Abstract
During the course of the development of nitrogen-fixing root nodules induced by Sinorhizobium meliloti on the model plant Medicago truncatula, tubules called infection threads are cooperatively constructed to deliver the bacterial symbiont from the root surface to cells in the interior of the root and developing nodule. Three-dimensional reconstructions of infection threads inside M. truncatula nodules showed that the threads formed relatively simple, tree-like networks. Some characteristics of thread networks, such as branch length, branch density, and branch surface-to-volume ratios, were remarkably constant across nodules in different stages of development. The overall direction of growth of the networks changed as nodules developed. In 5-d-old nodules, the overall growth of the network was directed inward toward the root. However, well-defined regions of these young networks displayed an outward growth bias, indicating that they were likely in the process of repolarizing their direction of development in response to the formation of the outward-growing nodule meristem. In 10- and 30-d-old nodules, the branches of the network grew outward toward the meristem and away from the roots on which the nodules developed.
The development of biological branching structures has been studied from theoretical, biomechanical, cellular, and molecular points of view, using model systems that include plant branches, plant roots, blood vessel networks, neuron dendrites, and tracheal development in invertebrates (Farnsworth and Niklas, 1995; Uylings and van Pelt, 2002; Uv et al., 2003; Masters, 2004; McColloh and Sperry, 2005; Wu et al., 2005). What these networks share are common solutions to the problems of optimizing resource uptake and waste dispersal. Many biological branching structures develop their final size and form in response to gradients in light, water, nutrients, or signaling molecules emanating from target and nontarget tissues. Work in this article describes the system of branched tubules, infection threads (ITs), that develop during symbiotic nodulation of certain plant roots. These branched networks share features in common with many other well-studied branched networks, including growth by tip extension and directed growth to deliver resources (the bacterial symbiont).
The symbiosis between Medicago truncatula, alfalfa (Medicago sativa), and their nodulating symbiont, Sinorhizobium meliloti, begins when the bacterium detects flavonoids and other compounds released by host plants. These activate S. meliloti transcriptional regulators, which then induce the expression of about 25 bacterial nod genes required for the biosynthesis of the lipooligosaccharide signaling molecule, Nod factor, which is obligatory for nodulation (Mulligan and Long, 1989; Brewin, 1991; Kondorosi et al., 1991; Denarie et al., 1996). An IT, a plant-derived tubule filled with dividing and growing bacteria, first develops in an infected root hair and then traverses several cell layers to deliver bacteria to root cells in the developing nodule (Brewin, 2004; Gage, 2004). Bacteria exit the ITs through infection droplets, thereby entering the cytoplasm of the nodule cells, and differentiate into nitrogen-fixing bacteroids (Rae et al., 1992; van Brussel et al., 1992; Gage, 2004).
The extension of root hairs is one of the few plant developmental processes that takes place by tip growth. Vesicles containing plant cell wall and cell membrane material travel in an actin- and microtubule-dependent fashion to the root hair tip where they fuse with the cell membrane, thus adding cell membrane to the tip and depositing cell wall material outside the membrane (Peterson and Farquhar, 1996; Miller et al., 2000; Hepler et al., 2001; Lhuisser et al., 2001). Successful infection by S. meliloti requires Nod factor-dependent reorientation of root hair growth, which results in root hair deformation and root hair curling. These major changes in growth direction correlate with, and are probably caused by, significant alterations in the plant cytoskeleton (Cardenas et al., 1998; Miller et al., 1999; Timmers et al., 1999; Catoira et al., 2001). It has been suggested that Nod factor-dependent reorganization of the root hair actin cytoskeleton redirects vesicle traffic from the root hair tip to new sites away from the center of the apical dome of the root hair, thereby causing root hair deformation and curling (Miller et al., 1999). The changes in actin-directed (and possibly microtubule-directed) vesicle deposition may eventually reorient tip growth to the extent that inward-directed tip growth of the IT results.
The IT itself, both in root hairs and in host cells that are infected at later stages, is a bacteria-filled invagination of the plant cell wall and its underlying plasma membrane. Thus, the lumen of the IT contains bacteria, secreted bacterial products such as exopolysaccharide and lipopolysaccharide, and material similar to that which makes up the plant extracellular matrix (Fraysse et al., 2003). In addition, IT lumens contain nodule-enriched extensins and other plant-derived glycoproteins that may modulate the physicochemical properties of the lumen matrix and extension of the thread (Rathbun et al., 2002; Brewin, 2004).
Early during infection, the growth of the IT network is highly polarized. Threads initiate in root hairs and then branch and ramify, to some degree, in both epidermal and cortical cells as they grow inward toward the developing nodule (Gage, 2004). In a variety of legumes that form indeterminate nodules, including M. truncatula, two series of Nod factor-dependent events take place concurrently. One begins on the surface, moves inward, and includes membrane depolarization of the root hair, Ca2+ oscillations in root hair cells, root hair curling, IT initiation, IT growth, and the reentry of outer cortical cells into the cell cycle (Libbenga and Harkes, 1973; Dudley et al., 1987; Ehrhardt et al., 1992, 1996; van Brussel et al., 1992; Mylona et al., 1995; Timmers et al., 1998, 1999). The other series of events begins near the center of the root and moves outward. This series of events starts with the activation of some pericycle cells next to protoxylem poles (Timmers et al., 1999). Following pericycle cell activation, cells of the inner cortex reenter the cell cycle and the resulting cell mass constitutes the nodule primordium. These cells are the first to receive the intracellular bacteria that differentiate into nitrogen-fixing bacteroids. The wave of cell activation continues to move outward, cells of the middle cortex become activated, and some of these will later form the nodule meristem.
Most of the volume of outer cortical cells is occupied by a large, centrally placed vacuole. Upon activation, cytoplasm moves from the cell periphery to a central position in those outer cortical cells located in columns above activated cells in the pericycle, inner, and middle cortex (van Brussel et al., 1992; Timmers et al., 1999). The cytoplasms in these cells align and are called preinfection threads (PIT). It is through these that inward-developing ITs are constructed. PITs have a polarized structure characterized by nuclei against the inner (proximal) wall and a wedge-shaped column of cytoplasm with its wider side against the outer (distal) wall (van Brussel et al., 1992). The polarized cytoplasm is probably responsible for the inward-directed construction of the IT network through these cells early during invasion. In addition, PITs contain bundled endoplasmic microtubules that run parallel to their long axes (van Brussel et al., 1992; Timmers et al., 1999). Thus, the future path of the developing IT through the outer cortex is marked by the polarized and aligned PITs. Micrographs show that when an IT leaves a root hair cell it fuses with the inner cell wall at a point that is marked by a wall weakening and a PIT in the underlying cell. The bacteria spill into the intercellular space and the weakened outer wall of the underlying cell invaginates and the IT propagates inward. By repeating this process, the IT eventually reaches the nodule primordium (van Brussel et al., 1992). After the thread network enters the nodule primordium, some of the uninfected, activated cells in the middle cortex organize into a nodule meristem (Timmers et al., 1999). At this time, the meristem is located on the distal side of the nodule primordium and is growing outward toward the root surface. The tips of the inward-developing IT network are located on the root (proximal) side of the outward-moving meristem. As the nodule grows, the ITs behind the meristem form a network in a region of the nodule termed the infection zone (Vasse et al., 1990). In indeterminate nodules, such as those of alfalfa and M. truncatula, the meristem continually grows away from the interior of the root. Clearly, as the meristem moves outward, the IT network must develop in the region behind the meristem that contains newly divided cells that need to be infected.
Once mature, indeterminate nodules contain at least five recognized zones (Vasse et al., 1990). Near the apex (distal end) of the nodule is the meristem (zone I), which gives rise to all tissues in the nodule. Cells here are small and cytoplasm rich and do not contain bacteria or ITs. Basal (proximal) to the meristem is the infection zone (zone II). Nodule cells in this zone are larger than the meristematic cells and they support the development of ITs. Bacteria are released in the distal part of this zone and small dividing bacteria can be found in the cytoplasm of zone II host cells. The nitrogen-fixation zone (zone III) is proximal to the infection zone. Here, nodule cells are large, contain a large, centrally placed vacuole, and have cytoplasm that is filled with large, elongated bacteroids capable of fixing atmospheric nitrogen. Zone III is generally eight to 12 cell layers wide in alfalfa. Between the infection and nitrogen-fixation zones lies interzone II-III. Here, nodule cells contain amyloplasts, and bacteroids are elongated and often fill nodule cells. A senescent zone (zone IV) is proximal to the nitrogen-fixation zone in older nodules. Here, the internal contents of nodule cells and their bacteroids undergo degradation.
Little is known about how ITs grow and develop in nodules. A fundamental question about the organization of IT networks inside nodules is whether, as a whole, they show growth toward the nodule meristem or whether their growth is random within the infection zone with only some branches developing toward the meristem. It is also unknown whether characteristics of IT networks remain fairly constant over time or change as nodules develop. Examination of the architecture of IT networks in nodules of different ages was thought likely to provide insight into how IT network architecture changes as nodules develop from immature to mature forms.
RESULTS
Overview of the Analyses
Three-dimensional (3-D) representations of IT networks were reconstructed from nodules harvested 5, 10, and 30 d after spot inoculation. These time points were chosen because they represent distinct stages of nodule development (Vasse et al., 1990). In alfalfa, and probably in M. truncatula, nodules 4 to 8 d old are not yet actively fixing nitrogen; they contain well-developed zones I, II, and sometimes interzone II-III. Ten-day-old nodules are newly mature, generally capable of fixing nitrogen, and contain zones I, II, II-III, and III. Thirty-day-old nodules are fully developed, contain zones I through IV, and represent the type of nodule present most often in mature alfalfa or M. truncatula plants. The growth characteristics of the IT networks were determined as described in “Materials and Methods.” The main processes used to generate data and perform growth analyses are outlined in Figure 1, A to D. In addition, network topology was analyzed by quantitating the degree of branching, average branch lengths, and branch density.
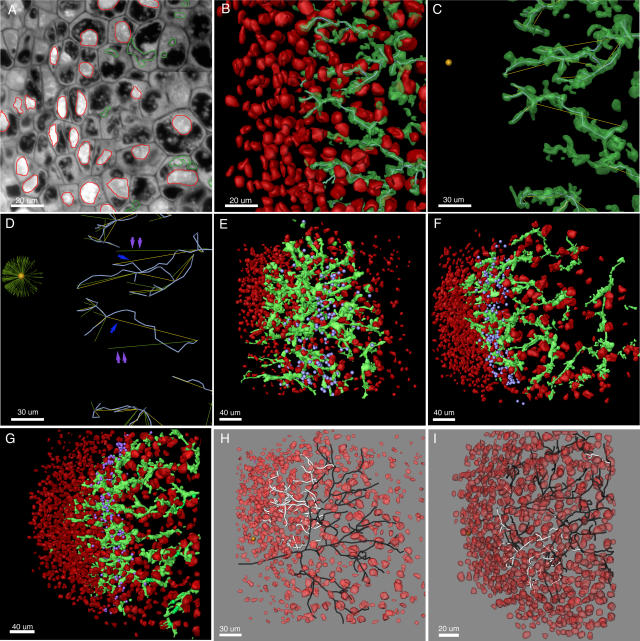
Figure 1.
Reconstruction of IT networks. Nodule apexes are to the left in all images. A to D, Region of the meristematic and infection zones of a 30-d-old alfalfa nodule. A, Stained 2-μm-thick section of an alfalfa nodule. ITs and nuclei are segmented and shown in green and red, respectively. B, Three-dimensional reconstruction of ITs and nuclei. Centerlines are shown threaded through the approximate center of volume of some of the ITs. C, Growth vectors (yellow), indicating the direction of IT growth, originate at branch points and extend to the next branch point or to ends of terminal branches. The center of the meristematic region is indicated by a gold sphere. D, Transformed growth vectors (green) show the direction of IT growth with respect to the meristem center. If a growth vector points directly toward the meristem center, it will point horizontally to the left following transformation. Blue arrows point to growth vectors that nearly point at the meristem center. Their transformed vectors are indicated with double violet arrows. Transformed growth vectors from the whole sample volume (much of which is not in the figure) are shown in the sunburst plot centered on the meristem center. E to G, Reconstructions of 5-, 10-, and 30-d-old M. truncatula nodules. ITs are green and nuclei are red. The location of the distal-most bacteria in the distal-most line of nodule cells of each stained section that contained released bacteria are indicated with violet spheres. H and I, Reconstructions of 5-d-old nodules showing nuclei (red) and skeletonized IT networks. Dark branches are inward growing and white branches are outward growing. The nodule shown in H is the same as the one shown in E. In H and I, only the main subtree is shown.
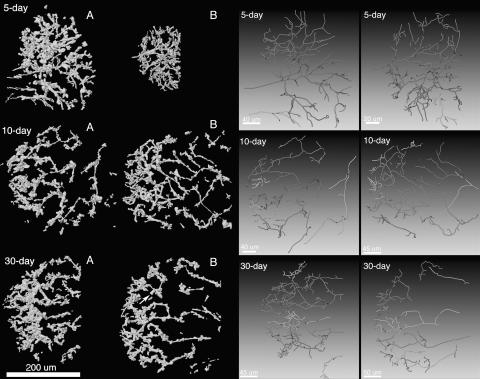
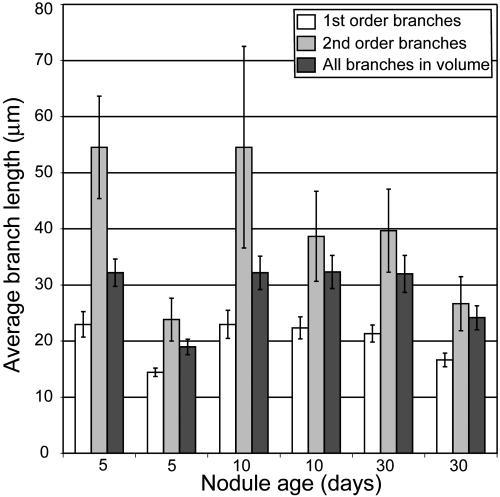
IT networks in 5-, 10-, and 30-d-old nodules shared many common structural features. They were generally made up of one or more fairly simple tree-like branching structures that occupied 1.5% to 3.0% of the volume of nodule tissue in zone II and interzone II-III (Table I). In some cases, network subtrees were likely connected by branches that were outside the nodule volume that had been reconstructed (Fig. 2). This was especially true for the larger 10- and 30-d-old nodules. One 5-d-old nodule had network subtrees that may have arisen from separate infection events (Fig. 2). Main ITs were generally oriented along the long axis of the nodule and exhibited lumps and protrusions, some of which were infection droplets, the sites of bacterial release. These could be discerned because they had a rather characteristic, wide, rectangular shape in the 3-D reconstructions, were contained within one nodule cell, and appeared to not be bounded by plant cell wall material in the stained sections (Supplemental Fig. 1). The average length of terminal (youngest) branches and the branches from which they arose (first and second order, respectively), and the average of all branches (including branches with orders >2) were similar among nodules of different ages (Fig. 3). The only major exceptions to this were the shorter branches found in one 5-d-old nodule that was relatively small (Figs. 2 and 3). Second-order branches were consistently longer than the terminal branches.
Table I.
Size of sampled nodule regions, IT network volumes, and IT network surface areas
| Nodule and Zonesa
|
Dimensions of Tissue Occupied by ITsb
|
Tissue Volume
|
IT Volume
|
IT Volume/Tissue Volume
|
IT Surface
|
IT Surface/IT Volume
|
||
|---|---|---|---|---|---|---|---|---|
| X | Y | Z | ||||||
| μm | μm3 | μm3 | μm2 | |||||
| Five-day A: I and II | 241 | 282 | 68 | 4,621,416b | 104,272 | 0.023 | 111,001 | 1.06 |
| Five-day B: I and II | 127 | 189 | 60 | 1,440,180 | 43,786 | 0.031 | 55,113 | 1.26 |
| Ten-day A: I, II, and II-III | 290 | 308 | 62 | 5,537,840 | 79,829 | 0.014 | 88,014 | 1.10 |
| Ten-day B: I, II, and II-III | 314 | 278 | 62 | 5,412,104 | 82,142 | 0.015 | 94,045 | 1.14 |
| Thirty-day A: I, II, and II-III | 228 | 304 | 62 | 4,297,344 | 87,590 | 0.020 | 96,873 | 1.11 |
| Thirty-day A: I, II, and II-III | 293 | 333 | 48 | 4,683,312 | 89,062 | 0.019 | 92,147 | 1.03 |
Nodules and their designations, A or B, are as shown in Figure 2. Zone numbers indicate which zones were present in the part of the nodule that was reconstructed. I, Meristem; II, infection zone; II-III, interzone II-III.
Dimensions of the smallest rectangular volume that included all of the ITs in the reconstructed dataset (e.g. the volume of the meristem is not included because it did not contain ITs).
Figure 2.
Reconstructed IT networks and network skeletons. The left half of the figure shows the six 3-D reconstructions used in this article. A and B, Nodules as presented in Table I. The arrow in the image of the 30-d-old nodule shows the infection droplet displayed in Supplemental Figure 1. The right half of the figure shows the centerlines for IT subtrees that contained two or more branches. Shades of gray are used to help visually separate overlapping subtrees. The order of the networks is the same on both sides of the figure.
Figure 3.
Branch lengths in IT networks in M. truncatula nodules of different ages. Average branch lengths are plotted for first- and second-order branches and for all branches completely contained in the sample volumes. Bars = se.
Structure of IT Networks in 5-d-Old M. truncatula Nodules
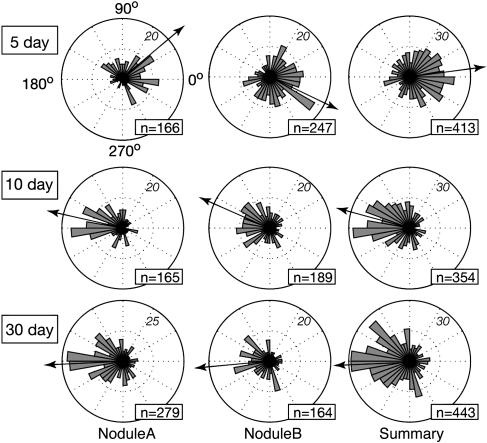
Five-day-old nodules were developmentally immature. Meristems had not yet developed to the point where they occupied a majority of the nodule apex. They contained developing zones I and II and relatively few nodule cells with released bacteria. Filled nodule cells with elongated bacteroids characteristic of interzone II-III had not yet developed. Network structure in the 5-d-old nodules was more complex than that seen in the older nodules in that the direction of branch growth was oriented in two main directions. Most of the branches of the IT networks were directed inward toward the root (Figs. 1 and 4), even though meristematic regions were developing near the nodule apexes. These inward-directed branches were not merely remnants of threads that developed in the root cortical layers, but likely developed in nodule primordium during the development of the outward-growing meristem. Both 5-d-old nodules showed well-defined regions where ITs were not growing inward, but rather were growing toward the nodule apex (Fig. 1, E, H, and I). In one case, the outward-growing branches were directed toward the only part of the nodule with obvious meristematic activity (Fig. 1H). While it is impossible to state for certain when, and by what mechanisms, the branches growing toward the nodule apex developed, their orientations and positions with respect to the branches from which they arose suggest that they originated by branching off of older ITs, rather than by changing their direction of growth from inward to outward during development.
Figure 4.
Direction of branch growth in IT networks from nodules of different ages. The directions of growth of branches of IT networks, with respect to center of the meristem (for 10- and 30-d-old nodules), or with respect to a point five to six cell layers behind the nodule apex (for 5-d-old nodules) are plotted as circular histograms. Branches growing directly toward the meristem have values of 180°, branches growing directly away have values of 0°. Italics indicate the number of branches in any bin that reaches the diagram edge; n values indicate the number of branches plotted in each diagram. Arrows show the mean growth direction of all branches and indicate overall growth direction of the IT networks. The right-most plot contains all data from nodule A and nodule B.
Bacterial release into nodule cells was seen over a region in the 5-d-old nodules that was broader than that seen in the older nodules (Fig. 1E). This zone was five to six cell layers wide in the 5-d-old nodules, two to four cell layers wide in the 10-d-old nodules, and two to three cell layers wide in the 30-d-old nodules. The zone seen in the 5-d-old nodules may have been relatively broad because bacteria were being released independently from putative infection droplets seen on both inward- and outward-directed branches.
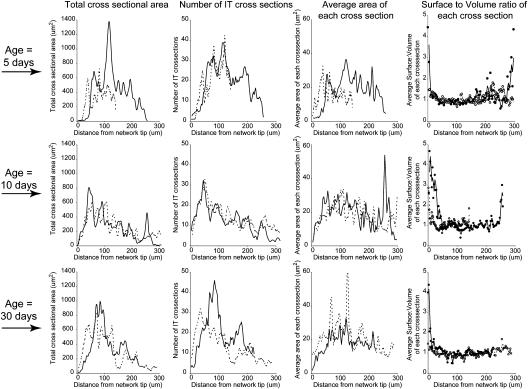
Surprisingly, the density, cross-sectional area, and surface-to-volume ratios of ITs in the 5-d-old nodules were not qualitatively different from those seen in the older, 10- and 30-d-old nodules (Table I; Fig. 5).
Figure 5.
Size of ITs and IT networks. Three-dimensional representations of IT networks were sampled by extracting 100 evenly spaced cross-sectional slices from the apexes of the networks to their bases. Cross sections were oriented perpendicular to the long axis of the nodules. Total IT area per slice, average number of IT cross sections per slice, average area of each IT cross section in each slice, and the average surface-to-volume ratio of each IT cross section in the slice, assuming a slice thickness of 0.1 μm, are shown. Top, middle, and bottom rows show data from 5-, 10-, and 30-d-old nodules, respectively.
Structure of IT Networks in 10- and 30-d-Old M. truncatula Nodules
Ten- and thirty-day-old nodules exhibited classic nodule structure with well-defined meristems, infection, and nitrogen-fixation zones. In these nodules, the IT network appeared to be growing toward the nodule apex and meristematic regions. This was confirmed by measuring the growth direction of the network branches with respect to the center of the meristematic region (Fig. 4). The density of ITs was greatest near the distal part of the infection zone, about 50 to 100 μm from the advancing tips of the IT network (Figs. 1, 2, and 5). This same IT-dense region held the youngest nodule cells containing intracellular S. meliloti (Fig. 1, E–G) and likely corresponded to the distal part of zone II described by Vasse et al. (1990), where bacteria are released from ITs. ITs were generally quite narrow at the advancing front of the network and often were wide enough to accommodate only two to three bacteria oriented side by side. For example, 25 μm from the distal edge of the networks, threads were on average 10 μm2 in cross section (Fig. 5). Generally, threads were widest about 50 to 100 μm from the advancing edge of the network where they had an average cross section of about 20 to 30 μm2. Proximal to this region, the ITs again became narrow, as can be seen from the decline in their average cross-sectional area (Fig. 5). It is possible that the decline in width was caused by the threads being pulled taut (lengthened and made narrower) as the host nodule cell to which the IT ends were anchored expanded in volume.
The surface-to-volume ratio of the threads in the network was close to 1.0 (Table I) and showed surprisingly little variation along the length of the nodules in spite of differences in cross-sectional area and network density (Fig. 5). An exception to this was near the advancing edge of the networks, where threads tended to have a higher surface-to-volume ratio than the rest of the threads in the network (Fig. 5).
DISCUSSION
Three-dimensional representations of IT networks were constructed from 2-μm-thick, stained, serial sections of M. truncatula nodules harvested 5, 10, and 30 d after inoculation with wild-type S. meliloti strain Rm1021. These revealed the structure of IT networks in nitrogen-fixing root nodules as well as information about their growth characteristics. While fragments of ITs can be seen in standard semithin sections of nodule tissue, these give a very limited view of the architecture of the IT networks because ITs occupy only 1.5% to 3.0% of a nodule's total volume (Table I). Thus, any single nodule cross section reveals little about the rest of the network.
In all nodules documented here, and in other nodules not presented, the IT networks exhibited simple, open tree-like structures in which fusion between separate branches was rare or nonexistent (Fig. 2; Supplemental Fig. 4). ITs in nodules displayed a preponderance of lumps and protrusions, some of which were infection droplets where bacteria were released from the threads into nodule cell cytoplasm (Rae et al., 1992; Supplemental Fig. 1). In older sections of the network, bacteria inside ITs often were haphazardly arrayed. In contrast, at the advancing edge of the network, ITs were thin and usually contained well-ordered columns of bacteria. The difference between the youngest part of the threads and their older counterparts suggests that ITs, and the bacterial populations inside, continue to grow for a period of time after a section of thread is synthesized. In the proximal part of zone II and in interzone II-III, ITs were on average thinner than they were in the more distal part of zone II where bacterial release was occurring. This may be because the walls of the ITs stopped expanding through the addition of cell wall material and were stretched by the expansion of the host cell in which they were housed. Such a stretching and thinning of tubes, with their entrance and exit openings anchored in the host cell wall, may explain the trumpet bell-like structures readily seen where ITs in interzone II-III and in zone III meet the host cell wall, as described and depicted by Ward (1887).
Bacterial release from ITs takes place in the infection zone (Vasse et al., 1990) and is known to require the products of the NIP and DMI2 genes in M. truncatula (Veereshlingam et al., 2004; Limpens et al., 2005). Release in this region was readily observed in the serial sections used for the 3-D reconstructions (Supplemental Fig. 1). Measurements indicated that the nodule cells containing newly released bacteria were 50 to 100 μm (two to four cell layers) from the distal, advancing edge of the IT network in the mature nodules. It was in this region that the IT networks were densest and where ITs were the widest (Fig. 5).
Analysis of the reconstructed IT networks showed that thread branch lengths, average cross-sectional areas, and average surface-to-volume ratios were remarkably similar in nodules of different ages. Such constancy may make future analysis of network development easier than it might have otherwise been if IT size, density, or surface area had changed radically with nodule age. Based on the data presented here, if a bacterial or plant mutant is shown to induce nodules that differ greatly from the wild type in one of the age-constant characteristics listed above, it is unlikely that the difference will be due to mutation-dependent delays or advances in nodule development, but rather because the mutation directly affects thread growth or development.
The work described in this article was initiated to characterize the structure of IT networks in nodules and determine whether IT networks displayed polarized development as they propagated through nodule tissue. It is known from the work of others that early during development ITs branch and display polarized growth as they develop in cortical tissue (Dudley et al., 1987; van Brussel et al., 1992; Timmers et al., 1999; Brewin, 2004). After reaching the primordium, the nodule meristem forms on the distal side of the primordium and the nodule begins to grow outward toward the root surface. It was not known how IT networks developed to deliver bacteria to the new, uninfected tissue laid down by the meristem. One possibility was that the inward-growing IT network repolarized, changed the direction of its development, and grew outward following the meristem. The other was that the network propagated in a directionally random fashion, with some of its branches delivering bacteria to the new, uninfected tissue derived from the meristem. The work described here clearly shows that by 10 d after inoculation, the IT networks in M. truncatula nodules had a strong growth bias toward the meristem and apex of the nodule. The story in 5-d-old nodules was more complex. In those nodules, the overall growth bias was toward the root interior. However, ITs in well-defined regions of these nodules showed local growth biases directed toward the nodule apex. These regions of the IT networks were likely in the process of changing their overall growth polarization from inward to outward. The outward-growing branches generally pointed toward the nodule apex from their inception and did not typically result from branches that turned or changed their growth from inward to outward. In one of the 5-d-old nodules, this region of newer, outward-growing branches developed such that they pointed toward the only well-developed meristematic region in the nodule (Fig. 1H). In the other 5-d-old nodule, the outward-growing branches were directed toward only one section of a well-developed meristematic region (Fig. 1I).
It is not known what is required for reorientation of IT networks in response to meristem development or for their progression through the infection zone. Outward-directed, polarized network growth could be supported if cells between the meristem and the infection zone were polarized, much like the cortical cells that support polarized IT growth early during infection (van Brussel et al., 1992; Timmers et al., 1999). Unlike IT progression through cortical tissue, there were no obvious visual markers indicating the future paths of ITs through the infection zone seen in this study. This may be because cortical cells contain large vacuoles, making it easy to discern PITs, whereas infection-zone cells in front of extending ITs are cytoplasm rich and have no large vacuoles to be displaced if polarized cytoplasm moves to a position indicative of the future paths of threads.
Alternatively, IT development may respond to a diffusible targeting signal. The hypothesis of response to a diffusible signal and the hypothesis of polarized cells regulating the direction of IT development in the nodule are not mutually exclusive because cells between the meristem and infection zone could be polarized in response to diffusible signals arising from the meristem or from signals arising from bacteria in ITs. It was recently shown that developing M. truncatula nodules form symplastic fields connected to phloem that are capable of trafficking large molecules. The location of these fields changes as nodules develop, and they correspond to regions of nodules that, in this study, supported IT growth. In young nodules, all internal regions formed a phloem-connected symplastic field, whereas only the meristem and infection zones did so in mature nodules (Complainville et al., 2003). Clearly, signals that influence IT targeting could be brought to their correct locations by such transport mechanisms (Crawford and Zambryski, 1999). Nod factor is also a candidate for a signal that might be needed for host cell polarization and IT propagation. It is clearly involved in these processes early in infection and the genes required for its synthesis and transport are known to be expressed by bacteria in the infection zone of nodules (Sharma and Signer, 1990).
CONCLUSION
ITs are composite structures, cooperatively built by both the symbiotic, nitrogen-fixing bacteria and their plant host. Their role is to deliver the bacteria to the interior of the developing nodule. ITs, and the bacterial populations inside them, have been studied in root hairs and in cortical cell layers near the surface of the root early during infection. To our knowledge, the networks that result from IT branching and development inside nodules have never been described, analyzed, or even visualized as a whole. We present 3-D reconstructions of IT networks in nodules of different developmental stages. This work shows how IT networks are structured inside nodules, the average size and surface-to-volume ratios of ITs in different parts of the nodule, where bacterial release occurs with respect to the whole IT network, and how the directional development of IT networks change as nodules mature. The analysis of network structure and development we present in this article is interesting in its own right, but it may also form a basis for the quantitative analysis of the development of the internal structure of nodules. In addition, the work presents techniques that can be used for future analyses of IT networks. The quantitative data and the introduced techniques will be important because large-scale genomics projects utilizing the model legume M. truncatula are starting to identify genes needed for proper IT growth, nodule formation, and nodule development.
MATERIALS AND METHODS
Nodule Fixation, Sectioning, and Staining
Wild-type Sinorhizobium meliloti strain Rm1021, grown in TY (Tryptone-Yeast) medium, was rinsed and suspended in Nod3 medium at 109 cells/mL, and spot inoculated onto Medicago truncatula roots of plants growing on slides covered with a layer of Nod3 medium (Bhuvaneswari et al., 1981; Gage et al., 1996). Slides were placed in loosely capped 50-mL polypropylene tubes containing 5 mL of liquid Nod3 medium in the bottom and incubated in a plant growth chamber (26°C, 16-h-light/8-h-dark period). Nodules were harvested 5, 10, and 30 d after inoculation, fixed in 4% paraformaldehyde in phosphate-buffered saline, dehydrated with an ethanol series, and embedded in JB4 plastic resin (PolySciences). Twenty to 35 serial, longitudinal, 2-μm-thick sections from near the central axis of each nodule were cut on a Shandon Finesse microtome, mounted on polylysine-coated slides, and stained with a mixture of 4′,6-diamino-phenylindole and acridine orange (Dudley et al., 1987), which allowed visualization of ITs, bacteria, plant cell nuclei, vacuoles, and cell walls. This was done on two nodules of each age.
Analysis of IT Networks
The main processes used to generate data and perform growth analyses are outlined in Figure 1, A to D. Images of serial sections were captured with a Qimaging Retiga 12-bit cooled CCD camera, using a 40× objective, which resulted in a resolution in the x and y planes of 0.162 μm/pixel. Images were aligned using ALIGN software (Fiala et al., 2002; Fiala and Harris, 2002). ITs, nuclei, and other structures in the sections were segmented and assembled into 3-D representations using Amira software (TGS; Fig. 1A). Surface rendering of nuclei and IT networks was done with Amira or Lightwave (NewTek) software. Following 3-D reconstruction, network skeletons were assembled from lines that went through the approximate center of IT volumes (Fig. 1B). The x, y, and z coordinates of points along the network skeletons were used for characterizing the topological and growth characteristics of the IT network. For topological analysis, a centripetal-segment numbering system was used (Berntson, 1997). In this system, terminal branches (youngest) were given an order of 1. The order of any interior branch was the same as the highest order of its two distal (younger), connected, branches. If the two distal branches were of equal order, the interior branch segment was given an order number of 1 greater than the order number of the distal two branches (Supplemental Fig. 2). Assigning the youngest branches an order number of 1 ensured that branches in the sample volume were compared with branches of the same developmental stage. Thus, the youngest terminal branches (order number 1) were compared with each other, and the branches from which they arose (order number 2) were compared with each other. This would not have been the case if the oldest branches were given an order number of 1 and their descendents higher order numbers, because the differing degrees of branching seen in different trees would lead to, for example, young terminal branches having different order numbers. For branch length calculations, contiguous groups of branches of the same order were counted as one branch, and its length was the sum of the lengths of its contiguous parts (Supplemental Fig. 2; Berntson, 1997). Only complete branches were used for topology analysis; any that left the sampled volume were disregarded.
Branch growth vectors were defined as beginning at a branch point and going to the next, more terminal branch point. Growth vectors of terminal branches went from their branch points to the ends of the branches (Fig. 1, C and D). Incomplete branches that left the sample volume were included in growth analysis because they retained directional information. ITs were 5.5 μm in width and 29 μm in length, on average. The error in assigning branch points and end points is expected to be no more than one-half of the diameter of the threads. For example, a maximum error in assigning an end point would be to place it on the surface of the thread rather than on the centerline of the thread. The assigned angle of an average branch, with a true branch angle of 90°, could be off by a value of 90° − arctan [(1/2 average length)/(1/2 average width)] = 11°. Average branches at other angles would also have a maximum possible error of 11°. The average error of the measured angles presented in the “Results” section is certainly much lower than 11° because care was taken to place branch and end points near the IT centerlines.
The growth vectors were transformed to reflect the direction each branch grew with respect to the nodule meristem (Supplemental Fig. 3). This was done by attaching the base of a new orientation vector to the base of the growth vector under consideration. The tip of the orientation vector was placed at the meristem center. The common base of the two vectors, and the attached vectors, were rotated around the tip of the orientation vector so that the y and z coordinates of the common base were the same as those of the meristem center. This resulted in an orientation vector that pointed horizontally (180°) at the meristem center. The altered growth vector, resulting from the rotations, was termed the transformed growth vector. If a growth vector had been pointing directly toward the meristem center from anywhere in the network, its transformed direction would point at 180° due west. Conversely, if it had been pointing directly away from the meristem center, its transformed direction would point at 0° due east. Growth vectors pointing neither at, nor away from, the meristem center had transformed vectors pointing in directions other than 180° or 0°. Transformed vectors were plotted with a common start point resulting in a sunburst plot (Fig. 1D) or their angles in the x-y plane were plotted in radial histograms. In cases where a well-defined meristem had not yet formed, vectors were transformed to show their growth with respect to a point five to six cell layers below the growing apex of the nodule (the approximate location of the meristem center in older nodules). In cases where a well-defined arc of meristematic cells had developed, transformation was done with respect to the meristem center. In these cases, transformed vectors from approximately 135° to 225° were growing toward meristematic tissue. The translations and rigid body rotations of datasets and their coordinate systems needed to generate transformed growth vectors, and generation of the radial histograms were done using MATLAB (Fisher, 1993; Angel, 2003).
To determine how IT number, area, and surface-to-volume ratio varied along the length of analyzed nodules, 3-D representations of IT networks were sampled by extracting 100 evenly spaced cross-sectional slices from the apexes of the networks to their bases. The cross sections were oriented perpendicular to the long axis of the nodules. Total IT area per slice, average number of IT cross sections per slice, average area of each IT cross section in each slice, and average surface-to-volume ratio of each IT cross section in the slice, assuming a slice thickness of 0.1 μm, were calculated. The error associated with these measurements is dependent on the accuracy of the segmentation. Segmentation was done by hand and care was taken to accurately demark objects of interest. We estimate that the maximum error associated with segmenting areas of interest was less than 25%.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Cynthia Jones and Dr. Marie Cantino for advice on tissue sectioning.
This work was supported by the National Science Foundation (grant no. IBN9974483) and by the University of Connecticut Research Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Daniel J. Gage (daniel.gage@uconn.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.072876.
References
- Angel E (2003) Interactive Computer Graphics: A Top-Down Approach with OpenGL. Addison Wesley, Boston
- Berntson GM (1997) Topological scaling and plant root system architecture: developmental and functional hierarchies. New Phytol 135: 621–634 [Google Scholar]
- Bhuvaneswari TV, Bhagwat AA, Bauer WD (1981) Transient susceptibility of root cells in four common legumes to nodulation by Rhizobia. Plant Physiol 68: 1144–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin NJ (1991) Development of the legume root nodule. Annu Rev Cell Biol 7: 191–226 [DOI] [PubMed] [Google Scholar]
- Brewin NJ (2004) Plant cell wall remodeling in the Rhizobium-legume symbiosis. CRC Crit Rev Plant Sci 23: 293–316 [Google Scholar]
- Cardenas L, Vidali L, Dominguez J, Perez H, Sanchez F, Hepler PK, Quinto C (1998) Rearrangement of actin microfilaments in plant root hairs responding to Rhizobium etli nodulation signals. Plant Physiol 116: 871–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Timmers ACJ, Maillet F, Galera C, Penmetsa RV, Cook D, Denarie J, Gough C (2001) The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128: 1507–1518 [DOI] [PubMed] [Google Scholar]
- Complainville A, Brocard L, Roberts I, Dax E, Sever N, Sauer N, Kondorosi A, Wolf S, Oparka K, Crespi M (2003) Nodule initiation involves the creation of a new symplasmic field in specific root cells of Medicago species. Plant Cell 15: 2778–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford KM, Zambryski PC (1999) Plasmodesmata signaling: many roles, sophisticated statutes. Curr Opin Plant Biol 2: 382–387 [DOI] [PubMed] [Google Scholar]
- Denarie J, Debelle F, Prome JC (1996) Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem 65: 503–535 [DOI] [PubMed] [Google Scholar]
- Dudley ME, Jacobs TW, Long SR (1987) Microscopic studies of cell divisions induced in alfalfa roots by Rhizobium meliloti. Planta 171: 289–301 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR (1992) Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256: 998–1000 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Farnsworth KD, Niklas KJ (1995) Theories of optimization, form and function in branching architecture in plants. Funct Ecol 9: 355–363 [Google Scholar]
- Fiala JC, Allwardt B, Harris KM (2002) Dendritic spines do not split during hippocampus LTP or maturation. Nat Neurosci 5: 297–298 [DOI] [PubMed] [Google Scholar]
- Fiala JC, Harris KM (2002) Computer-based alignment and reconstruction of serial sections. Microscopy Anal 52: 5–7 [Google Scholar]
- Fisher NI (1993) Statistical Analysis of Circular Data. Cambridge University Press, Cambridge, UK
- Fraysse N, Couderc F, Poinsot V (2003) Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur J Biochem 270: 1365–1380 [DOI] [PubMed] [Google Scholar]
- Gage DJ (2004) Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68: 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ, Bobo T, Long SR (1996) Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J Bacteriol 178: 7159–7166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Kondorosi A, Kondorosi E, John M, Schmidt J, Schell J (1991) The role of nodulation genes in bacterium-plant communication. Genet Eng 13: 115–136 [DOI] [PubMed] [Google Scholar]
- Lhuisser FGP, De Ruijter NCA, Sieberer BJ, Esseling JJ, Emons AMC (2001) Time course of cell biological events evoked in legume root hairs by Rhizobium Nod factors: state of the art. Ann Bot (Lond) 87: 289–302 [Google Scholar]
- Libbenga KR, Harkes PAA (1973) Initial proliferation of cortical cells in the formation of root nodules in Pisum sativum. Planta 114: 17–28 [DOI] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R (2005) Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA 102: 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters BR (2004) Fractal analysis of the vascular tree in the human retina. Annu Rev Biomed Eng 6: 427–452 [DOI] [PubMed] [Google Scholar]
- McColloh KA, Sperry JS (2005) Patterns in hydraulic architecture and their implications for transport efficiency. Tree Physiol 25: 257–267 [DOI] [PubMed] [Google Scholar]
- Miller D, De Ruijter NCA, Bisseling T, Emons AMC (1999) The role of actin in root hair morphogenesis: studies with lipochitooligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J 17: 141–154 [Google Scholar]
- Miller DD, Leferink-ten Klooster HB, Emons AMC (2000) Lipochito-oligosaccharide nodulation factors stimulate cytoplasmic polarity with longitudinal endoplasmic reticulum and vesicles at the tip in vetch root hairs. Mol Plant Microbe Interact 13: 1385–1390 [DOI] [PubMed] [Google Scholar]
- Mulligan JT, Long SR (1989) A family of activator genes regulates expression of Rhizobium meliloti nodulation genes. Genetics 122: 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona P, Pawlowski K, Bisseling T (1995) Symbiotic nitrogen fixation. Plant Cell 7: 869–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RL, Farquhar ML (1996) Root hairs: specialized tubular cells extending root surfaces. Bot Rev 62: 1–40 [Google Scholar]
- Rae AL, Bonfante-Fasolo P, Brewin NJ (1992) Structure and growth of infection threads in the legume symbiosis with Rhizobium leguminosarum. Plant J 2: 385–395 [Google Scholar]
- Rathbun EA, Naldrett MJ, Brewin NJ (2002) Identification of a family of extensin-like glycoproteins in the lumen of Rhizobium-induced infection threads in pea root nodules. Mol Plant-Microbe Interact 15: 350–359 [DOI] [PubMed] [Google Scholar]
- Sharma SB, Signer ER (1990) Temporal and spatial regulation of the symbiotic genes of Rhizobium meliloti in planta revealed by transposon Tn5-gusA. Genes Dev 4: 344–356 [DOI] [PubMed] [Google Scholar]
- Timmers AC, Auriac MC, de Billy F, Truchet G (1998) Nod factor internalization and microtubular cytoskeleton changes occur concomitantly during nodule differentiation in alfalfa. Development 125: 339–349 [DOI] [PubMed] [Google Scholar]
- Timmers AC, Auriac MC, Truchet G (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628 [DOI] [PubMed] [Google Scholar]
- Uv A, Cantera R, Samakovlis C (2003) Drosophila tracheal morphogenesis: intricate cellular solutions to basic plumbing problems. Trends Neurosci 26: 301–309 [DOI] [PubMed] [Google Scholar]
- Uylings HBM, van Pelt J (2002) Measures for quantifying dendritic arborizations. Network 13: 397–414 [PubMed] [Google Scholar]
- van Brussel AAN, Bakhuizen R, van Spronsen PC, Spaink HP, Tak T, Lugtenberg BJJ, Kijne JW (1992) Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharidess of Rhizobium. Science 257: 70–72 [DOI] [PubMed] [Google Scholar]
- Vasse J, de Billy F, Camut S, Truchet G (1990) Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol 172: 4295–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veereshlingam H, Haynes JG, Penmetsa RV, Cook DR, Sherrier DJ, Dickstein R (2004) nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol 136: 3692–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward HM (1887) On the tubercular swellings on the roots of Vicia fava. Philos Trans R Soc London Ser B 178: 539–562 [Google Scholar]
- Wu L, McGechan MB, Watson CA, Baddeley JA (2005) Developing existing plant root system architecture models to meet future, agricultural challenges. Adv Agron 85: 181–219 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.