Abstract
The v-rel oncogene encoded by reticuloendotheliosis virus strain T is the acutely transforming member of the Rel/NF-κB family of transcription factors. In v-Rel-transformed cells, v-Rel exists as homodimers or heterodimers with the endogenous Rel/NF-κB proteins c-Rel, NF-κB1, NF-κB2, and RelA. To examine the contribution of these complexes to v-Rel-mediated transformation, mutations were introduced into the dimerization interface of v-Rel to generate v-Rel mutants with selective dimerization properties. Nine mutants are described in this study that are defective in homodimer and/or heterodimer formation with specific Rel/NF-κB family members. Viruses expressing mutants that failed to homodimerize but were able to form heterodimeric complexes were unable to transform splenic lymphocytes in vitro, indicating that the dimerization of v-Rel with endogenously expressed Rel/NF-κB proteins is not in itself sufficient for transformation. In addition, two partially transforming mutants were identified that exhibited an impaired ability to form homodimers. Sequence analysis of the proviral DNA from cells transformed by these mutants revealed the presence of multiple secondary mutations in sequences responsible for dimerization and DNA binding. Two of these mutations either enhanced or restored the ability of these proteins to bind DNA as a homodimer. Viruses expressing these proteins transformed cells at levels comparable to or slightly less than v-Rel, suggesting that a threshold level of DNA binding by v-Rel homodimers is required for transformation.
The Rel/NF-κB family of transcription factors is highly conserved, and members are found in species ranging from insects to humans. The vertebrate members of this family include v-Rel, c-Rel, NF-κB1 (p105/p50), NF-κB2 (p100/p52), RelA, and RelB. Characteristic of this family is a highly conserved 300-amino-acid (aa) region in the N termini of these proteins termed the Rel homology domain (RHD). Crystal structures of several Rel/NF-κB dimers have identified the sequences within the RHD that allow these proteins to form homo- and heterodimers and bind DNA (9, 11, 22, 28, 29, 43, 55). Rel/NF-κB proteins are normally sequestered in inactive complexes in the cytoplasm of cells by a family of inhibitory proteins (IκBs) (60). Upon proper extracellular stimulation, IκBs are phosphorylated, ubiquitinated, and ultimately degraded, allowing for the nuclear translocation of active Rel/NF-κB complexes (31). In the nuclei of cells, Rel/NF-κB dimers bind to a 9- or 10-bp DNA sequence (κB site) to regulate the expression of genes involved in the immune response, differentiation, proliferation, and stress responses (28, 47).
The v-rel oncogene is the only acutely transforming Rel/NF-κB family member. v-rel was acquired as a result of a recombination event between the envelope sequences of the replication-competent reticuloendotheliosis-associated virus (REV-A) and turkey c-rel sequences (23). This transduction event resulted in the removal of sequences encoding 2 N-terminal and 118 C-terminal amino acids of c-Rel. REV-A envelope-derived sequences encode 11 N-terminal amino acids and 18 out-of-frame amino acids at the C terminus. In addition, v-Rel has acquired a number of amino acid substitutions and deletions, relative to turkey c-Rel. The structural changes acquired by v-Rel alter the DNA binding, dimerization, and transactivation properties of the protein and contribute to the high transforming potential of v-Rel (23).
Viruses expressing v-Rel induce fatal lymphomas in young birds 7 to 10 days after infection (4). v-Rel transforms cells of various lymphoid lineages, including B cells and T cells (2, 3, 36, 62). Although not a target cell in vivo, chicken embryo fibroblasts (CEFs) are also transformed by v-Rel (17, 39). In transformed cells, v-Rel is found in the cytoplasm in complexes with the endogenous Rel/NF-κB/IκB family members c-Rel, NF-κB1 (p105), NF-κB2 (p100), RelA, and IκBα (6, 13, 32, 37, 40, 56). Approximately 25% of v-Rel is associated with endogenous Rel/NF-κB proteins (12). Analysis of nuclear v-Rel DNA-binding complexes revealed the presence of v-Rel homodimers and v-Rel heterodimers containing c-Rel, NF-κB1 (p50), and NF-κB2 (p52) (26, 53).
The ability of v-Rel to form dimers and bind DNA to regulate gene expression is required for its transforming ability (23). A number of genes, including ikba, c-jun, and ch-IAP1, are directly regulated by v-Rel and c-Rel, and the altered regulation of these gene products by v-Rel has been shown to be important for v-Rel-mediated transformation (14, 18, 19, 33, 34, 51). These findings support a model in which v-Rel complexes directly regulate genes normally under the control of Rel/NF-κB family members, leading to transformation.
While previous studies have attempted to define the role of v-Rel complexes in transformation, the contribution of individual complexes remains largely undefined. A number of transformation-defective v-Rel mutants have been described, although the dimerization and DNA-binding properties of many of these mutants have only been partially characterized (20, 24, 35, 42, 58). One of the better-characterized mutants, v-Rel-SPW, contains a 2-aa insertion in the conserved protein kinase A site of v-Rel. This mutant failed to form homodimers or heterodimers with c-Rel (59). v-Rel-SPW did form heterodimers with NF-κB1 and NF-κB2, although at lower levels than v-Rel (6, 59). The expression of this mutant alone was unable to transform splenic lymphocytes in vitro, although transformation was observed when it was coexpressed with NF-κB2 (p52). Transformation by viruses coexpressing v-Rel-SPW and NF-κB2 (p52) was much weaker than by those expressing v-Rel alone, since these viruses failed to transform cells in soft agar. Nevertheless, these results suggest a potential role for v-Rel/NF-κB2 heterodimers in transformation.
In order to address the role of individual v-Rel complexes in transformation, we have identified v-Rel mutants defective in the formation of homodimers and/or selective heterodimeric complexes. A comprehensive analysis of the dimerization, DNA-binding, and transforming properties of these mutants is presented in this report. The results of these experiments indicate that the ability of v-Rel to dimerize and bind DNA with endogenously expressed Rel/NF-κB proteins is not in itself sufficient for transformation and suggest that a threshold level of DNA binding by v-Rel homodimers is required for transformation.
MATERIALS AND METHODS
Construction of a library of v-Rel mutants.
Based on the crystal structure of mammalian Rel/NF-κB proteins, a 151-nucleotide region of v-rel (nucleotides 933 to 1084) encoding the potential dimerization domain was targeted for mutagenesis. To facilitate mutagenesis of this region, two unique restriction endonuclease sites were introduced into v-rel that had been previously cloned in pBluescript (Stratagene, La Jolla, Calif.). Two oligonucleotides were used for the creation of pv-rel(B/A) which contained a BsmBI site (5′-CAGAAGAAAAATTTCgTCTCCTCCCTTTACACT-3′) at nucleotide 933 and an AvrII site (5′-GGGTTCTGTGATGTCTCCtAGGAACGGCGG-3′) at nucleotide 1084 by using the Transformer Site Directed Mutagenesis System according to the manufacturer's protocol (Clontech, Palo Alto, Calif.). Lowercase letters represent mutagenic nucleotides, and restriction sites are underlined. All oligonucleotides described here were manufactured by Integrated DNA Technologies (Coralville, Iowa).
Due to the size of the region targeted for mutagenesis, two parts of this region were mutagenized separately using partially degenerate mutagenic oligonucleotides that were 85 (5′-GGGAGGAGACGaaatTttTctTctgtgTgaCAaagtTCaaAaagaTgaCaTaGaggtCAgattTgtCtTgggCaaCTGGGAGGCA-3′) and 66 (5′-AAGGGCtcCttCtcCCaagcTgaTgtTcaTcgCCaggtCGcaatTGtattTAgaaCaCCGCCGTTC-3′) nucleotides in length. Lowercase letters represent the positions of the degenerate nucleotides. The oligonucleotides were synthesized with a 97:1:1:1 ratio of the wild-type nucleotides, relative to the remaining nucleotides at each degenerate position. Theoretically, this synthesis should yield an average of one mutation per oligonucleotide synthesized. The nucleotides represented by capital letters indicate locations where only the wild-type sequence was inserted. These positions were not used for mutagenesis, since they might interfere with cloning or potentially introduce translation termination codons. The corresponding nonmutagenic oligonucleotides were also synthesized. In addition, oligonucleotides complementary to the 85-mer (5′-CCTTTGCCTCCCAGTTGCCCAAGACAAATCTGACCTCTATGTCATCTTTTTGAACTTTGTCACACAGAAGAAAAATTTCGTCTCC-3′) or 66-mer (5′-CTAGGAACGGCGGTGTTCTAAATACAATTGCGACCTGGCGATGAACATCAGCTTGGGAGAAGGAGC-3′) were synthesized. These oligonucleotides were designed to create two overhangs when either of the mutagenic oligonucleotides (or the corresponding wild-type oligonucleotides) was annealed to the oligonucleotide containing its complementary sequences. The 5′ end of the annealed 85-mer oligonucleotides contained a 5′ overhang with sequences complementary to the BsmBI restriction endonuclease site. The 3′ end contained a 5′ overhang complementary to the 5′ overhang of the annealed 66-mer oligonucleotides. The 3′ end of this double-stranded 66-mer oligonucleotide contained a 5′ overhang complementary to the AvrII restriction endonuclease site.
Four pairs of these oligonucleotides were annealed to create two mutagenic and two wild-type double-stranded linkers. Pairs of oligonucleotides were annealed in a 50-μl reaction volume (20 mM Tris-Cl [pH 7.4], 2 mM MgCl2, 50 mM NaCl) by incubating 50 pmol of each oligonucleotide for 5 min at 75°C, followed by cooling to 20°C over 30 min in a thermocycler. To prepare these linkers for cloning, the 5′ ends were phosphorylated by using T4 polynucleotide kinase according to the manufacturer's directions (Life Technologies, Gaithersburg, Md.). The phosphorylated linkers were resolved on a nondenaturing 10% acrylamide (29:1 [wt/wt] acrylamide-bisacrylamide) gel, visualized by ethidium bromide staining, and eluted (52). Pairs of these four purified linkers were ligated to form two larger linkers for insertion into v-rel. Double-stranded 85-nucleotide linkers created with mutagenic oligonucleotides were ligated to wild-type 66-nucleotide linkers. Likewise, 66-nucleotide linkers created with mutagenic oligonucleotides were ligated to wild-type 85-nucleotide linkers. Ligation reactions were performed at room temperature for 10 min, resolved on a nondenaturing 10% acrylamide gel, and purified as described above. The purified linkers were ligated with pv-rel(B/A) that had been digested with BsmBI and AvrII and purified from the excised 151-nucleotide DNA fragment. The ligation mixture was electroporated directly into BMH-71-18 mutS cells. Colonies were selected and DNA was isolated by using a Qiagen Maxi Prep kit (Qiagen, Valencia, Calif.).
Two-hybrid screen for v-Rel dimerization mutants.
The LexA-based two-hybrid system used here included the expression plasmids pEG202 and pJG4-5 that encode the bacterial DNA-binding protein LexA and the acidic peptide B42, respectively (25). The reporter plasmid employed in these studies, pSH18-34, contains eight LexA DNA-binding sites upstream of lacZ. Yeast was cultured by using standard methods (1). Plasmids containing potential v-rel mutants in pEG202 were isolated with a modified boiling lysis protocol (49).
Sequences encoding aa 1 to 292 of v-Rel were excised from the v-rel mutant libraries described above and cloned into pEG202. Two different two-hybrid screens were employed to identify v-Rel dimerization mutants. The initial screen for v-Rel dimerization mutants scored for mutants that failed to interact with a specific Rel/NF-κB family member. Mutants identified in this manner were then scored for their ability to interact with full-length c-Rel (aa 1 to 598) and RelA (aa 1 to 558) and the p50 (aa 1 to 401) and p52 (aa 1 to 438) forms of NF-κB1 and NF-κB2, respectively. For these screens the pEG202-based library of v-rel mutants spanning nucleotides 1025 to 1075 was employed. This library and pJG4-5 expressing a Rel/NF-κB family member were cotransformed into the EGY48 yeast strain expressing pSH18-34 and plated on selective medium in the presence of glucose. Approximately 2,000 transformants were replica plated onto selective media in the absence of glucose (in the presence of galactose and raffinose) to induce the expression of the Rel/NF-κB family member. Cells were grown for 2 days at 30°C and screened for β-galactosidase activity by using a filter assay (1). Colonies that did not exhibit any β-galactosidase activity were picked and streaked onto selective media containing glucose to isolate a pure population of cells. These cells were replica plated and screened for β-galactosidase activity as described above. Colonies (180 to 250) that still failed to exhibit β-galactosidase activity were grown in liquid culture, and plasmid DNA was isolated. To select for the DNA-binding domain plasmid expressing the potential v-Rel dimerization mutant, DNA was transformed into Escherichia coli (strain KC8) and plated on M9 medium lacking uracil. DNA was isolated from these transformants, and the ability of the encoded v-Rel mutant to interact with Rel/NF-κB family members was evaluated once again. EGY48 yeast cells expressing these mutants were patch mated with yeast (strain RFY206) expressing pJG4-5, pJG4-5/c-Rel, pJG4-5/NF-κB1, pJG4-5/NF-κB2, or pJG4-5/RelA and then scored for β-galactosidase activity.
A second screen allowed for potential v-Rel mutants to be scored for interaction with c-Rel, NF-κB1, NF-κB2, and RelA at the same time. For these screens, the pEG202-based library of v-rel mutants spanning nucleotides 950 to 1006 was employed. This library was transformed into the EGY48 yeast strain expressing pSH18-34 and plated on selective medium in the presence of glucose. Approximately 1,056 transformants were picked and grown overnight in Cluster Tubes (Costar, Bedford, Mass.) containing 0.8 ml of media. A 96-prong replica plater (Sigma, St. Louis, Mo.) was used to plate overnight cultures on dishes containing selective media. Overnight cultures of the yeast (strain RFY206) expressing pJG4-5, pJG4-5/c-Rel, pJG4-5/NF-κB1, pJG4-5/NF-κB2, or pJG4-5/RelA were plated in a similar fashion. After 2 days of growth, cells were replica plated onto yeast extract-peptone-dextrose plates by using velveteen squares, allowed to mate, and replica plated onto selective media lacking glucose. After 5 days of growth, cells were assayed for β-galactosidase activity as described above. The final dimerization phenotype of v-Rel mutants was determined by quantitative liquid β-galactosidase assays (1).
In vitro translations.
In vitro translations were performed by using the T7 polymerase-based TNT-coupled transcription-translation wheat germ extract system according to the manufacturer's directions (Promega, Madison, Wis.). Genes encoding v-Rel, v-RelΔ (aa 1 to 292), c-Rel (aa 1 to 283), NF-κB1 (aa 1 to 401), and NF-κB2 (aa 1 to 438) were cloned in the T7 orientation of pTZ18R-based vectors (Bio-Rad, Hercules, Calif.) for these studies (45). Plasmids were linearized with a restriction endonuclease that cut a unique site in the polylinker 3′ to the expressed gene. Phosphorimager analysis of [35S]methionine-labeled products revealed that the cotranslated proteins were expressed in approximately equal amounts.
Immunoprecipitations.
Immunoprecipitations were performed with ca. 5 μl of in vitro-translated protein, 2.5 μl of antisera, and 200 μl of ice-cold low immunoprecipitation buffer IPB (25 mM Tris [pH 7.5], 150 mM NaCl, 2 mM EDTA, 0.5% NP-40). Reaction mixtures were rotated at 4°C for 1 h. A 50% slurry of protein A-Sepharose CL4B (25 μl) was added and incubated for 1 h at 4°C. Precipitates were collected by centrifugation for 30 s and washed three times in 1 ml of ice-cold IPB. Pellets were boiled in 25 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (250 mM Tris [pH 6.8], 20% glycerol, 2% SDS, 5% β-mercaptoethanol, bromophenol blue to color), and proteins were analyzed by electrophoresis with 10% polyacrylamide gels.
General cell culture conditions.
CEF cultures were prepared from embryonated SPAFAS (Charles River Laboratories, Preston, Conn.) or SC (Hyline International Hatcheries, Dallas Center, Iowa) eggs and grown in Dulbecco modified Eagle medium (DMEM) supplemented with 5% chicken serum (Life Technologies), 5% bovine calf serum (Atlanta Biologicals, Norcross, Ga.), penicillin (105 U/ml), and streptomycin (50 μg/ml). Primary cell lines transformed by v-Rel or v-Rel dimerization mutants were grown in DMEM supplemented with either 15 or 10% fetal bovine serum (Summit Biotechnology, Fort Collins, Colo.), 5% chicken serum, penicillin (105 U/ml), and streptomycin (50 μg/ml).
Spleen cell transformation assays.
Retroviral stocks were prepared as previously described (44). Titers of viral stocks were determined by dot blot analysis, and infectious units were determined by comparison to a viral stock of REV-TW, for which the titer had been determined by using an immunohistochemical assay (45, 46). For standard transformation assays, splenic lymphocytes were purified from 23- to 28-day-old chickens and resuspended in serum- and antibiotic-free DMEM (57). Purified lymphocytes (5 × 107 cells) were infected with 5 × 105 infectious units of virus for 24 h at 37°C in 8% CO2. Cells were collected by centrifugation and resuspended in 10 ml of plating medium (0.32% Noble agar, 15% fetal bovine serum, and 5% chicken serum in DMEM containing penicillin and streptomycin) and plated in two 60-mm petri dishes. Plates were incubated for 10 days at 37°C in 8% CO2 and scored for colony formation by microscopic analysis.
For transformation assays with concanavalin A (ConA)-stimulated lymphocytes, purified lymphocytes were plated at 107 cells/ml in serum-free DMEM containing ConA (250 μg/ml) and incubated at 37°C and 8% CO2 for 24 h. Cells were collected by centrifugation and infected with virus as described above. Infections were allowed to proceed overnight at 37°C in 8% CO2. Cells were then collected by centrifugation and grown for an additional 2 days in DMEM supplemented with 15% fetal bovine serum, 5% chicken serum, and ConA (250 μg/ml). Cells were again collected by centrifugation plated in soft agar as described above.
For liquid transformation assays, ConA-stimulated lymphocytes were infected as described above. After the overnight infection, cells were collected by centrifugation, resuspended in DMEM (supplemented with 15% fetal bovine serum, 5% chicken serum, and ConA), and grown for an additional 3 days. At least eight 1:2 serial dilutions of each infection were made. Each dilution was aliquoted (160 μl) into 12 wells in a 96-well plate. Plates were incubated for 10 days at 37°C and 8% CO2 and scored for growth by microscopic analysis. Transformation efficiency was calculated by multiplying the reciprocal of the highest dilution giving visible growth by the number of wells showing growth in that dilution.
EMSAs.
A double-stranded DNA probe containing a palindromic κB site was prepared for electrophoretic mobility shift assays (EMSAs). The probe was prepared by annealing a short oligonucleotide primer (5′-AGCTCAAGC-3′) to a longer template (5′-AATTCAGGGGAATTCCCCTAAGCTTGAGCT-3′) and extending the primer with Klenow in the presence of [32P]dCTP. The underlined sequences indicate the location of the κB site. The double-stranded DNA probe was purified from unincorporated [32P]dCTP by using a Tris-buffered Micro Bio-Spin 30 Chromatography Column according to the manufacturer's directions (Bio-Rad). EMSAs were performed with 50,000 cpm of probe and 4 μl of proteins translated in vitro in the absence of radioactive amino acids (44). The amount of protein in each DNA-binding reaction was normalized by comparing the incorporation of [35S]methionine into proteins translated in parallel reactions.
Isolation of genomic DNA from transformed cell lines.
Genomic DNA was isolated from transformed cells (5 × 106) for PCR amplification of proviral DNA (52). Primers complementary to REV-0 sequences flanking the cloning site for v-rel and v-rel mutants (5′-CCCTACACTGTAGTCCTCAGTG-3′ and 5′-CCAACAAGGGTAGCAAATATGG-3′) were used. Integrated proviral DNA was amplified with 45 μl of PCR Supermix (Life Technologies), 50 pmol of each of the REV-0 specific primers, and 1 μg of genomic DNA. Thermocycler settings were as follows: 94°C for 1 min plus 50°C for 2 min plus 72°C for 2 min for 25 cycles, followed by incubation at 72°C for 10 min.
Northern blot analysis.
Total RNA was isolated from CEF cultures or transformed cell lines by using RNAwiz according to the manufacturer's directions (Ambion, Austin, Tex.). RNA was electrophoretically separated by formaldehyde-agarose gel electrophoresis, transferred to Hybond N+ (Amersham Pharmacia Biotech, Piscataway, N.J.) by capillary transfer, and stained with methylene blue to confirm equal loading of the RNA (52). Membranes were hybridized with v-rel probes by using ULTRAhyb according to the manufacturer's directions (Ambion). Probes encoding the entire cDNA of v-rel were labeled with [32P]dCTP by using the Prime-A-Gene Labeling System (Promega).
Western blot analysis.
Cell lysates (40 μg) for Western blot analysis were resolved by SDS-PAGE and electrophoretically transferred to Optitran BA-reinforced nitrocellulose (Schleicher & Schuell, Keene, N.H.). Membranes were stained with Ponceau S to determine the quality of transfer and the equivalence of protein loading. Immunoblotting was performed with a monoclonal antibody (HY87) specific for v-Rel and c-Rel and a horseradish peroxidase-conjugated donkey anti-mouse antibody (Jackson ImmunoResearch, West Grove, Pa.) (27). Proteins were detected by using the Renaissance Western blot Chemiluminescence Reagent (Perkin-Elmer Life Sciences, Boston, Mass.).
Half-life analysis.
For half-life analysis, CEF cultures were plated at a density of 106 cells per 100-mm dish. After 16 h, cultures were treated with cycloheximide (50 μg/ml). At 2, 4, 6, 8, and 10 h after the addition of cycloheximide, cells were harvested by trypsinization, washed in phosphate-buffered saline, and lysed in 100 μl of loading buffer. Extracts (20 μl) were analyzed by Western blot analysis with the monoclonal antibody HY87. The expression levels of v-Rel and v-Rel dimerization mutants were quantitated with NIH Image version 1.59.
RESULTS
Mutations in the dimerization interface of v-Rel allow for selective dimerization and DNA binding by v-Rel.
v-Rel exists as homodimers or heterodimers with c-Rel, NF-κB1, NF-κB2, and RelA in v-Rel-transformed cells, and the formation of these dimers is believed to contribute to v-Rel-mediated transformation. Dimerization mutants of v-Rel were constructed to determine which complexes contribute to the transforming potential of v-Rel. To identify dimerization mutants of v-Rel, sequences corresponding to those that encode the dimerization interface of other Rel/NF-κB proteins were targeted for mutagenesis. Sequences encoding aa 208 to 254 were replaced with partially degenerate mutagenic linkers. This region contains 12 of the 16 aa positions shown to be involved in protein-protein contacts in the dimerization interface of other Rel/NF-κB family members, including 9 of the 12 aa involved in formation of c-Rel homodimers (9-11, 22, 28, 29, 43, 55). Mutant v-rel libraries were used in reverse two-hybrid screens to identify v-Rel mutants that failed to interact with specific Rel/NF-κB proteins while maintaining their interaction with others. The assignment of a dimerization phenotype to a mutant was based on the results of quantitative liquid β-galactosidase assays. A mutant was classified as defective in dimerization with a specific Rel/NF-κB family member if the β-galactosidase level observed when coexpressed with a Rel/NF-κB family member was less than twofold the level observed with a control plasmid.
A total of 16 mutants with altered dimerization properties were identified. Nine characteristic mutants were selected for this study. The location and nature of the amino acid changes in these mutants are shown in Table 1. These v-Rel mutants fell into five classes, based on their inability to interact with various Rel/NF-κB family members (Table 2). The class I mutant (M1) was defective in dimerization with RelA. The class II mutant (M2) failed to form homodimers, while maintaining the ability to dimerize with c-Rel, NF-κB1, NF-κB2, and RelA. This mutant induced high levels of β-galactosidase activity when expressed with NF-κB1 or NF-κB2 and low levels when expressed with c-Rel or RelA. The class III mutant (M3) was defective in dimerization with c-Rel and RelA and yet induced high levels of β-galactosidase activity when assayed for dimerization with either NF-κB1 or NF-κB2. However, when assayed for homodimer formation, it induced low levels of β-galactosidase activity relative to wild-type v-Rel. Class IV mutants were defective in homodimer formation and interaction with c-Rel. Both class IV mutants (M4 and M5) induced low levels of β-galactosidase activity when assayed for dimerization with RelA. The fifth class of mutants failed to form homodimers or heterodimers with c-Rel and RelA. Class V mutants (M6, M7, M8, and M9) all induced high levels of β-galactosidase activity when assayed for dimerization with NF-κB1 and NF-κB2, with the exception of M9, which induced low levels with NF-κB2.
TABLE 1.
Nature of amino acid changes in v-Rel mutants
| Protein | Mutation(s) | Relative location of mutationsa (aa 210-254) |
|---|---|---|
| • | ||
| ⧫⧫⧫⧫⧫•••⧫⧫⧫⧫•⧫⧫⧫ | ||
| v-Rel | DEIFLLCDKVQKDDIEVRFVLGNWEAKGSFSQADVHRQVAIVFRT | |
| M1 | I224L, E225D, E234D | --------------LD--------D-------------------- |
| M2 | E211K, I212T, D222A | -KT---------A-------------------------------- |
| M3 | F213V, V226F | ---V------------F---------------------------- |
| M4 | V226G | ----------------G---------------------------- |
| M5 | K218I, D223V | --------I----V------------------------------- |
| M6 | F213L, D223A, L230M, N232S | ---L---------A------M-S---------------------- |
| M7 | I250S | ----------------------------------------S---- |
| M8 | I212D | --D------------------------------------------ |
| M9 | V251L | -----------------------------------------L--- |
TABLE 2.
Dimerization and DNA-binding characteristics of v-Rel mutants
| Protein | Classa | Dimerizationb with:
|
DNA bindingc with:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| v-Reld | c-Rel | NF-κB1 | NF-κB2 | RelA | v-Reld | c-Rel | NF-κB1 | NF-κB2 | ||
| v-Rel | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | +++ | ++++ | ++++ | |
| M1 | I | +++ | ++++ | +++ | ++++ | − | +/− | ++ | +++ | +++ |
| M2 | II | − | + | +++ | ++++ | + | − | + | ++ | + |
| M3 | III | + | − | ++++ | ++++ | − | − | − | ++ | + |
| M4 | IV | − | − | + | ++ | + | − | − | + | + |
| M5 | IV | − | − | ++++ | ++++ | + | − | − | + | + |
| M6 | V | − | − | +++ | ++++ | − | − | − | ++ | + |
| M7 | V | − | − | +++ | ++++ | − | − | − | +++ | +++ |
| M8 | V | − | − | ++ | ++++ | − | − | − | + | + |
| M9 | V | − | − | ++++ | + | − | − | − | +++ | +++ |
Each dimerization mutant was assigned to a particular class based on the inability to interact with specific Rel/NF-κB family members.
Quantitative liquid β-galactosidase assays were performed to evaluate the formation of homodimers (23.2 ± 1.2) and heterodimers with c-Rel (23.6 ± 1.3), NF-κB1 (135.3 ± 5.1), NF-κB2 (60.7 ± 13.8), and RelA (61.4 ± 1.3). Dimerization: ++++, >75%; +++, 50 to 75%; ++, 25 to 50%; +, <25%; −, none detected. These values represent the β-galactosidase activity relative to that observed for v-Rel.
Qualitative assessment of DNA-binding activity relative to v-Rel.
Assayed for homodimer formation.
Since amino acids within the dimerization interface of Rel/NF-κB proteins contact the DNA backbone of κB sites, mutations within the dimerization interface may prevent a mutant from binding DNA without affecting its dimerization with a Rel/NF-κB family member. Therefore, the DNA-binding properties of the v-Rel dimerization mutants were analyzed. v-Rel dimerization mutants were either in vitro translated alone or cotranslated with c-Rel, NF-κB1, or NF-κB2 and used in EMSAs with a palindromic κB site as a probe. Under the conditions of these assays, v-Rel was able to bind DNA as a homodimer or heterodimer with each of these Rel/NF-κB proteins. The ability of mutants to bind DNA with RelA was not evaluated, since RelA was not found in the nuclei of v-Rel-transformed cells (W. Bargmann and H. R. Bose, Jr., unpublished results). The results of these experiments are summarized in Table 2. Eight mutants (M1, M2, M4, M5, M6, M7, M8, and M9) bound DNA with each of their dimerization partners identified in the two hybrid studies. One mutant, M3, failed to bind DNA as a homodimer despite the ability to weakly form homodimers in the two-hybrid system. In addition, a number of mutants (M2, M3, M5, M6, and M8) exhibited lower DNA-binding activity when expressed with NF-κB1 or NF-κB2 than expected based on their apparent ability to interact with these proteins in the two-hybrid system. One of these mutants (M5) contained an amino acid substitution at a position shown to be involved in protein-DNA interactions in other Rel/NF-κB proteins. The ability of the v-Rel dimerization mutants described above to selectively dimerize and bind DNA with Rel/NF-κB proteins makes them ideally suited to analyze the role of v-Rel complexes in transformation.
Dimerization of v-Rel enhances its stability.
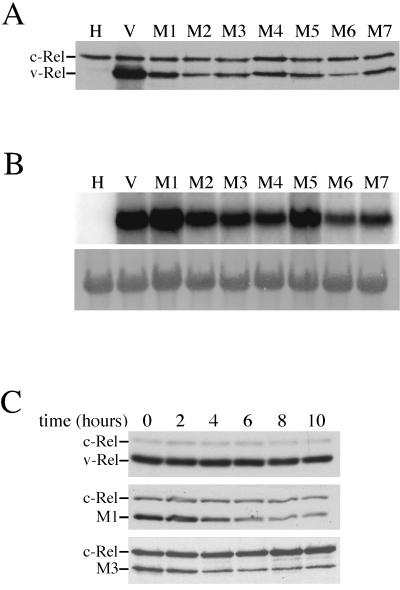
Seven v-Rel dimerization mutants were chosen for further characterization in vivo. The mutants were cloned into a reticuloendotheliosis virus-based retroviral vector (REV-0), and viral stocks were prepared. When these viruses were used to infect CEF cultures, mutant v-Rel proteins were expressed, although at reduced levels relative to v-Rel (Fig. 1A). The lower levels of protein expression could not be solely explained by differences in RNA levels. Cells infected with viruses expressing M1, M2, and M5 expressed comparable amounts of viral RNA as cells infected with viruses expressing wild-type v-Rel but exhibited diminished steady-state levels of protein (compare lane 2 with lanes 3, 4, and 7 in Fig. 1B).
FIG. 1.
Expression of v-Rel dimerization mutants in CEF cultures. (A) Western blot analysis of CEFs expressing v-Rel dimerization mutants. Protein (40 μg) in lysates from cells expressing CSV, v-Rel, or v-Rel dimerization mutants was resolved by SDS-PAGE, transferred to nitrocellulose, and analyzed by Western blotting with the anti-Rel monoclonal antibody HY87. The migration of v-Rel and c-Rel is indicated. (B) Northern blot analysis of CEFs expressing v-Rel dimerization mutants. Total RNA (10 μg) from the cultures expressing CSV, v-Rel, or v-Rel dimerization mutants in panel A was analyzed for the expression of viral RNA. The hybridization of the viral genomic RNA with a v-rel probe is shown in the top panel. The lower panel shows the methylene blue staining of the 26S rRNA after transfer to the membrane. (C) Half-life analysis of v-Rel mutants in CEF cultures. CEF cultures expressing v-Rel (top panel), M1 (middle panel), or M3 (lower panel) were treated with cycloheximide and whole-cell extracts prepared at various times after treatment. The expression of v-Rel and v-Rel dimerization mutants was evaluated by Western blot analysis. The location of v-Rel, M1, M3, or c-Rel is indicated on the left side of each panel. The length (in hours) of cycloheximide treatment is indicated on the top.
Protein half-life experiments were performed with mutants that were expressed at high (M1) and low (M3) levels to evaluate the mechanism for the reduced protein levels of the v-Rel dimerization mutants in these cells. CEF cultures expressing v-Rel, M1, or M3 were treated with the protein synthesis inhibitor cycloheximide, and whole-cell extracts were prepared at various times after treatment. The expression of v-Rel and v-Rel mutants was evaluated by Western blot analysis (Fig. 1C). The amount of v-Rel remained relatively constant in cells treated with cycloheximide, even after 10 h of treatment. In contrast, the levels of M1 and M3 decreased significantly after cycloheximide treatment. The level of M1 decreased approximately 30% after 4 h of cycloheximide treatment, and by 8 h the level of M1 decreased by approximately 40%. The level of M3 decreased more rapidly and to a greater extent than M1. After just 4 h a 50% reduction in M3 was observed, which then decreased an additional 10% between 4 and 8 h. Although this analysis is limited to two mutants, the results indicate that the lower levels of the mutant v-Rel proteins in cells are due to a reduction in their stability.
v-Rel heterodimers are not sufficient for transformation.
To elucidate the role of specific Rel/NF-κB family members in v-Rel-mediated transformation, v-Rel dimerization mutants were evaluated for their ability to transform splenic lymphocytes. Initial experiments revealed that infection with viruses expressing the v-Rel dimerization mutants failed to transform splenic lymphocytes of 3-week-old chickens under conventional soft agar conditions, although wild-type v-Rel routinely produced 50 to 100 colonies under these conditions. To enhance the sensitivity of this assay, splenic lymphocytes were first stimulated with ConA prior to infection. Under these conditions, colonies transformed by v-Rel were too dense to precisely quantitate, although they were transformed ca. 1,000 times more efficiently than untreated lymphocytes. Among the mutant v-Rel-expressing viruses tested, only M1 and M3 were able to induce colony formation in soft agar (Table 3). Levels of colony formation by these mutants were significantly lower than that induced by v-Rel. All other mutants tested were nontransforming under these assay conditions.
TABLE 3.
Transformation of splenic lymphocytes by v-Rel dimerization mutants
| Virusa | No. of colonies in soft agarb
|
Growth in liquidc
|
||||||
|---|---|---|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Avg | Expt 1 | Expt 2 | Expt 3 | Avg | |
| CSV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TW | TMTC | TMTC | TMTC | TMTC | 262,144 | 131,072 | 131,072 | 174,763 |
| TM1 | 36 | 79 | 103 | 73 | 64 | 16 | 64 | 48 |
| TM2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TM3 | 27 | 0 | 33 | 20 | 32 | 0 | 0 | 11 |
| TM4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TM5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TM6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TM7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Helper virus (CSV) or REV-0-based retroviruses expressing v-Rel (REV-TW) or v-Rel dimerization mutants (REV-TM) were used to infect ConA-stimulated splenic lymphocytes.
Infected splenic lymphocytes were plated in soft agar, and colonies were scored microscopically after 10 days.
Infected splenic lymphocytes were serially diluted in 96-well plates, and growth was scored microscopically after 10 days. Transformation efficiency was calculated by multiplying the reciprocal of the highest dilution giving visible growth by the number of wells showing growth in that dilution.
Since soft agar colony formation is one of the more stringent measurements of transformation, a less-growth-restrictive liquid transformation assay was employed to determine if any of the remaining mutants could transform lymphocytes (Table 3). Even under these more favorable conditions, only the mutants that induced colony formation in soft agar (M1 and M3) exhibited transforming properties in the liquid assays. Mutants that were unable to form homodimers (M2 and M4 to M9) failed to transform cells under either transformation conditions.
Proviral copies of v-rel mutants contain additional mutations.
M1 and M3 transformed splenic lymphocytes at a much-reduced rate relative to v-Rel. Furthermore, M3 failed to transform cells in half the assays performed. Given these observations and the error-prone nature of retroviral reverse transcriptase, the possibility of secondary mutations influencing the transformation potential of these mutants was investigated. Genomic DNA was isolated from at least three cell lines transformed by retroviruses expressing M1 and M3 and used as a template in PCRs to amplify viral DNA containing these mutants. These PCR products were cloned into pGEM-T, and three separate isolates from each cell line were sequenced.
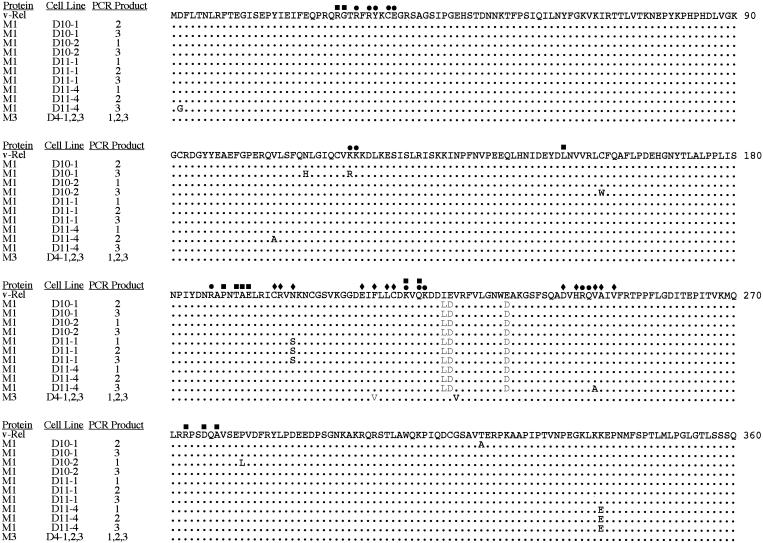
Proviral copies of M3 did not retain their original genotype. In each of the three cell lines analyzed, an identical change occurred that resulted in the restoration of the phenylalanine to the wild-type valine at aa 224 (Fig. 2). The phenylalanine-to-valine mutation at aa 213 remained unaltered in these proviral copies. Since all three of the cell lines transformed by M3 were derived from colonies from the same transformation assay and the proviral DNA from each cell line had the same reversion at the DNA level, it is likely these cell lines originated from a single transformation event.
FIG. 2.
Mutations in proviral DNA from cells transformed by v-Rel dimerization mutants. Genomic DNA was isolated from four cell lines (D10-1, D10-2, D11-1, and D11-4) transformed by viruses expressing M1 and three cell lines (D4-1, D4-2, and D4-3) transformed by viruses expressing M3. Proviral DNA was amplified with virus-specific primers and cloned into pGEM-T. Three separate clones of the PCR products from each cell line were sequenced over the entire length of v-rel. The amino acids from 1 to 360 of v-Rel and the location of amino acids involved in dimerization (⧫), DNA binding (•), and intramolecular interactions (▪) of chicken c-Rel are indicated. Only PCR products containing mutations that altered the amino acid sequence of the protein are shown. The gray letters represent the original amino acid differences found in M1 and M3 relative to v-Rel. The black letters represent secondary amino acid changes resulting from mutations in the proviral DNA.
Proviral DNA amplified from cells transformed by M1 contained numerous secondary mutations. All three PCR products from one cell line (D11-1) contained an asparagine-to-serine mutation at aa 202. This amino acid position has been shown to participate in direct protein-protein interactions at the dimerization interface of other Rel/NF-κB proteins, including chicken c-Rel. In addition, a lysine-to-glutamic acid change at aa 339 was observed in all three clones of the proviral PCR product from a second cell line (D11-4). This mutation is located in a region of v-Rel that does not appear to be directly involved in dimerization or DNA binding. Furthermore, point mutations were observed in one clone of PCR-amplified proviral DNA from a number of cell lines. Since cells transformed by REV-based retroviruses contain, on average, two to three proviral insertions, PCR clones having different sequences likely represent different proviral insertions (61). It is unlikely that these secondary mutations are due to errors during PCR amplification since no mutations were observed in PCR-amplified proviral DNA from a v-Rel-transformed cell line. The frequency of secondary mutations in the proviral copies suggests that they contribute to the transformed state of these cells.
Secondary mutations found in proviral DNA alter the dimerization, DNA-binding, and transforming properties of M1 and M3.
Since many of the mutations observed in the proviral DNA of cells transformed by M1 and M3 are found in regions important for dimerization and DNA binding, it is likely that these properties are altered. To test this possibility, immunoprecipitation analysis and EMSAs were performed with proteins expressed from the cloned proviral DNA. Clones tested included M1 with an asparagine-to-serine mutation at aa 202 (M1M) and M3 containing the phenylalanine-to-valine reversion at aa 224 (M3R).
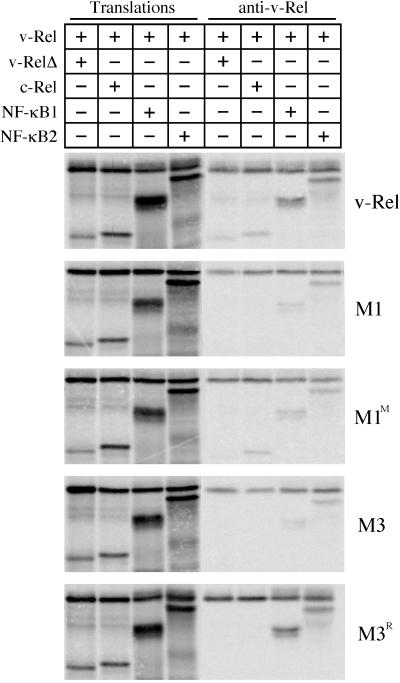
v-Rel or v-Rel mutants were coexpressed with c-Rel, NF-κB1, or NF-κB2 by using a coupled in vitro transcription-translation system, and v-Rel immunoprecipitates were analyzed. To evaluate homodimer formation, v-Rel and v-Rel mutants were also coexpressed with a truncated version of themselves (v-RelΔ) to provide distinguishable products in the coimmunoprecipitation experiments. Proteins were immunoprecipitated with antisera specific to the C terminus of v-Rel, resolved by SDS-PAGE, and visualized by phosphorimager analysis (Fig. 3) (26). Under these conditions v-RelΔ, c-Rel, NF-κB1, and NF-κB2 efficiently coimmunoprecipitated with wild-type v-Rel. Consistent with the two-hybrid results (Table 2), each of the cotranslated proteins coprecipitated with M1, although truncated M1 and c-Rel did so poorly. While M1M associated with NF-κB1 and NF-κB2 at levels similar to M1, it showed an increased ability to bind truncated M1M and c-Rel. These results suggest that the arginine-to-serine change at position 202 enhanced its ability to form homodimers and heterodimers with c-Rel. M3 coprecipitated both NF-κB1 and NF-κB2 and, as expected, failed to coprecipitate c-Rel. However, while M3 was able to weakly form homodimers in the two-hybrid system, M3 failed to coprecipitate with truncated M3, a result consistent with its inability to bind DNA alone (Table 2 and Fig. 4). Interestingly, M3R exhibited a stronger interaction with NF-κB1 than did M3 or v-Rel, but the reversion did not restore its ability to form homodimers or heterodimers with c-Rel. These results suggest that the single amino acid difference between v-Rel and M3R (Phe213→Val) is sufficient to increase the strength of association of M3R with NF-κB1 and, at the same time, decrease the formation of homodimers and heterodimers with c-Rel.
FIG. 3.
Association of v-Rel dimerization mutants with Rel/NF-κB proteins. v-Rel proteins were cotranslated in an in vitro system with truncated v-Rel proteins (v-RelΔ), c-Rel (aa 1 to 284), NF-κB1 (p50), and NF-κB2 (p52) in the presence of [35S]methionine. All cotranslations contained approximately equal amounts of each protein. Translated products were directly analyzed by SDS-PAGE (first four lanes) or were subjected to immunoprecipitation with an antiserum specific to the C terminus of v-Rel and then resolved by SDS-PAGE (last four lanes). Proteins were visualized by phosphorimager analysis. The identity of the v-Rel protein studied is indicated to the right of each panel. The Rel/NF-κB proteins present in each reaction are indicated on the top of each panel.
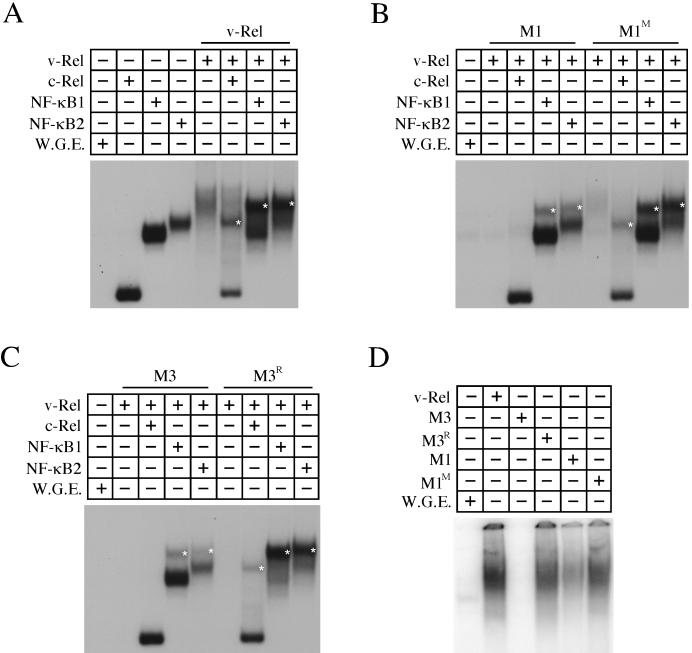
FIG. 4.
DNA binding of v-Rel dimerization mutants. v-Rel proteins were translated alone or cotranslated with c-Rel (aa 1 to 284), NF-κB1 (p50), and NF-κB2 (p52) and used in EMSAs with a 32P-labeled palindromic κB site probe (GGGGAATTCCCC). The Rel/NF-κB proteins in each reaction are indicated on the top of each panel. Reactions with translation mixtures containing v-Rel (A), M1 and M1M (B), and M3 and M3R (C) are indicated above each panel. The locations of heterodimeric complexes are indicated by asterisks. All cotranslations contained approximately equal amounts of each protein. The DNA-binding activity of v-Rel, M1, M1M, M3, and M3R homodimers when ca. 15-fold more protein is used are shown in panel D.
EMSAs were performed to test whether the altered dimerization properties of M1M and M3R affected their DNA-binding properties. v-Rel and v-Rel mutants were translated in vitro alone or cotranslated with c-Rel, NF-κB1, or NF-κB2. The translated proteins were used in EMSAs with a palindromic κB site. A diffuse band was observed when full-length c-Rel was tested under these conditions, making it difficult to distinguish between homodimeric and heterodimeric complexes. Therefore, a C-terminally truncated c-Rel was employed in these studies. Wild-type v-Rel homodimers, as well as v-Rel heterodimers containing c-Rel, NF-κB1, or NF-κB2, were able to bind DNA under these conditions (Fig. 4A). The mobility of each heterodimeric v-Rel complex (denoted by asterisks in Fig. 4) was distinct from the homodimeric DNA-binding complexes. M1 bound DNA as a heterodimer with NF-κB1 or NF-κB2, but at reduced levels relative to v-Rel (Fig. 4B). In addition, M1 poorly bound DNA as a heterodimer with c-Rel. The DNA binding of M1 homodimers was only observed when relatively large amounts of protein were used in the binding reaction (Fig. 4D). In contrast, M1M bound DNA as a heterodimer with NF-κB1 or NF-κB2 at levels comparable to that of v-Rel (Fig. 4B). In addition, M1M exhibited a significant increase in the ability to bind DNA as a heterodimer with c-Rel. While M1M bound DNA as a homodimer with greater efficiency than M1, it still did so at levels lower than v-Rel (Fig. 4D).
M3 bound DNA as a heterodimer with NF-κB1 or NF-κB2 and failed to bind DNA as a homodimer, even when large amounts of protein were used (Fig. 4C and D). By comparison, M3R exhibited strong DNA binding as a heterodimer with NF-κB1 or NF-κB2. Furthermore, M3R exhibited an ability to bind DNA as a homodimer and as a heterodimer with c-Rel. This result was unexpected since the results of the immunoprecipitation experiments suggested that M3R does not form homodimers or heterodimers with c-Rel. This difference may reflect the greater sensitivity of the EMSAs for examining complex formation. Alternatively, the binding of these complexes to DNA may stabilize their associations. Despite these differences, these results demonstrate that M3R forms homodimers and heterodimers with c-Rel at reduced levels relative to v-Rel.
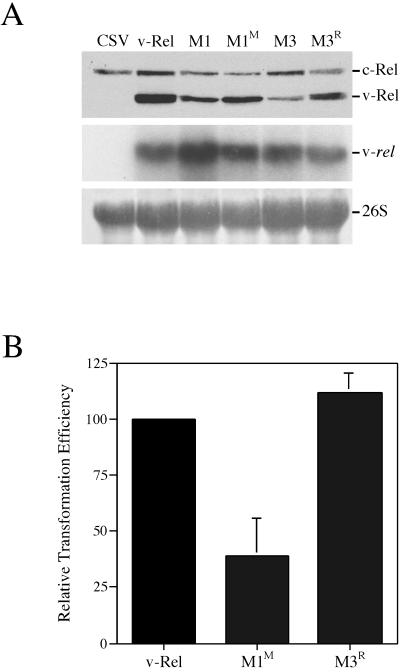
To evaluate whether the enhanced dimerization and DNA binding of M1M and M3R correlates with a greater transformation potential than M1 or M3, retroviruses expressing these mutants were prepared. CEF cultures transfected with these viral constructs expressed higher levels of M1M and M3R relative to M1 or M3, suggesting that M1M and M3R are more stable than M1 and M3 (Fig. 5A). Moreover, while viruses expressing M1 and M3 only induced transformation of lymphocytes stimulated with ConA, M1M and M3R were able to induce transformation of unstimulated splenic lymphocytes (Fig. 5B). M1M transformed cells at about 40% the efficiency of v-Rel, while M3R exhibited a transforming ability comparable to that of v-Rel. The combined results of the immunoprecipitation, DNA-binding, and transformation studies indicate that the secondary mutations acquired by M1 and M3 alter their biochemical and biological activities to more closely resemble v-Rel.
FIG. 5.
Transformation potential of v-Rel dimerization mutants M1M and M3R. (A) Expression of v-Rel dimerization mutants in CEFs. Proteins in lysates (40 μg) from CEFs expressing CSV, v-Rel, M3R, and M1M were resolved by SDS-PAGE and analyzed by Western blotting with monoclonal antibody HY87 (top panel). The location of v-Rel and c-Rel is indicated. Northern blot analysis of CEFs expressing v-Rel dimerization mutants is shown in the middle panel. Total RNA (10 μg) from the CEFs expressing v-Rel or v-Rel dimerization mutants described above was analyzed for the expression of viral RNA. The hybridization of the viral genomic RNA is shown. The lower panel shows the methylene blue staining of the 26S rRNA after transfer to the membrane. (B) Transformation potential of v-Rel, M3R, and M1M. Splenic lymphocytes were infected with 5 × 105 infectious units of a REV-based retrovirus expressing v-Rel, M3R, or M1M. Infected lymphocytes were plated in soft agar, and colonies were scored microscopically after 10 days. The relative transforming potential of each protein is indicated.
DISCUSSION
To define the roles of specific v-Rel complexes in transformation, we isolated and characterized v-Rel mutants with defects in the ability to form homodimers and/or heterodimers with selective Rel/NF-κB family members. Given the high degree of sequence homology in the dimerization interfaces of v-Rel and c-Rel or RelA of avian and mammalian species, it is likely that similar mutations in these Rel/NF-κB family members would confer analogous dimerization defects. Nine mutants described here exhibited one of five general dimerization defects: (i) failure to form heterodimers with RelA, (ii) failure to form homodimers, (iii) failure to form heterodimers with c-Rel and RelA, (iv) failure to form homodimers or heterodimers with c-Rel, or (v) failure to form homodimers and heterodimers with c-Rel and RelA. Mutants that failed to associate with NF-κB1 or NF-κB2 while maintaining their association with other Rel/NF-κB proteins were not identified. In fact, all mutants identified in two-hybrid screens based on the inability to dimerize with NF-κB1 failed to associate with c-Rel, NF-κB2, or RelA. NF-κB1 and NF-κB2 also exhibited the strongest associations with wild-type v-Rel in immunoprecipitation experiments and in the yeast two-hybrid system. The interaction of v-Rel with NF-κB1 and NF-κB2 may be strengthened by the presence of additional hydrogen bonds within the dimerization interface of these heterodimers that are not found in the homodimers. The crystal structure of a NF-κB1/RelA heterodimer revealed a hydrogen bond between equivalent positions in NF-κB1 (Asp 254) and RelA (Asn 200) that was not observed in either homodimer (9). A similar bond may form in v-Rel/NF-κB1 and v-Rel/NF-κB2 heterodimers, since v-Rel contains an Asn at the same position as RelA and since avian NF-κB1 and NF-κB2 contain an Asp at the same positions in the dimerization interface. Although the mutagenesis procedure employed here was not exhaustive, it remains likely that mutations that disrupt the formation of v-Rel/NF-κB1 and v-Rel/NF-κB2 heterodimers may disrupt all dimer formation by v-Rel.
The steady-state protein levels of the dimerization mutants were lower than those observed for v-Rel. The lower levels of these proteins were due to their reduced stability (Fig. 1A and C). The treatment of cells with cycloheximide resulted in a rapid turnover of M1 and M3 within the first 4 to 6 h after treatment, and then the levels of these proteins remained relatively constant until 10 h after treatment. This suggests that a portion of M1 and M3 existed in stable complexes, whereas monomeric v-Rel mutants were rapidly degraded. Furthermore, both the steady-state levels of these proteins and their rate of degradation after cycloheximide treatment correlated with their number of Rel/NF-κB dimerization partners. M3, which weakly forms homodimers and forms heterodimers only with NF-κB1 and NF-κB2, was expressed at lower levels and exhibited a more rapid degradation than M1, which efficiently formed homodimers and heterodimers with c-Rel, NF-κB1, and NF-κB2. Although a correlation was established between the stability of these mutants and their number of dimerization partners, there did not appear to be a direct correlation between their level of expression in cells and their apparent strength of dimer formation in vitro (Fig. 3 and 4). In cells, v-Rel mutants may be stabilized by association with other proteins such as IκBα. While these experiments suggest that the ability of v-Rel mutants to form homodimers and heterodimers with endogenous Rel/NF-κB proteins plays a key role in determining their stability, they do not rule out the possibility that other, undefined factors may contribute to their instability in cells.
Retroviruses encoding M1 and M3 formed a limited number of colonies in soft agar and liquid transformation assays with ConA-stimulated lymphocytes (Table 3). The use of ConA-stimulated lymphocytes enhanced the transforming efficiency of v-Rel by ca. 1,000-fold. Although the mechanism(s) that accounts for this is not known, ConA treatment may provide a higher percentage of proliferating target cells that can be productively infected by these retroviruses (50). Alternatively, ConA may activate genes that complement those regulated by v-Rel or induce the degradation of IκBα, resulting in a more efficient nuclear localization of v-Rel complexes (48). Analysis of proviral DNA from cell lines transformed by M1 and M3 revealed the accumulation of secondary mutations, the majority of which were found in regions known to be important for dimerization or DNA binding (Fig. 2). Additional mutations in the C-terminal transactivation domain and the N-terminal env-derived sequences were also observed in proviral DNA isolated from cell line D11-4. The ability of these regions to confer transactivation potential to v-Rel has been shown to be important for transformation (15, 16, 21). In addition, two proviral clones from cell line D10-2 contained mutations in amino acids located in close proximity to those found to participate in the intramolecular interactions between the N- and C-terminal halves of the RHD of c-Rel (28). One of these secondary mutations (Pro282→Leu) is located near an oncogenic point mutation found in v-Rel (Ala 278). Since mutations were found in the proviral DNA from all of the cell lines analyzed and these mutations are found in locations likely to alter the biochemical properties of v-Rel, it is possible that M1 and M3 are actually nontransforming v-Rel mutants. The increased transformation potential afforded by ConA stimulation may have allowed for the selection of rare secondary mutations that restored the transforming potential of these proteins.
The characterization of a secondary mutation found in the proviral DNA of a cell line (D11-1) transformed by M1 revealed a strong correlation between the ability of this protein (M1M) to bind DNA as a homodimer and its ability to transform cells. The asparagine-to-serine mutation at aa 202 in M1M allowed for an enhanced formation of homodimers and heterodimers with c-Rel, relative to M1 (Fig. 3). Furthermore, M1M bound DNA as heterodimers with c-Rel, NF-κB1, and NF-κB2 at comparable levels to v-Rel but exhibited reduced DNA binding as a homodimer (Fig. 4). M1M was able to transform normal splenic lymphocytes, although at ca. 40% the efficiency of wild-type v-Rel (Fig. 5B). The reduced transformation potential of M1M strongly supports a model in which the DNA binding by v-Rel homodimers is the major mediator of transformation in avian cells. Support for this model comes from the identification of seven nontransforming v-Rel mutants (M2 and M4 to M9) that failed to form homodimers but retained the ability to form heterodimers with endogenous Rel/NF-κB proteins (Tables 1 and 2). In particular, the inability of M2 to form homodimers, while retaining the ability to form heterodimers with c-Rel, NF-κB1, NF-κB2, and RelA, further supports the model that v-Rel homodimers are the critical complex in transformation. While this model has previously been proposed by others (23, 59), it was based largely on the phenotypes of v-Rel mutants that were characterized prior to the cloning of avian NF-κB1, NF-κB2, and RelA (30, 41, 42, 58). Only one v-Rel mutant defective in homodimer formation has been previously shown to form heterodimers with any of the avian Rel/NF-κB family members (6, 59). The thorough analysis of the biochemical and transforming properties of a large number of v-Rel dimerization mutants described in this study provides, for the first time, strong evidence to support a model in which DNA binding by v-Rel homodimers is critical for transformation and the ability of v-Rel to form heterodimers and bind DNA with endogenous Rel/NF-κB proteins is, in itself, not sufficient for transformation of avian cells.
Previous studies have demonstrated that the appearance of v-Rel heterodimeric complexes in the nuclei of v-Rel-expressing cells correlated with an increased proliferation of these cells (26, 45). Heterodimers of v-Rel with endogenous Rel/NF-κB proteins may therefore play an important role in the transformation process. The characterization of a secondary mutation found in the proviral DNA of cells transformed by M3 suggests a role for v-Rel/NF-κB1 heterodimers in transformation. The proline-to-valine substitution at aa 224 in M3R restored the transforming potential of M3, since M3R transformed normal splenic lymphocytes at levels comparable to v-Rel (Fig. 5B). Unlike M3, M3R demonstrated the ability to bind DNA as a homodimer and heterodimer with c-Rel, although it did not exhibit as high a level of homodimer DNA binding as did v-Rel (Fig. 4). However, M3R formed heterodimers with NF-κB1 more efficiently than v-Rel (Fig. 3). The enhanced interaction of M3R with NF-κB1 may compensate for the lower levels of homodimer formation, thereby allowing for the high transforming potential of this mutant. Previous studies demonstrated that the overexpression of NF-κB2 allowed for the transformation of lymphocytes by an otherwise nontransforming v-Rel dimerization mutant (v-Rel-SPW), suggesting a role for v-Rel/NF-κB2 heterodimers in transformation (59). Therefore, whereas DNA binding by v-Rel homodimers is likely a key factor for transformation, heterodimerization with endogenous Rel/NF-κB family members may also contribute to the ability of v-Rel to transform cells.
Although the results described in this study indicate that v-Rel homodimers are critical for transformation, the results do not establish that these homodimers are sufficient for this process. Previous studies employing v-rel transgenic mice suggested that v-Rel homodimers were sufficient for transformation in the murine system (7, 8). Murine cells transformed by v-Rel, however, contained a reduced assortment of v-Rel DNA-binding complexes, consisting only of v-Rel homodimers and heterodimers with NF-κB1. In addition, in contrast to the avian system, additional Rel/NF-κB DNA-binding complexes that did not contain v-Rel were also present in the nuclei of these cells and, therefore, may contribute to the transformed phenotype. There are also significant biological differences between the murine and avian systems which may preclude the conclusion that v-Rel homodimers would be sufficient for transformation of avian cells. While transgenic mice expressing v-rel driven by a murine T-cell-specific promoter develop T-cell tumors, they do so only after a protracted latent period (4 to 10 months). Furthermore, retroviruses expressing v-Rel fail to transform murine fibroblasts in culture or induce tumors in mice (54). Future experiments that reduce the activity of endogenous Rel/NF-κB proteins may define whether v-Rel heterodimers are required for transformation in the avian system. Previous studies demonstrated the ability of NF-κB1 mutants defective in DNA binding to function as transdominant-negative mutants and suggest the potential of the dimerization mutants described in this study to function in a similar manner (5, 38). The availability of mutants able to selectively compete with v-Rel for the limited supply of endogenous Rel/NF-κB proteins may provide additional insight into the role of v-Rel heterodimeric complexes in transformation.
Acknowledgments
We thank William Bargmann, Jarmila Kralova, and Emin Ulug for critical reading of the manuscript.
This work was supported by Public Health Service grants CA33192 from the National Cancer Institute. Andrew Liss was supported as an NIH predoctoral fellow on grant T32 CA09583 from the National Cancer Institute.
REFERENCES
- 1.Bartel, P. L., and S. Fields. 1997. The yeast two-hybrid system. Oxford University Press, New York, N.Y.
- 2.Barth, C. F., D. L. Ewert, W. C. Olson, and E. H. Humphries. 1990. Reticuloendotheliosis virus REV-T(REV-A)-induced neoplasia: development of tumors within the T-lymphoid and myeloid lineages. J. Virol. 64:6054-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehmelt, G., J. Madruga, P. Dorfler, K. Briegel, H. Schwarz, P. J. Enrietto, and M. Zenke. 1995. Dendritic cell progenitor is transformed by a conditional v-Rel estrogen receptor fusion protein v-RelER. Cell 80:341-352. [DOI] [PubMed] [Google Scholar]
- 4.Bose, H. R., Jr. 1992. The Rel family: models for transcriptional regulation and oncogenic transformation. Biochim. Biophys. Acta 1114:1-17. [DOI] [PubMed] [Google Scholar]
- 5.Bressler, P., K. Brown, W. Timmer, V. Bours, U. Siebenlist, and A. S. Fauci. 1993. Mutational analysis of the p50 subunit of NF-κB and inhibition of NF-κB activity by trans-dominant p50 mutants. J. Virol. 67:288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capobianco, A. J., D. Chang, G. Mosialos, and T. D. Gilmore. 1992. p105, the NF-κB p50 precursor protein, is one of the cellular proteins complexed with v-Rel oncoprotein in transformed chicken spleen cells. J. Virol. 66:3758-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrasco, D., P. Perez, A. Lewin, and R. Bravo. 1997. IκBα overexpression delays tumor formation in v-rel transgenic mice. J. Exp. Med. 186:279-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasco, D., C. A. Rizzo, K. Dorfman, and R. Bravo. 1996. The v-rel oncogene promotes malignant T-cell leukemia/lymphoma in transgenic mice. EMBO J. 15:3640-3650. [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, F. E., D. B. Huang, Y. Q. Chen, and G. Ghosh. 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature 391:410-413. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y.-Q., S. Ghosh, and G. Ghosh. 1998. A novel DNA recognition mode by the NF-κB p65 homodimer. Nat. Struct. Biol. 5:67-73. [DOI] [PubMed] [Google Scholar]
- 11.Cramer, P., C. J. Larson, G. L. Verdine, and C. W. Muller. 1997. Structure of the human NF-κB p52 homodimer-DNA complex at 2.1 Å resolution. EMBO J. 16:7078-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, N., W. Bargmann, M.-Y. Lim, and H. Bose, Jr. 1990. Avian reticuloendotheliosis virus-transformed lymphoid cells contain multiple pp59v-rel complexes. J. Virol. 64:584-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, N., S. Ghosh, D. L. Simmons, P. Tempst, H. C. Liou, D. Baltimore, and H. R. Bose, Jr. 1991. Rel-associated pp40: an inhibitor of the Rel family of transcription factors. Science 253:1268-1271. [DOI] [PubMed] [Google Scholar]
- 14.Diehl, J. A., T. A. McKinsey, and M. Hannink. 1993. Differential pp40IκBβ inhibition of DNA binding by rel proteins. Mol. Cell. Biol. 13:1769-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epinat, J. C., E. L. Dvorin, and T. D. Gilmore. 2000. Envelope-dependent transactivation by the retroviral oncoprotein v-Rel is required for efficient malignant transformation of chicken spleen cells. Oncogene 19:3131-3137. [DOI] [PubMed] [Google Scholar]
- 16.Epinat, J. C., D. Kazandjian, D. D. Harkness, S. Petros, J. Dave, D. W. White, and T. D. Gilmore. 2000. Mutant envelope residues confer a transactivation function onto N-terminal sequences of the v-Rel oncoprotein. Oncogene 19:599-607. [DOI] [PubMed] [Google Scholar]
- 17.Franklin, R. B., C.-Y. Kang, K. M.-M. Wan, and J. H. R. Bose. 1977. Transformation of chick embryo fibroblasts by reticuloendotheliosis virus. Virology 83:313-321. [DOI] [PubMed] [Google Scholar]
- 18.Fujii, M., T. Minamino, M. Nomura, K. I. Miyamoto, J. Tanaka, and M. Seiki. 1996. Selective activation of the proto-oncogene c-jun promoter by the transforming protein v-Rel. Oncogene 12:2193-2202. [PubMed] [Google Scholar]
- 19.Fujii, M., T. Minamino, M. Nomura, K. I. Miyamoto, J. Tanaka, and M. Seiki. 1997. v-Rel activates the proto-oncogene c-Jun promoter: a correlation with its transforming activity. Leukemia 11:402-404. [PubMed] [Google Scholar]
- 20.Garson, K., and C.-Y. Kang. 1990. Mapping of the functional domains of the v-rel oncogene. Oncogene 5:1431-1434. [PubMed] [Google Scholar]
- 21.Garson, K., H. Percival, and C. Y. Kang. 1990. The N-terminal env-derived amino acids of v-rel are required for full transforming activity. Virology 177:106-115. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh, G., G. Van Duyne, S. Ghosh, and P. B. Sigler. 1995. Structure of NF-κB p50 homodimer bound to a κB site. Nature 373:303-310. [DOI] [PubMed] [Google Scholar]
- 23.Gilmore, T. D. 1999. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene 18:6925-6937. [DOI] [PubMed] [Google Scholar]
- 24.Gilmore, T. D., and H. M. Temin. 1988. v-rel oncoproteins in the nucleus and in the cytoplasm transform chicken spleen cells. J. Virol. 62:703-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 26.Hrdlicková, R., J. Nehyba, and H. R. Bose, Jr. 1995. Mutations in the DNA-binding and dimerization domains of v-Rel are responsible for altered κB DNA-binding complexes in transformed cells. J. Virol. 69:3369-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hrdlicková, R., J. Nehyba, and E. H. Humphries. 1994. v-rel induces expression of three avian immunoregulatory surface receptors more efficiently than c-rel. J. Virol. 68:308-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, D., Y. Chen, M. Ruetsche, C. B. Phelps, and G. Ghosh. 2001. X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2. Structure 9:669-678. [DOI] [PubMed] [Google Scholar]
- 29.Huang, D.-B., T. Huxford, Y.-Q. Chen, and G. Ghosh. 1997. The role of DNA in the mechanism of NF-κB dimer formation: crystal structures of the dimerization domains of the p50 and p65 subunits. Structure 5:1427-1436. [DOI] [PubMed] [Google Scholar]
- 30.Kamens, J., P. Richardson, G. Mosialos, R. Brent, and T. Gilmore. 1990. Oncogenic transformation by v-Rel requires an amino-terminal activation domain. Mol. Cell. Biol. 10:2840-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 32.Kochel, T., J. F. Mushinski, and N. R. Rice. 1991. The v-rel and c-rel proteins exist in high molecular weight complexes in avian and murine cells. Oncogene 6:615-626. [PubMed] [Google Scholar]
- 33.Kralova, J., J. D. Schatzle, A. S. Liss, W. Bargman, and H. R. Bose, Jr. 1996. Synergistic stimulation of avian IκBα transcription by rel and fos/jun factors. Oncogene 12:2595-2604. [PubMed] [Google Scholar]
- 34.Kralova, J., A. S. Liss, W. Bargmann, and H. R. Bose, Jr. 1998. AP-1 factors play an important role in transformation induced by the v-rel oncogene. Mol. Cell. Biol. 18:2997-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar, S., A. B. Rabson, and C. Gélinas. 1992. The RxxRxRxxC motif conserved in all Rel/κB proteins is essential for the DNA-binding activity and redox regulation of the v-Rel oncoprotein. Mol. Cell. Biol. 12:3094-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis, R. B., J. McClure, B. Rup, D. W. Niesel, R. F. Garry, J. D. Hoelzer, K. Nazerian, and H. R. Bose, Jr. 1981. Avian reticuloendotheliosis virus: identification of the hematopoietic target cell for transformation. Cell 25:421-431. [DOI] [PubMed] [Google Scholar]
- 37.Lim, M. Y., N. Davis, J. Zhang, and H. R. Bose, Jr. 1990. The v-rel oncogene product is complexed with cellular proteins including its proto-oncogene product and heat shock protein 70. Virology 175:149-160. [DOI] [PubMed] [Google Scholar]
- 38.Logeat, F., N. Israël, R. Ten, V. Blank, O. Le Bail, P. Kourilsky, and A. Israël. 1991. Inhibition of transcription factors belonging to the rel/NF-κB family by a transdominant negative mutant. EMBO J. 10:1827-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore, B. E., and H. R. Bose, Jr. 1988. Expression of the v-rel oncogene in reticuloendotheliosis virus-transformed fibroblasts. Virology 162:377-387. [DOI] [PubMed] [Google Scholar]
- 40.Morrison, L. E., N. Kabrun, S. Mudri, M. J. Hayman, and P. J. Enrietto. 1989. Viral rel and cellular rel associate with cellular proteins in transformed and normal cells. Oncogene 4:677-683. [PubMed] [Google Scholar]
- 41.Mosialos, G., and T. D. Gilmore. 1993. v-Rel and c-Rel are differentially affected by mutations at a consensus protein kinase recognition site. Oncogene 8:721-729. [PubMed] [Google Scholar]
- 42.Mosialos, G., P. Hamer, A. J. Capobianco, R. A. Laursen, and T. D. Gilmore. 1991. A protein kinase A recognition sequence is structurally linked to transformation by p59v-rel and cytoplasmic retention of p68c-rel. Mol. Cell. Biol. 11:5867-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller, C. W., F. A. Rey, M. Sodeoka, G. L. Verdine, and S. C. Harrison. 1995. Structure of the NF-κB p50 homodimer bound to DNA. Nature 373:311-317. [DOI] [PubMed] [Google Scholar]
- 44.Nehyba, J., R. Hrdlicková, and H. R. Bose, Jr. 1997. Differences in κB DNA-binding properties of v-Rel and c-Rel are the result of oncogenic mutations in three distinct functional regions of the Rel protein. Oncogene 14:2881-2897. [DOI] [PubMed] [Google Scholar]
- 45.Nehyba, J., R. Hrdlicková, and E. H. Humphries. 1994. Evolution of the oncogenic potential of v-rel: rel-induced expression of immunoregulatory receptors correlates with tumor development and in vitro transformation. J. Virol. 68:2039-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson, D. M., J. J. Wahlfors, L. Chen, M. Onodera, and R. A. Morgan. 1998. Characterization of diverse viral vector preparations, using a simple and rapid whole-virion dot blot method. Hum. Gene Ther. 9:2881-2897. [DOI] [PubMed] [Google Scholar]
- 47.Pahl, H. L. 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 48.Rattner, A., M. Korner, H. Rosen, P. A. Baeuerle, and Y. Citri. 1991. Nuclear factor κB activates proenkephalin transcription in T lymphocytes. Mol. Cell. Biol. 11:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robzyk, K., and Y. Kassir. 1992. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 20:3790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sachdev, S., and M. Hannink. 1998. Loss of IκBα-mediated control over nuclear import and DNA binding enables oncogenic activation of c-Rel. Mol. Cell. Biol. 18:5445-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Schatzle, J. D., J. Kralova, and H. R. Bose, Jr. 1995. Avian IκBα is transcriptionally induced by c-Rel and v-Rel with different kinetics. J. Virol. 69:5383-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz, R. C., and O. N. Witte. 1988. A recombinant murine retrovirus expressing v-rel is cytopathic. Virology 165:182-190. [DOI] [PubMed] [Google Scholar]
- 55.Sengchanthalangsy, L. L., S. Datta, D. B. Huang, E. Anderson, E. H. Braswell, and G. Ghosh. 1999. Characterization of the dimer interface of transcription factor NFκB p50 homodimer. J. Mol. Biol. 289:1029-1040. [DOI] [PubMed] [Google Scholar]
- 56.Sif, S., and T. D. Gilmore. 1993. NF-κB p100 is one of the high-molecular-weight proteins complexed with the v-Rel oncoprotein in transformed chicken spleen cells. J. Virol. 67:7612-7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spector, D. L., R. D. Goldman, and L. A. Leinwand. 1998. Cells: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 58.Walker, W. H., B. Stein, P. A. Ganchi, J. A. Hoffman, P. A. Kaufman, D. W. Ballard, M. Hannink, and W. C. Greene. 1992. The v-rel oncogene: insights into the mechanism of transcriptional activation, repression, and transformation. J. Virol. 66:5018-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White, D. W., G. A. Pitoc, and T. D. Gilmore. 1996. Interaction of the v-Rel oncoprotein with NF-κB and IκB proteins: heterodimers of a transformation-defective v-Rel mutant and NF-κB p52 are functional in vitro and in vivo. Mol. Cell. Biol. 16:1169-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whiteside, S., and A. Israel. 1997. IkB proteins: structure, function and regulation. Semin. Cancer Biol. 8:75-82. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, J. Y., and H. R. Bose, Jr. 1989. Acquisition of new proviral copies in avian lymphoid cells transformed by reticuloendotheliosis virus. J. Virol. 63:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, J. Y., W. Olson, D. Ewert, W. Bargmann, and H. R. Bose, Jr. 1991. The v-rel oncogene of avian reticuloendotheliosis virus transforms immature and mature lymphoid cells of the B-cell lineage in vitro. Virology 183:457-466. [DOI] [PubMed] [Google Scholar]