Abstract
Sugars act as signaling molecules, whose signal transduction pathways may lead to the activation or inactivation of gene expression. Whole-genome transcript profiling reveals that the flavonoid and anthocyanin biosynthetic pathways are strongly up-regulated following sucrose (Suc) treatment. Besides mRNA accumulation, Suc affects both flavonoid and anthocyanin contents. We investigated the effects of sugars (Suc, glucose, and fructose) on genes coding for flavonoid and anthocyanin biosynthetic enzymes in Arabidopsis (Arabidopsis thaliana). The results indicate that the sugar-dependent up-regulation of the anthocyanin synthesis pathway is Suc specific. An altered induction of several anthocyanin biosynthetic genes, consistent with in vivo sugar modulation of mRNA accumulation, is observed in the phosphoglucomutase Arabidopsis mutant accumulating high levels of soluble sugars.
Anthocyanins are plant secondary metabolites playing a key role as flower pigments in signaling between plants and microbes, in responses related to nutrient availability, in male fertility of some species, in defense as antimicrobial agents and feeding deterrents, in the modulation of auxin transport, and in UV protection (Winkel-Shirley, 2001).
The anthocyanin biosynthetic pathway was described in different plants (Holton and Cornish, 1995), including Arabidopsis (Arabidopsis thaliana; Shirley et al., 1995; Bharti and Khurana, 1997), and several transcription factors regulating the anthocyanin biosynthetic pathway have been identified (Nesi et al., 2000; Vom Endt et al., 2002; Davies and Schwinn, 2003; Mathews et al., 2003; Broun, 2004; Matsui et al., 2004; Park et al., 2004).
The interrelationships between developmental, environmental, and metabolic signal transduction pathways control the production of flavonoids. Anthocyanin biosynthesis was often observed in plants germinated or grown on a sugar-containing medium (Mita et al., 1997; Németh et al., 1998; Baier et al., 2004). The chalcone synthase (CHS) gene derived from petunia (Petunia hybrida) petals in transgenic Arabidopsis leaves was induced by sugars (Tsukaya et al., 1991), and petunia corollas cultured in vitro without Suc do not show any pigmentation (Weiss, 2000). Petunia and Arabidopsis CHS genes are indeed characterized by the presence of Suc boxes in the 5′-flanking regions that may be also found in the Suc-inducible sporamin and amylase genes (Tsukaya et al., 1991). Arabidopsis grown on a Suc-containing medium shows high levels of anthocyanins (Tsukaya et al., 1991; Ohto et al., 2001).
Genes coding for dihydroflavonol reductase (DFR) and anthocyanidin synthase (ANS), also known as leucoanthocyanidin dioxygenase (LDOX), were up-regulated and the accumulation of anthocyanins was strongly increased by Suc in grape (Vitis vinifera) cells (Gollop et al., 2001, 2002), and signal transducers, such as Ca2+ and protein kinases/phosphatases, were shown to be involved in this process (Vitrac et al., 2000).
The Arabidopsis pho3 mutant, which has a defective copy of the Suc transporter 2 (SUC2) gene (encoding a phloem-loading Suc-proton symporter) leading to accumulation of soluble sugars and starch, showed growth retardation and anthocyanin accumulation (Lloyd and Zakhleniuk, 2004). The microarray analysis of pho3 adult leaves evidenced an enhanced expression of PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1), PRODUCTION OF ANTHOCYANIN PIGMENT 2 (PAP2), and TRANSPARENT TESTA 8 (TT8) transcription factors, as well as of genes coding for anthocyanin biosynthesis enzymes, suggesting that sugars are in vivo triggers of the anthocyanin biosynthesis (Lloyd and Zakhleniuk, 2004).
We investigated whether sugars coordinately induce most of the genes involved in the anthocyanin biosynthesis or if only a few genes play a pivotal role, and we studied the sugar specificity for the anthocyanin biosynthesis induction in Arabidopsis. In this article, we show evidence of the coordinated, Suc-specific modulation of most of the genes involved in the anthocyanin biosynthesis. Furthermore, induction of several anthocyanin biosynthetic genes in the phosphoglucomutase (pgm) Arabidopsis mutant accumulating high levels of soluble sugars is described.
RESULTS
Flavonols and Anthocyanins Are Inversely Modulated by Suc
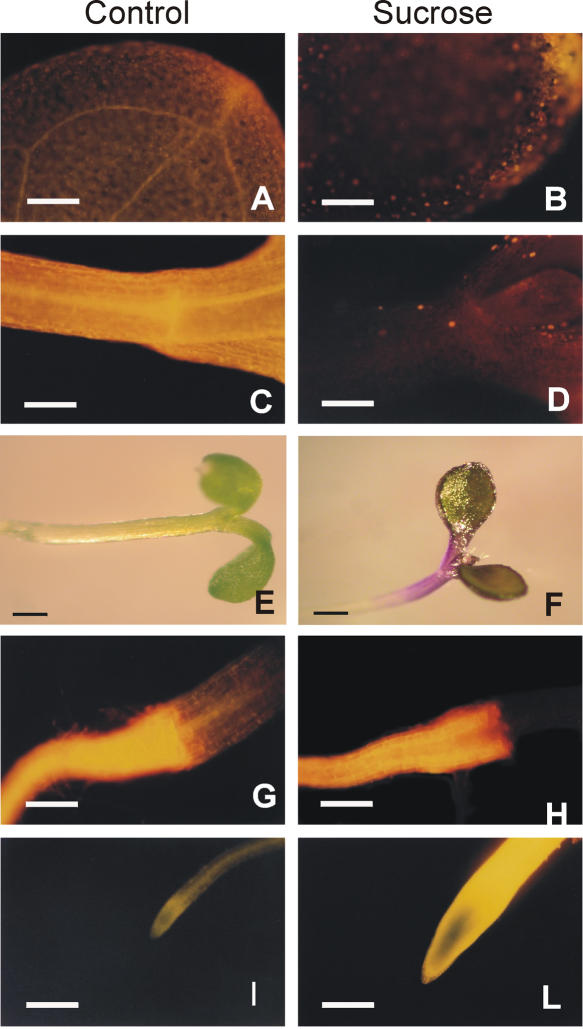
We analyzed the anthocyanin and flavonol content in Arabidopsis seedlings grown in the absence or presence of exogenous Suc. The histochemical analysis of flavonols (Fig. 1) shows the fluorescence that is characteristic of chlorophyll (red fluorescence), along with a limited orange fluorescence, which is typical of quercetin in cotyledons (Fig. 1A). The upper hypocotyl area exhibits a strong bright-yellow fluorescence (chalcone-naringenin; Fig. 1C). Whereas Suc does not affect the presence and distribution of flavonols in the roots (compare Fig. 1, G and I, with Fig. 1, H and L), this disaccharide leads to a decreased presence of flavonols (mainly represented by chalcone-naringenin) in the hypocotyl and cotyledons (Fig. 1, B and D). Interestingly, these tissues coincide with the anthocyanin accumulation site following Suc feeding of Arabidopsis seedlings (Fig. 1F).
Figure 1.
Effect of Suc on flavonoid and anthocyanin content and distribution in Arabidopsis seedlings. A to D and G to L, Effects of exogenous Suc on flavonoid content and distribution by DPBA staining of 3-d-old Arabidopsis seedlings, viewed through a fluorescein isothiocyanate filter. When reacting with flavonoids, DPBA emits orange fluorescence (quercetin), bright-yellow fluorescence (naringenin-chalcone), and bright-green fluorescence (kaempferol); chlorophylls exhibit a red autofluorescence. A, C, G, and I, Histochemical results of 3-d-old seedlings grown for an additional 72 h on control medium; the cotyledons (shown in A) display the fluorescence characteristic of quercetin; the cotyledonary node (shown in C) shows the bright-yellow fluorescence of chalcone-naringenin; the root (shown in G) contains mostly chalcone-naringenin; and the root tip (shown in I) contains mostly kaempferol. B, D, H, and L, Three-day-old seedlings grown for an additional 72 h on 90 mm Suc. Seedlings show a reduced bright-yellow fluorescence and the chlorophyll red fluorescence in cotyledons (shown in B). The cotyledonary node (shown in D) shows only the chlorophyll red fluorescence. Suc does not affect the flavonols' presence and distribution in the roots (compare G and I with H and L). E to F, Anthocyanin accumulation in Arabidopsis seedlings grown on control medium (shown in E) or grown for an additional 72 h on 90 mm Suc (shown in F). Bars in A to D, G to I, and L = 500 μm; bars in E and F = 200 μm.
Suc Affects the mRNA Level in Anthocyanin Biosynthetic Pathway Genes
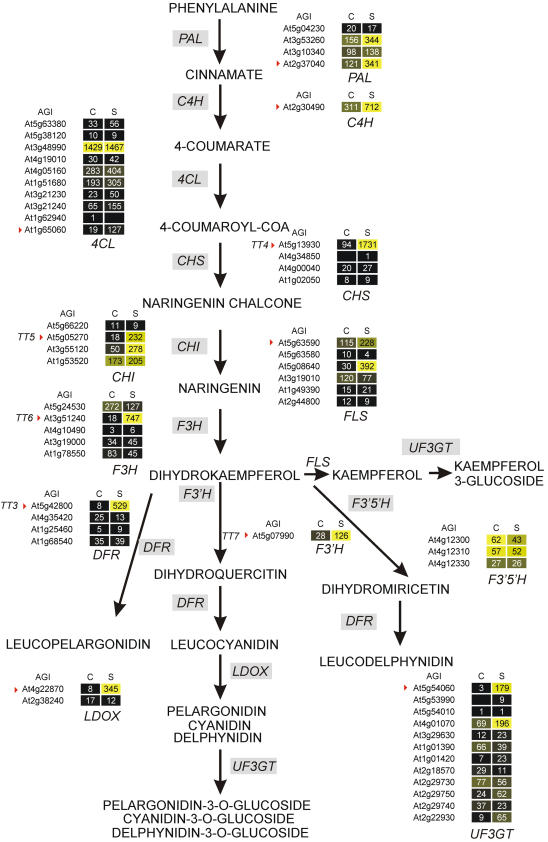
To identify the genes involved in the flavonol and anthocyanin biosynthesis that are regulated by Suc, we performed a microarray experiment with seedlings treated with Suc, compared to control seedlings. The rationale behind this experiment is that, besides the known genes involved in this pathway, there appears to be some gene redundancy for this cluster of genes (e.g. four 4-coumarate:CoA ligase [4CL] genes and six 4CL-like genes are represented in the Arabidopsis genome; http://www.arabidopsis.org/). The results of the microarray experiment are summarized in Figure 2. Interestingly, at least one gene up-regulated by Suc was detected in each step of the biosynthetic pathway, with the exception of the flavonoid 3′5′-hydroxylase (F3′5′H), which is expressed at a very low level. This genome-wide overview of the effects of Suc on the genes involved, or putatively involved, in the flavonoid and anthocyanin biosynthesis allowed us to select the genes to be further characterized in their response to sugar. One Suc-induced gene was selected (Arabidopsis Genome Initiative [AGI] codes marked with a red triangle in Fig. 2) for each biosynthetic step, giving preference to well-characterized genes when more than one gene was up-regulated by Suc (e.g. the TRANSPARENT TESTA 5 gene [TT5] corresponding to chalcone isomerase [CHI] was chosen among three Suc-induced CHI genes). As far as the transcription factors are involved in the regulation of the anthocyanin synthesis pathway, the PAP1 (At1g56650) transcript was 29-fold up-regulated by Suc, whereas PAP2 (At1g66390), TT8 (At4g09820), TRANSPARENT TESTA 2 (TT2; At5g35550), ANTHOCYANINLESS 2 (ANL2; At4g00730), and MYB family transcription factor 4 (AtMYB4; At4g38620) mRNA levels were unaffected by the treatment with Suc, or the induction of Suc was not confirmed by the biological replicate (TT8, ANL2; see Supplemental Table I).
Figure 2.
Effects of Suc on the mRNA accumulation for genes coding for flavonoid and anthocyanin biosynthetic enzymes in Arabidopsis seedlings. Arabidopsis seeds were germinated for 3 d and subsequently treated without (C) or with (S) 90 mm Suc for 6 h. Genes coding for enzymes involved in the flavonoid and anthocyanin pathways were identified by searching the Arabidopsis annotation in The Arabidopsis Information Resource (http://www.arabidopsis.org/). Microarray data (averaged transcript level from two biological replicates) were visualized using Heatmapper Plus software (http://bbc.botany.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi). The output of the software is shown, with the genes involved in each metabolic step represented by their respective AGI codes. A black square (and black shades) indicates a gene whose transcript level is low. A yellow square (and yellow shades) indicates a gene whose transcript level is relatively high within the group of genes putatively coding for the same function. AGI codes marked with a red triangle indicate the genes chosen for further studies. Arabidopsis mutants have been isolated on the basis of modified seed pigmentation and are therefore known as tt (for transparent testa) mutants (Koornneef, 1981, 1990), and TT loci identified have been characterized. When available, the TT gene codes are reported in the figure.
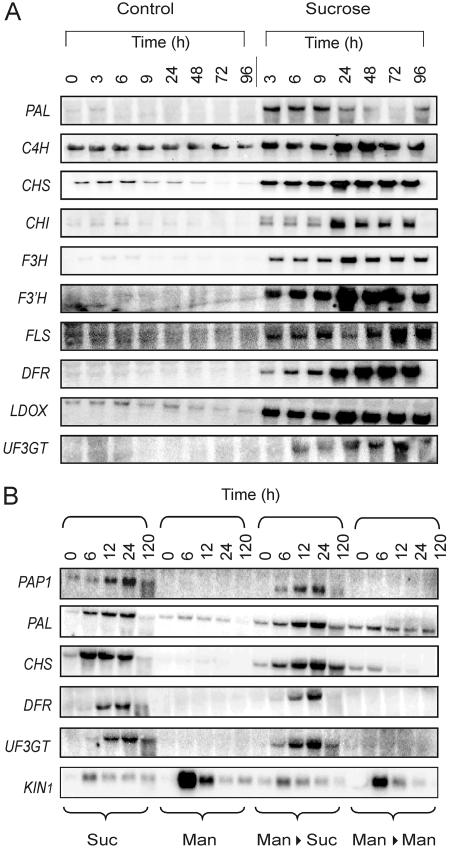
We analyzed the pattern of mRNA accumulation of the selected transcripts coding for proteins involved in the flavonoid biosynthesis pathway, selected on the basis of the microarray experiment results (Fig. 2). The results indicate that the mRNA level of several genes increases after the treatment with Suc (Fig. 3A). The induction is particularly evident for those genes coding for enzymes that act at the level and downstream of CHS, namely CHS, CHI, flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), flavonol synthase (FLS), DFR, LDOX, and UDP-Glc:flavonoid 3-O-glucosyltransferase (UF3GT). The cinnamate 4-hydroxylase (C4H) mRNA level is barely affected by Suc, whereas a transient induction of Phe ammonia-lyase (PAL) by Suc is observed (Fig. 3A). The 4CL mRNA level was below the detection threshold.
Figure 3.
A, Pattern of mRNA accumulation for genes coding for flavonoid and anthocyanin biosynthesis enzymes in Arabidopsis seedlings. Three-day-old Arabidopsis seedlings were grown for an additional 0 to 96 h on a Murashige and Skoog medium (Control) or on a Suc-enriched medium (Suc). RNA was extracted, electrophoresed, and northern analysis carried out using gene-specific probes. Equal loading was checked by reprobing with an rRNA probe (data not shown). A representative experiment is shown. B, Evaluation of the osmotic effect on the mRNA level of several anthocyanins biosynthetic genes and on the stress-induced KIN1 gene. Three-day-old Arabidopsis seedlings were treated for 0 to 120 h with Suc (Suc), mannitol (Man), pretreated (24 h) with mannitol followed by Suc for 0 to 120 h (Man→Suc), or pretreated (24 h) with mannitol followed by mannitol for 0 to 120 h (Man→Man). RNA was extracted, electrophoresed, and northern analysis carried out using gene specific probes. Equal loading was checked by reprobing with an rRNA probe (data not shown). A representative experiment is shown.
To evaluate whether the observed effects of Suc could be ascribed to an osmotic effect, we verified the effects of mannitol on some of the genes studied. Arabidopsis seedlings were treated with Suc (Fig. 3B, Suc), mannitol (Fig. 3B, Man), pretreated with mannitol (24 h) followed by Suc (Fig. 3B, Man→Suc), and pretreated with mannitol (24 h) followed by mannitol (Fig. 3B, Man→Man). The expression of several genes involved in the flavonoid/anthocyanin biosynthesis was analyzed, and the mRNA level of protein kinase 1 (KIN1), a stress-induced gene, was monitored (Kurkela and Franck, 1990). Genes involved in the flavonoid/anthocyanin synthesis were induced by Suc. The expression of KIN1 is transient and strongly influenced by mannitol, whereas Suc is unable to strongly induce this stress-related gene. A pretreatment with mannitol (Fig. 3B, Man→Suc) mitigates the perception of stress, leading to an increase in the level of KIN1 mRNA (compare Fig. 3B, Man, with Fig. 3B, Man→Man). This effect is not observed in the seedlings, which were pretreated with mannitol and subsequently exposed to Suc (Fig. 3B, Man→Suc): The Suc induction of flavonoid/anthocyanin genes is retained and is undistinguishable from the Suc-alone treatment (compare with Fig. 3B, Suc), ruling out a stress response, such as the triggering of flavonoid gene induction by Suc. These results suggest that the induction of flavonoid/anthocyanin synthesis genes is sugar specific and unlikely to be stress mediated.
A Specific Suc-Signaling Mechanism Requiring Low Sugar Concentrations Induces Anthocyanin Synthesis Genes
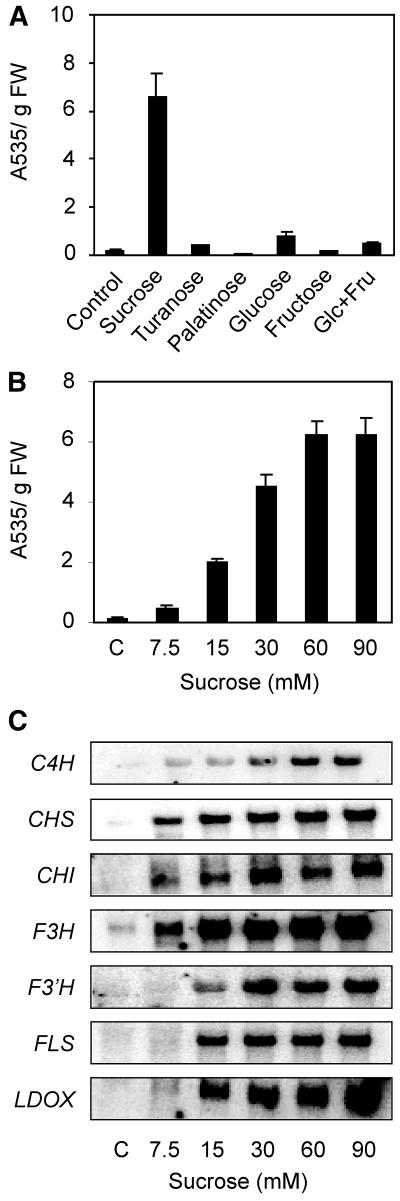
We investigated the Suc specificity of the anthocyanin biosynthesis by testing the effects of a set of metabolic sugars (Suc, Glc, Fru, and a 1:1 mixture of Glc + Fru) and nonmetabolic sugars (Suc analogs: turanose and palatinose; Loreti et al., 2000). The anthocyanin synthesis induction is Suc specific, with a strong accumulation of these pigments in Suc-treated seedlings only (Fig. 4A). The mRNA accumulation of transcripts related to flavonoid/anthocyanin synthesis is high in Suc-enriched media, whereas neither turanose nor palatinose were able to induce flavonoid/anthocyanin genes (data not shown). The threshold for the induction of the genes involved in the flavonoid/anthocyanin synthesis was investigated by testing different Suc concentrations. Three-day-old seedlings were treated with Suc concentrations ranging from 7.5 to 90 mm for 24 h, and the results show that 15 mm Suc is enough to enhance anthocyanin levels (Fig. 4B), although higher Suc concentrations lead to a more marked anthocyanin accumulation, reaching a plateau between 60 and 90 mm. The mRNA levels of CHS, CHI, and F3H readily increase when seedlings are treated with 7.5 mm Suc (Fig. 4C). Increasing the Suc concentration up to 15 mm results in an increased mRNA level of F3′H, FLS, and LDOX, whereas 30 to 60 mm is required to significantly increase the mRNA level of C4H (Fig. 4C).
Figure 4.
A, Effects of a set of metabolic sugars (Suc 90 mm, Glc 90 mm, Fru 90 mm, 1:1 mixture of Glc 45 mm + Fru 45 mm) and of nonmetabolic sugars (turanose and palatinose, 90 mm) on anthocyanin accumulation. B, Effect of Suc concentrations ranging from 7.5 to 90 mm on anthocyanin accumulation. C, mRNA accumulation for genes coding for flavonoids and anthocyanin biosynthesis enzymes in Arabidopsis seedlings. Seedlings were sugar treated for 48 h (A) or 24 h (B and C). RNA was extracted, electrophoresed, and northern analysis carried out using gene-specific probes. Equal loading was checked by reprobing with an rRNA probe (data not shown). A representative experiment is shown.
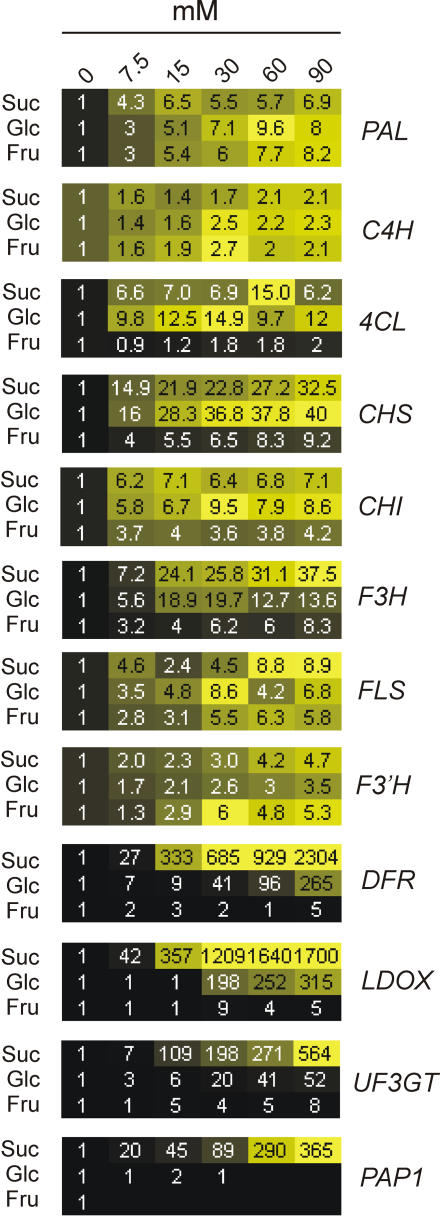
Northern analysis provides a semiquantitative profile of expression, and a more accurate quantitation can be obtained by means of real-time reverse transcription (RT)-PCR. Three-day-old, light-grown seedlings were fed for 12 h with Suc, Glc, or Fru in concentrations ranging from 7.5 to 90 mm. The real-time RT-PCR results indicate that Suc is the most efficient trigger of mRNA accumulation for genes, whose products act downstream along the anthocyanin biosynthetic pathway (DFR, LDOX, UF3GT), as well as for PAP1 (Fig. 5). These genes are induced by Suc several hundred-fold, whereas genes upstream of DFR show a lower induction by Suc and can also be induced by Glc and, to a minor extent, by Fru (Fig. 5).
Figure 5.
Effect of different metabolic sugars on the expression of genes involved in flavonoid/anthocyanin synthesis. Three-day-old Arabidopsis seedlings were grown for 12 h on a Murashige and Skoog standard medium (control) or standard medium supplemented with Suc, Glc, or Fru, at concentrations ranging from 7.5 to 90 mm. mRNA accumulation has been analyzed by real-time RT-PCR. Data (averaged transcript level from two biological replicates) were visualized using Heatmapper Plus software (http://bbc.botany.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi). Each treatment is represented by a row of colored boxes, and each sugar concentration is represented by a single column. Data are expressed as fold change (1 = control). Effects of sugars on gene expression range from pale to saturated yellow. Black indicates no change in gene expression. The value for image contrast was set to automatic for inductions lower than 500-fold, while it was fixed at 500 for induction higher than 500-fold.
In Vivo Sugar Modulation of Flavonoid and Anthocyanin Synthesis Genes
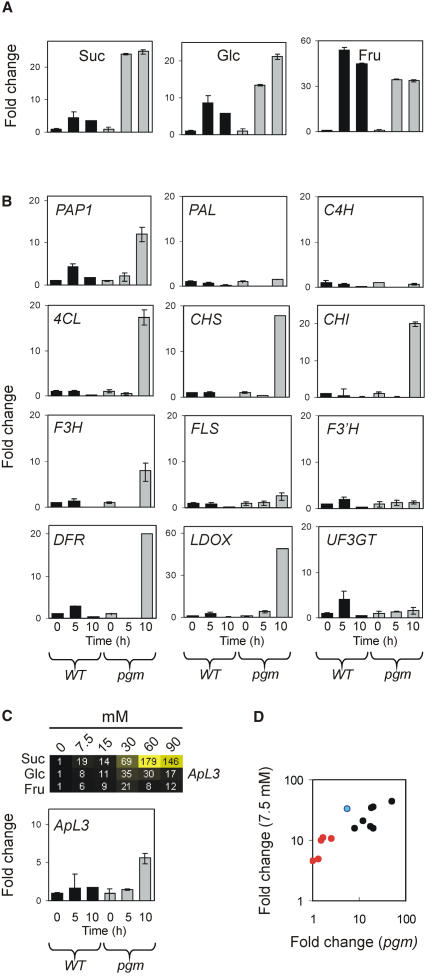
The pgm Arabidopsis mutant has a defect in the plastidial pgm gene hampering starch synthesis in the chloroplasts. Therefore, the mutant is starchless but accumulates high levels of soluble sugars as a consequence of its photosynthetic activity during the day (Caspar et al., 1985). Assuming that the experiments performed by treating seedlings with exogenous sugar reflect the ability of the plant to sense the endogenous sugars level, one would expect that the pgm mutant would show a sugar-controlled induction of genes, as soon as the endogenous sugar level increases beyond a certain threshold. Leaves were collected from plants at the rosette stage before beginning the light treatment (0 h), after 5 h under light (5 h), and after 10 h under light (10 h). The pgm mutant exhibits a clear increase in its Suc and Glc levels (up to 20-fold), whereas the wild type shows a lower diurnal fluctuation in Suc and Glc levels (Fig. 6A). The level of Fru increases almost equally in both the wild type and pgm mutant (Fig. 6A). The highest sugar level was measured in the leaves of the pgm mutant, 10 h after the beginning of the light treatment, with a measured concentration of about 10 mm soluble sugars (Suc + Glc + Fru). The expression of several of the flavonoid and anthocyanin synthesis genes increased about 10- to 20-fold after the 10-h light treatment in the pgm leaves but not in the wild-type leaves (Fig. 6B), suggesting that the in vivo expression of PAP1, 4CL, CHS, CHI, F3H, DFR, and LDOX is modulated by the rapid increase in sugar concentration observed in the pgm mutant (Fig. 6A). To confirm that the increases observed in the mRNA level of several of the genes studied is indeed the consequence of in vivo sugar sensing, we used the expression of the ApL3 gene, known to be sugar modulated (Sokolov et al., 1998), as a marker of sugar signaling occurring in leaves. The ApL3 mRNA level increases in response to Suc, with a limited induction by Glc and Fru (Fig. 6C, heatmap), and is induced in vivo only in the pgm mutant (Fig. 6C, pgm, time 10 h).
Figure 6.
Expression of flavonoid and anthocyanin synthesis genes in leaves of Arabidopsis wild type (WT) and pgm mutant. Leaves were collected from plants at the rosette stage before the beginning of the light treatment (0 h), after 5 h under light (5 h), and after 10 h under light (10 h). The results are means ± sd (n = 3). A, Changes in sugar concentration in leaves collected from wild-type and pgm mutant plants. Suc, Glc, and Fru were quantified, and changes in their amount were calculated (fold change 1 = time 0 h). B, mRNA accumulation was analyzed by real-time RT-PCR, and changes (fold change) in their amount were calculated (fold change 1 = time 0 h). C, Effect of sugars on ApL3 mRNA level. Arabidopsis seedlings were grown and mRNA analyzed by real-time RT-PCR as described in Figure 5. Data are expressed as fold change (fold change 1 = control). Effects of sugars on gene expression range from pale to saturated yellow (heatmap; see Fig. 5 legend for details). Black indicates no change in gene expression. The value for image contrast was set to automatic. The histogram shows ApL3 mRNA accumulation in leaves of Arabidopsis wild type and pgm mutant. mRNA was analyzed by real-time RT-PCR, and changes (fold change) in their amount were calculated (fold change 1 = time 0 h). D, Scatter plot showing the induction observed in pgm (data from Fig. 6, B and C) plotted against the induction by 7.5 mm sugars (data from Fig. 5). Red dots identify PAL, C4H, FLS, F3′H, and UF3GT. The blue dot identifies ApL3. Black dots identify PAP1, 4CL, CHS, CHI, F3H, DFR, and LDOX.
The comparison of the response of flavonoid and anthocyanin synthesis genes to low sugar concentration (7.5 mm; see Fig. 5) with their induction in the pgm mutant (Fig. 6B) indicates that the genes, which do not show any induction in the pgm mutant (PAL, C4H, FLS, F3′H, and UF3GT), are those showing a lower induction by the lowest sugar concentration tested (7.5 mm; Fig. 5). When the induction observed in pgm is plotted against the induction by 7.5 mm sugars (data from Fig. 5), it is possible to observe that all the genes lacking induction in the pgm mutant are grouped together and are those showing a lower induction by 7.5 mm exogenous sugars (Fig. 6D, red dots).
DISCUSSION
The expression of anthocyanin biosynthetic genes in grape berry skin appears to be highly coordinated during berry development (Boss et al., 1996), and the expression of grape DFR is responsive to Suc (Gollop et al., 2002). Since sugars accumulate during grape berry development (Boss et al., 1996), it is tempting to speculate that sugars are endogenous triggers modulating the expression of anthocyanin biosynthetic genes, possibly through the involvement of sugar-modulated regulatory genes. The ectopic expression of the transcription factor PAP1 (also AtMYB75) and of the related gene PAP2 results in an enhanced expression of the flavonoid biosynthetic genes PAL, CHS, and DFR (Borevits et al., 2000). The DFR expression is also under the control of TT2, interacting with TT8 (Nesi et al., 2001). Furthermore, the functional TRANSPARENT TESTA GLABROUS 1 (TTG1), encoding a WD40 repeat protein, is required for the normal expression of the DFR anthocyanin gene (Walker et al., 1999). AtMYB4 is another player in the biosynthesis of anthocyanins down-regulating C4H and 4CL genes (Jin et al., 2000), and ANTHOCYANINLESS 2 (ANL2) is involved in the accumulation of anthocyanins in subepidermal tissues (Kubo et al., 1999).
Our results suggest that sugars act as signaling molecules, activating the PAP1 gene by means of a Suc-specific signaling pathway. This is supported by the following experimental evidences. (1) Only PAP1 mRNA level was strongly Suc inducible, as previously suggested by Kranz et al. (1998), whereas other players involved in the anthocyanin biosynthesis regulation were unaffected by the treatment with Suc (see Supplemental Table I). (2) Suc, but neither Glc nor Fru, increases the mRNA level of PAP1 and of anthocyanin biosynthetic genes (Fig. 5, DFR, LDOX, UF3GT) and triggers an increased anthocyanin synthesis (Fig. 4A). (3) The effects of Suc on PAP1 mRNA level are unrelated to a generic osmotic response (Fig. 3B). We cannot, however, rule out the possibility of an involvement of other regulatory genes, since the sugar response expression pattern (Fig. 5) reveals that the response of genes upstream of DFR is clearly distinct from the response of PAP1.
A metabolite could control a pathway through selected steps. Regarding sugar regulation of the anthocyanin pathway, the effect is likely achieved through the last few steps, which are the most sensitive to sugar and show selective response to Suc (Fig. 5). Only Suc can elicit a clear increase in the anthocyanin content of Arabidopsis seedlings (Fig. 4A), and this is likely due to the induction, which is highly specific for Suc, of DFR, LDOX, and UF3GT (Fig. 5). This correlates nicely with the disappearance of naringenin chalcone from the seedling hypocotyl and its replacement with anthocyanins (Fig. 1). Gollop et al. (2001, 2002) reported that the induction of anthocyanin biosynthesis genes by sugars is rather unspecific, and Glc, Fru, and Suc treatments result in the induction of DFR and ANS (LDOX) in grape. The sugar-sensing mechanisms operating in grape are poorly studied, and it is reasonable to assume that they might be distinct from the sugar-sensing mechanisms active in Arabidopsis. Seedlings younger than 2 d show some degree of sensing unspecificity, and also Glc is able to induce DFR, LDOX, and UF3GT (data not shown), though not with the hundred-fold induction observed in the 3-d-old seedlings used in our experiments. Furthermore, it is worth outlining that the effects of Glc treatments longer than 12 h partly mirror the effects obtained with Suc, and feeding Arabidopsis seedlings with radiolabeled Glc reveals that this monosaccharide is readily converted into Suc within a few hours (data not shown).
As low as 7.5 mm Suc is enough to induce the mRNA accumulation of most of the genes studied, indicating that the sensing mechanism is compatible with physiological Suc concentrations and is likely to be operating in vivo. The expression of PAP1 shows a characteristic fluctuation during dark/light periods, the higher expression being reached around midday (Harmer et al., 2000), which is confirmed by our analysis of wild-type Arabidopsis leaves (Fig. 6B, PAP1; also observed in the Benshime ecotype, data not shown). The increase in sugar content during the light treatment peaks at 5 h in the wild type, and it is tempting to speculate that the PAP1 expression is sugar modulated in vivo. The expression of most of the genes involved in the flavonoid and anthocyanin biosynthesis is, however, unaffected in wild-type Arabidopsis leaves during the light treatment, a likely consequence of the limited fluctuation in the leaf content of Suc and Glc (Fig. 6A). The level of soluble sugars in wild-type Arabidopsis leaves increases from about 1 mm at time = 0 h to a maximum of 6 mm (reached at time = 5 h), whereas the pgm mutant shows a rise in soluble sugars from the very low level of 0.4 mm (time = 0) to a maximum of 10 mm (time = 10 h). These values are compatible with the increase observed in the mRNA level of several genes in the pgm mutant after 10 h of growth under light (Fig. 6B), as well as with their threshold for induction by sugars (Fig. 5). Indeed, the induction of genes in the pgm mutant correlates with gene sensitivity to exogenous sugars (Fig. 6D), and the pattern of expression of the ApL3 gene, known to be sugar modulated, mirrors that of anthocyanin-related genes, by showing an up-regulation in the pgm mutant (Fig. 6, B and C). The suggestion that sugars play a role in the modulation of the anthocyanin synthesis pathway in leaves is consistent with the microarray analysis of pho3 adult leaves accumulating high levels of soluble sugars and showing higher levels of expression of genes coding for anthocyanin biosynthesis enzymes (Lloyd and Zakhleniuk, 2004); these genes exhibit a very low expression level in sugar-depleted pgm leaves at the end of the night (Thimm et al., 2004).
The description of the anthocyanin biosynthesis as specifically responsive to Suc acting as a signaling molecule allows us to include this physiological process among the other Suc-specific processes that have been described up to now (Dijkwel et al., 1996; Smeekens and Rook, 1997; Chiou and Bush, 1998; Lalonde et al., 1999; Rolland et al., 2002; Vaughn et al., 2002; Koch, 2004). Furthermore, these results provide evidence for the occurrence of an in vivo Suc-sensing mechanism, which modulates the anthocyanin biosynthesis in Arabidopsis. Further work is needed to elucidate the molecular basis of this process and the possible interactions with the hormonal signaling network.
MATERIALS AND METHODS
Plant and Growth Condition
Arabidopsis (Arabidopsis thaliana) seeds were sterilized for 7 min in 1.7% (v/v) bleach solution, incubated overnight in 4% plant preservative mixture (PPM; Plant Cell Technology). PPM contains two isothiazolone class biocides, namely, methylchloroisothiazolinone and methylisothiazolinone (Paul et al., 2001) in full-strength sterilized Murashige and Skoog salt solution with gentle shaking, abundantly rinsed in sterile water, and transferred into 2.5 mL of liquid growing media (Murashige and Skoog half-strength solution ± sugars) with 0.05% PPM in six-well plates. Plates were incubated in the darkness at 4°C for 2 d and finally transferred to continuous light (90 μm photons m−2) with gentle swirling for experiment time in a plant growth chamber at 22°C. Treatments were performed by adding sugar solution to selected wells and water to the control wells.
Probe Design and Preparation
PCR primers were designed to amplify the most specific region inside the Affymetrix target region (sequence alignment was checked by the ClustalW multiple sequence alignment program (version 1.7, June 1997; http://www.ebi.ac.uk/clustalw/). For the design of the primers, we used the free Web-interfaced software Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Oligonucleotides were purchased from MWG-Biotech. Primer sequences for each gene are listed in Supplemental Table II. Poly(A+) RNA was purified by Oligotex (Qiagen) from total RNA extracted from 4-d-old Arabidopsis seedling incubated for 6 h in the presence of 90 mm Suc. About 150 ng of purified poly(A+) RNA was reverse transcribed with random primers by Improm-II (Promega) for 1 h at 42°C. PCR amplification on 15 ng cDNA (or 150 ng genomic DNA for intronless probed region) was performed with 400 nm specific primers and 2× PCR MIX (Promega). PCR conditions were as follows: 94°C for 2 min, 30 PCR cycles; 94°C 45 s, primers annealing temperature (see Supplemental Table II) 45 s; and 72°C for 45 s with a final extension of 8 min at 72°C.
RNA Isolation and Gel Blots
RNA extraction was performed using the aurintricarboxylic acid method as described previously (Perata et al., 1997). The amount of total RNA loaded per lane for electrophoresis was 20 μg. RNA was electrophoresed on 1% (w/v) agarose glyoxal gels, and blotted on nylon membrane (BrightStar-Plus) using the procedure suggested by the manufacturer. Membranes were prehybridized and hybridized using the NorthernMax-Gly kit (Ambion). Radiolabeled probes were prepared from gel-purified cDNAs by random primer labeling (Takara Chemicals) with [α32P]dCTP. Equal loading was checked by reprobing with an rRNA cDNA probe (data not shown). RNA blots were scanned using a Cyclone Phosphoimager (Packard Bioscience, Perkin Elmer). mRNA level was quantified using the Optiquant software (Packard Bioscience, Perkin Elmer).
RNA Isolation, cRNA Synthesis, and Hybridization to Affymetrix GeneChips
Total RNA was extracted from the seedling samples using the Ambion RNAqueous kit (Ambion). RNA quality was assessed by agarose gel electrophoresis and spectrophotometry. RNA was processed for use on Affymetrix Arabidopsis ATH1 GeneChip arrays as described previously (Loreti et al., 2005). Hybridization, washing, staining, and scanning procedures were performed by Biopolo (University of Milano Bicocca, Italy) as described in the Affymetrix technical manual. Expression analysis via the Affymetrix Microarray Suite software (version 5.0) was performed with standard parameters. Two independent, replicated experiments were performed for each experimental condition, and the output of the Affymetrix Microarray Suite software for each independent experiment was subjected to further analysis by using Microsoft Excel. Signal values (indicating the relative abundance of a particular transcript) and detection call values (indicating the probability that a particular transcript is present) were generated by Microarray Analysis Suite 5.0 software. Probe pair sets (genes) called Absent were removed from subsequent analyses. Furthermore, genes with Absent for the detection value in the baseline data and Decrease for the change call were excluded from the list. Similarly, genes with Absent for the detection call in the experimental data and Increase for the change value were also excluded from the list. Differences in transcript abundance, expressed as signal log ratio, were calculated using the Microarray Analysis Suite 5.0 software change algorithm. Signal log ratio was assumed to be correct only if the corresponding change call indicated a significant change (I, Increase; D, Decrease; generated by Microarray Analysis Suite 5.0 software). Expression data were filtered to select only genes showing a coinciding change call in the two biological replicates samples for each experimental condition.
Real-Time RT-PCR
RNA was extracted from seedlings grown on Murashige and Skoog 0.5× solution (control) or in the same medium supplemented with 90 mm sugars as indicated in figure legends. Total RNA, extracted with the RNAqueous kit (Ambion) according to the manufacturer's instruction, was subjected to a DNase treatment using the TURBO DNA free kit (Ambion). Two micrograms of each sample were reverse transcribed into cDNA with the high-capacity cDNA archive kit (Applied Biosystems). Real-time PCR amplification was carried out with the ABI Prism 7000 sequence detection system (Applied Biosystems), using the primers described in Supplemental Table III. Ubiquitin10 (UBQ10) was used as endogenous control. Taqman probes specific for each gene were used. Probe sequences are reported in Supplemental Table III. PCR reactions were carried out using 50 ng of cDNA and TaqMan Universal PCR master mix (Applied Biosystems) following the manufacturer's protocol. Relative quantitation of each single gene expression was performed using the comparative CT method as described in the ABI PRISM 7700 Sequence Detection System User Bulletin #2 (Applied Biosystems).
Anthocyanins Quantitation
Arabidopsis seedling extraction was performed as described by Ronchi et al. (1997) with minor modifications. In brief, seedlings were ground in 1 volume HCl 1% (v/v) in methanol with the addition of two-thirds volume of distilled water. Extracts were recovered, and 1 volume of chloroform was added to remove chlorophylls through mixing and centrifugation (1 min at 14,000g). Anthocyanins containing aqueous phase were recovered and absorption was determined spectrophotometrically (A535). Mean values were obtained from three independent replicates.
Flavonoid Staining
Flavonoid staining was performed as described by Peer et al. (2001). Three-day-old Arabidopsis seedlings, treated for 72 h with 90 mm Suc or minimal Murashige and Skoog medium (control), were stained for 5 to 15 min using saturated (0.25%, w/v) diphenylboric acid-2-aminoethyl ester (DPBA) with 0.005% Triton X-100, and were visualized with an epifluorescence microscope equipped with an fluorescein isothiocyanate filter (excitation 450–490 nm, suppression LP 515 nm). Photographs of seedlings were taken using color slide film (Kodak Elite, ASA 400) after 5 min of staining.
Sugar Analysis
Samples were rapidly frozen in liquid nitrogen and ground to a powder. Samples were then extracted and assayed by coupled-enzymatic assay methods measuring the increase in A340 as described by Guglielminetti et al. (1995).
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Silvia Gonzali for performing the sugar analysis and for helpful discussion.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Pierdomenico Perata (p.perata@sssup.it).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.072579.
References
- Baier M, Hemmann G, Holman R, Corke F, Card R, Smith C, Rook F, Bevan MW (2004) Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol 134: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti AK, Khurana JP (1997) Mutants of Arabidopsis as tools to understand the regulation of phenylpropanoid pathway and UVB protection mechanisms. Photochem Photobiol 65: 765–776 [DOI] [PubMed] [Google Scholar]
- Borevits JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P (2004) Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol 7: 202–209 [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C (1985) Alterations in growth, photosynthesis and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in a chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, Bush DR (1998) Sucrose is a signal molecule in assimilate partitioning. Proc Natl Acad Sci USA 95: 4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Schwinn KE (2003) Transcriptional regulation of secondary metabolism. Funct Plant Biol 30: 913–925 [DOI] [PubMed] [Google Scholar]
- Dijkwel PP, Kock P, Bezemer R, Weisbeek PJ, Smeekens SCM (1996) Sucrose represses the developmentally controlled transient activation of the plastocyanin gene in Arabidopsis thaliana seedlings. Plant Physiol 110: 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollop R, Even S, Colova-Tsolova V, Peri A (2002) Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. J Exp Bot 53: 1397–1409 [PubMed] [Google Scholar]
- Gollop R, Farhi S, Peri A (2001) Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Sci 161: 579–588 [Google Scholar]
- Guglielminetti L, Perata P, Alpi A (1995) Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiol 108: 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch LB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cornineill E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Koornneef M (1981) The complex syndrome of ttg mutants. Arabidopsis Inf Serv 18: 45–51 [Google Scholar]
- Koornneef M (1990) Mutations affecting the testa colour in Arabidopsis. Arabidopsis Inf Serv 27: 1–4 [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16: 263–276 [DOI] [PubMed] [Google Scholar]
- Kubo H, Peeters AJM, Aarts MGM, Pereira A, Koornneef M (1999) ANTHOCYANINLESS2, a homeobox gene affecting anthocyanins distribution and root development in Arabidopsis. Plant Cell 11: 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Franck M (1990) Cloning and characterization of a cold-inducible and ABA-inducible Arabidopsis gene. Plant Mol Biol 15: 137–144 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Boles E, Hellmann H, Barker L, Patrick JW, Frommer WB, Ward JM (1999) The dual function of sugar carriers: transport and sugar sensing. Plant Cell 11: 707–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Alpi A, Perata P (2000) Glucose and disaccharide-sensing mechanisms modulate the expression of α-amylase in barley embryos. Plant Physiol 123: 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of Arabidopsis mutant, pho3. J Exp Bot 55: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, et al (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanins biosynthesis, modification, and transport. Plant Cell 15: 1689–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Tanaka H, Ohme-Takagi M (2004) Suppression of the biosynthesis of proanthocyanidin in Arabidopsis by a chimeric PAP1 repressor. Plant Biotechnol J 2: 487–493 [DOI] [PubMed] [Google Scholar]
- Mita S, Hirano H, Nakamura K (1997) Negative regulation in the expression of a sugar-inducible gene in Arabidopsis thaliana. Plant Physiol 114: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kàlmàn Z, Stankovic-Stangeland B, Bakò L, Mathur J, Ökrész L, Stabel S, et al (1998) Pleiotropic control of glucose response by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev 12: 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon L, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes a R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M-A, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K (2001) Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol 127: 252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KI, Choi JD, Hoshino A, Morita Y, Lida S (2004) An intragenic tandem duplication in a transcriptional regulatory gene for anthocyanins biosynthesis confers pale-colored flowers and seeds with fine spots in Ipomea tricolor. Plant J 38: 840–849 [DOI] [PubMed] [Google Scholar]
- Paul A-L, Semer C, Kucharek T, Ferl RJ (2001) The fungicidal and phytotoxic properties of benomyl and PPM in supplemented agar media supporting transgenic Arabidopsis plants for a space shuttle flight experiment. Appl Microbiol Biotechnol 55: 480–485 [DOI] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol 126: 536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perata P, Matsukura C, Vernieri P, Yamaguchi J (1997) Sugar repression of a gibberellin-dependent signaling pathway in barley embryos. Plant Cell 9: 2197–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchi A, Farina G, Gozzo F, Tonelli C (1997) Effects of a triazolic fungicide on maize plant metabolism: modifications of transcript abundance in resistance-related pathways. Plant Sci 130: 51–62 [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Smeekens S, Rook F (1997) Sugar sensing and sugar-mediated signal transduction in plants. Plant Physiol 115: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov LN, Déjardin A, Kleczkowski LA (1998) Sugars and light/dark exposure trigger differential regulation of ADP-glucose pyrophosphorylase genes in Arabidopsis thaliana (thale cress). Biochem J 336: 681–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Ohshima T, Naito S, Chino M, Komeda Y (1991) Sugar-dependent expression of the CHS-A gene for chalcone synthase from petunia in transgenic Arabidopsis. Plant Physiol 97: 1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn MW, Harrington GN, Bush DR (2002) Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc Natl Acad Sci USA 99: 10876–10880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitrac X, Larronde F, Krisa S, Decendit A, Deffieux G, Merillon JM (2000) Sugar sensing and Ca2+-calmodulin requirement in Vitis vinifera cells producing anthocyanins. Phytochemistry 53: 659–665 [DOI] [PubMed] [Google Scholar]
- Vom Endt D, Kijne JW, Memelink J (2002) Transcription factors controlling plant secondary metabolism: What regulates the regulators? Phytochemistry 61: 107–114 [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray CJ (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D (2000) Regulation of flower pigmentation and growth: multiple signalling pathways control anthocyanin synthesis in expanding petals. Physiol Plant 110: 152–157 [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.