Abstract
We have previously shown that a human small GTPase Rac homolog, OsRac1, from rice (Oryza sativa) induces cascades of defense responses in rice plants and cultured cells. Sphingolipid elicitors (SEs) have been similarly shown to activate defense responses in rice. Therefore, to systematically analyze proteins whose expression levels are altered by OsRac1 and/or SE treatment, we performed a differential display analysis of proteins by the use of two-dimensional gel electrophoresis and mass spectrometry. A total of 271 proteins whose expression levels were altered by constitutively active (CA)-OsRac1 or SE were identified. Interestingly, of 100 proteins that were up-regulated by a SE, 87 were also induced by CA-OsRac1, suggesting that OsRac1 plays a pivotal role in defense responses induced by SE in cultured rice cells. In addition, CA-OsRac1 induces the expression of 119 proteins. Many proteins, such as pathogenesis-related proteins, SGT1, and prohibitin, which are known to be involved in the defense response, were found among these proteins. Proteins involved in redox regulation, chaperones such as heat shock proteins, BiP, and chaperonin 60, proteases and protease inhibitors, cytoskeletal proteins, subunits of proteasomes, and enzymes involved in the phenylpropanoid and ethylene biosynthesis pathways were found to be induced by CA-OsRac1 or SE. Results of our proteomic analysis revealed that OsRac1 is able to induce many proteins in various signaling and metabolic pathways and plays a predominant role in the defense response in cultured rice cells.
Plant proteomics has rapidly advanced over the last several years, and a number of studies have been undertaken in various species (van Wijk, 2001; Rose et al., 2004). So far, the most successful studies are those of subcellular compartments, since they contain a limited number of proteins. Such studies include the proteomics of the chloroplast (Peltier et al., 2000; Friso et al., 2004; Kleffmann et al., 2004; Lonosky et al., 2004; Rose et al., 2004), mitochondria (Kruft et al., 2001; Millar et al., 2001; Bardel et al., 2002), endoplasmic reticulum (Maltman et al., 2002), peroxisome (Fukao et al., 2002), and vacuole (Carter et al., 2004). Proteomic studies of specific stages in development or physiological conditions have been also performed with various plants. They include the proteomics of seed maturation (Finnie et al., 2002; Hajduch et al., 2005), seed germination (Gallardo et al., 2002), senescence (Wilson et al., 2002), nitrogen mobilization (Schiltz et al., 2004), salinity tolerance (Liska et al., 2004), and somatic embryogenesis (Imin et al., 2005). These studies mainly employ two-dimensional gel electrophoresis (2-DE) coupled with mass spectrometric analysis using matrix-assisted laser-desorption ionization time of flight (TOF) mass spectrometry (MS) or electrospray ionization-TOF to identify proteins.
One possible limitation in the application of proteomics in plant biology is the lack of complete genome data in most species (van Wijk, 2001; Rose et al., 2004). Since Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) are the only two plant species whose genomes have been almost completely analyzed, proteomics should be more easily applied to these species than to other plants. As expected, most extensive studies on differential protein expression in various tissues have been performed in rice (Koller et al., 2002; Komatsu and Tanaka, 2005) and Arabidopsis (Giavalisco et al., 2005).
Although proteomics is a promising technique that can be used for the study of signal transduction in plants, so far this method has not been extensively used in such studies in plants. In rice, proteomics has been utilized for studies of defensive responses, such as the elicitor response (Rakwal and Komatsu, 2000) and the identification of novel proteins induced by a pathogen attack (Hashimoto et al., 2004). A proteomics study of Arabidopsis proteins induced by infection with bacterial pathogens has been recently reported (Jones et al., 2004). Proteomics has also been used to identify a protein whose phosphorylation level is increased in a lesion-mimic mutant of rice, which confers resistance to the blast fungus infection (Takahashi et al., 2003). A general protein expression study was performed by using a lesion-mimic mutant of rice, cdr2, which shows resistance to the blast fungus infection (Tsunezuka et al., 2005). In Arabidopsis, a protein that is phosphorylated by an elicitor treatment has been identified by a proteome analysis (Peck et al., 2001), and a systematic analysis of phospholylation sites of proteins has been performed (Nuhse et al., 2004).
Rac GTPase belongs to a family of Rac/Rop GTPases in plants, which are closely related to the Rho-type GTPase of animals, and the Rac/Rop GTPases have been shown to play key roles in a number of cellular activities such as growth and differentiation, hormone signaling, defense signaling against pathogens, and responses to various stresses in plants (Valster et al., 2000; Yang, 2002; Gu et al., 2004). It has been previously shown that Rac GTPase of rice, OsRac1, is a molecular switch in the defense response of rice (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002). Constitutively active (CA)-OsRac1 induces the generation of reactive oxygen species (ROS), the activation of defense genes, and the production of phytoalexin, leading to disease resistance. Furthermore, the heterotrimeric G protein α-subunit has been shown to act upstream of OsRac1 and regulate ROS production and defense gene activation (Suharsono et al., 2002). More recently, OsRac1 and Gα were shown to positively regulate OsMAPK6, a rice homolog of SIPK and AtMAPK6, at the protein level (Lieberherr et al., 2005). It was also shown that OsRac1 transiently suppresses the mRNA levels of metallothionein, OsMT2b, which functions as a ROS scavenger, thus, leading to enhance ROS signaling in elicitor-induced rice cells (Wong et al., 2004). The dominant-negative (DN)-OsRac1 was shown to decrease ROS production and reduce the level of resistance to Tobacco mosaic virus in tobacco (Nicotiana tabacum) plants carrying the N resistance gene (Moeder et al., 2005). The regulation of ROS production by Rac GTPase has been demonstrated in other plant species (Potikha et al., 1999; Park et al., 2000). Therefore, these studies strongly suggest that Rac GTPase plays a pivotal role in the defense response in plants. However, its signaling cascade during the defense response remains to be studied.
Sphingolipid elicitors (SEs) were isolated from membranes of the rice blast fungus and shown to cause the accumulation of phytoalexins, cell death, increased resistance to infection by compatible pathogens, and induction of PR (pathogenesis-related) gene expression (Koga et al., 1998; Umemura et al., 2002). These results indicated that the SEs induce a cascade of responses normally induced by pathogens. We have shown that the elicitors require both OsRac1 and Gα, suggesting that they are involved in the elicitor's signal transduction pathway (Suharsono et al., 2002).
Therefore, to systematically analyze proteins whose expression levels are constitutively activated in rice cells expressing CA-OsRac1 or induced by a SE, we employed a proteomics approach by using 2-DE and mass spectrometric analysis. We identified 271 proteins whose expression levels were altered by elicitor or CA-OsRac1. The analysis of these proteins revealed that close to 90% of the proteins, which were induced by the SE, were constitutively activated in rice cells expressing CA-OsRac1. In addition to those proteins induced by elicitor and CA-OsRac1, CA-OsRac1 induces many other proteins that may be involved in defensive responses. These results strongly suggested that OsRac1 plays a pivotal role in the elicitor-induced defense response in cultured rice cells.
RESULTS AND DISCUSSION
The Expression Levels of 258 Proteins Are Altered by CA-OsRac1
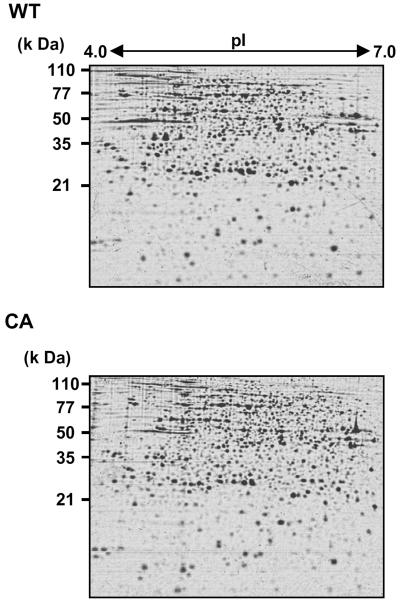
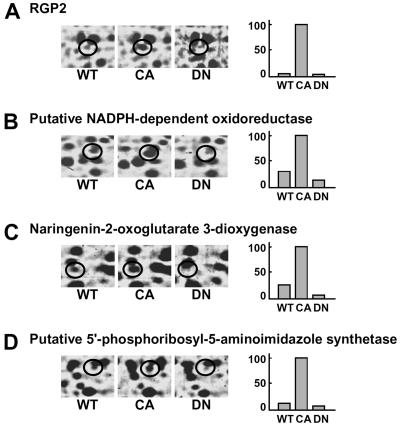
To investigate alterations in protein expression induced by CA-OsRac1 expression, we used a combination of 2-DE and MS. For the analysis of cultured rice cells transformed with CA-OsRac1 against the wild type (Kinmaze), we extracted total proteins from rice cells with a urea/thio-urea buffer. The total proteins from cultured rice cells were resolved into approximately 1,500 spots in reproducible SDS-polyacrylamide gels (isoelectric focusing pH range; 4–7 size; 24 cm, SDS-PAGE gel size; 26 × 20 cm). Protein spots were visualized by staining with colloidal Coomassie Brilliant Blue. Figure 1 shows the entire image of the Coomassie Brilliant Blue-stained 2-D gels of total extracted proteins from wild-type and CA-OsRac1 cultured rice cells. The image analysis of these gels was carried out with PDQuest. Overall, the protein levels of 258 spots were found to be altered by CA-OsRac1. Of 258 proteins, 206 were up-regulated, while 52 were down-regulated. Representative samples of proteins that were up-regulated by CA-OsRac1 are shown in Figure 2. They are RGP2 (reverse-glycosylating protein 2), putative NADPH-dependent oxidoreductase, naringenin-2-oxoglutarate 3-dioxygenase, and putative 5′-phosphoribosyl-5-aminoimidazole synthetase, and their protein levels were increased 17-fold, 3-fold, 4-fold, and 7.5-fold, respectively (Table I; Supplemental Table I). Their levels in DN-OsRac1-expressing cells were decreased as shown in Figure 2. Those proteins that showed more than 3-fold changes in all of three or more replicate gels from the independent protein extractions were chosen to be differentially regulated proteins (see “Materials and Methods”). These spots were excised from the gels, digested with trypsin, and analyzed by Q-TOF mass spectrometer. Protein identification was performed with the nonredundant database National Center for Biotechnology Information using the MASCOT search program (Perkins et al., 1999). Proteins were subsequently identified by a database search.
Figure 1.
2-DE of proteins isolated from rice cell cultures expressing CA-OsRac1. Proteins isolated from the wild-type (WT) and CA-OsRac1-expressing cells (CA) are shown.
Figure 2.
Examples of rice proteins whose levels were increased by CA-OsRac1 and decreased by DN-OsRac1. A, RGP2. B, Putative NADPH-dependent oxidoreductase. C, Naringenin-2-oxoglutarate 3-dioxygenase. D, Putative 5′-phosphoribosyl-5-aminoimidazole synthetase. Detailed information for proteins shown in A to C is provided in Table I and that of D is shown in Supplemental Table I. The level of increase found with each spot is shown in bars on the right. The highest level found among various treatments is shown as 100.
Table I.
Proteins that are induced by SE treatment and CA-OsRac1
These proteins belong to class I in Figure 4.
| Identified Protein | Accession No. | Mr (Da) | pI | Fold Increase |
|---|---|---|---|---|
| PR proteins | ||||
| Putative β-1,3-glucanase precursor | gi|33146691 | 54,631 | 5.68 | 69.0 |
| Sgt1 | gi|6581058 | 41,041 | 4.97 | 3.9 |
| Tau class GST protein 4 | gi|33304610 | 25,689 | 4.97 | 3.1 |
| Putative GST OsGSTF5 | gi|11177841 | 25,053 | 5.67 | 3.4 |
| Salt stress-induced protein | gi|134190 | 15,188 | 5.19 | 188.9 |
| Redox regulation | ||||
| Putative NADPH-dependent oxidoreductase | gi|37530522 | 36,032 | 5.32 | 3.2 |
| Cytochrome p450 isoform PM17 | gi|5524157 | 57,020 | 6.59 | 3.5 |
| Putative NADPH-thioredoxin reductase | gi|50912077 | 34,940 | 6.19 | 3.2 |
| NADP malic enzyme | gi|37694731 | 63,294 | 5.52 | 3.3 |
| Protein folding and homeostasis | ||||
| Proteasome subunit α-type 1 (20S proteasome α-subunit F) | gi|1709758 | 29,897 | 5.37 | 174.8 |
| 26S protease regulatory subunit 6A homolog | gi|1174613 | 47,995 | 4.97 | 4.3 |
| 26S proteasome regulatory particle non-ATPase subunit 12 | gi|17297985 | 31,095 | 4.99 | 3.5 |
| Hsp82 | gi|417154 | 80,429 | 5.00 | 3.5 |
| Hsp22 precursor, mitochondrial | gi|7441311 | 23,801 | 6.47 | 3.1 |
| Putative chaperonin CPN60-2, mitochondrial precursor | gi|46063348 | 72,531 | 7.06 | 252.5 |
| DnaK-type molecular chaperone BiP | gi|7441868 | 73,666 | 5.30 | 8.0 |
| Putative TCP-1/cpn60 chaperonin family protein | gi|50428682 | 57,592 | 5.56 | 4.0 |
| Cys proteinase inhibitor | gi|34903176 | 27,252 | 6.07 | 3.2 |
| Leucyl aminopeptidase | gi|7488956 | 31,173 | 5.33 | 4.6 |
| Phenylpropanoid biosynthesis pathway | ||||
| Naringenin-2-oxoglutarate 3-dioxygenase | gi|38344477 | 32,481 | 5.51 | 4.0 |
| Polyamine and ethylene biosynthesis pathway | ||||
| Spermidine synthase 1 | gi|12229998 | 35,523 | 5.23 | 3.4 |
| Arginase | gi|32488415 | 37,149 | 5.90 | 3.2 |
| Alcoholic fermentation-associated proteins | ||||
| Mitochondrial ALDH ALDH2a | gi|8574429 | 59,151 | 6.24 | 3.1 |
| ALDH | gi|8163730 | 59,626 | 6.33 | 25.5 |
| PDC isozyme 1 | gi|1706325 | 65,978 | 5.89 | 3.1 |
| Cytoskeletal proteins | ||||
| Tubulin β-chain | gi|629806 | 50,703 | 4.68 | 18.6 |
| Tubulin β-4 chain | gi|34902500 | 50,937 | 4.73 | 7.5 |
| Actin | gi|28301928 | 42,014 | 5.30 | 4.9 |
| Actin 82 | gi|3219766 | 37,323 | 5.54 | 3.5 |
| Putative actin | gi|34905850 | 41,929 | 5.23 | 3.1 |
| Transcription- and translation-related proteins | ||||
| Translational elongation factor Tu | gi|18001149 | 48,564 | 6.04 | 4.2 |
| Putative translational initiation factor eIF-4A | gi|45736055 | 47,393 | 5.43 | 3.8 |
| Eukaryotic initiation factor 4A | gi|547712 | 47,187 | 5.29 | 5.0 |
| Elongation factor 1-β | gi|232031 | 23,815 | 4.86 | 3.2 |
| Putative asparaginyl-tRNA synthetase | gi|34914792 | 62,948 | 5.68 | 4.5 |
| Importin | gi|33337497 | 58,197 | 5.06 | 96.5 |
| Putative Gly-rich protein 2 | gi|29467522 | 19,024 | 6.28 | 24.3 |
| Putative nucleic acid binding protein | gi|37536904 | 48,438 | 5.21 | 66.8 |
| 60S acidic ribosomal protein P0 | gi|730580 | 34,470 | 5.38 | 49.6 |
| Pentose phosphate pathway | ||||
| Putative Fru-bisphosphate aldolase | gi|34895322 | 42,253 | 8.81 | 8.2 |
| Fru-bisphosphate aldolase, cytoplasmic isozyme | gi|113622 | 39,185 | 8.50 | 37.7 |
| Probable phosphogluconate dehydrogenase (decarboxylating) | gi|7431262 | 8,580 | 10.00 | 79.2 |
| Cytosolic 6-phosphogluconate dehydrogenase | gi|38426301 | 51,784 | 6.58 | 32.9 |
| Phosphoglucomutase | gi|13324798 | 63,138 | 5.40 | 3.4 |
| Glc-6-P isomerase | gi|41529312 | 68,765 | 5.88 | 3.9 |
| Enolase (2-phosphoglycerate dehydratase) | gi|3023713 | 48,299 | 5.42 | 4.4 |
| Glycolysis/gluconeogenesis | ||||
| PDC | gi|7436705 | 65,964 | 5.89 | 35.9 |
| Pyruvate kinase isozyme G, chloroplast | gi|2497540 | 46,149 | 6.12 | 26.0 |
| Putative lipoamide dehydrogenase | gi|34894958 | 59,094 | 6.51 | 52.6 |
| Triosephosphate isomerase | gi|553107 | 27,816 | 6.60 | 95.3 |
| Glyceraldehyde 3-P dehydrogenase, cytosolic | gi|42407702 | 36,561 | 6.61 | 3.6 |
| Nucleic acid and amino acid metabolism | ||||
| Aspartate carbamoyl transferase | gi|9453916 | 39,981 | 6.95 | 3.3 |
| Cys synthase | gi|11131901 | 34,399 | 5.35 | 4.3 |
| Probable nitrilase | gi|14289301 | 33,670 | 5.28 | 5.1 |
| UDP-Glc pyrophosphorylase | gi|15823775 | 51,821 | 5.43 | 4.6 |
| Putative phosphoribosylaminoimidazole-carboxamide formyltransferase | gi|32974354 | 64,669 | 6.64 | 46.2 |
| Putative Asp-semi-ALDH | gi|14488363 | 40,438 | 6.73 | 2.7 |
| Wheat (Triticum aestivum) adenosylhomocysteinase-like protein | gi|29367605 | 53,860 | 5.62 | 3.6 |
| Putative 3-isopropylmalate dehydrogenase | gi|31415971 | 43,515 | 5.85 | 3.8 |
| Probable methylmalonate-semi-ALDH | gi|7431455 | 57,514 | 5.99 | 12.8 |
| Signaling | ||||
| Mitogen-activated protein kinase | gi|29500879 | 45,172 | 5.45 | 30.6 |
| Putative Ser/Thr protein kinase | gi|37536404 | 44,096 | 5.76 | 3.2 |
| Ser/Thr protein phosphatase PP2A-2 catalytic subunit | gi|11134218 | 35,847 | 5.16 | 3.1 |
| Glycerol kinase | gi|5852172 | 83,966 | 8.88 | 30.5 |
| Nucleoside diphosphate kinase I | gi|2498077 | 16,591 | 6.30 | 3.6 |
| Others | ||||
| RGP2 | gi|3646375 | 39,532 | 8.08 | 17.0 |
| Putative Glu-1-semialdehyde 2,1-aminomutase, chloroplast precursor | gi|42761379 | 50,434 | 6.48 | 3.8 |
| Betaine ALDH | gi|30698520 | 55,446 | 5.36 | 40.7 |
| Putative enoyl-acyl-carrier protein reductase | gi|38637198 | 39,277 | 8.81 | 5.1 |
| MTHFR, 3-partial | gi|37718877 | 42,245 | 6.10 | 3.1 |
| Glc-1-P adenylyltransferase precursor | gi|100675 | 53,747 | 5.65 | 27.7 |
| Glc-1-P adenylyltransferase small subunit, chloroplast precursor | gi|121289 | 53,764 | 5.65 | 10.2 |
| Delta-coat protein | gi|7677262 | 57,633 | 5.54 | 35.7 |
| Putative mitotic control protein dis3 | gi|23306125 | 10,101 | 6.36 | 3.0 |
| Putative iron deficiency protein Ids3 | gi|34395014 | 38,018 | 5.33 | 4.9 |
| Putative Xyl isomerase | gi|34395196 | 53,803 | 5.42 | 5.1 |
| Putative succinyl-CoA ligase (GDP forming) | gi|47847769 | 45,405 | 5.98 | 5.0 |
| Putative carboxymethylenebutenolidase | gi|34913460 | 30,505 | 6.31 | 3.1 |
| Unknown protein | gi|31415917 | 34,496 | 9.07 | 4.9 |
| Unnamed protein product | gi|34902832 | 58,162 | 5.10 | 71.7 |
| OSJNBa0020P07.11 | gi|38344868 | 46,571 | 6.16 | 5.9 |
| OSJNBa0032I19.9 | gi|38345357 | 38,591 | 5.66 | 6.3 |
| OSJNBa0044K18.22 | gi|38605779 | 36,882 | 5.77 | 3.9 |
| OSJNBa0085I10.14 | gi|38345797 | 44,129 | 6.23 | 3.3 |
| OSJNBb0059K02.15 | gi|32488924 | 64,153 | 6.83 | 90.8 |
| P0489A05.7 | gi|34903688 | 66,122 | 6.69 | 3.1 |
| P0665D10.11 | gi|34904026 | 55,518 | 8.33 | 3.1 |
A Total of 100 Proteins Induced by SE Are Identified
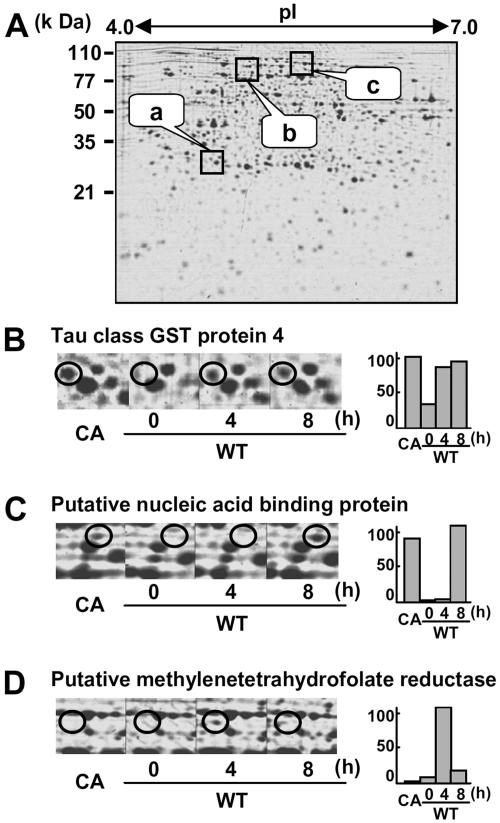
To analyze proteins whose expression levels are altered by SE, alterations in protein levels were examined at 4 and 8 h after initiation of the SE treatment (Fig. 3A). For instance, the tau class glutathione S-transferase (GST) protein 4 was not visible in the wild-type cells, while it was clearly induced at 4 and 8 h after elicitor treatment and in CA-OsRac1-expressing cells (Fig. 3B). A putative nucleic acid-binding protein was induced only at 8 h after the elicitor treatment and expressed in CA-OsRac1-expressing cells (Fig. 3C). Putative methylenetetrahydrofolate reductase (MTHFR) was induced at 4 h after elicitor treatment but disappeared at 8 h after treatment. This protein was not expressed in CA-OsRac1-expressing cells (Fig. 3D). We have identified a total of 100 proteins that were induced by SE in cultured rice cells (Fig. 4). Those proteins that were found only at either 4 h or 8 h after elicitor treatment were included in this category. We did not find any protein spots that were clearly down-regulated by SE. This is in contrast to CA-OsRac1-expressing cells, in which a number of proteins were down-regulated as shown below.
Figure 3.
2-DE of proteins isolated from rice cell cultures treated with SE. A, The entire image of the Coomassie Brilliant Blue-stained gel; a, b, and c are the regions enlarged in B, C, and D, respectively. Wild-type cells were treated with SE for 4 and 8 h. The level of the protein spot with varying times of SE treatment and that found in CA-OsRac1-expressing cells is shown in bars on the right. CA indicates cultured rice cells in which CA-OsRac1 was expressed. The highest level found among various treatments is shown as 100.
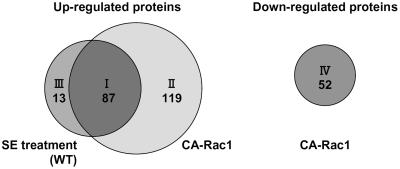
Figure 4.
Four categories of proteins identified by the proteome analysis of CA-OsRac1-expressing cells and wild-type cells treated with SE. Proteins grouped in class I (87 proteins; Table I) were those increased by both CA-OsRac1 and SE treatment. Class II (119 proteins; Supplemental Table I) includes proteins that were up-regulated by CA-OsRac1 but not increased by SE treatment. Class III (13 proteins; Supplemental Table II) includes those that were induced by SE treatment but not induced by CA-OsRac1. Class IV (52 proteins; Supplemental Table III) includes those whose expression levels were decreased by CA-OsRac1.
Proteins Whose Abundance Is Altered by CA-OsRac1 or SE Are Grouped into Four Categories
Proteins whose expression levels were altered by CA-OsRac1 or SE were grouped into four categories (Fig. 4). Proteins grouped in class I (87 proteins; Table I) were those whose abundance was increased by both CA-OsRac1 and SE treatment. Those in class II (119 proteins; Supplemental Table I) were proteins that were up-regulated by CA-OsRac1 but not increased by SE treatment. Class III (13 proteins; Supplemental Table II) included those that were induced by SE treatment but not by CA-OsRac1. Class IV (52 proteins; Supplemental Table III) included those whose expression levels were decreased by CA-OsRac1. For each protein listed in the tables an average fold induction or fold repression is shown at the right end of the column. For SE-induced proteins fold increase was calculated based on the higher values obtained at 4 or 8 h treatment.
The Majority of SE-Induced Proteins Are Constitutively Expressed in CA-OsRac1-Expressing Rice Cells
Surprisingly, 87 of 100 SE-inducible proteins (87%) were constitutively expressed in the cultured rice cells expressing CA-OsRac1 (class I). A total of 206 proteins that were up-regulated by CA-OsRac1 (classes I and II) were identified. Therefore, 42% of CA-OsRac1-induced proteins were similarly up-regulated by SE. These results suggest that OsRac1 could induce most SE-inducible proteins and that OsRac1 is likely to be placed very close to SE reception in the same signaling pathway. This conclusion is consistent with the previous findings that CA-OsRac1 could induce a variety of responses in rice cells and plants and leads to resistance against pathogens (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002; Wong et al., 2004; Lieberherr et al., 2005).
Proteins Whose Abundance Is Altered by CA-OsRac1 or SE Treatment
A large number of proteins were identified in the differential display analysis, and they could be classified into several distinct categories based on their functions. They will be discussed separately below.
Defense-Related Proteins
Proteins that are known to be induced during defense responses were identified in SE-treated cells (Table I) and CA-OsRac1-expressing cells (Supplemental Table I). We identified PR proteins, such as β-1, 3-glucanase and root-specific PR-10, and stress-induced proteins, such as salt tolerance protein 5, putative glucosyltransferase IS5a (salicylate induced), and salt stress-induced protein. In addition, SGT1, which has been shown to be a key regulator of R-gene-mediated disease resistance, was found to be induced by CA-OsRac1. SGT1 mRNA was previously shown to be up-regulated by infection by rice blast fungus in rice (Cooper et al., 2003).
Prohibitin, a mitochondrial protein, was previously shown to be phosphorylated in rice lesion-mimic mutants (Takahashi et al., 2003). It is implicated in cell death, and its protein level was increased by CA-OsRac1. We identified a rice mitogen-activated protein kinase (MAPK), OsMAPK6, which is an ortholog of tobacco SIPK and Arabidopsis MPK6; this protein was accumulated in CA-OsRac1-expressing cells and up-regulated by SE treatment. OsMAPK6 activity was recently shown to be induced by SE (Lieberherr et al., 2005). It was also shown that silencing of OsRac1 by RNAi strongly reduced OsMAPK6 at the protein level (Lieberherr et al., 2005). Therefore, it is likely that the protein spot whose level was increased by CA-OsRac1 and SE was the phosphorylated OsMAPK protein.
Redox Regulation
ROS have signaling roles in the induction of changes in gene expression associated with a number of physiological and developmental processes, including defense responses (Lamb and Dixon, 1997; Desikan et al., 2000; Dorey et al., 1999; Clarke et al., 2000; Grant and Loake, 2000; Rao et al., 2000; Apel and Hirt, 2004). Since we have shown previously that CA-OsRac1 can induce ROS production in cultured rice cells (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002), we assumed that proteins related to redox regulation were altered in cells expressing CA-OsRac1. As expected, we identified a number of oxidative stress-related proteins, including glyceraldehyde-3-P dehydrogenase and NADPH-thioredoxin reductase (Table I).
Ferredoxin-NADP(H) reductase, putative NADPH-dependent oxidoreductase, Gln synthetase, putative quinine oxidoreductase, putative quinine oxidoreductase QR2, and putative NADPH-thioredoxin reductase were constitutively up-regulated in CA-OsRac1-expressing cells. Putative MTHFR was induced by SE in the wild-type cells. GSTs can detoxify lipid peroxidation products to prevent ROS toxicity (Gronwald and Plaisance, 1998). Expression of GST1 transcripts in Arabidopsis is induced by oxidative stress (Reuber et al., 1998; Kliebenstein et al., 1999), and induced mRNA levels of GST1 are used as early markers of defense responses in this species (Rate and Greenberg, 2001).
Chaperones
Heat-shock proteins (Hsps) and chaperonin were identified in CA-OsRac1-expressing cells. We have previously identified four molecular chaperones that were up-regulated during the appearance of cell death in the rice lesion-mimic mutant, cdr2 (Tsunezuka et al., 2005). These proteins likely play a role in the stabilization and refolding of proteins that have been denatured during exposure to various stresses (Bukau and Horwich, 1998; Fink, 1999). Hsp70 and Hsp60 have been found to functionally cooperate in vitro and are regulated for the maintenance of proteins in cells (Langer et al., 1992). Additionally, in Escherichia coli, the DnaK and GroE systems cooperate for the preservation of aggregation in the folding of ferredoxin-NADP(H) reductase (Dionisi et al., 1998). We identified the Hsp22 precursor, which was up-regulated in CA-OsRac1-expressing cells and control cells after a 4-h treatment with SE. The results indicate that CA-OsRac1 strongly induces the protein expression of various molecular chaperones, which are probably required for the maintenance of cell functions under stress conditions.
Proteases and Protease Inhibitors
We found that a Cys protease was down-regulated, whereas a Cys protease inhibitor was increased by CA-OsRac1. Both proteins are shown to be important for the regulation of programmed cell death. The Cys protease inhibitor exhibits high homology to cyctatin, which is probably involved in the regulation of protein turnover, and plays an important role in resistance against insects and pathogens (Danon et al., 2004; Han et al., 2004). In soybean (Glycine max L. Merr.), transcripts of a Cys protease inhibitor are induced by wounding or methyl jasmonate treatment in local and systemic leaves coincident with increased papain inhibitory activity (Botella et al., 1996). Moreover, soybean cyctatin and the Arabidopsis homolog of this protein, AtCYS1, have been reported to effectively block cell death triggered by either oxidative stress or avirulent pathogens when transiently expressed in cultured cells (Solomon et al., 1999; Belenghi et al., 2003). Subunits of 20S and 26S proteasomes were found to be up-regulated in SE-treated wild-type cells and CA-OsRac1-expressing cells.
Phenylpropanoid Biosynthesis Pathway
We identified several proteins associated with the phenylpropanoid pathway in CA-OsRac1-induced proteins (Table I). They include the NADP-dependent malic enzyme and pyruvate decarboxylase (PDC), caffeic acid 3-o-methyltransferase, and putative isoflavone reductase. These proteins are enzymes involved in the lignin and phytoalexin biosynthesis of the phenylpropanoid pathway and known to be highly induced after infection of pathogens (Dumas et al., 1988; Jaeck et al., 1992; Pellegrini et al., 1993, 1994). We have recently found that PAL transcripts were decreased in rice cell cultures in which OsMAPK6 expression was silenced by RNAi (Lieberherr et al., 2005).
Polyamine and Ethylene Biosynthesis Pathway
We identified Met synthase and S-adenosyl-Met (SAM) synthase as CA-OsRac1-inducible proteins. SAM is involved in the biosynthesis of ethylene and polyamine (Even-Chen et al., 1982) and serves as a precursor of ethylene, which is induced by wounding, drought, and pathogen invasion (Arimura et al., 2002). In our previous study on the proteomics of a rice lesion-mimic mutant, cdr2, SAM synthase was found to accumulate during lesion formation, suggesting that ethylene may be involved in cell death in rice (Tsunezuka et al., 2005).
Many enzymes in the polyamine biosynthetic pathway were found to be induced by CA-OsRac1. Decarboxylated SAM, which is a SAM decarboxylation form, was catalyzed by SAM decarboxylase. Thus, this compound links the ethylene biosynthesis pathway with the polyamine biosynthesis pathway. Decarboxylated SAM is used as a substrate for spermidine synthase and spermine synthase, and these enzymes convert from putrescine to spermidine and from spermidine to spermine, respectively. Arginase is an enzyme in the polyamine biosynthesis pathway and is shown to be induced by wounding, jasmonic acid treatment, and bacterial pathogen attack (Chen et al., 2004).
Alcoholic Fermentation-Associated Proteins
We identified several proteins associated with alcoholic fermentation, PDC, alcohol dehydrogenase, and aldehyde dehydrogenase (ALDH), among proteins that were induced by CA-OsRac1. These enzymes are necessary for energy production of plants in anaerobic environments (Nakazono et al., 2000; Kirch et al., 2004). It was also reported that ALDH has a role in plant defense and abiotic stress tolerance (Sunkar et al., 2003; Kirch et al., 2004). It has been reported that anoxic pretreatment protects soybean cells from hydrogen peroxide-induced cell death (Amora et al., 2000). It is possible that the accumulation of alcoholic fermentation-associated proteins in CA-OsRac1-expressing cells may protect the cells from toxic effects of hydrogen peroxide. It was reported that in Arabidopsis, a Rop GTPase, corresponding to Rac GTPase in rice, regulates balance between ethanolic fermentation and survival (Baxter-Burrell et al., 2002).
Cytoskeletal Proteins
Rho GTPase is known to regulate cytoskeleton organization in animal cells (Etienne-Manneville and Hall, 2002). Cytoskeleton reorganization, which occurs during infection with pathogens, has been shown to be important for resistance to pathogens (Kobayashi et al., 1997; Opalski et al., 2005). The protein levels of various tubulins and actins were shown to be increased by SE or CA-OsRac1 (Table I; Supplemental Table II), suggesting the possibility that OsRac1 may regulate defense responses through control of cytoskeletal proteins.
Proteins Down-Regulated by CA-OsRac1
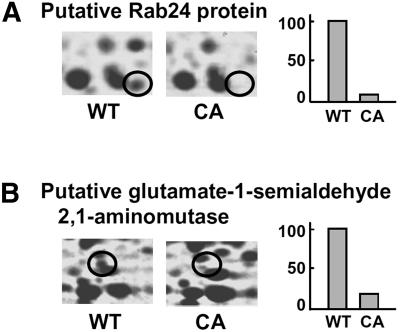
We identified 52 proteins that were down-regulated by CA-OsRac1. Although proteins grouped into this category do not clearly show a particular tendency, they might have negative regulators of defense responses (Fig. 5; Supplemental Table III). Calreticulin and the calreticulin precursor were down-regulated in CA-OsRac1. Calreticulin, a Ca2+-binding protein associated with several functions, including Ca2+ homeostasis and molecular chaperoning, is required for the stress response (Simpson et al., 1997; Park et al., 2001). In a proteomics study of rice cells in culture, calreticulin was identified as one of the phosphorylated proteins and shifted to an insoluble fraction with cold treatment (Li et al., 2003). We identified Rab24, which is one of the Rab family small G proteins. It is known that the Rab GTPase family regulates intracellular protein trafficking, but the function of Rab24 remains unknown.
Figure 5.
Examples of protein spots that were down-regulated by CA-OsRac1. A, Putative Rab24 protein. B, Putative Glu-1-semialdehyde 2,1-aminomutase. Levels of the protein spot in the wild-type (WT) and CA-OsRac1-expressing cells (CA) are shown. The highest level found among various treatments is shown as 100.
Are the Changes at the Protein Level Correlated with Those at the mRNA Level?
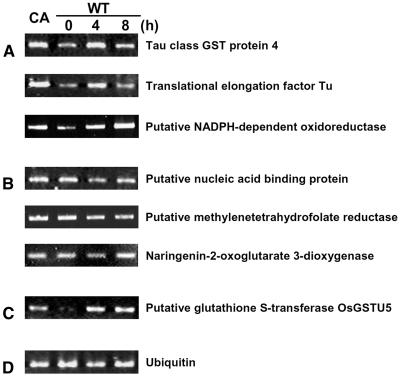
One important question originating from our proteomic study was whether the changes at the protein level caused by CA-OsRac1 expression or SE treatment were similar at the mRNA level. We have performed a preliminary analysis of global mRNA expression by the use of a rice microarray carrying 22,000 genes (T. Togashi, T. Kawasaki, and K. Shimamoto, unpublished data). Results showed that, for 87 class I proteins, which were up-regulated by both CA-OsRac1 and SE, 69 (79.3%) did not show a clear increase at the mRNA level. For 119 class II proteins, which were up-regulated by CA-OsRac1 but not by SE treatment, 52 (43.7%) exhibited no increase at the mRNA level, whereas 44 (37.0%) showed a similar increase at the mRNA level. Therefore, in these two classes, 121 of 206 proteins (58.7%) showed no clear increase at the mRNA level. These results indicate that almost 60% of the proteins whose levels were increased by either CA-OsRac1 or SE treatment did not show a similar increase at the mRNA level, suggesting that changes at the mRNA level do not necessarily show changes at the protein level under the conditions used in this study. It is possible that various modifications of proteins or changes in protein stability cause increased protein levels without a concomitant increase in mRNA levels. Results of reverse transcription-PCR analysis for some examples are shown in Figure 6. Six proteins whose mRNA levels were analyzed were class I proteins whose protein levels were increased by CA-OsRac1 and the SE treatment (Fig. 6, A and B). Three class I proteins whose abundance was essentially parallel with that of RNA are shown in Figure 6A while three other class I proteins whose mRNA levels did not greatly change by CA-OsRac1 or SE treatment are shown in Figure 6B. Figure 6C shows an example whose protein level was increased by CA-OsRac1 but not changed by SE. However, its RNA level was increased by CA-OsRac1 and SE. This kind of information can be obtained only after having conducted extensive studies on both functional proteomics and transcriptome analysis. Although this is preliminary information at the moment, our results on the functional proteomics of defense signaling in cultured rice cells suggest the importance of proteomics in plant cell and molecular biology.
Figure 6.
RNA analysis of proteins whose levels were increased by CA-OsRac1 and/or SE treatment. A, For these three proteins their RNA levels were similarly increased by CA-OsRac1 and SE treatment. Information on these proteins is shown in Table I. B, For these three proteins their RNA levels were not greatly increased by CA-OsRac1 or SE treatment. For their protein levels they were increased by CA-OsRac1 and SE treatment. Information on these proteins is shown in Table I. C, A protein whose RNA level was increased by SE treatment but its protein level was not. Both RNA and protein levels were increased by CA-OsRac1. Information on this protein is shown in Supplemental Table I. D, Ubiquitin RNA as a control.
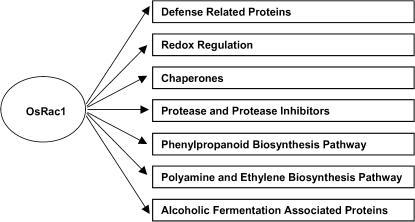
The Role of OsRac1 in Signaling in Cultured Rice Cells
From our proteomic study of OsRac1 it is clear that it plays an important role in various signaling pathways in rice cells as shown in Figure 7. Current study seems to indicate that OsRac1 regulates signaling pathways in various stress responses including defense. Increased levels of proteins involved in redox regulation by CA-OsRac1 may be the results of increased ROS production (Kawasaki et al., 1999), which in turn caused up-regulation of chaperones. The lack of clear changes in cell morphology might indicate that OsRac1 may not be involved in actin regulation as has been shown for some Rop proteins in Arabidopsis (Gu et al., 2004). Rice contains seven Rac genes (Miki et al., 2005), and various cellular functions of Rac genes have not been assigned to members of OsRac gene family. RNAi was recently used to specifically silence expression of each OsRac gene (Miki et al., 2005), thus they will be useful tools to identify signaling roles of individual OsRac proteins in the near future.
Figure 7.
Summary of signal transduction pathways that are activated by OsRac1 revealed in this study.
CONCLUSION
A systematic analysis of proteins whose expression was altered by CA-OsRac1 and/or a SE by 2-DE and mass spectrometric analysis identified 271 proteins. Although a large-scale proteome analysis of various tissues in rice identified more than 2,500 proteins (Koller et al., 2002), extensive studies on the proteome analysis of functionally separated proteins involved in particular signal transduction pathways have not been performed. Therefore, this study may represent one of the most extensive analyses of proteins involved in the defense signaling pathway, at least in rice. The most interesting observation in this study is that the majority of SE-induced proteins were also activated by CA-OsRac1. Considering the previous findings that a SE is able to elicit almost all the cellular responses induced by the rice blast fungus, these results might indicate that OsRac1 is indeed a molecular switch of defense in rice. Furthermore, the observation that CA-OsRac1 is able to induce many other proteins, which were not induced by a SE, indicates that OsRac1 is also involved in other signaling pathways. They may be other branches in the defense pathway and others in various signaling pathways. Our preliminary comparison of microarray data on global mRNA expression with the protein data shown here indicates that, for a large fraction of proteins whose expression levels were increased by CA-OsRac1 or SE treatment in cultured rice cells, their mRNA levels did not appreciably change. These results suggest the importance of proteomics in the analysis of actual modifications in protein expression that occur during responses to various internal and external stimuli.
MATERIALS AND METHODS
Rice Cell Cultures and Elicitor Treatment
Rice (Oryza sativa) suspension cultured cells expressing the CA and DN-OsRac1 have been described previously (Kawasaki et al., 1999; Ono et al., 2001). These transgenic cells in a 20-mL medium were subcultured every week and incubated on a rotary shaker (50 rpm) at 30°C. The SE treatment was performed according to Suharsono et al. (2002).
Protein Extraction
Rice cells in suspension cultures were harvested either before or after SE treatment and ground in a protein extraction buffer (7 m urea/2 m thio-urea, 4% w/v CHAPS, 20 mm dithiothreitol [DTT], 2% v/v, pharmalyte [Amersham Biosciences]; pH 3–10). Crude homogenates were sonicated and centrifuged at 4°C (9,000g for 30 min) to remove cellular debris. The supernatants were centrifuged again (20,000g for 30 min) and kept frozen at −80°C. The protein contents were determined by the Bradford (1976) method using bovine serum albumin as a standard.
2-DE and Gel Image Analysis
Isoelectric focusing was performed for 90 kVh at 20°C with a Multiphor II unit (Amersham Biosciences). For analytical separations, precast 24-cm immobilized pH gradient strips (pH 4–7; Immobiline DryStrip; Amersham Biosciences) were rehydrated overnight with 500 μg of sample proteins/450 μL of a rehydration buffer (7 m urea/2 m thio-urea, 4% w/v CHAPS, 20 mm DTT, 2% v/v, IPG buffer [Amersham Biosciences]; pH 4–7, bromphenol blue). The strips were placed in a reduction buffer (1% DTT, 6 m urea, 2% SDS, 30% glycerol, 50 mm Tris-HCl, pH 8.8) for 20 min with gentle shaking. The strips were then transferred to an alkylation buffer (2.5% iodoacetamide, 6 m Urea, 2% SDS, 30% glycerol, 50 mm Tris-HCl, pH 8.8) and shaken for 20 min. SDS-PAGE was carried out using an Ettan DALT twelve 2-D electrophoresis unit (Amersham Biosciences). The SDS polyacrylamide gels used contained 12.5% polyacrylamide gel (260 × 200 × 1.5 mm) and run in a Tris-Gly buffer (25 mm Tris, 192 mm Gly, 0.1% SDS) at a current setting of 10 mA/gel at 25°C.
The 2-DE gels were stained with Coomassie Brilliant Blue R-350 (Amersham Biosciences) or SYPRO Ruby protein gel stain (Invitrogen). Gel images were scanned using the GS-800 Calibrated Imaging Densitometer (Bio-Rad Laboratories). Noise reduction, background subtraction, spot detection, quantification, gel-to-gel matching, and differential protein display analysis were carried out using the PDQuest software (Bio-Rad Laboratories). After normalization of the spot intensities on each gel, the spot intensities of each protein were quantified using at least three gels from the independent extractions. High reproducibility of spot intensities was obtained among the replicates. The average of spot intensities of each protein was calculated and used to compare the protein levels among the samples. Those proteins exhibiting 3-fold changes in all of replicate gels were chosen to be differentially regulated proteins. Selected spots were excised and stored at 4°C in deionized water.
Peptide Preparation and Mass Spectrometric Analysis
Protein spots excised from the Coomassie Brilliant Blue-stained gels were washed twice with HPLC grade water containing 30% acetonitrile (Wako), washed with 100% of acetonitrile, and dried in a vacuum concentrator. The dried gel pieces were absorbed with 2 μL of a 0.5 μg/μL trypsin (Promega)/50 mm ammonium bicarbonate (Shevchenko and Shevchenko, 2001) and incubated at 37°C for 16 h. The digested peptides in the gel pieces were extracted twice with 20 μL of 5% formic acid (HCOOH)/50% acetonitrile. Finally, combined extracts were dried in a Speedvac evaporator. Trypsin-digested peptides were loaded on the column (PEPMAPC18, 5 μm, 75 μm internal diameter, 15 cm; Dionex) using the CapLC system (Waters). Buffers were 0.1% HCOOH in water (A) and 0.1% HCOOH in acetonitrile (B). A linear gradient from 5% to 45% B for 25 min was applied, and peptides eluted from the column were introduced directly into a Q-TOF mass spectrometer (Waters) with a flow rate of 100 nL/min. Ionization was performed with a PicoTip nanospray source (New Objective). MS/MS spectrums were subjected to the MASCOT server against a protein database from the National Center for Biotechnology Information.
Reverse Transcription-PCR
Total RNA was extracted using EASYPrep RNA (TaKaRa) according to the manufacturer's protocol. One microgram of total RNA was reverse transcribed into first-strand cDNA with an oligo poly-T primer (21-mer) and SuperScript II reverse transcriptase (Invitrogen) in 20 μL total volume. One microliter of the synthesized first-strand cDNA was used for PCR analysis with different sets of gene-specific primers. The primer's sequences, annealing temperatures, and number of cycles are shown in Supplemental Table IV.
Supplementary Material
Acknowledgments
We thank the members of the Laboratory of Plant Molecular Genetics at Nara Institute of Science and Technology for technical assistance, comments, and participation in discussions.
This work was supported by the Research for the Future Program of the Japan Society for Promotion of Science (JSPS; grant no. JSPS–RFTF 00L01604) and the Ministry of Agriculture, Forestry, and Fisheries of Japan, Rice Genome Project.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ko Shimamoto (simamoto@bs.naist.jp).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.068395.
References
- Amora Y, Chevionb M, Levinea A (2000) Anoxia pretreatment protects soybean cells against H2O2-induced cell death: possible involvement of peroxidases and of alternative oxidase. FEBS Lett 477: 175–180 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Arimura G, Ozawa R, Nishioka T, Boland W, Koch T, Kuhnemann F, Takabayashi J (2002) Herbivore-induced volatiles induce the emission of ethylene in neighboring lima bean plants. Plant J 29: 87–98 [DOI] [PubMed] [Google Scholar]
- Bardel J, Louwagie M, Jaquinod M, Jourdain A, Luche S, Rabilloud T, Macherel D, Garin J, Bourguignon J (2002) A survey of the plant mitochondrial proteome in relation to development. Proteomics 2: 880–898 [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J (2002) RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen-deprivation tolerance. Science 296: 2026–2028 [DOI] [PubMed] [Google Scholar]
- Belenghi B, Acconcia F, Trovato M, Perazzolli M, Bocedi A, Polticelli F, Ascenzi P, Delledonne M (2003) AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur J Biochem 270: 2593–2604 [DOI] [PubMed] [Google Scholar]
- Botella MA, Xu Y, Prabha TN, Zhao Y, Narasimhan ML, Wilson KA, Nielsen SS, Bressan RA, Hasegawa PM (1996) Differential expression of soybean cysteine proteinase inhibitor genes during development and in response to wounding and methyl jasmonate. Plant Physiol 112: 1201–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92: 351–366 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaig BC, Melotto M, He SY, Howe GA (2004) Regulation of plant arginase by wounding, jasmonate, and the phytotoxin coronatine. J Biol Chem 279: 45998–46007 [DOI] [PubMed] [Google Scholar]
- Clarke A, Desikan R, Hurst RD, Hancock JT, Neill SJ (2000) NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J 24: 667–677 [DOI] [PubMed] [Google Scholar]
- Cooper B, Clarke JD, Budworth P, Kreps J, Hutchison D, Park S, Guimil S, Dunn M, Luginbuhl P, Ellero C, et al (2003) A network of rice genes associated with stress response and seed development. Proc Natl Acad Sci USA 100: 4945–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Rotari VI, Gordon A, Mailhac N, Gallois P (2004) Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and defender against apoptotic death. J Biol Chem 279: 779–787 [DOI] [PubMed] [Google Scholar]
- Desikan R, Neill SJ, Hancock JT (2000) Hydrogen peroxide-induced gene expression in Arabidopsis thaliana. Free Radic Biol Med 28: 773–778 [DOI] [PubMed] [Google Scholar]
- Dionisi H, Checa S, Krapp A, Arakaki A, Ceccarelli E, Carrillo N, Viale A (1998) Cooperation of the DnaK and GroE chaperone systems in the folding pathway of plant ferredoxin-NADP+ reductase expressed in Escherichia coli. Eur J Biochem 251: 724–728 [DOI] [PubMed] [Google Scholar]
- Dorey S, Kopp M, Geoffroy P, Fritig B, Kauffmann S (1999) Hydrogen peroxide from the oxidative burst is neither necessary nor sufficient for hypersensitive cell death induction, phenylalanine ammonia lyase stimulation, salicylic acid accumulation, or scopoletin consumption in cultured tobacco cells treated with elicitin. Plant Physiol 121: 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas B, van Doorsselaere J, Geoffrey P, Fritig B (1988) Purification of tobacco o-methyltransferases by affinity chromatography and estimation of the rate of synthesis of the enzyme during hypersensitive reaction to virus infection. Planta 176: 36–41 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420: 629–635 [DOI] [PubMed] [Google Scholar]
- Even-Chen Z, Mattoo A, Goren R (1982) Inhibition of ethylene biosynthesis by aminoethoxyvinylglycine and by polyamines shunts label from 3,4-[14C]methionine into spermidine in aged orange peel disc. Plant Physiol 69: 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AL (1999) Chaperone-mediated protein folding. Physiol Rev 79: 425–449 [DOI] [PubMed] [Google Scholar]
- Finnie C, Melchior S, Roepstorff P, Svensson B (2002) Proteome analysis of grain filling and seed maturation in barley. Plant Physiol 129: 1308–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Nishimura M (2002) Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol 43: 689–696 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002) Proteomics of Arabidopsis seed germination: a comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol 129: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco P, Nordhoff E, Kreitler T, Kloppel KD, Lehrach H, Klose J, Gobom J (2005) Proteome analysis of Arabidopsis thaliana by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Proteomics 5: 1902–1913 [DOI] [PubMed] [Google Scholar]
- Grant JJ, Loake GJ (2000) Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol 124: 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronwald JW, Plaisance KL (1998) Isolation and characterization of glutathione S-transferase isozymes from sorghum. Plant Physiol 117: 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Wang Z, Yang Z (2004) ROP/RAC GTPase: an old new master regulator for plant signaling. Curr Opin Plant Biol 7: 527–536 [DOI] [PubMed] [Google Scholar]
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ (2005) A systematic proteomic study of seed filling in soybean: establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol 137: 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CU, Lee CH, Jang KS, Choi GJ, Lim HK, Kim JC, Ahn SN, Choi JE, Cha JS, Kim HT, et al (2004) Identification of rice genes induced in a rice blast-resistant mutant. Mol Cells 17: 462–468 [PubMed] [Google Scholar]
- Hashimoto M, Kisseleva L, Sawa S, Furukawa T, Komatsu S, Koshiba T (2004) A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid-signaling pathway. Plant Cell Physiol 45: 550–559 [DOI] [PubMed] [Google Scholar]
- Imin N, Nizamidin M, Daniher D, Nolan KE, Rose RJ, Rolfe BG (2005) Proteomic analysis of somatic embryogenesis in Medicago truncatula: explant cultures grown under 6-benzylaminopurine and 1-naphthaleneacetic acid treatments. Plant Physiol 137: 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeck E, Dumas B, Geoffroy P, Favet N, Inze D, Van Montagu M, Fritig B, Legrand M (1992) Regulation of enzymes involved in lignin biosynthesis: induction of O-methyltransferase mRNAs during the hypersensitive reaction of tobacco to tobacco mosaic virus. Mol Plant Microbe Interact 5: 294–300 [DOI] [PubMed] [Google Scholar]
- Jones AM, Thomas V, Truman B, Lilley K, Mansfield J, Grant M (2004) Specific changes in the Arabidopsis proteome in response to bacterial challenge: differentiating basal and gene mediated resistance. Phytochemistry 65: 1805–1816 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K (1999) The small GTP-binding protein Rac is a regulator of cell death in plants. Proc Natl Acad Sci USA 96: 10922–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch H-H, Bartels D, Wei Y, Schnable PS, Wood AJ (2004) The ALDH gene superfamily of Arabidopsis. Trends Plant Sci 9: 371–377 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjolander K, Gruissem W, Baginsky S (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14: 354–362 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Dietrich RA, Martin AC, Last RL, Dangl JL (1999) LSD1 regulates salycilic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol Plant Microbe Interact 12: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kobayashi I, Funaki Y, Fujimoto S, Takemoto T, Kunoh H (1997) Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non-host resistance in barley coleoptile cells. Plant J 11: 525–537 [Google Scholar]
- Koga J, Yamauchi T, Shimura M, Ogawa N, Oshima K, Umemura K, Kikuchi M, Ogasawara N (1998) Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J Biol Chem 273: 31985–31991 [DOI] [PubMed] [Google Scholar]
- Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, Haynes PA, Hays L, Schieltz D, Ulaszek R, Wei J, et al (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci USA 99: 11969–11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu S, Tanaka N (2005) Rice proteome analysis: a step toward functional analysis of the rice genome. Proteomics 5: 938–949 [DOI] [PubMed] [Google Scholar]
- Kruft V, Eubel H, Jansch L, Werhahn W, Braun H-P (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol 127: 1694–1710 [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer M, Hartl F (1992) Successive action of DnaK, DnaJ, and GroEL along the pathway of chaperone-mediated protein folding. Nature 356: 683–689 [DOI] [PubMed] [Google Scholar]
- Li Z, Onodera H, Ugaki M, Tanaka H, Komatsu S (2003) Characterization of calreticulin as a phosphoprotein interacting with cold-induced protein kinase in rice. Biol Pharm Bull 26: 256–261 [DOI] [PubMed] [Google Scholar]
- Lieberherr D, Thao NP, Nakashima A, Umemura K, Kawasaki T, Shimamoto K (2005) A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice. Plant Physiol 138: 1644–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liska AJ, Shevchenko A, Pick U, Katz A (2004) Enhanced photosynthesis and redox energy production contribute to salinity tolerance in Dunaliella, as revealed by homology-based proteomics. Plant Physiol 136: 2806–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonosky PM, Zhang X, Honavar VG, Dobbs DL, Fu A, Rodermel SR (2004) A proteomic analysis of maize chloroplast biogenesis. Plant Physiol 134: 560–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltman DJ, Simon WJ, Wheeler CH, Dunn MJ, Wait R, Slabas AR (2002) Proteomic analysis of the endoplasmic reticulum from developing and germinating seed of castor (Ricinus communis). Electrophoresis 23: 626–639 [DOI] [PubMed] [Google Scholar]
- Miki D, Itoh R, Shimamoto K (2005) RNA silencing of single and multiple members of a gene family in rice. Plant Physiol 138: 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giege P, Leaver CJ (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Moeder W, Yoshioka K, Klessig DF (2005) Involvement of the small GTPase Rac in the defense responses of tobacco to pathogens. Mol Plant Microbe Interact 18: 116–124 [DOI] [PubMed] [Google Scholar]
- Nakazono M, Tsuji H, Li Y, Saisho D, Arimura S, Tsutsumi N, Hirai A (2000) Expression of a gene encoding mitochondrial aldehyde dehydrogenase in rice increases under submerged conditions. Plant Physiol 124: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuhse TS, Stensballe A, Jensen ON, Peck SC (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalski KS, Schultheiss H, Kogel KH, Huckelhoven R (2005) The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J 41: 291–303 [DOI] [PubMed] [Google Scholar]
- Park BJ, Lee DG, Yu JR, Jung SK, Choi K, Lee J, Kim YS, Lee JI, Kwon JY, Singson A, et al (2001) Calreticulin, a calcium-binding molecular chaperone, is required for stress response and fertility in Caenorhabditis elegans. Mol Biol Cell 12: 2835–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Choi HJ, Lee S, Lee T, Yang Z, Lee Y (2000) Rac-related GTP-binding protein in elicitor-induced reactive oxygen generation by suspension-cultured soybean cells. Plant Physiol 124: 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck SC, Nuhse TS, Hess D, Iglesias A, Meins F, Boller T (2001) Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell 13: 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Geoffroy P, Fritig B, Legrand M (1993) Molecular cloning and expression of a new class of ortho-diphenol-O-methyltransferases induced in tobacco (Nicotiana tabacum L.) leaves by infection or elicitor treatment. Plant Physiol 103: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Rohfritsch O, Fritig B, Legrand M (1994) Phenylalanine ammonia-lyase in tobacco (molecular cloning and gene expression during the hypersensitive reaction to tobacco mosaic virus and the response to a fungal elicitor). Plant Physiol 106: 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J-B, Friso G, Kalume DE, Roepstorff P, Nilsson F, Adamska I, van Wijk KJ (2000) Proteomics of the chloroplast: systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12: 319–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Potikha TS, Collins CC, Johnson DI, Delmer DP, Levine A (1999) The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol 119: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakwal R, Komatsu S (2000) Role of jasmonate in the rice (Oryza sativa L.) self-defense mechanism using proteome analysis. Electrophoresis 21: 2492–2500 [DOI] [PubMed] [Google Scholar]
- Rao MV, Lee H-i, Creelman RA, Mullet JE, Davis KR (2000) Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate DN, Greenberg JT (2001) The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J 27: 203–211 [DOI] [PubMed] [Google Scholar]
- Reuber TL, Plotnikova JM, Dewdney J, Rogers EE, Wood W, Ausubel FM (1998) Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis-enhanced disease susceptibility mutants. Plant J 16: 473–485 [DOI] [PubMed] [Google Scholar]
- Rose JK, Bashir S, Giovannoni JJ, Jahn MM, Saravanan RS (2004) Tackling the plant proteome: practical approaches, hurdles, and experimental tools. Plant J 39: 715–733 [DOI] [PubMed] [Google Scholar]
- Schiltz S, Gallardo K, Huart M, Negroni L, Sommerer N, Burstin J (2004) Proteome reference maps of vegetative tissues in pea: an investigation of nitrogen mobilization from leaves during seed filling. Plant Physiol 135: 2241–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Shevchenko A (2001) Evaluation of the efficiency of in-gel digestion of proteins by peptide isotopic labeling and MALDI mass spectrometry. Anal Biochem 296: 279–283 [DOI] [PubMed] [Google Scholar]
- Simpson PB, Mehotra S, Lange GD, Russell JT (1997) High-density distribution of endoplasmic reticulum proteins and mitochondria at specialized Ca2+ release sites in oligodendrocyte processes. J Biol Chem 272: 22654–22661 [DOI] [PubMed] [Google Scholar]
- Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A (1999) The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suharsono U, Fujisawa Y, Kawasaki T, Iwasaki Y, Satoh H, Shimamoto K (2002) The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 99: 13307–13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Bartels D, Kirch H (2003) Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J 35: 452–464 [DOI] [PubMed] [Google Scholar]
- Takahashi A, Kawasaki T, Wong H, Suharsono U, Hirano H, Shimamoto K (2003) Hyperphosphorylation of a mitochondrial protein, prohibitin, is induced by calyculin A in a rice lesion-mimic mutant cdr1. Plant Physiol 132: 1861–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunezuka H, Fujiwara M, Kawasaki T, Shimamoto K (2005) Proteome analysis of programmed cell death and defense signaling using the rice lesion mimic mutant cdr2. Mol Plant Microbe Interact 18: 52–59 [DOI] [PubMed] [Google Scholar]
- Umemura K, Ogawa N, Koga J, Iwata M, Usami H (2002) Elicitor activity of cerebroside, a sphingolipid elicitor, in cell suspension cultures of rice. Plant Cell Physiol 43: 778–784 [DOI] [PubMed] [Google Scholar]
- Valster AH, Hepler PK, Chernoff J (2000) Plant GTPases: the Rhos in bloom. Trends Cell Biol 10: 141–146 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ (2001) Challenges and prospects of plant proteomics. Plant Physiol 126: 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KA, McManus MT, Gordon ME, Jordan TW (2002) The proteomics of senescence in leaves of white clover, Trifolium repens (L.). Proteomics 2: 1114–1122 [DOI] [PubMed] [Google Scholar]
- Wong HL, Sakamoto T, Kawasaki T, Umemura K, Shimamoto K (2004) Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol 135: 1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (2002) Small GTPases: versatile signaling switches in plants. Plant Cell (Suppl) 14: S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.