Abstract
Pepper (Capsicum annuum) serotonin N-hydroxycinnamoyltransferase (SHT) catalyzes the synthesis of N-hydroxycinnamic acid amides of serotonin, including feruloylserotonin and p-coumaroylserotonin. To elucidate the domain or the key amino acid that determines the amine substrate specificity, we isolated a tyramine N-hydroxycinnamoyltransferase (THT) gene from pepper. Purified recombinant THT protein catalyzed the synthesis of N-hydroxycinnamic acid amides of tyramine, including feruloyltyramine and p-coumaroyltyramine, but did not accept serotonin as a substrate. Both the SHT and THT mRNAs were found to be expressed constitutively in all pepper organs. Pepper SHT and THT, which have primary sequences that are 78% identical, were used as models to investigate the structural determinants responsible for their distinct substrate specificities and other enzymatic properties. A series of chimeric genes was constructed by reciprocal exchange of DNA segments between the SHT and THT cDNAs. Functional characterization of the recombinant chimeric proteins revealed that the amino acid residues 129 to 165 of SHT and the corresponding residues 125 to 160 in THT are critical structural determinants for amine substrate specificity. Several amino acids are strongly implicated in the determination of amine substrate specificity, in which glycine-158 is involved in catalysis and amine substrate binding and tyrosine-149 plays a pivotal role in controlling amine substrate specificity between serotonin and tyramine in SHT. Furthermore, the indisputable role of tyrosine is corroborated by the THT-F145Y mutant that uses serotonin as the acyl acceptor. The results from the chimeras and the kinetic measurements will direct the creation of additional novel N-hydroxycinnamoyltransferases from the various N-hydroxycinnamoyltransferases found in nature.
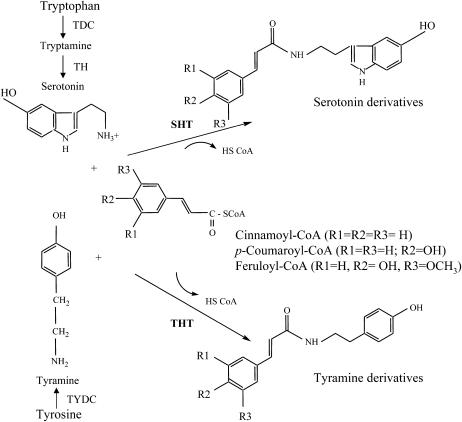
N-Hydroxycinnamic acid amides (HCAAs), which are present in a diverse array of plant species, have important roles in plant-plant (Martin-Tanguy and Negrel, 1987; Cutillo et al., 2003), plant-pathogen (Clarke, 1982), plant-insect (Lajide et al., 1995), and plant-environment (Negrel et al., 1993; Schraudner et al., 1993) interactions. HCAAs are synthesized by the condensation of hydroxycinnamoyl-CoA thioesters and aromatic amines (Fig. 1). The hydroxycinnamoyl-CoA thioesters include cinnamoyl-CoA, p-coumaroyl-CoA, caffeoyl-CoA, feruloyl-CoA, and sinapoyl-CoA, and are synthesized from cinnamic acid by a series of enzymes, including cinnamate-4-hydroxylase, coumarate-3-hydroxylase, caffeic acid O-methyltransferase, ferulate-5-hydroxylase, and hydroxycinnamate:CoA ligase (Douglas, 1996). Aromatic amines are generated from Tyr, Trp, and dihydroxyphenylalanine by decarboxylation or hydroxylation and include tyramine, octopamine, tryptamine, serotonin, dopamine, and noradrenaline (Wink, 1997). HCAAs represent an important class of antioxidant and chemotherapeutic agents (Zhang et al., 1996; Kawashima et al., 1998; Nagatsu et al., 2000; Park and Schoene, 2002). The biosynthesis of the HCAAs of tyramine is catalyzed by tyramine N-hydroxycinnamoyltransferase (THT; EC 2.3.1.110). This reaction has been well characterized in investigations of the mechanisms that regulate HCAA synthesis, THT activity, and the absolute levels of enzymes, in comparison with other classes of N-hydroxycinnamoyltransferases, such as anthranilate N-hydroxycinnamoyl/benzoyltransferase (EC 2.3.1.144; Yang et al., 1997) and agmatine N-hydroxycinnamoyltransferase (EC 2.3.1.64; Burhenne et al., 2003). Alignments of the amino acid sequences of these N-hydroxycinnamoyltransferases have revealed no significant similarities.
Figure 1.
Schematic diagram of the enzymatic reaction catalyzed by THT and SHT. Biosynthesis from Trp through serotonin is catalyzed by two enzymes, Trp decarboxylase (TDC) and tryptamine hydroxylase (TH), respectively. Tyrosine decarboxylase (TYDC) is responsible for the biosynthesis of tyramine.
Numerous THT enzymes from plant sources have been partially or completely purified and characterized (Farmer et al., 1999; Schmidt et al., 1999; Yu and Facchini, 1999; Ishihara et al., 2000). These enzymes behave as soluble proteins with a molecular mass of 26 to 28 kD. The deduced THT amino acid sequences, derived from cDNA sequences from potato (Solanum tuberosum), tobacco (Nicotiana tabacum), and tomato (Lycopersicon esculentum), show a high degree of amino acid sequence identity, ranging from 73% to 76%. Mechanistic studies have shown that all of the THT enzymes exhibit similar substrate affinity patterns, especially at the level of amine substrates, with the exception of the maize (Zea mays) enzyme (Ishihara et al., 2000). THT enzymes isolated from potato, tobacco, and tomato show the highest affinity for tyramine and octopamine (β-hydroxytyramine). Recently, a closely related N-hydroxycinnamoyltransferase gene was isolated from pepper (Capsicum annuum; Jang et al., 2004). This N-hydroxycinnamoyltransferase fused with a His tag was expressed in Escherichia coli and purified to homogeneity. The enzyme has a 16-fold lower Km for serotonin (73 μm) than for tyramine (1,165 μm) with feruloyl-CoA as the acyl donor, revealing serotonin N-hydroxycinnamoyltransferase activity (SHT). The subsequent ectopic expression of pepper SHT in rice (Oryza sativa) induced the production of large amounts of HCAAs of serotonin, such as feruloylserotonin and p-coumaroylserotonin, in transgenic rice, verifying in vivo that SHT is a true serotonin N-hydroxycinnamoyltransferase (Jang et al., 2004; Kang et al., 2005). In spite of considerable progress in the study of N-hydroxycinnamoyltransferases, at both the molecular and substrate specificity analysis levels, a more detailed study of the residues and domains required for substrate binding and enzyme activity has not been conducted.
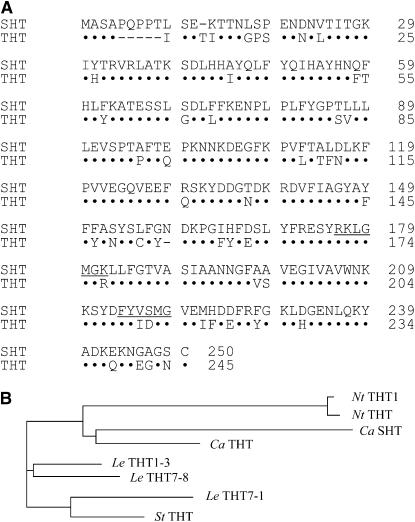
In this study, we attempted to determine the regions or the key residues of N-hydroxycinnamoyltransferase that are required for amine binding and enzyme activity, using pepper N-hydroxycinnamoyltransferases. We first isolated from pepper a THT that is not able to use serotonin as an acyl acceptor. Because of both the high level of amino acid identity between SHT and THT (78%) and a more or less equal distribution of amino acid substitutions and mismatches throughout the deduced amino acid sequences (Fig. 2A), we were not able to readily identify domains that might contribute to the amine-binding specificity domain. Therefore, a functional analysis using chimeric proteins constructed from the SHT and THT sequences was performed to identify a domain responsible for the amine-binding specificity, as well as key amino acids in pepper SHT and THT proteins.
Figure 2.
Comparison of the deduced amino acid sequences of SHT (GenBank accession no. AF329463) and THT (GenBank accession no. AY819700), and a phylogenetic tree of THTs and SHT. A, Identical amino acids are denoted as dots, and gaps are indicated by dashes. The conserved acyl-CoA binding sites are underlined (RxxxGXG and FYxxxG represent domains I and II, respectively). B, Phylogenetic analysis was performed using the PHYLODRAW program (http://pearl.cs.pusan.ac.kr/phylodraw). NtTHT1 and NtTHT represent THTs from tobacco (AJ131768 and AJ005062); LeTHT1-3, LeTHT7-8, and LeTHT7-1 are THTs from tomato (AY081905, AY081907, and AY081906); StTHT denotes a THT from potato (AB061243); and CaSHT is from pepper (AF329463).
RESULTS
Isolation and Characterization of a cDNA Clone Encoding THT
Based on the sequence of a pepper expressed sequence tag (EST; http://plant.pdrc.re.kr/ks200201/pepper.html), an EST clone (KS01044B06) highly homologous to tobacco THT was isolated and sequenced. The pepper cDNA clone contains a 956-bp fragment with a 50-bp 5′-untranslated region, a 738-bp open reading frame, and a 168-bp 3′-untranslated region (GenBank accession no. AY819700). The open reading frame in the clone encodes a 245-amino acid protein with a predicted molecular mass of 28,221 D. A comparison of this polypeptide with that of the previously reported pepper SHT (GenBank accession no. AF329463) revealed 78% identity between the proteins. The protein has a high level of homology with THTs from other plants, including 83%, 80%, and 71% amino acid identity with tomato, potato, and tobacco THT proteins, respectively (Farmer et al., 1999; Schmidt et al., 1999; Von Roepenack-Lahaye et al., 2003). In the phylogenetic tree based on amino acid sequence of N-hydroxycinnamoyltransferases from four plant species, pepper SHT showed a greater evolutionary distance from pepper THT than other THTs within species (Fig. 2B).
Enzymatic Properties of the Recombinant THT Enzyme
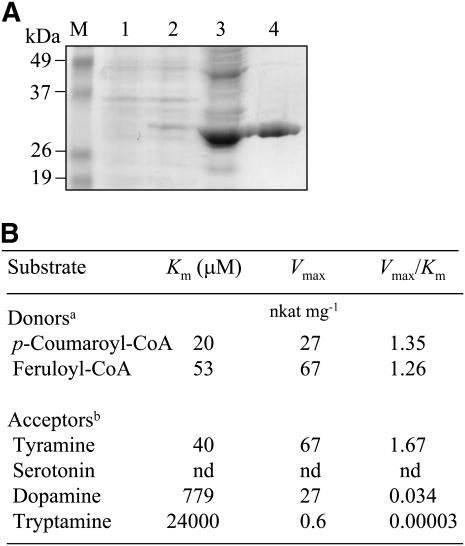
To verify that the pepper cDNA encodes a protein with THT activity, a His-tagged recombinant form of pepper THT was produced in E. coli, purified using affinity chromatography, and characterized (Fig. 3A). The enzyme kinetics of the purified His-tagged pepper THT were investigated using two cinnamoyl-CoA esters as acyl donors and a series of amines as acceptors (Fig. 3B). The reaction velocity (Vmax) of the recombinant pepper THT was highest for feruloyl-CoA (67 nkat mg−1), followed by p-coumaroyl-CoA (27 nkat mg−1). Of the acceptor amines tested, tyramine showed the maximum Vmax value (67 nkat mg−1), followed by dopamine (27 nkat mg−1), when feruloyl-CoA was used as the acyl donor. Tryptamine had a negligible Vmax of 1 nkat mg−1, and no serotonin conversion was detected. p-Coumaroyl-CoA had a Km value 2.5-fold less than that for feruloyl-CoA when tyramine was used as the acyl acceptor. Based on the Vmax/Km values, both acyl donors appear to be equally converted. In contrast to the acyl donors, tyramine was the preferred substrate, with a Km of 40 μm, which is approximately 20 times less than that of dopamine, 779 μm. Other acyl acceptors, such as tryptamine or serotonin, were not accepted as THT substrates. A preference for tyramine as the acyl acceptor was clearly shown by the Vmax/Km values, which for tyramine was 56 times higher than for dopamine. These substrate specificity data clearly show that the enzyme encoded by the pepper cDNA clone (KS01044B06) has THT enzyme activity.
Figure 3.
Affinity purification and substrate specificity analysis of the E. coli-expressed His-tagged THT protein. A, Expression of the pepper THT gene (AY819700) in E. coli. Protein samples were separated by SDS-PAGE and stained using Coomassie Blue. Lane M, Molecular standard; lane 1, total proteins in a 10-μL aliquot of bacterial cells grown without IPTG; lane 2, total proteins in a 10-μL aliquot of bacterial cells after IPTG treatment; lane 3, 20 μg of soluble proteins; lane 4, the THT protein (10 μg) purified by affinity (Ni-NTA) chromatography. B, Substrate specificity of the purified recombinant THT protein. Footnote a, Tyramine (1 mm) was used as the acyl acceptor. Footnote b, Feruloyl-CoA (250 μm) was used as the acyl donor. nd, Not detected.
Differential Expression of SHT and THT in Pepper Tissues
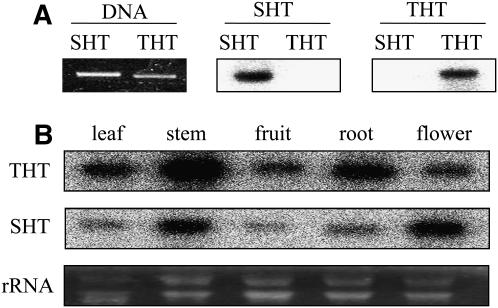
Full-length SHT and THT cDNA fragments were separated by agarose gel electrophoresis, and two identical DNA blots were hybridized independently with 32P-labeled SHT and THT cDNA clones (Fig. 4A). Under high-stringency hybridization and washing conditions, each DNA showed specific hybridization to its complementary sequence, despite the 77% identity of the SHT and THT nucleotide sequences. The differential and organ-specific expression of SHT and THT genes in mature pepper plants is shown in Figure 4B. Both the SHT and THT mRNAs were expressed constitutively in all tissues analyzed, but their expression levels in the different tissues varied. The highest amounts of THT transcripts were present in young stems, followed by roots, whereas peak levels of the SHT mRNA were observed in flowers and stems. Even though both the SHT and THT mRNAs were abundant in young stems, the THT mRNA was preferentially detectable in roots as compared to the SHT mRNA, which was present at higher levels in flowers. This result may suggest that THT and SHT genes have different biological functions in pepper plants.
Figure 4.
Specificity of N-hydroxycinnamoyltransferase probes and differential expression of the SHT and THT genes in pepper plants. A, Samples (100 ng) of the SHT and THT cDNA fragments were separated on a 1.5% agarose gel, stained with ethidium bromide, and blotted to a membrane. The blots were hybridized with the 32P-labeled probes denoted above the images. B, Northern-blot analysis of N-hydroxycinnamoyltransferase transcript levels in various organs from mature (2-month-old) pepper plants. The RNAs (20 μg) were hybridized with 32P-labeled SHT or THT probes at high stringency. Equal loading of the total RNA was demonstrated by ethidium bromide staining.
Determination of the Amine-Binding Specificity Domain
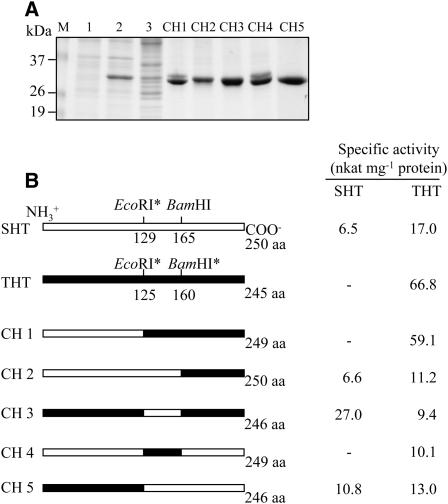
Pepper THT and SHT are responsible for the production of the HCAAs of tyramine and serotonin, respectively. Although the kinetics of these enzymes have been well documented, no information is currently available concerning their substrate specificity or catalytic domains. SHT prefers serotonin as an acyl acceptor and also has a low substrate specificity affinity toward tyramine, but we found that THT only uses tyramine as an acyl acceptor, with no affinity for serotonin. Both the high amino acid identity and the distinct substrate specificities of the two proteins prompted us to generate chimeric proteins to identify domains that might contribute to the amine-binding specificities for tyramine or serotonin. Chimeric genes were generated by swapping corresponding domains between the SHT and THT genes, after which the chimeric genes were expressed in bacteria as described in “Materials and Methods.” The recombinant chimeric proteins, each fused with a hexa-His tag, were affinity purified (Fig. 5A) and assayed for SHT and THT activity (Fig. 5B). Five chimeric constructs were generated and assayed. The specific activities of SHT, which has bifunctional enzyme activity, were 6.5 and 17 nkat mg−1 protein for SHT and THT, respectively. In contrast, THT exhibited only THT-specific activity, with 67 nkat mg−1 protein. Chimera 1 (CH1) showed only THT activity, with a specific activity similar to the original THT. This result suggests that the carboxy-terminal halves of the two proteins contribute to the specificity of the amine-binding reaction. CH5 is the converse construct of CH1, containing the THT amino terminus and the SHT carboxy terminus, and the specificity would be expected to be that of SHT. A comparison of CH1 and CH2 provides evidence that the amine-binding specificity resides within a domain of approximately 37 amino acids corresponding to amino acids 129 to 165 of the SHT sequence. This interpretation was further confirmed by the results from constructs CH3 and CH4. CH3 represents a substitution of amino acids 125 to 160 from THT with the corresponding amino acids of SHT and exhibits SHT activity. CH4 is a substitution of amino acids 129 to 165 of SHT with the corresponding THT sequence and results in THT-specific activity. This generation and characterization of a series of chimeric genes has demonstrated unequivocally that the amine-binding specificity domain resides within amino acid residues 129 to 165 of SHT and the counterpart residues 125 to 160 of THT.
Figure 5.
Chimeric constructs used to map the catalytic domains of the N-hydroxycinnamoyltransferases and specific enzyme activities. A, Expression of the chimeric genes in E. coli and affinity purification of His-tagged chimeric constructs. Lane M, Molecular standard; lane 1, total proteins in 10 μL of bacterial cells grown without IPTG; lane 2, total proteins in 10 μL of bacterial cells after IPTG treatment; lane 3, 20 μg of soluble proteins; lane 4, chimeric proteins (5–10 μg) purified by affinity (Ni-NTA) chromatography. B, Schematic diagrams of chimeric constructs. Specific enzyme activities were derived from kinetic analyses.
Catalytic Specificities of the Recombinant Chimeric Proteins
To examine more closely the mechanistic roles of the amine substrate specificity domains in these chimeric proteins, their kinetic constants were determined. As shown in Table I, the kinetic parameters of the different proteins spanned a remarkably wide range. The affinities for tyramine and serotonin varied 416- and 32-fold, respectively, whereas the corresponding maximal velocities varied 7- and 4-fold. Wild-type SHT and THT exhibited distinct substrate specificities, with the former having a higher Vmax/Km for serotonin and the latter having a Vmax and Km exclusive for tyramine. Of the five chimeric genes, the two chimeras (CH1 and CH4) containing the THT amine specificity region (amino acids 125–160) displayed THT-type kinetics, characterized by the exclusive use of tyramine as an acyl acceptor, but the Km and Vmax values of the chimeras differed from those of wild-type THT. The Km value of CH1 was 18-fold greater than that of wild-type THT, but its Vmax value was comparable to that of wild-type THT. CH4 showed a 266-fold increase in the Km for tyramine and an approximately 7-fold decrease in the Vmax, leading to an overall 1,861-fold decrease in the Vmax/Km. These results suggest that both the N-terminal and C-terminal portions of THT outside the amine-specificity domain (amino acids 125–160) are also involved in the catalytic process and the efficient binding of substrate. Of the five chimeras, three (CH2, CH3, and CH5) exhibited SHT-type kinetics because of the presence of the SHT amine-specificity region (amino acids 129–165). The affinity for tyramine (Km) was reduced by 6-, 14-, and 10-fold in CH2, CH3, and CH5, respectively, compared with that of wild-type SHT. In addition, these SHT-like chimeras showed a 1.3- to 1.8-fold decrease in the Vmax for tyramine. The Vmax for serotonin in the SHT-type chimeras increased from 1.2- to 4.2-fold, but the affinities for serotonin decreased by approximately 3-, 32-, and 13-fold for CH2, CH3, and CH5, respectively. These results suggest that the SHT specificity domain plays a role in increasing the Vmax for serotonin, as shown with the SHT-type chimeras (CH2, CH3, and CH5), but that the THT specificity domain tended to lower the Vmax values for tyramine (as seen with CH1 and CH4). Unlike the Vmax values, all of the chimeras had high Km values for both tyramine and serotonin.
Table I.
Kinetic constants of wild-type SHT, THT, and chimeric proteins with tyramine and serotonin as amine substrates
Feruloyl-CoA (250 μm) was used as the acyl donor. ND, Not detected.
| Enzyme
|
Tyramine
|
Serotonin
|
||||
|---|---|---|---|---|---|---|
| Km | Vmax | Vmax/Km | Km | Vmax | Vmax/Km | |
| μm | nkat mg−1 | μm | nkat mg−1 | |||
| SHT | 1,165 | 17.0 | 0.0146 | 73 | 6.5 | 0.0890 |
| THT | 40 | 66.8 | 1.6750 | ND | ND | ND |
| CH1 | 720 | 59.1 | 0.0820 | ND | ND | ND |
| CH2 | 7,400 | 11.2 | 0.0015 | 250 | 6.6 | 0.0264 |
| CH3 | 16,658 | 9.4 | 0.0005 | 2,360 | 27.0 | 0.0114 |
| CH4 | 10,650 | 10.1 | 0.0009 | ND | ND | ND |
| CH5 | 11,486 | 13.0 | 0.0011 | 942 | 10.8 | 0.0115 |
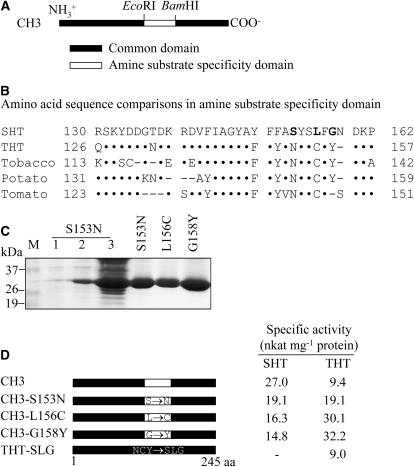
Catalytic Specificities of Point-Mutated Chimeric CH3 Proteins
Through the generation and characterization of a series of chimeric genes, we identified the amine substrate specificity domain of the SHT and THT proteins (Fig. 6A). An alignment of the amino acid sequences of these domains is shown in Figure 6B. The amine substrate specificity domains consist of 33 and 32 amino acids in SHT and THT, respectively, differing by seven amino acids. Two (Tyr or Phe at 149 and Phe or Tyr at 151, based on SHT numbering) of these seven amino acids were substituted by similar amino acids, and two others (Arg at 130 and Thr at 137) were variable between the pepper SHT and the various THTs. The amino acid residues that are distinctly different between SHT and THT are Ser, Leu, and Gly at 153, 156, and 158, respectively, of the SHT sequence. To verify the roles of these amino acid residues, we first employed a CH3 chimeric gene as a template for point mutation. The point-mutated CH3 chimeric genes were expressed in E. coli and affinity purified (Fig. 6C). To characterize the point-mutated CH3s, we assayed the N-hydroxycinnamoyltransferase activities of the mutated CH3s using tyramine and serotonin as acyl acceptors. All three of the mutated CH3 genes containing mutations within the amine substrate specificity domain (S153N, L156C, and G158Y) displayed lower SHT activity (Fig. 6D). However, the THT-specific activity of these mutated CH3 proteins increased accordingly. For example, in G158Y, in which the Tyr of THT was substituted for the Gly of SHT at position 158, the SHT activity was inhibited by 45%, but the THT activity was increased by 342%. Similarly, L156C, in which the Leu at 156 of SHT was replaced with the Cys of THT, exhibited a 41% lower SHT activity, whereas the THT activity increased by 320%. These results suggest that Ser-153, Leu-156, and Gly-158 have important roles in regulating the amine substrate specificity of SHT. Next, we investigated the kinetic constants of the mutated CH3 proteins to determine whether their specific activities are associated with more profound effects on the substrate specificity kinetics. As shown in Table II, the CH3 chimera displayed bifunctional substrate specificities with a Vmax/Km value for serotonin that was higher than that for tyramine. In agreement with the specific activity results obtained from the three mutated CH3 proteins, all three, in which residues present in the amine substrate specificity domain were mutated, exhibited different Vmax/Km values with either tyramine or serotonin as substrate because of variations in the Vmax and Km values, as compared to those of the unmutated CH3. All three of the mutated CH3 proteins exhibited decreased Vmax/Km values for serotonin, ranging from 18% to 60% of that of the original CH3, whereas the catalytic efficiency with tyramine as the substrate, as reflected by the Vmax/Km, increased toward tyramine by an average of 2-fold. It is particularly notable that the G158Y mutation resulted in a 3-fold increase in the Km for serotonin, whereas the Vmax decreased by 2-fold, leading to a 5-fold decrease in the Vmax/Km values. In contrast to the results for serotonin, G158Y exhibited Km and Vmax values for tyramine that were increased by 1.5- and 3.4-fold, respectively. This resulted in a greater than 2-fold increase in the Vmax/Km with tyramine as substrate. Similar to the G158Y mutant, mutations at S153N and L156C resulted in an increase in the Vmax/Km with tyramine and a decrease in the Vmax/Km with serotonin. These results indicate that these mutations at the three amino acids are sufficient to cause significant changes in the substrate specificity from serotonin to tyramine. However, none of these mutations was able to abolish the SHT activity of the bifunctional CH3 chimera. This result was further confirmed by constructing THT-SLG, in which Asn-149, Cys-152, and Tyr-154 of THT were replaced with the Ser-153, Leu-156, and Gly-158 of SHT. The NCY/SLG-mutated THT exhibited a significant decrease in the specific activity as compared to that of wild-type THT (Fig. 6D). The reduced specific activity was accompanied by a significant decrease in the substrate affinity and catalytic efficiency, with an approximately 425-fold increase in the Km and a 7-fold decrease in the Vmax. The concerted action of the three mutated residues (THT-SLG) resulted in a substantial reduction in the THT catalytic efficiency rather than a gain in the SHT activity (Table II).
Figure 6.
Comparison of amino acid sequences in the amine substrate specificity domain and specific activities of mutated CH3 proteins. A, Schematic diagram of the amine substrate specificity domain. B, Comparison of the amino acid sequences of the N-hydroxycinnamoyltransferases. The sequence of pepper SHT (AF329463) was aligned with the THT sequences from pepper (AY819700), tobacco (AJ131768), potato (AB061243), and tomato (AY081907). Identical amino acids are denoted as dots, and gaps are indicated by dashes. C, Expression of the mutated CH3 genes in E. coli and affinity purification of His-tagged mutated CH3 constructs. Lane M, Molecular standard; lane 1, total proteins in 10 μL of bacterial cells of the CH3-S153N strain grown without IPTG; lane 2, total proteins in 10 μL of bacterial cells of the CH3-S153N strain after IPTG treatment; lane 3, 20 μg of soluble proteins from the CH3-S153N strain; right lanes (S153N, L156C, and G158Y), mutated CH3 proteins (10 μg) purified by affinity (Ni-NTA) chromatography. D, Constructs of point-mutated CH3 chimeras and measurements of their specific activities. THT-SLG is a THT protein containing three mutated residues in which Asn-149, Cys-152, and Tyr-154 were replaced with the corresponding Ser-153, Leu-156, and Gly-158 of SHT, respectively. Enzyme assays were performed as described in Figure 5B.
Table II.
Kinetic constants of mutated CH3 and SHT proteins with tyramine and serotonin as amine substrates
Feruloyl-CoA (250 μm) was used as the acyl donor. ND, Not detected.
| Enzyme
|
Tyramine
|
Serotonin
|
||||
|---|---|---|---|---|---|---|
| Km | Vmax | Vmax/Km | Km | Vmax | Vmax/Km | |
| μm | nkat mg−1 | μm | nkat mg−1 | |||
| CH3 | 16,658 | 9.4 | 0.0005 | 2,360 | 27.0 | 0.0114 |
| CH3-S153N | 28,000 | 19.1 | 0.0007 | 3,789 | 19.1 | 0.0050 |
| CH3-L156C | 27,900 | 30.1 | 0.0011 | 2,382 | 16.3 | 0.0068 |
| CH3-G158Y | 25,800 | 32.2 | 0.0012 | 7,006 | 14.8 | 0.0021 |
| THT | 40 | 66.8 | 1.6750 | ND | ND | ND |
| THT-SLG | 17,000 | 9.0 | 0.0005 | ND | ND | ND |
| SHT | 1,165 | 17.0 | 0.0146 | 73 | 6.5 | 0.0890 |
| SHT-S153N | 1,325 | 55.3 | 0.0417 | 129 | 9.1 | 0.0705 |
| SHT-L156C | 1,739 | 40.8 | 0.0234 | 44 | 6.6 | 0.1500 |
| SHT-G158Y | 14,951 | 5.1 | 0.0003 | 19,876 | 29.4 | 0.0015 |
| SHT-NCY | 31,541 | 7.2 | 0.0002 | 7,546 | 15.7 | 0.0020 |
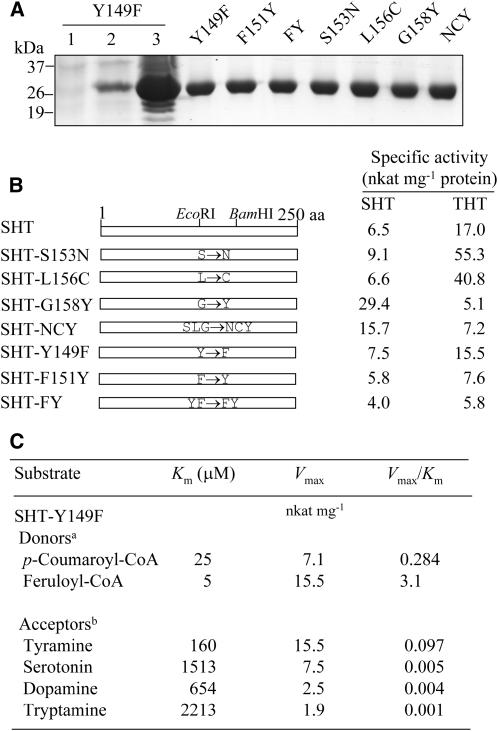
| SHT-Y149F | 160 | 15.5 | 0.0968 | 1,513 | 7.5 | 0.0049 |
| SHT-F151Y | 783 | 7.6 | 0.0097 | 450 | 5.8 | 0.0128 |
| SHT-FY | 1,370 | 5.8 | 0.0042 | 4,051 | 4.0 | 0.0010 |
Catalytic Specificities of Point-Mutated SHT Proteins
To confirm that the three amino acid residues mentioned above also play a significant role in regulating the catalytic activity of SHT protein, we also point mutated and affinity purified SHT proteins, as was done with the chimeric CH3 proteins. Furthermore, we point mutated two more amino acids, Tyr-149 and Phe-151, which were substituted by similar amino acids between SHT and THT. With the single point-mutated SHT proteins purified as above (Fig. 7A), the SHT- and THT-specific activities showed significant dissimilarities compared to those of the wild-type SHT protein (Fig. 7B). For example, the replacement of either Ser with Asn at 153 (SHT-S153N) or Leu with Cys at 156 (SHT-L156C) had an additive effect on the specific activities of THT by 2.8- and 2.1-fold, respectively. However, when Gly was replaced with Tyr at 158 (SHT-G158Y), the enzyme activity of THT decreased about 2.9-fold, whereas the SHT activity increased by about 4-fold, suggesting a crucial role of Gly for THT activity in the SHT enzyme. As for the three-point-mutated SHT (SHT-NCY), the specific activity of SHT increased 2-fold, but THT activity decreased about 2.3-fold. As compared to SHT-G158Y, SHT-NCY had 25% higher specific activity for THT, suggesting that these two amino acids (i.e. Ser-153 and Leu-156) are necessary to sustain a high rate of THT activity. Other mutant constructs, such as SHT-Y149F, SHT-F151Y, and SHT-FY, showed low THT and SHT specific activities relative to wild-type SHT, except the SHT-Y149F with a 1.2-fold increase of SHT activity.
Figure 7.
Specific activities of point-mutated SHT proteins and kinetic analysis of SHT-Y149F protein. A, Expression of the mutated SHT genes in E. coli and affinity purification of His-tagged mutated SHT constructs. Lane 1, Total proteins in 10 μL of bacterial cells of the SHT-Y149F strain grown without IPTG; lane 2, total proteins in 10 μL of bacterial cells of the SHT-Y149F strain after IPTG treatment; lane 3, 20 μg of soluble proteins from the SHT-Y149F strain; right lanes (Y149F through NCY), mutated SHT proteins (10 μg) purified by affinity (Ni-NTA) chromatography. B, Constructs of point-mutated SHT chimeras and measurements of their specific activities. SHT-FY is a SHT protein containing two mutated residues in which Tyr-149 and Phe-151 were replaced with the corresponding Phe-145 and Tyr-147 of THT. SHT-NCY is a SHT protein containing three mutated residues in which Ser-153, Leu-156, and Gly-158 were replaced with the corresponding Asn-149, Cys-152, and Tyr-154 of THT, respectively. Enzyme assays were performed as described above. C, Substrate specificity analysis of the SHT-Y149F protein. Footnote a, Tyramine (1 mm) was used as the acyl acceptor. Footnote b, Feruloyl-CoA (250 μm) was used as the acyl donor.
The pivotal role of these amino acids was clearly evident when judged using kinetic analyses (Table II). For example, the catalytic efficiency (Vmax/Km) with tyramine increased 2.8-fold in SHT-S153N as compared to wild-type SHT. The Vmax/Km value of SHT-L156C increased in a manner similar to that of SHT-S153N, implicating Ser-153 and Leu-156 in THT catalysis, rather than SHT catalysis. Additionally, the SHT-L156C exhibited the decrease in Km for serotonin by 1.7-fold, resulting in a new type of bifunctional SHT enzyme with a higher catalytic efficiency toward both tyramine and serotonin substrates relative to wild-type SHT. Unlike SHT-S153N and SHT-L156C, SHT-G158Y resulted in a dramatic decrease in Vmax/Km with tyramine because of a 13-fold increase in Km and a 3-fold decrease in Vmax as compared to wild-type SHT. This is in contrast with the Vmax value with serotonin, which increased 4-fold compared to wild-type SHT. Again, these data suggest that Gly-158 is essential for catalysis and substrate binding, especially with tyramine. SHT-NCY, which contained three amino acid substitutions (S153N, L156C, G158Y), showed a similar catalytic efficiency as compared to SHT-G158Y. Contrary to the specific activity data, SHT-Y149F transformed SHT enzymes into the THT type by reversing the substrate affinity toward serotonin and tyramine. The Km for serotonin in the SHT-Y149F was increased by 21-fold, but the Km for tyramine was reduced by 7-fold, with a marginal change of corresponding Vmax values resulting in a higher catalytic efficiency for tyramine (Vmax/Km = 0.0968) than for serotonin (Vmax/Km = 0.0049). This phenomenon is corroborated by the SHT-FY double mutant in which the catalytic efficiency for tyramine is 4-fold higher than for serotonin. SHT-F151Y plays a role in modulating substrate affinity, but its affinity to serotonin is still higher (Km = 450 μm) than tyramine (Km = 783 μm). The determining role of Tyr-149 in wild-type SHT for regulating a substrate preference between serotonin and tyramine was further confirmed on the kinetics and substrate specificity (Fig. 7C). The Km values in the SHT-Y149F for two acyl donors, p-coumaroyl-CoA and feruloyl-CoA, were 25 and 5 μm, respectively, which are comparable to those measured in wild-type SHT (Jang et al., 2004). This indicates that the mutation in Tyr-149 into Phe-149 does not alter acyl donor substrate binding or catalysis. The Vmax for various acyl acceptors differed within the range of 160 to 2,213 μm in the SHT-Y149F with a higher affinity for tyramine (Km = 160 μm) than for serotonin (Km = 1,513 μm) when feruloyl-CoA was used as the acyl donor. Thus, Km for tyramine is 9-fold lower than for serotonin, suggesting that the SHT-Y149F mutant behaves like a THT enzyme. In contrast to wild-type THT, the SHT-Y149F mutant has an additional ability to accept serotonin and tryptamine as acyl acceptors. Collectively, this result clearly reveals that Tyr-149 is a critical amino acid residue for enabling wild-type SHT to accept serotonin with a high affinity.
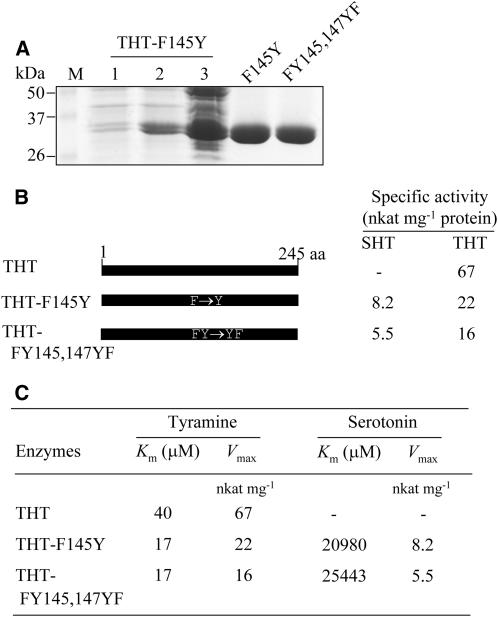
Conversion from THT to SHT by Mutating Phe-145 to Tyr in THT
Given the key role played by Tyr-149 in SHT in regulating the serotonin specificity, it is of great interest to examine whether the mutant THT enzyme, in which a Phe residue (F145Y) is mutated to a Tyr residue, gains SHT activity by accepting serotonin as the acyl acceptor. As shown in Figure 8, THT-F145Y is capable of utilizing serotonin as a substrate, showing SHT activity in addition to THT activity. Hence, the replacement of Phe-145 with Tyr in THT contributes pivotally to the change in accommodating serotonin as a substrate, resulting in a novel bifunctional THT enzyme like SHT. The kinetic constants of THT-F145Y differed from those of wild-type THT (Fig. 8, B and C). THT-F145Y possessed approximately 3-fold less THT-specific activity than wild-type THT, but acquired a novel SHT activity of 8 nkat mg−1 protein. Interestingly, THT-F145Y had increased affinity for tyramine (Km = 17 μm), and its Vmax value decreased by approximately 3-fold relative to that of wild-type THT. In addition, THT-F145Y had a Km for serotonin of 20,980 μm and a Vmax of 8.2 nkat mg−1. The double mutant THT-FY145,147YF showed kinetic values similar to those of THT-F145Y, indicating that Phe-145 in THT is primarily involved in regulating amine-binding specificity.
Figure 8.
Specific activities and kinetic constants of point-mutated THT-F145Y and THT-FY145,147YF proteins. A, Expression of the mutated THT genes in E. coli and affinity purification of His-tagged mutated THT constructs as described in Figure 7. B, Constructs of point-mutated THT and their specific activities. C, Kinetic constants of the mutated proteins. Enzyme assay and kinetic analysis were performed as described in Figure 7.
DISCUSSION
It has been well documented that THT enzymes have a broad range of substrate specificities and are encoded by a small gene family (Farmer et al., 1999; Schmidt et al., 1999). Tobacco and potato contain at least three THT isogenes, as shown by genomic Southern analyses. It would be intriguing to know whether all of the isogenes encode functional THT enzymes with HCAA synthesis activity and whether they display similar substrate specificities. With this aim, Von Roepenack-Lahaye et al. (2003) isolated and characterized four THT isogenes from tomato. The deduced amino acid sequences exhibited 93% similarity. However, only three of the isogenes encoded functional THT enzymes, each with the highest affinity toward tyramine as the acyl acceptor. Interestingly, the second best acyl acceptor varied among the three THT enzymes, suggesting that the THT isogenes encode proteins with different substrate specificities and physiological roles. The presence of a small gene family with the distinct characteristics of the THT isogenes was demonstrated in pepper. The two highly similar THT isogenes identified have been demonstrated to encode two different enzymes, THT and SHT. In addition to its presence in the Solanaceae, THT activity has been observed in other plant families, such as the Papaveraceae (Yu and Facchini, 1999) and Gramineae (Ishihara et al., 2000; Jang et al., 2004), but no gene structure data have been reported as yet.
SHT and THT belong to the GCN5-related family of structurally similar N-acetyltransferases, which includes streptothricin acetyltransferase, gentamicin 3′-acetyltransferase, aminoglycoside 6′-N-acetyltransferase, spermidine/spermine N-acetyltransferase, and serotonin N-acetyltransferase, although SHT and THT do not share extensive homology with these proteins (Lu et al., 1996). Nevertheless, all of these enzymes have molecular masses of around 20 to 25 kD and contain two conserved domains responsible for acetyl-CoA binding and enzyme activity. The domains I and II of the N-acetyltransferases, which have the amino acid sequences RGFGIG and FYXRXG, are well conserved, as compared to the RKLGMG and FYXXXG sequences of SHT and the THTs, respectively. Interestingly, the distance between domains I and II and their locations in the C-terminal portion of the N-acetyltransferase proteins are also well conserved. The pivotal roles of these domains were further confirmed by the determination of the three-dimensional structure of serotonin N-acetyltransferase (Hickman et al., 1999), which showed that domain I is involved in acetyl-CoA binding and domain II is associated with substrate binding. Furthermore, this study indicates that Tyr-168 may play a role in the reprotonation of the thiolate leaving group as a general acid. In contrast, our study of chimeras of SHT and THT showed that domain II is not implicated in amine substrate binding but contains the common domain for enzyme activity. The domain for amine substrate specificity is located within amino acids 129 to 165, a sequence that is upstream of domain I by only 10 amino acids. The amino acid sequences of the amine substrate domains of pepper SHT and pepper THT have 76% identity and 82% similarity. However, whether the entire stretch of amino acid residues is necessary for determination of the substrate by the SHT enzyme, controlling the accessibility of the bulky substrate serotonin versus the smaller substrate tyramine, is unknown. To clarify this issue, we generated several mutated SHT proteins to determine whether a single point mutation could affect the substrate specificity of SHT. Of the five mutants in wild-type SHT, mutation of Tyr-149 to Phe (SHT-Y149F) dramatically reverses the substrate affinity from serotonin to tyramine, indicating that this residue plays a key role in serotonin substrate binding and catalysis. The essential role of Tyr-149 in SHT seems to be different from that of Tyr-168 as a general acid in the serotonin N-acetyltransferase, whose mutation of Tyr-168 to Phe significantly reduces its catalysis (Hickman et al., 1999). In contrast, mutation of Tyr-149 to Phe in the SHT did not affect Vmax either for serotonin or tyramine, suggesting that Tyr-149 may not function as a general acid for amine substrates. It is therefore probable that Tyr-149 could serve to accurately place a serotonin substrate with an unknown Tyr residue played as a general acid in the active pocket.
Other mutant constructs were also capable of modulating the specific activity ratio in keeping with varying Km and Vmax, but none of the mutations, including Y149F, completely abolished the serotonin or tyramine substrate specificity in SHT. Thus, it is highly likely that the mutant THT enzyme, in which a Phe residue (F145Y) is mutated by a Tyr, will be a bifunctional enzyme accepting both serotonin and tyramine as the acyl acceptors as shown in the chimeric CH3. This hypothesis was further tested and confirmed by constructing the THT-F145Y in which the mutant THT-F145Y turned out a bifunctional enzyme catalyzing serotonin and tyramine as the acyl acceptors.
In addition, it is worth noting that SHT-L156C had 1.6-fold higher catalytic efficiencies with both serotonin and tyramine, respectively, as compared to wild-type SHT. This suggests that it is possible to generate either THT with greater catalytic efficiency for a tyramine substrate only or a bifunctional enzyme with a higher substrate affinity for the specific amine substrates (i.e. tyramine and serotonin) through a series of site-directed mutageneses within various N-hydroxycinnamoyltransferases found in nature.
MATERIALS AND METHODS
Isolation and Sequence Identification of a THT Clone
The EST clones KS01044B06 and KS01062D08 from pepper (Capsicum annuum) were provided by the Laboratory of Plant Functional Genomics (Korea Research Institute of Bioscience and Biotechnology). An EST database was then generated by single-pass sequencing of 5′-cDNA termini (http://plant.pdrc.re.kr/ks200201/pepper.html). Two EST cDNAs homologous to THT were identified in the EST database. The sequence of one cDNA clone (KS01062D08) was identical to that of a previously reported pepper SHT (GenBank accession no. AF329463) in the 3′-untranslated and coding regions. The other cDNA clone harbored a full-length THT-like insert, but differed from the previously described SHT gene in both the open reading frame and the 3′-untranslated region. The KS01044B06 cDNA was analyzed further and characterized. The DNA sequence was determined using the dideoxynucleotide chain termination method, according to the manufacturer's instructions (Sequenase). Northern-blot analysis was performed as described previously (Jang et al., 2004).
Bacterial Expression and Enzyme Purification
The plasmid pET-28(b) (Novagen) was employed to develop an Escherichia coli expression system for enzyme purification. The full-length THT gene was amplified by PCR using the primer F1003 5′-d(GCCATGGCTTCAGCTATA)-3′ as the forward primer (the NcoI restriction site is underlined and the translation start codon is in bold), the primer F1004 5′-d(CCCCTCGAGACAGTTTCCTCCTTC)-3′ (the XhoI site is underlined) as the reverse primer, and the cDNA harboring the THT gene as the template. The PCR product was digested with NcoI and XhoI, gel purified, and ligated in-frame into pET-28(b) between the same restriction sites. E. coli BL21(DE3) was used as the host strain for the construct containing the THT gene. Other chimeric constructs were fused into pET-28(b), as described above. The cell culture and purification procedures were as described previously (Mathis et al., 1997; Back et al., 2001).
Substrate Specificity Analyses
Substrate specificity analyses of the purified His-tagged proteins were performed as described by Jang et al. (2004). In brief, unless otherwise indicated, the purified recombinant enzymes were assayed in a mixture of 10 μL of 1 mm feruloyl-CoA, 10 μL of 800 mm tyramine (or serotonin for the SHT assay), and 70 μL of 100 mm Tris-HCl (pH 8.5). After incubation for 10 min at 30°C, the reaction was stopped with 20 μL of acetic acid. The mixture was diluted to 500 μL with methanol, and a 10-μL aliquot was subjected to HPLC analysis, as described previously (Jang et al., 2004). To determine the Km and Vmax, varying substrate and enzyme concentrations were used, depending on the recombinant enzymes tested. The Km and Vmax values were calculated from Lineweaver-Burk plots. Protein concentrations were determined by the Bradford method, using the Bio-Rad protein assay.
Construction of Chimeric Genes
The full-length SHT and THT cDNAs were initially fused between the NcoI and XhoI sites of pET-28(b), resulting in the constructs pET28(B)-SHT and pET-28(b)-THT. The chimeric constructs CH1, CH2, CH3, CH4, and CH5 were generated by introducing EcoRI and/or BamHI restriction sites without changing the encoded amino acids. For example, CH1 [pET28(b)-CH1] was constructed as follows. The NcoI-EcoRI fragment of CH1 was PCR amplified using B0301 [5′-d(CATACCATGGCTTCTGCTCCTCAA)-3′] as the forward primer (the NcoI restriction site is underlined and the translational start codon is in bold), G0401 [5′-d(CCGGAATTCCTCAACTTGTCCTTCCAC)-3′] as the reverse primer (the EcoRI site is underlined), and the pET28(b)-SHT gene as the template. The resulting PCR product was digested with EcoRI and gel purified. The carboxy-terminal EcoRI-XhoI fragment of CH1 was PCR amplified with the primers G0402 [5′-d(GAGGAATTCCAGTCCAAATACGATGAT)-3′; the underlined EcoRI site was introduced] and F1004 with pET28(b)-THT as the template. The resulting PCR product was digested with EcoRI and gel purified. The two EcoRI fragments of CH1 were joined in the presence of ligase (5 units) and ligase buffer (Invitrogen) at 21°C for 90 min. The CH1 gene construct was then amplified by PCR using the primers B0301 and F1004 with the ligated DNA fragments as the template. The resulting PCR product was digested with NcoI and XhoI, gel purified, and fused between the same restriction sites in pET-28(b), creating pET28(b)-CH1. For the CH2 gene construct, the NcoI-BamHI fragment of SHT cDNA was purified from a preparation of digested pET28(b)-SHT. The carboxy portion of the BamHI-XhoI fragment was amplified using the primers G0406 [5′-d(CCGGGGATCCATTTCGAGAGCCTTTACTTC)-3′; BamHI site underlined] and F1004 with pET28(b)-THT as the template. The resulting PCR product was digested with BamHI and XhoI. Ligation of the NcoI-BamHI fragment of the SHT gene with the BamHI-XhoI fragment of THT generated CH2. The CH3 gene construct was generated by ligating the NcoI-EcoRI fragment of THT and the EcoRI-XhoI fragment of CH2. The NcoI-EcoRI fragment of THT was PCR amplified with the primers F1003 and G0621 [5′-d(CTGGAATTCCTCAACTTGTCCTTCCAC)-3′; EcoRI site underlined], and gel purified after digestion with NcoI and EcoRI. The EcoRI-XhoI fragment was PCR amplified with the primers G0622 [5′-d(GAGGAATTCCGGTCCAAATATGATGAT)-3′; EcoRI site underlined] and F1004 using the CH2 gene as the template, and gel purified after digestion with EcoRI and XhoI. CH4 was constructed in a similar manner. The NcoI-BamHI fragment was PCR amplified with the primers B0301 and G0623 [5′-d(GAAATGGATCCCCGGTTTATCATAGAA)-3′; BamHI site underlined] using the CH1 gene as the template, and gel purified after digestion with NcoI and BamHI. The BamHI-XhoI fragment of SHT was gel purified after digestion of pET28(b)-SHT with BamHI and XhoI. Ligation of the NcoI-BamHI fragment of CH1 and the BamHI-XhoI fragment of SHT generated CH4. To generate the CH5 gene construct, the NcoI-EcoRI fragment of THT and the EcoRI-XhoI fragment of SHT were ligated into pET-28(b) predigested with NcoI and XhoI.
Construction and Site-Directed Mutagenesis of CH3 Chimeric Genes
Site-directed mutagenesis of the CH3 chimeric gene was performed using in vitro DNA synthesis and PCR with mutagenic primers. First, a point-mutated amine-binding domain representing amino acids 125 to 161 in the CH3 chimeric gene was synthesized in vitro with the mutagenic primers. The forward primer was 66 bp in length and had the sequence 5′-d(GAATTCCGGTCCAAATATGATGATGGAACCGATAAACGTGATGTGTTCATCGCGGGATATGCTTAC)-3′. The mutated reverse primers were 60 bp in length and harbored point-mutated nucleotides (underlined), as follows: 5′-d(ATGGATCCCCGGTTTGTCGTTCCCGAAGAGTGAATAATTAGCAAAAAAGTAAGCATATCC)-3′, the Ser codon 153 (AGT) was changed to an Asn codon (AAT); 5′-d(ATGGATCCCCGGTTTGTCGTTCCCGAAGCATGAATAACTAGCAAAAAAGTAAGCATATCC)-3′, the Leu codon 156 (CTC) was changed to a Cys codon (TGC); and 5′-d(ATGGATCCCCGGTTTGTCGTTATAGAAGAGTGAATAACTAGCAAAAAAGTAAGCATATCC)-3′, the Gly codon 165 (GGG) was changed to a Tyr codon (TAT). Both the forward and reverse primers were designed to contain overlapping and complementary regions of 12 bp at their 3′ ends. Three primer sets with the forward primer and one of the three reverse primers were prepared to 200 nm in 5 mm MgCl2 and 5 mm dNTPs. The primers were annealed by heating at 85°C for 2 min, followed by gradual cooling to room temperature over 25 min. After annealing, DNA polymerase I (Invitrogen) and the appropriate buffer were added, and the reaction was incubated at 37°C for 30 min. Following synthesis of the complementary strand, the mutated DNA fragments were amplified by PCR using the primer 5′-d(GAGGAATTCCGGTCCAAATATGATGAT)-3′ as the forward primer (EcoRI site underlined), 5′-d(AATGGATCCCCGGTTTGTC)-3′ (BamHI site underlined) as the reverse primer, and the in vitro-synthesized DNA fragments as templates. The PCR products were digested with EcoRI and BamHI, gel purified, and fused into pBluescript SK+ between the same restriction sites. Next, the EcoRI and BamHI fragments of the pET28b-CH3 plasmid were replaced with the point-mutated EcoRI and BamHI fragments, resulting in pET28(b)-CH3:S153N, -L156C, and -G158Y.
Construction and Site-Directed Mutagenesis of SHT Genes
Sited-directed mutagenesis of the SHT gene was performed as follows. SHT-S153N was created by replacing the EcoRI-BamHI fragment of CH4 with the EcoRI-BamHI fragment of the CH3-S153N fragment. Similarly, SHT-L156C and SHT-G158Y were constructed by ligating the EcoRI-BamHI fragments of CH3-L156C and CH3-G158Y into the corresponding CH4 restriction endonuclease sites. The three-point-mutated SHT-NCY was generated in vitro with mutagenic primers. The forward primer was 66 bp long and had the sequence 5′-d(GAATTCCGGTCCAAATATGATGATGGAACCGATAAACGTGATGTGTTCATCGCGGGATATGCTTAC)-3′. The mutated reverse primer was 60 bp long and had the following sequence, which harbored three-point-mutated nucleotides (underlined): 5′-d(ATGGATCCCCGGTTTGTCGTTATAGAAGCATGAATAATTAGCAAAAAAGTAAGCATATCC)-3′. Both primers were annealed and synthesized in vitro as described above. The in vitro-synthesized DNA fragment was digested with EcoRI and BamHI and ligated into the corresponding sites of the CH4 chimera leading to SHT-NCY. SHT-Y149F, SHT-F151Y, and SHT-FY were also generated in vitro with mutagenic primers in the same way as employed in the SHT-NCY.
Construction and Site-Directed Mutagenesis of THT Genes
pET28(b)-THT:NCY/SLG was constructed by PCR amplification using the three-point-mutated primers G0810 [5′-d(ATAGCGGGATACGCTTTCTTTTACGCGTCTTATTCACTCTTCGGGGATAAACCGGGGTTCTATTTCGAGA)-3′] and G0811 [5′-d(ATAAGACGCGTAAAAGAAAGCGTATCCCGCTAT)-3′]. An MluI site (underlined) was introduced to facilitate construction of the gene, and the point-mutated nucleotides are shown in bold. The PCR product generated from primers F1003 and G0811 using pET28(b)-THT as the template was digested with NcoI and MluI and gel purified. The PCR product amplified using the primers G0810 and F1004 was digested with MluI and XhoI and gel purified. Ligation of the NcoI-MluI and MluI-XhoI fragments into pET-28(b) predigested with NcoI and XhoI generated pET28(b)-THT-SLG. pET28(b)-THT-F145Y and pET28(b)-THT-FY145,147YF were also constructed by PCR amplification as above. The point-mutated primers used were H0816 [5′-d(GTAGCCGGCTATGAAAACATCACGCTT)-3′], H0815 [5′-d(ATAGCCGGCTACGCTTACTTTTATGCGAATTATTCA)-3′], and H0817 [5′-d(ATAGCCGGCTACGCTTACTTTTTCGCGAATTATTCATGCTTC)-3′]. An NaeI site (underlined) was introduced to facilitate construction of the gene and the point-mutated nucleotides are shown in bold. The same approach was employed as described above.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AY819700.
This work was supported by a grant from the Science Research Center program of the Ministry of Science and Technology/Korea Science and Engineering Foundation (R11–2001–09203001–0) to the Agricultural Plant Stress Research Center of Chonnam National University.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kyoungwhan Back (kback@chonnam.ac.kr).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.071514.
References
- Back K, Jang SM, Lee BC, Schmidt A, Strack D, Kim KM (2001) Cloning and characterization of a hydroxycinnamoyl-CoA:tyramine N-(hydroxycinnamoyl) transferase induced in response to UV-C and wounding from Capsicum annuum. Plant Cell Physiol 42: 475–481 [DOI] [PubMed] [Google Scholar]
- Burhenne K, Kristensen BK, Rasmussen SK (2003) A new class of N-hydroxycinnamoyltransferases. Purification, cloning, and expression of a barley agmatine coumaroyltransferase (EC 2.3.1.64). J Biol Chem 278: 13919–13927 [DOI] [PubMed] [Google Scholar]
- Clarke DD (1982) The accumulation of cinnamic acid amides in the cell walls of potato tissue as an early response to fungal attacks. In RKS Wood, ed, Active Defense Mechanisms in Plants. Plenum, New York, pp 321–332
- Cutillo F, D'Abrosca B, DellaGreca M, Marino CD, Golino A, Previtera L, Zarrelli A (2003) Cinnamic acid amides from Chenopodium album: effects on seeds germination and plant growth. Phytochemistry 64: 1381–1387 [DOI] [PubMed] [Google Scholar]
- Douglas CJ (1996) Phenylpropanoid metabolism and lignin biosynthesis: from weeds to trees. Trends Plant Sci 1: 171–178 [Google Scholar]
- Farmer MJ, Czernic P, Michael A, Negrel J (1999) Identification of cDNA clones encoding hydroxycinnamoyl-CoA:tyramine N-hydroxycinnamoyltransferase from tobacco. Eur J Biochem 263: 686–694 [DOI] [PubMed] [Google Scholar]
- Hickman A, Namboodiri M, Klein D, Dyda F (1999) The structural basis of ordered substrate binding by serotonin N-acetyltransferase: enzyme complex at 1.8 Å resolution with a bisubstrate analog. Cell 97: 361–369 [DOI] [PubMed] [Google Scholar]
- Ishihara A, Kawata N, Matsukawa T, Iwamura H (2000) Induction of N-hydroxycinnamoyltyramine synthesis and tyramine N-hydroxycinnamoyltransferase (THT) activity by wounding in maize leaves. Biosci Biotechnol Biochem 64: 1025–1031 [DOI] [PubMed] [Google Scholar]
- Jang SM, Ishihara A, Back K (2004) Production of coumaroylserotonin and feruloylserotonin in transgenic rice expressing pepper hydroxycinnamoyl-coenzyme A:serotonin N-(hydroxycinnamoyl)transferase. Plant Physiol 135: 346–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Jang S-M, Kang S, Back K (2005) Enhanced neutraceutical serotonin derivatives of rice seed by hydroxycinnamoyl-CoA:serotonin N-(hydroxycinnamoyl)transferase. Plant Sci 168: 783–788 [Google Scholar]
- Kawashima S, Hayashi M, Takii T, Kimura H, Zhang HL, Nagatsu A, Sakakibara J, Murata K, Oomoto Y, Onozaki K (1998) Serotonin derivative, N-(p-coumaroyl) serotonin, inhibits the production of TNF-alpha, IL-1 alpha, IL-1 beta, and IL-6 endotoxin-stimulated human blood monocytes. J Interferon Cytokine Res 18: 423–428 [DOI] [PubMed] [Google Scholar]
- Lajide L, Escoubas P, Mizutani J (1995) Termite antifeedant activity in Xylopia aethiopica. Phytochemistry 40: 1105–1112 [Google Scholar]
- Lu L, Berkey KA, Casero RA (1996) RGFGIGS is an amino acid sequence required for acetyl coenzyme A binding and activity of human spermidine/spermine N1 acetyltransferase. J Biol Chem 271: 18920–18924 [DOI] [PubMed] [Google Scholar]
- Martin-Tanguy J, Negrel J (1987) Hydroxycinnamic acid amides, hypersensitivity, flowering and sexual organogenesis in plants. In D von Wettstein, NH Chua, eds, Plant Molecular Biology. Plenum, New York, pp 253–263
- Mathis JR, Back K, Starks C, Noel J, Poulter CD, Chappell J (1997) Pre-steady-state study of recombinant sesquiterpene cyclases. Biochemistry 36: 8340–8348 [DOI] [PubMed] [Google Scholar]
- Nagatsu A, Zhang HL, Mizukami H, Okuyama H, Sakakibara J, Tokuda H, Nishino H (2000) Tyrosinase inhibitory and anti-tumor promoting activities of compounds isolated from safflower (Carthamus tinctorius L.) and cotton (Gossypium hirsutum L.) oil cakes. Nat Prod Lett 14: 153–158 [Google Scholar]
- Negrel J, Javelle F, Paynot M (1993) Wound-induced tyramine hydroxycinnamoyltransferase in potato (Solanum tuberosum) tuber discs. J Plant Physiol 142: 518–524 [Google Scholar]
- Park JB, Schoene N (2002) Synthesis and characterization of N-coumaroyltyramine as a potent phytochemical which arrests human transformed cells via inhibiting protein tyrosine kinases. Biochem Biophys Res Commun 292: 1104–1110 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Grimm R, Schmidt J, Scheel D, Strack D, Rosahl S (1999) Cloning and expression of a potato cDNA encoding hydroxycinnamoyl-CoA:tyramine N-(hydroxycinnamoyl)transferase. J Biol Chem 274: 4273–4280 [DOI] [PubMed] [Google Scholar]
- Schraudner M, Langebartels C, Negrel J, Sanderman H (1993) Plant defense reactions induced in tobacco by the air pollutant ozone. In B Fritig, M Legrand, eds, Mechanisms of Plant Defense Responses. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 286–290
- Von Roepenack-Lahaye E, Newman MA, Schornack S, Hammond-Kosack KE, Lahaye T, Jones JDG, Daniels MJ, Dow JM (2003) p-Coumaroylnoradrenaline, a novel plant metabolite implicated in tomato defense against pathogens. J Biol Chem 278: 43373–43383 [DOI] [PubMed] [Google Scholar]
- Wink M (1997) Special nitrogen metabolism. In PM Dey, JB Harborne, eds, Plant Biochemistry. Academic Press, London, pp 439–486
- Yang Q, Reinhard K, Schiltz E, Matern U (1997) Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol Biol 35: 777–789 [DOI] [PubMed] [Google Scholar]
- Yu M, Facchini PJ (1999) Purification, characterization, and immunolocalization of hydroxycinnamoyl-CoA: tyramine N-(hydroxycinnamoyl) transferase from opium poppy. Planta 209: 33–44 [DOI] [PubMed] [Google Scholar]
- Zhang HL, Nagatsu A, Sakakibara J (1996) Antioxidative compounds isolated from safflower (Carthamus tinctorius L.) oil cake. Chem Pharm Bull (Tokyo) 44: 874–876 [DOI] [PubMed] [Google Scholar]