Abstract
Enzymatic removal of the methoxycarbonyl group of pheophorbide (Pheid) a in chlorophyll degradation was investigated in cotyledons of radish (Raphanus sativus). The enzyme pheophorbidase (PPD) catalyzes the conversion of Pheid a to a precursor of pyropheophorbide (PyroPheid), C-132-carboxylPyroPheid a, by demethylation, and then the precursor is decarboxylated nonenzymatically to yield PyroPheid a. PPD activity sharply increased with the progression of senescence in radish, suggesting de novo synthesis of PPD. The enzyme activity was separated into two peaks in anion-exchange and hydrophobic chromatography; the terms type 1 and type 2 were applied according to the order of elution of these enzymes in anion-exchange chromatography. PPD types 1 and 2 were purified 9,999- and 6,476-fold, with a yield of 0.703% and 2.73%, respectively. Among 12 substrates tested, both enzymes were extremely specific for Pheids of the dihydroporphyrin and tetrahydroporphyrin types, indicating that they are responsible for the formation of these PyroPheids. Both PPDs had molecular masses of 113,000 kD on gel filtration and showed three bands of 16.8, 15.9, and 11.8 kD by SDS-PAGE. The partial N-terminal amino acid sequences for these bands of PPD (type 2) were determined. Based on their N-terminal amino acid sequences, a full-length cDNA of PPD was cloned. The molecular structure of PPD, particularly the molecular mass and subunit structure, is discussed in relation to the results of SDS-PAGE.
Changes in the color of leaves and the ripening of fruits are visible results of the breakdown of chlorophylls (Chls) due to senescence or maturation. The pathway for breakdown of Chls consists of several reaction steps (for reviews, see Hendry et al., 1987; Brown et al., 1991; Hörtensteiner, 1999; Kräutler and Matile, 1999; Matile et al., 1999; Takamiya et al., 2000), and the pathway is operationally divided into three stages. The early stage includes modification of the side chains of the tetrapyrrole macrocyclic and isocyclic rings. The middle stage involves cleavage of the macrocyclic ring by pheophorbide (Pheid) a oxygenase (PaO) and its successive modifications (Rodoni et al., 1997; Wüthrich et al., 2000; Pružinská et al., 2003). The last stage is the subsequent degradation of an open tetrapyrrole to smaller carbon- and nitrogen-containing fragments, such as organic acids, via monopyrroles (Suzuki and Shioi, 1999; Losey and Engel, 2001).
Information concerning the enzymes involved in the early stage modification of macrocylic and isocyclic rings has gradually accumulated in recent years. The first step in the degradation of Chl a is hydrolysis of the phytyl ester linkage catalyzed by chlorophyllase (EC 3.1.1.14), which forms chlorophyllide (Chlid) a and phytol. Although the activity of chlorophyllase was revealed about 90 years ago (Willstätter and Stoll, 1913), molecular cloning of the gene was accomplished only recently (Jacob-Wilk et al., 1999; Tsuchiya et al., 1999). Based on homology searching of sequences and expression, it was determined that the coronatine-induced gene ATHCOR1 was the gene encoding chlorophyllase (Tsuchiya et al., 1999).
A release of magnesium (Mg) from the macrocyclic ring causes the formation of Pheid a. An activity catalyzing this reaction has been reported in photosynthetic bacteria, algae, and higher plants and is considered to be due to an enzyme that has been designated Mg-dechelatase (Owens and Falkowski, 1982; Ziegler et al., 1988; Shioi et al., 1991; Vicentini et al., 1995). Previously, we demonstrated that the release of Mg2+ from Chlid a is not due to an enzyme, but to a low-molecular-mass, heat-stable substance that has been designated Mg-dechelating substance (Shioi et al., 1996a). The highly purified substance is, however, specific not only for Mg2+ but also for divalent cations. Therefore, the substance was renamed metal-chelating substance (Suzuki and Shioi, 2002). Recent studies confirmed these results, and metal-chelating substance is a possible candidate for the substance that catalyzes the Mg-dechelating reaction (Kunieda et al., 2005; Suzuki et al., 2005).
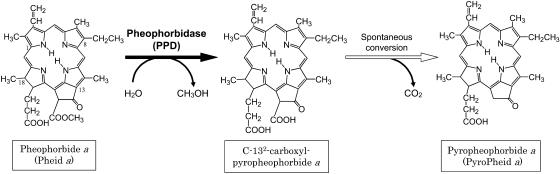
The final step of macrocyclic ring modification is the conversion of Pheid a to pyropheophorbide (PyroPheid) a. Two types of enzymes that catalyze alternative reactions in the formation of PyroPheid a were found (Shioi et al., 1996b; Watanabe et al., 1999; Doi et al., 2001; Suzuki et al., 2002). As shown in Figure 1, one route consists of two reactions: first, enzymatic conversion of Pheid a to a precursor of PyroPheid a, identified as C-132-carboxylPyroPheid a, and, next, spontaneous conversion of the precursor to PyroPheid a (Shioi et al., 1996b; Watanabe et al., 1999). This enzyme was designated pheophorbidase (PPD). Its activity is widely distributed but confined to some orders of higher plants, and, thus, this reaction may be specific for certain orders of plants (Suzuki et al., 2002). The other enzyme, termed Pheid demethoxycarbonylase, was partially purified from the Chl b-less mutant NL-105 of Chlamydomonas reinhardtii (Doi et al., 2001). This enzyme produced no intermediate, as shown in the PPD reaction, indicating that it converts Pheid a directly into PyroPheid a, probably by an acetyl transferase reaction.
Figure 1.
Demethoxycarbonyl reaction of Pheid a to PyroPheid a catalyzed by PPD. The reaction is composed of two steps: enzymatic conversion of Pheid a to a precursor, C-132-carboxylPyroPheid a, followed by spontaneous conversion of the precursor to PyroPheid a. The site of the enzymatic reaction is indicated by a black arrow.
Previously, we purified PPD from Chenopodium album and the N-terminal sequence was determined (Watanabe et al., 1999); however, there is no information concerning the substrate specificity of this enzyme or the molecular structure, including gene cloning. Furthermore, little is known about the significance of this reaction step, including whether this is a true auxiliary step in vivo or whether it is a side reaction of the esterase. In this study, we purified two types of PPD from the senescent cotyledons of radish (Raphanus sativus) to homogeneity and examined their substrate specificity in a variety of Chl derivatives; we also determined N-terminal and internal amino acid sequences and the cDNA sequence. Based on the results of substrate specificity, we discuss the role of this enzyme in vivo in relation to the formation of Chl catabolites.
RESULTS
Enzyme Activities and Chl Breakdown during Senescence
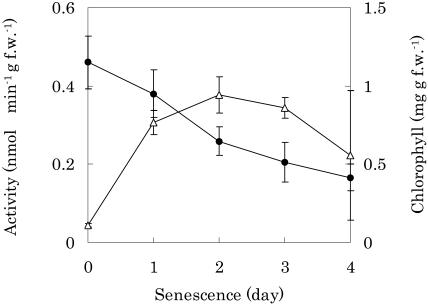
The concentrations of Chl in the senescent cotyledons of radish were followed spectrophotometrically after extraction (Fig. 2). Chl concentrations gradually decreased as the duration of senescence increased and reached 44% after 3 d. In contrast, PPD activity sharply increased at 1 d, was maintained at a stationary level up to 3 d, and then decreased. The activity in senescent leaves at the first day increased about 7 times compared to that of nonsenescent leaves. This was not due to an increase in the activity itself, but to de novo synthesis of the enzyme in accordance with the results of inhibitor studies using cyclohexamide. This finding is also confirmed by immunoblot analysis using a polyclonal antibody raised against a purified recombinant PPD, although several minor faint bands appeared (data not shown). Pigment analysis using HPLC showed that the substrate and product of this enzyme, Pheid and PyroPheid species, were not detected in the senescent cotyledons of radish (Suzuki and Shioi, 2004). This finding suggests that they are further metabolized in vivo.
Figure 2.
Changes in Chl concentrations and specific activity of PPD during senescence. Shoots were excised and placed in 50-mL flasks containing 15 mL of distilled water. The cotyledons were allowed to reach senescence in complete darkness at 25°C. Chls were extracted from cotyledons with 80% (v/v) acetone on the indicated day of senescence and measured spectrophotometrically. The enzyme activity was determined for the crude extracts as described in the text. PPD activity is expressed by the amount of C-132-carboxylPyroPheid a calculated from HPLC detection. •, Chls; ▵, PPD activity.
Purification of Type 1 and Type 2 PPDs
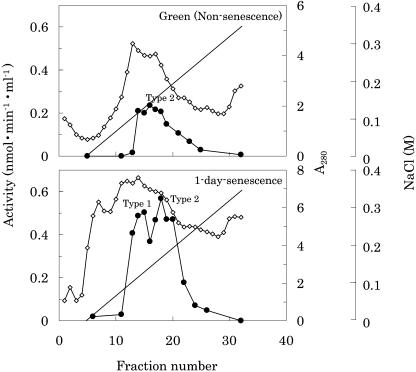
The crude enzyme, extracted from about 5 kg (fresh weight) of 1-d-senescent cotyledons of radish, was purified by four steps of successive chromatography (Table I). The enzyme activity was separated into two peaks by chromatography on DEAE-Toyopearl and butyl-Toyopearl. These peaks are not artifacts during purification, but probably isomers, because they were reproducibly observed in one-step purification with a simple extraction followed by chromatography. The PPDs were operationally termed type 1 and type 2 according to their order of elution in DEAE-Toyopearl chromatography (Fig. 3). These enzymes increased to high levels during senescence, as seen from the results of the time-dependent study (Fig. 2), indicating that these PPDs are induced in parallel by senescence. In comparison with the activity at 1-d senescence (Fig. 3), the total activity of type 1 enzyme was about 5 times higher than that of nonsenescence, although type 2 stayed about 3 times higher. These enzymes were purified separately using chromatography. PPD types 1 and 2 were purified 9,999- and 6,476-fold with a yield of 0.703% and 2.73%, respectively. A summary of the purification of the two PPDs is presented in Table I. Some enzymatic properties (e.g. optimal pH and the effect of reaction products) were reported previously (Suzuki et al., 2002).
Table I.
Purification of PPD types 1 and 2 from cotyledons of radish
Purification was done from 4,937 g (fresh weight) of 1-d-senescent cotyledons.
| Purification Step | Protein | Total Activity | Specific Activity | Yield | Purification |
|---|---|---|---|---|---|
| mg | nmol min−1 | pmol min−1 mg−1 | % | -fold | |
| Crude | 121,940 | 258.4 | 2.119 | 100 | 1 |
| Heat treatment | 110,675 | 240.7 | 2.175 | 93.1 | 1.03 |
| Acetone fractionation | 3,255 | 78.08 | 23.99 | 30.2 | 11.3 |
| Type 1 | |||||
| DEAE Toyopearl | 163.9 | 64.60 | 394.1 | 25.0 | 186 |
| Butyl Toyopearl | 7.483 | 29.86 | 3,990 | 11.6 | 1,883 |
| Mono Q | 0.6890 | 4.464 | 6,479 | 1.73 | 3,057 |
| Superdex 200 | 0.08568 | 1.816 | 21,190 | 0.703 | 9,999 |
| Type 2 | |||||
| DEAE Toyopearl | 126.6 | 44.44 | 351.0 | 17.2 | 166 |
| Butyl Toyopearl | 26.94 | 37.39 | 1,388 | 14.5 | 655 |
| Mono Q | 1.362 | 17.69 | 12,990 | 6.84 | 6,128 |
| Superdex 200 | 0.5145 | 7.062 | 13,730 | 2.73 | 6,476 |
Figure 3.
Elution profile of DEAE-Toyopearl chromatography of PPD. Enzyme extracts were used after ammonium sulfate precipitation (30%–70% saturation). The column was equilibrated with 20 mm Tris-HCl buffer (pH 7.5) and eluted with the same buffer containing a linear gradient of NaCl (0–0.35 m) at a flow rate of 1 mL/min. Three-milliliter fractions were collected. The enzyme assay and conditions were as described in Figure 2. ⋄, Protein concentrations were estimated with A280; •, PPD activity; —, NaCl concentration. The peak fractions were designated type 1 (front peak) and type 2 (back peak) by the order of elution.
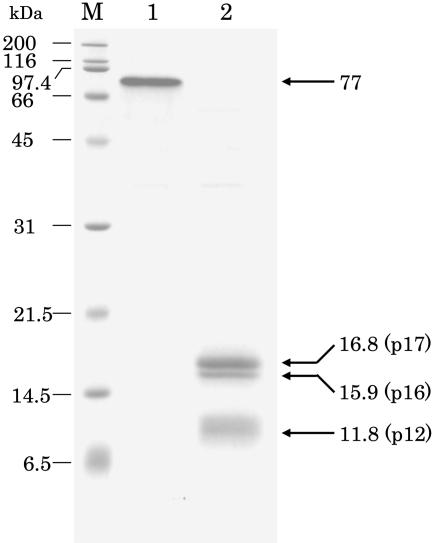
SDS-PAGE was performed with purified PPD type 2 (Fig. 4). In non-heat-treated samples, one major band whose size was about 77 kD appeared in 15% polyacrylamide gel (Fig. 4, lane 1), while on 12% gel a single band emerged at the size of 61.3 kD. Even after 1 min of heat treatment of the enzyme in the sample buffer containing SDS and 2-mercaptoethanol, PPD was separated into three bands with molecular masses of 16.8, 15.9, and 11.8 kD (Fig. 4, lane 2). The pattern of SDS-PAGE was not changed after heating for 1 to 20 min. Similar results were also obtained for PPD type 1.
Figure 4.
SDS-PAGE on 15% polyacrylamide gel of purified PPD (type 2) from radish cotyledons. Peptides were stained by Coomassie Brilliant Blue R-250. In the nonheated sample (lane 1), only one band was stained. After heating at 95°C for 3 min (lane 2), PPD was separated into three bands. Numerals under kD denote the molecular mass of marker proteins. Lane M, Molecular markers.
Substrate Specificity of PPD
The Km for Pheid a of the two enzymes was determined in the concentration range from 1 to 40 μm. The values obtained were 14.1 and 15.1 μm for types 1 and 2, respectively (see Table II).
Table II.
Substrate specificity of PPD types 1 and 2 from cotyledons of radish
The C-position number refers to the structure formula of Pheid a in Figure 1. Preparation of Chl derivatives and the enzyme assay were performed as described in the text.
| Chl Species
|
Metal (Mg)
|
C-Position
|
Km
|
||||
|---|---|---|---|---|---|---|---|
| C17a | C7 | C17-C18b | C7-C8b | Type 1 | Type 2 | ||
| μm | |||||||
| Porphyrins | |||||||
| ProtoChlid a | + | Propionyl | CH3 | Double | Double | – | – |
| ProtoPheid a | − | Propionyl | CH3 | Double | Double | – | – |
| Pheophytin c | − | Acryl | CH3 | Double | Double | – | – |
| Dihydroporphyrins | |||||||
| Chl a | + | Phytyl | CH3 | Single | Double | – | – |
| Chl b | + | Phytyl | CHO | Single | Double | – | – |
| Chlid a | + | Propionyl | CH3 | Single | Double | – | – |
| Pheophytin a | − | Phytyl | CH3 | Single | Double | – | – |
| Pheid a | − | Propionyl | CH3 | Single | Double | 14 | 15 |
| Pheid a′ | − | Propionyl | CH3 | Single | Double | – | – |
| Pheid b | − | Propionyl | CHO | Single | Double | 243 | 232 |
| Tetrahydroporphyrins | |||||||
| Bacterio-Chl a | + | Phytyl | CH3 | Single | Single | – | – |
| Bacterio-Pheid a | − | Propionyl | CH3 | Single | Single | 39 | 37 |
Phytyl, -CH2-CH2-COOC20H30; propionyl, -CH2-CH2-COOH; acryl, -CH = CH-COOH.
Single, single bond; Double, double bond.
To determine the substrate specificity of the enzyme, several Chl derivatives were examined under standard assay conditions. As shown in Table II, both enzymes used Pheid a/b and bacterio-Pheid a as substrates, but not other Chl derivatives, such as protoChlid a, pheophytin c, Chl a/b, Chlid a, pheophytin a, and bacterio-Chl a. These results indicate that the enzymes are specific for the structure of the substrate, (e.g. the absence of Mg and a phytol chain) and also the existence of a single bond at the position of C17-C18. The Km values of the two enzymes as active substrates were calculated to be about 240 and 40 μm for Pheid b and bacterio-Pheid a, respectively. The Km value for Pheid b was the highest among the three Pheid species, suggesting that a methyl group at C7 confers a greater affinity than a formyl group.
Furthermore, the Chl catabolite, nonfluorescent Chl catabolite (NCC), was examined to determine whether PPD uses it as a substrate or not. Cj-NCC, prepared from Cercidiphyllum japonicum, was kindly provided by Professor Bernhard Kräutler. No change in the HPLC peak of Cj-NCC at a retention time of 70.2 min was observed, indicating that PPD did not recognize Cj-NCC as a substrate.
To test the esterase activity of this enzyme, several nitrophenyl compounds, such as nitrophenyl acetate and nitrophenyl butylate, which are used for general esterase assays, were used as the substrates. However, little or no activity was observed for these substrates, indicating that the PPD activity is not a side reaction of an esterase activity as far as the substrates tested.
Identification of the N-Terminal Sequence and Cloning of cDNA for PPD
Analyses of the N-terminal sequences were performed with the three peptides separated by SDS-PAGE after heat treatment of the purified PPD type 2 (Fig. 4). The amino acid sequences of three peptides were determined: 16.8 kD, EEDIWEYIYGEGADKPPTGVLMKEEFFRRY; 15.9 kD, EDIWEYIYGEGADKPPTGVLMKEEFFRHYY; and 11.8 kD, DDSVVHFVFFHGASHGAAWYYKPTTTLV (Fig. 5A). In the 15.9-kD peptide, only one amino acid of the N-terminal sequence was missing as compared to that of the 16.8 kD, although the C terminus is unknown. Two different partial amino acid sequences were thus revealed from these analyses.
Figure 5.
A, Partial N-terminal amino acid sequences of the peptide from SDS-PAGE. The number corresponds to the peptide size of SDS-PAGE (Fig. 4). B, Nucleotide sequence of a full-length cDNA encoding PPD of radish. The deduced amino acid sequence is denoted below in the standard one-letter code. The translation termination codon is designated with an asterisk (*). The lipase domain is underlined. Dashed lines indicate the results of N-terminal sequence analyses.
A BLAST search (BLASTp; Altschul et al., 1997) was performed against the National Center for Biotechnology Information (NCBI) databases with the two amino acid sequences. The results showed that the protein NP_193402 of Arabidopsis (Arabidopsis thaliana) had homologies of 77% and 64% to PPD peptides, 16.8 and 11.8 kD, respectively. From the comparison with NP_193402, the amino acid sequence of 11.8 kD was located upstream from those of 16.8 and 15.9 kD. Sense and antisense degenerate primers for reverse transcription (RT)-PCR were therefore designed from the amino acid sequence of PPD, which had been constructed based on the sequences of the 11.8- and 16.8-kD peptides.
From the results of RT-PCR and 3′-RACE, the partial cDNA of PPD that corresponds to the 862 bp containing the untranslated region was sequenced. The translated region of PPD had 84% identity to At4g16690, which is the cDNA of NP_193402. We assumed that the 5′ ends of these mRNAs are conserved between PPD and At4g16690. To obtain the full sequence-coding region of PPD, an At4g16690-specific sense primer and a PPD-specific antisense primer containing the initiation and termination codons, respectively, were then designed.
Finally, the 792-bp cDNA fragment from the initiation codon, ATG, to the termination codon, TGA, was cloned by a combination of PCRs with the template synthesized from radish RNA (Fig. 5B). We named this gene RsPPD. The nucleotide sequence reported in this article has been submitted to the DNA Data Bank of Japan (DDBJ; accession no. AB218276). Genomic Southern analysis for RsPPD showed a major band with one or two minor bands, depending on the restriction enzymes used, suggesting that RsPPD exists as multiple copies on the Raphanus genome.
The amino acid sequence deduced from the nucleic acid sequence was 263 amino acid residues corresponding to a Mr of 28,974. The Mr and subunit structure are discussed below. The deduced polypeptide has a lipase domain (Fig. 5B, underline) that was presumed to be an active site of esterases, as in the case of chlorophyllase (Tsuchiya et al., 2003). The recombinant PPD that had been expressed using the glutathione S-transferase (GST) fusion system (data not shown) had comparable activity to the native purified PPDs for the formation of C-132-carboxylPyroPheid a from Pheid a, confirming that the cloned gene is PPD. For the type 1 enzyme, no clone was obtained in spite of several screenings. A reason for this might be that the population of transcripts for type 1 is much lower than that of type 2, despite the large amount of proteins.
Deduced PPD Sequence and Its Homologs
Homology searching against the deduced amino acid sequence of PPD was performed with BLAST programs using the NCBI databases GenomeNet BLAST2 and The Arabidopsis Information Resource (TAIR) BLAST 2.0. We operationally named the homologs of PPD using the prefix of their species name to distinguish the sources (i.e. RsPPD [R. sativus PPD]). From the results of the homology search, two homologs, NP_193402 from Arabidopsis (GI: 15235844, AtPPD) and CAC82615 from Capsella rubella (GI: 15866583, CrPPD), were found by BLASTp, and four homologs, CD819739 and CD818940 from Brassica napus (GI: 32501679 and GI: 32500880, BnPPD1 and BnPPD2) and BH478656 and BH720243 from Brassica oleracea (GI: 17686760 and GI: 18820838, BoPPD1 and BoPPD2), were found by BLASTn. The multiple alignments of the deduced PPD sequences and its homologs using ClustalW (Higgins et al., 1996) are shown in Figure 6. BnPPDs and BoPPDs were translated in the deduced peptides. All homologs were highly conserved. AtPPD is an esterase/lipase/thioesterase family protein and is a cyanohydrin lyase-like protein of Arabidopsis consisting of 262 amino acids (Mr = 28,996). The gene of AtPPD, At4g16690, is located on chromosome 4 of Arabidopsis and contains two introns. CrPPD is a hypothetical protein of C. rubella consisting of 265 amino acids (Mr = 29,347). AtPPD and CrPPD had high levels of amino acid sequence conservation to the deduced amino acid of RsPPD at 84% and 77% identity, respectively. All peptides had a lipase domain (Hasslacher et al., 1995) whose pattern is [LIV]-x-[LIVFY]-[LIVMST]-G-[HYWV]-S-x-G-[GSTAC] (underlined in Fig. 6). When the amino acid sequences were aligned with CrPPD, it was found that RsPPD, AtPPD, BnPPDs, and BoPPDs have gaps. In particular, the gap shown by an arrowhead in Figure 6 corresponds to the position of RsPPD cleaved by heating, as shown in SDS-PAGE (Fig. 4, lane 2). The homologs of PPD were thus found mainly in Brassicaceae. This finding is in agreement with the patterns of distribution of PPD activity in plants reported previously (Suzuki et al., 2002).
Figure 6.
Comparison of amino acid sequences among PPDs from radish and its homologs. Homology searching was performed with BLAST programs. The alignments of the sequences were obtained using ClustalW. The lipase domain is underlined. The arrowhead indicates the position that was cleaved by heating at 95°C. Identical residues in over four proteins are boxed in black, while gray boxes indicate similar amino acids. Gaps introduced are shown by hyphens. AtPPD, NP_193402 from Arabidopsis (GI: 15235844); CrPPD, CAC82615 from C. rubella (GI: 15866583); BnPPD1, deduced sequence of CD819739 from B. napus (GI: 32501679); BnPPD2, deduced sequence of CD818940 from B. napus (GI: 32500880); BoPPD1, deduced sequence of BH478656 from B. oleracea (GI: 17686760); BoPPD2, deduced sequence of BH720243 from B. oleracea (GI: 18820838).
Subsequently, homologs of RsPPD were searched against the C. reinhardtii expressed sequence tag (EST) index of the Kazusa DNA Research Institute because C. reinhardtii has an enzyme that catalyzes the conversion of Pheid to PyroPheid (Doi et al., 2001). However, no homologs were found in the C. reinhardtii EST (http://www.kazusa.or.jp/en/plant/chlamy/EST) or in the CyanoBase (http://www.kazusa.or.jp/cyano/cyano.html). A homology search at the C. reinhardtii genomic database Chlamy Center (http://www.chlamy.org/chlamydb.html) was also done, but no significant homolog was found. This indicates that Pheid demethoxycarbonylase from C. reinhardtii and PPD from radish are genetically different enzymes, which was expected because their reaction mechanisms differ, although they are both involved in the reaction for synthesizing PyroPheid.
DISCUSSION
In this study, senescence-induced RsPPDs were purified from cotyledons of radish to homogeneity and their amino acid sequences were deduced along with the full cDNA sequence using RT-PCR and RACE. A substrate specificity study showed that these enzymes were extremely specific for Pheids of the dihydroporphyrin and tetrahydroporphyrin types, indicating that they are responsible for the reaction of those structures.
Total RsPPD activity markedly increased with senescence. This phenomenon is predominantly due to up-regulation of proteins by senescence. In this sense, this enzyme, PaO, is one of the senescence-induced genes as well as a porphyrin-opening enzyme in the Chl degradation pathway (Matile et al., 1999). Type 1 and type 2 RsPPDs could not be separated by gel filtration, but they could be reproducibly separated using ion-exchange and hydrophobic chromatography, indicating that they are isomers. After six steps of purification to homogeneity, 86 and 514 μg of types 1 and 2 enzymes, respectively, were finally obtained from approximately 5 kg of radish cotyledons.
The molecular mass of non-heat-treated, denatured samples of RsPPD was estimated to be in the range of 77 to 61.3 kD, depending on the concentration of polyacrylamide gel used (see Fig. 4). Combined with the results of the deduced amino acid (28,000) and gel filtration (113,000), RsPPD is probably a tetramer. Moreover, the peptide, with a molecular mass of 77 to 61.3 kD, seems to maintain its dimeric form after being denatured, but not in non-heat-treated conditions, although it had a somewhat large molecular mass. This suggests RsPPD is a strongly associated dimer. After heat treatment of RsPPD, three bands, 16.8, 15.9, and 11.8 kD, appeared on SDS-PAGE, but the 16.8- and 15.9-kD peptides overlapped some of the amino acid sequences. Excluding the overlapped peptide, the sum of the molecular mass of the other bands was 28.6 kD. The sizes of the deduced peptide fragments, N-terminal and C-terminal fragments, from the cleavage site were 12,800 and 15,100, respectively, corresponding to the approximate band sizes. As shown in Figure 6, RsPPD was cleaved at a nonconserved amino acid site (gap structure). Based on the strong dimer formation and the fragile primary structure, it is likely that RsPPD forms a dimer by three-dimensional domain swapping (Jaskólski, 2001) in which a structural element of a monomeric protein is replaced by the same element from another subunit.
In the degradation pathway of Chl, Pheid is catabolized by PaO via red Chl catabolites to NCC and further modified (Matile et al., 1999). Besides Pheid, the formation of PyroPheid was reported in a variety of photosynthetic organisms, such as higher plants, algae, and bacterial cells (Owens and Falkowski, 1982; Ziegler et al., 1988; Shimokawa et al., 1990; Shioi et al., 1991; Langmeier et al., 1993). In higher plants, however, the distribution of the enzyme catalyzing this reaction is confined to certain plant orders, and this reaction is thus considered to be an auxiliary reaction. The significance of this reaction is still unknown, although enzyme formation is clear, as presented in this study. Moreover, little attention has been paid to the enzymatic catabolism of PyroPheid. Our recent analysis showed that Pheid and PyroPheid species were not detected in the senescent cotyledons of radish (Suzuki and Shioi, 2004). This finding suggests that PyroPheid is also further metabolized in vivo, probably as a substrate for PaO, as is Pheid. This is supported by the result that these compounds accumulated simultaneously in certain conditions, such as anaerobic or reduced states that inhibit PaO (Shioi et al., 1995; Doi et al., 2001).
Several NCC derivatives were found in different plant species (Matile et al., 1999). Interestingly, the presence of NCC derivatives lacking a methoxycarbonyl group was consistent with the distribution of the enzyme PPD in plant species. Therefore, the question arises whether PPD actually acts for NCC. To investigate this question, the interaction between PPD and NCC was verified by the use of Cj-NCC instead of Pheid in a standard assay mixture. Judging from HPLC analyses (Oberhuber et al., 2001), Cj-NCC was, however, not changed, indicating that NCC was not recognized as the substrate by PPD. At present, we have not completely excluded the possibility that PPD is involved in the Chl degradation pathway, particularly for the modification of open tetrapyrrole products of Chl catabolites, because there are several intermediates from red Chl catabolite to NCC at different steps of Chl degradation. Additional experiments are necessary to clarify this question.
It is considered that Chl degradation proceeds only in thylakoids and inner envelope membranes (Matile et al., 1999). Recent investigations, however, suggest the possibility that Chl is degraded not only inside, but also outside, the chloroplast. Guiamét et al. (1999) reported that numerous large plastoglobuli containing Chls and Chl protein complexes were extruded into the cytosol through the senescent chloroplast envelope membrane. In addition, enzymes involved in the degradation of Chls are localized outside the chloroplasts. For instance, chlorophyllase has a signal peptide for the endoplasmic reticulum in addition to a transit peptide for plastids (Tsuchiya et al., 1999). In the case of PPDs, it has been determined that CaPPD is located outside the chloroplast in C. album (Watanabe et al., 1999). RsPPD and its homologs are also estimated to be soluble proteins that are localized to the cytosol by analysis based on amino acid sequences using the SOSUI system (http://sosui.proteome.bio.tuat.ac.jp/cgi-bin/sosui.cgi?/sosui_submit.html). These facts indicate that PPDs are involved in the demethoxycarbonylation reaction in the cytosol. This finding suggests that PPDs actually function in the modification of open tetrapyrrole products of Chl catabolites rather than in the formation of PyroPheid, since Pheid is the preceding and direct substrate for PaO in the Chl degradation pathway. Determination of the functional significance of this enzyme awaits further study.
MATERIALS AND METHODS
Plant Materials
Cotyledons of radish (Raphanus sativus) were purchased from a local market. Induction of senescence was performed according to the method described previously (Suzuki and Shioi, 1999). Briefly, shoots were excised and placed in 50-mL flasks containing 15 mL of distilled water. The cotyledons were allowed to reach senescence in complete darkness at 25°C.
Chls
Chl a, Pheid a, and PyroPheid a were purchased from Wako Pure Chemical Industries. The concentrations of Chls and their derivatives were determined spectrophotometrically as described previously (Shioi et al., 1991; Doi et al., 2001).
Preparation of Chl Derivatives
Chl a/b was extracted from the leaves of spinach (Spinacia oleracea) with acetone and partially purified by precipitation with dioxane (Iriyama et al., 1974). The dioxane precipitation was repeated once, and Chls obtained were separated and further purified by Suc-column chromatography (Perkins and Roberts, 1962). Bacterio-Chl a was extracted from the cells of Rhodobacter sulfidophilus. ProtoChlid a was obtained with acetone from 6-d-old etiolated leaves of barley (Hordeum vulgare). Chl c (c1 and c2 mixture) was extracted from the thalli of Undaria pinnatifida. Pheophytins a/c, Pheid b, and bacterio-Pheid a were prepared by acid treatment of pure respective Chls, according to the method of Perkins and Roberts (1962) and Hynninen (1973).
Chlid species were prepared from pure respective Chls by the action of chlorophyllase, which catalyzes the hydrolysis of esterified alcohols. Chlorophyllase was obtained from mature leaves of Chenopodium album as described previously (Tsuchiya et al., 1997). Each pigment was purified further by DEAE-Toyopearl (Tosoh; acetate form) chromatography (Omata and Murata, 1980) or by Suc-column chromatography.
Enzyme Assay
The activity of PPD was assayed basically according to the methods described by Shioi et al. (1996b). The reaction mixture consisted of 20 mm phosphate buffer (pH 7.0), 160 μm Pheid a in acetone (final 20%, v/v), and PPD preparation in a total volume of 100 μL. After incubation in darkness at 30°C for 10 min, the reaction was terminated by adding 200 μL of acetone. The amount of C-132-carboxylPyroPheid a formed was assayed using the HPLC system described below. HPLC analysis of the pigments was carried out according to our previous method with a slight modification (Shioi et al., 1996b). Briefly, HPLC was performed using a Zorbax octadecyl silica column (250 × 4.6 mm; Agilent Technologies) or Cosmosil 5C18-MS column (250 × 4.6 mm; Nacalai Tesque). Pigments were eluted isocratically with methanol-2 m ammonium acetate (95:5, v/v) at a flow rate of 1.0 mL/min at 30°C. The pigments were monitored spectrophotometrically at 410 nm and quantified by an integrator. Peak areas were used for the calculation of the enzyme activity. The concentration of C-132-carboxylPyroPheid a was tentatively calculated using the standard curve for Pheid a because isolated C-132-carboxylPyroPheid a rapidly changed to PyroPheid a spontaneously (Shioi et al., 1996b).
Purification of PPD
Senescent cotyledons of radish (approximately 5,000 g) were homogenized with 20 mm phosphate buffer (pH 7.0) at 4°C in a blender. The homogenate was filtered through six layers of cotton gauze and centrifuged at 17,000g for 30 min at 4°C. The cell-free extract was incubated at 60°C for 10 min and centrifuged at 12,000g for 15 min. The supernatant was fractionated with 60% acetone. The precipitate was dissolved in a small volume of 20 mm Tris-HCl (pH 7.5). After clarifying the solution by centrifugation at 12,000g for 15 min, the enzyme solution was applied to a column of DEAE-Toyopearl 650 m (2.5 × 8 cm; Tosoh) previously equilibrated with 20 mm Tris-HCl (pH 7.5) and eluted with the same buffer containing a linear gradient of NaCl (0–0.35 m). Fractions eluted at 0.1 to 0.14 m NaCl were classified as type 1 and at 0.15 to 0.2 m as type 2 and were pooled separately (see Fig. 3).
The following purification procedures were carried out separately for types 1 and 2, but the procedures used for both types 1 and 2 are described together. After adding ammonium sulfate to 30% saturation, the enzyme solution was applied to a butyl-Toyopearl 650 m column (2.5 × 8 cm; Tosoh) previously equilibrated with 20 mm Tris-HCl (pH 7.5) containing 30% saturation of ammonium sulfate and eluted with the same buffer containing a reverse-linear gradient of ammonium sulfate (30%–0% saturation). Fractions eluted at about 0% to 6% saturation for type 1 and 7% to 12% saturation for type 2 were pooled. The active fraction was dialyzed against 20 mm Tris-HCl (pH 7.2) and charged onto a column of Mono-Q (Amersham Biosciences) that had been equilibrated with the same buffer using the ÄKTAFPLC system (Amersham Biosciences). The column was eluted with the same buffer containing a linear gradient of NaCl (0–0.45 m). Fractions eluted at 0.22 m NaCl for type 1 and 0.27 m NaCl for type 2 were pooled. The pooled enzyme solution was concentrated by Centriflo CF25 Membrane Cones (Millipore) and applied to a column of Superdex 200 (Amersham Biosciences) equilibrated with 20 mm Tris-HCl (pH 7.5) containing 0.15 m NaCl and eluted with the same buffer. Active fractions were collected and used for the subsequent experiments.
Detection of NCC
Cj-NCC prepared from Cercidiphyllum japonicum was kindly provided by Professor Bernhard Kräutler of Leopold-Franzens-Universität, Innsbruck, Austria. HPLC was carried out with a model LC-10AT (Shimadzu) equipped with a column-temperature controller, using 5-μm Hypersil octadecyl silica (250 × 4.6 mm; Agilent Technologies). Analysis of NCC was performed according to the method of Oberhuber et al. (2001). Pigments were eluted at a flow rate of 0.5 mL/min at 25°C with a programmed binary gradient elution system. The solvents used were 100 mm potassium phosphate buffer (pH 7.0) for A and methanol for B. Separation was performed with a gradient containing the break points of 0 min (80:20, v/v), 10 min (80:20), 70 min (40:60), and 80 min (40:60). Separated pigments were detected spectrophotometrically with a photodiode array detector (Shimadzu SPD-M10A), measuring from 280 to 500 nm and monitoring at 320 nm.
Analysis of the N-Terminal Amino Acid Sequence
Purified PPD was electrophoresed on a 15% SDS-polyacrylamide gel and electrotransferred to a polyvinylidene difluoride membrane. The protein bands visualized with Coomassie Brilliant Blue were cut out and subjected to amino acid sequencing on a protein sequencer PPSQ-21A (Shimadzu) according to the procedure provided by the manufacturer.
RNA Extraction, PCR Amplification, and Molecular Cloning
Total RNA was isolated from 1-d-senescent cotyledons of radish by the guanidinium thiocyanate method (Chomczynski and Sacchi, 1987). Total RNA concentrations were determined by UV spectrophotometry.
The cloning of the PPD gene (PPD) was achieved in several steps. The first step was performed on template cDNA synthesis using ReverTra Ace-α (Toyobo) according to the manufacturer's instructions. Total RNA from 1-d-senescent radish leaves was used as a template with oligo(dT) primer to produce single-strand cDNA. For the next PCR, which was RT-PCR (Frohman et al., 1988) based on the partial amino acid sequences, degenerate primers were designed as follows: a forward primer, 5′-CAYTTYGTNTTYGTNCAYGGNGC-3′, and a reverse primer, 5′-ATRTAYTCCCTDATRTCYTCYTC-3′, where D indicates not C, N indicates any, R indicates a purine (G + A), and Y indicates a pyrimidine (T + C). A partial cDNA (375 bp) was amplified using TaKaRa Taq (TaKaRa) with primers for 40 cycles at 97°C for 0.5 min and 47°C and 72°C for 1 min each. To identify the 3′-end, 3′-RACE was carried out. Sequence-specific primers were designed from the results of RT-PCR as follows: 5′-GGTATCAACCTCACCGACTCTAA-3′. A partial cDNA (550 bp) was amplified using TaKaRa Taq with a sequence-specific primer and oligo(dT) for 40 cycles at 97°C for 0.5 min and 60°C and 72°C for 1 min each.
From a BLAST search using the results of RT-RCR and 3′-RACE, the Arabidopsis (Arabidopsis thaliana) homolog At4g16690, which shows 84% identity, was found. Full-length cDNA was amplified using primers containing the following restriction enzyme site: a forward primer based on a homolog sequence, 5′-TCCCCGGGAATGGGAGGAGAAGGTGGTGCTGA-3′, and a reverse primer based on the specific sequence 5′-ATGAATTCTTATAGTTGAAGAAAAGATACAGCAC-3′. PCR was performed using TaKaRa Taq and the template cDNA with primers for 40 cycles at 97°C for 0.5 min and 45°C and 72°C for 1 min each. All PCR products were cloned into pT7-Blue T-Vector (Novagen) and sequenced.
BLAST Search
The analogous genomic and amino acidic sequences of PPD were obtained by searching against nonredundant NCBI databases (http://www.ncbi.nlm.nih.gov/BLAST), GenomeNet BLAST2 (http://blast.genome.jp), and TAIR BLAST 2.0 (http://www. arabidopsis.org/Blast). Homology searches at the Chlamydomonas reinhardtii EST index of the Kazusa DNA Research Institute (http://www.kazusa.or.jp/en/plant/chlamy/EST), the C. reinhardtii genomic database, Chlamy Center (http://www.chlamy.org/chlamydb.html), and the CyanoBase (http://www.kazusa.or.jp/cyano/cyano.html) were also done.
Expression of PPD-GST Fusion Protein and Preparation of Antibody
The PPD coding region was amplified by RT-PCR using total RNA isolated from 1-d-senescent cotyledons of radish as a template. The 5′ primer was 5′-TCCCCGGGAATGGGAGGAGAAGGTGGTGCTGA-3′ and the 3′ primer was 5′-ATGAATTCTTATAGTTGAAGAAAAGATACAGCAC-3′. The PPD fragment was digested with SmaI and EcoRI restriction enzymes (Toyobo) and cloned into the SmaI and EcoRI sites of the expression vector pGEX-2T. The plasmid pGEX-2T-PPD was used to transform the Escherichia coli BL21(DE3)pLysS strain. Expression of recombinant PPD protein was induced by addition of isopropyl β-d-thiogalactopyranoside and cultivation for 2 h. Bacteria were collected by centrifugation at 5,500g for 5 min.
The GST fusion protein was purified using GSTrap FF (1 mL; Amersham Biosciences) and a template program for GSTrap of an ÄKTAprime (Amersham Biosciences) liquid chromatography system according to the manufacturer's protocol. To yield the native protein from GST-PPD, the fusion protein was digested with thrombin. The purified PPD was used as an antigen to raise polyclonal antisera in guinea pigs. Polyclonal antibody was prepared by standard protocol and used without further purification.
Electrophoresis
SDS-PAGE was performed by the method of Laemmli (1970) using 15% or 12% polyacrylamide gel under reducing conditions. Heat treatment was performed at approximately 95°C for 1 to 20 min. Fixing and staining were done in aqueous methanol (25%, v/v) containing acetic acid (7.5%, v/v) and Coomassie Brilliant Blue R-250, respectively.
Protein Determination
Protein concentrations were determined using a bicinchoninic acid protein assay kit (Pierce) with bovine serum albumin as a standard. For column chromatographic fractions, protein was assayed spectrophotometrically by measuring at 280 nm at room temperature.
Sequence data from this article can be found in the GenBank/EMBL/DDBJ data libraries under accession number AB218276.
Acknowledgments
We thank Professor Bernhard Kräutler (Leopold-Franzens-Universität, Innsbruck, Austria) for providing the Cj-NCC used in this study. We also thank Keiko Fukuda, Keiko Soga, and Keiko Furuya for technical assistance.
This work was supported by the Ministry of Education, Science, Sports and Culture of Japan (grant nos. 12640631 and 07856).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yuzo Shioi (sbysioi@ipc.shizuoka.ac.jp).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.071290.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SB, Houghton JD, Hendry GAF (1991) Chlorophyll breakdown. In H Scheer, ed, Chlorophylls. CRC Press, Boca Raton, FL, pp 465–489
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenolchloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Doi M, Inage T, Shioi Y (2001) Chlorophyll degradation in a Chlamydomonas reinhardtii mutant: an accumulation of pyropheophorbide a by anaerobiosis. Plant Cell Physiol 42: 469–474 [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin G (1988) Rapid production of full-length cDNAs from rare transcripts; amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85: 8998–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiamét JJ, Pichersky E, Noodén LD (1999) Mass exodus from senescing soybean chloroplasts. Plant Cell Physiol 40: 986–992 [Google Scholar]
- Hasslacher M, Schall M, Hayn M, Griengl H, Kohlwein SD, Schwab H (1995) Molecular cloning of the full-length cDNA of (S)-hydroxynitrile lyase from Hevea brasiliensis: functional expression in Escherichia coli and Saccharomyces cerevisiae and identification of an active residue. J Biol Chem 271: 5884–5891 [DOI] [PubMed] [Google Scholar]
- Hendry GAF, Houghton JD, Brown SB (1987) The degradation of chlorophyll—a biological enigma. New Phytol 107: 255–302 [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266: 383–402 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S (1999) Chlorophyll breakdown in higher plants and algae. Cell Mol Life Sci 56: 330–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynninen PH (1973) Chlorophylls. IV. Preparation and purification of some derivatives of chlorophylls a and b. Acta Chem Scand 27: 1771–1780 [Google Scholar]
- Iriyama K, Ogura N, Takamiya A (1974) A simple method for extraction and partial purification of chlorophyll from plant material, using dioxane. J Biochem (Tokyo) 76: 901–904 [PubMed] [Google Scholar]
- Jacob-Wilk D, Holland D, Goldschmidt EE, Riov J, Eyal Y (1999) Chlorophyll breakdown by chlorophyllase: isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J 20: 653–661 [DOI] [PubMed] [Google Scholar]
- Jaskólski M (2001) 3D domain swapping, protein oligomerization, and amyloid formation. Acta Biochim Pol 48: 807–827 [PubMed] [Google Scholar]
- Kräutler B, Matile P (1999) Solving the riddle of chlorophyll breakdown. Acc Chem Res 32: 35–43 [Google Scholar]
- Kunieda T, Amano T, Shioi Y (2005) Search for chlorophyll degradation enzyme, Mg-dechelatase, from extracts of Chenopodium album with native and artificial substrates. Plant Sci 169: 177–183 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Langmeier M, Ginsburg S, Matile P (1993) Chlorophyll breakdown in senescent leaves: demonstration of magnesium-dechelatase activity. Physiol Plant 89: 347–353 [Google Scholar]
- Losey FG, Engel N (2001) Isolation and characterization of a urobilinogenoidic chlorophyll catabolite from Hordeum vulgare L. J Biol Chem 276: 8643–8647 [DOI] [PubMed] [Google Scholar]
- Matile P, Hörtensteiner S, Thomas H (1999) Chlorophyll degradation. Annu Rev Plant Physiol Plant Mol Biol 50: 67–95 [DOI] [PubMed] [Google Scholar]
- Oberhuber M, Berghold J, Mühlecker W, Hörtensteiner S, Kräutler B (2001) Chlorophyll breakdown—on a nonfluorescent chlorophyll catabolite from spinach. Helv Chim Acta 84: 2615–2627 [Google Scholar]
- Omata T, Murata N (1980) A rapid and efficient method to prepare chlorophyll a and b from leaves. Photochem Photobiol 31: 183–185 [Google Scholar]
- Owens TG, Falkowski PG (1982) Enzymatic degradation of chlorophyll a by marine phytoplankton in vitro. Phytochemistry 21: 979–984 [Google Scholar]
- Perkins HJ, Roberts DWA (1962) Purification of chlorophylls, pheophytins and pheophorbides for specific activity determinations. Biochim Biophys Acta 58: 486–498 [DOI] [PubMed] [Google Scholar]
- Pružinská A, Tanner G, Anders I, Roca M, Hörtensteiner S (2003) Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA 100: 15259–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodoni S, Mühlecker W, Anderl M, Kräutler B, Moser D, Thomas H, Matile P, Hörtensteiner S (1997) Chlorophyll breakdown in senescent chloroplasts. Cleavage of pheophorbide a in two enzymic steps. Plant Physiol 115: 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa K, Hashizume A, Shioi Y (1990) Pyropheophorbide a, a catabolite of ethylene-induced chlorophyll a degradation. Phytochemistry 29: 2105–2106 [Google Scholar]
- Shioi Y, Masuda T, Takamiya K, Shimokawa K (1995) Breakdown of chlorophylls by soluble proteins extracted from leaves of Chenopodium album. J Plant Physiol 145: 416–421 [Google Scholar]
- Shioi Y, Tatsumi Y, Shimokawa K (1991) Enzymatic degradation of chlorophyll in Chenopodium album. Plant Cell Physiol 32: 87–93 [Google Scholar]
- Shioi Y, Tomita N, Tsuchiya T, Takamiya K (1996. a) Conversion of chlorophyllide to pheophorbide by Mg-dechelating substance in extracts of Chenopodium album. Plant Physiol Biochem 34: 41–47 [Google Scholar]
- Shioi Y, Watanabe K, Takamiya K (1996. b) Enzymatic conversion of pheophorbide a to the precursor of pyropheophorbide a in leaves of Chenopodium album. Plant Cell Physiol 37: 1143–1149 [Google Scholar]
- Suzuki T, Kunieda T, Murai F, Morioka S, Shioi Y (2005) Mg-dechelation activity in radish cotyledons with artificial and native substrates, Mg-chlorophyllin a and chlorophyllide a. Plant Physiol Biochem 43: 459–464 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Shioi Y (2002) Two enzymatic reaction pathways in the formation of pyropheophorbide a. Photosynth Res 74: 225–233 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Doi M, Shioi Y (2002) Two enzymatic reaction pathways in the formation of pyropheophorbide a. Photosynth Res 74: 225–233 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Shioi Y (1999) Detection of chlorophyll breakdown products in the senescent leaves of higher plants. Plant Cell Physiol 40: 909–915 [Google Scholar]
- Suzuki Y, Shioi Y (2004) Chlorophyll and carotenoid changes during the senescence of cotyledons of Raphanus sativus L.: simultaneous and systemical analyses. Physiol Plant 122: 291–296 [Google Scholar]
- Takamiya K, Tsuchiya T, Ohta H (2000) Degradation pathway(s) of chlorophyll: What has gene cloning revealed? Trends Plant Sci 5: 426–431 [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Ohta H, Masuda T, Mikami B, Kita N, Shioi Y, Takamiya K (1997) Purification and characterization of two isozymes of chlorophyllase from mature leaves of Chenopodium album. Plant Cell Physiol 38: 1026–1031 [Google Scholar]
- Tsuchiya T, Ohta H, Okawa K, Iwamatsu A, Shimada H, Masuda T, Takamiya K (1999) Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA 96: 15362–15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Suzuki T, Yamada T, Shimada H, Masuda T, Ohta H, Takamiya K (2003) Chlorophyllase as a serine hydrolase: identification of a putative catalytic triad. Plant Cell Physiol 44: 96–101 [DOI] [PubMed] [Google Scholar]
- Vicentini F, Iten F, Matile P (1995) Development of an assay for Mg-dechelatase of oilseed rape cotyledons, using chlorophyllin as the substrate. Physiol Plant 94: 57–63 [Google Scholar]
- Watanabe K, Ohta H, Tsuchiya T, Mikami B, Masuda T, Shioi Y, Takamiya K (1999) Purification and some properties of pheophorbidase in Chenopodium album. Plant Cell Physiol 40: 104–108 [Google Scholar]
- Willstätter R, Stoll A (1913) Die Wirkungen der Chlorophyllase. In Untersuchungen über Chlorophyll. Springer, Berlin, pp 172–187
- Wüthrich KL, Bovet L, Hunziker PE, Donnison IS, Hörtensteiner S (2000) Molecular cloning, functional expression and characterization of RCC reductase involved in chlorophyll catabolism. Plant J 21: 189–198 [DOI] [PubMed] [Google Scholar]
- Ziegler R, Blaheta A, Guha N, Schönegge B (1988) Enzymatic formation of pheophorbide and pyropheophorbide during chlorophyll degradation in a mutant of Chlorella fusca Shihira et Kraus. J Plant Physiol 132: 327–332 [Google Scholar]