Abstract
Spleen necrosis virus (SNV) proteins can package RNA from distantly related murine leukemia virus (MLV), whereas MLV proteins cannot package SNV RNA efficiently. We used this nonreciprocal recognition to investigate regions of packaging signals that influence viral RNA encapsidation specificity. Although the MLV and SNV packaging signals (Ψ and E, respectively) do not contain significant sequence homology, they both contain a pair of hairpins. This hairpin pair was previously proposed to be the core element in MLV Ψ. In the present study, MLV-based vectors were generated to contain chimeric SNV/MLV packaging signals in which the hairpins were replaced with the heterologous counterpart. The interactions between these chimeras and MLV or SNV proteins were examined by virus replication and RNA analyses. SNV proteins recognized all of the chimeras, indicating that these chimeras were functional. We found that replacing the hairpin pair did not drastically alter the ability of MLV proteins to package these chimeras. These results indicate that, despite the important role of the hairpin pair in RNA packaging, it is not the major motif responsible for the ability of MLV proteins to discriminate between the MLV and SNV packaging signals. To determine the role of sequences flanking the hairpins in RNA packaging specificity, vectors with swapped flanking regions were generated and evaluated. SNV proteins packaged all of these chimeras efficiently. In contrast, MLV proteins strongly favored chimeras with the MLV 5′-flanking regions. These data indicated that MLV Gag recognizes multiple elements in the viral packaging signal, including the hairpin structure and flanking regions.
Retroviruses package two copies of viral RNA into virions as genetic material (19, 34, 35). Viral RNAs are specifically selected to be packaged during virus assembly; the specificity of RNA packaging is determined by the interactions between the packaging signal in the viral RNA and the viral polyprotein Gag (8, 9, 38, 58). Deletion of the packaging signal can drastically reduce viral RNA packaging (3, 36, 41, 61), whereas mutations in the Gag polyprotein can affect packaging specificity. The Gag polyproteins of all retroviruses have three common domains: matrix (MA), capsid (CA), and nucleocapsid (NC) (58). Among these domains, NC plays an important role in RNA recognition and packaging, although other domains in Gag have also been suggested previously to contribute to RNA packaging specificity (23, 30, 53, 57). Mutations in NC can result in decreased viral RNA packaging (2-4, 20, 24, 25, 27, 43, 44, 50, 65, 66), whereas swapping the NC domain from different viruses can alter packaging specificity (10, 13, 21, 32, 66).
Packaging signals in many retroviruses have been identified; the major packaging signals are generally located in the 5′ untranslated regions of the viral RNA, with some signals extending into the group-specific antigen gene (gag) (8, 37). Sequences elsewhere in the viral genome have also been suggested previously to affect RNA packaging (5, 49, 64). Packaging signals are generally mapped by using deletion analyses to identify elements necessary for RNA packaging or by determining which elements are sufficient to allow the encapsidation of a heterologous RNA (8, 37).
One of the first packaging signals identified in retroviruses was that in reticuloendotheliosis virus, an avian pathogen closely related to spleen necrosis virus (SNV) (41, 56, 61). This packaging signal was first defined by deletion studies as a 190-nucleotide (nt) sequence located in the 5′ untranslated region of the viral genome. This region was termed the encapsidation sequence (E). Deletion of E or part of E resulted in decreased SNV RNA being packaged into the viral particles (60-62). Relocating SNV E to the 3′ untranslated region of the viral vector still allowed efficient packaging of the vector RNA (28). The defined 190-nt SNV E was shown elsewhere to allow the packaging of heterologous RNAs (16).
The packaging signal of murine leukemia virus (MLV) was defined both by deletion studies and by demonstrating its ability to allow the encapsidation of heterologous RNA (1, 41). A 350-nt sequence in the 5′ untranslated region was first defined by restriction enzyme mapping to be important for viral RNA packaging; this sequence was termed Ψ (41). It was later shown that a longer sequence, including Ψ and the 5′ portion of gag (approximately 420 nt), allowed more efficient packaging of the vector RNA (7). This extended packaging signal was termed Ψ+. Abolishing the gag start codon did not interfere with the enhancement of RNA packaging, indicating a cis-acting role for the gag sequences (7). Deletion of Ψ or portions of Ψ resulted in decreases in the amount of viral RNA that was encapsidated into retroviral particles (41, 47, 48). Ψ can be moved to the 3′ end of the viral vector genome and still allow efficient vector RNA packaging (40). Ψ and Ψ+ can also be inserted into nonretroviral RNAs to allow the packaging of these RNAs into viral particles (1).
With a few reported exceptions, packaging specificity is generally limited to the same virus or closely related viruses (12, 32, 52, 58). One of the exceptions is that SNV Gag polyprotein can efficiently package MLV RNA (22). However, this recognition is nonreciprocal: MLV Gag cannot package SNV RNA efficiently (14). Furthermore, replacing the NC domain of SNV Gag with MLV NC drastically reduces the ability of this chimeric Gag to recognize SNV vector RNA, although it can package MLV RNA efficiently (13). This finding indicates that MLV Gag cannot recognize a certain RNA motif(s) in SNV E. Such nonreciprocal RNA packaging between SNV and MLV provides us with a model system to analyze RNA motifs involved in the interactions between Gag and packaging signals.
The primary sequences of MLV Ψ and SNV E do not contain significant homology (Fig. 1) (33, 41, 61). However, they share a similar RNA secondary structure: a pair of hairpins demonstrated previously to be essential for RNA packaging (33, 62). In both MLV and SNV, deletions or mutations that destabilized the hairpin pair drastically reduced the efficiencies of RNA packaging (47, 48, 62). Based on its important role in RNA packaging, the hairpin pair was proposed previously to be the core encapsidation signal of MLV (47). Interestingly, the MLV hairpin pair can functionally replace the SNV hairpin pair, since an SNV E-based packaging signal containing the MLV hairpin pair can be recognized by SNV Gag efficiently (62). Because SNV Gag recognizes both packaging signals, it remains unclear whether the hairpin pair motif plays a role in the nonreciprocal recognition between MLV and SNV. It is possible that MLV Gag can discriminate between the SNV and MLV hairpin pairs, whereas SNV Gag can recognize both hairpin pairs. It is also possible that, despite the importance of the hairpin pair, other RNA regions in MLV Ψ that are necessary for MLV Gag-RNA interaction are lacking or different in SNV E.
FIG. 1.
Comparison of SNV (A) and MLV (B) packaging signals. The sequences of SNV E and MLV Ψ are shown. Hairpin pair sequences are shown in boldface; in addition, the two hairpins are shown by dashed lines and the word “hairpin.” The restriction enzyme sites are shown in italics. The proposed hairpins A and B are underlined.
In this report, we sought to delineate the motifs in the two packaging signals that lead to the nonreciprocal packaging of MLV and SNV. We took advantage of the conserved hairpin pair structure and constructed vectors that contained chimeric packaging signals, replacing either the hairpin pair or one of the regions flanking the hairpin pair. We then examined the abilities of these chimeric packaging signals to be encapsidated by MLV Gag and SNV Gag independently. With this approach, we evaluated the contribution of different RNA regions in the viral packaging signals that determine the specificity of viral RNA encapsidation.
MATERIALS AND METHODS
Plasmid construction and definitions.
All of the retroviral vectors were constructed using standard techniques (54). In this report, the “p” in front of the vector name (e.g., pBB4-SE) refers to the plasmid, whereas the vector name without the “p” refers to virus derived from this plasmid (e.g., BB4-SE).
Plasmid pAR2 is an MLV vector (63) that encodes the hygromycin phosphotransferase B gene (hygro) (26). Plasmid pAR3 was derived from pAR2 by deleting the entire MLV Ψ+ in the 5′ untranslated region (7). Plasmid pAR2 was digested with MscI plus SalI, treated with the Escherichia coli DNA polymerase I large fragment (Klenow), and religated. This deletion was confirmed by restriction enzyme mapping and DNA sequencing. DNA sequencing analyses revealed that the ligation junction contained a 1-bp deletion (G) and a 2-bp insertion (CA), presumably generated during the cloning procedure. This minor variation was not expected to affect viral replication and RNA packaging.
Plasmid pAR3L was derived from pAR3 by insertion of a linker (annealed DNA oligonucleotides BENLINK1 and 2KNILNEB [Table 1]) into the ClaI site of the viral 3′ untranslated regions. This linker contained the BsrGI and SalI restriction enzyme sites followed by stop codons in three reading frames. The previously described vectors pSY223, pSY254, pSY281, and pSY280 (62) (kind gifts from Shiaolan Yang) were digested with Asp718 plus SalI to yield DNA fragments that were ligated to the BsrGI-plus-SalI-digested pAR3L to generate pBB4-SNVE, pBB5-MLVΨ+, pBB6-SEMhp, and pBB7-MLVΨ, respectively. The Ψ and Ψ+ sequences from pSY281 and pSY280 were derived from pLN, which contains a hybrid signal with the 5′ portion from Moloney murine sarcoma virus and the 3′ portion from Moloney MLV (46). The packaging signals from Moloney MLV and Moloney murine sarcoma virus contain minor differences; hybrid packaging signals, especially this set, have been used frequently in previous studies. For convenience, we use MLV Ψ or Ψ+ to designate these packaging signals.
TABLE 1.
Primers used to construct vectors and generate probes
| Primer | Sequence |
|---|---|
| BENLINK1 | 5′ CGATAGCTAAGTGATGTACAAGCTTGTCGAC 3′ |
| 2KNILNEB | 5′ CGGTCGACAAGCTTGTACATCACTTAGCTAT 3′ |
| P1MLVP5 | 5′ CAGTAGGCCTGGTACCTGGCCAGCAACTTATCTG 3′ |
| 4P3-PVLM | 5′ TATTTTATCGGTCGACGAATTCCGGCGCC 3′ |
| 2P5PHVNS | 5′ TCCCCTGCTGAGCGGACCTGGAGCTTCCCGGCAGCGTCCCGCCACTCCGATCC TCCCAGATACAGAGCTAGTTAGC 3′ |
| P3SNVHP3 | 5′ CGGGACGCTGCCGGGAAGCTCCACCTCCGCTCAGCAGGGGACGCCCTGGTCT GAGCACTTTGGGGGCCGTTTTTGTGG 3′ |
| MLVHP5′ | 5′ GCGGACCCGTGGTGGAACTGAC 3′ |
| 3′HYGRO | 5′ TCCGGATCAGCTTGGCACTGGC 3′ |
| 3′PHVLM | 5′ ACGTCTCCCAGGGTTGCGGCC 3′ |
| TPPVLM | 5′ TTTCATTCCCCCCTTTTTCTGG 3′ |
| SNVHP5′ | 5′ GAGGATCGGAGTGGCGGGAC 3′ |
| PHVNS3′ | 5′ GCTCAGACCAGGGCGTCCCC 3′ |
| ORGYH7T | 5′ TAATACGACTCACTATAGGGAGGCTTGCAACGTGACACCCTGTG 3′ |
| SV40PEND | 5′ AGGCTTTTTTGGAGGCCTAGGC 3′ |
| RD5′toSMA | 5′ GTCCACTCCCAGGTCCAACCG 3′ |
| AMVNS7T | 5′ TAATACGACTCACTATAGGGAGGCCTTCCTTCGGCCACCCTACG 3′ |
| SNVU3R | 5′ GTCGCCGTCCTACACATTGTTG 3′ |
| JD2143′U5 | 5′ TAATACGACTCACTATAGGGAGGCGGGCTTGTCTGCTCCCGGC 3′ |
The chimeric packaging signal sequences located in plasmids pBB8-MΨ+Shp, pBB10-MShpS, pBB11-SShpM, pBB12-SMhpM, and pBB13-MMhpS were generated by overlapping extension PCR using the primers listed in Table 1. This was accomplished by first amplifying a DNA fragment containing the upstream flanking region plus the hairpin pair (5′ fragment) and a DNA fragment containing the hairpin pair plus the downstream flanking region (3′ fragment), followed by joining of the two fragments by PCR. Vector pBB8-MΨ+Shp was generated using the following procedures. The 5′ fragment was amplified from pBB5-MLVΨ+ with primers P1MLVP5 and 2P5PHVNS. The 3′ fragment was amplified from pBB5-MLVΨ+ with primers 4P3-PVLM and P3SNVHP3. These two DNA fragments were joined by PCR with primers P1MLVP5 and 4P3-PVLM, digested with Asp718 plus SalI, and inserted into BsrGI-plus-SalI-digested pAR3L. Plasmid pBB9-MΨShp was generated by digesting pBB8-MΨ+Shp with Eco47III plus SalI, treating the DNA with the Klenow fragment, and self-ligating the DNA. MLV Ψ is defined as the region between the MscI and PstI sites in the MLV 5′ untranslated region (41); the Eco47III site is immediately downstream of the PstI site.
Plasmid pBB10-MShpS was generated by the following steps. The 5′ fragment was amplified from pBB9-MΨShp with primers 3′HYGRO and PHVNS3′. The 3′ fragment was amplified from pBB4-SNVE with primers TPPVLM and SNVHP5′. These two DNA fragments were joined by PCR with primers 3′HYGRO and TPPVLM, digested with ClaI plus SalI, and inserted into ClaI-plus-SalI-digested pAR3L.
Plasmid pBB11-SShpM was generated in the following manner. The 5′ fragment was amplified from pBB4-SNVE with primers 3′HYGRO and PHVNS3′. The 3′ fragment was amplified from pBB9-MΨShp with primers TPPVLM and SNVHP5′. These two DNA fragments were joined by PCR with primers 3′HYGRO and TPPVLM, digested with ClaI plus SalI, and inserted into ClaI-plus-SalI-digested pAR3L.
Plasmid pBB12-SMhpM was generated in the following manner. The 5′ fragment was amplified from pBB6-SEMhp with primers 3′HYGRO and 3′PHVLM. The 3′ fragment was amplified from pBB7-MLVΨ with primers MLVHP5′ and TPPVLM. These two DNA fragments were joined by PCR with primers 3′HYGRO and TPPVLM, digested with ClaI and SalI, and inserted into ClaI-plus-SalI-digested pAR3L.
Plasmid pBB13-MMhpS was generated by the following procedure. The 5′ fragment was amplified from pBB7-MLVΨ with primers 3′HYGRO and 3′PHVLM. The 3′ fragment was amplified from pBB6-SEMhp with primers TPPVLM and MLVHP5′. These two DNA fragments were joined by PCR with primers 3′HYGRO and TPPVLM, digested with ClaI and SalI, and inserted into ClaI-plus-SalI-digested pAR3L.
The general structures of all the plasmids were verified by restriction enzyme analyses. To ensure that inadvertent mutations were not introduced during PCR or the cloning procedures, all of the packaging signals and neighboring sequences were characterized by DNA sequencing (55) (AutoRead kit [Pharmacia]).
Cell lines, transfection, and infection.
PG13 and D17 cell lines were obtained from the American Type Culture Collection. Cell line PG13 was derived from murine NIH 3T3 TK− cells and expresses MLV Gag/Gag-Pol and gibbon ape leukemia virus Env (45). D17 is a dog osteosarcoma cell line that is permissive for infection by SNV Env- and gibbon ape leukemia virus Env-pseudotyped virus (51). Cell line DSH134G was derived from D17 and expresses SNV Gag/Gag-Pol and Env (42) (a kind gift from Ralph Dornburg). All cells were maintained in Dulbecco's modified Eagle's medium supplemented with either 6% (D17 and DSH134G) or 10% (PG13) calf serum (HyClone Laboratories, Inc.) with penicillin (50 U/ml; Gibco) plus streptomycin (50 μg/ml; Gibco). Cells were maintained at 37°C with 5% CO2.
Plasmids were introduced into PG13 and DSH134G cells by calcium phosphate transfection (54) and dimethyl sulfoxide-Polybrene transfection (31), respectively. Hygromycin-resistant helper cell colonies were pooled; each transfected cell pool contained at least 500 colonies. Transfected DSH134G cell pools were propagated in the presence of 3′-azido-3′-deoxythymidine (1 μM) to prevent potential vector reinfection (29). 3′-Azido-3′-deoxythymidine was removed from the cell culture before plating of the transfected cells for virus harvest.
Transfected cells were plated at a density of 5 × 106 cells per 100-mm-diameter dish, fresh medium was added 24 h later, and another 24 h later supernatants were collected. Virus-containing cell culture medium was centrifuged at 3,000 × g for 10 min to pellet cellular debris. The supernatant was then aliquoted for use in infection, reverse transcriptase (RT) assay, and isolation of cell-free virion RNA.
Tenfold serial dilutions of each virus were used to infect D17 target cells for 4 h in the presence of Polybrene (50 μg/ml). The infected D17 cells were placed under hygromycin selection (120 μg/ml). Viral titers were determined by the numbers of hygromycin-resistant D17 cell colonies.
RNA isolation and analysis.
Total cellular RNAs were isolated by using either Trizol (Life Technologies, Inc.) or the AquaPure RNA isolation kit (Bio-Rad) as specified by the manufacturers. The integrity of the cellular RNA was verified by gel electrophoresis and inspection of the rRNA bands, whereas the amount of total cellular RNA was quantified by spectrophotometric analyses. Cell-free virions were concentrated by centrifugation at 25,000 rpm for 90 min in a Beckman SW28 rotor. Viral pellets were resuspended in Dulbecco's modified Eagle's medium; a portion was removed from each sample for the RT assay, and the remaining sample was used for RNA analysis. Prior to RNA isolation, a portion of wild-type SNV was added to each virus preparation as an internal standard to monitor the loss of RNA during the isolation procedure. RNA was isolated by lysing the viral preparation with 0.1% sodium dodecyl sulfate in the presence of 200 μg of tRNA/ml followed by phenol-chloroform extraction and ethanol precipitation. Alternatively, cell-free virion RNA was isolated using the QIAamp viral RNA isolation kit (Qiagen). All RNA samples were resuspended in diethyl pyrocarbonate-treated water.
DNA fragments used to generate RNA probes were amplified by PCR; T7 promoters were introduced into the DNA fragments during PCR by using primers containing the promoter sequences. Antisense RNA probes were generated from these PCR DNA fragments by using the Maxiscript T7 kit (Ambion) with the addition of [α-32P]UTP (ICN).
Vector RNAs were detected using a 385-nt probe containing a portion of hygro (hygro probe). A 325-nt sequence in this probe is complementary to the hygro sequences in all of the vectors containing various packaging signals. This probe was generated by transcribing a DNA fragment amplified with primers ORGYH7T and SV40PEND (Table 1), with pJD220SVhy as a template (17). Two probes were used to detect wild-type SNV RNA. The first was a 223-nt probe containing a 180-nt sequence complementary to the gag sequences of SNV RNA (gag probe). This probe was generated by transcribing a DNA fragment amplified with primers RD5′toSMA and AMVNS7T, with pRD136 as a template (42). The second probe was a 250-nt fragment containing a 146-nt sequence complementary to R-U5 of SNV viral RNA (R-U5 probe). This probe was generated by transcribing a DNA fragment amplified with primers SNVU3R and JD2143′U5, with pJD214 as a template (17).
RNase protection assays were performed using the RPAII kit (Ambion) as specified by the manufacturer. For the detection of cellular RNAs, 20 μg of total cellular RNA was used in each sample. Equal volumes of RNA obtained from viral pellets were analyzed under the same conditions, with the hygro, gag, or R-U5 probe in separate assays. RNA fragments were resolved on 5% Tris-boric acid-EDTA-urea polyacrylamide Ready Gels (Bio-Rad) and exposed to X-OMAT film (Kodak) and a PhosphorImager cassette (Bio-Rad). Quantification of the RNA fragments was performed using the Quantity One software program (version 4.10) on a PhosphorImager (Bio-Rad).
RT assay.
The RT activity of each viral sample was determined using an equal volume of the concentrated virus by modification of the standard method (6, 59), which has also been described recently (15). In general, samples from a single set of transfection experiments generated similar RT activities, with less than twofold variation.
RESULTS
System used to examine the role of the hairpin pair motif in specific RNA recognition by SNV and MLV proteins.
We hypothesized that the differences between the hairpin pair motifs of MLV and SNV are responsible for the inability of MLV Gag proteins to recognize SNV E, based on the proposed central role of this motif in the function of MLV Ψ (47). To test this hypothesis, we examined the abilities of MLV and SNV proteins to recognize vectors containing SNV E, MLV Ψ, or chimeric packaging signals with heterologous hairpin pairs.
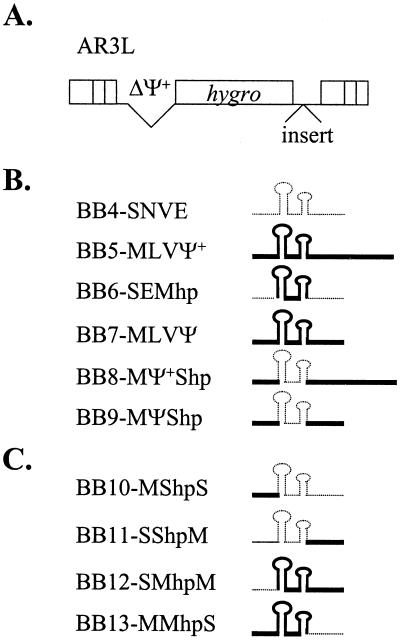
A series of vectors containing different packaging signals was constructed. All of these vectors were derived from an MLV-based vector, pAR3L (Fig. 2A). pAR3L contains hygro and MLV cis-acting elements except Ψ+, which was completely deleted. Various packaging signals were inserted into the 3′ untranslated region of pAR3L between hygro and the 3′ long terminal repeat (LTR). These sequences were inserted into the 3′ untranslated region instead of the natural 5′ untranslated region to avoid the possible effects of these packaging signals on reverse transcription and gene expression (11, 48).
FIG. 2.
Schematic representation of vectors containing canonical or chimeric packaging signals. (A) Structure of vector AR3L. AR3L expresses the hygromycin phosphotransferase B gene (hygro) and contains MLV-derived cis-acting elements. The MLV extended packaging signal is deleted in AR3L (ΔΨ+). Various packaging signals were inserted into the 3′ untranslated region of AR3L (“insert”). Open boxes at the two ends of AR3L represent LTRs. (B) Structures of canonical packaging signals and chimeric packaging signals with heterologous hairpin pairs. (C) Structures of chimeric packaging signals with one of the regions flanking the hairpin pair replaced with the heterologous counterpart. Structures of the packaging signals are shown on the right, whereas names of resulting vectors are shown on the left. Thick solid lines indicate MLV-derived sequences; thin dotted lines denote SNV-derived sequences.
The structures of six vectors containing different packaging signals are shown in Fig. 2B; three of these vectors have canonical packaging signals (SNV E, MLV Ψ+, and MLV Ψ), whereas the other three vectors contain chimeric sequences (SNV E with MLV hairpin pair [SEMhp], MLV Ψ+ with SNV hairpin pair [MΨ+Shp], and MLV Ψ with SNV hairpin pair [MΨShp]). All of the chimeric packaging signals contained the exact replacement of the hairpin pair sequences (Fig. 1).
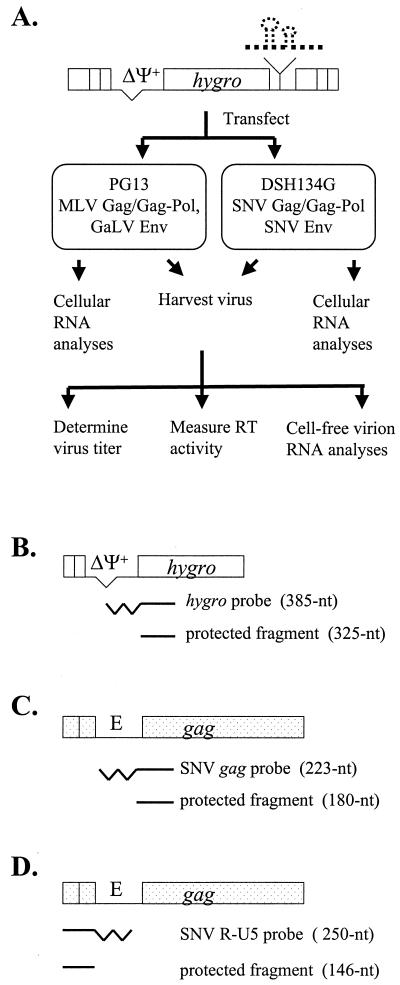
The abilities of MLV and SNV proteins to recognize these different chimeric packaging signals were examined by using the protocol illustrated in Fig. 3A. These vectors were introduced into helper cells expressing MLV Gag/Gag-Pol (PG13) or SNV Gag/Gag-Pol (DSH134G) by DNA transfection. Hygromycin-resistant colonies from each DNA sample were pooled; in general, each pool contained more than 500 colonies. An equal number of cells was plated from each cell pool, and virus was harvested from these cells 48 h later. For each virus sample, one portion was used to measure RT activity and isolate cell-free virion RNA, whereas the other portion was used to infect D17 target cells. Infected D17 target cells were subjected to hygromycin selection, and the number of resistant colonies was used to calculate viral titer. In addition to the cell-free virion RNA, total cellular RNA was isolated from each pool to directly examine the level of viral RNA expression. This protocol was designed to allow only one round of retroviral replication to ensure that the differences observed between these vectors were not amplified by multiple rounds of replication.
FIG. 3.
Experimental protocol and probes used in RNase protection assay. (A) Experimental protocol. Various vectors were separately transfected into PG13 or DSH134G cells. Cellular RNAs were isolated from stably transfected cell pools, and viruses were harvested from these pools. For each viral sample, viral titer and RT activity were measured, and cell-free virion RNA analysis was performed. (B) hygro probe used to detect vector RNA. The partial structure of AR3L-derived vectors is shown on the top, the full-length hygro probe is shown in the middle, and the expected protected fragment is shown on the bottom. (C and D) gag probe and R-U5 probes, respectively, used to detect wild-type SNV RNA as an internal control. Thick straightline, protected portion of the probe; thick zigzag line, unprotected portion of the probe; E, SNV packaging signal. Other abbreviations are the same as in Fig. 2.
Viral titers generated by vectors with different packaging signals.
Although all of the chimeric packaging signals were constructed by precise swapping of the motifs, it is still possible that these signals may not form correct RNA structures, therefore losing their ability to be recognized by Gag polyprotein. SNV Gag can recognize both SNV E and MLV Ψ; therefore, if these chimeric packaging signals retain their functions, they should be recognized by SNV Gag. A comparison of viral titers generated from 6 or 10 independent experiments using SNV-based DSH134G helper cells is shown in Table 2. Vector BB4-SNVE contains the SNV packaging signal and was used as a standard for comparison of viral titers propagated by SNV Gag. In each set of experiments, virus titers generated from various vectors were first standardized to the RT activities and then compared to the RT-standardized BB4-SNVE titer. BB4-SNVE generated titers with a mean of 3.3 × 104 CFU/ml (standard error of 0.6 × 104 CFU/ml).
TABLE 2.
Virus titers generated by vectors containing canonical and hairpin pair chimeric packaging signals
| Vector | Data for cell line:
|
|||
|---|---|---|---|---|
| DSH134G (SNV Gag/Gag-Pol)
|
PG13 (MLV Gag/Gag-Pol)
|
|||
| No. of expts | Viral titer ratioa | No. of expts | Viral titer ratiob | |
| AR3L | 10 | 0.026 ± 0.006 | 7 | 0.0019 ± 0.0005 |
| BB4-SNVE | 10 | 1 | 11 | 0.014 ± 0.003 |
| BB5-MLVψ+ | 6 | 3.3 ± 0.9 | 7 | 1.2 ± 0.2 |
| BB6-SEMhp | 10 | 0.48 ± 0.09 | 7 | 0.019 ± 0.007 |
| BB7-MLVψ | 6 | 1.8 ± 0.4 | 11 | 1 |
| BB8-Mψ+Shp | 6 | 1.6 ± 0.4 | 7 | 0.70 ± 0.09 |
| BB9-MψShp | 6 | 1.9 ± 0.3 | 11 | 0.35 ± 0.06 |
Virus titers were first standardized to RT activities and then standardized to the BB4-SNVE titer.
Virus titers were first standardized to RT activities and then standardized to the BB7-MLVψ titer.
Vector AR3L generated titers dramatically lower than those from the other six vectors, with all differences being statistically significant (P < 0.001). These data indicated that the deletion of the packaging signal had a dramatic impact on the viral titer. All statistical analyses were performed using paired t tests with a modified Bonferroni approach to control the type 1 error rate (per comparison alpha set at 0.01). As previously shown, a chimeric packaging signal with SNV E and the MLV hairpin was functional (62); however, vector BB6-SEMhp generated titers slightly lower than (48%) but significantly different from those of BB4-SNVE (P < 0.001). Vector BB7-MLVΨ generated titers that were not significantly different from those of BB4-SNVE (P = 0.274), whereas vector BB5-MLVΨ+ generated titers slightly higher (3.3-fold) than those of BB4-SNVE (P = 0.005). Vectors containing chimeric MLV packaging signals with SNV hairpins replicated efficiently; BB8-MΨ+Shp and BB9-MΨShp generated titers not statistically different from those of BB4-SNVE (P = 0.299 and 0.031, respectively). These data indicated that, although there were subtle differences, all of these packaging signals could be recognized efficiently by SNV Gag; therefore, the three chimeric packaging signals with swapped hairpin pairs were functional.
If the presence of the MLV hairpin pair motif was necessary for recognition by the MLV protein, then the MLV protein should not recognize the chimeric MLV packaging signals with replaced SNV hairpin pair motifs. Similarly, if the presence of the MLV hairpin pair motif was sufficient for the recognition, then the MLV protein should recognize the chimeric SNV packaging signal with the replaced MLV hairpin pair motif.
A comparison of the titers generated by viruses obtained from PG13 cells is shown in Table 2. In each set of experiments, which were performed 7 or 11 times, all of the vector titers were standardized to the RT activities and compared with the titer of standardized BB7-MLVΨ. BB7-MLVΨ generated titers with a mean of 10 × 104 CFU/ml (standard error of 2.7 × 104 CFU/ml). As expected, AR3L and BB4-SNVE produced very low titers (approximately 0.2 and 1.4% of the titers generated by BB7-MLVΨ, respectively); these differences were statistically significant (P < 0.001 for both vectors). BB5-MLVΨ+ generated titers similar to those of BB7-MLVΨ (P = 0.386). Vectors containing a chimeric MLV packaging signal with the SNV hairpin pair generated viral titers that were 70 and 35% of the BB7-MLVΨ titers; this result is in sharp contrast to the BB4-SNVE titers (1.4% of BB7-MLVΨ titer). These data indicate that the SNV hairpin pair structure functionally replaces the MLV hairpin pair and is recognized by MLV Gag. However, the interaction between the SNV hairpin and MLV Gag may not have been as efficient, because BB9-MΨShp titers are significantly lower than those of BB7-MLVΨ (P < 0.001). BB6-SEMhp generated low viral titers that were significantly different from those of BB7-MLVΨ (P < 0.001) and not significantly different from those of BB4-SNVE (P = 0.027). This finding indicates that the presence of the MLV hairpin pair in the SNV packaging signal does not result in a significant rescue of RNA packaging by the MLV protein. Together, these data demonstrate that the MLV and SNV hairpin pair motifs can functionally replace each other with only minor effects on recognition by MLV or SNV proteins. Therefore, the hairpin pair motif is not the major component in SNV E that prevents MLV Gag from recognizing SNV RNA.
Cellular and viral RNA analyses of the vectors containing hairpin pair chimeric packaging signals.
RNase protection assays were performed to directly examine the expression of vector RNA in transfected helper cells and the amount of vector RNA encapsidated in cell-free virions. An RNA probe containing sequences from the simian virus 40 promoter and hygro was used in these assays to detect vector RNA. Of the 385-nt probe, only a 325-nt portion hybridized to the RNA of AR3L and all AR3L-derived vectors and was protected from RNase treatment (Fig. 3B).
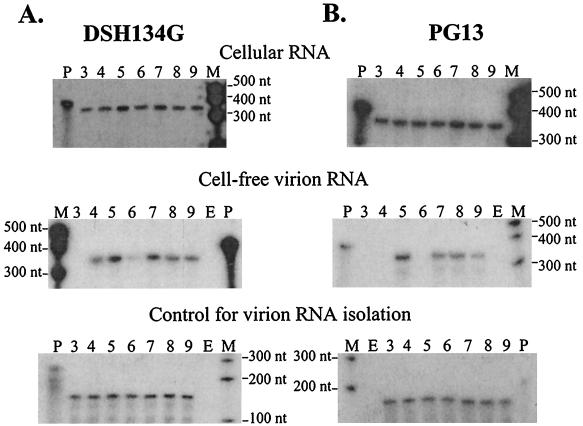
Cellular RNAs were isolated from the transfected cell pools, and an equal amount of total cellular RNA from each transfected cell pool was used to determine the levels of vector RNA expression by RNase protection assays. The top panels of Fig. 4A and B show examples of RNase protection assays with cellular RNA isolated from transfected DSH134G and PG13 cell pools, respectively. All of the cell pools within each set of transfections generally had similar levels of vector RNA expression (with less than twofold variation) as quantified by a PhosphorImager.
FIG. 4.
RNase protection analyses of vectors containing canonical or hairpin pair chimeric packaging signals. (A) RNA samples derived from transfected DSH134G cell pools. (B) RNA samples derived from transfected PG13 cell pools. Top panels, vector expression in transfected cells (cellular RNA); middle panels, cell-free viral RNA released from transfected cells; bottom panels, wild-type SNV RNA added to cell-free viruses as an internal control. Lanes: P, full-length probe; M, molecular size marker; E, empty lane; 3, AR3L; 4, BB4-SNVE; 5, BB5-MLVΨ+; 6, BB6-SEMhp; 7, BB7-MLVΨ; 8, BB8-MΨ+Shp; 9, BB9-MΨShp.
The amounts of cell-free viral RNAs released from transfected helper cell pools were determined, and examples of the RNase protection assays are shown in the middle panels of Fig. 4. In cell-free viral RNAs isolated from transfected DSH134G cells, viral RNAs were detected in samples from all vectors except the AR3L sample (lane 3). RNA quantification indicated that the amounts of RNA packaged in cell-free virions closely followed the viral titers (data not shown). In sharp contrast, not all the vectors with packaging signals were efficiently encapsidated in cell-free viral RNAs produced from transfected PG13 cells. As expected, viral RNAs were detected in BB5-MLVΨ+ and BB7-MLVΨ samples (lanes 5 and 7, respectively) but not in AR3L and BB4-SNVE samples (lanes 3 and 4, respectively). Viral RNA was not detected in BB6-SEMhp (lane 6), indicating inefficient RNA packaging by MLV Gag and providing an explanation for the low viral titers generated by these vectors in PG13 cells. RNA analyses also indicated that BB8-MΨ+Shp and BB9-MΨShp cell-free viral RNAs (lanes 8 and 9, respectively) were packaged by MLV Gag with a two- to threefold reduction in efficiency relative to that of BB7-MLVΨ (lane 7). These results are consistent with the viral titer data.
To demonstrate that the differences detected in the amounts of cell-free viral RNAs reflected packaging efficiencies and not loss of RNA during isolation and RNase protection procedures, these experiments were repeated at least four times. In addition, an aliquot of replication-competent wild-type SNV was added to each viral sample as an internal control prior to viral RNA isolation to monitor the loss of RNA during this procedure. The amount of SNV in each sample was also examined by the RNase protection assay using RNA probes that hybridized to SNV gag (gag probe) or the R-U5 region of SNV RNA (R-U5 probe) (Fig. 3C and D). DSH134G cells express SNV gag-pol from a helper construct lacking E and SNV LTR sequences. Although RNA from this helper construct should not be packaged into virions, to avoid any potential complications, the R-U5 probe was used to detect wild-type SNV RNA added to the viral samples isolated from DSH134G cells. Two representative RNase protection assays of the internal control SNV RNA from various samples are shown in the lower panels of Fig. 4. Within each experiment, all samples had similar amounts of internal control SNV RNA, demonstrating that the viral RNAs were recovered at similar efficiencies. These RNA analyses indicated that the amounts of RNA that were packaged into cell-free virions generated from both sets of cells were proportional to the viral titers.
The role of the flanking region(s) in nonreciprocal RNA packaging between MLV and SNV.
To further dissect the regions that affect specific RNA-protein interactions, we generated additional pAR3L-based vectors that contain packaging signals with one of the flanking regions replaced by the heterologous counterparts. The structures of the four vectors containing chimeric packaging signals are shown in Fig. 2C. The origin of the sequence is reflected in the vector name. For example, pBB10-MShpS contains a chimeric packaging signal consisting of the MLV upstream flanking region, SNV hairpin pair, and SNV downstream flanking region; therefore, the abbreviation MShpS is used.
These vectors were introduced into DSH134G and PG13 cells, viral titers were determined, and RNA analyses were performed as described above. Comparisons of viral titers generated by these vectors in DSH134G cells from 6 to 10 experiments are summarized in Table 3. All of the vectors containing chimeric packaging signals with swapped flanking regions replicated well in DSH134G cells. None of these chimeras generated titers significantly different from the titers of BB4-SNVE (P = 0.691, 0.591, 0.04, and 0.25 for BB10-MShpS, BB11-SShpM, BB12-SMhpM, and BB13-MMhpS, respectively). This result indicated that these chimeric packaging signals were functional. However, vectors containing heterologous flanking regions did not generate similar titers in PG13 cells. BB10-MShpS and BB11-SShpM contained SNV E with swapped MLV upstream and downstream flanking sequences, respectively. Both vectors generated significantly lower viral titers than those of BB7-MLVΨ (P < 0.001 for both vectors), indicating that neither region could fully rescue RNA packaging by MLV Gag. However, compared with BB4-SNVE, BB10-MShpS had a fivefold increase in the viral titer, which was statistically significant (P = 0.008). In contrast, the viral titers generated by BB11-SShpM were not significantly different from those of BB4-SNVE (P = 0.245). These data indicated that the MLV upstream flanking region could partially rescue packaging by MLV Gag, whereas the MLV downstream flanking region did not have this effect.
TABLE 3.
Virus titers generated by vectors containing canonical and flanking region chimeric packaging signals
| Vector | Data for cell line:
|
|||
|---|---|---|---|---|
| DSH134G (SNV Gag/Gag-Pol)
|
PG13 (MLV Gag/Gag-Pol)
|
|||
| No. of expts | Viral titer ratioa | No. of expts | Viral titer ratiob | |
| BB4-SNVE | 10 | 1 | 11 | 0.014 ± 0.003 |
| BB7-MLVψ | 6 | 1.8 ± 0.4 | 11 | 1 |
| BB10-MShpS | 6 | 1.0 ± 0.3 | 7 | 0.073 ± 0.01 |
| BB11-SShpM | 6 | 1.0 ± 0.4 | 7 | 0.011 ± 0.003 |
| BB12-SMhpM | 6 | 0.5 ± 0.1 | 7 | 0.069 ± 0.02 |
| BB13-MMhpS | 6 | 0.8 ± 0.3 | 7 | 0.36 ± 0.08 |
Virus titers were first standardized to RT activities and then standardized to the BB4-SNVE titer.
Virus titers were first standardized to RT activities and then standardized to the BB7-MLVψ titer.
If replacing the SNV upstream flanking region with that of MLV resulted in a gain of RNA packaging by MLV Gag, then it is likely that replacing the MLV upstream flanking region with that of SNV would cause a reduction in RNA packaging. BB12-SMhpM contained MLV Ψ with a swapped SNV upstream flanking region. This vector generated titers that were 14-fold lower than those of BB7-MLVΨ; the difference between these two sets of viral titers was statistically significant (P < 0.001).
BB13-MMhpS contained MLV Ψ with a swapped SNV downstream flanking region. This vector generated titers that were threefold lower than those of BB7-MLVΨ; the difference between these two sets of viral titers was statistically significant (P < 0.001).
RNA analyses of vectors with chimeric packaging signals containing swapped flanking regions.
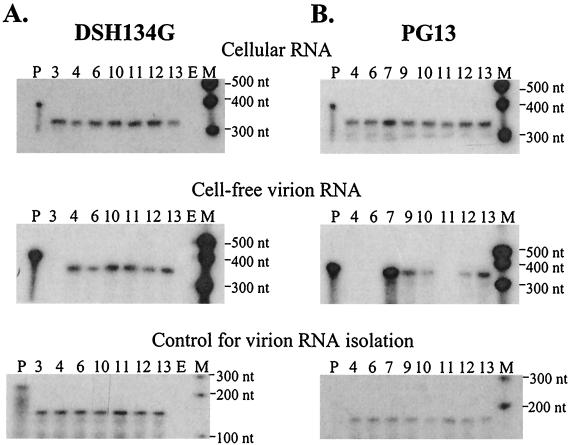
RNase protection assays were performed to monitor vector RNA gene expression in transfected DSH134G and PG13 cells; representative assays are shown in the top panels of Fig. 5A and B, respectively. Cellular vector RNA expression in each set of transfections was generally within a twofold variation among the different vectors. Cell-free viral RNAs released from DSH134G cells that were transfected with different vectors contained amounts of RNA similar to those of BB4-SNVE with the exception of AR3L, which did not contain a detectable amount of RNA. Cell-free viral RNAs released from transfected PG13 cells reflected the viral titers generated from these cells. The two vectors containing SNV E with the replaced MLV upstream or downstream flanking regions were packaged by MLV Gag at different efficiencies. BB11-SShpM resembled BB4-SNVE: neither vector generated a sufficient amount of cell-free viral RNA to be detected in the assay (lanes 11 and 4, respectively). In contrast, BB10-MShpS generated a detectable amount of cell-free viral RNA, although the level was approximately 13-fold lower than that of BB7-MLV Ψ (lanes 10 and 13, respectively). Both vectors containing MLV Ψ with swapped flanking upstream or downstream regions generated detectable amounts of cell-free viral RNA. BB12-SMhpM RNA was packaged 13-fold less efficiently, whereas BB13-MMhpS RNA was packaged threefold less efficiently, than BB7-MLVΨ RNA. The different levels of cell-free viral RNA were not caused by loss of RNA during the isolation and analysis procedures, because all of these samples had similar amounts of the internal control wild-type SNV RNA (bottom panels, Fig. 5A and B).
FIG. 5.
RNase protection analyses of vectors containing canonical and flanking region chimeric packaging signals. (A) RNA samples generated from DSH134G cells. (B) RNA samples generated from PG13 cells. Panel descriptions are the same as those for Fig. 4. Lanes: P, full-length probe; M, molecular size marker; E, empty lane; 3, AR3L; 4, BB4-SNVE; 6, BB6-SEMhp; 7, BB7-MLVΨ; 9, BB9-MΨShp; 10, BB10-MShpS; 11, BB11-SShpM; 12, BB12-SMhpM; 13, BB13-MMhpS.
Taken together, the viral titers and RNA analyses indicated that both the upstream and downstream flanking sequences affected the recognition by MLV Gag; however, the upstream flanking region had a much more significant role than did the downstream sequence. The presence of the MLV upstream flanking region could partially rescue RNA packaging by MLV Gag, and when it was replaced with the SNV upstream flanking region, a dramatic reduction of viral RNA packaging was observed. The MLV downstream flanking region had a lower effect on the efficiency of MLV Gag-mediated RNA packaging. The presence of the MLV downstream flanking region alone could not rescue RNA packaging by MLV Gag; however, replacing this region with the comparable SNV sequence resulted in only a threefold reduction in RNA packaging.
DISCUSSION
Use of chimeric RNA to study protein-RNA interactions.
In this report, we determined the ability of chimeric packaging signals to be recognized by two different Gag polyproteins. Using this strategy, we delineated the contribution of the different RNA motifs in the nonreciprocal packaging between SNV and MLV. We were also able to determine that although MLV Gag cannot efficiently package vectors containing SNV E, MLV Gag can interact with some elements in SNV E. Therefore, chimeric RNA can be used as a powerful tool to study specific viral protein-RNA motif interactions. In addition, this study demonstrates the fluidity of the RNA sequences in packaging signals. SNV E and MLV Ψ do not contain significant homology, and yet all of the seven chimeras are functional. This study also emphasizes the importance of the structure of various RNA motifs during the interaction of Gag and packaging signal.
Mechanistically similar and distinct interactions of SNV Gag and MLV Gag with RNA motifs.
The structurally conserved hairpin pair motif plays an important role in specific RNA packaging of MLV-related retroviruses. In SNV, deletion of one of the hairpins in the hairpin pair resulted in at least a 50-fold decrease in the viral titer (62). In MLV, deletion of the hairpin pair resulted in a 250-fold decrease in viral titer (48). Our data indicated that, despite the importance of the hairpin pair motif in RNA packaging, this motif was not the major element that distinguished between the SNV and MLV packaging signals during MLV Gag-RNA interactions. In contrast to the previously observed severe effect from deletion of the hairpin pairs, replacement of the hairpin pair with the heterologous counterpart had only a slight effect in viral titers (up to threefold reduction). MLV Gag and SNV Gag contain limited identity (35%) in protein sequences, and the two hairpin pairs also do not contain significant homology. Therefore, these results suggest that, despite the limited identity between MLV Gag and SNV Gag, these two polyproteins interact with the hairpin pair RNA structure in a mechanistically similar manner.
Despite the similarity in Gag-hairpin pair interactions, MLV and SNV Gag polyproteins differ significantly in their requirements for other packaging signal motifs. MLV Gag strongly prefers the MLV upstream flanking region (Table 3 and Fig. 5). In contrast, SNV Gag does not significantly distinguish between upstream sequences from MLV and SNV. Pairs of vectors that differed only in the upstream flanking regions generated similar viral titers, e.g., BB4-SNVE and BB10MShpS (P = 0.691) and BB7-MLVΨ and BB12-SMhpM (P = 0.566). These results indicated that SNV Gag and MLV Gag have different requirements for the upstream sequences that are essential for efficient RNA packaging.
Comparison of the upstream flanking region in MLV Ψ and SNV E.
These studies indicate that elements in the upstream flanking region of MLV Ψ are required for optimal MLV Gag-mediated RNA packaging. The sequence length from splice donor to hairpin pair is 102 nt in MLV and 30 nt in SNV (Fig. 1). The defined Ψ includes 97 of the 102 nt (39, 41, 46), whereas the defined E contains 18 of the 30 nt (61, 62). In MLV Ψ, two secondary structures, termed hairpins A and B, were proposed to be present upstream of the hairpin pair described in this report. The canonical Ψ contains only half of hairpin A but has the complete hairpin B, which is located immediately upstream of the hairpin pair (Fig. 1).
In a recent binding study using purified MLV NC protein and short RNA, it was found that NC bound with high affinity to RNA containing hairpin B plus the hairpin pair, but not to RNA consisting of only the hairpin pair (18). This observation is in complete agreement with our findings that the upstream sequence from MLV is critical to RNA packaging by MLV Gag. In a different study, MLV Ψ was replaced by human immunodeficiency virus type 1 (HIV-1) Ψ in an MLV-based vector; because MLV Gag could not recognize HIV-1 RNA, this RNA could not be packaged by MLV Gag at a detectable level (47). When most of hairpin B, the hairpin pair, and the 46-nt downstream sequence were inserted into the vector, packaging of vector RNA was partially rescued. Although we could not directly compare our data with this previous study because our chimeras were constructed with precise replacement of hairpins or flanking regions, all these studies are in general agreement that the upstream sequence plays an important role in RNA-MLV Gag interaction.
In a deletion analysis, it was shown previously that deletion of hairpin A or hairpin B resulted in a seven- or sixfold decrease in the level of RNA encapsidation, respectively (48). Interestingly, double deletion did not have an additive effect: deleting both hairpin A and hairpin B but retaining the 54 nt between the hairpins resulted in only a fourfold reduction in RNA packaging. These data indicated that there might be complex interactions between different motifs in the upstream sequence. Therefore, despite strong evidence of the importance of hairpin B, we cannot eliminate the possibility that other sequences in the upstream region also have an impact on RNA-protein interactions.
In this study, the canonical SNV E was inserted into the 3′ untranslated region of an MLV vector. One possible argument is that the canonical SNV E has a portion of the sequence missing in the upstream region, which leads to the inability of MLV Gag to recognize SNV RNA. In other experiments, we have used SNV vectors with the defined E as well as all of the sequences upstream from E and have shown that MLV Gag cannot package these RNAs efficiently (13, 14). Therefore, the inability of the SNV E upstream region to replace the MLV counterpart is not due to incomplete sequences being present in the vectors.
Cooperative effect of the three regions in MLV Ψ that alter MLV Gag-mediated RNA packaging.
The most optimal MLV Gag-mediated RNA packaging involves the presence of all three regions from MLV Ψ. When a single region of SNV E was replaced by the corresponding Ψ region, only the presence of the Ψ upstream region yielded a detectable increase (fivefold) in RNA packaging by MLV Gag. Although neither the hairpin pair nor the downstream region by itself could rescue RNA packaging by MLV Gag, the presence of both regions resulted in a fivefold increase in RNA packaging by MLV Gag (BB6-SEMhp and BB11-SShpM versus BB12-SMhpM). Similarly, either hairpin pair or downstream region in the presence of the upstream region could further enhance RNA packaging 25-fold by MLV Gag (BB9-MΨShp and BB13-MMhpS versus BB10-MShpS). MLV Gag packaged a Ψ-containing vector 70-fold more efficiently than it packaged an E-containing vector. Therefore, increasing the number of MLV-derived regions improves MLV Gag-mediated RNA packaging in a nonlinear, nonadditive manner, indicating that there is a cooperative effect among these regions during RNA packaging. This effect is likely to exist at the level of RNA-Gag interaction; for example, it is possible that Gag recognizes more than one region, and having these sequences together increases the binding affinity or binding efficiency of Gag.
Weak recognition of MLV Gag with SNV E and other RNA elements important for RNA packaging.
The viral titers generated from transfected PG13 cells by BB4-SNVE were significantly higher than those generated by AR3L (P < 0.001; 2 × 103 and 3 × 102 CFU/ml, respectively). We therefore conclude that MLV Gag can interact with SNV E, albeit at a very weak level.
Besides MLV Ψ or Ψ+, sequences located elsewhere in the viral genome have been proposed to impact RNA packaging. These sequences include U5 and a 21-nt segment in the 3′ untranslated region (49, 64). Because all packaging signals were inserted into AR3L, which contain U5 and the 21-nt segment of the 3′ untranslated region, the roles of these sequences were not addressed in this study. However, without any canonical packaging signals, AR3L was able to replicate at a 0.2 to 3% level compared with the vectors that had the canonical packaging signal (Table 2). It is possible that this level of replication is sustained by vector sequences that allow a low level of RNA packaging. The ability of AR3L to replicate at a low level also led us to speculate on the evolutionary role of minor RNA packaging sequences. One possible explanation is that the presence of these sequences allows low-level rescue of a virus that suffered a detrimental mutation in the major packaging signal. Interestingly, the low level of replication of AR3L was observed in both SNV- and MLV-based helper cells, suggesting that SNV Gag can also recognize the minor packaging signals from MLV. Therefore, the presence of minor RNA packaging signals may be common among retroviruses.
Acknowledgments
We thank Douglas Powell for multiple discussions and his expert help with the statistical analyses of this study, Shiaolan Yang for her generous gift of plasmids, and Dusty Miller and Ralph Dornburg for sharing cell lines. We thank Vinay K. Pathak for encouragement, intellectual input, and critical reading of the manuscript and Alan Rein for encouragement, helpful discussions, and critical reading of the manuscript. We thank Anne Arthur for expert editorial help with the manuscript and Sara Cheslock, Que Dang, William Fu, Sook-Kyung Lee, Dexter Poon, Terence Rhodes, and Heather Wargo for critical reading of the manuscript. We also thank Mike Summers for communicating data prior to publication.
B.E.B. was partially supported by the medical scientist training program at West Virginia University. This work is supported by the HIV Drug Resistance Program, NCI, NIH.
REFERENCES
- 1.Adam, M. A., and A. D. Miller. 1988. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J. Virol. 62:3802-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini, A., and R. A. Young. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 64:1920-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. J., P. Lee, K. L. Levine, J. S. Sang, S. A. Shah, O. O. Yang, P. R. Shank, and M. L. Linial. 1992. Molecular cloning and characterization of the RNA packaging-defective retrovirus SE21Q1b. J. Virol. 66:204-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, D. J., J. Stone, R. Lum, and M. L. Linial. 1995. The packaging phenotype of the SE21Q1b provirus is related to high proviral expression and not trans-acting factors. J. Virol. 69:7319-7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aschoff, J. M., D. Foster, and J. M. Coffin. 1999. Point mutations in the avian sarcoma/leukosis virus 3′ untranslated region result in a packaging defect. J. Virol. 73:7421-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baltimore, D. 1970. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 226:1209-1211. [DOI] [PubMed] [Google Scholar]
- 7.Bender, M. A., T. D. Palmer, R. E. Gelinas, and A. D. Miller. 1987. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J. Virol. 61:1639-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 9.Berkowitz, R. D., J. Luban, and S. P. Goff. 1993. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol. 67:7190-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkowitz, R. D., A. Ohagen, S. Hoglund, and S. P. Goff. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69:6445-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berlioz, C., and J. L. Darlix. 1995. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J. Virol. 69:2214-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Browning, M. T., R. D. Schmidt, K. A. Lew, and T. A. Rizvi. 2001. Primate and feline lentivirus vector RNA packaging and propagation by heterologous lentivirus virions. J. Virol. 75:5129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Certo, J. L., T. O. Kabdulov, M. L. Paulson, J. A. Anderson, and W. S. Hu. 1999. The nucleocapsid domain is responsible for the ability of spleen necrosis virus (SNV) Gag polyprotein to package both SNV and murine leukemia virus RNA. J. Virol. 73:9170-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Certo, J. L., B. F. Shook, P. D. Yin, J. T. Snider, and W. S. Hu. 1998. Nonreciprocal pseudotyping: murine leukemia virus proteins cannot efficiently package spleen necrosis virus-based vector RNA. J. Virol. 72:5408-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang, Q., and W. S. Hu. 2001. Effects of homology length in the repeat region on minus-strand DNA transfer and retroviral replication. J. Virol. 75:809-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dornburg, R., and H. M. Temin. 1990. Presence of a retroviral encapsidation sequence in nonretroviral RNA increases the efficiency of formation of cDNA genes. J. Virol. 64:886-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dougherty, J. P., and H. M. Temin. 1987. A promoterless retroviral vector indicates that there are sequences in U3 required for 3′ RNA processing. Proc. Natl. Acad. Sci. USA 84:1197-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Souza, V., J. Melamed, D. Habib, K. Pullen, K. Wallace, and M. F. Summers. 2001. Identification of a high affinity nucleocapsid protein binding element within the Moloney murine leukemia virus Ψ-RNA packaging signal: implications for genome recognition. J. Mol. Biol. 314:217-232. [DOI] [PubMed] [Google Scholar]
- 19.Duesberg, P. H. 1968. Physical properties of Rous sarcoma virus RNA. Proc. Natl. Acad. Sci. USA 60:1511-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupraz, P., S. Oertle, C. Meric, P. Damay, and P. F. Spahr. 1990. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J. Virol. 64:4978-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupraz, P., and P. F. Spahr. 1992. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J. Virol. 66:4662-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Embretson, J. E., and H. M. Temin. 1987. Lack of competition results in efficient packaging of heterologous murine retroviral RNAs and reticuloendotheliosis virus encapsidation-minus RNAs by the reticuloendotheliosis virus helper cell line. J. Virol. 61:2675-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geigenmuller, U., and M. L. Linial. 1996. Specific binding of human immunodeficiency virus type 1 (HIV-1) Gag-derived proteins to a 5′ HIV-1 genomic RNA sequence. J. Virol. 70:667-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelick, R. J., S. M. Nigida, Jr., J. W. Bess, Jr., L. O. Arthur, L. E. Henderson, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gritz, L., and J. Davies. 1983. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25:179-188. [DOI] [PubMed] [Google Scholar]
- 27.Housset, V., H. De Rocquigny, B. P. Roques, and J. L. Darlix. 1993. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J. Virol. 67:2537-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, J. S., R. W. Allan, and H. M. Temin. 1993. Alteration of location of dimer linkage sequence in retroviral RNA: little effect on replication or homologous recombination. J. Virol. 67:3151-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julias, J. G., T. Kim, G. Arnold, and V. K. Pathak. 1997. The antiretrovirus drug 3′-azido-3′-deoxythymidine increases the retrovirus mutation rate. J. Virol. 71:4254-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh, I., H. Kyushiki, Y. Sakamoto, Y. Ikawa, and Y. Yoshinaka. 1991. Bovine leukemia virus matrix-associated protein MA(p15): further processing and formation of a specific complex with the dimer of the 5′-terminal genomic RNA fragment. J. Virol. 65:6845-6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai, S., and M. Nishizawa. 1984. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol. Cell. Biol. 4:1172-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaye, J. F., and A. M. Lever. 1998. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J. Virol. 72:5877-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konings, D. A., M. A. Nash, J. V. Maizel, and R. B. Arlinghaus. 1992. Novel GACG-hairpin pair motif in the 5′ untranslated region of type C retroviruses related to murine leukemia virus. J. Virol. 66:632-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kung, H. J., J. M. Bailey, N. Davidson, M. O. Nicolson, and R. M. McAllister. 1975. Structure, subunit composition, and molecular weight of RD-114 RNA. J. Virol. 16:397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kung, H. J., J. M. Bailey, N. Davidson, P. K. Vogt, M. O. Nicolson, and R. M. McAllister. 1975. Electron microscope studies of tumor virus RNA. Cold Spring Harbor Symp. Quant. Biol. 39:827-834. [DOI] [PubMed] [Google Scholar]
- 36.Lever, A., H. Gottlinger, W. Haseltine, and J. Sodroski. 1989. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 63:4085-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lever, A. M. 2000. HIV RNA packaging and lentivirus-based vectors. Adv. Pharmacol. 48:1-28. [DOI] [PubMed] [Google Scholar]
- 38.Luban, J., and S. P. Goff. 1991. Binding of human immunodeficiency virus type 1 (HIV-1) RNA to recombinant HIV-1 gag polyprotein. J. Virol. 65:3203-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ly, H., D. P. Nierlich, J. C. Olsen, and A. H. Kaplan. 1999. Moloney murine sarcoma virus genomic RNAs dimerize via a two-step process: a concentration-dependent kissing-loop interaction is driven by initial contact between consecutive guanines. J. Virol. 73:7255-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mann, R., and D. Baltimore. 1985. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNAs. J. Virol. 54:401-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mann, R., R. C. Mulligan, and D. Baltimore. 1983. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell 33:153-159. [DOI] [PubMed] [Google Scholar]
- 42.Martinez, I., and R. Dornburg. 1995. Improved retroviral packaging lines derived from spleen necrosis virus. Virology 208:234-241. [DOI] [PubMed] [Google Scholar]
- 43.Meric, C., E. Gouilloud, and P. F. Spahr. 1988. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. J. Virol. 62:3328-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meric, C., and P. F. Spahr. 1986. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J. Virol. 60:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7:980-982, 984-986, 989-990. [PMC free article] [PubMed] [Google Scholar]
- 47.Mougel, M., and E. Barklis. 1997. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 71:8061-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mougel, M., Y. Zhang, and E. Barklis. 1996. cis-active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J. Virol. 70:5043-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy, J. E., and S. P. Goff. 1989. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: effects on RNA packaging and reverse transcription. J. Virol. 63:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poon, D. T., J. Wu, and A. Aldovini. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 70:6607-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riggs, J. L., R. M. McAllister, and E. H. Lennette. 1974. Immunofluorescent studies of RD-114 virus replication in cell culture. J. Gen. Virol. 25:21-29. [DOI] [PubMed] [Google Scholar]
- 52.Rizvi, T. A., and A. T. Panganiban. 1993. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J. Virol. 67:2681-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakalian, M., J. W. Wills, and V. M. Vogt. 1994. Efficiency and selectivity of RNA packaging by Rous sarcoma virus Gag deletion mutants. J. Virol. 68:5969-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shank, P. R., and M. Linial. 1980. Avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: characterization of a deletion in the provirus. J. Virol. 36:450-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steeg, C. M., and V. M. Vogt. 1990. RNA-binding properties of the matrix protein (p19gag) of avian sarcoma and leukemia viruses. J. Virol. 64:847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, Plainview, N.Y. [PubMed]
- 59.Temin, H. M., and S. Mizutani. 1970. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226:1211-1213. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe, S., and H. M. Temin. 1983. Construction of a helper cell line for avian reticuloendotheliosis virus cloning vectors. Mol. Cell. Biol. 3:2241-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe, S., and H. M. Temin. 1982. Encapsidation sequences for spleen necrosis virus, an avian retrovirus, are between the 5′ long terminal repeat and the start of the gag gene. Proc. Natl. Acad. Sci. USA 79:5986-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, S., and H. M. Temin. 1994. A double hairpin structure is necessary for the efficient encapsidation of spleen necrosis virus retroviral RNA. EMBO J. 13:713-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yin, P. D., V. K. Pathak, A. E. Rowan, R. J. Teufel II, and W. S. Hu. 1997. Utilization of nonhomologous minus-strand DNA transfer to generate recombinant retroviruses. J. Virol. 71:2487-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, S. S., J. M. Kim, and S. Kim. 2000. The 17 nucleotides downstream from the env gene stop codon are important for murine leukemia virus packaging. J. Virol. 74:8775-8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71:6765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, Y., and E. Barklis. 1995. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 69:5716-5722. (Erratum, 71:5712, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]