Abstract
Root elongation and root hair formation are important in nutrient absorption. We found that two Arabidopsis (Arabidopsis thaliana) phospholipase Ds (PLDs), PLDζ1 and PLDζ2, were involved in root elongation during phosphate limitation. PLDζ1 and PLDζ2 are structurally different from the majority of plant PLDs by having phox and pleckstrin homology domains. Both PLDζs were expressed more in roots than in other tissues. It was reported previously that inducible suppression or inducible overexpression of PLDζ1 affected root hair patterning. However, gene knockouts of PLDζ1, PLDζ2, or the double knockout of PLDζ1 and PLDζ2 showed no effect on root hair formation. The expression of PLDζs increased in response to phosphate limitation. The elongation of primary roots in PLDζ1 and PLDζ2 double knockout mutants was slower than that of wild type and single knockout mutants. The loss of PLDζ2, but not PLDζ1, led to a decreased accumulation of phosphatidic acid in roots under phosphate-limited conditions. These results indicate that PLDζ1 and PLDζ2 play a role in regulating root development in response to nutrient limitation.
Plants encounter heterogeneous, constantly fluctuating environments and need many essential nutrients for survival and growth (Schachtman et al., 1998). Of them, phosphate plays essential roles in metabolic and regulatory reactions. Lack of phosphate in soil has profound effects on plant growth and development (Bieleski and Ferguson, 1983); for example, phosphate deprivation leads to root growth and architecture modifications that enable interactions with an increased volume of soil (Williamson et al., 2001; Ma et al., 2003).
Changes in lipid metabolism are involved in response to phosphate limitation. Upon phosphate starvation, plants increase levels of digalactosyldiacylglycerol in roots; this lipid class is believed to functionally replace acidic phospholipids (Härtel et al., 2000). A nonspecific phospholipase C, which hydrolyzes common membrane phospholipids, such as phosphatidylcholine to diacylglycerol, has been shown to increase in expression and activity during phosphate deprivation (Nakamura et al., 2005). This phospholipase C is hypothesized to provide diacylglycerol as a substrate for the synthesis for digalactosyldiacylglycerol (Nakamura et al., 2005).
Phospholipase D (PLD), which cleaves phospholipids to produce phosphatidic acid (PA) and a free head group such as choline, has been implicated in root hair growth and patterning (Ohashi et al., 2003). Ohashi and coworkers also imply that PLD and its product, PA, play a role in root elongation (Ohashi et al., 2003). In this work, we investigate the role of PLD in plant response to phosphate limitation. The PLD gene family has 12 members in Arabidopsis (Arabidopsis thaliana), designated as PLDα1, PLDα2, PLDα3, PLDβ1, PLDβ2, PLDγ1, PLDγ2, PLDγ3, PLDδ, PLDɛ, PLDζ1, and PLDζ2 (Zhang et al., 2005). PLD previously has been proposed to play a pivotal role in many cellular processes, including signal transduction, membrane trafficking, cytoskeletal rearrangements, and membrane degradation (Wang, 2002). PLDα1 and PLDδ were demonstrated to function in freezing and drought tolerance (Sang et al., 2001; Welti et al., 2002; Li et al., 2004; Zhang et al., 2004). PLDζ1 and PLDζ2 are distinctively different from other PLDs; they have phox homology (PX) and pleckstrin homology (PH) domains that are also found in mammalian PLDs (Qin and Wang, 2002). PLDζ1 uses phosphatidylcholine selectively as a substrate (Qin and Wang, 2002), and PLDζ1 gene function is implicated in mediating initiation and maintenance of root hairs (Ohashi et al., 2003). On the other hand, there is no detailed understanding of the function of PLDζ1 in plant growth and development, particularly under stress conditions, nor have the functions of PLDζ2 been identified. To understand the in planta roles of these PLD gene products, we isolated mutants defective in the expression of PLDζ1 and PLDζ2, as well as double mutants lacking both PLDs. Analyses of the mutants revealed that PLDζ1 and PLDζ2 play a role in primary root elongation under low-phosphate conditions.
RESULTS
PLDζ1 and PLDζ2 Are Expressed Highly in Roots
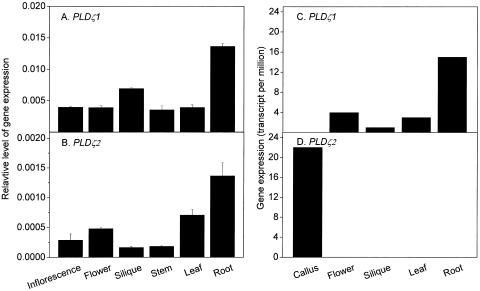
The steady-state levels of PLDζ1 and PLDζ2 transcripts in various organs of 6-week-old soil-grown Arabidopsis plants were determined by real-time PCR (Fig. 1, A and B). Both PLDζ1 and PLDζ2 transcripts were detectable in inflorescences, flowers, siliques, stems, leaves, and roots, but the expression of PLDζ1 was about 10-fold higher than that of PLDζ2. Both genes were expressed most highly in roots, but relative levels of PLDζ1 and PLDζ2 transcripts varied in siliques, flowers, stems, and leaves. Arabidopsis massively parallel signature sequencing (MPSS) data for PLDζ expression was obtained from http://mpss.udel.edu/at (Meyers et al., 2004). For PLDζ1, the MPSS expression profile was in general agreement with the real-time PCR data; PLDζ1 expression level in roots was 5 times higher than that in leaves (Fig. 1C). PLDζ2 was undetectable in normal organs, such as flowers, siliques, leaves, and roots, again consistent with the PCR data, indicating that PLDζ2 expression was much lower than that of PLDζ1. Interestingly, the PLDζ2 transcript accumulated to a high level in callus, but expression of PLDζ1 was not detected in this undifferentiated tissue. Data from both the quantitative PCR and MPSS measurements indicate that the expression of PLDζ1 and PLDζ2 is differentially regulated.
Figure 1.
Expression levels of PLDζ1 and PLDζ2 in Arabidopsis tissues. Total RNA from tissues of 6-week-old, soil-grown plants were used for real-time PCR. A and B, PCR quantification of PLDζ1 and PLDζ2 transcripts, respectively. Transcript levels of each gene were expressed in relation to the UBQ10 gene. Experiments for A and B were repeated with similar results. The bar represents se (n = 3). C and D, Expression levels of PLDζ1 and PLDζ2 as detected by MPSS. The MPSS data were obtained from the Arabidopsis MPSS Web site (http://mpss.udel.edu/at; Meyers et al., 2004).
Mutants Ablate the Expression of One or Both PLDζs
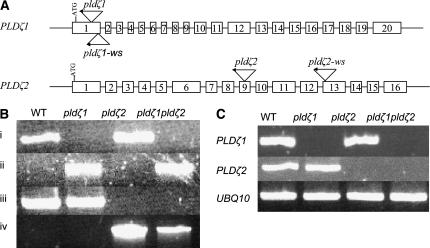
To determine the physiological function of PLDζ1 and PLDζ2 in Arabidopsis, we isolated two sets of T-DNA insertional mutants for PLDζ1 and PLDζ2: pldζ1 and pldζ2 in the Columbia (Col-0) ecotype, and pldζ1-ws and pldζ2-ws in the Wassilewskija (Ws) ecotype. pldζ1 has the T-DNA inserted at the first exon and 231 nucleotides downstream of the ATG codon of PLDζ1, and pldζ1-ws has an insertion near the end of the first exon (Fig. 2A). The T-DNA insertions in pldζ2 and pldζ2-ws are at the ninth exon (2,416 nucleotides downstream of the ATG codon) and the thirteenth exon, respectively (Fig. 2A). In each case, the presence and location of the T-DNA insertion were verified by junction PCR and sequencing, and homozygous lines were identified by PCR (Fig. 2B). In each line, the F2 progeny segregated 3:1 for resistance to kanamycin, indicating a single insertion in the genome. The presence of a single insertion in each genome was further demonstrated by DNA gel-blot analyses of pldζ1-ws and pldζ2-ws (data not shown).
Figure 2.
T-DNA insertion positions and loss of gene expression in PLDζ single and double mutants. A, PLDζ1 and PLDζ2 genes with the sites of the T-DNA insertions in Col-0 ecotype, pldζ1 and pldζ2, and Ws ecotype, pldζ1-ws and pldζ2-ws, indicated. Exons are shown as white boxes, not drawn to scale. B, PCR confirmation of T-DNA insertions in PLDζ genes and of the homozygosity of the mutations. Genomic DNA was isolated from soil-grown plant leaves. i, Products produced using two PLDζ1-specific primers, 1a and 1b. The lack of the PLDζ1 DNA band in pldζ1 and pldζ1pldζ2 demonstrates that the mutant is homozygous; the presence of T-DNA made the fragment too large to be amplified under these PCR conditions. The presence of the T-DNA insert is confirmed using a left-border (LB) primer and a PLDζ1 primer (1a) as shown in ii. iii and iv, Primer combinations 2a and 2b, and 2a and LB, respectively, are used to confirm the T-DNA insertion in pldζ2 and pldζ1pldζ2. C, Verification of the loss of PLDζ transcripts in pldζ1, pldζ2, and pldζ1pldζ2 by RT-PCR. Total RNA was isolated from leaves of soil-grown plants. RT-PCR analyses were carried out with primers specific for PLDζ1 and PLDζ2 genes. Gene-specific primers, 1c and 1d, were used to detect PLDζ1 mRNA, and primers 2c and 2d were used to detect PLDζ2 mRNA. PLDζ1 and PLDζ2 gene-specific primers failed to amplify a band in pldζ1 and pldζ2 mutants. The sequence of the gene-specific primers, 1a, 1b, 1c, 1d, 2a, 2b, 2c, and 2d, is listed in Table I.
Double mutants in both Col-0 and Ws ecotypes were generated by crossing single PLDζ1 and PLDζ2 knockout mutants. Homozygous double mutants, pldζ1pldζ2, were identified in the F3 generation. Loss of PLDζ1 and PLDζ2 gene expression in each single or double mutant was verified by reverse transcription (RT)-PCR (Fig. 2C). Arabidopsis plants deficient in PLDζ1, PLDζ2, or both PLDζs grew and developed normally under regular laboratory growth conditions. These mutants were also compared with wild-type plants in response to hormones abscisic acid and auxin indole-3-acetic acid, and to salt (NaCl) and osmotic (mannitol) stresses, and no apparent difference in plant growth and development was observed between these genotypes.
Loss of PLDζ1 and PLDζ2 Reduces Primary Root Elongation, But Promotes Lateral Root Elongation under Low-Phosphate Conditions
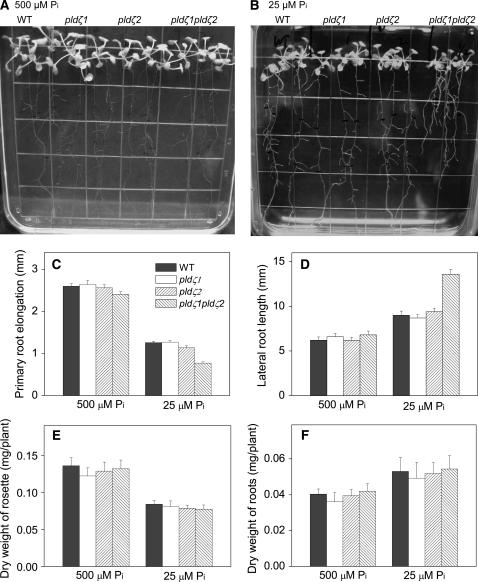
Because PLDζ1 and PA have been implicated as having roles in root function (Ohashi et al., 2003), we investigated the potential effect of PLDζ mutations on roots. Seeds were germinated on Murashige and Skoog agar media, and 3-d-old seedlings were transferred to fresh agar media that either contained the standard amount of phosphate (500 μm) or low phosphate (25 μm). Root growth patterns and elongation were examined. In 500 μm phosphate, there was no difference in primary root elongation among wild-type, pldζ1, pldζ2, and pldζ1pldζ2 plants (Fig. 3A). With limited phosphate, wild type and the single mutants pldζ1 and pldζ2 still displayed no difference in primary root growth (Fig. 3B). However, primary root elongation of the double mutant pldζ1pldζ2 was retarded; the primary root elongation between the fourth and seventh days after transfer was reduced 33% compared to wild type (Fig. 3, B and C). In contrast, lateral roots grew longer in the double mutant than in wild-type plants in low phosphate (Fig. 3, B and D). There was no significant difference in lateral root length among wild type and single mutants at either low or standard phosphate levels (Fig. 3, B and D), and the numbers of lateral roots were also similar among wild type, single, and double mutants (data not shown). The phenotypes of single and double pldζ mutants in the Col-0 and Ws backgrounds were the same, so only data obtained from pldζ1 and pldζ2 (Col-0 background) are presented hereafter. Taken together, the results suggest that PLDζ1 and PLDζ2 promote primary root growth but inhibit lateral root elongation when plants are challenged with low-phosphate levels. The fact that the root alteration occurs only in double, but not in single, mutants indicates that the PLDζs have overlapping functions in primary root elongation.
Figure 3.
Effect of PLDζ mutants on root growth under two phosphate conditions. A and B, Seedlings of wild type, pldζ1, pldζ2, and pldζ1pldζ2 grown for 7 d in 500 and 25 μm phosphate agar media, respectively. Seedlings were transferred to the indicated media 3 d after germination. C, Elongation of primary roots from the fourth to the seventh day (mm/3 d) after transfer onto 500 or 25 μm phosphate agar plates. D, Elongation of lateral roots from the first to the seventh day (mm/7 d) after transfer. Data for primary roots were obtained from 20 seedlings in five agar culture plates. Data for lateral roots were obtained from 20 seedlings, four lateral roots per seedling for each genotype. Values are means ± se. E and F, Dry weights of rosettes and roots, respectively, of wild type, pldζ1, pldζ2, and pldζ1pldζ2 under the two phosphate conditions. Seedlings were grown on 500 μm phosphate agar plates for 3 d and then transferred to agar plates containing 500 and 25 μm phosphate for an additional 7 d before harvesting for dry weight measurement. Values are means ± se (n = 5); each was performed on 20 seedlings of each genotype per treatment.
In addition to alterations in root elongation, a decrease in the dry weight of shoots has been observed in response to phosphate starvation (Williamson et al., 2001). Plants at lower phosphate showed lower rosette growth, but higher root growth, than those grown on standard Murashige and Skoog medium (Fig. 3, E and F). The root-to-rosette growth ratios, as measured by the dry weights, were greater in all genotypes in 25 μm than in 500 μm phosphate (Fig. 3, E and F). However, there was no difference in total rosette and root weights among wild type, single mutants, and double mutants, indicating that the total root volume for the nutrient absorption is not impaired by the loss of PLDζs. The pldζ1pldζ2 plants had shorter primary roots, but longer lateral roots, in 25 μm phosphate.
Ablation of PLDζ1 and PLDζ2 Does Not Affect Root Hair Patterning
Elongation of root hairs is a response to a low-phosphate environment (Williamson et al., 2001), and PLDζ1 was implicated in root hair initiation and patterning (Ohashi et al., 2003). In Arabidopsis, the root epidermis is composed of two types of cell files, hair cell files and hairless cell files, of which only hair cell files produce root hairs (Dolan et al., 1993). Mutation of GLABRA2 (GL2) and TRANSPARENT TESTA GLABRA1 (TTG1) caused hairless cell files in the root epidermis to produce root hairs (Cristina et al., 1996). TTG1 was suggested to regulate GL2 positively, and GL2 regulates PLDζ1 negatively by binding to its promoter region (Ohashi et al., 2003). When expression of PLDζ1 was suppressed by an inducible promoter, root hairs developed from random positions and appeared to be expanded and globular (Ohashi et al., 2003). On the other hand, when PLDζ1 expression was increased, root hairs were initiated from both types of cell files, and the hairs were frequently branched and swollen (Ohashi et al., 2003). These results led to the hypothesis that PLDζ1 is involved in both initiation and maintenance of root hair development.
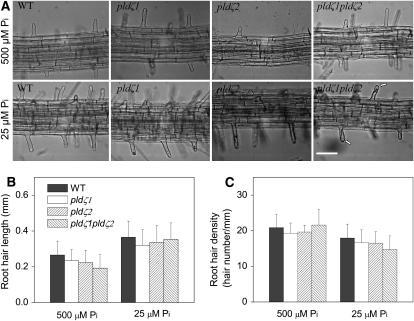
To investigate whether the loss of PLDζs affected root hairs, 3-d-old seedlings in standard agar plates were transferred to fresh standard and low-phosphate media. The root hair pattern, root hair length, and root hair density were recorded (Fig. 4). Neither single mutants pldζ1 and pldζ2 nor double mutant pldζ1pldζ2 exhibited root hair patterning (Fig. 4A) or root hair growth that differed from wild type under standard phosphate conditions (Fig. 4, B and C). The pattern of root hair initiation and growth was examined from seedling to flowering stages, and no apparent difference was observed between wild type and PLDζ mutants under normal growth conditions. At the low-phosphate level, some root hairs of the pldζ1pldζ2 double mutants appeared slightly globular at the tip (Fig. 4A), but the overall root hair pattern in low-phosphate conditions was not affected by the loss of the PLDζs. The same subtle phenotype was observed in PLDζ single and double mutants in the Ws background (data not shown).
Figure 4.
Morphology, length, and density of root hairs from wild type, pldζ1, pldζ2, and pldζ1pldζ2 under two phosphate conditions. A, Top images are root hairs of wild type, pldζ1, pldζ2, and pldζ1pldζ2, respectively, under standard phosphate (500 μm) conditions. Bottom images are root hairs of wild type, pldζ1, pldζ2, and pldζ1pldζ2, respectively, under low-phosphate (25 μm) conditions. Arrows indicate slightly globular root hair tips. Scale bars represent 100 μm. B and C, Root hair length and root hair density of the PLDζ mutant lines were similar to wild type. Plants were grown for 3 d on agar plates containing 500 μm phosphate and then transferred to agar plates containing either 500 or 25 μm phosphate. Twenty-four hours after the transfer, images of the root hairs were captured. Root hairs growing in the region 3 to 4 mm above the root tips were measured (B) and root hair numbers within the region 3 to 4 mm from the root tips were counted (C). Data are based on the measurements of root hairs on 20 seedlings; values are means ± se.
Transcripts of PLDζs Increase during Phosphate Deprivation
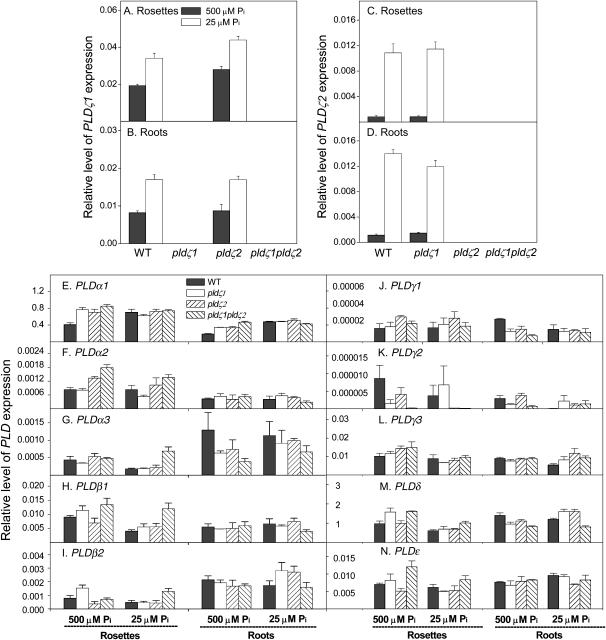
One potential explanation for the discrepancy between the lack of changes in root hair patterning that we observed in the PLDζ knockout lines and the much more dramatic phenotype affecting root hair patterning, observed by Ohashi et al. (2003) when expression of PLDζ1 was suppressed by an inducible promoter, is that other PLDs in Arabidopsis may compensate when PLDζ1, PLDζ2, or both are permanently lost. Therefore, expression of other PLD genes was quantified with real-time PCR in wild type and single and double mutants to determine whether there was any compensation at the transcriptional level. First, we examined whether loss of one PLDζ affected the expression of the other PLDζ or other PLDs. Loss of PLDζ2 did not affect the expression of PLDζ1 in roots (Fig. 5B), but the PLDζ1 transcript level was about 20% higher in pldζ2 than in wild-type rosettes under either standard or low-phosphate conditions (Fig. 5A). Loss of PLDζ1 did not affect the expression of PLDζ2 in roots and rosettes at either phosphate level (Fig. 5, C and D). The level of PLDζ1 expression was higher than that of PLDζ2 in young seedlings on agar plates, and this result is consistent with that of plants grown in soil (Figs. 1 and 5). However, PLDζ1 expression is highest in roots of 6-week-old, soil-grown plants (Fig. 1), but this is not the case in 10-d-old, agar plate-grown seedlings (Fig. 5). The difference in growth conditions and developmental stage might cause this discrepancy. In addition, rosettes used in Figure 5 contained all parts above roots, in particular the apical meristematic tissues that were not included in the soil-grown plants (Fig. 1). Thus, the difference in tissue types might also account for the discrepancy.
Figure 5.
Effect of phosphate levels on the expression of 12 PLD genes in Arabidopsis. Primers specific to each PLD gene (Table II) were used for real-time PCR to determine the expression levels in wild type, pldζ1, pldζ2, and pldζ1pldζ2 under the two phosphate conditions. Three-day-old seedlings were transferred to fresh 500 or 25 μm phosphate agar media, and, after 7 d, total RNA was isolated from rosettes and roots. The levels of expression are expressed relative to the expression of UBQ10. Values are the mean of three replicates ± se from one of the two independent experiments that gave similar results.
Next, the transcript levels of the other 10 PLD genes were monitored in pldζ1, pldζ2, and pldζ1pldζ2 mutants to determine whether their expression in roots and rosettes was significantly altered relative to wild-type plants. At the standard phosphate level, the transcript level of PLDα1 was slightly higher in the single and double knockout mutants than in wild type in both roots and rosettes (Fig. 5E). The PLDα2 transcript level in double mutants was higher than in wild type in rosettes, but not in roots (Fig. 5F). The transcript levels of the rest, PLDα3, PLDβ1, PLDβ2, PLDγ1, PLDγ2, PLDγ3, PLDδ, and PLDɛ, were not significantly different from wild-type levels or the difference was very subtle (Fig. 5, G–N). With low phosphate, the PLDα1 transcript levels of wild type and mutants were similar in rosettes and roots, and PLDα2, PLDα3, PLDβ1, and PLDβ2 expression levels were higher than wild-type levels in rosettes, but not in roots (Fig. 5, F–I). However, the other five PLDs, γ1, γ2, γ3, δ, and ɛ, were expressed in both rosettes and roots, at levels similar to wild type in low-phosphate medium (Fig. 5, J–N).
When the expression of all 12 PLDs was compared under standard and phosphate-limited conditions, the expression of PLDζ2 showed the most prominent increase in low phosphate. The levels of PLDζ2 transcript in roots and rosettes were more than 10-fold higher under phosphate-limited than under standard conditions (Fig. 5, C and D). The expression of PLDζ1 increased about 2-fold (Fig. 5, A and B), as did PLDβ2, but the expression of other PLDs exhibited no major changes in response to low-phosphate conditions (Fig. 5, E–N). The PLD expression patterns are consistent with the observation that PLDζs are involved in root elongation during phosphate limitation. The results also suggest that PLDζ2 plays a more important role than PLDζ1 in the ability of a plant to cope with phosphate deficiency.
PLDζ2 Is Involved in PA Production in Roots under Phosphate Limitation
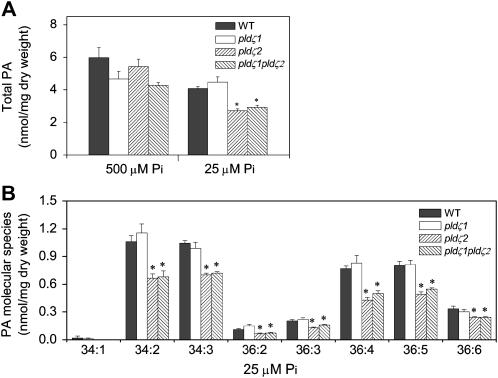
To investigate the metabolic consequences resulting from the loss of the single or double PLDζs, we analyzed the level and composition of PA, the direct lipid product of PLD activity. Under standard phosphate conditions, there was no significant difference in PA levels in roots in single pldζ1 or pldζ2 mutants, but the PA level tended to be lower in the pldζ1pldζ2 double mutant than in roots of wild-type plants (Fig. 6A). In low-phosphate medium, however, pldζ2 had only 70% as much PA as in wild-type plants, but pldζ1 had a similar level of PA compared to wild type (Fig. 6A). The PA level of pldζ1pldζ2 was the same as that of pldζ2, implying that the decrease in the double mutant resulted from the loss of pldζ2. This difference was also observed when the data were expressed as mol% of total phospholipids. This was because the absolute amounts of total phospholipids were not significantly different between wild type and mutants in roots under growth conditions, and also because the amount of PA accounted for only a small fraction of total phospholipids. Thus, the decrease in PA in the double mutant was not due to an alteration in content of other phospholipids. These results show that PLDζ2, but not PLDζ1, plays a major role in PA production during phosphate deprivation, and that PLDζ1 does not compensate for PLDζ2 function in pldζ2. This metabolic activity of PLDζ2 is consistent with gene expression data showing that transcript levels of PLDζ2 were induced most by phosphate deficiency (Fig. 5, C and D). All PA molecular species were lower in roots of pldζ2 and the double mutant than in wild type or pldζ1 (Fig. 6B), suggesting that PLDζ2 produces a variety of PA molecular species.
Figure 6.
Total PA and PA molecular species in wild type, pldζ1, pldζ2, and pldζ1pldζ2. PA was analyzed in roots of wild type and PLDζ mutant lines. The content of total PA and PA molecular species was determined by electrospray ionization/tandem mass spectrometry. A, Total PA content in roots in standard and low-phosphate conditions is shown. B, PA molecular species in roots of wild type, pldζ1, pldζ2, and pldζ1pldζ2 under low-phosphate conditions. Values are means ± se (n = 5). The stars indicate that mutant and wild-type values are significantly different (P < 0.05).
DISCUSSION
Phosphorus is an essential element for plant growth, development, and reproduction. It plays an important role not only in regulation of various enzymes but also as a constituent of components such as membrane phospholipids and nucleic acids (Schachtman et al., 1998). Plants have developed distinct systems to cope with phosphate deficiency. When a plant grows under low-phosphate conditions, highly integrated systems in plants are activated (Lynch, 1995; Vance et al., 2003) to increase the assimilation of free inorganic phosphate. One alteration is a morphological modification of root architecture upon phosphate limitation, which presumably facilitates phosphate uptake by enlargement of absorptive root surface areas (Lynch, 1995; Vance et al., 2003). We show here that PLDζ1 and PLDζ2 are involved in the root growth modification and that these two PLD gene products have both overlapping and unique functions.
These data show that the loss of PLDζ1 and PLDζ2 decreases primary root growth and that loss of PLDζ2 results in lower PA content under a low-phosphate condition. Root elongation is regulated by many factors. A recent study indicates that PA interacts with AtPDK1, stimulates a protein kinase cascade, and promotes root apical growth and initiation (Anthony et al., 2004). However, the in vivo source of PA is unclear. This is because, in addition to PLD, signaling PA can be generated from other routes, such as diacylglycerol kinase-mediated phosphorylation of diacylglycerol. The decrease in PA formation in the PLDζ2 mutant raises the possibility that PA generated by PLDζs may promote root elongation and that AtPDK1 might be the target for PA under low-phosphate conditions. However, there is a discrepancy between the metabolic alteration and morphological phenotype: Decreased apical growth in primary roots occurred only in plants lacking both PLDζs, indicating that either PLDζ1 or PLDζ2 can function to promote apical growth. However, PLDζ2 is sufficient to produce PA in the absence of PLDζ1, while PLDζ1 does not seem to produce PA in the absence of PLDζ2. The production of PA by the PLDζ2 gene product occurs in concert with the induction of PLDζ2 during phosphate deprivation. These results suggest that PLDζ2-derived PA is not the only mediator of root elongation and that PLDζ1 plays a significant role, uncorrelated with the production of measurable levels of PA.
Interestingly, the consequence of loss of PLDζs is different in the elongation of primary roots from that of lateral roots. Recently, it was reported that phosphate transport double mutants, plt1;1 and plt1;4, exhibited lower plant growth, including that of primary roots (Shin et al., 2004). However, the plt1;1plt1;4 double mutant showed faster lateral root growth than wild type when high levels of phosphate were supplied to phosphate-starved roots (Shin et al., 2004). These results support the notion that the response to low phosphate could be different in primary as compared to lateral roots. Our data indicate that PLDζs are involved in these different responses to low phosphate between primary roots and lateral roots; they promote primary root elongation but inhibit lateral root elongation under low-phosphate conditions (Fig. 3, B and D).
The lack of dramatic differences in root hair growth and patterning in the single and double PLDζ mutants is perplexing because a previous study showed that PLDζ1 was involved in root hair initiation and patterning. GL2 is a key component of a regulatory circuit mediating root hair patterning in Arabidopsis (Masucci et al., 1996). PLDζ1 is a direct target of GL2 as GL2 binds to its promoter region of PLDζ1 (Ohashi et al., 2003). Inducible expression of PLDζ1 promoted ectopic root hair initiation, whereas inducible suppression inhibited root hair initiation (Ohashi et al., 2003). These results suggest that GL2 regulates root hair development through modulation of PLDζ1 and lead to the prediction that loss of PLDζ1 would affect root hair patterning. However, our data show that knockout of PLDζ1 and PLDζ2 in both the Col-0 and Ws backgrounds had no effect on root hair patterning and only a very subtle phenotype of slightly globular root tips under low-phosphate conditions. One potential cause of the apparent discrepancy is the difference in experimental approaches: The previous studies were done with an inducible PLDζ1 suppression/overexpression system, not gene knockouts. It might be possible that the antisense suppression was not specific to PLDζ1, and other PLDs might also be suppressed. Another potential explanation is that the threshold of PLDζ1 expression and protein levels might be important in root hair initiation and growth because the antisense suppression might not completely ablate the proteins and expression of PLDζ1, and the alterations on root hairs might result from the suboptimal PLDζ1 levels in the cell. Taken together, our results indicate that more than one PLD is involved in root hair initiation and patterning, that PLDζ1 and PLDζ2 may have overlapping functions, and that PLDζs promote primary root growth when plants are challenged with low-phosphate levels.
MATERIALS AND METHODS
Mutant Isolation
Two Arabidopsis (Arabidopsis thaliana) ecotypes, Col-0 and Ws, were employed. Potential pldζ1 (Salk_083090) and pldζ2 (Salk_094369) mutant lines, which are in Col-0, were identified from the Salk T-DNA lines (Alonso et al., 2003) through the analysis of the SiGnAL database (http://www.signal.salk.edu/cgi-bin/tdnaexpress), and seeds were obtained from the Ohio State University Arabidopsis Biological Resource Center (ABRC). Homozygous mutant plants were isolated using the T-DNA left-border primer and gene-specific primers: Primers 1a and 1b were used to identify pldζ1 and primers 2a and 2b to identify pldζ2 (Table I). pldζ1-ws and pldζ2-ws insertion mutants are in the Ws background, and mutants were identified by screening pools of T-DNA insertion lines according to the protocol of the Arabidopsis Gene Knockout Research Facility at the University of Wisconsin (Sussman et al., 2000). PCR was carried out on pooled genomic DNA with a T-DNA left-border primer, JL202, and gene-specific primers, 1e and 1f for the PLDζ1 gene and 2e and 2f for the PLDζ2 gene. Pools were deconvoluted and individual lines identified by PCR. The number of single T-DNA inserts in pldζ1-ws and pldζ2-ws knockout lines was assessed by Southern-blot analysis.
Table I.
Primers used for identification and verification of T-DNA insertions in Figure 2
| Gene | Primer Names and Sequences |
|---|---|
| PLDζ1 | 1a 5′-CGCTACTTTCAGATGCAGCCTGAGCAATT-3′ |
| 1b 5′-ACTGATATGGACTGCTGTCTTCCCA-3′ | |
| 1c 5′-CGGCGATATTAGCCCTGTACTCTT-3′ | |
| 1d 5′-GCCATTCTTTAACCTGTTCCTGCT-3′ | |
| 1e 5′-CAAGATGAAGGCTGAGAAGGCTAGAGAGA-3′ | |
| 1f 5′-ATCAGAGAAATGGCATCTGAGCAGTTGAT-3′ | |
| PLDζ2 | 2a 5′-CAGGTGAAGGAACACAAC-3′ |
| 2b 5′-CAGACCATTGGCTAACAC-3′ | |
| 2c 5′-AGACAGGAGAAAGTACCCGCGAAT-3′ | |
| 2d 5′-TGTGGTGGTGAGGCATCAACAATG-3′ | |
| 2e 5′-GGATTTTTGATATGCAAGGATCAGTTGTG-3′ | |
| 2f 5′-GTCTCGTGAACCTAGTAAGCTCCTATCGT-3′ | |
| T-DNA left border
|
JL202 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′ |
| LB 5′-GCGTGGACCGCTTGCTGCAACT-3′ |
To generate pldζ1pldζ2 double mutants in both ecotypes, homozygous single mutants in PLDζ1 were crossed with mutants in PLDζ2. F1 plants were self-pollinated and individual F2 plants were screened for homozygous double mutants. Because PLDζ1 and PLDζ2 genes are both located on chromosome III, a large population of plants was screened to obtain homozygous double mutants. Of 200 individual F2 plants screened by PCR, eight plants were found to be homozygous for PLDζ1 and heterozygous for PLDζ2. These plants were self-pollinated and double homozygous pldζ1pldζ2 plants were obtained in the F3 generation.
Plant Growth and Phosphate Treatments
For mutant isolation and routine plant growth, seeds were sown in soil and treated at 4°C for 2 d. Plants were grown in growth chambers under a 12-h-day/12-h-night at 23°C/18°C cycle. For phosphate treatments, seeds were surface sterilized and germinated on modified Murashige and Skoog medium under a 12-h-day/12-h-night at 22°C cycle. The modified medium contained 1.25 mm KNO3, 1.5 mm Ca(NO3)2, 0.75 mm MgSO4, 0.5 mm KH2PO4, 75 μm FeEDTA, 50 μm H3BO3, 10 μm MnCl, 2 μm ZnSO4, 1.5 μm CuSO4, and 0.075 μm (NH4)6Mo7O24. Three-day-old seedlings on standard 500 μm phosphate medium were transferred onto either 500 or 25 μm phosphate medium and grown for an additional 7 d. The seedlings grown on both media were used to determine dry weight, lipid composition, and RNA isolation.
Measurements of Roots and Root Hairs
Seedlings were grown for 3 d on agar plates containing 500 μm phosphate and then transferred to agar plates containing either 500 or 25 μm phosphate agar plates. The replacement of the root tip was measured between the fourth and seventh days after transfer, and the data are based on 20 seedlings per genotype. The lateral root length was measured at the seventh day after transferring, and data are based on four lateral roots per seedling and 20 seedlings per genotype. Forty-eight hours after transfer, images of root hair patterns in the region 3 to 4 mm above the root tips were captured using a MagnaFire (Optronics) system. Root hair length was determined by measuring 100 root hairs located 10 mm from the root tip in 12 individual plants from each line.
Lipid Profiling
The processes of lipid extraction, analysis, and quantification were performed as described (Wanjie et al., 2005). Briefly, rosettes or roots from 25 seedlings were collected at the sampling time and immersed immediately into 3 mL hot isopropanol with 0.01% butylated hydroxytoluene at 75°C to inhibit lipolytic activities. The tissues were extracted with chloroform-methanol five times with 30 min agitation each time. The remaining plant tissues were dried in an oven at 105°C overnight and then weighed. The weights of these dried, extracted tissues are the dry weights of the samples. Lipid samples were analyzed on an electrospray ionization triple quadrupole mass spectrometer (API 4000). The molecular species of PA were quantified in comparison to the two internal standards using a correction curve determined between standards. Five replicates of each treatment for each phenotype were processed and analyzed. The Q-test for discordant data was done on the replicates of the total lipid. Paired values were subjected to Student's t test to determine statistical significance.
Real-Time PCR
Total RNA was isolated using a rapid cetyl-trimethyl-ammonium bromide method (Stewart and Via, 1993), and RNA was precipitated using 2 m LiCl overnight under 4°C. RNA integrity was checked by 1% (w/v) agarose gel electrophoresis prior to DNase I digestion. Eight micrograms of total RNA were digested with RNase-free DNase I according to the manufacturer's instructions (Ambion). The absence of genomic DNA contamination was subsequently confirmed by PCR, using RNA without RT. For RT, first-strand cDNA was synthesized from 1 μg of total RNA using an iScript cDNA synthesis kit (Bio-Rad) in a total reaction volume of 20 μL according to the manufacturer's instructions. The efficiency of the cDNA synthesis was assessed by real-time PCR amplification of a control gene encoding UBQ10 (At4g05320), and the UBQ10 gene threshold cycle (Ct) value was 20 ± 0.5. Only cDNA preparations that yielded similar Ct values for the control genes were used for determination of PLD gene expression. The level of PLD expression was normalized to that of UBQ10 by subtracting the Ct value of UBQ10 from the Ct value of PLD genes. PCR was performed with a MyiQ sequence detection system (Bio-Rad) using SYBR Green to monitor double-stranded DNA synthesis. Each reaction contained 7.5 μL 2× SYBR Green master mix reagent (Bio-Rad), 1.0 ng cDNA, and 200 nm of each gene-specific primer in a final volume of 15 μL. The following standard thermal profile was used for all PCRs: 95°C for 3 min; and 50 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s.
Table II.
Primers used for real-time PCR in Figure 1, A and B, and Figure 5
| Gene | Primer Sequences | Product Size | |
|---|---|---|---|
| PLDα1 | At3g15730 | Forward 5′-TCTCTGCTTTGCTGCTGTTGTAGC-3′ | 150 bp |
| Reverse 5′-CACAAAGCTACATTCTCTCACCACGTC-3′ | |||
| PLDα2 | At1g52570 | Forward 5′-GAGGATCCGCGAGATTATCTGACA-3′ | 116 bp |
| Reverse 5′-TGTGCTCGGATATAGTCAGTGTCC-3′ | |||
| PLDα3 | At5g25370 | Forward 5′-ATGGTTAATGCAACGGCAGACGAG-3′ | 77 bp |
| Reverse 5′-CCCGGTAAATCGTCATTTCGAGGA-3′ | |||
| PLDβ1 | At2g42010 | Forward 5′-AGGAAGGATCCAGTGGCACACTTT-3′ | 82 bp |
| Reverse 5′-TCTCCCACCCTCTTATTATCAACTCTCA-3′ | |||
| PLDβ2 | At4g00240 | Forward 5′-CTGCTAGATGATTGCTTTGTGGAGC-3′ | 108 bp |
| Reverse 5′-CTCCGACACTTCCTCACTCCT-3′ | |||
| PLDγ1 | At4g11850 | Forward 5′-ACTTTTTCTGTCTTGGAACCAGAG-3′ | 97 bp |
| Reverse 5′-GCATTTGCATTTGCGTTTGGCTGA-3′ | |||
| PLDγ2 | At4g11830 | Forward 5′-TGCTCCCTTTGCGTCTAGGTTTCT-3′ | 78 bp |
| Reverse 5′-TCTATTCCAGCAGCAACACCACGA-3′ | |||
| PLDγ3 | At4g11840 | Forward 5′-GGTTTCCATAACACCCTTGTTGGT-3′ | 73 bp |
| Reverse 5′-TTCTCACCATCCACTTTATGATTCCT-3′ | |||
| PLDδ | At4g35790 | Forward 5′-TGGGCGCATACCAACCTAATCA-3′ | 128 bp |
| Reverse 5′-TGGCTCCACAAACTCATCTCCA-3′ | |||
| PLDɛ | At1g55180 | Forward 5′-GGGACACCGAGATTGCAATAGGTT-3′ | 79 bp |
| Reverse 5′-AGCGATAACCGGTAAGCTTGGA-3′ | |||
| PLDζ1 | At3g16785 | Forward 5′-TGGATGGCAACCGCAAAGACAA-3′ | 137 bp |
| Reverse 5′-ATCGTTGTGTGTCCCAGCTTCT-3′ | |||
| PLDζ2 | At3g05630 | Forward 5′-TTTGAGGACGGTCCAATTGCCA-3′ | 143 bp |
| Reverse 5′-ACAACACCGATCTCAGAGTCTCGT-3′ | |||
|
UBQ10
|
At4g05320
|
Forward 5′-CACACTCCACTTGGTCTTGCGT-3′ | 71 bp
|
| Reverse 5′-TGGTCTTTCCGGTGAGAGTCTTCA-3′ |
This work was supported by the National Science Foundation (NSF; grant nos. MCB–0416839 and INB–0454866) and the U.S. Department of Agriculture (grant no. 2005–35818–15253). The Kansas Lipidomics Research Center Analytical Laboratory received support from the NSF EPSCoR program (grant no. EPS–0236913) with matching support from the State of Kansas through Kansas Technology Enterprise Corporation and Kansas State University. The Kansas Lipidomics Research Center also received Core Facility support from K-IDeA Networks of Biomedical Research Excellence (INBRE) through the National Institutes of Health (grant no. P20 RR16475 from the INBRE program of the National Center for Research Resources).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Xuemin Wang (wangxue@umsl.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.070995.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anthony RG, Henriques R, Helfer A, Meszaros T, Rios G, Testerink C, Munnik T, Deak M, Koncz C, Bogre L (2004) A protein kinase target of a PDK1 signaling pathway is involved in root hair growth in Arabidopsis. EMBO J 23: 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski RL, Ferguson IB (1983) Physiology and metabolism of phosphate and its compounds. In A Lauchi, RL Bieleski, eds, Encyclopedia of Plant Physiology. Springer-Verlag, New York, pp 422–449
- Cristina MD, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G (1996) The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J 10: 393–402 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organization of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Härtel H, Dormann P, Benning C (2000) DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA 97: 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li M, Zhang W, Welti R, Wang X (2004) The plasma membrane-bound phospholipase Dδ enhances freezing tolerance in Arabidopsis thaliana. Nat Biotechnol 22: 427–433 [DOI] [PubMed] [Google Scholar]
- Lynch J (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP (2003) Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol 131: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Lee DK, Vu TH, Tej SS, Edberg SB, Matvienko M, Tindell LD (2004) Arabidopsis MPSS. An online resource for quantitative expression analysis. Plant Physiol 135: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya KI, Ohta H (2005) A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J Biol Chem 280: 7469–7476 [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T (2003) Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Qin C, Wang X (2002) The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol 128: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Cui D, Wang X (2001) Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol 126: 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman D, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Stewart CN Jr, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14: 748–758 [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a non-renewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Wang X (2002) Phospholipase D in hormonal and stress signaling. Curr Opin Plant Biol 5: 408–414 [DOI] [PubMed] [Google Scholar]
- Wanjie SW, Welti R, Moreau RA, Chapman KD (2005) Identification and quantification of glycerolipids in cotton fibers: reconciliation with metabolic pathway predictions from DNA databases. Lipids 40: 773–785 [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X (2002) Profiling membrane lipids in plant stress response. Role of phospholipidase D in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SPCP, Fitter AH, Ottoline Leyser HM (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X (2004) Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yu L, Zhang Y, Wang X (2005) Phospholipase D in the signaling networks of plant response to abscisic acid and reactive oxygen species. Biochim Biophys Acta 1736: 1–9 [DOI] [PubMed] [Google Scholar]