SUMMARY
Acid resistance (AR) for Escherichia coli is important for its survival in the human gastrointestinal tract and involves three systems. The first AR system is dependent on the sigma factor RpoS. The second system (GAD system) requires glutamate decarboxylase isoforms encoded by the gadA and gadB genes. The third system (ARG system) requires arginine decarboxylase encoded by adiA. Loss of topoisomerase I function from topA deletion or Tn10 insertion mutations lowered the resistance to killing by pH 2 or 2.5 treatment by 10 to >100 fold. The RpoS and GAD systems were both affected by the topA mutation but the ARG system of acid resistance was not affected. Northern blot analysis showed that induction of gadA and gadB transcription in stationary phase and at pH 5.5 was decreased in the topA mutant. Western blot analysis showed that the topA mutation did not affect accumulation of RpoS, GadX or GadW proteins. Topoisomerase I could have a direct influence on transcription of acid resistance genes. This influence did not involve R-loop formation as the overexpression of RNase H did not alleviate the decrease of acid resistance from the topA mutation. The effect of the topA mutation could be suppressed by the hns mutation so topoisomerase I might be required to counteract the effect of H-NS protein on gene expression in addition to its influence on RpoS-dependent transcription.
INTRODUCTION
For pathogenic or non-pathogenic Escherichia coli to colonize human gastroinestinal tracts, it has to survive the challenge of extreme acid pH (Giannella et al., 1972; Gordon & Small, 1973). Enterohemorrhagic E. coli strains require a low infectious dose (ID) to cause disease due partly to their high degree of acid resistance (AR) to pH of 2.5 or lower (Benjamin & Datta, 1995; Conner & Kotrola, 1995; Lin et al., 1996, Price et al., 2004). Three distinct acid resistance systems are responsible for protecting stationary phase E. coli cells from killing by the extreme low pH (Castanie-Cornet et al., 1999; Foster, 2004). The oxidative or glucose-repressed system (System 1) is dependent on the stationary-phase sigma factor RpoS (Small et al., 1994). The GAD system requires glutamate during acid challenge and involves glutamate decaroxylase activity (Lin et al., 1995; Hersh et al., 1996; De Biase et al., 1999). The ARG system requires arginine in the acid medium and arginine decarboxylase activity (Lin et al., 1995; Castanie-Cornet et al., 1999). RpoS is also required for expression of gadA and gadBC, the structural genes for glutamate decarboxylase isozymes, at neutral pH in the stationary phase (Castanie-Cornet et al., 1999; Castanie-Cornet & Foster, 2001).
Topoisomerase I is the major activity for removal of negative supercoils from DNA in E. coli, and along with DNA gyrase and topoisomerase IV, plays an important role in regulation of both global and localized supercoiling (Zechiedrich et al., 2000; Champoux, 2001). Four promoters have been characterized for the E. coli topA gene encoding topoisomerase I (Qi et al., 1997). Promoter P1 is recognized by the heat shock responsive sigma factor σ32 (Lesley et al., 1990; Qi et al., 1996) while promoter Px1 can be recognized by the stationary phase and stress responsive sigma factor σs (RpoS). The transcription of topA by these sigma factors in response to environmental challenge suggests a role of topoisomerase I function in adaptation and survival of E. coli. This hypothesis is supported by the lower survival rates of the topA deletion mutants versus topA+ isogenic strains after challenges with high temperature or oxidative stress (Qi et al., 1999; Tse-Dinh, 2000). Loss of topoisomerase I activity can affect transcription initiation at promoters due to its influence on DNA topology (Steck et al., 1993; Wang & Lynch, 1993), and is needed during transcription elongation to suppress hypernegative supercoiling and R-loop formation (Massé & Drolet, 1999; Hraiky et al., 2000). Efficient transcription of genes required for acid resistance may thus require topoisomerase I activity to be present in the cell. In Helicobacter pylori, topA was found by differential display PCR to be one of the genes induced by prolonged acid exposure (Dong et al., 2001), suggesting the involvement of topoisomerase I in adaptation to low pH by this organism. We now present results of experiments that demonstrate the effect of ΔtopA mutation on E. coli acid resistance, and on the expression of genes involved in the oxidative and GAD acid resistance mechanisms.
METHODS
Bacterial strains, plasmids and culture media
The bacterial strains and plasmids used in this study are described in Table 1. P1 transduction was carried out according to standard protocols (Miller, 1992). The insertion mutation in rpoS or topA mutant strains selected by Tetr was confirmed by Western blotting with antibodies against the targeted protein. E. coli was grown at 37°C in Luria Broth (LB), LB supplemented with 0.4% glucose (LBG), or LB buffered with either 100 mM morpholinepropanesulfonic acid (MES; pH 5.5) or with 100 mM morpholinepropanesulfonic acid (MOPS; pH 8). Overall acid resistance (AR) is measured by diluting the culture into LB pH 2.0 or pH 2.5 after overnight growth in LB buffered at pH 5.5 or pH 8.0 (Castanie-Cornet et al., 1999). To test the individual acid resistance system, survival rates were measured after treatment in E minimal medium (Vogel & Bonner, 1956) containing 0.4% glucose (EG) at pH 2.5 after different overnight growth conditions as described (Castanie-Cornet et al., 1999). For System 1 (oxidative system), overnight growth was in LB pH 5.5, followed by treatment in EG pH2.5 with no supplement. For the GAD acid resistance system, overnight culture in LBG was diluted into EG pH 2.5 supplemented with 1.5 mM glutamate. For the ARG acid resistance system, overnight growth was in brain heart infusion medium supplemented with glucose (BHIG), followed by treatment in EG pH 2.5 supplemented with 0.6 mM arginine. In the experiments involving hns mutants, M9 medium at pH 2.5 with 0.4% glucose was used as the acid challenge medium (Hommals et al., 2001). After challenge for the indicated period of time, the viable counts were determined by dilutions in M9 minimal medium (Sambrook et al., 1989) and plating on LB plates, supplemented by ampicillin (100 μg ml−1), tetracycline (15 μg ml−1), or kanamycin (25 μg ml−1) when appropriate.
Table 1.
E. coli strains and plasmids used in this study.
| Strain or plasmid | Relevant genotype and characteristics | Reference or Source |
|---|---|---|
| E. coli strains | ||
| RFM443 | rpsL galK2, Δlac74 | Drolet et al., 1995 |
| RFM445 | rpsL galK2, Δlac74, gyrB221(couR), gyrB203(Ts) | Drolet et al., 1995 |
| RFM475 | rpsL galK2, Δlac74, gyrB221(couR), gyrB203(Ts) Δ(topAcysB)204 | Drolet et al., 1995 |
| 2111 | W3110, gyrB(Ts, cou R),topA20::Tn10 | Zechiedrich et al., 2000 |
| YT481 | RFM445 topA20::Tn10 | P1(2111)x(RFM445), tetr |
| GY12 | rpoS::Tn10 | Yamashino et al., 1995 |
| YT445R | RFM445 rpoS::Tn10 | P1(GY12)x(RFM445), tetr |
| YT475R | RFM475 rpoS::Tn10 | P1(GY12)x(RFM475), tetr |
| FB20271 | MG1655 hns::Tn5 | F. Blattner Laboratory |
| YT445H | RFM445 hns::Tn5 | P1(FB20271)x(RFM445), kanr |
| YT475H | RFM475 hns::Tn5 | P1(FB20271)x(RFM475), kanr |
| Plasmids | ||
| pSK760 | pBR322 derivative with the rnhA gene expressing RNase H | Kanaya & Crouch, 1983 |
| pSK762c | pBR322 derivative with mutated rnhA gene expressing inactive RNase H | Drolet et al., 1995 |
Northern blotting and real-time PCR
Total RNA was isolated from cells grown in buffered LB to OD600 0.4–0.5 (EP, exponential phase) or 1.2–1.5 (SP, early stationary phase) using previously published procedures (Hraiky et al., 2000). The 1.4 kb gad probe for detection of both gadA and gadBC was generated by PCR as described previously (Castanie-Cornet & Foster, 2001) and labeled with [α-32P]dCTP using the Mega Prime kit from Amersham. Electrophoresis and transfer of RNA samples (5 μg) were carried out as described (Sambrook et al., 1989). The PerfectHyb Plus solution (from Sigma) was used for hybridization. For real-time PCR analysis, RNA was treated with DNase I (Promega) prior to cDNA synthesis with random primers and AMV reverse transcripase (Promega). Primers 5′-GAACTGGCCACTTTCTGC-3′ and 5′-GCGCATAGGGATCTCACG-3′ were used to amplify a 376 bp fragment from both gadA and gadB cDNA using the Roche Life-Cycler.
Western blotting
Cells from exponential phase or overnight cultures were collected by centrifugation and lyzed by boiling in the sample buffer for SDS gel electrophoresis for 5 min. The total lysates with volumes adjusted for cell densities were electrophoresed in 10% SDS polyacrylamide gels. The proteins in the gel were either stained with coomassie blue to check for equal loading or transferred onto nitrocellulose filters for Western blotting using the ECL Plus reagents (Amersham). The antibodies against E. coli topoisomerase I (Qi et al., 1996) and GAD (Castanie-Cornet et al., 1999) were described previously. Mouse monoclonal antibodies against E. coli RpoS was purchased from Neoclone. Antibodies against GadW were raised in rat using the peptide sequence YHEQQKISLHNESILC. Antibodies against GadX (Shin et al., 2001) were kindly provided by Dr. J. B. Kaper. The coomassie stained gels and exposed films were analyzed by the Alpha-Imager for quantitation.
RESULTS
Loss of topoisomerase I function leads to reduced acid resistance
The increased sensitivity of the topA deletion mutant strain RFM475 to high temperature and oxidative stress challenge when compared to its isogenic topA+ counterpart, RFM445, has been reported (Qi et al., 1999; Tse-Dinh, 2000). The effect of loss of topoisomerase I function on acid resistance was investigated here. After overnight growth in either LB buffered at pH 8.0 or 5.5, RFM475 was found to have 20–30 fold lower acid resistance than RFM445 (Fig. 1). The cysB gene function, also missing in RFM475 due to the topA-cysB deletion, is required for the induction of the arginine decarboxylase activity in the ARG acid resistance mechanism (Shi & Bennett, 1994; Lin et al., 1996). This loss of cysB function is expected to lead to decreased arginine decarboxylase-dependent acid resistance in RFM475. We therefore also tested a topA20::Tn10 insertion mutant (YT481) that is wild-type for cysB function for acid resistance and found similar degree of sensitivity due to the topA mutation in the absence of the cysB mutation (Fig. 1). This indicates that topA mutations affects acid resistance in mechanisms independent of the arginine decarboxylase activity. Topoisomerase I protein was not detected in the total protein of YT481 by Western blot analysis (data not shown).
Fig. 1.
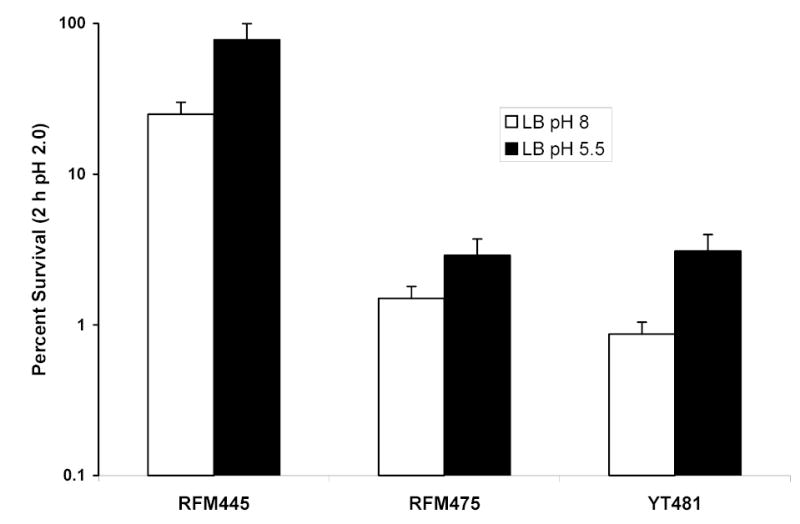
Effect of topA mutation on overall acid resistance as measured by survival rates after overnight growth in LB pH 5.5 or 8.0, and challenge in LB pH 2.0 for 2 h. The results represent the average from three independent experiments.
Loss of topoisomerase I function affects the oxidative and GAD systems of acid resistance
To demonstrate directly which of the three acid resistance mechanisms was affected by the loss of topoisomerase I function, the three AR systems were tested individually and the results are shown in Fig. 2. As expected from the loss of the cysB function, RFM475 was defective in arginine-dependent AR. However, the arginine-dependent AR was not affected in YT481 with the topA20::Tn10 mutation, indicating that topoisomerase I function is not required for the arginine-dependent acid resistance mechanism. The oxidative system and glutamate-dependent systems were both affected by the loss of topoisomerase I function in the two topA mutants. However, the two topA mutants differed in the relative degree of sensitivity. Mutant RFM475 appeared to be more severely affected in the oxidative system while the glutamate-dependent system was more severely affected in mutant YT481. The partial loss of oxidative AR in mutant YT481 was confirmed by longer exposure to the pH 2.5 EG medium. After 6 h of exposure, the survival rate of YT481 was 0.046% versus 7.1% for RFM445.
Fig. 2.
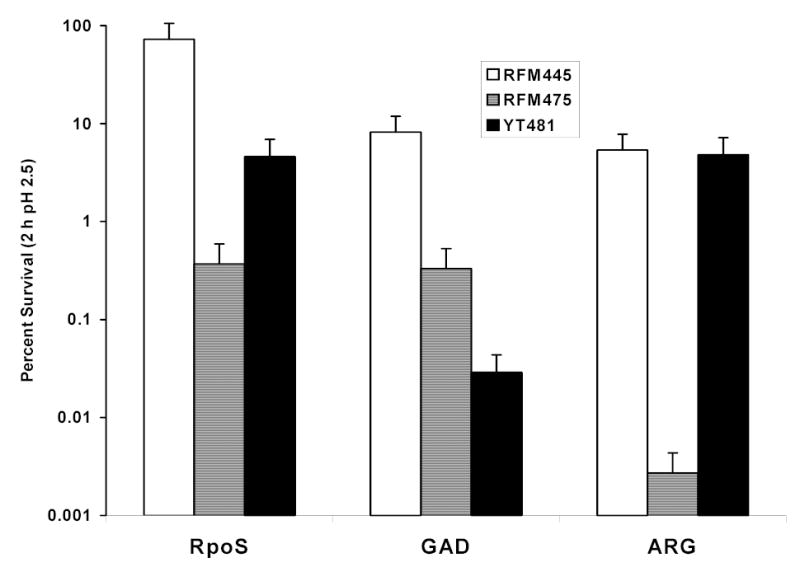
Effect if topA mutation on the three different mechanisms of acid resistance. The test for the oxidative system (RpoS) involved overnight growth in LB-MES (pH5.5) followed by dilution into EG (pH 2.5). Survival (viable counts) was measured after 2 h at 37°C. The glutamate (GAD) and arginine (ARG) systems required overnight LBG (glutamate system) or BHIG (arginine system) cultures which were diluted into EG (pH 2.5) containing 1.5 mM glutamate or 0.6 mM arginine respectively. The deletion of cysB gene in RFM475 (ΔtopAcysB) accounts for the low acid resistance from the arginine-dependent mechanism. Strain YT481 (topA20::Tn10) has wild-type cysB gene. The results represent the average from four independent experiments.
Overexpression of RNAse H does not improve the AR of topA mutants
In topA mutants, the formation of R-loops during transcription elongation is driven by the accumulation of hypernegative supercoiling. The blocking of RNA polymerase by these R-loops can account at least partially the growth defect of topA mutants (Drolet et al., 1995; Massé & Drolet, 1999; Baaklini et al., 2004). Overexpression of RNase H in topA mutants including RFM475 has been shown to correct this defect of R-loop formation (Drolet et al., 1995; Hraikly et al., 2000). The overexpression of RNase H from plasmid pSK760 has also been shown to partially alleviate the effect of topA deletion on the survival rate of RFM475 after high temperature and oxidative challenge (Cheng et al., 2003). We therefore compared the survival rates of topA mutants transformed with either pSK760 expressing wild type RNAse H or pSK762c expressing inactive RNAse H mutant. The results (Table 2) showed that there was no significant increase in the overall AR (left column) or GAD-dependent AR (right column) of RFM475 or YT481 that could be attributed to the overexpression of RNAse H when the pSK760 plasmid was present.
Table 2. Effect of RNase overexpression from plasmid pSK760 on acid resistance of topA mutants.
Each experiment was carried out four times. Mean results and standard deviations are shown.
| Overall Acid Resistance | GAD Acid Resistance | ||
|---|---|---|---|
| Adaptation: Overnight in LB pH 5.5 | Adaptation: Overnight in LBG | ||
| Acid challenge: EG pH 2.5, 2 h | Acid challenge: EG pH 2.5 + Glutamate, 2 h | ||
| Strain | Percent Survival | Strain | Percent Survival |
| RFM445 | 73±14 | RFM445 | 8.2±3.2 |
| RFM475 Δ(topAcysB) | 0.37±0.18 | YT481 topA20::Tn10 | 0.029±0.011 |
| RFM475/pSK760 | 0.15±0.03 | YT481/pSK760 | 0.030±0.012 |
| RFM475/pSK762c | 0.66±0.30 | YT481/pSK762c | 0.043±0.014 |
Effect of Topoisomerase I function on the level of glutamate decarboxylase isoforms
The glutamate decarboxylase activity encoded by gadA and gadB consumes an intracellular proton for the decarboxylation of each molecule of glutamate. It thus plays a major role in acid resistance. Transcription of both gadA and gadBC is positively controlled by the transcriptional activator GadX (Shin et al., 2001; Tramonti et al., 2002). RpoS is also involved in the induction of gad and gadB during the stationary phase of cell growth (Castanie-Cornet & Foster, 2001). The total level of GadA and GadB proteins in RFM445 and the topA mutants RFM475, YT481 was measured by Western blotting. The results (Fig. 3) showed that the GadA and GadB proteins were present in significantly lower level in the topA mutants after overnight growth in both neutral (pH 8) and acidic pH (pH 5.5). The decrease of GadA/B resulting from the topA mutation was also seen under conditions with active oxidative system (during stationary phase in LB or EG pH 5.5 medium) or repressed RpoS activity (during exponential phase in EG medium or during stationary phase in LBG medium). This decrease in GadA and GadB proteins in the topA mutants would account for the diminished acid resistance observed under these different growth conditions.
Fig. 3.
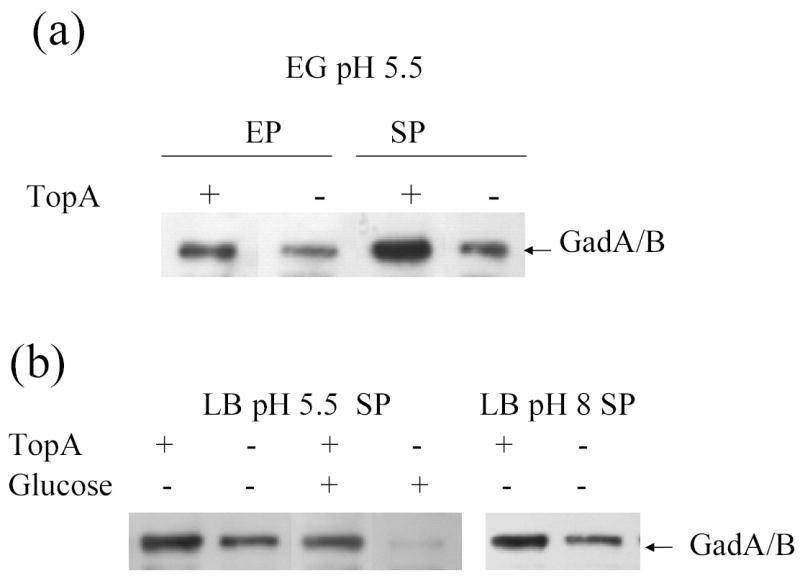
Effect of topA mutation on the GadA and GadB protein levels determined by western blot analysis. (a) RFM445 (topA+) and YT481(topA20::Tn10) were grown in EG pH5.5 medium to EP (OD600=0.5) and SP (18 h). (b) RFM445 and RFM475 (ΔtopA) were grown in buffered LB for 18 h with either no glucose or 0.4% glucose added.
TopA mutation diminished the induction of gadA and gadBC genes in acidic LB during stationary phase
In order to determine if transcription of gadA and gadBC genes was affected by the topA mutation, total RNA was prepared and used for Northern blotting. The results (Fig. 4) showed that the induction of the gadA and gadBC message level during entry into stationary phase (OD600 = 1.4) at acidic pH was drastically decreased in the topA mutants RFM475 and YT481 versus RFM445. The difference in gadA and gadB message levels in pH5.5 stationary phase LB cultures was quantified by real-time PCR. The ratio measured for YT481 versus RFM445 was found to be 1:60 + 10 (average of results from two independent RNA preparations in three separate experiments).
Fig. 4.
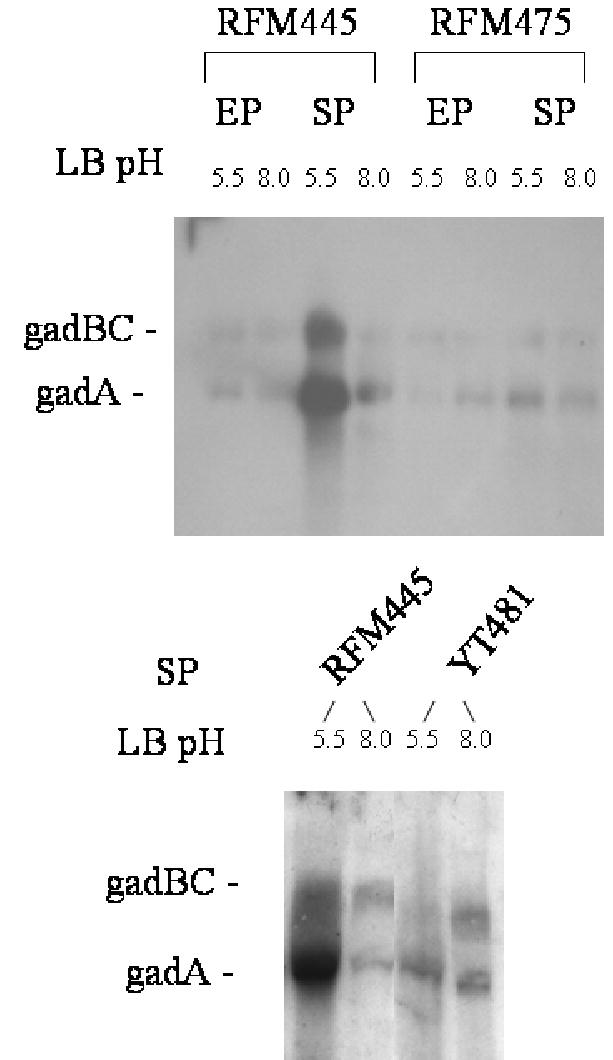
Effect of topA mutation on the gadA and gadBC mRNA levels determined by Northern blot analysis. RNA was extracted from RFM445 (topA+), RFM475 (ΔtopA) and YT481(topA20::Tn10) grown in LB at pH 5.5 or pH 8.0 to exponential phase (EP) or early stationary phase (SP).
The levels of RpoS, GadX and GadW were not altered by the topA mutation
The sigma factor RpoS is central to the resistance to acidic pH in the glucose-repressed mechanism in complex medium. The AraC-like regulators GadX and GadW have also been implicated in the complex regulation of glutamate decarboxylase system (Shin et al., 2001; Tramonti et al., 2002; Ma et al., 2002, 2003). Western blotting was used to compare the amounts of RpoS, GadX and GadW in the topA+ and topA mutant cultures to determine if change in levels of these regulators accounted for the decreased gadA and gadBC expression in the topA mutants. The results showed that the levels of RpoS accumulated in overnight stationary phase cultures of the topA mutants RFM475 and YT481 were similar to those found in RFM445 (Fig. 5). There was also no difference observed for the level of GadX and GadW due to the topA mutation under different growth conditions (Fig. 5). Therefore topoisomerase I function had no significant effect on the accumulated levels of these regulators.
Fig. 5.
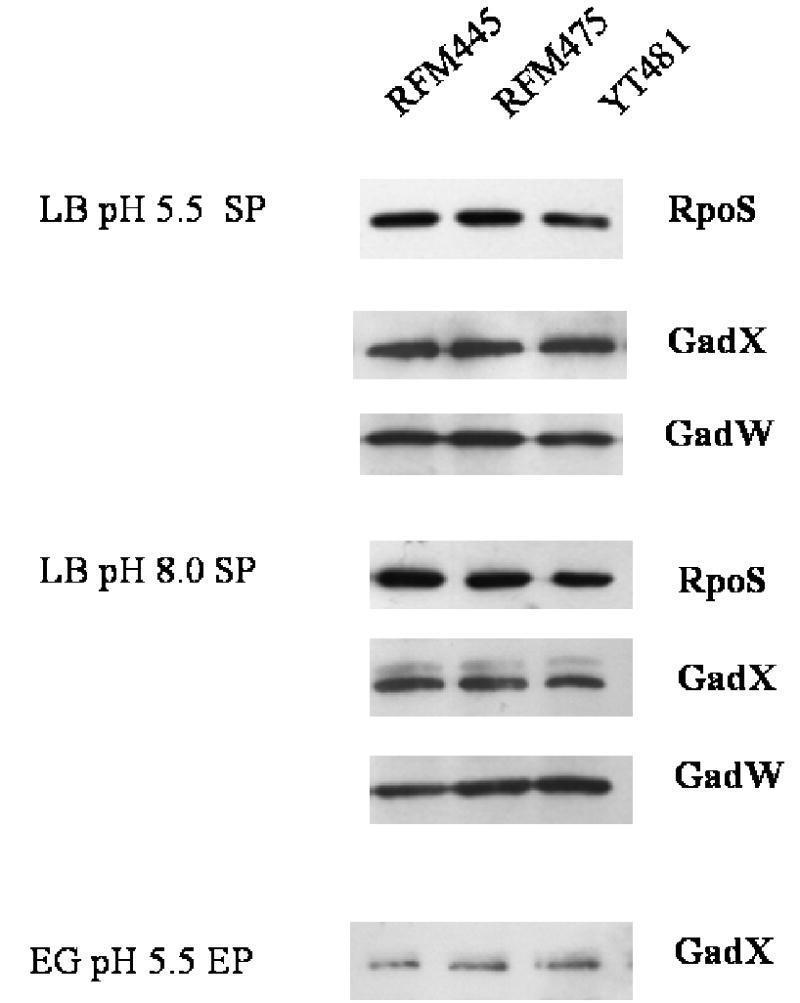
Western blot analysis to compare levels of RpoS, GadX and GadW proteins in RFM445 and its topA mutant derivatives.
RpoS-independent mechanism also accounts for some of the effect of topA mutation
A RpoS dependent promoter is involved in the transcription of topA (Qi et al., 1997), suggesting that topoisomerase I function may be important for gene expression directed by RpoS. The rpoS::Tn10 mutation was introduced into the strains RFM445 and RFM475 to determine if the effect of topA mutation on acid resistance was entirely dependent on transcription directed by RpoS. Survival in the absence of glutamate at pH 2.5 for 1 hour was 3.5% for YT445R (rpoS) and 0.098% for YT475R (rpoS topA). In the presence of glutamate at pH 2.5 for 2 hours, survival was 0.1% vs 0.0029%, respectively. These results showed that the presence of topA mutation in the rpoS mutant background further decreased the survival rate. Therefore the effect of the topA mutation on acid resistance was not entirely due to RpoS-dependent transcription. Measurement of the level of GadA and GadB proteins by Western blot also showed that there was a further decrease of expression from the topA mutation in the rpoS null background (Fig 6a).
Fig. 6.
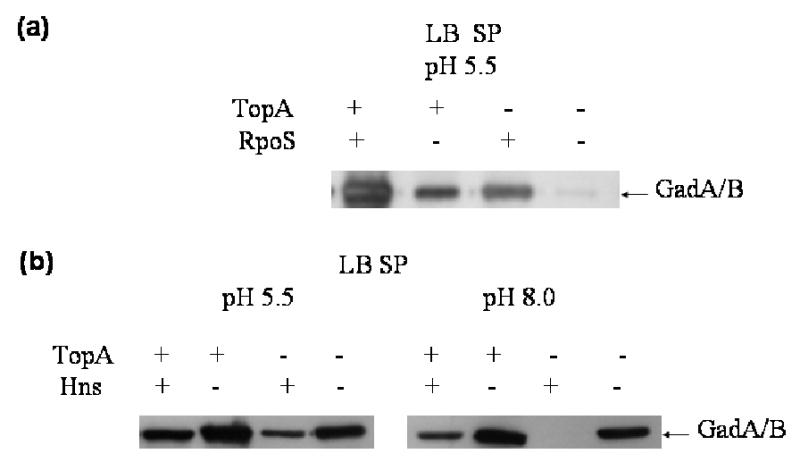
Western blot analysis of the effect of topA mutation on the GadA and GadB protein levels in the presence of background mutation of either (a) rpoS (b) hns. Mutations of rpoS or hns were introduced by transduction into RFM445 (topA+) and RFM475 (topA−)
The effect of topA deletion can be suppressed by hns mutation
H-NS protein is a negative regulator of acid resistance (Foster, 2004). The hns mutation affecting the nucleoid protein H-NS has been shown to increase expression of the gadA and gadBC genes (Yoshia et al., 1993a,b; De Biase et al., 1999; Hommals et al., 2001; Tramonti et al., 2002; Ma et al., 2002; Waterman & Small, 2003). The hns::Tn5 mutation was introduced into RFM445 and RFM475. Acid resistance of the resulting strains YT445H and YT475H were compared after using conditions previously used to demonstrate the increase in acid resistance from hns mutation (Hommals et al., 2001). The effect of the topA mutation in RFM475 versus RFM445 under these experimental conditions was >100 fold reduction in acid resistance (Table 3). There was a modest increase in acid resistance when the hns mutation was introduced into RFM445. The increase in acid resistance from the hns mutation was much greater in RFM475, restoring it close to the level observed for RFM445. The increase in acid resistance in the hns mutants correlates to the increase in expression of GadA and Gad B proteins as analyzed by Western blots (Fig. 6b).
Table 3. hns mutation can suppress the effect of the topA mutation on acid resistance.
Each experiment was carried out four times. Mean results and standard deviations are shown.
| Adaptation: Overnight in M9 pH 5.5 | |
|---|---|
| Acid challenge: M9 pH 2.5 + Glutamate, 2 h | |
| Strain | Percent Survival |
| RFM445 | 47 ±9 |
| YT445H hns::Tn5 | 59 ±9 |
| RFM475 | 0.25±0.09 |
| YT475H hns::Tn5 | 44 ±5 |
Effect of gyrB mutations and gyrase inhibition on acid resistance
The topA+ strain RFM445 contains gyrase mutations necessary for survival of RFM475 and YT481. The acid resistance of the gyr+ strain RFM443 was also measured to determine if the gyrB221(couR), gyrB203(Ts) mutations in RFM445 had any effect. Overnight growth of RFM443 was also carried out in the presence of sub-lethal concentrations of novobiocin to determine if inhibition of gyrase activity influenced acid resistance. Besides the conditions of adaptation in pH 5.5 M9 medium used previously to test the effect of the hns mutation (Hommals et al., 2001), the RpoS suppressed condition (overnight growth in LBG) with the relatively lower survival rates for the topA+ strains was also tested, so that any positive effect from the reduced gyrase activity could potentially be detected. The results (Table 4) showed that the gyrB mutations in RFM445 resulted in a small increase (~2 fold) of the RpoS-independent and glutamate-dependent acid resistance after overnight growth in LBG. This could be due to the moderate level of reduction of gyrase activity in RFM445, compared to topoisomerase I activity missing totally in the topA mutants. However, the gyrase mutations had the opposite effect when acid resistance was measured after overnight growth in M9 medium at pH 5.5. The presence of novobiocin at 15 and 30 μg/ml inhibited overnight growth of RFM443 by about ten fold (data not shown). There was a small decrease in acid resistance after adaptation in LBG (~2–3 fold) and a more significant decrease in acid resistance after adaptation in pH5.5 M9 medium (up to ten fold).
Table 4. Effect of the gyrB mutations in RFM445 and inhibition of gyrase activity in RFM443 by novobiocin on acid resistance.
Each experiment was carried out four times. Mean results and standard deviations are shown.
| Percent Survival | |||
|---|---|---|---|
| RFM445
gyrB221(couR), gyrB203(Ts) |
RFM443 | RFM443
15 μg/ml novobiocin |
RFM443
30 μg/ml novobiocin |
| Overnight in M9 pH5.5, challenged for 2 hr in M9 pH 2.5 + glutamate | |||
| 47±9 | 90±10 | 25±6 | 8.0 ±1.0 |
| Overnight in LBG, challenged for 2 hr in EG pH 2.5 + glutamate | |||
| 8.2±3.2 | 4.4±2.2 | 2.6±1.4 | 1.4±0.6 |
DISCUSSION
Glutamate decarboxylase activity plays an important role in the acid resistance of E. coli. Data presented here showed that a topA mutation resulted in significantly lower transcription of gadA and gadBC, accounting at least in part for the decreased acid resistance of the topA mutants. Unlike other growth defects of topA mutants including increased sensitivity to high temperature and oxidative stress, the reduced acid resistance could not be rescued by the overexpression of RNase H. Therefore it is unlikely for R-loop accumulation during transcription elongation to be the cause of the reduced acid resistance even though it could conceivably be argued that degradation of the mRNA required for acid resistance by the overexpressed RNase H led to the low level of survival.
Expression of gadA and gadBC in stationary phase has been shown to be largely dependent on RpoS (Castanie-Cornet et al., 1999; De Biase et al., 1999). This is mediated by the RpoS-dependent transcription of GadX, an activator of gadA and gadBC (Ma et al, 2000). Our results showed that the decreased survival of the topA mutants under the RpoS-dependent mechanism of acid resistance was not due to a reduction in level of RpoS or GadX protein. The loss of topoisomerase I function could have affected the transcription of other RpoS target promoters directly, since high level of negative supercoiling has been reported to inhibit transcription by RpoS (Bordes et al., 2003), and localized hypernegative supercoiling might be present in the topA mutants. It was found that the gyrase inhibitor novobiocin induced the RpoS-directed transcription initiation from the osmEp gene without affecting the level of RpoS accumulation (Bordes et al., 2003). However, in this study the introduction of gyrase mutations or the addition of novobiocin to RFM443 resulted in only small changes in acid resistance, and not always in the opposite direction of topA mutation. In addition, the data from the rpoS mutants indicated that topoisomerase I function was also involved in an RpoS-independent mechanism of acid resistance.
The nucleoid protein H-NS has been shown to act as a negative regulator on E. coli genes required for acid resistance (Yoshida et al., 1993a,b; Hommals et al., 2001). The repression of gene expression by H-NS may be due to its effect on DNA supercoiling (Tupper et al., 1994) or its binding to specific curved DNA sites in the promoter regions of these genes. The large effect of the hns mutation on strain RFM475 relative to RFM445 suggested that topoisomerase I function may be required to overcome the repression of acid resistance genes by H-NS. It has been shown that H-NS can repress gadA and gadBC expression independent of GadX (Ma et al., 2002). The effect of hns mutation on gadA and gadBC transcription remained highly significant even in a rpoS mutant background (Waterman & Small, 2003), so this might account for the observation that the topA effect could still be seen in the rpoS mutants studied here. Topoisomerase I function may be needed to counteract the H-NS repression on gadA and gadBC expression. The role of topoisomerase I at such loci is not simply due to its relaxation activity, since the addition of novobiocin resulted in decrease of acid resistance.
A previous study with Salmonella typhimurium established that DNA topology can affect the environmental regulation of the pH-regulated locus aniG (Karem & Foster, 1993). Expression of aniG in mannose media is decreased in a topA mutant, but increased in two hns mutants at pH 6.0–7.0 (Karem & Foster, 1993). This is also in agreement with a possible role of topoisomerase I in relieving repression of aniG by H-NS. Many other bacterial genes including some of the genes involved in virulence and osmotic control are also known to be regulated by H-NS (Hommais et al., 2001; Madrid et al., 2002). The virF gene of Shigella required for invasion functions is suppressed by interaction of H-NS with its promoter, and its thermal induction was found to be inhibited in a topA mutant (Ni Bhriain & Dorman, 1993). This is also similar to the effect of topA on the induction gad expression during stationary phase observed here. The effective DNA superhelicity at any gene promoter region is a result of the local dynamic competition between structural constraining proteins, including H-NS, RNA polymerase and the topoisomerases (Travers & Muskhelishvili, 2005). This dynamic competition changes during growth-phase transition and adaptative response to challenge. Topoisomerase I function could have a direct role in relieving repression of H-NS at specific gene loci, but it also cannot be ruled out that topoisomerase I may be counteracting the general effect of H-NS on DNA topology in the other cases (Mojica & Higgins, 1997). In a study of E. coli protein-protein interaction network, topoisomerase I was found to interact H-NS (Butland et al., 2005), suggesting a functional link between the two proteins. We have demonstrated previously that E. coli topoisomerase I function was required for R-loop suppression during transcription elongation of stress genes during adaptation to high temperature and oxidative stress (Cheng et al., 2003). In this study, it was found that E. coli topoisomerase I played a different role in gene expression for acid resistance that could involve the influence of DNA topology on the action of the positive and negative regulators of transcription. This further supports the importance of type IA topoisomerase function during environmental adaptation and its conservation in evolution (Wang, 2002). Furthermore inhibitors of bacterial topoisomerase I activity may be useful for general suppression of adaptation of pathogenic bacteria to host environments.
Acknowledgments
This work was supported by National Institutes of Health award R01-GM54226 (to Y.T.) and R01-GM61147 (to J.W.F.). Technical assistance was provided by Kenneth Montini Shikha Shukla and Molly Boyd.
References
- Baaklini I, Hraiky C, Rallu F, Tse-Dinh YC, Drolet M. RNase H1 overproduction is required for efficient full-length RNA synthesis in the absence of topoisomerase I in Escherichia coli. Mol Microbiol. 2004;54:198–211. doi: 10.1111/j.1365-2958.2004.04258.x. [DOI] [PubMed] [Google Scholar]
- Benjamin MM, Datta AR. Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1995;61:1669–1672. doi: 10.1128/aem.61.4.1669-1672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordes P, Conter A, Morales V, Bouvier J, Kolb A, Gutierrez C. DNA supercoiling contributes to disconnect σs accumulation from σs-dependent transcription in Escherichia coli. Mol Microbiol. 2003;48:561–571. doi: 10.1046/j.1365-2958.2003.03461.x. [DOI] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Foster JW. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiol. 2001;147:709–715. doi: 10.1099/00221287-147-3-709. [DOI] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Cheng B, Rui S, Ji C, Gong VW, Van Dyk TK, Drolet M, Tse-Dinh YC. RNase H overproduction allows the expression of stress-induced genes in the absence of topoisomerase I. FEMS Microbiol Letters. 2003;221:237–242. doi: 10.1016/S0378-1097(03)00209-X. [DOI] [PubMed] [Google Scholar]
- Conner DE, Kotrolla JS. Growth and survival of Escherichia coli O157:H7 under acidic conditions. Appl Environ Microbiol. 1995;61:382–385. doi: 10.1128/aem.61.1.382-385.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase D, Tramonti A, Bossa F, Visca P. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol. 1999;32:1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- Dong Q, Hyde H, Herra C, Kean C, Murphy P, O’Moran CA, Buckley M. Identification of genes regulated by prolonged acid exposure in Helicobacter pylori. FEMS Microbiol Letters. 2001;196:245–249. doi: 10.1111/j.1574-6968.2001.tb10572.x. [DOI] [PubMed] [Google Scholar]
- Drolet M, Phoenix P, Menzel R, Massé E, Liu LF, Crouch RJ. Overexpression of RNase H partially complements the growth defect of an Escherichia coli topA mutants: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JW. Escherichia coli acid resistance: Tales of an amateur acidophile. Nature Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- Giannella RA, Broitman SA, Zamcheck N. Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro. Gut. 1993;13:251–256. doi: 10.1136/gut.13.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J, Smith PLC. Acid resistance in enteric bacteria. Infect Immun. 1993;61:364–367. doi: 10.1128/iai.61.1.364-367.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh BM, Farooq FT, Barstad DN, Blankenhorn DL, Slonczewski JL. A gluatmate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178:3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, Danchin A, Bertin P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- Hraiky C, Raymond MA, Drolet M. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J Biol Chem. 2000;275:11257–11263. doi: 10.1074/jbc.275.15.11257. [DOI] [PubMed] [Google Scholar]
- Kanya S, Crouch RJ. DNA seqence of the gene coding for Escherichia coli ribonuclease H. J Biol Chem. 1983;258:1276–1281. [PubMed] [Google Scholar]
- Karem K, Foster JW. The influence of DNA topology on the environmental regulation of a pH-regulated locus in Salmonella typhimurium. Mol Microbiol. 1993;10:75–86. doi: 10.1111/j.1365-2958.1993.tb00905.x. [DOI] [PubMed] [Google Scholar]
- Lesley SA, Jovanovich SB, Tse-Dinh YC, Burgess RR. Identification of a heat shock promoter in the topA gene of Escherichia coli. J Bacteriol. 1990;172:6871–6874. doi: 10.1128/jb.172.12.6871-6874.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Lee IS, Frey J, Slonzewski JL, Foster JW. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri and Escherichia coli. J Bacteriol. 1995;177:4097–4105. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Richard H, Tucker DL, Conway T, Foster JW. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators. GadX and GadW (YhiW) J Bacteriol. 2002;184:7001–7002. doi: 10.1128/JB.184.24.7001-7012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Richard H, Foster JW. pH-Dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of the GadX regulator and Escherichia coli acid resistance. J Bacteriol. 2003;185:6852–6859. doi: 10.1128/JB.185.23.6852-6859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid C, Nieto JM, Paytubi S, Falconi M, Gualerzi CO, Juárez A. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J Bacteriol. 2002;184:5058–5066. doi: 10.1128/JB.184.18.5058-5066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Drolet M. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J Biol Chem. 1999;274:16659–16684. doi: 10.1074/jbc.274.23.16659. [DOI] [PubMed] [Google Scholar]
- Miller, J. H. (1992). A Short Course in Bacterial Genetics: a Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Mojica FJM, Higgins CF. In vivo supercoiling of plasmid and chromosomal DNA in an Escherichia coli hns mutant. J Bacteriol. 1997;179:3528–3533. doi: 10.1128/jb.179.11.3528-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Bhriain N, Dorman CJ. Isolation and characterization of a topA mutant of Shigella flexneri. Mol Microbiol. 1993;7:351–358. doi: 10.1111/j.1365-2958.1993.tb01127.x. [DOI] [PubMed] [Google Scholar]
- Price SB, Wright JC, DeGraves FJ, Castanie-Corner MP, Foster JW. Acid resistance systems required for survival of Escherichia coli O157:H7 in the bovine gastrointestinal tract and in apple cider are different. App Env Microbiol. 2004;70:4792–4799. doi: 10.1128/AEM.70.8.4792-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Menzel R, Tse-Dinh YC. Effect of the deletion of the σ32-dependent promoter (P1) of the Escherichia coli topoisomerase I gene on thermotolerance. Mol Microbiol. 1996;21:703–711. doi: 10.1046/j.1365-2958.1996.241390.x. [DOI] [PubMed] [Google Scholar]
- Qi H, Menzel R, Tse-Dinh YC. Regulation of Escherichia coli topA gene transcription: involvement of a σs-dependent promoter. J Mol Biol. 1997;267:481–489. doi: 10.1006/jmbi.1997.0901. [DOI] [PubMed] [Google Scholar]
- Qi H, Menzel R, Tse-Dinh YC. Increased thermosensitivity associated with topoisomerase I deletion and promoter mutations in Escherichia coli. FEMS Microbiol Letters. 1999;178:141–146. doi: 10.1111/j.1574-6968.1999.tb13770.x. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Shi X, Bennett GN. Effects of rpoA and cysB mutations on acid induction of biodegradative arginine decarboxylase in Escherichia coli. J Bacteriol. 1994;176:7071–7023. doi: 10.1128/jb.176.22.7017-7023.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Castanie-Cornet MP, Foster JW, Crawford JA, Brinkley C, Kaper JB. An activator of glutamate decarboxylase gene regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol Microbiol. 2001;41:1133–1150. doi: 10.1046/j.1365-2958.2001.02570.x. [DOI] [PubMed] [Google Scholar]
- Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramonti A, Visca P, De Canio M, Falconi M, De Biase D. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J Bacteriol. 2002;184:2603–2613. doi: 10.1128/JB.184.10.2603-2613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A, Muskhelishvili G. DNA supercoiling – A global transcriptional regulator for enterobacterial growth? Nature Rev Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- Tse-Dinh YC. Increased sensitivity to oxidative challenges associated with topA deletion in Escherichia coli. J Bacteriol. 2000;182:829–832. doi: 10.1128/jb.182.3.829-832.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupper AE, Owen-Hughes TA, Ussery DW, Santos DS, Ferguson DJP, Sidebotham JM, Hinton JCD, Higgins CF. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 1994;13:258–278. doi: 10.1002/j.1460-2075.1994.tb06256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Waterman SR, Small PLC. Transcriptional expression of Escherichia coli glutamate-dependent acid resistance gene gadA and gadBC in an hns rpoS mutant. J Bacteriol. 2003;185:4644–4647. doi: 10.1128/JB.185.15.4644-4647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T, Ueguchi C, Mizuno T. Quantitative control of the stationary phase-specific sigma factor, σs, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J. 1995;14:594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Ueguchi C, Yamada H, Mizuno T. Function of the Escherichia coli nucleoid protein, H-NS: molecular analysis of a subset of proteins whose expression is enhanced in a hns deletion mutant. Mol Gen Genetic. 1993a;237:113–122. doi: 10.1007/BF00282791. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Yamashimo T, Ueguchi C, Mizuno T. Expression of the Escherichia coli dimorphic glutamic acid decarboxylases is regulated by the nucleoid protein H-NS. Biosci Biotechnol Biochem. 1993b;57:1568–1569. doi: 10.1271/bbb.57.1568. [DOI] [PubMed] [Google Scholar]
- Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]


