Abstract
A series of recombinant viruses with either site-specific mutations or various deletions of the early UL4 promoter of human cytomegalovirus were used to determine the roles of regulatory elements and the effects of the mitogen-activated protein kinase (MAPK) pathways. Viral gene expression was regulated by upstream cis-acting sites and by basic promoter elements that respond to the MAPK signal transduction pathways. Inhibitors of either the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) pathway or the p38 MAPK pathway affected expression equally with either wild-type or mutant early UL4 promoters in the viral genome, indicating that the effects of the inhibitors are not exclusive for a single transcription factor. The minimal responsive element is the TATA box-containing early viral promoter.
Human cytomegalovirus (HCMV), a member of the betaherpesvirus family, is a ubiquitous pathogen that infects only humans. In healthy individuals, primary infection is normally asymptomatic and persistent. In immunocompromised adults, such as transplantation recipients or human immunodeficiency virus-infected patients, pneumonitis, hepatitis, retinitis, gastrointestinal diseases, and other severe medical problems can develop. Congenital birth defects, which include sight and hearing loss and brain damage, can occur (3). HCMV replicates productively in terminally differentiated cells, including fibroblasts, epithelial, endothelial, smooth muscle, and microglial cells and monocyte-derived macrophages (64, 66). HCMV latent viral DNA can be detected in macrophage-granulocyte progenitors in the bone marrow and in the peripheral blood monocytes (25, 44, 52, 57, 67).
During productive infection, HCMV gene expression can be divided into three categories designated immediate-early (IE, α), early (β), and late (γ). The IE genes include major IE genes UL123/122 (IE1/IE2) and auxiliary IE genes, such as UL36 to UL38, UL115 to UL119, IRS1/TRS1, and US3. The IE proteins are required for subsequent early viral gene expression. The early genes code for proteins required for viral DNA replication. After viral DNA replication, the transcription of late genes occurs, the products of which are involved in virion assembly and maturation (17, 19, 68).
Two major IE proteins, IE72 and IE86, which are encoded by the IE1 (UL123) and IE2 (UL122) genes, respectively, are key transactivators for HCMV early gene expression and other heterologous viral promoters as demonstrated by transient-transfection assays (17, 68). Compared to the IE86 protein, the IE72 protein alone is a weak transactivator, but it is capable of synergistically activating viral promoters with IE86. Although the IE72 protein is not essential for early viral gene expression after a high multiplicity of infection (MOI), it is required after a low MOI (23). In contrast, the IE86 protein is essential for early viral gene expression (14, 53). The IE86 protein upregulates early viral promoters in transient-transfection experiments in a TATA box-dependent manner. The TATA box and the transcription initiation complex are assumed to be critical for viral gene expression, because the IE86 fusion protein produced in bacteria interacts in vitro with members of the basal transcription machinery, which include TATA-binding protein (TBP), TFIIB, TAFII-130, and TFIID (9, 18, 24, 39, 40, 49). The IE86 protein also interacts with other transcription factors, such as Tef-1, Sp1, c-Jun, JunB, ATF-2, CREB, histone acetyltransferase CREB-binding protein (CBP)-associated factor (P/CAF), and p53 (7, 47, 50, 65, 69). cis-acting regulatory elements upstream of the early viral promoters have a role in maximum viral gene expression. For example, the USF/MLTF, AP-1, NF-Y, Sp1, and CREB/ATF cis-acting regulatory elements have an effect on the TRL4, TRL6, UL4, UL54, and UL112-113 promoters in transient transfection assays, respectively (30, 41-43, 47, 51, 70, 79, 83).
The mitogen-activated protein kinase (MAPK) signal transduction pathways respond to various extracellular stimuli, ranging from growth factors and cytokines to cellular stress (60, 63, 82). The MAPK signal transduction cascade consists of a three-component module consisting of MAPK, MAPK kinase (MEK, MAPKK, or MKK), and MAPK kinase kinase (MEKK, MAPKKK, or MKKK), which is conserved from yeasts to humans. The MAPK cascade is activated by sequential phosphorylation. The MAPKs are activated by dual phosphorylation of the Thr-X-Tyr motifs. In mammalian cells, there are five subfamilies known as the extracellular signal-regulated kinase (MAPK/ERK or ERK1/2), p38 MAPK (p38α, p38β, p38γ, and p38δ), stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK)(1/2/3), ERK5, and ERK3/4 (78, 82). Activation of MAPK/ERK and p38 MAPK has been reported to be important for productive HCMV replication (34-36, 61). The MAPK/ERK pathway is activated at 4 h postinfection (hpi), and the p38 MAPK pathway is activated at 8 hpi. Both early viral gene expression and late viral gene expression are enhanced by the MAPK/ERK and p38 MAPK pathways (35, 36, 61).
The MAPK/ERK pathway activates nuclear transcription factors, such as Elk-1, c-fos, c-myc, Sap-1, and Tal (13, 20, 33, 82). The p38 MAPK pathway activates nuclear transcription factors, such as Elk-1, Sap-1, ATF-2, CREB, CHOP, and Max (54, 59, 62, 80-82). The phosphorylation of these cellular transcription factors increases their DNA binding or protein interaction capabilities. The MAPKs also activate viral transactivators, which are important in the activation of early viral promoters. For example, ERK phosphorylates both IE72 and IE86 proteins in vitro and in vivo (17, 27). In addition, MAPK/ERK kinase (MEK1/2) inhibitors UO126 and PD98059 repress phosphorylation of IE72 and IE86 proteins in infected cells without decreasing the level of IE proteins (36, 61).
The viral UL4 gene codes for a virion-associated glycoprotein of unknown function (11). The early UL4 gene of HCMV has been used as a model system for studying the multiple mechanisms of activation of viral gene expression. Expression of the UL4 gene is regulated at both the transcriptional and translational levels (2, 8, 12, 30, 31). To further understand the mechanisms of UL4 gene expression in the context of the viral genome, we constructed a series of recombinant HCMVs with either wild-type or mutated upstream sequence elements of the UL4 promoter. We determined the effect of the MAPK/ERK and p38 MAPK pathways on early viral gene expression. We report that the Elk-1 binding site, the sequences between −102 and −50 upstream of the transcription start site, and the TATA box all have a role in the activation of the UL4 promoter in the context of the viral genome. All elements were affected by the MAPK/ERK and p38 MAPK pathways.
MATERIALS AND METHODS
Virus and cell culture.
The maintenance of primary human foreskin fibroblasts (HFFs) was described previously (72). The maintenance and propagation of HCMV AD169 and AD169 strain-derived recombinant viruses were described previously (38, 71).
Enzymes.
Restriction endonucleases were purchased from New England Biolabs, Inc. (Beverly, Mass.). T4 DNA ligase, the Klenow fragment of Escherichia coli DNA polymerase I, and calf intestinal alkaline phosphatase were obtained from Boehringer Mannheim Biochemicals (Indianapolis, Ind.). Vent and Taq DNA polymerases were purchased from New England Biolabs, Inc., and Fisher Scientific (Pittsburgh, Pa.), respectively. RNasin and RNase-free DNase were purchased from Promega (Madison, Wis.). The enzymes were used according to the manufacturers' instructions.
Plasmids.
Plasmid pwt-xs has been described previously (12). pwt-xs contains the UL4 (E1) promoter and 220 bp upstream of the transcription start site and the downstream chloramphenicol acetyltransferase (CAT) reporter gene. pwt-xs has a 1,411-bp XbaI-SacI DNA fragment (bp 200392 to 201803) of HCMV Towne strain containing the US12 and US13 genes and a 1,205-bp HindIII-BamHI DNA fragment (bp 195,838 to 197,043) containing the US6 and US7 genes flanking the promoter and CAT gene (12). To generate pwt-AatII/S, a 76-bp XbaI-to-AatII DNA fragment (bp 200392 to 200468) containing the US11 TATA box was deleted from pwt-xs. Plasmid pwt-xs was double digested with XbaI and AatII, and then the digested plasmid was religated back with an 8-mer linker oligonucleotide fragment, 5′-CTAGACGT-3′, with the 5′ end phosphorylated.
To generate pdlElk-1-AatII/S, two fragments were generated by PCR with two sets of primers: primer 5′-GTAGCacactaGtcaTGGAATCGTTCGGCT-3′ with primer 5′-GCCATACGGAATTCCGGATGAGCA-3′ and primer UL4 SEQ 5′-GCCCCCATCTGGTATCCAA-3′ with primer 5′-GATTCCAtgaCtagtgtGCTACATACCTGCCA-3′. (Mutant bases are indicated by lowercase letters.) Plasmid pwt-AatII/S was used as the template. The PCR fragments of 440 and 221 bp, respectively, were digested with SpeI to generate subfragments of 433 and 210 bp, respectively, and then ligated at room temperature overnight. A 643-bp DNA fragment containing the two subfragments was gel purified and double digested with XbaI and BspEI. The resulting 484-bp fragment was isolated and replaced the 484-bp XbaI-BspEI DNA fragment of pwt-AatII/S to generate plasmid pdlElk-1-AatII/S. Plasmid pdlElk-1-AatII/S contains a mutation in the consensus Elk-1 binding site, which changes the Elk-1 consensus from wild-type GTATCCGGTT to mutant acactaGtca, but leaves intact the potential IE86 protein binding site within the site 2 region (12).
For construction of truncation mutations of the UL4 promoter, a series of oligonucleotides were employed in PCR-directed mutagenesis with pwt-AatII/S as the template. The sense primers for p-137, p-102, p-50, and p-23 are 5′-CGGCTTCTAGACGGGGGATAGTGAG-3′, 5′-GGGATTCTAGAGGACCCAATCACT-3′, 5′-GATGATTCTAGAATCACATGAGGTCTGG-3′, and 5′-GGATATGTCTAGATGAGGAGTGAA-3′, respectively. The antisense primer for these plasmids was 5′-GCCATACGGAATTCCGGATGAGCA-3′. DNA fragments of 422, 387, 336, and 308 bp were generated by PCR, isolated by low-melting-point agarose gel electrophoresis, and digested with XbaI and BspEI. The resulting 399-, 364-, 312-, and 285-bp fragments replaced the same site in pwt-AatII/S to generate plasmids p-137, p-102, p-50, and p-23, respectively. All primers were purchased from Life Technologies (Grand Island, N.Y.) unless otherwise specified. All mutations were confirmed by automated dideoxynucleotide sequencing (University of Iowa DNA Core).
HCMV recombination and plaque purification.
The isolation of recombinant virus RVUL4CATgpt, which has a 2,115-bp BsrGI-BamHI DNA fragment containing the guanine phosphoribosyltransferase gene (gpt) under the control of a minimal simian virus 40 (SV40) promoter and the CAT gene inserted between the downstream US6/7 and upstream US12/13 genes, has been described previously (12). Recombinant viruses RVwt, RVdlElk-1, RV-137, RV-102, RV-50, and RV-23 were generated by the back-selection method of Greaves et al. (22). HFFs were cotransfected with infectious RVUL4CATgpt viral DNA and plasmid pwt-AatII/S, pdlElk-1-AatII/S, p-137, p-102, p-50, or p-23. After 100% cytopathic effect (CPE), virus was harvested and diluted 1:10 or 1:20. Lesch-Nyhan cells GM02291 (Coriell Cell Repository, Camden, N.J.) in 100-mm-diameter plates in medium containing 6-thioguanine (50 μg/ml; Sigma, St. Louis, Mo.) were infected. Virus plaques were picked and transferred to HFFs grown in 24-well plates. Cell-associated viral DNA was screened by dot blot hybridization, and cell lysates were screened by CAT assay as described previously (12, 21, 56). Positive plaques were subjected to two additional rounds of plaque purifications. Two or three recombinant virus clones were obtained from independent transfection-recombination procedures to control for spurious genomic mutations. All recombinant viruses had the same growth characteristics and were propagated as described previously (71).
Southern blot analysis.
Southern blot analysis of the recombinant virus genome structure was performed essentially as described previously with minor modifications (12). Four days after 100% CPE, viral supernatant was collected, and virus particles were pelleted by centrifugation as described previously (71). The viral DNA was purified from the viral pellet as described previously (56). Viral DNAs were digested with restriction endonuclease HindIII or double digested with HindIII and SpeI. DNAs were fractionated in a 3% agarose gel with a 1:4 ratio of agarose to NuSieve GTG agarose (BioWhittaker Molecular Applications, Rockland, Maine). HindIII and SpeI double-digested viral DNAs were loaded onto a 2% gel (1:3 ratio of regular agarose to GTG agarose) and subjected to electrophoresis at 40 V for 6 h at 4°C. The DNA was transferred to Maximum Strength Nytran (Schleicher & Schuell, Keene, N.H.), and Southern blot analysis was performed as described previously (4, 56). All probes used in dot blotting as well as Southern blot analysis were prepared by randomly labeling gel-purified DNA fragments with [32P]dCTP (Amersham, Arlington Heights, Ill.) and Ready-To-Go DNA labeling beads from Amersham Pharmacia Biotech, Inc. (Piscataway, N.J.). Unincorporated nucleotides were removed by a Nuctrap purification column (Stratagene, Cedar Creek, Tex.). The 32P-HindIII X probe was prepared with the 5,019-bp HindIII-HindIII DNA fragment of pMSDT DG (75). The 32P-AA probe was prepared by labeling the 110-bp AatII-AvrII fragment isolated from pwt-xs (12). The 32P-gpt probe was generated by labeling the 262-bp KpnI-EcoRV fragment isolated from p dlMSVgpt (56).
RNase protection assays.
The methods used to generate the plasmid DNA templates for synthesis of antisense CAT, actin, and IE1 riboprobes have been described previously (28, 48, 76). The riboprobes were synthesized with [32P]UTP (Amersham) as described previously (45).
Cytoplasmic RNA was harvested from four 100-mm-diameter plates of HFFs either mock infected or infected with HCMV recombinant viruses at an MOI of 5 PFU/cell at various times after infection. Twenty micrograms of RNA was hybridized to 32P-labeled antisense CAT, IE1, or actin probes at room temperature overnight before digestion with RNase T1 (150 U) as described previously (48). The protected RNA fragments were subjected to electrophoresis in denaturing 6% polyacrylamide gels, followed by autoradiography on Hyperfilm MP (Amersham). Signals were quantitated by an electronic Autoradiographic Instant Imager (Packard Instant Imager, Meridan, Conn.).
CAT assays.
HFFs seeded in 100-mm-diameter plates were infected with recombinant viruses in quadruplicate. At indicated time points after infection, cell lysates were harvested and subjected to CAT assays as described previously (21). Acetylated derivatives were separated from nonacetylated 14C-chloramphenicol by thin-layer chromatography. The percent conversion of 14C-chloramphenicol to acetylated derivatives was determined by image acquisition analysis and normalized to the amount of cell lysate protein in each reaction. The protein concentration was measured by the Bradford method (Bio-Rad Laboratories, Hercules, Calif.).
For infection of HFFs in the presence of the MEK inhibitor UO126 (Promega) or the p38 MAPK inhibitor SB202190 (FHPI) (Calbiochem Corp., San Diego, Calif.), the drugs were dissolved in dimethyl sulfoxide (DMSO) to make a 10 mM stock as recommended by the manufacturer. Various amounts of the 10 mM stock drug were diluted in serum-free medium to be used in subsequent treatment. Confluent HFFs in 100-mm-diameter plates in quadruplicate were serum starved for 24 h and then preincubated with various concentrations of the inhibitors in serum-free medium at 37°C for 1 h before infection with recombinant viruses in the presence of the inhibitors. The desired concentration of drug was present throughout the experiment. The amount of DMSO applied on each plate was balanced and did not exceed 0.2%. Cells were harvested at the indicated time points after infection (8 hpi for UO126 and 14 hpi for FHPI), and CAT assays were carried out as described previously (21).
Northern blot analysis.
HFFs were washed with phosphate-buffered saline and serum starved for 24 h. Before infection with RVwt, HFFs were either mock treated or treated with UO126 or FHPI as described above. At indicated time points after infection, cytoplasmic RNAs from mock-treated or inhibitor-treated HFFs were isolated as described previously (10, 29). Ten micrograms of cytoplasmic RNA was subjected to electrophoresis in a 1% agarose gel containing 2.2 M formaldehyde at 5 V/cm and transferred to Nytran Super Charge (Schleicher & Schuell). Equal loading of the RNAs was confirmed by visualization of ethidium bromide staining of the 28S and 18S rRNAs. Northern blot analysis was performed as described previously (12, 56). UL4-, CAT-, and actin-specific DNA probes were derived from the 230-bp AvaII-DraI DNA fragment of pEgp48, corresponding to 13,630 to 13,860 bp of the HCMV genome (10, 12), the 403-bp PvuII-NcoI DNA fragment of p-220CAT, and the 810-bp PstI-PstI fragment of pBR-actin (26, 56), respectively. All DNA probes were labeled with the Ready-To-Go DNA labeling beads and [32P]dCTP (Amersham). The same blot was serially stripped in 0.1× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) and 0.5% sodium dodecyl sulfate and rehybridized with different probes. All hybridization was performed overnight at 68°C.
Cell toxicity assays.
To measure cytotoxic effects of the MEK inhibitor UO126 or the p38 MAPK inhibitor FHPI, the CytoTox 96 assay kit (Promega) was used according to the manufacturer's instructions. The CytoTox 96 assay kit quantitatively measures a cytosolic enzyme, lactate dehydrogenase (LDH), which is released upon cell lysis. A total of 10,000 cells were plated in a 96-well tissue culture plate in quadruplicate and serum starved for 24 h, then serum-free medium containing various concentrations of drug was present for either 9 h for UO126 or 15 h for FHPI before the cytotoxicity assays. The released LDH is able to convert the substrate tetrazolium salt into a red formazan product, which can be measured at 492 nm in a 96-well tissue culture plate. The value of LDH released by cells after drug treatment divided by the value of LDH released after complete cell lysis was used to determine the percent cytotoxicity.
RESULTS
Recombinant viruses with UL4 promoter mutations.
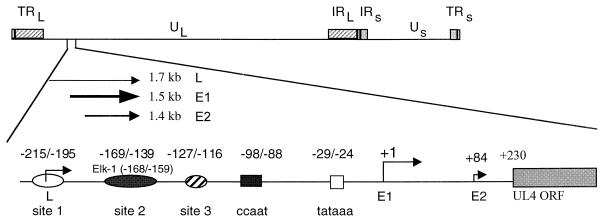
DNase I footprinting analysis demonstrated three cellular protein binding sites, designated sites 1, 2, and 3, upstream of the UL4 transcription start site of HCMV (31) (Fig. 1). Site 2 (−169/−139) and an NF-Y binding site (−98/−88) were important for promoter activation in transient-transfection assays (30, 31). In addition, recombinant HCMVs with mutations in an Elk-1 site plus a potential IE86 protein binding site within the site 2 region also decreased expression from the UL4 promoter in the context of the viral genome (12). However, mutations in the NF-Y binding site had no effect in the context of the viral genome. To determine the role of upstream cis-acting sites, we performed site-directed mutagenesis of the Elk-1 binding site and constructed a series of deletion mutations upstream of the UL4 transcription start site. The recombinant viruses were derived from RVUL4CATgpt by the method of back-selection against gpt (22). The UL4 promoter drives transcription of the CAT gene (Fig. 2A). The UL4 promoter-CAT constructs were inserted by homologous recombination between the US7 to US12 region of the viral genome, which has been shown to be dispensable for viral replication in tissue culture (37, 38).
FIG. 1.
Schematic representation of the early viral UL4 transcription unit. The UL4 transcription unit in its natural locus drives the synthesis of three unspliced RNAs. Three upstream cellular protein binding sites, 1, 2, and 3, are shown by open, solid, and hatched ovals, respectively. For the E1 (early) viral UL4 promoter, the imperfect dyad NF-Y binding site (CCAAT box) and the TATA box are designated by the solid and open boxes, respectively. The numbers above the symbols are relative to the transcription start site (arrow) of E1.
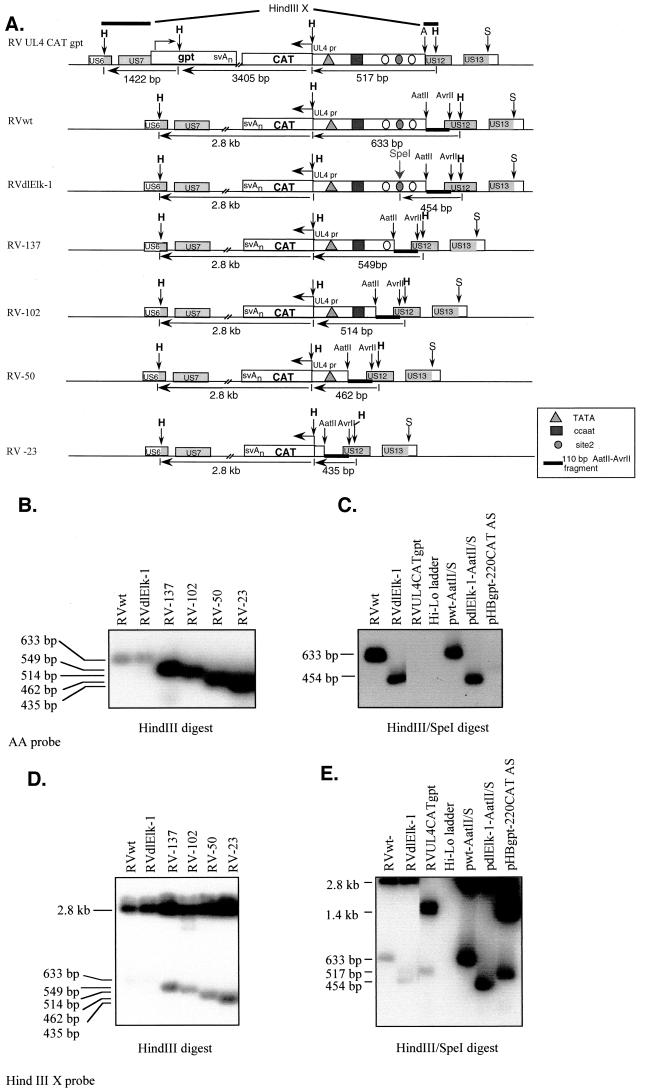
FIG.2.
Southern blot analysis of recombinant viruses. (A) Schematic genome maps of RVUL4CATgpt, RVwt, RVdlElk-1, RV-137, RV-102, RV-50, and RV-23. The UL4-CAT in its ectopic location is oriented according to the prototype position of the US genes, where transcription is from right to left. The sizes of the DNA fragments resulting from HindIII and SpeI restriction endonuclease digestion are indicated in base pairs. The genes involved in homologous recombination in shuttle vectors are shown in shaded boxes. A, S, and H represent restriction endonuclease sites AvrII, SacI, and HindIII, respectively. (B to E) Autoradiograms of Southern blots to identify the recombinant viruses by using either the 32P-labeled 110-bp AatII-Avr II DNA (AA probe [B and C]) or HindIII X probe (D and E). Lanes containing viral DNA fragments from different recombinant viruses were spliced together from the same gel. Shuttle vectors pwt-AatII/S, pdlElk-1-AatII/S, and pHBgpt-220CAT AS were used as controls. Panels D and E result from stripping the AA probe from panels B and C, respectively, and then hybridization with the HindIII X probe.
The genome structures of the recombinant viruses were analyzed by HindIII restriction endonuclease digestion of viral DNAs followed by Southern blot hybridization with a 110-bp 32P-labeled AatII-AvrII DNA (AA) probe (Fig. 2A). The predicted sizes of viral DNA fragments are indicated in Fig. 2A. The resulting recombinant viruses differ from the parental RVUL4CATgpt by the presence of the AatII-AvrII DNA fragment and the absence of the gpt gene. Site specific mutations within the site 2 region of the UL4 promoter were confirmed by DNA sequencing of the shuttle vector and by double restriction endonuclease digestion of the recombinant viral DNAs as shown in Fig. 2B and C. The flanking regions of the recombinant viruses were determined to be intact by using the HindIII X probe (Fig. 2A), which spans part of the US6 and US12 in the recombinant viruses (Fig. 2D and E).
In addition to analyzing the genome structure of the recombinant viruses by Southern blotting, we determined that the CAT open reading frame (ORF) of each recombinant virus does not contain functionally deleterious mutations. All recombinant viruses analyzed had a functional CAT ORF determined by CAT assays as described in Materials and Methods (data not shown).
Effect of the Elk-1 binding element.
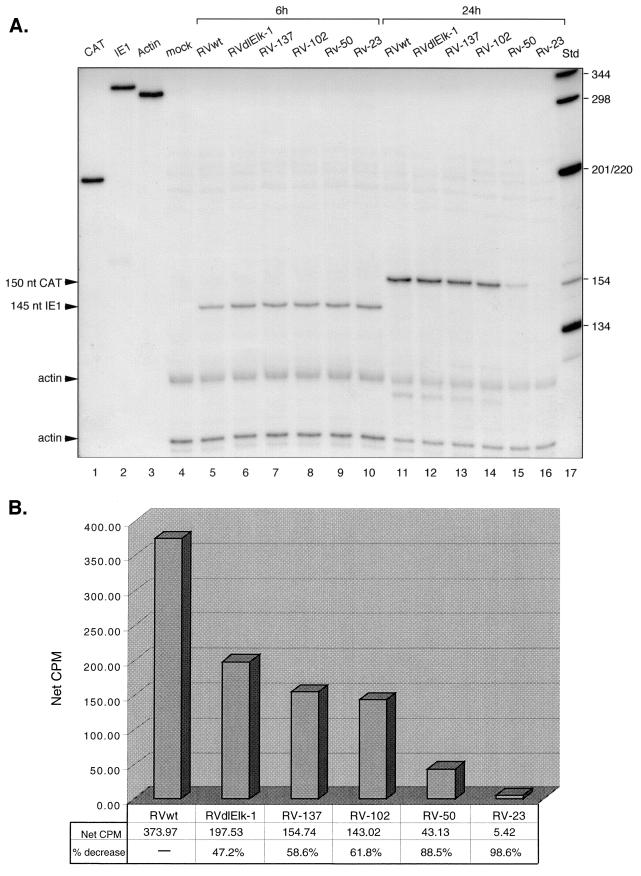
We previously reported that mutations in the Elk-1 site plus a proximal putative IE86 binding site (between −148 and −136 relative to the E1 transcription start site) within the site 2 region upstream of the UL4 promoter decreased promoter activity in HCMV-infected cells (12). To determine whether the Elk-1 binding site itself was responsible for the reduction of the UL4 promoter activity, HFF cells were infected at 5 PFU/cell with either wild-type (RVwt) or mutant (RVdlElk-1) virus and then analyzed for steady-state levels of RNA transcribed from the UL4 promoter. Cytoplasmic RNAs were isolated from infected cells at very early (6 hpi) or early (24 hpi) times after infection and subjected to RNase protection assays as described in Materials and Methods. The internal control for multiplicity of viral infection is the protected IE1 RNA at 6 hpi (Fig. 3). The protected CAT RNA level from RVwt-infected cells was higher than that from RVdlElk-1 infected cells at 24 hpi. The level of protected CAT RNA of RVdlElk-1 exhibited an approximately 50% reduction relative to wild-type virus, as shown in Fig. 3B. A 50% reduction in the level of UL4 protein, which is nonessential for viral replication in cell culture, may be important for viral pathogenesis. We conclude that the Elk-1 binding site alone plays a role in UL4 promoter activation in the context of the viral genome.
FIG. 3.
Steady-state RNA levels transcribed from the UL4-CAT promoter with either the wild type or various mutations at early times after infection. Cytoplasmic RNA was isolated at the indicated time points after HFFs were infected with RVwt, RVdlElk-1, RV-137, RV-102, RV-50, or RV-23 at approximately 5 PFU/cell and then subjected to the RNase protection assay as described in Materials and Methods. (A) Autoradiogram of RNase protection assay. Lanes: 1 to 3, 32P-labeled CAT, IE1, and actin riboprobes not treated with RNase, respectively; 4, mock infected; 5 to 10, RVwt, RVdlElk-1, RV-137, RV-102, RV-50, and RV-23 at 6 hpi, respectively; 11 to 16, RVwt, RVdlElk-1, RV-137, RV-102, RV-50, and RV-23 at 24 hpi, respectively; 17, 32P-labeled DNA standard molecular weight markers (Std). For lanes 4 to 10, 150 cpm of both 32P-labeled IE1 and actin antisense probe was used. For lanes 11 to 16, 150 cpm of both 32P-labeled actin and CAT antisense probe was used. The sizes of the protected CAT and IE1 RNAs are indicated. nt, nucleotides. (B) Image acquisition analysis of steady-state levels of CAT RNA. The IE1 signal at 6 hpi or the CAT RNA signals at 24 hpi were first normalized to the corresponding actin signals. The resulting numbers were used to determine the final normalized CAT RNA signal. Net cpm, total cpm for an equal area.
Effect of deletion mutations upstream of the UL4 promoter.
To test for other cis-acting elements that control the expression from the early viral UL4 promoter, we constructed a series of recombinant HCMVs that contain various deletions. RV-137, RV-102, and RV-50, respectively, contain UL4 promoter with sites 1 and 2 deleted, with sites 1, 2, and 3 deleted, and with sites 1, 2, and 3 plus the NF-Y binding site deleted but with an intact TATA box (Fig. 1 and 2A). RV-23 has only 23 bp upstream of the transcription start site and is a recombinant virus without the TATA box upstream of the UL4 early promoter. We determined the steady-state level of RNA transcribed from the UL4-CAT promoter in HFFs infected at 5 PFU/cell with either the wild type (RVwt) or the various mutants. Cytoplasmic RNAs were isolated from infected cells at very early (6 hpi) or early (24 hpi) times after infection and RNase protection assays were performed. The steady-state level of CAT RNA from RV-137 or RV-102 was reduced approximately 50 to 60% compared to that of the wild type (Fig. 3A, lanes 13 and 14, and B). This level of reduction was similar to that of RVdlElk-1 when compared to the wild type (Fig. 3B).
With the three upstream sites deleted plus the NF-Y binding site (−98/−88) deleted in recombinant virus RV-50, the steady-state level of CAT RNA was decreased by 88.5% (Fig. 3A, lane 15, and B). These results indicated that the upstream sequences from −102 to −50 have an important regulatory element. Although RV-50 contains only the minimal promoter region (TATA box containing) without known upstream elements, it still had basal transcriptional activity. These results suggested that the basal transcription machinery was able to bind to the TATA box to initiate transcription from the UL4-CAT promoter in the absence of known upstream activators.
RV-23, which has the TATA box deleted and has the sequence up to −23 relative to the transcription start site, had a 98.6% reduction in the steady-state CAT RNA level in virus-infected cells (Fig. 3A, lane 16, and B). Although previous transient-transfection assays have demonstrated that activation of early HCMV promoters is TATA box dependent, this is the first report that demonstrated this notion in the context of the HCMV genome.
Cumulative effects on CAT gene expression.
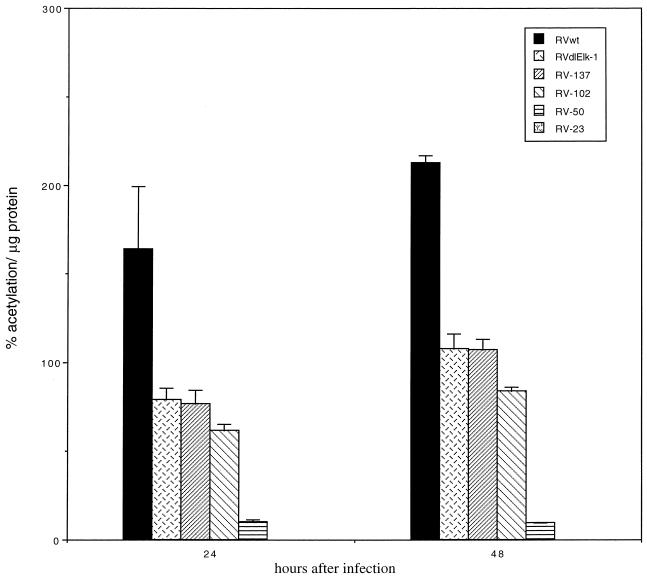
Since the CAT RNA turns over rapidly and the CAT protein is very stable in the mammalian cell, we assayed the cumulative effects of the various mutations upstream of the UL4-CAT promoter by determining the CAT activity. HFFs were infected with RVwt, RVdlElk-1, RV-137, RV-102, RV-50, or RV-23 at approximately 5 PFU/cell. Equal infectivity was established by RNase protection assays for the IE1 RNA in HFFs as shown in Fig. 3. CAT assays were performed to determine CAT activity as described in Materials and Methods.
At both early (24 hpi) and late (48 hpi) times after infection, the CAT activity of RVwt was the highest. When the Elk-1 binding site was mutated, the CAT activity was reduced compared to that of the wild type at both 24 and 48 hpi (Fig. 4 [see RVdlElk-1]). After considering the IE1 RNA signal from the RNase protection assays to normalize for MOI difference, the CAT activity of RVdlElk-1 repeatedly exhibited a 50% reduction compared to that of the wild type. These results were consistent with the RNase protection data (Fig. 3A and B) and indicated that the Elk-1 binding site of the UL4-CAT promoter is a positive element in the context of the viral genome. Mutations in both RV-137 and RV-102 decreased the CAT activity to 50 to 60% of the wild-type level. This reduction could be due to the deletion of the Elk-1 binding element, as demonstrated by RVdlElk-1 described above. The deletion of sequences between −220 and −50 had approximately a 95% reduction compared to the wild type (Fig. 4 [see RV-50]). RV-23, which has the TATA box deleted, had little to no CAT activity. These data support the results of the RNase protection assays (Fig. 3). Taken together, these results demonstrate that both the Elk-1 binding element and the sequences between −102 and −50 relative to the transcription start site were upstream cis-acting sites for the activation of the UL4-CAT promoter in virus-infected cells. The activation of the UL4-CAT promoter in the context of the viral genome was also TATA box dependent.
FIG. 4.
Effect of deletion mutations on CAT expression from the UL4-CAT promoter in infected HFFs. HFFs were infected with recombinant viruses at 5 PFU/cell in quadruplicate as described in the legend to Fig 3. The infection was performed in parallel with that of the RNase protection assay as described in Fig. 3. Equal MOI was confirmed by RNase protection assay of protected IE1 mRNA. The CAT activity per microgram of protein was normalized to IE1 mRNA for each recombinant virus at different time points after infection as described in Materials and Methods. The results of the CAT assays were averaged, and the standard deviations were calculated.
Effect of the MAPK/ERK and p38 MAPK signal transduction pathways on UL4-CAT expression.
Since HCMV infection activates both the MAPK/ERK and p38 MAPK pathways, but not the SAPK/JNK pathway (34, 36, 61), we tested the effect of the MAPK/ERK and p38 MAPK pathways by using inhibitors UO126 and SB202190 (FHPI), respectively. UO126 inhibits the (MAPK/ERK kinase) MEK activity by preventing the activation of MEK by upstream Raf kinase and also blocks the catalytic activity of preexisting activated MEK to activate downstream MAPK/ERK (15). FHPI inhibits the α and β isoforms of the p38 kinase activity by competing for the ATP binding site of p38 (58, 84).
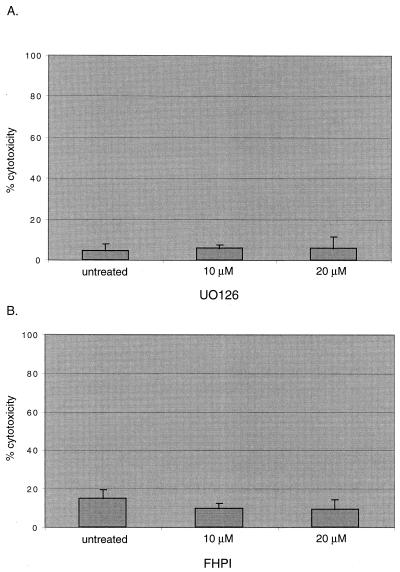
First, we determined the cytotoxic effect of the kinase inhibitors on HFF cells. To eliminate the stimulatory effects of serum in the media, HFFs were serum starved and either mock treated or treated with various concentrations of the inhibitors. Cytotoxic effects were determined by measuring the release of LDH as described in Materials and Methods. Both UO126 and FHPI had similar cytotoxic effects compared to untreated cells (Fig. 5). It was expected that the longer incubation time with FHPI would result in a higher cytotoxic effect. It was also expected that the two different drugs together would have an additive cytotoxic effect.
FIG. 5.
Cytotoxicity assays of MAPK inhibitor-treated cells. HFFs in 96-well plates were serum starved for 24 h before treatment with either UO126 for 9 h or FHPI for 15 h in quadruplicate. The cytotoxicity effect of the drug treatment was determined as described in Materials and Methods. (A) Effect of UO126 treatment. (B) Effect of FHPI treatment.
To determine whether the MAPK/ERK pathway or the p38 MAPK pathway has an effect on the activation of the UL4-CAT promoter, we pretreated serum-starved HFFs with the kinase inhibitors for 1 h before infection. The cells were infected with 5 PFU/cell in the presence of the inhibitor. MAPK/ERK is phosphorylated at 4 hpi and reaches high levels of phosphorylation at 8 to 24 hpi (61), but the p38 MAPK is phosphorylated at 10 hpi, and levels of phosphorylation decrease between 14 and 24 hpi (34, 36, 61). Therefore, cell lysates were harvested at either 8 or 14 h after infection to determine the early effects of the MEK inhibitor or the p38 inhibitor, respectively. CAT assays were performed as described in Materials and Methods. The percent acetylation per microgram of protein was compared between untreated and drug-treated cells.
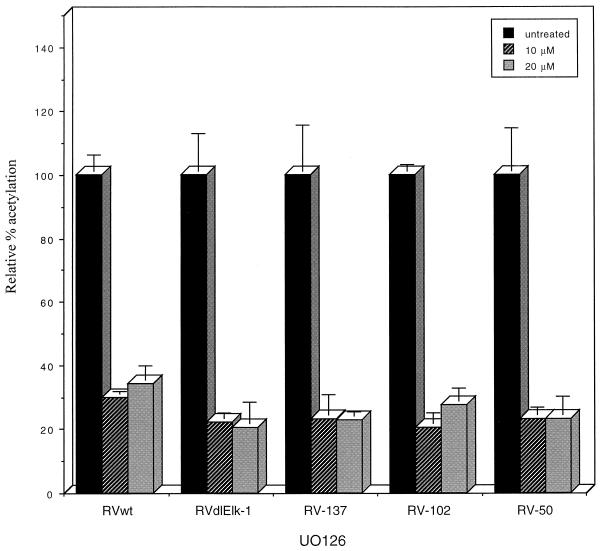
In both cells treated with 10 μM UO126 and those treated with 20 μM UO126, there was an approximately 70 to 80% reduction in UL4 promoter activity for all recombinant viruses compared to recombinant virus-infected cells without drug treatment (Fig. 6). Another MEK inhibitor, PD98059, also decreased UL4-CAT expression by approximately 50% (data not shown). Western blot analysis confirmed previous reports demonstrating that the MEK inhibitors did not affect the level of expression of the viral IE72 and IE86 proteins (36, 61; data not shown).
FIG. 6.
Effect of a MEK inhibitor, UO126, on CAT expression from the UL4-CAT promoter in recombinant virus-infected HFFs. Confluent HFFs were serum starved for 24 h and then treated with UO126 for 1 h before infection with recombinant viruses in the presence of UO126 in quadruplicate as described in Materials and Methods. Cells were harvested at 8 h after infection and subjected to CAT assays. The results of the CAT assays were averaged, and the standard deviations were calculated. The CAT activity per microgram of protein at various concentrations of the drug is relative to that of the nontreated recombinant viruses.
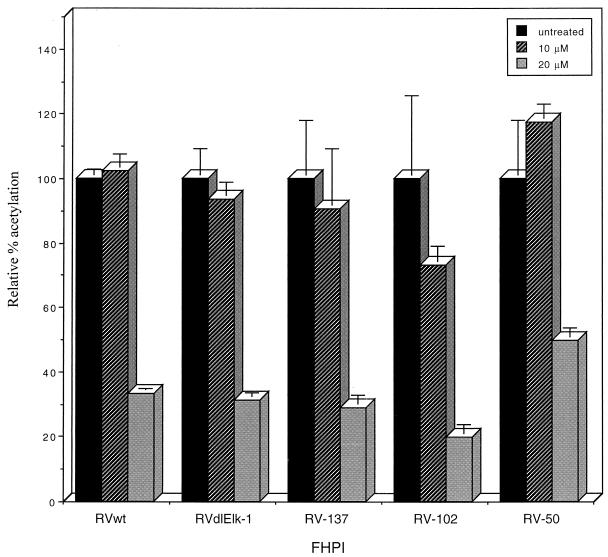
In 10 μM FHPI-treated cells, there was little to no significant reduction in CAT activity. In 20 μM FHPI-treated cells, all recombinant viruses exhibited a 50 to 80% reduction compared to recombinant virus-infected cells without drug treatment (Fig. 7). The UL4-CAT expression decreased almost equally upon drug treatment for recombinant viruses with either wild-type or mutant UL4 promoters, indicating that the effects of the MAPK/ERK and p38 MAPK are not exclusive for a single transcription factor. We conclude that the UL4-CAT promoter is a MAPK-responsive promoter and the minimal responsive element is the TATA box-containing promoter.
FIG. 7.
Effect of p38 MAPK inhibitor SB202190 (FHPI) on CAT expression from the UL4-CAT promoter in recombinant virus-infected HFFs. Confluent HFFs were serum starved for 24 h and then treated with FHPI for 1 h before infection with various recombinant viruses in the presence of FHPI in quadruplicate as described in Materials and Methods. Cells were harvested at 14 h after infection and subjected to CAT assays. The results of the CAT assays were averaged, and the standard deviations were calculated. The CAT activity per microgram of protein at various concentrations of the drug is relative to that of the nontreated recombinant viruses.
Effect of MAPK signal transduction pathway on UL4 promoter in both ectopic and natural positions.
The UL4-CAT promoter recombined into the US component of the viral genome has upstream sequence to only −220 bp relative to the E1 transcription start site. It is possible that additional upstream sequence elements affect the UL4 promoter in the UL component of the viral genome. To determine whether the expression pattern of the ectopic UL4-CAT promoter correlates with that of UL4 promoter in the UL component of the viral genome, serum-starved HFFs were pretreated with U0126 or FHPI for 1 h. After infection with RVwt at 5 PFU/cell in the presence of the inhibitor, as described in Materials and Methods, cytoplasmic RNAs were isolated at 8 hpi and subjected to Northern blot analysis with 32P-labeled DNA probes.
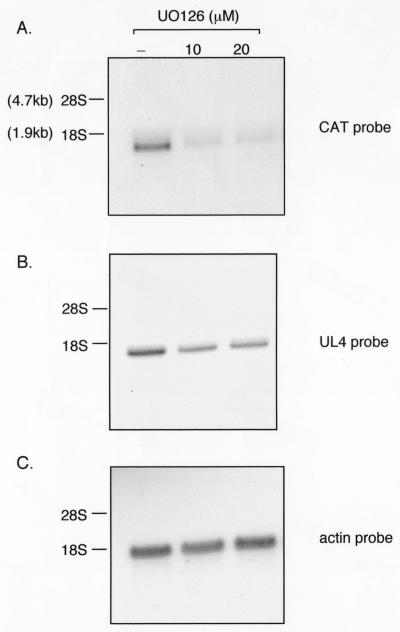
Treatment with UO126 or FHPI had a significant negative effect on the steady-state level of CAT RNA (approximately 66 to 75%). Treatment with U0126 had an effect on UL4 RNA (approximately 22 to 33% reduction) compared to untreated cells (Fig. 8A and B, compare lane 1 with lanes 2 and 3), but the effect of FHPI was less. The inhibitory effects were more pronounced on the ectopic UL4-CAT promoter. UO126 had no effect on actin RNA levels (Fig. 8C). Differences with the UL4 promoter in the ectopic location of the viral genome compared to the natural position may be due to the additional upstream sequences in the natural position. We conclude that the UL4-CAT promoter expression pattern correlates with the UL4 promoter in the UL component of the viral genome. While the viral promoters were affected by inhibitors of the MAPK signal transduction pathways, effects on the actin promoter were not detected.
FIG. 8.
Northern blot analysis of UL4-CAT or UL4 RNA expression in HFFs infected with RVwt in the presence of the MEK inhibitor UO126. Confluent HFFs were serum starved and pretreated with UO126 for 1 h before infection with RVwt as described in the legend to Fig. 6. The drug was present throughout the experiment. Cytoplasmic RNA was harvested at 8 hpi and analyzed by Northern blot hybridization with either the CAT (A), UL4 (B), or actin (C) probe as described in Materials and Methods. The blot was serially stripped and hybridized with different probes. Lanes: 1, untreated; 2, treated with 10 μM UO126; 3, treated with 20 μM UO126.
DISCUSSION
The early viral UL4 promoter (E1), which drives the synthesis of a 1.5-kb mRNA during early times after HCMV infection, has been studied extensively in our laboratory as a model system for early viral gene expression (10, 11, 30, 31). Within the early UL4 promoter, there are an Elk-1 binding site and an NF-Y binding site, as demonstrated by in vitro binding assays (12, 30). Recombinant viruses with a mutation of the Elk-1 element located between −168 and −159 (−168/−159) relative to the transcription start site and the proximal putative IE86 protein binding site had decreased levels of the UL4 promoter activity by approximately 50%. Mutation of the NF-Y binding site had no effect (12). To extend our understanding of how the UL4 promoter is regulated, we isolated recombinant viruses with site-specific mutations solely in the Elk-1 binding site. We also constructed a series of recombinant HCMVs with either wild-type or deletion mutations in the UL4 promoter and tested the effect of inhibitors of the MAPK signal transduction pathways.
RNase protection assays and CAT assays demonstrated that the Elk-1 site and the sequences between −102 and −50 relative to the transcription start of site of the UL4 promoter are two of the major upstream regulatory elements in the context of the viral genome. The Elk-1 site alone, but not the proximal potential IE86 binding site, is a regulatory element of the UL4 promoter in virus-infected cells. A transcription factor binding site database search (http://molsun1.cbrc.aist.go.jp/research/db/TFSEARCH.html) revealed that the sequences between −102 and −50 have potential transcription factor binding sites for Evi-1, c-rel, and NF-Y. Among these sites, the NF-Y binding site had the highest score (data not shown). However, site-specific mutations in the NF-Y binding site in recombinant virus RVdlNF-Y-xs did not induce a reduction in the UL4-CAT expression (12). Even though the NF-Y binding was abolished, other transcription factors might be able to compensate. There is a potential Evi-1 binding site. Evi-1 is a 145-kDa nuclear zinc finger protein (46, 74). The Evi-1 binding site (−88/−79) overlaps the NF-Y binding site (−98/−88) by one nucleotide. In RVdlNF-Y-xs, the nucleotide at −88 was changed from a wild-type G to a mutant C, which changed the potential consensus binding site for Evi-1 by only one nucleotide at the 5′ end. Since this change did not induce a phenotype, the role of the Evi-1 site in the regulation of the UL4 promoter requires further investigation. To determine the exact location of the positive element, a series of recombinant viruses with site-specific mutations between −102 and −50 would be necessary.
Recombinant virus RV-50, which has 50 bp upstream of the transcription start site and an intact TATA box, had basal transcription activity. The viral IE proteins were able to activate this minimal promoter by forming a preinitiation complex in the absence of known upstream activator binding sites. Recombinant virus RV-23, which has no TATA box, had little to no activity. Therefore, activation of the early viral UL4 promoter is TATA box dependent in the context of the viral genome. These results indicated that the Elk-1 site, the sequences between −102 and −50 relative to the transcription start site, and the TATA box constitute cis-regulatory elements for activation of transcription from the UL4 promoter in the context of the viral genome.
The results obtained with recombinant virus-infected cells are different from those of the transient-transfection assays as described previously (30, 31). The NF-Y binding site did not play a significant role in the activation of the UL4 early promoter in the context of the viral genome, whereas in transient-transfection assays, it was a positive element. The deletion of site 2 had an increase in UL4 promoter activity in the absence of the IE86 protein, but exhibited little decrease compared to the wild type in the presence of the IE86 protein in transient-transfection assays. In virus-infected cells, a condition in which the IE86 protein was naturally expressed, site 2 was a positive element. The results from recombinant virus-infected cells might mimic naturally infected cells, since the templates from which viral genes are expressed are similar.
One of the mechanisms for activation of the UL4 promoter is through the MAPKs. HCMV infection activates at least two MAPK pathways, which include the MAPK/ERK and the p38 MAPK signal transduction pathways (34, 36, 61). Activation of the MAPK/ERK and the p38 MAPK pathways requires de novo viral protein synthesis, and activation of these pathways is essential for DNA replication (34, 61). The MEK inhibitor UO126 reduced UL4 promoter activity by 70 to 80% in HFFs infected with recombinant viruses at 8 h after infection. One explanation for the remaining activity could be due to activation of MAPKs other than ERKs. Alternatively, the remaining activity might be due to incomplete inhibition of the MEK activity by the inhibitor even at high concentrations or might be due to a MAPK-independent component that could contribute to the UL4 promoter activation. The p38 MAPK inhibitor FHPI reduced the promoter activity at higher concentrations, but not at lower concentrations. FHPI inhibits the α and β isoforms of p38 kinase. Whether the other two isoforms, p38γ and p38δ, contributed to UL4 promoter activation is not known.
The reduction of the UL4 promoter activity by the MAPK inhibitors was demonstrated to be qualitatively similar in both the ectopic position of the viral genome and the natural position in the viral genome. However, the inhibitory effect of UO126 and FHPI was more dramatic in the ectopic position of the viral genome. This may be due to the differences in the context of the viral genome between an ectopic position and its corresponding natural locus. In all recombinant viruses, the UL4 promoter contained only 220 bp upstream of the E1 transcription start site. Therefore, the strength of transcription could be different. The difference could also be due to the stability of the CAT RNA versus the UL4 RNA. We conclude that the early viral UL4 promoter is a MAPK-responsive promoter.
Our original rationale was that site-specific mutations or deletions of responsive elements in the UL4 promoter might eliminate or decrease the effect of the drug compared to that of the wild type. Instead, all recombinant viruses containing mutations in the UL4 promoter-regulatory region were affected by the drug treatments. Recombinant virus RV-50 with a minimal promoter containing an intact TATA box was affected by the drug treatment, which suggests that a responsive element lies between −50 and the transcription start site of the UL4 promoter. Recombinant virus RV-23, which lacks a TATA box, had little to no activity. Phosphorylation affects the ability of transcription factors to interact with other proteins (32). Since phosphorylation of the IE86 protein changes its ability to transactivate promoters in transient-transfection assays (27), inhibition of the MAPKs might affect the activation of the UL4 promoter by affecting the activity of the IE86 protein. It is possible that phosphorylation of IE86 increases its ability to interact with transcription factors that bind to upstream cis-acting elements and with the basal transcription machinery to form the preinitiation complex at the TATA box. Since recombinant HCMVs with either wild-type or mutant UL4 promoters are equally affected by MAPK inhibitors, the MAPKs might also affect nuclear matrix-bound transcription complexes. Finally, it is possible that MAPKs also affect transcript stability. One advantage of drug inhibitor studies is that they provide results rapidly and help to determine research directions. However, drug inhibitors can have general specificity problems. Confirmation of the data presented above by expression of dominant-negative forms of kinase is currently being investigated.
Other viruses have evolved to take advantage of the cellular MAPK/ERK and p38 MAPK signal transduction pathways to upregulate their gene expression. Among the alphaherpesvirus family, HSV-1 infection induces the activation of the p38 MAPK and SAPK/JNK signal transduction pathways, but the MAPK/ERK pathway is not affected (55, 85). Latency established by gammaherpesvirus Epstein-Barr virus is disrupted by activation of the MAPK/ERK and p38 MAPK pathways (1, 16). Other DNA viruses, such as adenovirus and hepatitis B virus, activate the MAPK/ERK and p38 MAPK pathways after infection (5, 6, 73). The activation of the MAPK/ERK and p38 MAPK pathways by these DNA viruses may contribute to the efficiency of viral replication (77).
In HCMV-infected cells, de novo viral protein synthesis is necessary to fully activate the MAPK/ERK or p38 MAPK pathways (34, 61). The viral proteins required for activation of the MAPKs at early times after infection are currently not known. We have used the early viral UL4 promoter as a model to study activation of HCMV early gene expression. The early UL4 promoter of HCMV requires the activation of the MAPK/ERK and p38 MAPK signal transduction pathways for full activation. These MAPK pathways play an important role in HCMV early promoter activation, which affects the efficiency of viral replication.
Acknowledgments
We thank members of the laboratory for helpful discussions and comments on the manuscript. We are grateful to Philip Lashmit for assistance and to Richard Roller for critical reading of the manuscript.
This work was supported by grant AI-13562 from the National Institutes of Health.
REFERENCES
- 1.Adamson, A. L., D. Darr, E. Holley-Guthrie, R. A. Johnson, A. Mauser, J. Swenson, and S. Kenney. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alderete, J. P., S. J. Child, and A. P. Geballe. 2001. Abundant early expression of gpUL4 from a human cytomegalovirus mutant lacking a repressive upstream open reading frame. J. Virol. 75:7188-7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alford, C. A., and W. J. Britt. 1990. Cytomegalovirus, p. 1981-2010. In B. N. Fields, D. M. Knipe et al (ed.), Virology. Raven Press, Ltd., New York, N.Y.
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology, vol. 1, p. 2.9.1-2.9.5. John Wiley & Sons, New York, N.Y.
- 5.Benn, J., F. Su, M. Doria, and R. J. Schneider. 1996. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J. Virol. 70:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruder, J. T., and I. Kovesdi. 1997. Adenovirus infection stimulates the Raf/MAPK signaling pathway and induces interleukin-8 expression. J. Virol. 71:398-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant, L. A., P. Mixon, M. Davidson, A. J. Bannister, T. Kouzarides, and, J. H. Sinclair. 2000. The human cytomegalovirus 86-kilodalton major immediate-early protein interacts physically and functionally with histone acetyltransferase P/CAF. J. Virol. 74:7230-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, J., and A. P. Geballe. 1996. Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell. Biol. 16:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caswell, R., C. Hagemeier, C.-J. Chiou, G. Hayward, T. Kouzarides, and J. Sinclair. 1993. The human cytomegalovirus 86K immediate early (IE2) protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via region of IE2 required for transcriptional regulation. J. Gen. Virol. 74:2691-2698. [DOI] [PubMed] [Google Scholar]
- 10.Chang, C.-P., C. L. Malone, and M. F. Stinski. 1989. A human cytomegalovirus early gene has three inducible promoters that are regulated differentially at various times after infection. J. Virol. 63:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, C.-P., D. H. Vesole, J. Nelson, M. B. A. Oldstone, and M. F. Stinski. 1989. Identification and expression of a human cytomegalovirus early glycoprotein. J. Virol. 63:3330-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J., and M. F. Stinski. 2000. Activation of transcription of the human cytomegalovirus early UL4 promoter by the Ets transcription factor binding element. J. Virol. 74:9845-9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, P. 1997. The search for physiological substrates of MAP and SAP kinases in mammalian cells. Trends Cell. Biol. 7:353-361. [DOI] [PubMed] [Google Scholar]
- 14.Dwarakanath, R. S., C. L. Clark, A. K. McElroy, and D. H. Spector. 2001. The use of recombinant baculoviruses for sustained expression of human cytomegalovirus immediate early proteins in fibroblasts. Virology 284:297-307. [DOI] [PubMed] [Google Scholar]
- 15.Favata, M. F., K. Y. Horiuchi, E. J. Manos, A. J. Daulerio, D. A. Stradley, W. S. Feeser, D. E. Van Dyk, W. J. Pitts, R. A. Earl, F. Hobbs, R. A. Copeland, R. L. Magolda, P. A. Scherle, and J. M. Trzaskos. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273:18623-18632. [DOI] [PubMed] [Google Scholar]
- 16.Fenton, M., and A. J. Sinclair. 1999. Divergent requirements for the MAPKERK signal transduction pathway during initial virus infection of quiescent primary B cells and disruption of Epstein-Barr virus latency by phorbol esters. J. Virol. 73:8913-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortunato, E. A., and D. H. Spector. 1999. Regulation of human cytomegalovirus gene expression. Adv. Virus Res. 54:61-128. [DOI] [PubMed] [Google Scholar]
- 18.Furnari, B. A., E. Poma, T. F. Kowalik, S.-M. Huong, and E.-S. Huang. 1993. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J. Virol. 67:4981-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 20.Gille, H., M. Kortenjann, O. Thomae, C. Moomaw, C. Slaughter, M. H. Cobb, and P. E. Shaw. 1995. ERK phosphorylation potentiates Elk-1 mediated ternary complex formation and transactivation. EMBO J. 14:951-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorman, C. M., L. F. Moffat, and B. H. Howard. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greaves, R. F., J. M. Brown, J. Vieira, and E. S. Mocarski. 1995. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the Escherichia coli guanosine phosphoribosyl transferase (gpt) gene. J. Gen. Virol. 76:2151-2160. [DOI] [PubMed] [Google Scholar]
- 23.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagemeier, C., S. Walker, R. Caswell, T. Kouzarides, and J. Sinclair. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J. Virol. 66:4452-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn, G., R. Jores, and E. S. Mocarski. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 95:3937-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanukoglu, I., N. Tanese, and E. Fuchs. 1983. Complementary DNA sequence of a human cytoplasmic actin. Interspecies divergence of 3′ non-coding regions. J. Mol. Biol. 163:673-678. [DOI] [PubMed] [Google Scholar]
- 27.Harel, N. Y., and J. C. Alwine. 1998. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2. J. Virol. 72:5481-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermiston, T. W., C. L. Malone, and M. F. Stinski. 1990. Human cytomegalovirus immediate-early two protein region involved in negative regulation of the major immediate-early promoter. J. Virol. 64:3532-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermiston, T. W., C. L. Malone, P. R. Witte, and M. F. Stinski. 1987. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J. Virol. 61:3214-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, L., C. L. Malone, and M. F. Stinski. 1994. A human cytomegalovirus early promoter with upstream negative and positive cis-acting elements: IE2 negates the effect of the negative element, and NF-Y binds to the positive element. J. Virol. 68:2108-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, L., and M. F. Stinski. 1995. Binding of cellular repressor protein or the IE2 protein to a cis-acting negative regulatory element upstream of a human cytomegalovirus early promoter. J. Virol. 69:7612-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter, T., and M. Karin. 1992. The regulation of transcription by phosphorylation. Cell 70:375-387. [DOI] [PubMed] [Google Scholar]
- 33.Janknecht, R., and T. Hunter. 1997. Convergence of MAP kinase pathways on the ternary complex factor Sap-1a. EMBO J. 16:1620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, R. A., S.-M. Huong, and E.-S. Huang. 2000. Activation of the mitogen-activated protein kinase p38 by human cytomegalovirus infection through two distinct pathways: a novel mechanism for activation of p38. J. Virol. 74:1158-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, R. A., S.-M. Huong, and E.-S. Huang. 1999. Inhibitory effect of 4-(4-fluorophenyl)-2-(4-hydroxyphenyl)-5-(4-pyridyl)1H-imidazole on HCMV DNA replication and permissive infection. Antivir. Res. 41:101-111. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, R. A., X.-L. Ma, A. D. Yurochko, and E.-S. Huang. 2001. The role of MKK1/2 kinase activity in human cytomegalovirus infection. J. Gen. Virol. 82:493-497. [DOI] [PubMed] [Google Scholar]
- 37.Jones, T. R., and V. P. Muzithras. 1992. A cluster of dispensable genes within the human cytomegalovirus genome short component: ISR1, US1 through US5, and the US6 family. J. Virol. 66:2541-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones, T. R., V. P. Muzithras, and Y. Gluzman. 1991. Replacement mutagenesis of the human cytomegalovirus genome: US10 and US11 gene products are nonessential. J. Virol. 65:5860-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jupp, R., S. Hoffmann, A. Depto, R. M. Stenberg, P. Ghazal, and J. A. Nelson. 1993. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J. Virol. 67:5595-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jupp, R., S. Hoffmann, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1993. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J. Virol. 67:7539-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerry, J. A., M. A. Priddy, T. Y. Jervey, C. P. Kohler, T. L. Staley, C. D. Vanson, T. R. Jones, A. C. Iskenderian, D. G. Anders, and R. M. Stenberg. 1996. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J. Virol. 70:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klucher, K. M., D. K. Rabert, and D. H. Spector. 1989. Sequences in the human cytomegalovirus 2.7-kilobase RNA promoter which mediate its regulation as an early gene. J. Virol. 63:5334-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klucher, K. M., and D. H. Spector. 1990. The human cytomegalovirus 2.7-kilobase RNA promoter contains a functional binding site for the adenovirus major late transcription factor. J. Virol. 64:4189-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondo, K., H. Kaneshima, and E. S. Mocarski. 1994. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc. Natl. Acad. Sci. USA 91:11879-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krieg, P. A., and D. A. Melton. 1987. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 155:397-414. [DOI] [PubMed] [Google Scholar]
- 46.Kurokawa, M., K. Mitani, T. Yamagata, T. Takahashi, K. Izutsu, S. Ogawa, T. Moriguchi, E. Nishida, Y. Yazaki, and H. Hirai. 2000. The Evi-1 oncoprotein inhibits c-Jun N-terminal kinase and prevents stress-induced cell death. EMBO J. 19:2958-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, B., T. W. Hermiston, and M. F. Stinski. 1991. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J. Virol. 65:897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukac, D. M., J. R. Manuppello, and J. C. Alwine. 1994. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J. Virol. 68:5184-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luu, P., and O. Flores. 1997. Binding of SP1 to the immediate-early protein-responsive element of the human cytomegalovirus DNA polymerase promoter. J. Virol. 71:6683-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maciejewski, J. P., E. E. Bruening, R. E. Donahue, E. S. Mocarski, N. S. Young, and S. C. St. Jeor. 1992. Infection of hematopoietic progenitor cells by human cytomegalovirus. Blood 80:170-178. [PubMed] [Google Scholar]
- 53.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin-Blanco, E. 2000. p38 MAPK signaling cascades: ancient roles and new functions. BioEssays 22:637-645. [DOI] [PubMed] [Google Scholar]
- 55.McLean, T. I., and S. L. Bachenheimer. 1999. Activation of cJUN N-terminal kinase by herpes simplex virus type 1 enhances viral replication. J. Virol. 73:8415-8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meier, J. L., and M. F. Stinski. 1997. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J. Virol. 71:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minton, E. J., C. Tysoe, J. H. Sinclair, and J. G. Sissons. 1994. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J. Virol. 68:4017-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nemoto, S., J. Xiang, S. Huang, and A. Lin. 1998. Induction of apoptosis by SB202190 through inhibition of p38 beta mitogen-activated protein kinase. J. Biol. Chem. 273:16415-16420. [DOI] [PubMed] [Google Scholar]
- 59.Price, M. A., F. H. Cruzalegui, and R. Treisman. 1996. The p38 and ERK MAP kinase pathways cooperate to activate ternary complex factors and c-fos transcription in response to UV light. EMBO J. 15:6552-6563. [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 61.Rodems, S. M., and D. H. Spector. 1998. Extracellular signal-regulated kinase activity is sustained early during human cytomegalovirus infection. J. Virol. 72:9173-9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rolli, M., A. Kotlyarov, K. M. Sakamoto, M. Gaestel, and A. Neininger. 1999. Stress-induced stimulation of early growth response gene-1 by p38/stress-activated protein kinase 2 is mediated by a cAMP-responsive promoter element in a MAPKAP kinase 2-independent manner. J. Biol. Chem. 274:19559-19564. [DOI] [PubMed] [Google Scholar]
- 63.Schaeffer, H. J., and M. J. Weber. 1999. Mitogen-activated protein kinase: specific messages from ubiquitous messengers. Mol. Cell. Biol. 19:2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidbauer, M., H. Budka, W. Ulrich, and P. Ambros. 1989. Cytomegalovirus (CMV) disease of the brain in AIDS and connatal infection: a comparative study by histology, immunocytochemistry, and in situ DNA hybridization. Acta Neuropathol. 79:286-293. [DOI] [PubMed] [Google Scholar]
- 65.Scully, A. L., M. H. Sommer, R. Schwartz, and D. H. Spector. 1995. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 69:6533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinzger, C., B. Plachter, A. Grefte, T. H. The, and G. Jahn. 1996. Tissue macrophages are infected by human cytomegalovirus. J. Infect. Dis. 173:240-245. [DOI] [PubMed] [Google Scholar]
- 67.Soderberg-Naucler, C., K. N. Fish, and J. A. Nelson. 1997. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91:119-126. [DOI] [PubMed] [Google Scholar]
- 68.Spector, D. H. 1996. Activation and regulation of human cytomegalovirus early genes. Intervirology 39:361-377. [DOI] [PubMed] [Google Scholar]
- 69.Speir, E., R. Modali, E. S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 70.Staprans, S. I., D. K. Rabert, and D. H. Spector. 1988. Identification of sequence requirements and trans-acting functions necessary for regulated expression of a human cytomegalovirus early gene. J. Virol. 62:3463-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stinski, M. F. 1976. Human cytomegalovirus: glycoproteins associated with virions and dense bodies. J. Virol. 19:594-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stinski, M. F. 1978. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J. Virol. 26:686-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suomalainen, M., M. Y. Nakano, K. Boucke, S. Keller, and U. F. Greber. 2001. Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J. 20:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanaka, T., K. Mitani, M. Kurokawa, S. Ogawa, K. Tanaka, J. Nishida, Y. Yazaki, Y. Shibata, and H. Hirai. 1995. Dual functions of the AML1/Evi-1 chimeric protein in the mechanism of leukemogenesis in t(3;21) leukemias. Mol. Cell. Biol. 15:2383-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thomsen, D. R., and M. F. Stinski. 1981. Cloning of the human cytomegalovirus genome as endonuclease XbaI fragments. Gene 16:207-216. [DOI] [PubMed] [Google Scholar]
- 76.Thrower, A. R., G. C. Bullock, J. E. Bissell, and M. F. Stinski. 1996. Regulation of a human cytomegalovirus immediate-early gene (US3) by a silencer-enhancer combination. J. Virol. 70:91-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tibbles, L. A., and J. R. Woodgett. 1999. The stress-activated protein kinase pathways. Cell. Mol. Life Sci. 55:1230-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Treisman, R. 1996. Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol. 8:205-215. [DOI] [PubMed] [Google Scholar]
- 79.Wade, E. J., K. M. Klucher, and D. H. Spector. 1992. An AP-1 binding site is the predominant cis-acting regulatory element in the 1.2-kilobase early RNA promoter of human cytomegalovirus. J. Virol. 66:2407-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whitmarsh, A. J., P. Shore, A. D. Sharrocks, and R. J. Davis. 1995. Integration of MAP kinase signal transduction pathways at the serum response element. Science 269:403-407. [DOI] [PubMed] [Google Scholar]
- 81.Whitmarsh, A. J., S.-H. Yang, M. S.-S. Su, A. D. Sharrocks, and R. J. Davis. 1997. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol. Cell. Biol. 17:2360-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Widmann, C., S. Gibson, M. B. Jarpe, and G. L. Johnson. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79:143-180. [DOI] [PubMed] [Google Scholar]
- 83.Wu, J., J. O'Neill, and M. S. Barbosa. 1998. Transcription factor Sp1 mediates cell-specific trans-activation of the human cytomegalovirus DNA polymerase gene promoter by immediate-early protein IE86 in glioblastoma U373MG cells. J. Virol. 72:236-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Young, P. R., M. M. McLaughlin, S. Kumar, S. Kassis, M. L. Doyle, D. McNulty, T. F. Gallagher, S. Fisher, P. C. McDonnell, S. A. Carr, M. J. Huddleston, G. Seibel, T. G. Porter, G. P. Livi, J. L. Adams, and J. C. Lee. 1997. Pyridinyl imidazole inhibitors of p38 mitogen-activated protein kinase bind in the ATP site. J. Biol. Chem. 272:12116-12121. [DOI] [PubMed] [Google Scholar]
- 85.Zachos, G., B. Clements, and J. Conner. 1999. Herpes simplex virus type 1 infection stimulates p38/c-Jun N-terminal mitogen-activated protein kinase pathways and activates transcription factor AP-1. J. Biol. Chem. 274:5097-5103. [DOI] [PubMed] [Google Scholar]