Abstract
The 14-3-3 family of proteins functions through protein:phosphoprotein interactions, the nature of which has been elucidated using x-ray crystallography. However, some key structural features in nonconserved regions have yet to be fully resolved, leaving open questions regarding the functional selectivity of 14-3-3 family members for diverse clients. In an effort to study surface accessible structural features in 14-3-3 containing macromolecular complexes and to illuminate important structure/function variations among the 14-3-3 isoforms, we determined the epitopes for three unique monoclonal antibodies (mAbs) developed against the Arabidopsis (Arabidopsis thaliana) G-box DNA:protein complex. The epitopes mapped to different loops in a phylogenetically important subset of the 13 14-3-3 family members. All three epitopes were on a common exposed face of complexed 14-3-3s. Two of the mAbs recognized linear sequences within loops 5 and 6, while the third mAb recognized 14-3-3 residues surrounding the pivotal medial Gly in the divalent cation-binding domain of loop 8, together with distal residue(s) in the putative dynamic 10th helix that has yet to be determined by crystallography. Gly at this loop 8 position is unique to nonepsilon 14-3-3 isoforms of the plant kingdom, suggesting that this region constitutes a plant-specific key functional 14-3-3 feature and highlighting that the loop 8 region is functionally significant. Mutagenesis of the medial amino acid in the loop 8 domain changed the flexibility of the C terminus and altered client peptide-binding selectivity, demonstrating the functional significance of the surface accessible, evolutionarily distinct loop 8 domain.
The key functional capacity of 14-3-3 proteins is their ability to bind to specifically phosphorylated target proteins, which enables 14-3-3 participation in many cellular-signaling roles. The general mechanism and structural basis of phospho-Ser/Thr binding has been described through x-ray crystallographic analyses of several diverse 14-3-3 isoforms, 14-3-3:phosphopeptide complexes, and a 14-3-3:target protein complex (Liu et al., 1995; Xiao et al., 1995; Yaffe et al., 1997; Petosa et al., 1998; Obsil et al., 2001; Wilker et al., 2005). However, in all of these analyses some structural features of the 14-3-3s, including divergent C-terminal and interhelical loop regions, have yet to be fully resolved, and little information is available regarding the structures of 14-3-3s in multimacromolecular complexes such as for DNA:protein associations.
The 14-3-3 families consist of groups of small, 30-kD, soluble, dimeric proteins separated into two major evolutionary groups, ɛ and non-ɛ. These groups are separated by a deep branch in the evolutionary tree, providing a basis for potential differences in fundamental structural or functional properties within the family (Ferl et al., 1994). Associated activities of 14-3-3 proteins range from organellar protein translocation to enzymatic regulation to transcriptional control (Pan et al., 1999; Sehnke et al., 2002a; Roberts, 2003). Analysis of the Arabidopsis (Arabidopsis thaliana) genome, for which the entire 14-3-3 family is known, predicts that potential plant 14-3-3 targets may account for up to 20% of the genome (Sehnke et al., 2002a) and may explain the large family size of plant 14-3-3s. Additionally, since the 14-3-3s bind phosphoproteins through a highly conserved core in the groove of the antiparallel helices and are experimentally interchangeable in several systems, it is believed that most 14-3-3 target specificity may reflect spatial and temporal 14-3-3 localization differences among isoforms. However, 14-3-3 isoform preference for specific target proteins both in vivo and in vitro (Bachmann et al., 1996b; Athwal et al., 2000; Rosenquist et al., 2000; Roberts and de Bruxelles, 2002; Sorrell et al., 2003; Alsterfjord et al., 2004; Bornke, 2005; Sinnige et al., 2005) argues that different 14-3-3s contain unique regions such as that of the highly variable termini and less conserved internal loops that may contribute to differentially regulate target binding of the various 14-3-3 isoforms. Recent in vivo 14-3-3 target peptide competition studies in Arabidopsis suggest 14-3-3 target specificity at the cellular level (Paul et al., 2005). Further, it has been established that the extreme C termini of 14-3-3s plays a role in client binding (Truong et al., 2002; Shen et al., 2003; Bornke, 2005; Wilker et al., 2005), although the exact mechanism is not fully understood. However, due to unresolved areas of published 14-3-3 crystal structures, the structural details of regions key to client interactions in general and to plant-specific interactions in particular remain unclear.
Some of the first identification of 14-3-3s in plants came about from their role as a partner in the G-box cis-acting DNA regulatory element-binding protein complex (Lu et al., 1992). Monoclonal antibodies (mAbs) generated against purified G-box complex (GBC) were tested in electrophoretic mobility supershift assays to provide probes for discovering the proteins present in the complex. These antibodies were used to immunoscreen cDNA expressing λ phage libraries, and 14-3-3 proteins were identified as the antigens (Lu et al., 1992). Since these antibodies recognized 14-3-3s complexed with other proteins and DNA (DeLisle and Ferl, 1990), they should cross-react with surface-accessible regions of the complexed 14-3-3s and therefore provide insight into 14-3-3 protein architecture while the 14-3-3s are part of a larger, multicomponent complex. In fact, preliminary characterization of a GBC mAb identified the trypsin-accessible C terminus as the general region recognized by that antibody (Lu et al., 1994b).
Using random peptide phage display, recombinantly expressed 14-3-3 truncations and site-directed mutagenesis, we have characterized the epitopes that are recognized by three supershifting GBC mAbs that recognize 14-3-3s. The epitopes are localized to three distinct regions of 14-3-3s and are present only in non-ɛ 14-3-3 family members. One particularly interesting epitope is central to the divalent cation-binding domain of 14-3-3s that is important for changing the structure of the C termini (Lu et al., 1994b; Athwal et al., 1998; Athwal and Huber, 2002) and represents a major divergence not only among plant 14-3-3s, but also between plant and animal 14-3-3s. Characterization of this difference that exists between 14-3-3s that are unique to plants and those that share similarities to animal isoforms is presented using site-directed mutagenesis, divalent cation-induced proteolytic protection experiments, and immobilized 14-3-3 binding experiments with phosphopeptides. This work provides evidence that all 14-3-3s bind divalent cations and in a way that affects the structure and function of the C terminus. But while all 14-3-3s bind cations to affect the C-terminal structure, those plant-specific isoforms recognized by the GBC mAb for loop 8 had an increased flexibility in the C terminus and an altered phosphopeptide-binding specificity.
RESULTS
Production of 13 Isoforms of Arabidopsis 14-3-3s for Testing Cross-Reactivity to GBC mAbs
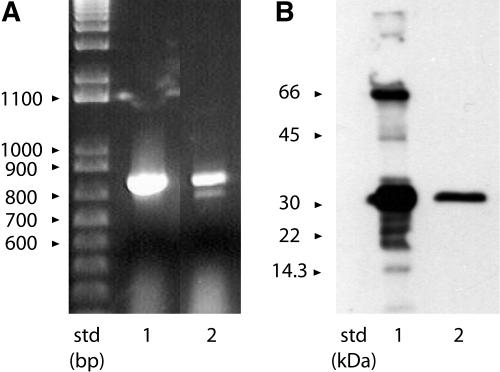
Since it is known that only one of the three GBC mAb cross-reacts with at least one Arabidopsis 14-3-3 (Lu et al., 1992), we wished to establish the cross-reactivity of all the mAbs against all of the Arabidopsis 14-3-3 isoforms. The Arabidopsis genome predicts 13 functional full-length 14-3-3 genes; however, to our knowledge, only 12 protein isoforms have been reported in the literature (Rosenquist et al., 2001; Sehnke et al., 2002b). We used reverse transcription (RT)-PCR with total RNA and specific primers to the predicted gene product to GF14π (GRF13), At1g78220.1 to search for the corresponding mRNA. The GF14π mRNA (GenBank accession no. AF543836) was present in both the leaves of 10-d-old plants and the green siliques of plants, grown under constant light (Fig. 1A). The sequence for GF14π matched the predicted protein coded for by At1g78220 locus and produced recombinant protein of a size consistent with that of other Arabidopsis 14-3-3s (Fig. 1B).
Figure 1.
Detection, isolation, and bacterial expression of Arabidopsis 14-3-3 GF14π. A, The cDNA for the Arabidopsis gene GRF13 (Rosenquist et al., 2001; Sehnke et al., 2002b) was isolated from leaf total RNA using RT-PCR (lane 1). GF14π transcript was also detected in RNA isolated from green siliques (lane 2). The cDNA sequence was deposited in GenBank with the accession number AF543836. B, Western analysis of E. coli expressed His-tagged GF14π. The coding region for GF14π was subcloned into pET15b and transformed into BL21 DE3 AI E. coli. The protein was induced with IPTG and purified with nickel-charged IMAC. Protein samples from the crude bacterial extract (lane 1) and the IMAC-purified GF14π (lane 2) were analyzed with SDS-PAGE/western analysis using antibodies to the His tag.
The cDNAs for ι and ο were produced from total RNA using RT-PCR with gene-specific primers corresponding to coding sequence identified previously (Rosenquist et al., 2001) and subcloned along with GF14π cDNA into the pET15b His-tag fusion expression vector for production of recombinant protein in Escherichia coli. Expression of GF14ι and ο was consistent with previous expression levels of the other Arabidopsis 14-3-3s (data not shown). GF14π was less soluble under standard expression conditions and required a modified induction protocol in BL21DE3 AI cells to produce sufficient protein that could be purified by immobilized metal affinity chromatography (IMAC; Fig. 1B). These proteins, in addition to Arabidopsis 14-3-3 GF14ω, φ, χ, ψ, ν, υ, κ, λ, ɛ, and μ also expressed as His-tagged fusion proteins (Wu et al., 1997a), purified easily using IMAC.
Specific Subsets of Arabidopsis 14-3-3 Isoforms Cross-React Differentially to GBC mAbs 2A3, 4B9, and 5D6
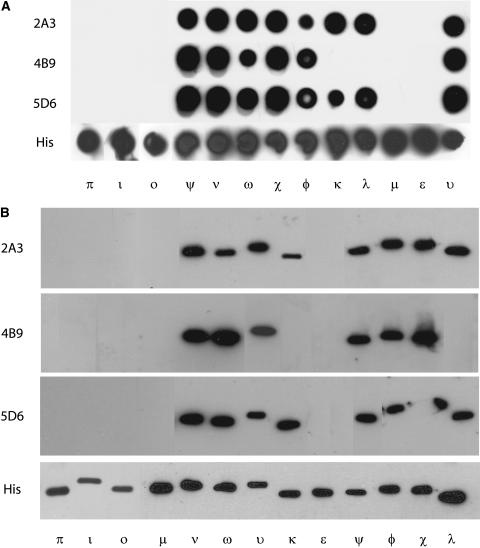
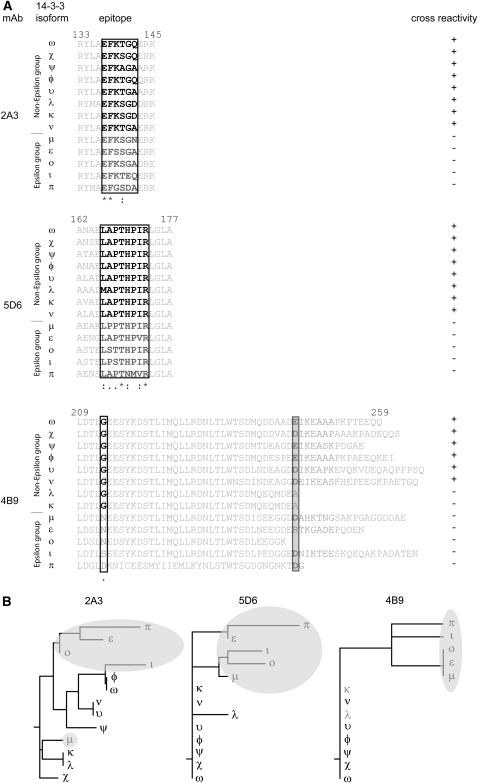
Samples of 1 μg of each isoform were transferred to nitrocellulose using a Minifold I Dot-Blot system (Schleicher & Schuell BioScience) and probed with GBC mAbs 2A3, 4B9, and 5D6 (Fig. 2A). None of the GBC mAbs recognized the ɛ group isoforms (ɛ, μ, ι, ο, and π); however, GBC mAbs 2A3 and 5D6 cross-reacted with the entire non-ɛ group (ω, φ, υ, ν, ψ, χ, κ, and λ). GBC mAb 4B9 recognized the non-ɛ members with the exception of κ and λ.
Figure 2.
Identification of 14-3-3 isoforms recognized by three mAbs generated from 14-3-3/G-box DNA-binding complexes using 13 recombinant Arabidopsis 14-3-3 proteins. A, GBC mAbs detect a subset of nondenatured recombinant Arabidopsis 14-3-3 proteins. Samples of each purified recombinant Arabidopsis 14-3-3 protein were transferred to nitrocellulose with a dot-blot apparatus. After blocking, the blot was probed with GBC mAb 2A3, 4B9, and 5D6. Cross-reactivity was detected with anti-mouse horseradish peroxidase conjugate and chemiluminescence. B, GBC mAbs detect a subset of denatured recombinant Arabidopsis 14-3-3 proteins. Aliquots of 10 μg of each recombinant Arabidopsis 14-3-3 proteins were denatured in reducing SDS sample buffer prior to electrophoresis. After transferring electrophoresed proteins to nitrocellulose and blocking, the blot was probed with GBC mAb 2A3, 4B9, and 5D6. Immuno-cross-reactivity was detected with anti-mouse horseradish peroxidase conjugate and chemiluminescence.
To test the ability of the antibodies to detect denatured 14-3-3s, we first denatured the samples of the recombinant 14-3-3s in reducing SDS sample buffer prior to electrophoresis and western analysis (Fig. 2B). Probing the blots with the GBC mAbs produced similar results as native dot-blot analysis, suggesting the epitopes are either not dependent upon structural conformation or that those regions of the proteins can refold sufficiently for antibody recognition.
Epitope Mapping Using Random Peptide Phage Display with GBC mAbs Identifies C-Terminal Regions as Epitopes for GBC mAbs 5D6 and 4B9
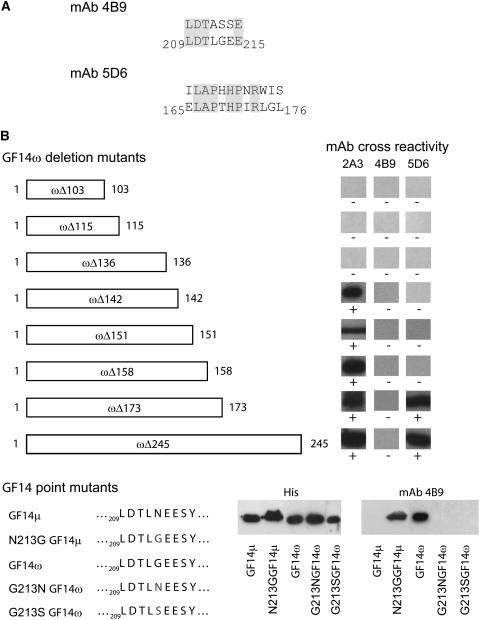
Screening commercially available random dodecamer, heptamer, and Cys cyclic heptamer peptide phage display libraries with GBC mAbs produced a consensus binding sequence for 4B9 and 5D6 (Fig. 3A). The GBC mAb 5D6 clone matched sequence with 75% identity over eight internal amino acids of a 12-amino acid display peptide that corresponded to GF14ω sequence Leu-166 to Arg-173. The GBC mAb 4B9 consensus binding sequence matched with 71% identity over seven amino acids of GF14ω sequence 209 to 215. The medial Gly is missing from the phage display peptide; however, it has been replaced by a Ser, and the following residue is also a Ser, thereby allowing for similar behavior as a Gly residue because of hydrogen binding (Adamian and Liang, 2002; Dawson et al., 2002).
Figure 3.
GBC mAbs epitopes map to three distinct discontinuous sequences within the Arabidopsis 14-3-3s. A, GBC mAbs were used to screen phage display heptamer and dodecamer peptide libraries. Screening of 1 × 1011 clones identified peptides with a consensus sequence corresponding to Arabidopsis GF14 sequences for two of the antibodies. B, Truncations of GF14ω were used to localize the 2A3 epitope and confirm the phage display identification of the 4B9 and 5D6 epitopes. PCR was used to generate C-terminal truncation constructs of GF14ω, which were examined by western analysis. C, Mutation of GF14μ and GF14ω were used to further confirm and establish a key residue for the 4B9 GBC mAb epitope. Strand overlap extension PCR was used with Arabidopsis GF14μ to produce an Asn to Gly mutation and GF14ω to produce a Gly to Ser or Asn mutation at residue 213 in the constructs, respectively.
Western Analysis with Truncated GF14ω Constructs and GBC mAbs Identifies Epitopes for GBC mAb 2A3 and Confirms Results of Phage Display for GBC mAbs 5D6 and 4B9
Since no consensus phage display sequences were obtained using GBC mAb 2A3, we used SDS-PAGE western analysis of recombinant GF14ω truncations (Fig. 3B). The GBC mAb 2A3 cross-reacted with the C-terminally truncated GF14ω stopping at residue 142 or longer but not with shorter proteins terminating at residues 136, 115, or 103. Therefore the epitope for GBC mAb 2A3 would be from Glu-136 to Gln-142 on GF14ω. Western analysis with GBC mAb 5D6 and the GF14ω truncations confirmed the results obtained with phage display. Similar analysis with GBC mAb 4B9 supports the phage display results in that Gly-213 was important for epitope formation. However, since 4B9 antigenicity was lost by the removal of the GF14ω C-terminal extension (GF14ωΔ245), an extension that is not present in the nonantigenic Gly-213-containing GF14λ and κ isoforms, the complete 4B9 epitope must rely upon residues in this C-terminal extension region as well as the divalent cation-binding domain.
Western Analysis with Mutated N213G GF14μ, G213N GF14ω, G213S GF14ω, and GBC mAb 4B9 Confirms Phage Display Results
The phage display results for GBC mAb 4B9 demonstrated that the sequence from Leu-209 to Glu-215 was important for antibody binding. Given that this mAb did not recognize ɛ group members but recognized only most of the non-ɛ members, and Gly-213 is unique to non-ɛ group members, it seemed likely that the conserved Gly-213 was important for binding of the actual 14-3-3. To test this theory we mutated GF14μ Asn-213 to Gly (N213G GF14μ) and checked the ability of GBC mAb 4B9 to detect the protein in an SDS western system (Fig. 3C). The N213G GF14μ mutant protein was detected by GBC mAb 4B9, indicating that Gly-213 is critical for formation of the epitope. To test if the conversion of Gly-213 from an antigenic isoform to that of Asn or Ser found in nonantigenic isoforms would abolish the epitope, we mutated GF14ω Gly-213 to Asn (G213N GF14ω) or Ser (G213S GF14ω) and again checked for GBC 4B9 SDS western cross-reactivity (Fig. 3C). Neither G213N GF14ω nor G213S GF14ω was recognized by the antibody.
Location of the Three GBC mAb Epitopes in Secondary Structural Elements of GF14ω
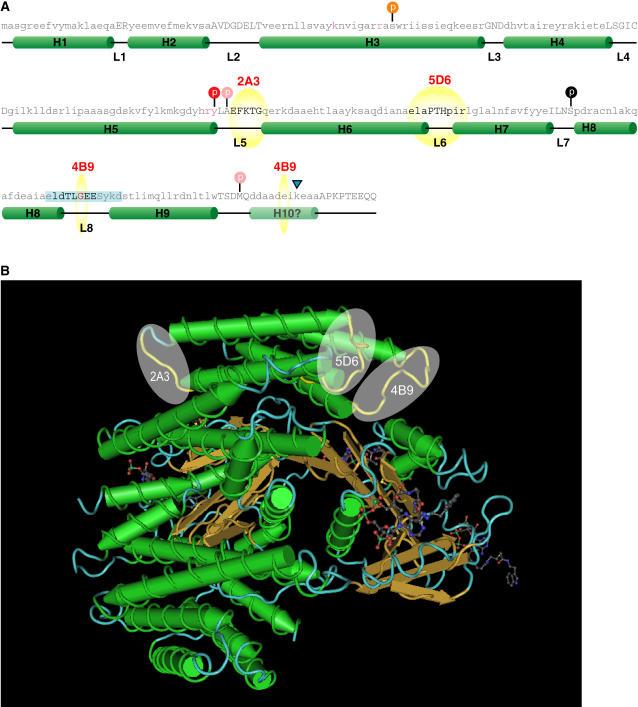
The sequence of the central core of 14-3-3s is highly conserved and as such allows for precise alignment of the Arabidopsis 14-3-3s with known 14-3-3 three-dimensional structures. This allows for mapping of the GBC mAb epitopes on the structural elements. All three GBC mAbs epitopes exist within loop regions of the 14-3-3s (Fig. 4A). The GBC mAb 2A3 epitope resides in loop 5, 5D6 in loop 6, and 4B9 in loop 8 that also contains the divalent cation-binding domain.
Figure 4.
GBC mAb epitopes map to loops residing on a common surface on 14-3-3s. A, Primary and secondary structural elements of GBC mAb epitopes on Arabidopsis 14-3-3 ω. The sequence of GF14ω was superimposed on the secondary structural elements from the crystal structures of tobacco and human 14-3-3s based on sequence alignment. Helices are represented by green cylinders with the helix number (Hn where n = 1–10) and loops as black lines with the designation L (loop number). The mapped GBC mAb epitopes are indicated by yellow spheres and bold text. The location of potential posttranslational events based upon the 14-3-3 literature and using sequence alignment is represented by a colored circle on top of a vertical black line. Red circles indicate a decrease in binding with the modification, while orange implies decrease in dimerization. Pink circles indicate that the specific residue is required for the modification and is not conserved in plant 14-3-3s. The residues important for phospho-Ser target ligand coordination are in red. Tryptic cleavage of GF14ω is indicated by the blue triangle. B, The location of the mAb epitopes cluster on the 14-3-3 three-dimensional structure. Using the x-ray diffraction derived three-dimensional model of the human 14-3-3 ζ complexed with AANAT PDB accession number 1IB1, the regions corresponding to the determined GBC mAb epitopes were mapped and are highlighted in yellow. The regions corresponding to mAb epitopes are represented as yellow spheres.
The only 14-3-3 target protein complex three-dimensional model that exists is the structure of the human 14-3-3 ζ and phosphorylated serotonin N-acetyltransferase (AANAT; Obsil et al., 2001). Superimposing the GBC mAb epitopes upon the structure maps to regions that are away from the complexed protein (Fig. 4B) and as such should not be hidden while the 14-3-3 is complexed.
GF14μ Displays Calcium-Induced Structural Changes
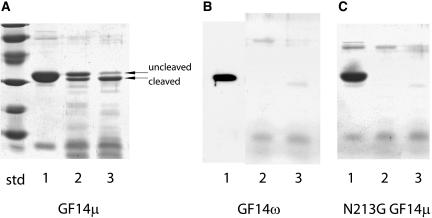
Calcium (Ca2+) and magnesium (Mg2+) cations are both bound by GF14ω and result in indistinguishable structural changes in the extreme C terminus (Lu et al., 1994b; Shen et al., 2003); however, to our knowledge this has not been demonstrated for any other 14-3-3s. While other non-ɛ plant 14-3-3s possessing conserved metal ligand residues and a medial Gly thought important for bending or kinking in this region should bind metal, it was not clear if 14-3-3s lacking this medial Gly would bind divalent cations nor possess C termini capable of structural changes. Proteolytic protection experiments with GF14μ and endoproteinase Lys-C in the presence of Ca2+ and without Ca2+ clearly demonstrate Ca2+-induced structural changes (Fig. 5A). Identical proteolytic conditions applied to GF14ω totally removed the C terminus with subsequent complete protein degradation (Fig. 5B), demonstrating a higher sensitivity to the protease by GF14ω as compared to GF14μ.
Figure 5.
Proteolytic analysis of GF14μ, GF14ω, and N213G GF14μ mutant structures demonstrates increased flexibility for the medial Gly containing divalent cation-binding domain. Aliquots of purified, bacterially expressed GF14μ (A), GF14ω (B), and N213G GF14μ (C) were digested at 25°C with endoproteinase Lys-C at an enzyme to substrate ratio of 1:100 in the presence (A, B, and C, lane 2) or absence (A, B, and C, lane 3) of 5 mm CaCl2. Denatured samples of 0 h digest (A, B, and C, lane 1), 4 h digest with CaCl2 (A, B, and C, lane 2), and 4 h digest without CaCl2 (A, B, and C, lane 3) were electrophoresed and visualized with Coomassie stain. Uncleaved and cleaved proteins are indicated. Divalent cation induced structural changes were observed; however, replacement of Gly for Asn in the divalent cation-binding loop increased sensitivity to proteolytic activity suggesting increased flexibility.
N213G GF14μ Displays Increased Protease Sensitivity
Treatment of N213G GF14μ with endoproteinase Lys-C protease with Ca2+ under the same conditions used for GF14μ and ω resulted in a dramatically increased sensitivity to the protease (Fig. 5C) relative to GF14μ and very similar results to that of GF14ω with endoproteinase Lys-C (Fig. 5B), namely cleavage of the C terminus followed by further protein degradation.
GF14μ and N213G GF14μ Preferentially Bind Different Phosphopeptide Targets
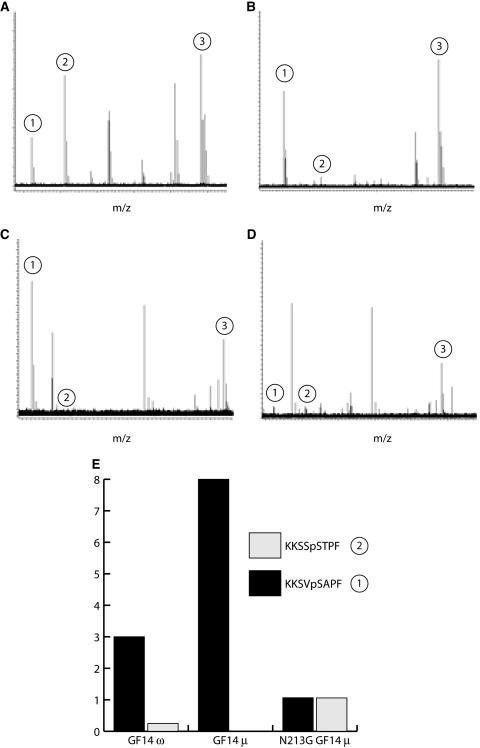
Isoform-specific binding by 14-3-3s is thought to depend upon unique regions of the 14-3-3s, namely extreme termini and nonconserved loops. The exact mechanism for binding is not known but may involve C-terminal autoregulation (Truong et al., 2002; Shen et al., 2003; Obsilova et al., 2004). Since non-ɛ group 14-3-3s and N213G GF14μ exhibit increased flexibility of this region in proteolytic assays, we tested whether these proteins could bind similar targets using a panning protocol that flowed degenerative octamer phosphopeptides derived from Arabidopsis nitrate reductase over ZipTip immobilized GF14ω, GF14μ, and N213G GF14μ followed by elution of bound peptides. The binding conditions employed Mg2+ cation concentrations as per previous 14-3-3/nitrate reductase interaction studies (Shen et al., 2003). Mass spectral analysis of three consistently bound peptides showed a dramatically different profile for N213G GF14μ compared to GF14μ or GF14ω (Fig. 6, A–D). To highlight the differential binding of N213G GF14μ, the peak heights of two of the bound peptides, KKSVpSAPF and KKSSpSTPF (Fig. 6), were normalized against the peak height for the third peptide KKSVpSYPF (Fig. 6), which remained fairly uniformly bound by all three 14-3-3s. The N213G mutation in GF14μ resulted in a reduced selection of peptide KKSVpSAPF and an increase in the selection of peptide KKSSpSTPF (Fig. 6).
Figure 6.
Mutation of medial divalent cation-binding residue Asn-213 to Gly alters the target-binding specificity of GF14μ. N213G GF14μ, GF14μ, or GF14ω were immobilized on ZipTips and used to pan a mixture of synthetic octamer phosphopeptides based on the Arabidopsis nitrate reductase target sequence. The peptide mixture and the 14-3-3 bound peptides were analyzed by direct infusion ESI-FTICR-mass spectrometry. The resultant profiles are presented with the mass-to-charge ratio plotted on the x axis and relative signal on the y axis. A, Complete phosphopeptide mixture. B to D, Preferential selection of three specific peptides for the GF14ω (B), GF14μ (C), and N213G GF14μ (D) that correlated to the presence or absence of a medial Gly in the divalent cation-binding domain in loop 8. The three peptides retained by all three 14-3-3s were KKSVpSAPF (1), KKSSpSTPF (2), and KKSVpSYPF (3). Peptides not retained by all samples were excluded from analysis. E, The binding of peptides KKSVpSAPF (1) and KKSSpSTPF (2) are compared relative to the binding of peptide KKSVpSYPF (3) to highlight the binding differential among isoforms.
DISCUSSION
Arabidopsis Has Thirteen Expressed 14-3-3 Isoforms
Previous studies had identified 12 expressed members of the Arabidopsis 14-3-3 family (Wu et al., 1997b; Rosenquist et al., 2001). However, genomic support for highly divergent or truncated 14-3-3 genes or pseudogenes exists. Isolation of the cDNA for the 13th isoform, GF14π, and initial biochemical characterization reveal that this isoform is rather rare and unique in the uncharacteristically low solubility for a normally extremely soluble family of proteins. The gene organization and primary sequence argue that GF14π is an ɛ member. However because of stretches of major sequence divergence, especially in regions such as the divalent cation-binding domain, further studies are required to determine if this isoform possesses 14-3-3 functional features such as the ability to dimerize and bind phosphoproteins.
An exhaustive search of all currently available cDNA and expressed sequence tag databases argues strongly that no other expressed Arabidopsis 14-3-3s exist beyond those described here, and as such GF14π completes the pool of 14-3-3 proteins available in Arabidopsis. Comparison of all 13 isoforms reveals several obvious shared structural features based upon sequence conservation, suggesting that little variation exists in the overall secondary structure of 14-3-3s and yet biological isoform-specific targeting exists (Bachmann et al., 1996a; Rosenquist et al., 2000; Alsterfjord et al., 2004), hinting that subtle divergences of the 14-3-3s are responsible for Arabidopsis isoform specificity.
14-3-3 Loops Are Exposed When the Proteins Are Complexed and Are Phylogenetically Diverse and Structurally Dissimilar
We have identified on Arabidopsis 14-3-3s three structural features that were accessible to mAbs while the 14-3-3s were present in a DNA-binding complex. The existence of the 14-3-3 in the complex was demonstrated by the ability of all three of these antibodies to recognize the GBC in plant cell extracts as well as recombinant 14-3-3 expressed in E. coli. The capacity of the antibodies to bind to the complexed 14-3-3 suggested that the antibodies recognized surface-accessible regions of the 14-3-3s. However the structural model of the 14-3-3s, based upon comparison with the three-dimensional model of the tobacco (Nicotiana tabacum) 14-3-3 C (Wurtele et al., 2003) or even mammalian ζ 14-3-3 complexed with serotonin AANAT (Obsil et al., 2001), does not immediately specify potential surface-accessible antigenic regions while the 14-3-3s are part of a DNA/protein macromolecular complex. The location of all three of the epitopes in loops between helices suggests that these loops must protrude from the 14-3-3 such that they do not interfere with client binding and can still be recognized by the antibody in either a complexed or free 14-3-3 state. These data also suggest that these epitopes maintain the same structural conformation regardless of the client-binding state. Also of interest is the fact that GBC mAb 5D6 epitope in loop 6 is in the same region that has been recognized as the site of Ser phosphorylation in mammalian 14-3-3s (Woodcock et al., 2003) and potential Tyr phosphorylation in plant 14-3-3s (Giacometti et al., 2004). This strengthens the argument that loop 6 is accessible at the surface of 14-3-3s, since it must be available to kinases and that loop 6 remains accessible in complexes.
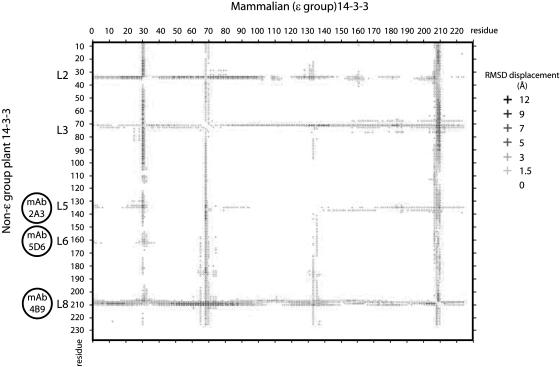
The surface-accessible features recognized by GBC mAb 5D6 and 2A3 epitopes were present in subsets of the 14-3-3s, fundamentally differentiating the non-ɛ group from the ɛ group (Fig. 7A). Since the epitope conservation relies upon amino acid conservation, this is not surprising, except that these epitopes are completely conserved for the entire non-ɛ group and divergent for the entire ɛ group in Arabidopsis, while being sequence divergent to varying degrees (Fig. 7B). This would suggest that two different structures exist for loops 5 and 6 (the GBC mAb 5D6 and 2A3 epitopes), one for ɛ group members and one for non-ɛ group members. Since the structure of any Arabidopsis 14-3-3 has yet to be solved, comparison of a best resolved, closest representative of the plant ɛ group (mammalian 14-3-3 ζ) and a representative of the non-ɛ group (tobacco 14-3-3 C) may present some clues as to the structural differences among these loop 5 and loop 6 epitopes. Using the protein superimposition server SuperPose 1.0 to superimpose the two 14-3-3 structures identifies these two loops as structurally dissimilar based upon the positional deviation from the similar helices (Fig. 8). These data support the hypothesis that loops 5 and 6 are different structurally between the non-ɛ and ɛ group as suggested by the antibody specificity. Additionally, since sequence conservation of the GBC mAb 2A3 epitope in loop 5 is limited to higher plants, while the GBC mAb 5D6 epitope in loop 6 is also found in lower plants such as Chlamydomonas reinhardtii, as well as yeast (Saccharomyces cerevisiae), fungi, insects, and vertebrates, shared functions among all 14-3-3s versus kingdom specific may also be reflected in these exposed loops.
Figure 7.
Specificity, alignment, and comparative analysis of Arabidopsis 14-3-3 family mAb epitopes. A, The location of the epitope areas for GBC mAb 2A3, 4B9, and 5D6 are presented by sequence alignment for each of the 13 Arabidopsis 14-3-3 isoforms. The epitope sequence and adjoining residues are presented, with the limits of the epitope area for each isoform boxed. The isoform immuno-cross-reactivity for each mAb is indicated by bold text and by a plus sign (+) in the cross-reactivity column. B, Phylogenetic analysis of the Arabidopsis 14-3-3 mAb epitopes areas. A rooted tree analysis of Arabidopsis 14-3-3 mAb epitope areas was performed with the boxed sequences from A using ClustalW at the San Diego super computer Biology Workbench. Cladograms for each mAb are presented.
Figure 8.
The epitope loop regions of 14-3-3s are the most variable structural elements between plant and animal 14-3-3s. The three-dimensional structures of a human (PDB accession no. 1IB1) and tobacco 14-3-3 (PDB accession no. 109C), an isoform with a medial Gly in the divalent cation-binding domain, were superimposed for identification of homologous and dissimilar structural elements using SuperPose 1.0 (Maiti et al., 2004). The difference distance plot identified several regions of the 14-3-3s that were structurally divergent at distances between 12 to 1.5 angstroms (Å). The GBC mAb epitopes mapped to three such regions, L5, L6, and L8.
The surface feature recognized by GBC mAb 4B9 is also absent in ɛ group members; however, non-ɛ group members GF14κ and λ are not recognized by this mAb. Conservation of a medial Gly that is specific to non-ɛ group members in the divalent cation-binding domain that makes up part of this epitope suggests that GF14κ and λ should cross-react with the mAb. This premise is further supported by the conversion of the nonantigenic GF14μ into a cross-reacting protein with the mutation of a single amino acid, the medial Asn in the divalent cation-binding loop into a Gly and the reciprocal conversion to non-cross-reactivity for the mutants G213N and G213S GF14ω. However, the truncated nature of the GF14κ and λ C termini, together with the prediction of this terminal region undergoing structural changes as identified by proteolytic protection assays, suggests that acidic residues in the region missing in these isoforms constitutes the remainder of the GBC mAb 4B9 epitope. Removal of this C-terminal extension by mutation of GF14ω (Δ245) abolishes the ability of the antibody to recognize the protein in a western analysis; however, the structural consequences of this mutation are not known. In fact these amino acids are present in GF14μ thereby allowing the recognition of the N213G GF14μ mutation. This missing region in GF14κ and λ C termini also contains nonacidic residues, but based on alignment between all cross-reacting isoforms (ω, φ, χ, υ, ν, and N213G GF14υ) the acidic residues are most conspicuously conserved, suggesting their inclusion in the epitope. This concept is further supported by the high incidence of acidic amino acids in the other phage display binding sequences for GBC mAb 4B9. Interestingly, this region, which is missing in GF14κ and λ, has been predicted to contain a 10th helix that becomes ordered upon binding to divalent cation (Shen et al., 2003), which occurs in the other portion of the GBC mAb 4B9 epitope. Thus the GBC mAb 4B9 epitope consists of residues surrounding the medial Gly of the cation-binding loop together with the portion of the 10th helix, whose structure is influenced by cation binding.
Comparative analysis of the GBC mAb 4B9 epitope in the structurally distinct ɛ and non-ɛ groups is important because of the fact that, largely, the loop 8 and C-terminal regions are not visible in the models referenced in this study presumably because those regions are disordered or take on multiple conformations. However, based upon the superimposition of the structures closest to those regions, the structural divergence of this loop between the non-ɛ and ɛ group is larger than the divergence that occurs in the loops for the two other epitopes (Fig. 8). This is consistent with altered flexibility in the loop 8 region induced by divalent cation binding.
ɛ 14-3-3 Members Possess a Functional yet Less Flexible Divalent Cation-Binding Domain
We had previously demonstrated that divalent cation binding occurs for the non-ɛ group 14-3-3s (Lu et al., 1994b) and this notion was further refined by Athwal and Huber (2002) and most recently by Shen et al. (2003). Cation binding also brings about an induced structural change as indicated by proteolytic digestion experiments and fluorescent probe experiments (Lu et al., 1994b; Athwal et al., 2000; Shen et al., 2003). Until this analysis, there were no data that ɛ group members demonstrated the associated structural change that occurs with cation binding. We have shown here that GF14μ, an ɛ group member, undergoes a structural change in the presence of divalent cations This effect, however, was not identical to that of the non-ɛ group members in that GF14μ was more resistant to the protease albeit at similar divalent cation levels. Conversion of the medial Asn of GF14μ in the divalent cation-binding domain into a Gly resulted in a dramatic increase in sensitivity to the protease. This suggests that the medial Gly allows for increased flexibility of the loop and thereby increased proteolytic sensitivity. This is consistent with increased flexibility with the presence of Gly in proteins and protein loops (Yan and Sun, 1997; Zeng et al., 2003). Again this argues that ɛ and non-ɛ groups have a different structure in this region of the molecule. However, the demonstration that 14-3-3s with medial Asn in loop 8 bind divalent cations argues that most 14-3-3s, including those from animals, bind divalent cations, but those with a medial Gly in loop 8 have an increased flexibility in the region.
Flexible Loop 8 Domains Containing a Medial Gly Are Plant Specific
Interestingly, only plant 14-3-3s possess the flexibility-enhancing medial Gly in loop 8, which may reflect a plant-evolved specific function (Fig. 9). Within plants, distribution of the medial Gly versus non-Gly containing plant 14-3-3s appears to be conserved and delimited, supporting the theory that there are plant-specific functions for those 14-3-3s with a medial Gly in loop 8. Since plant 14-3-3s can be substituted for yeast and mammalian 14-3-3 isoforms in vivo and in vitro, respectively (Lu et al., 1994a; van Heusden et al., 1996), there is an implied conservation of some basic function for all 14-3-3s, but such basic functions would still permit potential specialization in the plant kingdom. Interestingly, in the case of C. reinhardtii, which has only two 14-3-3 isoforms, both contain medial Gly residues in the divalent cation-binding loop supporting the theory that medial Gly containing 14-3-3s have additional capabilities. Of course the ultimate test for the theory that the loop 8 medial Gly-containing plant 14-3-3s have specialized or added function would be to utilize a nonplant 14-3-3 in a plant-specific assay to determine if plant-specific 14-3-3 functions truly exist.
Figure 9.
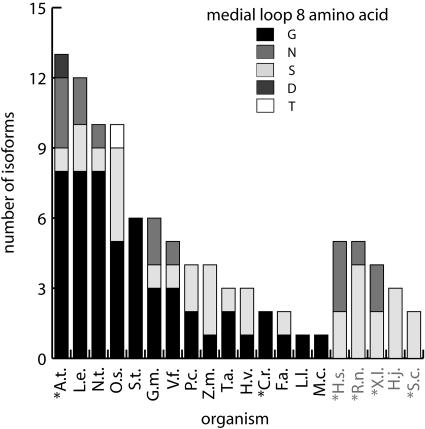
Plants and photosynthetic organisms uniquely contain 14-3-3s isoforms with a medial Gly residue in the divalent cation-binding domain. A sequence comparison of plant and nonplant families of 14-3-3s available from EMBL Interpro release 8.0 was used to score the structurally important medial amino acid in the divalent cation-binding domain (between the Z and −X position) of 14-3-3s. The isoforms are grouped by host. Arabidopsis, A.t.; C. reinhardtii, C.r.; Fritillaria species, F.a.; Glycine max, G.m.; Hypocrea jecorina, H.j.; Homo sapiens, H.s.; Hordeum vulgare, H.v.; Lilium longiflorum, L.l.; Lycopersicon esculentum, L.e.; Mesembryanthemum crystallinum, M.c.; tobacco, N.t.; Oryza sativa, O.s.; Populus canescens, P.c.; Rattus norvegicus, R.n.; Saccharomyces cerevisiae, S.c.; Solanum tuberosum, S.t.; Triticum aestivum, T.a.; Vicia faba, V.f.; and Xenopus laevis, X.l. Completely sequenced genomes are marked by an asterisk. The total number of isoforms and occurrence of amino acids are presented for each family as indicated in the legend.
Flexibility of the Divalent Cation-Binding Domain Alters Target Binding
Since the presence of Gly at the medial position in the divalent cation-binding loop alters at least the flexibility of the C-terminal region, the immediate question was what this might be doing to the binding specificity of the 14-3-3s to phosphotargets. Recent attention has been given to the position of the C terminus and phosphotarget binding (Truong et al., 2002; Shen et al., 2003; Obsilova et al., 2004; Bornke, 2005). A second phospho-independent ligand-binding site in loop 8 of the human 14-3-3 isoform σ has also recently been suggested (Wilker et al., 2005), providing further belief for this region's involvement in target binding. Using GF14μ and N213G GF14μ as probes for phosphotarget selection, we demonstrated that the characteristics of binding were strongly influenced and that the simple conversion of a Gly at the loop 8 medial position altered the binding preference among ɛ-like and non-ɛ isoforms. This suggests that in nature the two groups have different preferential targets, preferences that may further be defined by additional residues that can contact the target protein.
In summary, the examination of three loops of the 14-3-3 molecule that are accessible while in a protein:DNA complex and uniquely limited to one of the major branches of the 14-3-3 phylogenetic tree has illuminated several specific and distinct relationships regarding 14-3-3 function and structure. One of these regions, loop 8, has a strong role in defining structural features based upon a single amino acid and corresponding selective functions that can be specifically assigned to certain 14-3-3s. Knowledge of the biochemical contributions of specific 14-3-3 features and the distribution of those features on the wider evolutionary tree is key to understanding the distribution of specific 14-3-3 functions.
MATERIALS AND METHODS
Materials
GBC mAbs were produced against Arabidopsis (Arabidopsis thaliana) G-Box binding complex as described previously (Lu et al., 1992). Ascites from three established hybridoma lines (2A3, 4B9, and 5D6) that supershifted Arabidopsis alcohol dehydrogenase G-Box probes and recognized Arabidopsis 14-3-3s were used in this experiment. Anti-mouse horseradish peroxidase or alkaline phosphatase antibodies were obtained commercially (Sigma). Anti-rabbit and anti-mouse horseradish peroxidase conjugates were purchased from Amersham (Amersham Biosciences).
Detection and Isolation of GF14π, ι, and ο cDNA
RT-PCR was performed with gene-specific primers and total RNA to detect and isolate Arabidopsis GF14π, ι, and ο. Samples of 2 μg of RNA, isolated from green siliques and leaves using the RNeasy Plant Mini RNA kit (Qiagen), were used as the template for PCR with the One Step RT-PCR kit (Qiagen) and primers corresponding to the coding region sequence with added terminal NdeI and BamHI restriction sites for subsequent cloning. Reactions were analyzed on agarose gels stained with ethidium bromide.
Expression Plasmid Constructions
The GF14π, ι, and ο cDNA PCR products were purified with the QiaQuick gel extraction kit (Qiagen), restricted, and subcloned into pET15b His tag fusion expression vector (Novagen). The vectors produced recombinant 14-3-3 proteins fused in frame to an amino-terminal His tag for detection and purification. Expression vectors for producing His-tagged GF14ɛ, μ, ω, χ, φ, ν, υ, ψ, κ, and λ were described previously (Lu et al., 1992; Wu et al., 1997a). The 14-3-3 point mutants were generated by strand overlap extension PCR using the corresponding 14-3-3 expression vectors as templates with internal mutant primers and subcloned into pET100, pET200 (Invitrogen), or Gateway expression vector pDEST17 (Invitrogen). Truncations were produced with PCR and gene-specific vectors to regions within the coding region and subcloned into pET200 or pDEST17. All resultant vectors were sequenced at the University of Florida (UF) Interdisciplinary Center for Biotechnology Research DNA sequencing core.
Recombinant 14-3-3 Protein Expression and Purification
The pET15b 14-3-3 expression vectors were transformed into Escherichia coli BL21DE3 or BL21-AI (Invitrogen). Bacteria were grown in a shaker at 250 rpm 37°C to a density of 0.6 optical density (OD) 600 nm. Proteins were induced with the addition of 1 mm isopropylthio-β-galactoside (IPTG) and grown for an additional 3 h. Soluble expression of GF14π required induction at lower temperatures (20°C) and reduced levels of IPTG (0.01 mm IPTG). Bacteria were pelleted by centrifugation and protein extracted using the Ni-NTA HisBind purification kit (Novagen). Proteins were dialyzed into phosphate buffered saline pH 7.4 for storage at 4°C. Protein concentrations were measured using a 280 nm absorbance with an extinction coefficient of 1 mg/mL per OD 280 nm.
Electrophoresis and Western Analysis
Proteins were analyzed using discontinuous SDS-PAGE. Protein samples were transferred to nitrocellulose using a Minifold I Dot-Blot system (Schleicher & Schuell BioScience).
Phage Display Analysis with GBC mAbs
The phage display combinatorial libraries of random peptides fused to the M13 PIII proteins Ph.D.-7, Ph.D.-C7C, and Ph.D.-12 used in this work were obtained commercially (New England Biolabs). The complexity for the heptapeptide, disulfide-constrained heptapeptide, and dodecapeptide library were 2.8 × 109, 1.2 × 109, and 2.7 × 109 sequences, respectively. Protein A agarose was washed with Tris buffered saline plus 0.1% Tween 20 (TBST) and resuspended in 0.1 m sodium carbonate pH 8.6, 5 mg/mL bovine serum albumin (BSA), and 0.02% sodium azide for 1 h. After blocking, the resin was washed with TBST plus 0.1% BSA. Approximately 1 × 1011 phage plaque-forming units from PhD-12 and 7 were incubated with 300 ng antibodies for 20 min at room temperature. Following the incubation, the antibody-phage reaction was transferred to a microcentrifuge tube containing preblocked resin. The slurry was mixed gently and incubated for 10 min at room temperature. The resin was pelleted, supernatant discarded, and pellet washed with TBST. The bound phage were eluted with 0.2 m Gly-HCl pH 2.0 containing 1 mg/mL BSA after a 10-min incubation at room temperature. The eluted phage solution was neutralized with 1 m Tris pH 9.1 and retitered using E. coli ER2738 strain. From isolated plaques phagemid DNA was amplified and extracted for sequencing using the manufacturer's protocols. DNA was sequenced with gIII primers at the UF DNA sequencing core. The second and third rounds of panning were performed as described above except the Tween 20 detergent concentration was increased to 0.5% as recommended by the phage display supplier. In addition, to rule out protein A binding peptides, protein G resin was substituted for the affinity resin in the second capture. Alignments against Arabidopsis 14-3-3 sequences were performed with Vector NTI 8 software to identify binding consensus sequences.
Proteolytic Protection Experiments of GF14ω, GF14μ, and N213G GF14μ in the Presence and Absence of Calcium
Endoproteinase Lys-C was purchased from Roche and used according to manufacturer's instructions. Proteins to be digested were first dialyzed against 25 mm Tris, 1 mm sodium azide pH 7.8 at 25°C. Digestions were performed at 25°C in the same buffer with and without 5 mm calcium chloride. Time course samples were taken and protease inhibitors (Protease Inhibitor Cocktail set III; Calbiochem) were added prior to heated 4× SDS-PAGE sample buffer.
Binding and Selection of Eight Synthesized Phosphopeptides by Microresin-Immobilized GF14μ, N213G GF14μ, and GF14ω
Samples of recombinant His-tagged GF14μ, N213G GF14μ, and GF14ω were diluted to 0.1 μg/μL in 10 mm Tris, 250 mm sodium chloride, and 2.5 mm imidazole prior to loading onto nickel-charged MC ZipTip pipette tips (Millipore). Unbound protein was removed by washing with the same buffer minus the protein. The protein-bound pipette tips were equilibrated with peptide-binding buffer (20 mm HEPES, 300 mm NaCl, 5 mm MgCl2 pH 7.2) prior to incubation at room temperature with a mixture of the synthesized phoshopeptides KKSYpSTPF, KKSFpSTPF, KKSSpSTPF, KKSTpSTPF, KKSVpSAPF, KKSVpSMPF, KKSVpSYPF, and KKSVpSIPF dissolved in the same buffer. The incubation was allowed to continue for 15 min and then unbound peptide was washed off rapidly by micropipetting 15 μL of peptide-binding buffer minus the peptides twice. The buffer was exchanged for a more mass spectrometry-compatible buffer of 10 mm ammonium bicarbonate pH 6.8 twice prior to protein and peptide elution with 0.1% acetic acid. Samples were analyzed via direct infusion Electrospray Ionization Fourier Transform Ion Cyclotron Resonance Mass Spectrometry (ESI-FTICR) using a Bruker APEXII, 4.7 Tesla (Bruker Daltonics) with relative changes in ESI peptide signal probes before and after binding were plotted. Only those peptides bound by all 14-3-3s tested were used for the evaluation in Figure 6.
Protein Alignments
Protein alignments for epitopes of 14-3-3s across different hosts were performed with ClustalX at the San Diego super computer.
Structure Comparisons
Three-dimensional structures of mammalian 14-3-3 ζ:AANAT protein database (PDB) accession number 1IB1, tobacco (Nicotiana tabacum) 14-3-3 protein C PDB accession number 109E were superimposed using the protein superposition server Superpose 1.0 for analysis comparison. Distance difference plots were produced for comparing structural deviations of specific regions of the structures.
Note Added in Proof
While this article was in press, Sinnige et al. (Sinnige MP, Roobeek I, Bunney TD, Visser AJ, Mol JN, de Boer AH [2005] Single amino acid variation in barley 14-3-3 proteins leads to functional isoform specificity in the regulation of nitrate reductase. Plant J 44: 1001–1009) reported that the central Gly of loop 8 in the barley 14-3-3 isoforms is critical in the definition of isoform functional specificity.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF543836.
Acknowledgments
We thank the UF Interdisciplinary Center for Biotechnology Research DNA sequencing core for sequencing the vector constructs and positive phage plasmid DNA. We also wish to thank the UF protein core for synthesis of phosphopeptides.
This work was supported by the U.S. Department of Agriculture (grant no. 00–35304–9601) and the National Science Foundation (grant no. MCB 0114501).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert J. Ferl (robferl@ufl.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.073916.
References
- Adamian L, Liang J (2002) Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins 47: 209–218 [DOI] [PubMed] [Google Scholar]
- Alsterfjord M, Sehnke PC, Arkell A, Larsson H, Svennelid F, Rosenquist M, Ferl RJ, Sommarin M, Larsson C (2004) Plasma membrane H+-ATPase and 14-3-3 isoforms of Arabidopsis leaves: evidence for isoform specificity in the 14-3-3/H+-ATPase interaction. Plant Cell Physiol 45: 1202–1220 [DOI] [PubMed] [Google Scholar]
- Athwal GS, Huber JL, Huber SC (1998) Biological significance of divalent metal ion binding to 14-3-3 proteins in relationship to nitrate reductase inactivation. Plant Cell Physiol 39: 1065–1072 [DOI] [PubMed] [Google Scholar]
- Athwal GS, Huber SC (2002) Divalent cations and polyamines bind to loop 8 of 14-3-3 proteins, modulating their interaction with phosphorylated nitrate reductase. Plant J 29: 119–129 [DOI] [PubMed] [Google Scholar]
- Athwal GS, Lombardo CR, Huber JL, Masters SC, Fu H, Huber SC (2000) Modulation of 14-3-3 protein interactions with target polypeptides by physical and metabolic effectors. Plant Cell Physiol 41: 523–533 [DOI] [PubMed] [Google Scholar]
- Bachmann M, Huber JL, Athwal GS, Wu K, Ferl RJ, Huber SC (1996. a) 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett 398: 26–30 [DOI] [PubMed] [Google Scholar]
- Bachmann M, Huber JL, Liao PC, Gage DA, Huber SC (1996. b) The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14-3-3 protein. FEBS Lett 387: 127–131 [DOI] [PubMed] [Google Scholar]
- Bornke F (2005) The variable C-terminus of 14-3-3 proteins mediates isoform-specific interaction with sucrose-phosphate synthase in the yeast two-hybrid system. J Plant Physiol 162: 161–168 [DOI] [PubMed] [Google Scholar]
- Dawson JP, Weinger JS, Engelman DM (2002) Motifs of serine and threonine can drive association of transmembrane helices. J Mol Biol 316: 799–805 [DOI] [PubMed] [Google Scholar]
- DeLisle AJ, Ferl RJ (1990) Characterization of the Arabidopsis Adh G-box binding factor. Plant Cell 2: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl RJ, Lu G, Bowen BW (1994) Evolutionary implications of the family of 14-3-3 brain protein homologs in Arabidopsis thaliana. Genetica 92: 129–138 [DOI] [PubMed] [Google Scholar]
- Giacometti S, Camoni L, Albumi C, Visconti S, De Michelis MI, Aducci P (2004) Tyrosine phosphorylation inhibits the interaction of 14-3-3 proteins with the plant plasma membrane H+-ATPase. Plant Biol (Stuttg) 6: 422–431 [DOI] [PubMed] [Google Scholar]
- Liu D, Bienkowska J, Petosa C, Collier RJ, Fu H, Liddington R (1995) Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376: 191–194 [DOI] [PubMed] [Google Scholar]
- Lu G, de Vetten NC, Sehnke PC, Isobe T, Ichimura T, Fu H, van Heusden GP, Ferl RJ (1994. a) A single Arabidopsis GF14 isoform possesses biochemical characteristics of diverse 14-3-3 homologues. Plant Mol Biol 25: 659–6678061318 [Google Scholar]
- Lu G, DeLisle AJ, de Vetten NC, Ferl RJ (1992) Brain proteins in plants: an Arabidopsis homolog to neurotransmitter pathway activators is part of a DNA binding complex. Proc Natl Acad Sci USA 89: 11490–11494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Sehnke PC, Ferl RJ (1994. b) Phosphorylation and calcium binding properties of an Arabidopsis GF14 brain protein homolog. Plant Cell 6: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti R, Van Domselaar GH, Zhang H, Wishart DS (2004) SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res 32: W590–W594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obsil T, Ghirlando R, Klein DC, Ganguly S, Dyda F (2001) Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex: a role for scaffolding in enzyme regulation. Cell 105: 257–267 [DOI] [PubMed] [Google Scholar]
- Obsilova V, Herman P, Vecer J, Sulc M, Teisinger J, Obsil T (2004) 14-3-3zeta C-terminal stretch changes its conformation upon ligand binding and phosphorylation at Thr232. J Biol Chem 279: 4531–4540 [DOI] [PubMed] [Google Scholar]
- Pan S, Sehnke PC, Ferl RJ, Gurley WB (1999) Specific interactions with TBP and TFIIB in vitro suggest that 14-3-3 proteins may participate in the regulation of transcription when part of a DNA binding complex. Plant Cell 11: 1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AL, Sehnke PC, Ferl RJ (2005) Isoform specific subcellular localization among 14-3-3 proteins in Arabidopsis appears to be driven by client interactions. Mol Biol Cell 16: 1735–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petosa C, Masters SC, Bankston LA, Pohl J, Wang B, Fu H, Liddington RC (1998) 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J Biol Chem 273: 16305–16310 [DOI] [PubMed] [Google Scholar]
- Roberts MR (2003) 14-3-3 proteins find new partners in plant cell signalling. Trends Plant Sci 8: 218–223 [DOI] [PubMed] [Google Scholar]
- Roberts MR, de Bruxelles GL (2002) Plant 14-3-3 protein families: evidence for isoform-specific functions? Biochem Soc Trans 30: 373–378 [DOI] [PubMed] [Google Scholar]
- Rosenquist M, Alsterfjord M, Larsson C, Sommarin M (2001) Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes: expression is demonstrated for two out of five novel genes. Plant Physiol 127: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist M, Sehnke P, Ferl RJ, Sommarin M, Larsson C (2000) Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? J Mol Evol 51: 446–458 [DOI] [PubMed] [Google Scholar]
- Sehnke PC, DeLille JM, Ferl RJ (2002. a) Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell (Suppl) 14: S339–S354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke PC, Rosenquist M, Alsterfjord M, DeLille J, Sommarin M, Larsson C, Ferl RJ (2002. b) Evolution and isoform specificity of plant 14-3-3 proteins. Plant Mol Biol 50: 1011–1018 [DOI] [PubMed] [Google Scholar]
- Shen W, Clark AC, Huber SC (2003) The C-terminal tail of Arabidopsis 14-3-3omega functions as an autoinhibitor and may contain a tenth alpha-helix. Plant J 34: 473–484 [DOI] [PubMed] [Google Scholar]
- Sinnige MP, ten Hoopen P, van den Wijngaard PWJ, Roobeek I, Schoonheim PJ, Mol JNM, de Boer AH (2005) The barley two-pore K+-channel HvKCO1 interacts with 14-3-3 proteins in an isoform specific manner. Plant Sci 169: 612–619 [Google Scholar]
- Sorrell DA, Marchbank AM, Chrimes DA, Dickinson JR, Rogers HJ, Francis D, Grierson CS, Halford NG (2003) The Arabidopsis 14-3-3 protein, GF14omega, binds to the Schizosaccharomyces pombe Cdc25 phosphatase and rescues checkpoint defects in the rad24-mutant. Planta 218: 50–57 [DOI] [PubMed] [Google Scholar]
- Truong AB, Masters SC, Yang H, Fu H (2002) Role of the 14-3-3 C-terminal loop in ligand interaction. Proteins 49: 321–325 [DOI] [PubMed] [Google Scholar]
- van Heusden GP, van der Zanden AL, Ferl RJ, Steensma HY (1996) Four Arabidopsis thaliana 14-3-3 protein isoforms can complement the lethal yeast bmh1 bmh2 double disruption. FEBS Lett 391: 252–256 [DOI] [PubMed] [Google Scholar]
- Wilker EW, Grant RA, Artim SC, Yaffe MB (2005) A structural basis for 14-3-3 sigma functional specificity. J Biol Chem 280: 18891–18898 [DOI] [PubMed] [Google Scholar]
- Woodcock JM, Murphy J, Stomski FC, Berndt MC, Lopez AF (2003) The dimeric versus monomeric status of 14-3-3zeta is controlled by phosphorylation of Ser58 at the dimer interface. J Biol Chem 278: 36323–36327 [DOI] [PubMed] [Google Scholar]
- Wu K, Lu G, Sehnke P, Ferl RJ (1997. a) The heterologous interactions among plant 14-3-3 proteins and identification of regions that are important for dimerization. Arch Biochem Biophys 339: 2–8 [DOI] [PubMed] [Google Scholar]
- Wu K, Rooney MF, Ferl RJ (1997. b) The Arabidopsis 14-3-3 multigene family. Plant Physiol 114: 1421–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele M, Jelich-Ottmann C, Wittinghofer A, Oecking C (2003) Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J 22: 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Smerdon SJ, Jones DH, Dodson GG, Soneji Y, Aitken A, Gamblin SJ (1995) Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376: 188–191 [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91: 961–971 [DOI] [PubMed] [Google Scholar]
- Yan BX, Sun YQ (1997) Glycine residues provide flexibility for enzyme active sites. J Biol Chem 272: 3190–3194 [DOI] [PubMed] [Google Scholar]
- Zeng WY, Wang YH, Zhang YC, Yang WL, Shi YY (2003) Functional significance of conserved glycine 127 in a human dual-specificity protein tyrosine phosphatase. Biochemistry (Mosc) 68: 634–638 [DOI] [PubMed] [Google Scholar]