects, see Fig. 4) was added into the equilibration buffer to give the medium-containing ABA, or distilled water containing the same amount of ethanol for solubilizing ABA was added in this equilibration buffer to give the control medium. The equilibrated discs were incubated in vivo in this ABA-containing medium or in the control medium at a ratio of tissue fresh weight to buffer volume being 1 to 3 (w/v) by gentle shaking at 20°C for 1 h (except when analyzing time course of ABA-induced effects, see Fig. 3). Following washes with double-distilled water, the treated tissues were used immediately for assays or frozen in liquid nitrogen and kept at −80°C until use.
ABA Analysis
Grape tissues were immediately frozen with liquid N2 and then homogenized in double-distilled water at a ratio of sample weight to double-distilled water volume being 1 to 3 at ice-cold temperature. Samples were extracted with oscillation at 4°C in dark for 24 h and then centrifuged for 25 min at 20,000g, and the supernatants were used for the ABA analysis. The ABA assay was carried out using the radioimmunoassay method as described by Quarrie et al. (1988) and used in our laboratory (Peng et al., 2003). The anti-ABA antibody was provided by Dr. Quarrie (Cambridge Laboratory, John Innes Centre, UK). The crude extract (50 μL) was mixed with 200 μL phosphate-buffered saline (PBS; pH 6.0), 100 μL diluted antibody solution, and 100 μL 3H-ABA solution. The reaction mixture was incubated at 4°C for 25 min, and the bound radioactivity was measured in 50%-saturated (NH4)2SO4 precipitated pellets with a liquid scintillation counter. ABA concentration was calculated according to the standard radioimmunoassay curve.
Purification and Sequencing of the 58-kD ACPK1
The microsomal proteins extracted from berry mesocarp were separated on SDS-polyacrylamide gels. The portions of the gels containing the ABA-stimulated 58-kD ACPK1, indicated by the markers of molecular mass and autophosphorylation and immunorecognized signals of this kinase, were as precisely as possible excised and crushed with pestles in microcentrifuge tubes. The elution buffer containing 4 mm HEPES-NaOH (pH 7.5), 0.45 mm EGTA, 0.55 mm CaCl2, 10 mm MgCl2, and 2 mm 1,4-DTT was added into the tubes to elute proteins from the gels. The collected eluate, considered as the primarily purified 58-kD kinase, was lyophilized for identification of 58-kD kinase and further purification of the kinase by IEF/SDS-PAGE.
For sequencing of the purified kinase, 58-kD kinase spots were excised from five two-dimensional gels (first dimension IEF followed by nondenaturing PAGE). The peptides generated by in-gel digestion with trypsin were sequenced by TMS on an electrospray ion trap mass spectrometer (LCQ Deca XPPlus spectrometer, Thermo Finnigan) in Beijing Proteome Research Center, The Chinese Academy of Medical Sciences.
Cloning of ACPK1 Gene
The data from the first sequencing by TMS indicated that the sequence of the purified 58-kD kinase matches that of plant CDPKs in their conserved domains, which formed the basis for the cloning of ACPK1 by PCR. For the cloning, single-stranded cDNA was synthesized from total RNA of grape berry using PowerScript reverse transcriptase and SMART III Oligonucleotide/CDSIII3′ as the primer (CLONTECH). Using the cDNA as the template, the main part of a CDPK cDNA was amplified in a PCR reaction with two degenerate primers [forward primer, 5′-GA(G/A) ATTCAGATAATGCAICAITTG-3′; reverse primer, 5′-TTTITTCATTGCTGAIAA(T/C) TGCTTC-3′] that correspond to two of the matched sequences of the 58-kD kinase and also to the conversed domain of most CDPKs (-EIQIMHHL- and -KQFSAMNK-). PCR products were cloned into pMD-18T plasmid (TAKARA, Dalian Division) and were confirmed by sequence analysis. 5′-RACE PCR was performed by using a nested PCR with gene-specific primers derived from the cloned CDPK 5′ end (reverse primer 1, 5′-CCAACAATGGTCTTGATCGGCTTT-3′; reverse primer 2, 5′-CCTCACACAACTCCATGACCAAAT-3′) and SMART III Oligonucleotide primer as forward primer. 3′-RACE PCR was done with gene-specific primers derived from the cloned CDPK 3′ end (forward primer1, 5′-CTATTGCGGAAAATGCTTGATCGG-3′; forward primer2, 5′-CCCATGGGTCGTAGATGATAGAAT-3′) and CDS III3′ as reverse primer. 5′-RACE and 3′-RACE products were cloned into pMD-18T plasmid (TAKARA, Dalian Division) and were sequenced. Splicing three fragments, and thus designing a pair of specific primer (forward primer, 5′-GCCATTACGGCCGGGAGAGAGAGAGTCTTCC-3′; reverse primer, 5′-ACGATCAAATTAATGCCGAAACCAGCTACAAG-3′), a full-length cDNA encoding a putative CDPK from grape berry was obtained by PCR amplification. The data of the second sequencing of the purified 58-kD kinase by TMS (see Fig. 6) demonstrated that the cloned putative CDPK cDNA indeed encodes ACPK1.
Northern- and Southern-Blot Analysis
Total RNA (20 μg) was subjected to electrophoresis on formaldehyde/agarose gels and transferred to nylon membranes (Hybond-N+, Amersham Pharmacia Biotech) according to standard protocols. For Southern-blot analysis, genomic DNA was extracted from young leaf tissue using the method of Doyle and Doyle (1990). Ten micrograms of DNA was digested to completion with BamHI, KpnI, EcoRI, and HindIII restriction enzymes, electrophoresed through 0.8% agarose, and blotted onto nylon membranes (Hybond-N+, Amersham Pharmacia Biotech). RNA and DNA blots were probed with the 32P-labeled full-length cDNA or a cDNA fragment corresponding to the variable domain of ACPK1. Hybridization was performed according to the method of Church and Gilbert (1984).
Expression, Purification, and Antiserum Preparation of Recombinant ACPK1L and ACPK1-N40
Two constructs were generated for the expression of ACPK1 in Escherichia coli as glutathione S-transferase (GST) fusion protein. Full-length ACPK1 (designated ACPK1L) corresponds to the protein encoded by the entire ORF and ACPK1-N40 represents the N-terminal fragment of 40 amino acids covering N-terminal variable domain and 11 amino acids in its adjacent kinase domain. The corresponding regions of the ACPK1 cDNA were amplified by PCR using pfu DNA polymerase (TAKARA, Dalian Division) and synthetic oligonucleotide primers (forward primer for both ACPK1L and ACPK1-N40, 5′-ATGAAGAAATCGTCCGCAGGAGC-3′; reverse primer for ACPK1L, 5′-GTTTGTCAAGCGCATATCTGGTA-3′; and that for ACPK1-N40, 5′-TGTCCCAAACTGCCCTTGAC-3′). The PCR products were cloned into the BamHI (5′ end)/XhoI (3′ end) sites of pGEX-4T-1 (Amersham Pharmacia Biotech) for the expression of N-terminal GST fusion proteins under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter. To ensure that no errors were introduced by PCR, each construct was checked by sequencing. To produce GST tag fusion proteins, DH5α cells transformed with pGEX-4T-1/ACPK1L and pGEX-4T-1/ACPK1-N40 were induced with 20 mm IPTG for 3 h. The fusion proteins were purified from IPTG-induced cultures by Glutathione Sepharose 4B column (Amersham Pharmacia Biotech) and analyzed by SDS-PAGE. The purified two fusion proteins were used for standard immunization protocols in rabbits. Polyclonal antisera were affinity purified first by HiTrap Protein-G HP (Amersham Pharmacia Biotech) and further by immunosorbent column coupled with GST-ACPK1-N40 or GST-ACPK1L fusion protein. The affinity-purified antisera were evaluated by western blotting and shown to be highly specific.
Phosphoamino Acid Analysis
Phosphoamino acids were analyzed essentially as described by Shi et al. (1999). In brief, after ACPK1L (10 μg) was autophosphorylated in 0.5 mL of 40 mm HEPES-NaOH buffer (pH 7.5) containing 10 mm MgCl2, 0.45 EGTA, 2 mm DTT, 0.55 mm CaCl2, 50 μm ATP, and 15 μCi/mL [γ32-P]-ATP (3,000 Ci/mmol), the protein was precipitated, and then hydrolyzed in 6 n HCl at 110°C for 1 h. After centrifugation, the hydrolysate was dried in vacuum and spiked with phosphoamino acids standards (12 μg each of l-phospho-Ser, phospho-Thr, and phospho-Tyr). The mixture of the samples and the standards was spotted onto two-dimensional thin-layer chromatography plates. The first-dimensional thin-layer electrophoresis was done at 1.5 kV for 20 min in the pH 1.9 electrophoresis buffer, and the second dimension at 1.3 kV for 16 min in pH 3.5 electrophoresis buffer. The plates were dried in an oven at 60°C. Phosphoamino acid standards were visualized by spraying with ninhydrin (0.25% [w/v] in acetone) followed by heat treatment, and the labeled amino acids were detected by autoradiography.
Immunoprecipitation
The microsomal proteins (50 μg) were resuspended in 0.5 mL immunoprecipitation buffer containing 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EGTA, 1 mm Na3VO4, 1 mm NaF, 10 mm β-glycerophosphate, 1 mm phenylmethylsulfonyl fluoride, 5 μg mL−1 antipain, 5 μg mL−1 aprotinin, 5 μg mL−1 leupeptin, and 0.5% Triton X-100. The mixture was incubated with either the purified anti-ACPK-N40 serum (about 3 μg protein) or the same amount of preimmune serum protein (as a control) at 4°C for 2 h. Then 25 μL protein A-agarose suspension was added to the mixture, and the mixture was incubated further for 2 h. Following a brief centrifugation, the immunoprecipitated proteins, after three washes with the immunoprecipitation buffer, were used for the assays of autophosphorylation, kinase activity, or immunoblotting.
Transient Expression of GFP-ACPK1 in Onion Epidermis and Arabidopsis Protoplasts
For observation of the subcellular localization of ACPK1, the full-length ORF of ACPK1 cDNA was PCR amplified by using primers 5′-GCCTCGAGATGAAGAAATCGTCCGCAGGAGCA-3′ (forward primer) and 5′-GCCGGATCCTTTGTCAAGCGCATATCTGGTATC-3′ (reverse primer). The PCR product was then fused to the upstream of the enhanced GFP (Cormack et al., 1996) at the XhoI (5′ end)/BamHI (3′ end) sites in the cauliflower mosaic virus 35S-EGFP-Ocs 3′ vector (p-EZS-NL vector, Dr. Ehrhardt, http://deepgreen.stanford.edu). This vector does not express GFP well without adding coding sequence to the 5′ end of the ORF of GFP, thus the control cells do not show fluorescence of GFP. Protoplasts were isolated from the leaves of 3- to 4-week-old plants of Arabidopsis (ecotype Columbia) and transiently transformed by using PEG essentially according to Ueda et al. (2001). The strips of onion (Allium cepa) bulb epidermis were bombarded essentially according to Scott et al. (1999) with gold particles coated with plasmids using a Bio-Rad PDS-1000/He particle delivery system. Fluorescence of GFP was observed by a confocal laser scanning microscope (Bio-Rad MRC 1024) after incubation at 23°C for 16 h.
Subcellular Immunogold Labeling
Subcellular immunogold labeling of ACPK1 was done essentially as described previously (Zhang et al., 2004). Briefly, the ultra-thin sections were prepared from Lowicryl K4M-embedded specimen. They were first blocked for 30 min at room temperature by floating the grids on droplets of a PBS containing 8 mm Na2HPO4, 1.5 mm KH2PO4, 3 mm KCl, and 500 mm NaCl (pH 7.4) supplemented with 50 mm Gly and continuously blocked with the PBS supplemented with 0.1% (w/v) gelatin, 0.5% (w/v) BSA, and 0.1% (v/v) Tween 20 (pH 7.4; PBGT). Without rinsing, the sections were incubated with rabbit antiserum directed against the ACPK1L or ACPK1-N40 or Arabidopsis PM H+-ATPase AHA3 (all diluted 1:100 in PBGT buffer) overnight at 4°C. Following extensive washes with PBGT buffer, the sections were incubated with secondary antibody (goat anti-rabbit IgG antibody conjugated with 10 nm gold) at 1:100 dilution in PBGT buffer for 2 h at room temperature. The sections were rinsed consecutively with PBGT and double-distilled water. The sections were then stained with 2% uranyl acetate in 50% ethanol for 25 min at 25°C and with alkaline lead citrate for 15 min. After extensively washing with double-distilled water, the ultra-thin sections were examined with an electron microscope.
The specificity and reliability of the immunogold labeling were tested by two negative controls. In the first one, the antiserum was omitted to test possible unspecific labeling of the goat anti-rabbit IgG antibody-gold conjugate. In the second one, rabbit preimmune serum was used instead of the rabbit antiserum before immunogold labeling to test the specificity of the antiserum. More than three repetitions of the control experiments were conducted for each sample.
Assays of Effects of ACPK1 on PM H+-ATPase Activity
PM H+-ATPase activity was assayed as described by Olivari et al. (2000) with modifications. The reaction medium (50 μL) contains 5 mm MgSO4, 0.1 mm Na2MoO4, 50 mm KNO3, 1 μg mL−1 oligomycin, 5 μm carbonyl cyanide p-(trifluoromethoxy) phenyl-hydrazone, 50 μg mL−1 polyoxyethylene-20-cetyl ether (Brij 58), 40 mm 1,3-bis[tris(hydroxymethyl) methylamino]-propane (BTP)-MES (pH 6.5) or BTP-HEPES (pH 7.0), and 50 μg microsomal protein. A given amount of EGTA or CaCl2 was added to the reaction medium as indicated (see text below). The reaction was started by adding 3 mm ATP plus or minus 200 μm vanadate, and the assay was performed at 30°C over 1 h of incubation. The PM H+-ATPase activity was evaluated as the activity inhibited by vanadate. Released inorganic phosphate produced from ATP hydrolysis was assayed as described by De Michelis and Spanswick (1986).
To assess the effect of ACPK1 on PM H+-ATPase activity and Ca2+ dependence of this effect (see Fig. 10D), the microsomal proteins only (50 μg, control 1) or the microsomal proteins (50 μg) together with either 10 μg denatured ACPK1L protein (boiling for 10 min, control 2) or 10 μg ACPK1L protein were added to the above-described reaction medium supplemented with 0.1 mm ATP and 0.1 mm CaCl2 or in the absence of 0.1 mm CaCl2 but with 0.3 mm EGTA, and the mixtures were subjected to a 15-min preincubation at 20°C before adding ATP and 0.3 mm EGTA to start the reaction at pH 6.5. The pH dependence of PM H+-ATPase activation (see Fig. 10E) was assayed essentially in the same way where the mixture of ACPK1 and microsomal proteins was preincubated in the presence of 0.1 mm CaCl2.
For the trypsin treatment assay (see Fig. 10F), microsomal proteins (2 mg mL−1) were incubated at 20°C with 100 μg mL−1 trypsin for 10 min in a medium containing 20 mm BTP-HEPES (pH 7.0), 5% glycerol, 0.1 mm CaCl2, 100 μg mL−1 Brij 58, 0.5 mm DTT, and 1 mm ATP. The reaction was blocked by addition of 0.5 mg mL−1 soybean trypsin inhibitor. The control was run in the absence of trypsin. The treated microsomal proteins were used for assaying PM H+-ATPase activity as described above in the medium at pH 7.0 with 0.3 mm EGTA after a preincubation of ACPK1 and microsomal proteins in the presence of 0.1 mm CaCl2.
To assay phosphorylation of PM H+-ATPase by ACPK1 (see Fig. 10G), we partially purified PM H+-ATPase essentially according to the technique described by Jahn et al. (1997). Briefly, plasma membrane proteins containing PM H+-ATPase solublized by dodecyl β-d-maltoside were precipitated in PEG 3350, and the precipitated proteins were subjected to a Sephacryl S-300 HR gel filtration column to obtain a partially purified PM H+-ATPase fraction. In vitro phosphorylation of the PM H+-ATPase fraction by ACPK1 was assayed as described above for “In Vitro Phosphorylation of Total Microsomal Proteins” with addition of 50 μg partially purified PM H+-ATPase and 50 μg ACPK1L protein to the buffer B (50 μL). The control was run in the absence of ACPK1L protein.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers or locus tag numbers (for Arabidopsis only) with some references related to the gene cloning or characterization being cited in parenthesis: ACPK1, AY394009; AtCPK4, At4g09570 (Hrabak et al., 1996); AtCPK10, At1g18890 (formerly AtCDPK1; Sheen, 1996); AtCPK11, At1g35670 (formerly AtCDPK2); AtCPK12, At5g23580 (formerly CDPK9); AtCPK30, At1g74740 (formerly AtCDPK1a; Sheen, 1996); GmCDPKα, M64987 (Harper et al., 1991); GmCDPKβ, U69173 (Lee et al., 1997); GmS-CDPKa, AY247754; GmS-CDPKb, AY247755; NtCDPK1, AF72908 (Yoon et al., 1999); OsCDPK, AC128673; StCDPK, AB051809; VfCDPK, AY753552; and ZmCDPK, AY301072.
Supplementary Material
Acknowledgments
We thank Dr. Alice Harmon (University of Florida, Gainesville, FL) for her generous gift of antibodies to the CaM-like domain of soybean CDPKα, and Dr. R. Serrano (Universidat Politecnica de Valencia, Valencia, Spain) for his kindly providing us with the antibodies against Arabidopsis PM H+-ATPase AHA3. We thank Dr. Dong-Tao Ren (China Agricultural University, Beijing) for his kind advice in kinase assays.
This research was supported by the China National Natural Science Foundation (grant nos. 30421002, 30330420, 30471193, and 30270919 to D.-P.Z.), the National Key Basic Research Program of China (grant no. 2003CB114302 to D.-P.Z.), and Research Programs of the China Ministry of Education (grant no. 03018 to D.-P.Z).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Da-Peng Zhang (zhangdp@sohu.net).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074971.
References
- Allen GJ, Kuchitsu K, Chu SP, Murata Y, Schroeder JI (1999) Arabidopsis abi1-1 and abi-2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11: 1785–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderberg RJ, Walker-Simmons MK (1992) Isolation of a wheat cDNA clone for an ABA-inducible transcript with homology to protein kinases. Proc Natl Acad Sci USA 89: 10183–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M, Shiraishi N, Campbell WH, Yoo BC, Harmon AC, Huber SC (1996) Identification of a Ser-543 as the major regulatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell 8: 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsevich JL, Cutler AJ, Lamb N, Friesen LOJ, Kurz EU, Perras MR, Abrams SR (1994) Response of cultured maize cells to (+)-abscisic acid (−)-abscisic acid, and their metabolites. Plant Physiol 106: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Burnett EC, Desikan R, Moser RC, Neil SJ (2000) ABA activation of an MBP kinase in Pisum sativum epidermal peels correlates with stomatal responses to ABA. J Exp Bot 51: 197–205 [DOI] [PubMed] [Google Scholar]
- Camoni L, Harper JF, Palmgren MG (1998) 14-3-3 protein activates a plant calcium-dependent protein kinase (CDPK). FEBS Lett 430: 381–384 [DOI] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J (2002) Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe BG (1992) Research on development and ripening of the grape berry. Am J Enol Vitic 43: 101–111 [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173: 33–38 [DOI] [PubMed] [Google Scholar]
- Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, Pickard BG, Harper JF (2003) Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol 132: 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Michelis MI, Spanswick RM (1986) H+-pumping driven by the vanadate-sensitive ATPase in membrane vesicles from corn roots. Plant Physiol 81: 542–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nisi P, Dell'Orto M, Pirovano L, Zocchi G (1999) Calcium-dependent phosphorylation regulates the plasma membrane H+-ATPase activity of maize (Zea mays L.) roots. Planta 209: 187–194 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15 [Google Scholar]
- D'Souza JS, Johri MM (2002) ABA and NaCl activate myelin basic protein kinase in the chloronema cells of the moss Funaria hygrometrica. Plant Sci 40: 17–24 [Google Scholar]
- Fan LM, Zhao ZX, Assmann SM (2004) Guard cells: a dynamic signaling model. Curr Opin Plant Biol 7: 537–546 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Gampala S, Rock C (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Rock C (2002) Abscisic acid biosynthesis and signaling. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD
- Gomez-Cadenas A, Verhey SD, Holappa LD, Shen Q, Ho THD, Walker-Simmons MK (1999) An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc Natl Acad Sci USA 96: 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas A, Zentella R, Walker-Simmons MK, Ho THD (2001) Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13: 667–679 [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233–244 [DOI] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Gubrium E, Harper JF (2001) The CDPK superfamily of protein kinases. New Phytol 151: 175–183 [DOI] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF (2000) CDPKs: a kinase for every Ca2+ signal? Trends Plant Sci 5: 154–159 [DOI] [PubMed] [Google Scholar]
- Harper JF, Huang JF, Lloyd SJ (1994) Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 33: 7267–7277 [DOI] [PubMed] [Google Scholar]
- Harper JF, Sussman MR, Schaller GE, Putnam-Evans C, Charbonneau H, Harmon AC (1991) A calcium-dependence protein kinase with a regulatory domain similar to calmodulin. Science 252: 951–954 [DOI] [PubMed] [Google Scholar]
- Hepler PK (2005) Calcium: a central regulator of plant growth and development. Plant Cell 17: 2142–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RD, Liu JH, Durnin D, Lamb N, Shaw A, Abrams SR (1995) Abscisic acid structure-activity relationships in barley aleurone layers and protoplasts. Plant Physiol 108: 573–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hőhener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling. Curr Opin Plant Biol 6: 470–479 [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Dickman LJ, Satterlee JS, Sussman MR (1996) Characterization of eight new members of the calmodulin-like domain protein kinase gene family from Arabidopsis thaliana. Plant Mol Biol 31: 405–412 [DOI] [PubMed] [Google Scholar]
- Jahn T, Fuglsang AT, Olsson A, Bruntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C (1997) The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 9: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK (2002) The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol 130: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameshita I, Fujisawa H (1989) A sensitive method for detection of calmodulin-dependent protein kinase II activity in sodium dodecyl sulfate-polyacrylamide gel. Anal Biochem 183: 139–143 [DOI] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15: 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18: 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knetsch MW, Wang M, Snaar-Jagalska BE, Heimovaara-Dijkstra S (1996) Abscisic acid induces mitogen-activated protein kinase activation in barley aleurone protoplasts. Plant Cell 8: 1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T (2004) Different activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16: 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Leon-Kloosterziel KM, Schwartz SH, Zeevaart JAD (1998) The genetic and molecular dissection of abscisic acid biosynthesis and signal transduction in Arabidopsis. Plant Physiol Biochem 36: 83–89 [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, Delong A, Schroeder JI (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lee JY, Roberts DM, Harmon AC (1997) Isolation of two new CDPK isoforms (accession nos. U69173 and U69174) from soybean (Glycine max L.) (PGR 97-128). Plant Physiol 115: 314 [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49: 199–222 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Assmann SM (1996) An abscisic acid-activated and calcium-independent protein kinase from guard cells of Fava bean. Plant Cell 8: 2359–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kinoshita T, Pandey S, Ng CKY, Gygi SP, Shimazaki K, Assmann SM (2002) Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418: 793–797 [DOI] [PubMed] [Google Scholar]
- Li J, Wang XQ, Watson MB, Assmann SM (2000) Regulation of abscisic acid stomatal closure and anion channels by guard cell AAPK kinase. Science 287: 300–303 [DOI] [PubMed] [Google Scholar]
- Li WG, Komatsu S (2000) Cold stress-induced calcium-dependent protein kinase(s) in rice (Oryza sativa L.) seedling stem tissues. Theor Appl Genet 101: 355–363 [Google Scholar]
- Lino B, Baizabal-Aguirre VM, Vara LEG (1998) The plasma membrane H+-ATPase from beet root is inhibited by a calcium-dependent phosphorylation. Planta 204: 352–359 [DOI] [PubMed] [Google Scholar]
- Lu C, Han MH, Guevara-Garcia A, Fedoroff NV (2002) Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci USA 99: 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Hrabak EM (2002) An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol 128: 1008–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell (Suppl) 14: S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig AA, Romeis T, Jones JDG (2004) CDPK-mediated signaling pathways: specificity and cross-talk. J Exp Bot 55: 181–188 [DOI] [PubMed] [Google Scholar]
- Martin M, Busconi L (2000) Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J 24: 429–435 [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signaling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Morsomme P, Boutry M (2000) The plasma membrane H+-ATPase: structure, function and regulation. Biochim Biophys Acta 1465: 1–16 [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari C, Albumi C, Pugliarello MC, De Michelis MI (2000) Phenylarsine oxide inhibits the fusicoccin-induced activation of plasma membrane H+-ATPase. Plant Physiol 122: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opaskornkul C, Lindberg S, Tiliberg JE (1999) Effect of ABA on the distribution of sucrose and protons across the plasmalemma of pea mesophyll protoplasts suggesting a sucrose/proton symport. J Plant Physiol 154: 447–453 [Google Scholar]
- Palmgren MG (2001) Plant plasma membrane H+-ATPase: powerhouse for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52: 817–845 [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Larsson C, Sommarin M (1990) Proteolytic activation of the plant plasma membrane H+-ATPase by removal of a terminal segment. J Biol Chem 265: 13423–13426 [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C (1991) Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J Biol Chem 266: 20470–20475 [PubMed] [Google Scholar]
- Pan QH, Li MJ, Peng CC, Zhang N, Zou X, Zou KQ, Wang XL, Yu XC, Wang XF, Zhang DP (2005) Abscisic acid activates acid invertases in developing grape berry. Physiol Plant 125: 157–170 [Google Scholar]
- Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D'Angelo C, Weinl S, Kudla J, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo JM, Serrano R (1989) Structure of a plasma membrane H+-ATPase gene from the plant Arabidopsis thaliana. J Biol Chem 264: 8557–8562 [PubMed] [Google Scholar]
- Peng YB, Lu YF, Zhang DP (2003) Abscisic acid activates ATPase in developing apple fruit especially in fruit phloem cells. Plant Cell Environ 26: 1329–1342 [Google Scholar]
- Quarrie SA, Whitford PN, Appleford NEJ, Wang TL, Cook SK, Henson IE, Loveys BR (1988) A monoclonal antibody to (S)-abscisic acid: its characterization and use in a radioimmunoassay for measuring abscisic acid in crude extracts of cereal and lupin leaves. Planta 173: 330–339 [DOI] [PubMed] [Google Scholar]
- Rasi-Caldognov F, Pugliarello MC, Olivari C, De Michelis MI (1993) Controlled proteolysis mimics the effect of fusicoccin on the plasma membrane H+-ATPase. Plant Physiol 103: 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg B, Villalba JM, Lanfermeijer FC, Palmgren MG (1995) C-terminal deletion analysis of plant plasma membrane H+-ATPase: yeast as a model system for solute transport across plant plasma membrane. Plant Cell 7: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DM, Harmon AC (1992) Calcium-modulated proteins: targets of intracellular calcium signals in higher plants. Annu Rev Plant Physiol Plant Mol Biol 43: 375–414 [Google Scholar]
- Rock CD, Quatrano RS (1995) The role of hormones during seed development. In PJ Davies, ed, Plant Hormones. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 671–697
- Romeis T, Ludwig AA, Martin R, Jones JDG (2001) Calcium-dependent protein kinase plays an essential role in a plant defense response. EMBO J 20: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd JJ, Franklin-Tong VE (2001) Unravelling response-specificity in Ca2+ signaling in plant cells. New Phytol 151: 7–33 [DOI] [PubMed] [Google Scholar]
- Rutschmann F, Stalder U, Piotrowski M, Oeking C, Schaller A (2002) LeCDPK, a calcium-dependent protein kinase from tomato: plasma membrane targeting and biochemical characterization. Plant Physiol 129: 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Brownlee C, Harper JF (1999) Communicating with calcium. Plant Cell 11: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Sussman MR (1988) Phosphorylation of the plasma membrane H+-ATPase of oat roots by a calcium-stimulated protein kinase. Planta 173: 509–518 [DOI] [PubMed] [Google Scholar]
- Scott AC, Wyatt S, Tsou P, Robertson D, Allen NS (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26: 1125–1132 [DOI] [PubMed] [Google Scholar]
- Sekler I, Weiss M, Pick U (1994) Activation of the Dunaliella acidophila plasma membrane H+-ATPase by trypsin cleavage of a fragment that contains a phosphorylation site. Plant Physiol 105: 1125–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (1996) Ca++-dependent protein kinases and stress signal transduction in plants. Science 274: 1900–1902 [DOI] [PubMed] [Google Scholar]
- Sheen J (1998) Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA 95: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YY, Duan CQ, Liang XE, Zhang DP (2004) Membrane-associated protein kinase activities in the developing mesocarp of grape berry. J Plant Physiol 161: 15–23 [DOI] [PubMed] [Google Scholar]
- Shi J, Kim KS, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinase associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11: 2392–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Fromm H (2001) Calmodulin as a versatile calcium signal transducer in plants. New Phytol 151: 35–66 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H, Li X, Palmgren MG (1999) Energization of plant cell membranes by H+-pumping ATPase: regulation and biosynthesis. Plant Cell 11: 677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Katagiri T, Mizoguchi T, Yamaguchi-Shinozaki K, Hayashida N, Shinozaki K (1994) Two genes that encode Ca2+-dependent protein kinases are induced by drought and high-salt stresses in Arabidopsis thaliana. Mol Gen Genet 244: 331–340 [DOI] [PubMed] [Google Scholar]
- Walker-Simmons MK, Holappa LD, Abrams GD, Abrams SR (1997) ABA metabolites induce group 3 LEA mRNA and inhibit germination in wheat. Physiol Plant 100: 474–480 [Google Scholar]
- Wayne HL, John DE (1996) Sugar alcohol metabolism in sinks and sources. In E Zamski, AA Schaffer, eds, Photoassimilate Distribution in Plants and Crops: Source-Sink Relationships. Marcel Dekker, New York, pp 185–207
- Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S, Asakura T (1991) Stimulation of the uptake of sorbitol into vacuoles from apple fruit flesh by abscisic acid and into protoplasts by indoleacetic acid. Plant Cell Physiol 32: 315–318 [Google Scholar]
- Yao B, Zhang Y, Delikat S, Mathias S, Basu S, Kolesnick R (1995) Phosphorylation of Raf by ceramide-activated protein kinase. Nature 378: 303–310 [DOI] [PubMed] [Google Scholar]
- Yoon GM, Cho HS, Ha HJ, Liu JR, Lee HP (1999) Characterization of NtCDPK1, a calcium-dependent protein kinase gene in Nicotiana tabacum, and the activity of its encoded protein. Plant Mol Biol 39: 991–1001 [DOI] [PubMed] [Google Scholar]
- Zhang DP, Chen SW, Peng YB, Shen YY (2001) Abscisic acid-specific binding sites in the flesh of developing apple fruit. J Exp Bot 52: 2097–2103 [DOI] [PubMed] [Google Scholar]
- Zhang DP, Li M, Wang Y (1997) Ultrastructural changes in the mesocarp cells of grape berry during its development. Acta Bot Sin 39: 389–396 [Google Scholar]
- Zhang DP, Wu ZY, Li XY, Zhao ZX (2002) Purification and identification of a 42-kilodalton abscisic acid-specific-binding protein from epidermis of broad bean leaves. Plant Physiol 128: 714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DP, Zhang ZL, Chen J, Jia WS (1999) Specific abscisic acid-binding sites in mesocarp of grape berry: properties and subcellular localization. J Plant Physiol 155: 324–331 [Google Scholar]
- Zhang LY, Peng YB, Pelleschi ST, Fan Y, Lu YF, Lu YM, Gao XP, Shen YY, Delrot S, Zhang DP (2004) Evidence for apoplasmic phloem unloading in developing apple fruit. Plant Physiol 135: 574–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski RE (1998) Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 697–725 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.