Abstract
Hypocotyl segments of Arabidopsis (Arabidopsis thaliana) produce adventitious roots in response to exogenously supplied auxin. root primordium defective 1 (rpd1) is a temperature-sensitive mutant isolated on the basis of impairment in this phenomenon. This study describes further phenotypic analysis of the rpd1 mutant and isolation of the RPD1 gene. When adventitious root formation was induced from the rpd1 explants at the restrictive temperature, cell proliferation leading to root promordia formation was initiated at the same time as in wild-type explants. However, development of the root primordia was arrested thereafter in the mutant. Temperature-shift experiments indicated that RPD1 exerts its function before any visible sign of root primordium formation. The expression patterns of the auxin-responsive gene DR5:β-glucuronidase and the cytodifferentiation marker gene SCARECROW suggest that the rpd1 mutation interferes with neither axis formation nor cellular patterning at the initial stage of root primordium development. Taken together with the effect of the rpd1 mutation on callus cell proliferation, these data imply a role for RPD1 in prearranging the maintenance of the active cell proliferation during root primordium development. Positional cloning of the RPD1 gene revealed that it encodes a member of a novel protein family specific to the plant kingdom. Disruption of the RPD1 gene by a T-DNA insertion caused embryogenesis arrest at the globular to transition stages. This phenotype is consistent with the hypothesized function of RPD1 in the maintenance of active cell proliferation.
The formation of adventitious and lateral roots is a typical example of de novo postembryonic organogenesis that is achieved through the elaborate regulation of cell proliferation. The process of morphogenesis common to adventitious and lateral roots can be summarized as follows. First, cell proliferation is initiated from quiescent cells in response to endogenous or external stimuli, including auxin (Boerjan et al., 1995; Celenza et al., 1995; Hobbie and Estelle, 1995; Reed et al., 1998; Tian and Reed, 1999; Xie et al., 2000; Malamy and Ryan, 2001; Nakazawa et al., 2001; Rogg et al., 2001; Fukaki et al., 2002). A rapid increase in cell number leads to the formation of the root primordium (MacLeod and Thompson, 1979; Laskowski et al., 1995). During the development of the root primordium, cellular patterning generates the structure of the root apical meristem (Malamy and Benfey, 1997). Cell proliferation then diminishes and the primordium emerges from the parental tissue mainly by cell elongation (Friedberg and Davidson, 1971; MacLeod and McLachlan, 1975; Malamy and Benfey, 1997). The reactivation of cell proliferation in the root apical meristem after root emergence is dependent on environmental conditions, and in some cases, arrest is maintained for a long time (Celenza et al., 1995; Zhang et al., 1999; Zhang and Forde, 2000; Signora et al., 2001; De Smet et al., 2003). Once the root apical meristem is activated, it becomes responsible for the subsequent growth of the root. Therefore, the regulation of cell proliferation is the central concern of postembryonic root formation.
Previously, we isolated temperature-sensitive mutants of Arabidopsis (Arabidopsis thaliana) that are defective in adventitious root formation (Konishi and Sugiyama, 2003; Sugiyama, 2003). Of these mutants, root primordium defective 1 (rpd1) was characterized by developmental arrest of the adventitious root primordia at the restrictive temperature. This mutant was also temperature sensitive for callus formation, suggesting a role of RPD1 in cell proliferation (Konishi and Sugiyama, 2003). Because there are no known mutants similar to rpd1, it may provide a unique tool for the study of the regulatory mechanisms of cell proliferation underlying root primordium development.
In this article, we report a detailed phenotypic analysis of the rpd1 mutant and the cloning of the RPD1 gene. We also describe the knockout phenotype of RPD1. Our work implicates the involvement of RPD1 in the maintenance of active cell proliferation and sheds light on the novel plant-specific protein family to which RPD1 belongs.
RESULTS
Effects of rpd1 Mutation on Adventitious Root Formation
When hypocotyl explants of wild-type Arabidopsis are cultured on root-inducing medium (RIM), they generate adventitious roots within several days. Two allelic mutants of the RPD1 locus, rpd1-1 and rpd1-2, were isolated as temperature sensitive for this process (Konishi and Sugiyama, 2003). Hypocotyl explants of the rpd1-1 mutant formed root primordium-like structures after 16 d of culture at the restrictive temperature (28°C) on RIM, whereas at the permissive temperature (22°C), they formed normal adventitious roots. The inhibitory effect of the rpd1 mutations on adventitious root formation was clearly detected at the primordial stage after 6 d of culture (Konishi and Sugiyama, 2003).
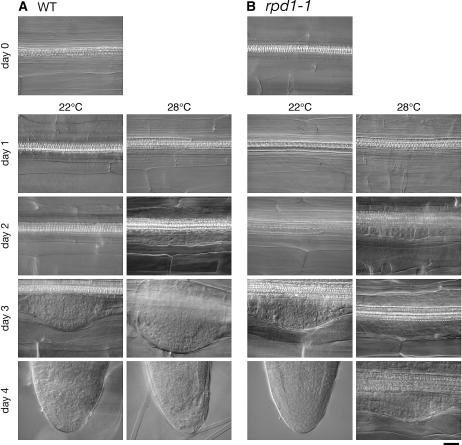
Closer observation of the time course of adventitious root development showed that the effect of the rpd1 mutation occurred in a stage-specific manner (Fig. 1). When cultured at 28°C, stele cells of wild-type hypocotyl explants initiated cell proliferation, giving rise to root primordia between day 1 and day 2. On day 2, adventitious root primordia reached the two-cell-layer stage. The root primordium formation of the rpd1-1 mutant proceeded normally until this stage. However, after 3 d of culture, developmental defects became evident in the mutant. On day 3, the wild-type primordium continued to develop, becoming hemispherical. In the rpd1-1 mutant, however, the development of the root primordium was arrested or strongly retarded at the two- to four-cell-layer stages. Therefore, the rpd1 mutation inhibited the development of adventitious root primordia beyond the two-cell-layer stage without affecting the earlier processes of primordium development, including the initiation of cell proliferation.
Figure 1.
Time course of adventitious root formation as influenced by the rpd1 mutation. Hypocotyl explants of the wild type (A) and rpd1-1 mutant (B) were cultured at 22°C or 28°C for the indicated days. Bar = 20 μm.
Because the hypocotyl segments used in the above experiments were excised from seedlings that had been grown at the permissive temperature (22°C), the normal development of root primordia at the very initial stage in the rpd1-1 explants cultured at 28°C might be attributable to the remnant of the RPD1 function active at 22°C. To test this possibility, we examined the temperature sensitivity of adventitious root formation using hypocotyl segments excised from seedlings that had been exposed to 28°C for the 4 d before tissue culture, which was expected to eliminate residual RPD1 activity. In this experiment, cell proliferation was initiated normally in the rpd1-1 explants in culture on RIM at either 22°C or 28°C (data not shown). This result corroborates the proposition that the RPD1 function is not required for the initiation of cell proliferation.
Temporal Requirement for RPD1 Activity during Adventitious Root Formation
To estimate the critical period for RPD1 activity during adventitious root formation, we performed temperature-shift experiments with rpd1-1 (Fig. 2). Hypocotyl explants of the wild-type and rpd1-1 mutant were cultured at 22°C for 1 to 3 d, after which the temperature was shifted up to 28°C. After a total 6 d in culture, the explants were examined for adventitious rooting phenotypes. When the temperature was maintained at 28°C throughout the culture (Fig. 2K) or shifted up on day 1 (Fig. 2J), the adventitious roots of the rpd1-1 explants showed developmental arrest at the primordial stage. In contrast, when the temperature was shifted up at day 3, all the rpd1-1 explants formed normal adventitious roots (Fig. 2H), as occurred in culture at 22°C (Fig. 2G). A temperature shift on day 2 induced variable phenotypes among the rpd1-1 explants, but about half of them formed normal adventitious roots (Fig. 2I). This indicates that 2 d of culture at 22°C is sufficient for subpopulation of rpd1-1 hypocotyl explants to complete adventitious root formation. As no cell proliferation was recognizable in any of the hypocotyl explants before temperature shift in this condition (Fig. 1), this suggests that RPD1 is required during the limited period before cell proliferation becomes visible.
Figure 2.
Effects of temperature shift on adventitious root formation in rpd1-1. Hypocotyl explants of the wild type (A–F) and rpd1-1 mutant (G–L) were cultured on RIM with the indicated temperature programs. A and G, Twenty-two degrees Celcius, 6 d. E and K, Twenty-eight degrees Celcius, 6 d. B to D and H to J, Temperature shift up. Hypocotyl explants were cultured at 22°C, followed by culture at 28°C. F and L, Temperature shift down. Hypocotyl explants were cultured at 28°C, followed by culture at 22°C. Bar = 200 μm.
When rpd1-1 explants were grown at 28°C for 3 d, after which the temperature was shifted down to 22°C, they formed adventitious roots that were not morphologically different from those of the wild-type explants (Fig. 2L). Therefore, the defect caused by the rpd1 mutation is reversible, and the rpd1 root primordia were readily released from developmental arrest by a temperature shift from 28°C to 22°C.
DR5 Reporter Expression in Adventitious Root Primordia of the rpd1 Mutant
During the early phase of root primordium development, auxin accumulates within the root primordium. An auxin gradient with the maximum at the primordium tip is then gradually established, which is considered to play an important role in the further development of the root primordium (Benková et al., 2003). This phenomenon raises the possibility that the developmental arrest in the adventitious root primordia of rpd1 mutants might be related to some aberration in auxin accumulation or in the generation of the auxin gradient. To address this issue, we examined the expression patterns of an auxin-responsive reporter, DR5:β-glucuronidase (DR5:GUS), during the development of adventitious root primordia in the rpd1-1 mutant at 28°C, and compared it with those of the wild type (Fig. 3A).
Figure 3.
Effects of the rpd1 mutations on expression patterns of DR5:GUS and SCR:GUS (END199) during adventitious root formation. Hypocotyl explants of the DR5:GUS (A) and END199 (B) reporter lines with or without the rpd1 mutations were cultured at 28°C on RIM for the indicated days and stained for GUS activity. Bars = 100 μm.
In the wild-type explants, DR5:GUS activity became detectable in association with the initiation of adventitious root primordia by day 2 (Fig. 3A). Thereafter, the GUS activity increased around the primordial tip, and a steep gradient was evident within the root primordium on day 4. This gradient of GUS activity was also established in the adventitious root primordium of the rpd1-1 mutant at days 3 and 4, but it was much more broadly localized than in the wild-type primordium. The altered pattern of DR5:GUS expression in the rpd1-1 primordium appeared to correlate with the developmental arrest of the primordium. Before the defect in the rpd1-1 primordium was morphologically detectable, however, no obvious difference in DR5:GUS expression was observed between the wild type and rpd1-1. These results suggest that the rpd1 mutation does not primarily affect auxin accumulation patterns.
SCARECROW Expression in Adventitious Root Primordia in the rpd1 Mutant
END199 is a SCARECROW (SCR) reporter line in which the uidA gene is inserted in the promoter region of the SCR gene. In this line, GUS activity is expressed under the control of the SCR promoter specifically in the endodermis, endodermis/cortex initials, and the quiescent center of mature roots (Malamy and Benfey, 1997). During lateral root development, GUS activity of END199 is localized to specific cell layers of the root primordium, which indicates the onset of cellular differentiation (Malamy and Benfey, 1997). When hypocotyl explants of END199 were cultured at 28°C on RIM, GUS activity appeared in association with the initiation of adventitious root primordia on day 2, and in developing root primordia at days 3 and 4, high activity of GUS was observed inside the outermost layer (Fig. 3B). END199 in the rpd1-2 background showed similar patterns of GUS expression until the developmental arrest of adventitious root primordia became obvious (Fig. 3B). This suggests that the rpd1 mutation does not impede the beginning of cellular differentiation during root primordium development.
Effect of the rpd1 Mutation on the Growth of Primary Roots
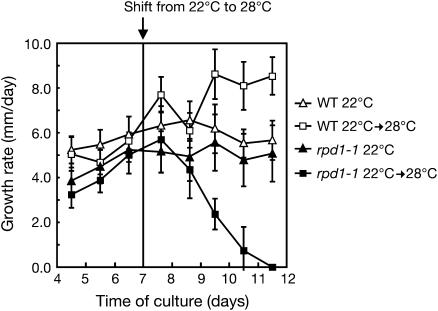
The effect of the rpd1 mutation on the activity of the root apical meristem was examined by monitoring the growth rates of primary roots. When rpd1-1 seedlings were first grown at 22°C and the temperature was then shifted up to 28°C, the growth rate decreased with a time lag of about 2 d after the temperature shift. It then dropped to zero within the next 3 d (Fig. 4). This result demonstrates that the rpd1 mutation interferes with the maintenance of meristematic activity, as well as with the development of the root primordium.
Figure 4.
Effect of temperature shift on the growth rate of the rpd1-1 primary root. Seedlings of the wild type and rpd1-1 were grown at 22°C on GMA. After 7 d of culture, half of them were transferred to 28°C. Growth rate was measured by marking the position of the root tip every day. Symbols represent averages of data from eight to 15 primary roots for each condition. Vertical lines indicate sds.
Effect of the rpd1 Mutation on Callus Formation
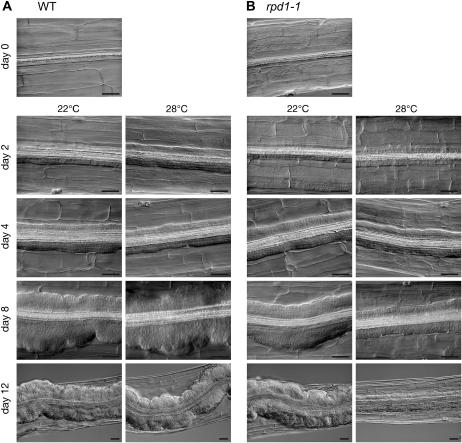
Hypocotyl explants of the rpd1-1 mutant were cultured on callus-inducing medium at 22°C or 28°C, and time course of callus formation was compared with that in the wild-type explants (Fig. 5). In the rpd1 explants cultured at 28°C, callus formation started at the same time as in the wild-type explants. During the first 4 d of culture, callus growth appeared not to be affected by the rpd1 mutation. After day 4, however, callus growth in the rpd1 explants was retarded severely at 28°C. Thus the rpd1 mutation inhibited not the initiation but the maintenance of callus cell proliferation.
Figure 5.
Time course of callus formation as influenced by the rpd1 mutation. Hypocotyl explants of the wild type (A) and rpd1-1 mutant (B) were cultured at 22°C or 28°C for the indicated days. Bars = 50 μm.
Cloning of the RPD1 Gene
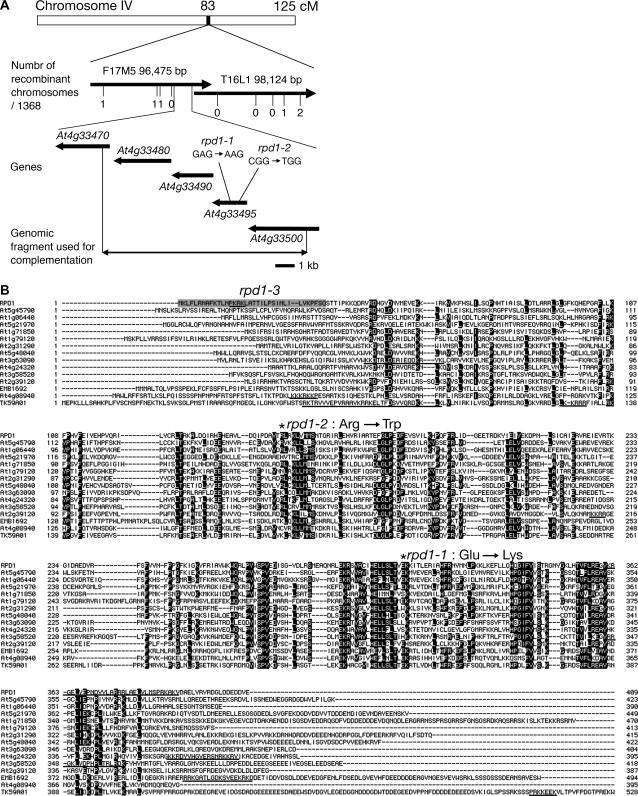
The rpd1-1 mutation was mapped within a 110-kb region at the 83-cm position of chromosome IV (Fig. 6A). Sequence analysis of genomic DNA in this region revealed that both rpd1-1 and rpd1-2 have a nucleotide substitution in At4g33495. A genomic fragment encompassing At4g33495 complemented the adventitious rooting phenotype of rpd1-1 at 28°C (data not shown). From these results, we concluded that At4g33495 corresponds to the RPD1 gene.
Figure 6.
Map-based cloning of RPD1. A, Chromosomal position of RPD1. B, Alignment of the amino acid sequences of full-length RPD1 and related proteins. Identical amino acid residues are highlighted on a black background. Positions of the rpd1-1 and rpd1-2 mutations are marked with asterisks. An N-terminal peptide retained in the T-DNA insertion mutant (rpd1-3) is shadowed. Predicted nuclear localization signals are underlined. The full-length amino acid sequence of TK59A01 is deduced from the genomic sequence around the EST, TK59A01 (accession no. CO048598; Katari et al., 2005), which is located on chromosome IV between At4g01030 and At4g01040.
Determination of the cDNA sequence of RPD1 and its comparison with the genomic sequence showed that the RPD1 gene is composed of two exons and one intron. The intron is located downstream from the stop codon (data not shown).
The predicted RPD1 protein consists of 409 amino acid residues. RPD1 shows no homology with any proteins of known function. A database search detected several sequences related to RPD1 among the expressed sequence tags (ESTs) of various plants, including the fern, Ceratopteris richardii (accession nos. BE643047 and BE643199), and the moss, Physcomitrella patens (BJ954318), but no such sequences were found of algal, animal, fungal, or bacterial origin (data not shown). In the Arabidopsis genome, there are 13 annotated genes and one EST encoding RPD1-like proteins (Fig. 6B). RPD1 and RPD1-like proteins share similarities throughout their sequences, except for the amino- and carboxy-terminal extensions (Fig. 6B). They are annotated to localize in mitochondrion or plastid except for At3g63090, although RPD1 and some of RPD1-like proteins have putative nuclear localization signals according to analysis with the PSORT prediction software (http://psort.ims.u-tokyo.ac.jp/; Fig. 6B).
Expression Profiles of the RPD1 Gene
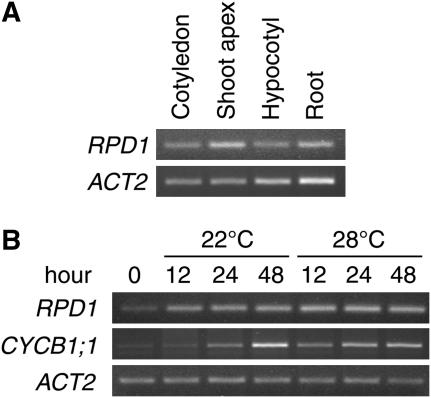
Expression profiles of the RPD1 gene were analyzed by semiquantitative reverse transcription (RT)-PCR (Fig. 7). RPD1 mRNA was expressed in all organs of 14-d-old seedlings (Fig. 7A). It was relatively abundant in the shoot apex and the root. In hypocotyl explants induced to form adventitious roots, RPD1 expression showed a 2- to 3-fold increase in the first 12 h of culture at either 22°C or 28°C (Fig. 7B). This was at least 1 d earlier than the first sign of adventitious root initiation detectable microscopically (Fig. 1).
Figure 7.
Semiquantitative RT-PCR analysis of RPD1 expression. A, Expression of RPD1 mRNA in various organs of 14-d-old wild-type seedlings. Seedlings were divided into cotyledons, hypocotyls, roots, and the remaining shoot apex parts with true leaves. B, Expression of RPD1 mRNA upon adventitious root induction in wild-type hypocotyl explants. ACT2 was used as the control.
As the sole intron of RPD1 is more than 360 bases distant from the rpd1-1 and rpd1-2 mutations, its splicing does not seem to be influenced by these mutations. In support of this, RT-PCR analysis indicated accumulation of the normally spliced RPD1 mRNA in the rpd1-1 explants cultured at 28°C at a similar level to that found in the wild type (data not shown).
Knockout Phenotypes of the RPD1 Gene
Two lines that harbor T-DNA insertions in the RPD1 gene were identified in the collection of SALK T-DNA insertion lines (Alonso et al., 2003; http://signal.salk.edu/). T-DNA was inserted at the same position in both these lines, 107 bp downstream from the start codon. The predicted fusion gene product resulting from the T-DNA insertion contains the first 36 amino acids of RPD1 followed by 36 amino acids derived from the T-DNA insertion (Fig. 6B). This fusion protein lacks any conserved regions of RPD1 and should be nonfunctional. Therefore, the T-DNA insertion in these lines caused a presumably null rpd1 mutation, which was designated rpd1-3.
The T4 population was produced from each T3 plant heterozygous for the rpd1-3 mutation through self reproduction. The T4 individuals were genotyped with respect to the T-DNA insert and the RPD1 gene. All the T4 individuals had the intact RPD1 gene, and two thirds of them had the T-DNA insertion (Table I), suggesting that homozygotes for the rpd1-3 mutation are embryo lethal. Introduction of the genomic fragment containing At4g33495, namely RPD1 into the RPD1/rpd1-3 heterozygotes resulted in the recovery of progeny homozygous for the rpd1-3 mutation in the presence of a transgene-derived copy of RPD1. This demonstrated that the embryo lethality in the rpd1-3 lines was caused by the knockout of the RPD1 gene.
Table I.
Segregation of genotypes among the T4 populations derived from self reproduction of T3 plants heterozygous for the rpd1-3 mutation
Numbers in the three middle columns represent the number of individuals with the indicated genotype. Numbers in the Total column represent the total number of plants examined.
| T4 Line
|
Genotype
|
Total
|
||
|---|---|---|---|---|
| RPD1/RPD1 | RPD1/rpd1-3 | rpd1-3/rpd1-3 | ||
| 123424-12 | 19 | 45 | 0 | 64 |
| 123424-67 | 18 | 38 | 0 | 56 |
| 123464-12 | 39 | 94 | 0 | 133 |
| 123464-13 | 48 | 89 | 0 | 137 |
| 123464-14 | 32 | 95 | 0 | 127 |
| 123464-19 | 52 | 98 | 0 | 150 |
| 123464-22 | 35 | 70 | 0 | 105 |
| 123464-24 | 29 | 58 | 0 | 87 |
| 123464-32 | 14 | 17 | 0 | 31 |
| 123464-37 | 9 | 20 | 0 | 29 |
| Total | 295 | 624 | 0 | 919 |
T4 plants genotyped as RPD1/rpd1-3 were indistinguishable in morphology and growth from RPD1/RPD1 plants of the same population (data not shown). However, heterozygous T4 plants produced both green and white seeds by self reproduction (Fig. 8A). In their mature siliques, embryos at the globular or transition stages were observed together with bent-cotyledon stage embryos (Fig. 8, C and D). The overall morphology of the arrested embryos appeared normal (Fig. 8E).
Figure 8.
Effect of RPD1 knockout on embryogenesis. A, Opened silique of RPD1/rpd1-3. B, Opened silique of RPD1/RPD1. C and D, Seeds in the mature RPD1/rpd1-3 silique. E, Higher magnification of embryo in D. Bars = 5 mm (A and B) or 50 μm (C–E).
To gain information about effects of disruption of RPD1-related genes on embryogenesis, we searched the database of SeedGenes project (http://www.seedgenes.org/; Tzafrir et al., 2004) and found one RPD1-related gene, At5g62990/EMB1692. According to the presented information, T-DNA insertion in At5g62990/EMB1692 causes embryogenesis arrest at the transition stage, which is similar to the case of the RPD1 knockout line.
Structural Characterization in Silico of RPD1 Family Proteins
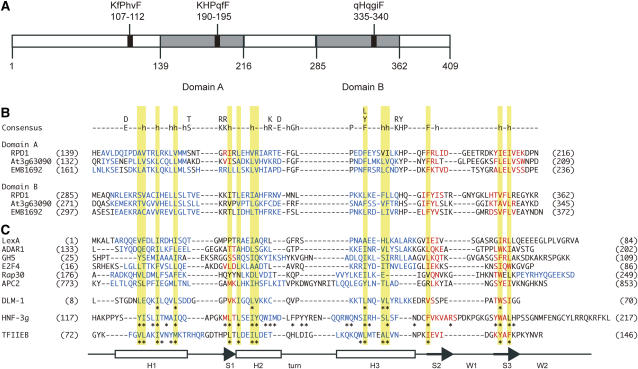
We characterized in silico the two conserved regions of RPD1 and RPD1-like proteins where the rpd1-1 and rpd1-2 mutations reside, and noticed that they are structurally related to each other (Fig. 9). These regions are designated domain A (residues 139–216 of RPD1) and domain B (residues 285–362 of RPD1).
Figure 9.
Possible structural features of RPD1 and related proteins. A, Putative structural units of RPD1. Positions of three K/RH/YPXXF motifs are shown by black boxes with corresponding amino acid sequences. Uppercase letters indicate residues that match to the motif consensus and lowercase letters indicate residues that differ from the consensus. Domains A and B are shown by gray boxes. B, Sequence alignment of domains A and B of RPD1 and related proteins. Residues predicted to form α helix and β strand are written in blue and red, respectively. Consensus between domain A and domain B is given above the domain A sequence of RPD1. Lowercase h indicates hydrophobic residues. C, Sequence alignment of winged helix proteins. Residues in α helix and β strand are written in blue and red, respectively. LexA, DNA-binding domain of Escherichia coli LexA repressor, Protein Data Bank (PDB) accession 1LEA (Fogh et al., 1994); ADAR1, Zα domain of human ADAR1, PDB accession 1QBJ (Schwartz et al., 1999); GH5, chicken linker histone H5, PDB accession 1HST (Ramakrishnan et al., 1993); E2F4, DNA-binding domain of human E2F4, PDB accession 1CF7 (Zheng et al., 1999); Rap30, DNA-binding domain of Rap30 subunit of human TFIIF, PDB accession 2BBY (Groft et al., 1998); APC2, WH-B domain of Saccharomyces cerevisiae APC2, PDB accession 1LDD (Zheng et al., 2002); DLM-1, Zα domain of mouse DLM-1, PDB accession 1J75 (Schwartz et al., 2001); HNF-3g, DNA-binding domain of rat HNF-3γ, Swissprot accession P32183 (Clark et al., 1993); TFIIEB, central core domain of human TFIIEβ, PDB accession 1D8K (Okuda et al., 2000). Asterisks indicate residues that constitute the hydrophobic cores of DLM-1, TFIIEβ, and HNF-3γ. Secondary structures are illustrated at the bottom. Yellow shadings show that the positions of conserved hydrophobic residues of RPD1 and related proteins correspond to those of the hydrophobic core residues of the winged helix fold.
As shown in Figure 9B, domains A and B share low but significant sequence similarity. Higher structures of these domains were compared using the three-dimensional position-specific scoring matrix (3D-PSSM) system (Kelly et al., 2000; http://www.sbg.bio.ic.ac.uk/∼3dpssm/), which returns secondary structure predictions based on the PSI-Pred program (Jones, 1999), in addition to the top 20 hits of potentially structurally related proteins. The secondary structures of domains A and B were predicted to be H-short S-H-H-S-S and H-H-H-S-S, respectively (H for α helix and S for β strand; Fig. 9B). Thus domains A and B are very similar in their overall secondary structures. Searches of 3D-PSSM with domains A and B as queries hit various helical proteins. Proteins that belong to the winged helix superfamily were most frequently found in the search results for both domains A and B. Winged helix consists of three α helices, three β strands, and two extended loops (wings; W) that are arranged in the order of H1-S1-H2-turn-H3-S1-W1-S2-W2 (Gajiwala and Burley, 2000). Alignment of the sequences of RPD1 and RPD1-like proteins with those of winged helix proteins indicated that conserved hydrophobic residues in domains A and B are located at the position of the hydrophobic core of winged helix proteins (Fig. 9, B and C; Clark et al., 1993; Gajiwala and Burley, 2000; Okuda et al., 2000; Schwartz et al., 2001). These results suggested a possible relationship of RPD1 and RPD1-like proteins with winged helix proteins.
Sequence comparison of RPD1 and RPD1-like proteins also detected three 6-amino acid motifs, K/RH/YPXXF (X denotes arbitrary amino acid; Figs. 6B and 9A). Domain A and domain B contain the second and the third K/RH/YPXXF motifs, respectively. The last residue Phe of each K/RH/YPXXF motif is completely conserved among RPD1-related proteins, suggesting its importance in this group of proteins.
DISCUSSION
RPD1 Is Required for the Maintenance of Active Cell Proliferation
The rpd1 mutants (rpd1-1 and rpd1-2) were initially characterized by their temperature-sensitive development of adventitious root primordia. In this study, we further analyzed the adventitious rooting phenotype of rpd1, taking advantage of its temperature sensitivity. Microscopic observation of the time course of adventitious root formation at the restrictive temperature showed that the rpd1 mutation does not affect the initial phase of development of adventitious root primordia, but retards their development beyond the two- to four-cell-layer stages (Fig. 1). Temperature-shift experiments localized the window of requirement for the RPD1 function to the period before cell proliferation becomes recognizable (Fig. 2). These findings suggest that the RPD1 function is related to stage-specific events involved in root primordium development, rather than to fundamentals required throughout organogenesis.
During the early development of root primordia, establishment of the apical-basal axis and cellular organization occur in correlation with changes in auxin accumulation patterns (Malamy and Benfey, 1997; Benková et al., 2003). This organization process was considered a possible target of the rpd1 mutation, but our results refute this possibility. Adventitious root primordia of the rpd1 mutant, arrested at the early stages under the restrictive conditions, were not deformed and could resume normal development after transfer to the permissive conditions (Fig. 2L). This means that the rpd1 mutation does not seriously damage the cellular patterning of the root primordium. The relatively normal organization of the rpd1 root primordium was also supported by expression analysis of an auxin-responsive reporter, DR5:GUS and a cytodifferentiation marker gene, SCR. The results indicated that the rpd1 mutant can generate the auxin gradient associated with axis formation and start proper cytodifferentiation in the root primordium at the restrictive temperature (Fig. 3).
The other notable feature of root primordium development is a rapid increase in cell number (MacLeod and Thompson, 1979; Laskowski et al., 1995). The above-described phenotypes of the rpd1 mutant imply that it has a primary defect in the maintenance of the highly active cell division characteristic of the early development of the root primordium, instead of in the cellular organization of the root primordium. Temperature sensitivity of unorganized cell proliferation of the rpd1 callus (Fig. 5) also suggests that the maintenance of active cell division requires the RPD1 function. This inference is in good agreement with the defect of the RPD1 knockout (rpd1-3) in embryogenesis at the globular to transition stages (Fig. 8), where rapid cycles of cell division are involved (Bowman and Mansfield, 1994).
As discussed above, the rpd1 mutations caused apparently stage-specific defects in root primordium development. However, in the apical growth that depends on meristematic activity, the effects of the rpd1 mutations seem to be cumulative. For example, exposure of the rpd1-1 mutant to the restrictive temperature did not halt primary root growth immediately, but decreased the growth rate gradually after a time lag of about 2 d (Fig. 4). These differential effects of the rpd1 mutations might reflect differences in cell-cycle duration. Estimates of 5 to 9 h have been reported for cell-cycle duration during the development of lateral root primordia and embryos in Arabidopsis (Bowman and Mansfield, 1994; Laskowski et al., 1995). These are much shorter than the estimates of 19 to 48 h for the duration of the meristematic cell cycle (Beemster et al., 2002). We speculate that the extraordinary shortening of the cell cycle may increase the requirement for the RPD1 function.
In summary, the RPD1 gene is inferred to have an important function in maintaining active cell proliferation, particularly when the cell-division cycle is very rapid. It should be noted again that the initial stages of adventitious root development and callus formation are quite normal in the rpd1-1 explants, even at the restrictive temperature. Therefore, we could say that the RPD1 function is required for the maintenance of active cell proliferation, but not for the reactivation of the cell cycle in quiescent cells or the progression of every cycle of cell division.
RPD1 Belongs to a Novel Protein Family Specific to the Plant Kingdom
RPD1 encodes a member of a novel protein family (Fig. 6). Database searches detected many sequences encoding RPD1-like proteins in ESTs of various plants, including monocots, a fern, and a moss. However, no related proteins were found in algae, animals, fungi, or bacteria. Therefore, this protein family is specific to plants. It is of interest to resolve how and when the RPD1 family originated in the history of plant evolution.
RPD1 and related proteins contain no known domains or motifs, except the putative transit peptide and nuclear localization signal. However, careful comparison of the sequences of these proteins and 3D-PSSM search led to the identification of two domains representing a large repeat conserved among RPD1-related proteins (Fig. 9). The rpd1-2 and rpd1-1 mutations occur in the first and second domains, respectively, suggesting that these domains are important for the function of RPD1. It is notable that these domains might be structurally related to the winged helix proteins, which are involved in various regulatory functions through DNA binding, RNA binding, or protein-protein interaction (Clark et al., 1993; Ramakrishnan et al., 1993; Fogh et al., 1994; Finnin et al., 1997; Groft et al., 1998; Schwartz et al., 1999; Zheng et al., 1999; Gajiwala and Burley, 2000; Okuda et al., 2000; Kamada et al., 2001; Schwartz et al., 2001; Wilce et al., 2001; Selmer and Su, 2002; Zheng et al., 2002; Alfano et al., 2004; Dong et al., 2004; Wei et al., 2004). Studies focusing on this unique structure should provide insight into the molecular function shared by the RPD1 family proteins.
MATERIALS AND METHODS
Plant Materials
Of two rpd1 mutants (rpd1-1 and rpd1-2) of the ecotype Landsberg erecta (Ler) origin, we used rpd1-1 as a representative of the rpd1 mutant in this study, unless otherwise indicated. An auxin-responsive reporter line of the Columbia origin carrying DR5:GUS (Ulmasov et al., 1997) was provided by Dr. Tom J. Guilfoyle (University of Missouri). A SCR reporter line END199 (Malamy and Benfey, 1997) was provided by Dr. Philip N. Benfey (New York University). These reporter lines were crossed with the rpd1-1 or rpd1-2 mutant. After two rounds of self reproduction, F3 lines homozygous for both the reporter gene and the rpd1 mutation were selected and used for the reporter analysis. F3 lines homozygous for the reporter gene and for the wild-type RPD1 allele were used as the wild-type control. T-DNA insertion lines SALK_123424 and SALK_123464 of the Columbia origin were obtained from The Arabidopsis Information Resource (Alonso et al., 2003; http://www.arabidopsis.org).
Tissue Culture
Tissue culture experiments were performed as described by Konishi and Sugiyama (2003). Hypocotyl segments of 5 mm in length were excised from 12- to 14-d-old seedlings and cultured under continuous light (15–25 μmol m−2 s−1) at either 22°C or 28°C. For the induction of adventitious roots, hypocotyl explants were cultured on RIM:B5 medium (Gamborg et al., 1968) supplemented with 20 g L−1 Glc and 0.5 mg L−1 indole-3-butyric acid. For the induction of callus, explants were cultured on callus-inducing medium:B5 medium supplemented with 20 g L−1 Glc, 0.5 mg L−1 2,4-dichlorophenoxyacetic acid, and 0.1 mg L−1 kinetin. These culture media were buffered at pH 5.7 with 0.5 g L−1 MES, and solidified with 2.5 g L−1 gellan gum.
Histological Observation
Hypocotyl explants were fixed at 4°C in a 9:1 mixture of ethanol and acetic acid, hydrated through a graded series of ethanol, and mounted with a drop of clearing solution (a mixture of 8 g chloral hydrate, 2 mL water, and 1 mL glycerol). Cleared samples were observed under a light microscope equipped with Nomarski optics (BX50-DIC; Olympus).
GUS Staining
Hypocotyl explants were fixed in 90% acetone at −20°C overnight and rinsed three times with 0.1 m sodium phosphate buffer (pH 7.4). They were incubated in X-Glc solution (0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide cyclohexylammonium salt, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 0.1 m sodium phosphate, pH 7.4) at 37°C for 3 h.
Root Growth Analysis
Seeds were surface sterilized and sown on germination medium A (GMA). GMA is Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 10 g L−1 Suc, buffered to pH 5.7 with 0.5 g L−1 2-(N-morpholino) ethanesulfonic acid, and solidified with 15 g L−1 agar. GMA plates were placed vertically under continuous light (30–50 μmol m−2 s−1). Primary roots grown on the surface of GMA were subjected to growth analysis.
Chromosome Mapping
Chromosome mapping was performed according to the standard procedure as described previously (Konishi and Sugiyama, 2003).
Sequence Analysis
Genomic DNA was extracted from the wild-type (Ler) and the rpd1 mutants with Isoplant II (Nippon Gene), and subjected to PCR amplification with gene-specific primer sets. The PCR product was treated with ExoSAP-IT (USB) and used as the template for cycle sequence reactions with BigDye Terminator Cycle Sequencing FS Ready Reaction kit (Applied Biosystems), according to the manufacturer's protocol. Sequencing was carried out with an ABI PRISM 377 DNA sequencer (Applied Biosystems). The sequence data were analyzed with Genetyx-Mac software version 10 (Genetyx).
Determination of the RPD1 cDNA Sequence
Total RNA was extracted from Ler hypocotyl explants that had been cultured on RIM for 1 d at 28°C and used as the template for the first-strand cDNA synthesis. cDNA synthesis and its amplification by nested PCR were performed with the SMART RACE cDNA amplification kit (BD Biosciences). The RPD1-specific primers used were: 5′-GSP (5′-AAGCCGAGACCTAGACGACGAGCGAGCG-3′) and 5′-GSP2 (5′-CTGTGAAATTTGACGACTTTGCGG-3′) to amplify the 5′ end of RPD1 cDNA; and 3′-GSP (5′-CGAGGAAATTACGGGAAGCTTCATACGG-3′) and 3′-GSP2 (5′-AAGAGAGGGGAATTGGTAGAACCG-3′) to amplify the 3′ end. The amplified DNA fragments were cloned into the pGEM-T Easy Vector (Promega) and subjected to sequence analysis.
Isolation of Genomic Clone
The genomic clone GL3631, which was used for the complementation test, was isolated from our transformation-competent genomic library. In this library, genomic fragments of Ler were inserted into the pNH01 binary vector, which had been constructed from pIG121-Hm (Akama et al., 1992) by removing the 35S:GUS reporter gene. Sequencing of the ends of the genomic fragment in GL3631 indicated that it encompassed a genomic span of 10.7 kb from residue 16105028 to residue 16115778 of chromosome IV (accession no. NC003075). The genomic clone GL3631 was introduced into Agrobacterium tumefaciens EHA101 by electroporation.
Transformation of Plants
A simplified version of the floral dip method was used for plant transformation (Clough and Bent, 1998). T1 seedlings derived from the infected plants were selected for hygromycin B resistance on GMA (solidified with 0.8% agar in some cases) containing 15 or 20 mg L−1 hygromycin B and 100 mg L−1 carbenicillin sodium salt.
Semiquantitative RT-PCR
RNA extraction was carried out as described by Ozeki et al. (1990). Total RNA was treated with DNase I (Sigma) and reverse transcribed with SuperScript II RNase H− Reverse Transcriptase (Invitrogen). The first-strand cDNA was used as template for PCR amplification of cDNA fragments. Primer sets used were 5′-TGATGTCGAATGAACATACG-3′ and 5′-TCACAATCTTCGATTCTTCG-3′ for RPD1; 5′-AGACGCCCCCACTACTTAGACTT-3′ and 5′-GGTTTAGCTCGAATCGGACATGC-3′ for CYCB1;1 (Richard et al., 2001); and 5′-GTTGCACCACCTGAAAGGAAG-3′ and 5′-CAATGGGACTAAAACGCAAAA-3′ for ACT2 (An et al., 1996; Himanen et al., 2002). PCR was performed for 27 cycles (RPD1), 29 cycles (CYCB1;1), or 19 cycles (ACT2), consisting of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s. The cycle numbers used here produced the exponential phase of PCR amplification, which was verified by performing the PCRs for various numbers of cycles.
Knockout Analysis
Insertion mutant information was obtained from the SIGnAL Web site at http://signal.salk.edu. For knockout analysis of the RPD1 gene, T3 plants heterozygous for the T-DNA insertion were selected by PCR-based screening of the SALK lines, SALK_123424 and SALK_123464. The presence of the RPD1-inserted T-DNA was checked by PCR amplification of it with a RPD1-specific primer and the LBb1 primer (http://signal.salk.edu/). The PCR product was sequenced to confirm the site of T-DNA insertion. The presence of the intact RPD1 gene was checked by PCR amplification with RPD1-specific primer sets. T4 lines obtained by self reproduction of each T3 plant were used for genotypic and phenotypic analyses.
For complementation analysis, the genomic fragment of the Ler origin containing RPD1 was introduced into plants heterozygous for the T-DNA insertion (Columbia background) via the transformation-competent genomic clone GL3631, and the resultant transformants were genotyped. The GL3631-derived copy of RPD1 (Ler origin) and the endogenous RPD1 (Columbia origin) were distinguished using a new CAPS marker, which was developed for a flanking single nucleotide polymorphism (residue 16113626 of chromosome IV) between Ler and Columbia.
The nucleotide sequence of RPD1 mRNA of Ler origin has been deposited with the DNA Data Bank of Japan/EMBL/GenBank databases under accession number AB189464.
Acknowledgments
We thank Dr. Tom. J. Guilfoyle (University of Missouri) for the generous gift of the DR5:GUS line seeds, Drs. Philip N. Benfey (New York University) and Hidehiro Fukaki (Nara Institute of Science and Technology) for the END199 seeds, and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants.
This work was supported by Grants-in-Aid from the Ministry of Education, Sports, Culture, Science and Technology of Japan (RFTF00L01605 and no.14036209). Funding for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Munetaka Sugiyama (sugiyama@ns.bg.s.u-tokyo.ac.jp).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074724.
References
- Akama K, Shiraishi H, Ohta S, Nakamura K, Okada K, Shimura Y (1992) Efficient transformation of Arabidopsis thaliana: comparison of the efficiencies with various organs, plant ecotypes and Agrobacterium strains. Plant Cell Rep 12: 7–11 [DOI] [PubMed] [Google Scholar]
- Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR (2004) Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 11: 323–329 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- An Y-Q, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Beemster GTS, De Vusser K, De Tavernier E, De Bock K, Inzé D (2002) Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol 129: 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera M-T, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D (1995) superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Mansfield SG (1994) Embryogenesis: introduction. In J Bowman, ed, Arabidopsis, an Atlas of Morphology and Development. Springer-Verlag, New York, pp 351–361
- Celenza JL Jr, Grisafi PL, Fink GR (1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK (1993) Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364: 412–420 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang H (2003) An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J 33: 543–555 [DOI] [PubMed] [Google Scholar]
- Dong G, Chakshusmathi G, Wolin SL, Reinisch KM (2004) Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J 23: 1000–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin MS, Cicero MP, Davies C, Porter SJ, White SW, Kreuzer KN (1997) The activation domain of the MotA transcription factor from bacteriophage T4. EMBO J 16: 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogh RH, Ottleben G, Rüterjans H, Schnarr M, Boelens R, Kaptein R (1994) Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy. EMBO J 13: 3936–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg SH, Davidson D (1971) Cell population studies in developing root primordia. Ann Bot (Lond) 35: 523–533 [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Gajiwala KS, Burley SK (2000) Winged helix proteins. Curr Opin Struct Biol 10: 110–116 [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirement of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Groft CM, Uljon SN, Wang R, Werner MH (1998) Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly. Proc Natl Acad Sci USA 95: 9117–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, Estelle M (1995) The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J 7: 211–220 [DOI] [PubMed] [Google Scholar]
- Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Kamada K, De Angelis J, Roeder RG, Burley SK (2001) Crystal structure of the C-terminal domain of the RAP74 subunit of human transcription factor IIF. Proc Natl Acad Sci USA 98: 3115–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari MS, Balija V, Wilson RK, Martienssen RA, McCombie WR (2005) Comparing low coverage random shotgun sequence data from Brassica oleracea and Oryza sativa genome sequence for their ability to add to the annotation of Arabidopsis thaliana. Genome Res 15: 496–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LA, MacCallum RM, Sternberg MJE (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol 299: 499–520 [DOI] [PubMed] [Google Scholar]
- Konishi M, Sugiyama M (2003) Genetic analysis of adventitious root formation with a novel series of temperature-sensitive mutants of Arabidopsis thaliana. Development 130: 5637–5647 [DOI] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM (1995) Formation of lateral root meristems is a two-stage process. Development 121: 3303–3310 [DOI] [PubMed] [Google Scholar]
- MacLeod RD, McLachlan SM (1975) The development of large primordia in Vicia faba L.: some cytological and anatomical changes. Protoplasma 85: 291–304 [Google Scholar]
- MacLeod RD, Thompson A (1979) Development of lateral root primordia in Vicia faba, Pisum sativum, Zea mays and Phaseolus vulgaris: rates of primordium formation and cell doubling times. Ann Bot (Lond) 44: 435–449 [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Ryan KS (2001) Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol 127: 899–909 [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25: 213–221 [DOI] [PubMed] [Google Scholar]
- Okuda M, Watanabe Y, Okamura H, Hanaoka F, Ohkuma Y, Nishimura Y (2000) Structure of the central core domain of TFIIEβ with a novel double-stranded DNA-binding surface. EMBO J 19: 1346–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki Y, Matsui K, Sakuta Y, Matsuoka M, Ohashi Y, Kano-Murakami Y, Yamamoto N, Tanaka Y (1990) Differential regulation of phenylalanine ammonia-lyase genes during anthocyanin synthesis and by transfer effect in carrot cell suspension cultures. Plant Physiol 80: 379–387 [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362: 219–223 [DOI] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK (1998) Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard C, Grainer C, Inzé D, De Veylder L (2001) Analysis of cell division parameters and cell cycle gene expression during the cultivation of Arabidopsis thaliana cell suspensions. J Exp Bot 52: 1625–1633 [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13: 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T, Behlke J, Lowenhaupt K, Heinemann U, Rich A (2001) DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat Struct Biol 8: 761–765 [DOI] [PubMed] [Google Scholar]
- Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A (1999) Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 284: 1841–1845 [DOI] [PubMed] [Google Scholar]
- Selmer M, Su X-D (2002) Crystal structure of an mRNA-binding fragment of Moorella thermoacetica elongation factor SelB. EMBO J 21: 4145–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Sugiyama M (2003) Isolation and initial characterization of temperature-sensitive mutants of Arabidopsis thaliana that are impaired in root redifferentiation. Plant Cell Physiol 44: 588–596 [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed JW (1999) Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Zhang P, Zhou Z, Cheng Z, Wan M, Gong W (2004) Crystal structure of human eIF3k, the first structure of eIF3 subunits. J Biol Chem 279: 34983–34990 [DOI] [PubMed] [Google Scholar]
- Wilce JA, Vivian JP, Hastings AF, Otting G, Folmer RHA, Duggin IG, Wake RG, Wilce MCJ (2001) Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest. Nat Struct Biol 8: 206–210 [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua N-H (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14: 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG (2000) Regulation of Arabidopsis root development by nitrate availability. J Exp Bot 51: 51–59 [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG (1999) Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA 96: 6529–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Fraenkel E, Pabo CO, Pavletich NP (1999) Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev 13: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]