Abstract
Phosphoprotein NSP5 is a component of replication intermediates that catalyze the synthesis of the segmented double-stranded RNA (dsRNA) rotavirus genome. To study the role of the protein in viral replication, His-tagged NSP5 was expressed in bacteria and purified by affinity chromatography. In vitro phosphorylation assays showed that NSP5 alone contains minimal autokinase activity but undergoes hyperphosphorylation when combined with the NTPase and helix-destabilizing protein NSP2. Hence, NSP2 mediates the hyperphosphorylation of NSP5 in the absence of other viral or cellular proteins. RNA-binding assays demonstrated that NSP5 has unique nonspecific RNA-binding activity, recognizing single-stranded RNA and dsRNA with similar affinities. The possible functions of the RNA-binding activities of NSP5 are to cooperate with NSP2 in the destabilization of RNA secondary structures and in the packaging of RNA and/or to prevent the interferon-induced dsRNA-dependent activation of the protein kinase PKR.
Rotaviruses, members of the family Reoviridae, are the major cause of acute severe gastroenteritis in infants and young children (18). The infectious virion is an icosahedron consisting of three concentric layers of protein and a genome of 11 segments of double-stranded RNA (dsRNA) (40). The innermost layer is a T=1 structure formed by VP2 (24). Associated with the 12 vertices of the VP2 layer are one copy each of the viral RNA polymerase VP1 (50) and the mRNA-capping enzyme VP3 (8, 25). Collectively, VP1, VP2, VP3, and the dsRNA genome make up the core of the virion. The middle protein layer of the virus is formed by VP6, and the outer protein layer is formed by the glycoprotein VP7, and the spike protein VP4 (40). The RNA polymerase activity of double-layered particles (cores surrounded by VP6) catalyzes the synthesis of the 11 viral mRNAs (23). During viral replication, the mRNAs not only direct the synthesis of six structural proteins (VP) and six nonstructural proteins (NSP) (11) but also serve as templates for the synthesis of minus-strand RNAs, forming the dsRNA genome (9).
The synthesis of viral dsRNA and the assembly of replication intermediates (RIs) with replicase activity occur in cytoplasmic inclusions termed viroplasms (37). Characterization of subviral particles from infected cells has indicated that core-like RIs (core RIs) are the simplest complexes that catalyze the synthesis of the dsRNA genome (14, 32). Core RIs, which are composed of VP1, VP2, and VP3 and nonstructural proteins NSP2 and NSP5, undergo maturation into double-layered RIs as they synthesize dsRNA. Both NP2 and NSP5 accumulate in viroplasms, and the expression of these proteins together, but not separately, generates viroplasm-like structures in uninfected cells (12). NSP2 has strong sequence-independent affinity for single-stranded RNA (ssRNA) (20), interacts with the viral RNA polymerase (19), and self-assembles into stable octamers (44). The NSP2 octamers have Mg2+-dependent NTPase activity (47) and Mg2+-independent helix-destabilizing activity (46). During hydrolysis of NTP by NSP2, the protein undergoes phosphorylation (47).
NSP5 is a dimeric (39), serine/threonine-rich protein of 198 amino acids that is slightly acidic and undergoes O-linked glycosylation (15) and phosphorylation in infected cells (2, 5). Due to variations in the extent of phosphorylation, several phosphorylated isomers of NSP5 have been identified (5, 39). One or more kinase activities may be involved in the phosphorylation of NSP5. Some studies have suggested that NSP5 is phosphorylated due to an intrinsic low-level protein kinase activity, and other studies have provided evidence that cellular proteins are involved in the hyperphosphorylation of NSP5 (2, 6, 39). When coexpressed in uninfected cells, NSP2 induces the hyperphosphorylation of NSP5, indicating that NSP2 may also play a role in the posttranslation modification of NSP5 (1). Because NSP2 and NSP5 (i) are common components of RIs containing replicase activity (3, 14), (ii) function cooperatively to form viroplasm-like structures (12), and (iii) interact to promote phosphorylation of NSP5 (1, 39), they are believed to be involved in RNA packaging and replication. In addition to NSP2, there is evidence that NSP5 interacts with NSP6, a small protein of unknown function that localizes to viroplasms in infected cell (49).
Proteins of core RIs support the packaging of the 11 viral mRNAs and the replication of the dsRNA genome. Because of their involvement in these processes, the discovery that most of the proteins of core RIs have associated RNA-binding activity (i.e., VP1 [30], VP2 [7], VP3 [31], and NSP2 [20]) is not unexpected. To gain further insight into its role in viral replication, recombinant NSP5 (rNSP5) containing an N-terminal His tag was expressed in bacteria and purified by affinity chromatography. Although rNSP5 possesses little or no kinase activity alone, its phosphorylation level was significantly enhanced when it was incubated with rNSP2. Further analysis of the properties of rNSP5 by UV cross-linking, filter-binding, and RNA capture assays revealed that the protein possesses nonspecific RNA-binding activity, recognizing both ssRNA and dsRNA with similar affinities. Analysis of infected-cell extracts confirmed that this is an activity of NSP5 in vivo. It is possible that NSP5 recruits helix-destabilizing protein NSP2 to secondary structures on the mRNA template during genome replication, thereby promoting removal of the RNA-RNA duplexes that may interfere with RNA packaging or that may activate the interferon-induced dsRNA-dependent protein kinase PKR (10).
Expression and purification of rNSP5
To produce rNSP5, a full-length cDNA of the strain SA11 gene encoding NSP5 (segment 11) was prepared from virion dsRNA by reverse transcription and PCR. The cDNA was ligated into pGEM-T-easy, generating the vector pGEM-Tg11. The gene 11 insert was released from the vector by digestion with BamHI and PstI and ligated into the same restriction sites of bacterial isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression vector pQE30 (Qiagen). The gene 11 cDNA was positioned in pQE30g11 such that the open reading frame (ORF) for NSP5 was located downstream of six in-frame His codons. Analysis of the nucleotide sequence of pQE30g11 showed that rNSP5 encoded by the vector was identical in sequence to that reported previously for SA11 NSP5 (28). Following expression of His-tagged rNSP5 by Escherichia coli M15 containing pQE30g11, the protein was recovered under native conditions from the soluble fraction of the bacterial lysate by Ni-nitrilotriacetic acid (NTA) affinity chromatography (47). The final eluate was dialyzed against low-salt buffer (LSB; 2 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 0.5 mM dithiothreitol) and stored at 4°C.
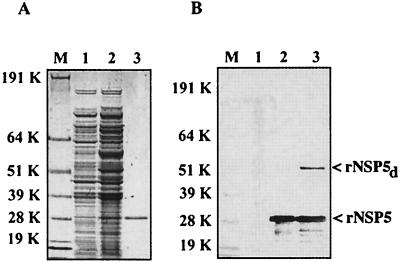
Electrophoresis demonstrated that IPTG induction resulted in the expression of soluble rNSP5 in E. coli that could be purified by Ni-NTA chromatography to near homogeneity (Fig. 1A). The identity of the putative rNSP5 protein was confirmed by Western blot analysis with anti-NSP5 polyclonal antisera (Fig. 1B). In addition to the expected 28-kDa rNSP5 protein, the antisera also recognized protein migrating with a molecular mass of ∼56 kDa, approximately twice the size of the rNSP5 monomer. This same ∼56-kDa material was also recognized by mouse anti-NSP5 monoclonal antibody 158G37 (data not shown). Based on its molecular mass, recognition by anti-NSP5 antibodies, and previous evidence for the homodimerization of NSP5 (39), it is presumed that the 56-kDa material probably represents dimers of rNSP5.
FIG. 1.
Expression and purification of rNSP5. Proteins were denatured in sample buffer containing LDS and dithiothreitol (NuPAGE LDS sample buffer; Invitrogen) and resolved by PAGE on NuPAGE 10% Bis-Tris polyacrylamide gels. Proteins were detected by staining the gel with Coomassie blue (A). To locate rNSP5, protein in the gel was transferred onto nitrocellulose and the blot was probed sequentially with guinea pig anti-NSP5 polyclonal antisera (1:750 dilution) and goat anti-guinea pig horseradish peroxidase-conjugated antibody (1:5,000 dilution) (47). The blot was developed with the Sigma Fast system (B). Lanes: M, protein molecular size standards; 1, bacterial lysate prior to induction; 2, bacterial lysate after induction with IPTG; 3, His-tagged rNSP5 eluted from Ni affinity column. K, kilodaltons.
In addition to the NSP5 ORF, the gene 11 cDNA in pQE30g11 contained the complete ORF for the 11-kDa protein NSP6. The initiation codon for NSP6 was located 58 bases downstream of the initiation codon for NSP5, and the NSP6 ORF was out of frame with respect to the NSP5 ORF and the His tag. As a consequence, any rNSP6 produced by pQE30g11 would lack a His tag and would be unable to directly bind the NTA matrix. Unlike the NSP5 ORF, the NSP6 ORF in pQE30g11 was not preceded by the prokaryotic ribosome-binding site and thus, its expression in bacteria would be expected to be inefficient, at best. To test whether rNSP5 purified by NTA affinity chromatography contained rNSP6, a large amount of an rNSP5 preparation was resolved by electrophoresis on a 10 to 20% polyacrylamide gel. Although staining of the gel with Coomassie blue revealed the presence of several minor bands in the range of 8 to 26 kDa, a Western blot assay with NSP5 polyclonal antisera established that the minor bands were fragments of rNSP5 (data not shown). The absence of bands of ∼11 kDa not reacting with the NSP5 antisera indicates that the rNSP5 preparation did not contain rNSP6.
rNSP2-mediated phosphorylation of rNSP5
To examine rNSP5 for intrinsic protein kinase activity, the purified protein was incubated with [γ-32P]ATP in an in vitro phosphorylation assay and the reaction mixture was analyzed for the production of 32P-labeled rNSP5 by polyacrylamide gel electrophoresis (PAGE) and autoradiography. Consistent with earlier reports (5, 39), the protein was only weakly autophosphorylated, even when large amounts of the protein were included in the assay (∼40 pmol), suggesting that, by itself, rNSP5 has little or no kinase activity (Fig. 2B, lane 2). Unexpectedly, a radiolabeled protein of ∼70 kDa was detected in the products of the rNSP5 phosphorylation assay (lane 2). The radiolabeled protein corresponded in size to a minor protein detectable upon electrophoresis and staining of large amounts of the purified rNSP5 preparation (Fig. 2A, lane 2). Microsequencing of the ∼70-kDa protein recovered from a polyacrylamide gel showed that its N-terminal sequence was SKIIGIDL (48). A search of the protein sequence data bank revealed that the ∼70-kDa protein was DnaK, a member of the Hsp70 family of molecular chaperones (4). Earlier studies of DnaK have shown that the protein has a strong affinity for ATP and a low level of intrinsic ATPase activity and is not a protein kinase (42). It is likely that DnaK became radiolabeled in the phosphorylation assay due to its affinity for [γ-32P]ATP.
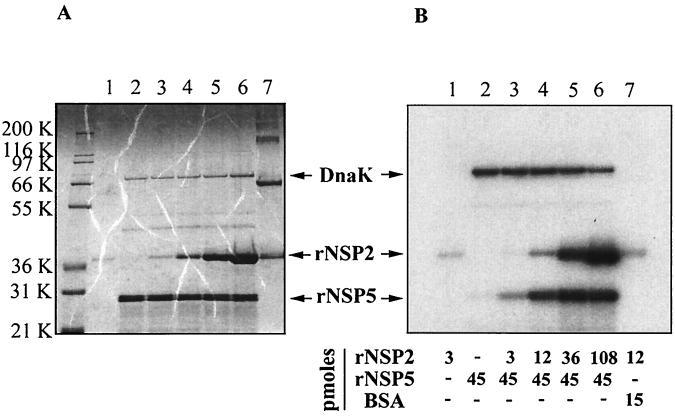
FIG. 2.
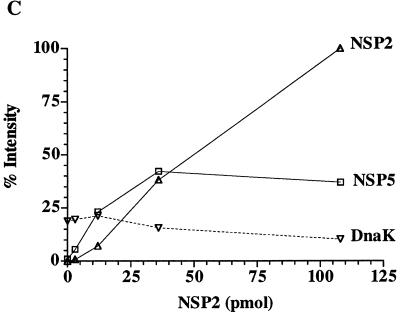
rNSP2-stimulated phosphorylation of rNSP5. Phosphorylation assay reaction mixtures contained rNSP2 (47), rNSP5 and/or BSA, 2 to 10 μCi of [γ-32P]ATP (30 to 3,000 Ci/mmol; NEN), 50 mM Tris-HCl (pH 7.5), and 5 mM MgCl2 in a final volume of 30 μl. After incubation for 2 h at 37°C, proteins in the mixtures were subjected to LDS-PAGE and detected by staining with Coomassie blue (A). Autoradiography was used to identify 32P-labeled proteins resolved on the gel (B). The intensities of radiolabeled rNSP2, rNSP5, and DnaK in panel B were quantified with a phosphorimager, and the values were adjusted relative to that obtained with rNSP2 in the reaction mixture containing 108 pmol of rNSP2 and 45 pmol of rNSP5, which was arbitrarily defined as 100%. The adjusted values were plotted as a function of the rNSP2 concentration in the reaction mixtures (C). K, kilodaltons.
As was found previously (47, 48), purified rNSP2 was phosphorylated in the in vitro phosphorylation assays even when only a relatively small amount of the protein (3 pmol) was present (Fig. 2B, lane 1). To assess the impact of rNSP2 on the extent of rNSP5 phosphorylation, reaction mixtures containing a constant amount of rNSP5 (45 pmol) were incubated with increasing amounts of rNSP2 (3 to 108 pmol). Electrophoretic analysis revealed that the presence of rNSP2 significantly enhanced the extent of rNSP5 phosphorylation even when only a small amount of rNSP2 relative to rNSP5 was present in the assay (Fig. 2B, lanes 3 and 4). Quantitation of the results showed that the degree of rNSP5 phosphorylation reached a maximum in reaction mixtures in which the molar ratio of rNSP2 to rNSP5 was close to 1:1 (Fig. 2C). At higher levels of rNSP2, the extent of rNSP5 phosphorylation was unchanged although the extent of rNSP2 phosphorylation increased as the amount of rNSP2 included in the reaction mixtures increased. The level of DnaK radiolabeling in the phosphorylation assays decreased slightly as the amount of added rNSP2 increased (Fig. 2C), possibly reflecting competition and hydrolysis of ATP by rNSP2.
Detection of the RNA-binding activity of rNSP5
Three approaches were used to analyze rNSP5 for associated RNA-binding activity: UV cross-linking of RNA to protein, filter binding of RNA-protein complexes, and RNA capture with immunoadsorbed protein (33). As detailed below, all of these approaches indicated that rNSP5 has affinity for RNA.
(i) UV cross-linking assay
32P-labeled gene 8 mRNA and v40 RNA were prepared by transcribing SacII-linearized vectors pSP65g8(+) and pSP72v40 with T7 RNA polymerase using Ambion MEGAscript and MEGAshortscript kits, respectively (30, 35). Reaction mixtures contained 100 μCi of [α-32P]UTP (800 Ci/mmol; NEN) and one-fourth of the recommended concentration of unlabeled UTP. 33P-labeled gene 8 mRNA was prepared by including 100 μCi of [α-33P]UTP (3,000 Ci/mmol; NEN) in reaction mixtures in place of [32P]UTP. To remove unincorporated nucleotides, the gene 8 mRNA was passed through NucAway spin columns (Ambion) and the v40 RNA was eluted from an 8% polyacrylamide gel containing 7 M urea (36). The gene 8 mRNA is 1,059 nucleotides in length and is identical in sequence to that of the SA11 strain of rotavirus (35). The 3′-terminal 40 nucleotides of the 72-nucleotide v40 RNA are the same as those found at the 3′ end of the SA11 gene 8 mRNA (30).
Purified rNSP5 was incubated with 32P-labeled gene 8 mRNA, and the mixtures were exposed to 254-nm UV light for 15 min at a distance of 3 to 4 cm to introduce covalent linkages between the RNA and any protein to which it was bound. Afterwards, some mixtures were treated with RNase T1, leaving only short stretches of the RNA linked to the protein. The samples were analyzed for 32P-labeled rNSP5 by PAGE and autoradiography. As shown in Fig. 3A, 32P-labeled rNSP5 was detected in the mixtures, indicating that the protein had affinity for ssRNA. Formation of 32P-labeled rNSP5 required exposure of the reaction mixtures to UV light (data not shown) and was not detected unless the UV cross-linked mixtures were treated with RNase (Fig. 3A). Due to its large size, the intact 32P-labeled gene 8 mRNA remained near the origin of the 10% polyacrylamide gel (lane 1). The 15-min UV exposure time used in these experiments was based on the observation that a nearly linear increase in the formation of 32P-labeled rNSP5 resulted as the light exposure time was increased from 0 to 20 min (data not shown). Under the conditions used in this assay, the amount of 32P-labeled rNSP5 formed in the reaction mixtures increased as the amount of protein in the reaction mixtures was increased to 20 pmol (data not shown).
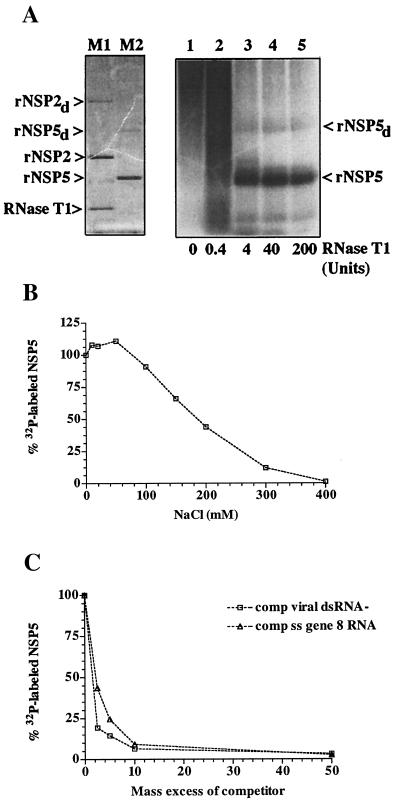
FIG.3.
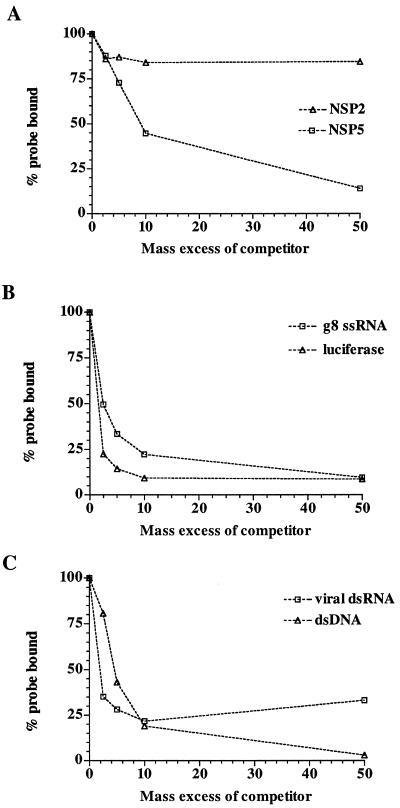
UV cross-linking of RNA to rNSP5. (A) Reaction mixtures containing 18 pmol of rNSP5 and 0.14 pm of 32P-labeled gene 8 mRNA were incubated in LSB for 15 min at room temperature and then exposed to UV light at room temperature. Afterwards, the mixtures were treated with 0, 0.4, 4, 40, or 200 U of RNase T1 for 30 min at 37°C and then analyzed by LDS-PAGE. Staining of the gel with Coomassie blue revealed the locations of rNSP2, rNSP2 dimers (rNSP2d), and RNase T1 (11 kDa) (lane M1) and rNSP5 and rNSP5 dimers (rNSP5d) (lane M2). 32P-labeled rNSP5 was located by autoradiography (lanes 1 to 5). (B) After incubation of reaction mixtures containing 9 pmol of rNSP5, 2 pmol of 32P-labeled v40, and 0 to 400 mM NaCl, the samples were exposed to UV light and digested with 40 U of RNase T1. 32P-labeled rNSP5 was detected in the samples by LDS-PAGE and quantified with a phosphorimager. The values were normalized to the intensity of the 32P-labeled rNSP5 produced in the reaction mixture containing 0 mM NaCl, which was assigned a value of 100%. The normalized values were plotted as a function of the NaCl concentration. (C) Reaction mixtures containing 16 pmol of rNSP5, 0.28 pmol of 32P-labeled gene 8 mRNA, and various amounts of unlabeled gene 8 mRNA or total virion dsRNA were incubated and exposed to UV light. After digestion with RNase T1, 32P-labeled rNSP5 in the mixtures was resolved by LDS-PAGE and quantified with a phosphorimager. The values were plotted as a function of the molar ratio of unlabeled to labeled RNA in the assays.
The stability of the interaction between rNSP5 and RNA was evaluated by analyzing the effect that inclusion of NaCl in UV cross-linking assays had on the formation of 32P-labeled rNSP5. As shown in Fig. 3B, the presence of 100 mM NaCl in the reaction mixtures reduced the formation of 32P-labeled rNSP5 by <20%. Given that the intracellular concentration of monovalent salt is ∼100 mM, the limited effect that 100 mM NaCl had on the formation of 32P-labeled-rNSP5 suggests that the protein possesses RNA-binding activity under physiological conditions. rNSP5 retained one-half of its RNA-binding activity in the UV cross-linking assay, even in the presence of ∼175 mM NaCl.
(ii) Filter-binding assay
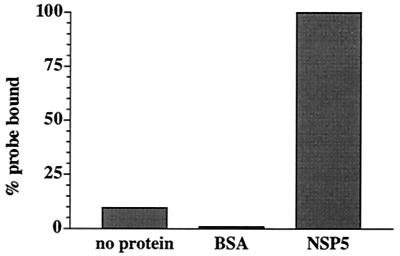
Reaction mixtures containing 33P-labeled gene 8 mRNA and no protein or the same RNA and either 75 ng of bovine serum albumin (BSA) or purified rNSP5 were incubated and then filtered through nitrocellulose membranes to recover protein and any RNA bound to the protein (Fig. 4). To increase the stringency of the assay for RNA-protein interactions, a 100 mM concentration of NaCl was also included in the reaction mixtures. The analysis showed that significant levels of 33P-labeled gene 8 mRNA bound to the membranes only when rNSP5 was present in the reaction mixture. Consistent with the results of the UV cross-linking assays, these findings suggest that rNSP5 has affinity for ssRNA. In the filter-binding assay, the amount of 33P-labeled RNA binding to the nitrocellulose filters increased as the amount of rNSP5 included in the reaction mixtures was increased to 75 ng.
FIG. 4.
Detection of rNSP5 RNA-binding activity by filter-binding assay. Reaction mixtures (25 μl) contained 0.07 pmol of 33P-labeled gene 8 mRNA, 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, and either no protein or 500 ng of BSA or rNSP5. After incubation for 30 min at room temperature, the mixtures were filtered through 25-mm-diameter nitrocellulose membranes (0.45-μm pore size) presoaked in TN buffer (50 mM Tris-HCL [pH 8.0], 100 mM NaCl). Subsequently, the filters were washed two or three times with TN buffer and air dried and the quantity of 33P-labeled RNA bound to the filter was determined with a liquid scintillation counter. All assays were performed in duplicate, and the values were averaged. The values were normalized to that of the reaction mixture containing rNSP5, which was assigned a value of 100%.
(iii) RNA capture assay
The His-tagged proteins rNSP2, rNSP5, and recombinant dihydrofolate reductase (rDHFR) were each incubated with 32P-labeled v40 RNA. Subsequently, the mixtures were incubated with anti-NSP2 or -NSP5 antiserum produced by immunizing guinea pigs with purified, His-tagged rNSP2 or rNSP5. Antibody-antigen complexes in the mixtures were recovered with protein A-Sepharose, and 32P-labeled v40 RNA bound to the beads was detected by gel electrophoresis and autoradiography (Fig. 5). The results showed that little or no v40 RNA was bound to beads recovered from mixtures containing rDHFR and NSP5 antiserum (lane 3). In contrast, 32P-labeled v40 RNA was bound to beads recovered from mixtures containing rNSP2 and either NSP2 (lane 6) or NSP5 antiserum (lanes 4). The reason that the use of NSP5 antiserum led to the recovery of 32P-labeled v40 is that this serum contains anti-His antibody, which is able to bind to His-tagged rNSP2. Although only a relatively low level of v40 RNA was associated with beads recovered from a mixture containing 10 pmol of rNSP5 and NSP5 antisera (lane 1), the level of v40 increased markedly when the amount of rNSP5 in the mixture was increased to 50 pmol (lane 2). The recovery of 32P-labeled v40 RNA with immunoadsorbed rNSP5 provided additional evidence that rNSP5 possesses RNA-binding activity.
FIG. 5.
RNA-binding activity of immunoadsorbed rNSP5. Reaction mixtures (50 μl) containing 2.5 pmol of 32P-labeled v40 RNA and 10 or 50 pmol of His-tagged DHFR (47), rNSP2, or rNSP5 in LSB were incubated for 30 min at room temperature. A 950-μl volume of LSB and 4 μl of guinea pig anti-NSP2 or anti-NSP5 antisera were then added, and the mixtures incubated overnight at 4°C with gentle mixing. After addition of 50 μl of a 50% slurry of protein A-Sepharose beads in LSB, the mixtures were incubated for 1 h. The beads were washed three times with LSB and resuspended in LDS sample buffer. 32P-labeled v40 RNA in the sample buffer was detected by PAGE and autoradiography and quantified with a phosphorimager.
Specificity of the RNA-binding activity of rNSP5
To test whether rNSP5 recognizes dsRNA in addition to ssRNA by the UV cross-linking assay, 32P-labeled gene 8 mRNA was combined with various amounts of unlabeled gene 8 mRNA or virion dsRNA to produce ratios of cold competitor RNA to radiolabeled RNA of 2- to 50-fold in reaction mixtures. After addition of the same amount of rNSP5 to each, the mixtures were exposed to UV light, treated with RNase, and analyzed by PAGE to detect the formation of 32P-labeled rNSP5. Quantitation of the results indicated that gene 8 mRNA and virion dsRNA competed similarly with 32P-labeled gene 8 mRNA for binding to rNSP5 (Fig. 3C). Indeed, when present in reaction mixtures at a 10-fold excess over the radiolabeled RNA, each nonradiolabeled competitor RNA reduced by ∼10-fold the amount of 32P-labeled rNSP5 produced in the assay. Thus, the results of UV cross-linking assays indicated that rNSP5 possesses affinity for both ssRNA and dsRNA.
The filter-binding assay was used to verify that rNSP5 recognizes dsRNA in addition to ssRNA. To accomplish this, 32P-labeled gene 8 mRNA was mixed with various amounts of unlabeled gene 8 dsRNA to yield ratios of nonradiolabeled dsRNA to radiolabeled ssRNA of 2- to 50-fold in reaction mixtures. Following addition of equivalent amounts of rNSP5 or rNSP2 to the mixtures and incubation, RNA-protein complexes were collected by filtration onto nitrocellulose membranes. rNSP2 was used as a control in this experiment since the protein is known to have strong affinity for ssRNA and to have poor affinity for dsRNA (47). Analysis of the amount of radiolabeled RNA-protein complexes that were formed indicated, as expected, that the presence of dsRNA in reaction mixtures had little effect on the binding of rNSP2 to the 32P-labeled gene 8 mRNA (Fig. 6A). In contrast, the presence of dsRNA in the reaction mixtures significantly reduced the amount of radiolabeled ssRNA that bound to rNSP5, consistent with the hypothesis that this protein possesses affinity for both ssRNA and dsRNA.
FIG. 6.
Specificity of the RNA-binding activity of rNSP5 measured by filter-binding assay. (A) Reaction mixtures containing 0.03 pmol of 33P-labeled gene 8 mRNA, various amounts of unlabeled gene 8 dsRNA (47), and 2.7 pmol of either rNSP2 or rNSP5 were incubated for 30 min at room temperature. 33P-labeled RNA bound to the protein was recovered by filtration through nitrocellulose membranes and quantified with a scintillation counter. The normalized values were plotted as a function of the molar ratio in mass of unlabeled to labeled nucleic acid in the reaction mixtures. (B and C) Same as in panel A except that the reaction mixtures contained 0.07 pmol of 33P-labeled gene 8 mRNA, various amounts of unlabeled gene 8 mRNA, luciferase RNA (Promega), rotavirus genomic dsRNA or SacII-linearized pSP65g8(+) dsDNA, and 0.9 pmol of rNSP5.
The RNAs used in the RNA-binding assays described above for rNSP5 all contained virus-specific sequences. To examine whether the ssRNA-binding activity of rNSP5 was sequence dependent or independent, the protein was added to mixtures containing 32P-labeled gene 8 mRNA and various concentrations of unlabeled gene 8 or luciferase mRNA. After incubation, RNA-protein complexes were collected by filtration and the amount of 32P-labeled RNA bound to the filters was determined. The results showed that both the unlabeled gene 8 and luciferase mRNAs interfered in a concentration-dependent manner with the interaction of rNSP5 and the 32P-labeled gene 8 mRNA (Fig. 6B). From this analysis, it was concluded that the RNA-binding activity of rNSP5 is sequence independent.
To evaluate whether rNSP5 can distinguish between dsRNA and dsDNA, 32P-labeled gene 8 mRNA was combined with various amounts of unlabeled virion dsRNA or linearized plasmid dsDNA and a constant amount of rNSP5. After incubation, RNA-protein complexes in the reaction mixtures were collected by filtration on nitrocellulose membranes and the amount of 32P-labeled RNA on the membranes was determined. The results showed that dsRNA and dsDNA interfered similarly with the binding of 32P-labeled RNA to rNSP5 (Fig. 6C). Hence, rNSP5 possesses affinity for dsDNA and the extent of that affinity is indistinguishable from that for dsRNA.
Cointeraction of rNSP2 and rNSP5 with RNA
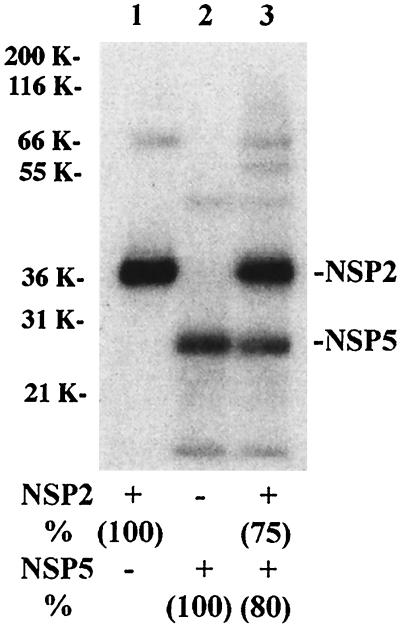
Our results demonstrated that rNSP2 interacts with rNSP5, causing its hyperphosphorylation, and that both rNSP2 and rNSP5 have nonspecific affinity for ssRNA. To assess whether these proteins affect one other's ability to bind ssRNA, reaction mixtures containing either rNSP2, rNSP5, or both proteins were incubated with 32P-labeled v40 RNA and then exposed to UV light to covalently link RNA-protein complexes. Following RNase treatment, the mixtures were resolved by PAGE and the levels of 32P-labeled rNSP2 and rNSP5 were determined with a phosphorimager (Fig. 7). The analysis revealed that there were similar reductions in the levels of 32P-labeled rNSP2 (20% reduction) and rNSP5 (25% reduction) formed in the mixture containing both proteins (lane 3) compared to the mixtures containing one of the proteins (lane 1 and 2). Thus, although these proteins can interact, neither exerted a trans-dominating activity that interfered with the RNA-binding activity of the other. The 20 to 25% reduction in the formation of 32P-labeled rNSP2 and rNSP5 when both proteins were present in the reaction mixture most likely was due to increased competition for the probe.
FIG. 7.
Cobinding of rNSP2 and rNSP5 to RNA. Reaction mixtures containing 8 pmol of rNSP2, 7 pmol of rNSP5, or both proteins were incubated with 6 pmol of 32P-labeled v40 RNA and then exposed to UV light. After RNase digestion, 32P-labeled rNSP2 and rNSP5 were detected in the mixtures by LDS-PAGE and autoradiography and quantified with a phosphorimager. The values for reaction mixtures containing only rNSP2 or rNSP5 were arbitrarily set at 100. The adjusted values are given in parentheses. K, kilodaltons.
Detection of RNA-NSP5 interactions in infected cells
To determine whether NSP5 was bound to RNA in rotavirus-infected cells, MA104 cell monolayers were infected with SA11-4F and, at 7 h postinfection, rinsed twice with TMN buffer (3 mM Tris-HCl [pH 8.1], 66 mM NH4Cl, 3 mM magnesium acetate, 14 mM potassium acetate, 1 mM dithioerythritol), and overlaid with TMN buffer containing antiprotease cocktail. The monolayers were then exposed to 254-nm UV light for 0, 1, or 2 min at a distance of 1 cm. The cells were scraped into the TMN buffer, pelleted by low-speed centrifugation, and lysed with TMN buffer containing 0.5% Triton X-100. After incubation on ice for 5 min with occasional gentle mixing, nuclei and large cellular debris were removed from the lysates by centrifugation at 13,000 × g for 10 min. One-half of each clarified lysate was adjusted to 30,000 U of RNase T1 and 10 μg of RNase A (Ambion) per ml and incubated for 30 min at 37°C, while the other half of each lysate was mock treated. To promote disruption of noncovalent interactions holding protein-protein and RNA-protein complexes together, the mock- and RNase-treated portions of the lysates were adjusted to 4 M urea-1% Triton X-100-0.25% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonic acid (CHAPS)-0.25% deoxycholate in a final volume of 4 ml of TMN and incubated for 15 min at 37°C and then for 10 min at 50°C. After addition of CsCl to a density of 1.45 g/ml, the samples were centrifuged at 35,000 rpm in a Beckman TI55 rotor for 18 h at 12°C. Due to their intrinsic densities, it is expected that RNA will pellet and protein will form a band of ∼1.30 g/ml in such CsCl gradients. Recovery of protein from the RNA pellet of the gradients indicates that the protein either has a strong affinity for RNA or is covalently linked to the RNA. The presence of NSP5 in the pellet fractions of the CsCl gradients was monitored by lithium dodecyl sulfate (LDS)-PAGE and Western blot assay using mouse anti-NSP5 monoclonal antibody 158G37 (39) and peroxidase-conjugated immunoglobulin G antibody. Blots were developed with Pierce SuperSignal chemiluminescent substrate and exposed to X-ray film
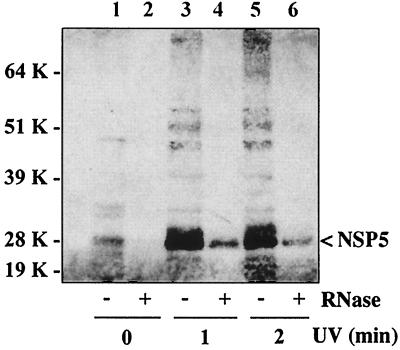
As shown in Fig. 8, little or no NSP5 pelleted in gradients containing RNase-digested lysates (lanes 2, 4, and 6). Similarly, little NSP5 pelleted in the gradient containing an undigested lysate derived from cells not exposed to UV light (lane 1). In contrast, large amounts of NSP5 pelleted in gradients containing undigested lysates derived from cells treated with UV light (lanes 3 and 5). Hence, recovery of large amounts of NSP5 in the pellet required UV cross-linking and that RNA in the lysate not be removed by RNase digestion prior to CsCl centrifugation. Collectively, the analyses indicated that exposure of infected cells to UV light results in the cross-linking of NSP5 to RNA to form complexes recoverable in the pellets of CsCl gradients.
FIG. 8.
UV cross-linking of NSP5 to RNA in vivo. At 7 h postinfection, SA11-4F-infected cells were exposed to UV light for 0, 1, or 2 min. Clarified lysates prepared from the cells were divided into two portions, and one portion was digested with RNase and the other portion was mock digested. The portions were incubated with denaturants and at elevated temperatures and then adjusted to ∼1.45 g/ml with CsCl, and the samples were centrifuged to equilibrium. Material in the pellet of each gradient was subjected to LDS-PAGE, and protein in the gel was transferred to nitrocellulose. NSP5 was located in the blot by incubation with anti-NSP5 antibody and peroxidase-conjugated immunoglobulin G antibody. The blot was developed with Pierce SuperSignal chemiluminescent substrate and exposed to X-ray film. K, kilodaltons.
Analysis of the pellet fractions of the CsCl gradients by PAGE and Western blot assay with NSP2 polyclonal antisera showed that RNA-binding protein NSP2 was also present (data not shown). In contrast, a similar analysis performed with VP6 polyclonal antisera showed that VP6, a protein that lacks RNA-binding activity, was not present in the pellets of the CsCl gradients. However, VP6 was detectable in the infected-cell lysate prior to centrifugation. Considered together, these data indicate that the detection of NSP5 in the pellets of the CsCl gradients stems from the affinity of the protein for RNA and not from nonspecific contamination of the pellet by proteins in the infected-cell lysate.
NSP5 in comparison to other RNA-binding proteins
This study has shown that NSP5 is a sequence-independent RNA-binding protein with affinity for both ssRNA and dsRNA. Our results are consistent with the results of preliminary experiments by Mattion et al. (26), who indicated that NSP5 has affinity for poly(U). The findings of the study described herein, combined with those of previous studies, establish that of the 12 proteins encoded by the group A rotaviruses, 7 have affinity for RNA. Of these seven, five are involved in RNA packaging or synthesis (VP1 [30], VP2 [7], VP3 [31], NSP2 [20], and NSP5), one is involved in viral mRNA translation (NSP3 [38]), and the function of one is unknown (NSP1 [17]). The only rotavirus protein besides NSP5 known to have affinity for dsRNA is VP2. However, while competition assays indicate that NSP5 has similar affinity for ssRNA and dsRNA (Fig. 3C), the affinity of VP2 for ssRNA is significantly higher than its affinity for dsRNA (7, 21). Hence, among the rotavirus RNA-binding proteins, the affinity of NSP5 is unique.
Analysis of the NSP5 sequence has indicated that the protein does not contain the classical dsRNA-binding motif found in PKR and many other dsRNA-binding proteins (13). Although members of the family Reoviridae in addition to the group A rotaviruses (e.g., bluetongue virus [BTV] and orthoreovirus) encode proteins with affinity for dsRNA, comparison of their sequences with that of NSP5 has not revealed similarities suggestive of functional homologs. Likewise, comparison of the biochemical and enzymatic properties of NSP5 with those of proteins of other viruses of the family have not suggested which, if any, are functional homologs. dsRNA-binding protein VP6 of BTV is similar to NSP5 in that it is a phosphoprotein and has nonspecific affinity for ssRNA and dsRNA and for DNA (45). However, BTV VP6 is a basic, glycine-rich structural protein residing inside the core of the virion. Also, BTV VP6 possesses RNA-RNA helicase activity, which we have not detected for NSP5 (data not shown). Multifunctional outer capsid protein σ3 of orthoreovirus (29, 43) and the protein products of the NSP3 gene of group C rotavirus (22) have dsRNA-binding activity, but the primary function of this activity appears to be to inhibit the RNA-dependent activation of PKR by sequestering RNA-RNA duplexes that form in viral RNA in vivo (22, 51). Suppression of PKR activation can be important for productive infection, as this enzyme inhibits protein synthesis by phosphorylating eukaryotic initiation factor 2α (10). Although σ3 and the group C NSP3 gene products have no similarity to NSP5, it remains possible that by interacting with RNA-RNA duplexes, NSP5 also inhibits PKR activation.
Importance of NSP2-NSP5 interactions
Since NSP2 and NSP5 physically interact with one another (1, 16, 39), cooperate in the formation of viroplasms, and are common components of RIs with replicase activity, these proteins likely work together during genome replication. Because the segmented dsRNA genomes of rotavirus and related viruses (e.g., orthoreovirus, BTV, and bacteriophage φ6) are made only following packaging of the mRNA templates into T=1 icosahedral procapsids, naked genomic dsRNA is not present in infected cells (34). As a consequence, the target of the dsRNA-binding activity of NSP5 in vivo is probably not genomic dsRNA, as access to this RNA by a nonstructural protein would be prevented by the shell of the procapsid. Instead, the target of the dsRNA-binding activity of NSP5 may include the secondary and tertiary structures of those mRNAs that are to be packaged into and replicated within the procapsid. From analysis of the replication of φ6 dsRNA in vitro, it is known that such higher-order structures in the mRNA template can impede packaging (27, 41). Although we have not been able to detect helicase activity for NSP5 (data not shown), the fact that NSP5 has affinity for NSP2, a protein with helix-destabilizing activity, raises the possibility that NSP5 recruits NSP2 to sites on the mRNA template containing higher-order structures. In turn, NSP2 may disrupt RNA-RNA duplexes at these sites and the ssRNA-binding activities of NSP2 and/or NSP5 may then serve to prevent re-formation of the higher-order structures. Similarly, NSP2 and NSP5 may work together to preclude inappropriate trans interactions among the 11 viral mRNAs that might interfere with packaging.
Phosphorylation assays indicate that NSP5, in the absence of other viral proteins or cellular components, contains little or no autophosphorylation activity (5, 39). In comparison, NSP2 contains an intrinsic NTPase activity that results in its autophosphorylation in vitro. NSP2 that is transiently expressed in MA104 cells in the absence of other rotavirus proteins is also phosphorylated, but a phosphorylated form of NSP2 has not been detected in rotavirus-infected cells (47). Consistent with a physical interaction between the proteins, inclusion of NSP2 with NSP5 in phosphorylation assays causes hyperphosphorylation of NSP5 via a mechanism that can occur in the absence of RNA. The source of the phosphate residues added to NSP5 in the presence of NSP2 is not known but could result from the induction of a dormant kinase activity associated with either NSP2 or NSP5. However, we favor the alternative hypothesis that the phosphate residues added to NSP5 originate from the transfer of phosphate groups that are generated by the NTPase activity of NSP2 and that are initially covalently linked to NSP2. If this hypothesis is correct, then the inability to detect phosphorylated NSP2 in the infected cell results from rapid and efficient transfer of phosphate groups from NSP2 to NSP5. Addition of phosphatase inhibitors to rotavirus-infected cells increases the level of hyperphosphorylated NSP5, implying that, in vivo, this protein is transiently hyperphosphorylated and that phosphate groups are removed from the protein via cellular phosphatases (6). Thus, it can be proposed that the NTPase activity of NSP2 generates phosphate residues that are transferred from NSP2 to NSP5 and subsequently removed from NSP5 by phosphatases. This pathway would allow continuing cycling of NSP2 and NSP5 between the phosphorylated and nonphosphorylated forms and provide a means by which to dispose of the phosphate residues generated by the NTPase activity of NSP2.
Acknowledgments
P.V. was supported in part by a grant from INRA, Jouy-en-Josas, France.
Mouse anti-NSP5 monoclonal antibody 158G37 was kindly provided by Didier Poncet (INRA, Jouy-en-Josas, France). We thank Jonas Chnaiderman for constructing pQE30g11.
REFERENCES
- 1.Afrikanova, I., E. Fabretti, M. C. Miozzo, and O. R. Burrone. 1998. Rotavirus NSP5 phosphorylation is up-regulated by interaction with NSP2. J. Gen. Virol. 79:2679-2686. [DOI] [PubMed] [Google Scholar]
- 2.Afrikanova, I., M. C. Miozzo, S. Giambiagi, and O. R. Burrone. 1996. Phosphorylation generates different forms of rotavirus NSP5. J. Gen. Virol. 77:2059-2065. [DOI] [PubMed] [Google Scholar]
- 3.Aponte, C., D. Poncet, and J. Cohen. 1996. Recovery and characterization of a replicase complex in rotavirus-infected cells using a monoclonal antibody against NSP2. J. Virol. 70:985-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Zvi, A. P., and P. Goloubinoff. 2001. Mechanisms of disaggregation and refolding of stable protein aggregates by molecular chaperones. J. Struct. Biol. 135:84-93. [DOI] [PubMed] [Google Scholar]
- 5.Blackhall, J., A. Fuentes, K. Hansen, and G. Magnusson. 1997. Serine protein kinase activity associated with rotavirus phosphoprotein NSP5. J. Virol. 71:138-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackhall, J., M. Munoz, A. Fuentes, and G. Magnusson. 1998. Analysis of rotavirus nonstructural protein NSP5 phosphorylation. J. Virol. 72:6398-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle, J. F., and K. V. Holmes. 1986. RNA-binding proteins of bovine rotavirus. J. Virol. 58:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, D., C. L. Luongo, M. L. Nibert, and J. T. Patton. 1999. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology 265:120-130. [DOI] [PubMed] [Google Scholar]
- 9.Chen, D., C. Y. Zeng, M. J. Wentz, M. Gorziglia, M. K. Estes, and R. F. Ramig. 1994. Template-dependent in vitro replication of rotavirus RNA. J. Virol. 68:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens, M. J. 1997. PKR—a protein kinase regulated by double-stranded RNA. Int. J. Biochem. Cell Biol. 29:945-949. [DOI] [PubMed] [Google Scholar]
- 11.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Fabbretti, E., L. Afrikanova, F. Vascotto, and O. R. Burrone. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333-339. [DOI] [PubMed] [Google Scholar]
- 13.Fierro-Monti, I., and M. B. Mathews. 2000. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci. 25:241-246. [DOI] [PubMed] [Google Scholar]
- 14.Gallegos, C. O., and J. T. Patton. 1989. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology 172:616-627. [DOI] [PubMed] [Google Scholar]
- 15.Gonzáles, S. A., and O. Burrone. 1991. Rotavirus NS26 is modified by addition of a single O-linked residue of N-acetylglucosamine. Virology 182:8-16. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez, R. A., M. A. Torres-Vega, S. Lopez, and C. F. Arias. 1998. In vivo interactions among rotavirus nonstructural proteins. Arch. Virol. 143:981-996. (Erratum, 143:2064.) [DOI] [PubMed] [Google Scholar]
- 17.Hua, J., X. Chen, and J. T. Patton. 1994. Deletion mapping of the rotavirus metalloprotein NS53 (NSP1): the conserved cysteine-rich region is essential for virus-specific RNA binding. J. Virol. 68:3990-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 19.Kattoura, M. D., X. Chen, and J. T. Patton. 1994. The rotavirus RNA binding protein NS35 (NSP2) forms 10S multimers and interacts with the viral RNA polymerase. Virology 202:803-813. [DOI] [PubMed] [Google Scholar]
- 20.Kattoura, M., L. L. Clapp, and J. T. Patton. 1992. The rotavirus non-structural protein, NS35, is a nonspecific RNA-binding protein. Virology 191:698-708. [DOI] [PubMed] [Google Scholar]
- 21.Labbe, M., P. Baudoux, A. Charpilienne, D. Poncet, and J. Cohen. 1994. Identification of the nucleic acid binding domain of the rotavirus VP2 protein. J. Gen. Virol. 75:3423-3430. [DOI] [PubMed] [Google Scholar]
- 22.Langland, J. O., S. Pettiford, B. Jiang, and B. L. Jacobs. 1994. Products of the porcine group C rotavirus NSP3 gene bind specifically to double-stranded RNA and inhibit activation of the interferon-induced protein kinase PKR. J. Virol. 68:3821-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawton, J. A., M. K. Estes, and B. V. V. Prasad. 1997. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 2:118-121. [DOI] [PubMed] [Google Scholar]
- 24.Lawton, J. A., C.Q.-Y. Zeng, S. K. Mukherjee, J. Cohen, M. K. Estes, and B. V. V. Prasad. 1997. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal deleted VP2: implications for the architecture of the VP2 capsid layer. J. Virol. 71:7353-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lui, M., N. M. Mattion, and M. K. Estes. 1992. Rotavirus VP3 expressed in insect cells possesses guanylyltransferase activity. Virology 188:77-84. [DOI] [PubMed] [Google Scholar]
- 26.Mattion, N. M., D. B. Mitchell, G. W. Both, and M. K. Estes. 1991. Expression of rotavirus proteins encoded by alternative open reading frames of genome segment 11. Virology 181:295-304. [DOI] [PubMed] [Google Scholar]
- 27.Mindich, L. 1999. Reverse genetics of dsRNA bacteriophage phi6. Adv. Virus Res. 53:341-353. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell, D. B., and G. W. Both. 1988. Simian rotavirus SA11 segment 11 contains overlapping reading frames. Nucleic Acids Res. 16:6244.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oiland, A. M., J. Jane-Valbuena, L. A. Schiff, M. L. Nibert, and S. C. Harrison. 2001. Structure of the reovirus outer capsid and dsRNA-binding protein σ3 at 1.8 Å resolution. EMBO J. 20:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patton, J. T. 1996. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA but the interaction is not sufficient to initiate minus-strand synthesis. J. Virol. 70:7940-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patton, J. T., and D. Chen. 1999. RNA-binding and capping activities of proteins in rotavirus open cores. J. Virol. 73:1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patton, J. T., and C. O. Gallegos. 1990. Rotavirus RNA replication: single-strand RNA extends from the replicase particle. J. Gen. Virol. 71:1087-1094. [DOI] [PubMed] [Google Scholar]
- 33.Patton, J. T., and J. Hua. 1995. Using the RNA-capture assay to assess the RNA-binding activity of viral proteins. Methods Mol. Genet. 7:373-387. [Google Scholar]
- 34.Patton, J. T., and E. Spencer. 2000. Genome replication and packaging of segmented double-stranded RNA viruses. Virology 277:217-225. [DOI] [PubMed] [Google Scholar]
- 35.Patton, J. T., M. Wentz, J. Xiaobo, and R. F. Ramig. 1996. cis-acting signals that promote genome replication in rotavirus mRNAs. J. Virol. 70:3961-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patton, J. T., Z. Taraporewala, V. Chizhikov, and D. Chen. 1999. Virus replication. Methods Mol. Med. 34:33-66. [DOI] [PubMed] [Google Scholar]
- 37.Petrie, B. L., H. B. Greenberg, D. Y. Graham, and M. K. Estes. 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1:133-152. [DOI] [PubMed] [Google Scholar]
- 38.Poncet, D., S. Laurent, and J. Cohen. 1994. Four nucleotides are the minimal requirement for RNA recognition by rotavirus non-structural protein NSP3. EMBO J. 13:4165-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poncet, D., P. Lindenbaum, R. L. Haridon, and J. Cohen. 1997. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J. Virol. 71:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasad, B. V. V., G. J. Wang, J. P. M. Clerx, and W. Chiu. 1988. Three-dimensional structure of rotavirus. J. Mol. Biol. 199:269-275. [DOI] [PubMed] [Google Scholar]
- 41.Qiao, X., J. Qiao, and L. Mindich. 1995. Interference with bacteriophage φ6 genomic RNA packaging by hairpin structures. J. Virol. 69:5502-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell, R., A. Wali Karzai, A. F. Mehl, and R. McMacken. 1999. DnaJ dramatically stimulates ATP hydrolysis by DnaK: insight into targeting of Hsp70 proteins to polypeptide substrates. Biochemistry 38:4165-4176. [DOI] [PubMed] [Google Scholar]
- 43.Schiff, L. A., M. L. Nibert, M. S. Co, E. G. Brown, and B. N. Fields. 1988. Distinct binding sites for zinc and double-stranded RNA in the reovirus outer capsid protein σ3. Mol. Cell. Biol. 8:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuck, P., Z. Taraporewala, P. McPhie, and J. T. Patton. 2000. Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes. J. Biol. Chem. 276:9679-9687. [DOI] [PubMed] [Google Scholar]
- 45.Stäuber, N., J. Martinez-Costas, G. Sutton, K. Monastyrskaya, and P. Roy. 1997. Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J. Virol. 71:7220-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taraporewala, Z., and J. T. Patton. 2001. Identification and characterization of the helix destabilizing activity of the rotavirus nonstructural protein NSP2. J. Virol. 75:4519-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taraporewala, Z., D. Chen, and J. T. Patton. 1999. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 73:9934-9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taraporewala, Z., D. Chen, and J. T. Patton. 2001. Multimers of the bluetongue virus nonstructural protein, NS2, possess nucleotidyl phosphatase activity: similarities between NS2 and rotavirus NSP2. Virology 280:221-231. [DOI] [PubMed] [Google Scholar]
- 49.Torres-Vega, M. A., R. A. González, M. Duarte, D. Poncet, S. López, and C. F. Arias. 2000. The C-terminal domain of rotavirus NSP5 is essential for its multimerization, hyperphosphorylation and interaction with NSP6. J. Gen. Virol. 81:821-830. [DOI] [PubMed] [Google Scholar]
- 50.Valenzuela, S., J. Pizarro, A. M. Sandino, M. Vasquez, J. Fernandez, O. Hernandez, J. Patton, and E. Spencer. 1991. Photoaffinity labeling of rotavirus VP1 with 8-azido-ATP: identification of the viral RNA polymerase. J. Virol. 65:3964-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yue, Z., and A. J. Shatkin. 1997. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology 234:364-371. [DOI] [PubMed] [Google Scholar]