Abstract
The coronavirus membrane (M) protein is the most abundant virion protein and the key component in viral assembly and morphogenesis. The M protein of mouse hepatitis virus (MHV) is an integral membrane protein with a short ectodomain, three transmembrane segments, and a large carboxy-terminal endodomain facing the interior of the viral envelope. The carboxy terminus of MHV M has previously been shown to be extremely sensitive to mutation, both in a virus-like particle expression system and in the intact virion. We have constructed a mutant, MΔ2, containing a two-amino-acid truncation of the M protein that was previously thought to be lethal. This mutant was isolated by means of targeted RNA recombination with a powerful host range-based selection allowed by the interspecies chimeric virus fMHV (MHV containing the ectodomain of the feline infectious peritonitis virus S protein). Analysis of multiple second-site revertants of the MΔ2 mutant has revealed changes in regions of both the M protein and the nucleocapsid (N) protein that can compensate for the loss of the last two residues of the M protein. Our data thus provide the first genetic evidence for a structural interaction between the carboxy termini of the M and N proteins of MHV. In addition, this work demonstrates the efficacy of targeted recombination with fMHV for the systematic genetic analysis of coronavirus structural protein interactions.
The assembly of progeny virions of coronaviruses is brought about by cooperative interactions among a minimal set of four structural proteins, the large positive-stranded RNA genome, and a membrane envelope acquired from the intermediate compartment between the endoplasmic reticulum and the Golgi complex (15, 17, 49). The viral nucleocapsid is formed by the packaging of the genome by the nucleocapsid (N) protein into a helical ribonucleoprotein structure (19) that is further compacted into a core possibly having icosahedral symmetry (37, 38). This nucleocapsid is incorporated into virions by intracellular budding through a membrane containing three envelope proteins: the membrane (M) glycoprotein, the small envelope (E) protein, and the spike (S) glycoprotein.
Although the details of the coronavirus assembly process are not well understood, in recent years tremendous progress has been made in elucidating the molecular interactions that determine the formation of the virion envelope, particularly for the prototype murine coronavirus mouse hepatitis virus (MHV). Much of this progress has been realized by the study of virus-like particles (VLPs) formed by coexpression of the coronavirus M and E proteins, which has defined the minimal molecular interactions required for production of particles resembling authentic virions (3, 52). This system has provided an invaluable tool leading to detailed exploration of the roles of individual proteins and intermolecular interactions in the coronavirus assembly process (2-4, 6-8, 11, 52).
One outcome of the MHV VLP work confirmed a previous conclusion, based on studies with the glycosylation inhibitor tunicamycin (12, 40) or with an S gene mutant (36), that the S protein, despite its essential role in virus infectivity, is not an obligatory participant in viral assembly. The S glycoprotein, which forms the peplomers characteristic of coronaviruses, is the structural protein involved in the binding of virions to host cell receptors and in mediating virus-cell and cell-cell fusion. Recent evidence supports the notion that incorporation of the S protein into virions is directed by specific heterotypic interactions with the M protein that are initiated after the folding of the S protein in the endoplasmic reticulum (7, 28, 30, 31). VLP studies have allowed the localization of the segment responsible for specific incorporation to the carboxy-terminal 64 residues of the S molecule, a region encompassing the transmembrane and endodomains (11). This point was most convincingly demonstrated by the construction, through targeted RNA recombination (18), of an MHV mutant in which the entire ectodomain of the MHV S protein was replaced by the equivalent, but highly divergent, ectodomain of the feline infectious peritonitis virus (FIPV) S protein. The resulting chimeric recombinant, fMHV, acquired the ability to infect feline cells, while it simultaneously lost the ability to grow in murine cells (18).
The E protein is a minor, yet critical, structural component in coronavirus assembly, as demonstrated for MHV by investigations with both the VLP system (52) and constructed viral mutants (10). However, the exact nature of the involvement of the E protein in the assembly process remains poorly characterized.
The M protein is the most abundant structural protein in virions and is the key player in MHV assembly. It spans the membrane bilayer three times, displaying a short amino-terminal domain on the virion exterior surface and a large carboxy-terminal tail in the virion interior (39). When expressed alone, M accumulates in the Golgi complex in homomultimeric complexes (15). However, in combination with the E protein, M is retained in the budding compartment and is incorporated into VLPs, which are exported from the cell (52). It has been shown that mutational changes in any of the domains of the M protein affect VLP formation, and in some cases, these conclusions were further supported by the incorporation of the same mutational changes into recombinant viruses (6).
Among the mutationally sensitive domains of the M protein, the extreme carboxy terminus is the most intriguing. Deletion of the last amino acid residue of M, T228 (the MΔ1 mutation), was sufficiently devastating to reduce VLP formation to minimally detectable levels when the MΔ1 protein was coexpressed with the wild-type E protein. In contrast, when the MΔ1 mutation was incorporated into the MHV genome, the resulting recombinant virus exhibited a phenotype indistinguishable from the wild type. This suggested that a third component was available in complete MΔ1 virions to stabilize structures that were weakened in MΔ1 VLPs. We speculated that the viral nucleocapsid, absent in the VLPs, was this stabilizing factor. Further truncation of the carboxy terminus of the M protein (the MΔ2 and larger mutations) completely abolished VLP formation, and we were unable to recover recombinant viruses containing any of these mutations (6). Thus, removal of as few as two amino acid residues from the carboxy-terminal tail of the M protein appeared to be lethal.
We were prompted to reexamine this conclusion following the recent isolation of the chimeric S protein mutant fMHV (18), which has enabled a much more powerful selection for MHV mutants. When fMHV was used as the recipient virus in the targeted RNA recombination scheme that is used to carry out reverse genetics of MHV (21), its stringent host cell species specificity allowed the exclusive selection of newly generated recombinants on the basis of their regaining the ability to grow in murine cells. In the present study, we successfully obtained the MΔ2 mutant, which, although viable, is drastically defective, yielding tiny plaques and growing to maximal titers more than 103-fold lower than those of wild-type MHV. Furthermore, we isolated second-site revertants of the MΔ2 mutant and the reverting mutations in these were individually mapped to the carboxy terminus of either the M protein or the N protein. Reconstruction of the reverting changes combined with the original MΔ2 mutation provided the first genetic evidence for a structural interaction between the carboxy termini of the M and N proteins of MHV. Our results complement recent biochemical studies that have demonstrated M protein-nucleocapsid interactions for MHV (24, 25) and transmissible gastroenteritis virus (TGEV) (9).
MATERIALS AND METHODS
Cells and viruses.
Wild-type MHV A59 and all MHV mutants containing the wild-type MHV S gene were propagated in mouse 17 clone 1 (17Cl1) or L2 cells, and plaque assays and purifications were carried out with mouse L2 cells. MHV mutant fMHV, which contains the ectodomain of the FIPV S protein, was maintained in feline FCWF cells or AK-D fetal lung cells (American Type Culture Collection).
Plasmid constructs.
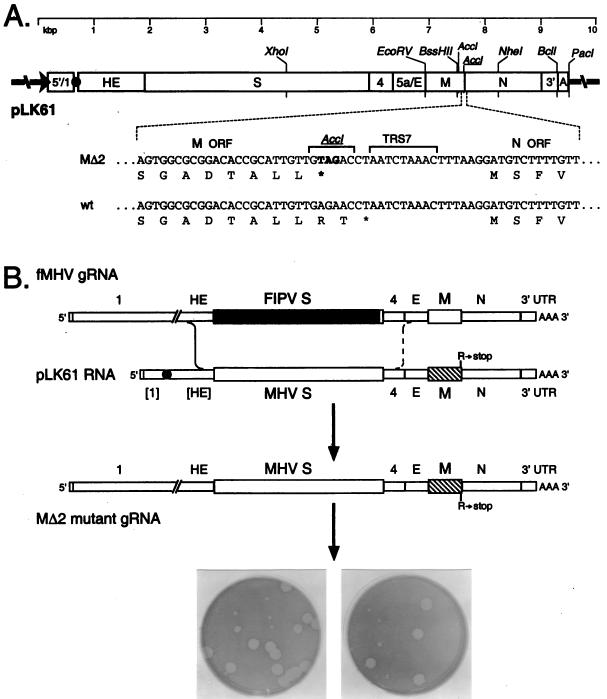
The parent plasmid used in this study was pMH54, which has been described in detail previously (18). It encodes a 9.1-kb T7 RNA polymerase transcript consisting of the 5′ 467 nucleotides (nt) of the MHV genome connected, via a 72-nt linker, to the entire 3′ 8.6 kb of the MHV genome, starting at codon 28 of the hemagglutinin-esterase (HE) pseudogene and proceeding through to a poly(A) tail of approximately 115 nt. The MΔ2 mutation, which truncates the M open reading frame (ORF) by substituting the stop codon TAG in place of the penultimate codon (AGA, coding for arginine), was first generated in plasmid pCFS8, as described in an earlier study (6). This substitution simultaneously created a new AccI site (Fig. 1A). The MΔ2 mutation was transferred into pMH54 by replacement of the XhoI-NheI fragment, resulting in plasmid pLK61 (Fig. 1A).
FIG. 1.
Selection of the MΔ2 mutant. (A) Construction of a transcription vector for synthesis of donor RNA. Plasmid pLK61 was derived from pMH54 (18) as detailed in Materials and Methods. The restriction sites shown are those relevant to plasmid construction (XhoI, EcoRV, BssHII, NheI, and BclI), in vitro transcription (PacI), or mutant analysis (AccI). The filled circle denotes the linker between the cDNA segments corresponding to the 5′ and 3′ ends of the MHV genome, and the arrow indicates the T7 promoter. The expanded region of sequence shows the three base changes made in pLK61 to create a stop codon at codon 227 of the M ORF and a new AccI site (underlined). The corresponding wild-type (wt) sequence from pMH54 is aligned for comparison. Also indicated is TRS7, the TRS governing synthesis of sgRNA7 (N mRNA). (B) Scheme for generation of the MΔ2 mutant by targeted RNA recombination between the interspecies chimera fMHV (18) and donor RNA transcribed from plasmid pLK61. fMHV contains the ectodomain-encoding region of the FIPV S gene (shaded rectangle) and is able to grow in feline cells but not in murine cells. A single crossover (solid line), within the HE gene, should generate a recombinant that has simultaneously reacquired the MHV S ectodomain and the ability to grow in murine cells and has also incorporated the mutant M gene (hatched rectangle). A potential second crossover (broken line), in gene 4 or the E gene, would exclude the mutant M gene from the recombinant. At the bottom is shown the mixed progeny of two independent targeted recombination experiments, forming tiny and large plaques on mouse L2 cells at 72 h postinfection.
For the purpose of reconstructing MΔ2 revertants, reverse transcription (RT)-PCR products generated from selected revertants were incorporated into pLK61 via unique restriction sites. For the T185I mutation in the M protein, an RT-PCR product was generated and cloned into the EcoRV and BssHII sites occurring at the 3′ end of the E gene and near the 3′ end of the M gene, respectively (Fig. 1A). For the M protein mutations D195G, D195N, G196S, and S206F and the N protein mutation S98L, RT-PCR products were inserted between the BssHII site in the M gene and the NheI site in the N gene. Similarly, the N protein mutations Q437L and Q437MMA were cloned between the NheI site in the N gene and the BclI site located in the 3′ untranslated region (Fig. 1A). All plasmid constructs were verified by restriction analysis; all PCR-generated segments and newly created junctions of each plasmid were confirmed by automated sequencing with an Applied Biosystems 3100 or 3700 DNA sequencer.
Targeted RNA recombination.
The incorporation of mutations into the MHV genome by targeted RNA recombination was carried out as previously described (10, 22), except that the interspecies chimeric virus fMHV was used as the recipient virus (13). Feline FCWF or AK-D cells in monolayers were infected with fMHV at a multiplicity of approximately 1 PFU per cell for 4 h at 37°C, after which the cells were suspended by limited trypsin treatment. Capped, runoff donor transcripts were synthesized from PacI-truncated pLK61 (or derivatives of pLK61) using a T7 RNA polymerase kit (Ambion) as specified by the manufacturer. Donor RNA (5 to 10 μg), without further purification, was transfected into 8 × 106 fMHV-infected cells by using two pulses at 960 μF and 0.3 kV with a Gene Pulser electroporation apparatus (Bio-Rad). Infected and transfected feline cells were then plated onto monolayers of murine 17Cl1 cells. At 48 h postinfection at 37°C, following the appearance of syncytia in the murine cells, progeny virus in the supernatant medium was harvested. All recombinant candidates were purified by two rounds of plaque titration on murine L2 cell monolayers at 37°C.
Genomic analysis of candidate recombinants.
Purified independent candidate recombinants, for the MΔ2 mutant and for the subsequently reconstructed revertants, were used to infect 25-cm2 monolayers of mouse L2 cells at 37°C, and total cellular RNA was harvested with the Ultraspec reagent (Biotecx) at 5 days postinfection for the MΔ2 mutant and the MΔ2 S98L double mutant or 24 to 48 h postinfection for all other revertants and controls. RNA was reverse transcribed under standard conditions (41) with a hexanucleotide random primer, p(dN)6 (Boehringer Mannheim), and avian myeloblastosis virus reverse transcriptase (Life Sciences). The cDNA was amplified by PCR with various primer pairs to characterize different genomic regions of candidate recombinants. PCR amplifications were run for 30 cycles of 1 min at 94°C, 1 min at 48°C, and 2 min at 72°C with AmpliTaq DNA polymerase (Perkin-Elmer). Products were directly analyzed by agarose gel electrophoresis or were purified with either preparative agarose gels or Quantum-prep columns (Bio-Rad). Purified PCR fragments were then analyzed by restriction digestion or automated sequencing.
RESULTS
Generation of the MΔ2 mutant.
In previous work done to study the structural requirements of the M protein in coronavirus assembly, MHV M gene mutants were successfully created by using targeted RNA recombination (6). Combined data from the VLP expression system and recombinant viruses indicated a major involvement of the carboxy-terminal residues of the M protein in the assembly process or in the stability of the resulting virions. Among the many mutants generated in the study, MΔ1 was highly interesting for the fact that truncation of the carboxy-terminal amino acid residue T228 almost entirely abolished VLP formation when MΔ1 was coexpressed with the wild-type E protein. Surprisingly, however, the isolated MΔ1 viral recombinant was indistinguishable from the wild type in all measurable aspects of growth, including plaque size, growth rate, and viral yield (6). This pointed to the existence of interactions in addition to those between the M and E proteins contributing to overall virion stability. Nucleocapsids, not present in VLPs, were the most likely candidate for filling this role.
Despite repeated efforts, we were not previously able to generate an MΔ2 recombinant in which the carboxy-terminal two amino acids of the M protein were truncated. This abrupt difference between MΔ1 and MΔ2 was very intriguing. It led us to conclude either that the MΔ2 mutation was lethal or that MΔ2 mutants were so highly defective that they could not be recovered by the targeted RNA recombination technique, which could only select for constructed recombinants that were more fit than the thermolabile N gene deletion recipient virus (10, 21, 22).
The creation of the FIPV-MHV chimeric S recombinant fMHV (18) allowed us to revisit this issue more decisively. By taking advantage of the stringent host cell species specificity of fMHV, we could now, in principle, completely eliminate the background of recipient virus in targeted RNA recombination. We expected that restoration of the MHV S gene to fMHV, via recombination with an engineered donor RNA, would allow selection for virtually any nonlethal cotransduced mutation.
For the purpose of generating an MΔ2 MHV recombinant, a vector, pLK61, was constructed from pMH54, which had been designed previously to allow the shuttling of different S genes into MHV (18, 26, 35). This transcription vector, in order to provide a sufficient flanking region for recombination, contains almost all of the HE pseudogene upstream of the S gene as well as the entire distal portion of the MHV genome thereafter (18). In pLK61, the penultimate codon (AGA) of the M ORF, encoding R227, was replaced with the stop codon TAG (Fig. 1A). This approach was taken to avoid unintended effects on the transcription of the downstream N gene that might have resulted from the actual deletion of the RNA sequence at the 3′ terminus of the M ORF adjacent to the transcription-regulating sequence (TRS) for the N gene. Donor RNA transcripts from pLK61 or from pMH54 as a control were transfected into feline cells that first had been infected with fMHV. The supernatant media from these infected and transfected cells were harvested and analyzed by plaque assay on murine L2 cells. For recombinants obtained with pLK61 donor RNA, two starkly different plaque sizes were clearly observed (Fig. 1B). One was very small and grew much more slowly than a wild-type MHV control, and the other was very similar to the wild type in both plaque size and growth rate. These dramatic differences were preserved through two subsequent rounds of plaque purification (Fig. 2A). By contrast, all recombinants obtained with the control pMH54 donor RNA gave rise to only wild-type-size plaques. Furthermore, as expected, no plaques were detected from samples that had been infected with fMHV and then mock transfected, proving the stringency of the host range-based selection.
FIG. 2.
Purification and analysis of targeted recombination progeny. (A) Appearance of purified plaques on mouse L2 cells at 72 h postinfection. Alb231 and Alb233 are tiny-plaque recombinants from two independent infection-transfection experiments, and Alb243 is a large-plaque recombinant that arose in the same infection-transfection as Alb231. Alb239 is a wild-type control recombinant reconstructed from fMHV and pMH54 donor RNA. A control mock-infected L2 cell monolayer is included for comparison. (B) RT-PCR analysis. RNA was isolated from infected cells, reverse transcribed with random primer p(N)6, and amplified with primers PM145 and PM159. PCR products were analyzed by electrophoresis in 1.2% agarose with (+) or without (−) prior digestion with AccI. Lane M, DNA fragment size markers. wt, wild type.
Genotypic confirmation and phenotypic description of the MΔ2 mutant.
Previous experience with targeted recombination has shown that it is possible to have multiple crossover events between the recipient virus genome and the donor RNA if there is a strong selective pressure to exclude a particular genetic marker (6, 13, 14). This suggested to us that the wild-type-size plaques obtained with pLK61 donor RNA probably resulted from two crossover events, one upstream of the S gene and one between the S gene and the MΔ2 mutation (Fig. 1B). The tiny plaques, on the other hand, were most likely to be the MΔ2 mutant. To test this notion, we purified RNA from cells infected with either small- or large-plaque recombinants isolated from independent infection-transfection samples. Random-primed RT was carried out followed by PCR with primers PM159 and PM145, which are specific for the upstream and downstream flanking genes E and N, respectively (Fig. 2B). Restriction digestion of the resulting 1.3-kb PCR product with AccI was expected to produce a fragment pattern of 1,029, 207, and 98 bp for the MΔ2 mutant, compared to fragments of 1,029 and 305 bp predicted for the wild type. All of the large plaques obtained from the targeted recombination with pLK61 donor RNA (one of which, Alb243, is shown in Fig. 2B) yielded exactly the same restriction digest pattern as that of an isogenic wild-type control, Alb239, that had been generated with pMH54 donor RNA (Fig. 2B). A number of other wild-type controls also showed identical restriction patterns (data not shown).
All of the small-plaque recombinants characterized contained the extra AccI site associated with the stop codon placed at codon 227. Data for two independent isolates, Alb231 and Alb233, are shown in Fig. 2B. The presence of the MΔ2 mutation was conclusively confirmed by direct sequencing of these PCR products. Moreover, there were no other changes found in the M gene for these recombinants. Although it was unlikely that the defective phenotype of the small-plaque mutants resulted from changes other than the MΔ2 mutation, PCR products encompassing the entire E and N genes were also obtained and directly sequenced. As expected, no other mutations were found in either of these genes. We therefore concluded that the MΔ2 mutation was solely responsible for the observed small-plaque phenotype of Alb231 and Alb233, consistent with previous results indicating a critical role for the carboxy terminus of the M protein in viral assembly (6).
The MΔ2 mutant exhibited an extremely defective phenotype in both murine L2 and 17Cl1 cells at 37°C. Additionally, it recovered no significant growth advantage relative to the wild type when grown at either 33 or 39°C. Therefore, the MΔ2 mutant was not temperature or cold sensitive. Besides its dramatically smaller plaque size, the MΔ2 mutant had infectious titers of less than 1 × 104 PFU/ml, over 3 orders of magnitude lower than those of wild-type controls (data not shown). With this level of mutant growth and viral yield it was possible to achieve 100% syncytium formation and cytopathic effect in L2 cell monolayers, albeit with greatly protracted kinetics compared to the wild-type virus. However, in the more slowly fusing 17Cl1 cell line, the rate of progeny viral production appeared never to be able to outrun the rate of cell growth; thus, MΔ2-infected monolayers of these cells were never entirely infected before being overtaken by uninfected cells.
Our success in obtaining the MΔ2 mutant strongly supported our initial expectations regarding the potential power of genetic selection exploiting the host cell species specificity of fMHV. MHV mutants harboring structural gene lesions detrimental to viral growth have previously been characterized by our laboratory, most notably the N gene mutants Alb4 (16, 22), Alb1 (22), and Alb25 (22), but all of these were more amenable to manipulation in tissue culture. To our knowledge, the MΔ2 mutant is the most severely impaired coronavirus mutant yet obtained by either classical or reverse genetic methods.
Isolation and analysis of revertants of the MΔ2 mutant.
A major consequence of the severity of the MΔ2 lesion was its instability. It was not possible to grow mutant viral stocks for more than two or three passages before an acceleration of viral growth was noted, which suggested to us that faster growing revertants had taken over the culture. Originally, this possibility was confirmed by plaque assay of passage 3 virus stock, which had a titer 2 to 3 orders of magnitude higher than the original MΔ2 mutant and consisted almost entirely of much larger plaques. This indicated that there was a strong selective pressure to remedy the MΔ2 defect. It was of great interest to identify any second-site mutations capable of compensating for the original mutation, since this would provide genetic information regarding interactions between the critical carboxy-terminal region of the M protein and other potential participants in the viral assembly process.
To pursue this, viral stocks begun from independent small plaques of the MΔ2 mutant were passaged multiple times at 37°C with daily examination of infected monolayers. Once the onset of accelerated syncytium formation and cytopathic effect was observed, the released virus in the supernatant media was assayed by plaque titration, with parallel wild-type and MΔ2 mutant controls. Larger-plaque-forming revertants from three independent searches were isolated and purified through two rounds of plaque titration (Fig. 3, sets 1 to 3) (revertant R1 was the first revertant, obtained from the original passage 3 stock of MΔ2). All revertants yielded plaques that were slightly smaller than those of the wild type but were significantly larger than those of the MΔ2 mutant.
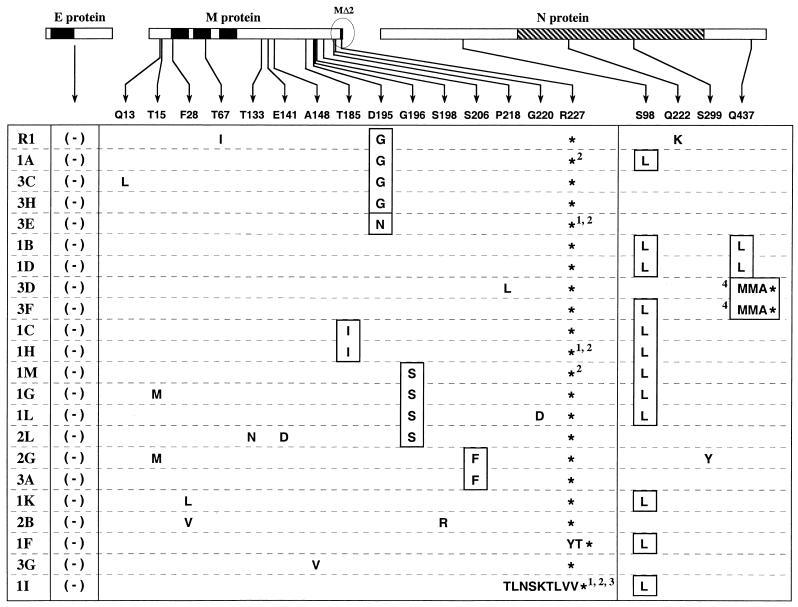
FIG. 3.
Sequence analysis summary for the E, M, and N genes of revertants of MΔ2 mutants Alb231 and Alb233. Revertant R1 and sets 1, 2, and 3 (left column) constitute four independent groups of revertants. R1 and sets 1 and 3 were obtained from Alb231; set 2 was from Alb233. The E, M, and N proteins are represented linearly at the top. The solid rectangles indicate the membrane-bound domains of the E and M proteins (10, 39); the hatched rectangle indicates the RNA-binding domain of the N protein (20, 27). Arrows show the positions within the M and N proteins at which potential reverting mutations were found, and amino acid changes are grouped under the corresponding wild-type residue at each of these positions. Asterisks indicate stop codons. No changes were found in the E gene in any of the revertants. Those amino acid residues or positions that were chosen for further analysis are boxed. Numbers denote the following: 1, mutation of the MΔ2 stop codon from UAG to UAA; 2, mutation of the wild-type codon 228 from ACC to AUC; 3, replacement of nt 650 to 678 of the MΔ2 gene by nt 31 to 56 of the MHV leader; and 4, a deletion of nt 1309 to 1318 of the N gene resulting in a frameshift and truncation of the 15 carboxy-terminal amino acids of the N protein.
In order to identify all potential changes that may be involved in compensation for the MΔ2 lesion, we sequenced RT-PCR products encompassing the entirety of the region most likely to genetically interact with this mutation: the E, M, and N genes. All changes found are listed in the compilation in Fig. 3, from which several important points emerge. First, no change was found in the E gene in any of the 22 independent revertants. This suggests either that there is no possible residue change in the E protein that can offset the loss of the two carboxy-terminal residues of M or that such changes are too rare to be detected because other reverting mutations arise with higher frequency. A second salient result from the revertant analysis was that, with the exception of a single case, 1F (Fig. 3), all revertants arose from second-site mutations and still retained the MΔ2 mutation. For revertant 1F, the stop codon UAG was changed to UAU, encoding tyrosine. Since this change restored the M ORF, the larger-plaque phenotype of revertant 1F may be a result of either the Y227 residue or the simultaneously reinstated T228 residue.
Most of the changes found in the revertants were in the M protein itself, with the majority of these clustered in the carboxy-terminal region of M (Fig. 3). In particular, changes at residues T185, D195, G196, and S206 appeared multiple times in revertants from separate searches. More interestingly, at any given one of these positions, mutational change was almost always to the same amino acid residue. For example, all four apparent reverting mutations at residue 196 (in revertants 1M, 1G, 1L, and 2L) were from G to S (Fig. 3). Similarly, in four of the five revertants containing a mutation at residue 195 (revertants R1, 1A, 3C, and 3H) the change was from D to G, and in the fifth (revertant 3E) it was from D to N (Fig. 3). These multiple occurrences of the same mutations led us to conclude that these were most likely the changes responsible for counteracting the effect of the MΔ2 lesion. However, many of these revertants also contained changes elsewhere, particularly in the ectodomain or transmembrane domains of the M protein. Most of these latter types of mutations each appeared only once among the 22 revertants.
Remarkably, for some of the revertants, changes were also found in the N protein. Among these, the S98L mutation, which was always due to the same C to U change at the RNA level, was the most frequent, appearing in 12 of the revertants (Fig. 3). Two other changes, Q222K and S299Y, each appeared only once (revertants R1 and 2G, respectively) and occurred in revertants that also contained one of the conserved M mutations. The most intriguing alteration in the N protein was at residue Q437, in the carboxy-terminal domain of the protein. It was either changed by a point mutation to leucine (revertants 1B and 1D) or frameshifted to methionine as the result of a 10-nt deletion (revertants 3D and 3F). In the latter case, the mutated N protein became truncated two residues later, making it 15 amino acids shorter than the wild-type N protein. It was somewhat surprising to us that the carboxy terminus of the N protein, which is highly conserved among different strains of MHV (33), was dispensable.
Revertants resulting from apparent nonhomologous recombination between genomic RNA and sgRNA.
In five of the revertants, 1A, 1H, 1M, 3E, and 1I (Fig. 3), sequence analysis also revealed a curious aberration at the 3′ end of the M gene. In the most extreme case, that of revertant 1I, 32 nt of the MΔ2 ORF were replaced by 29 heterologous nt (Fig. 4A). This resulted in the replacement of the 10 carboxy-terminal amino acids of the MΔ2 protein by 9 amino acids having limited homology to the same region. In addition, the stop codon of the MΔ2 ORF was changed from UAG to UAA, and the codon immediately downstream of this (which had been codon 228 in the original M ORF) was changed from ACC to AUC. Strikingly, the source of the heterologous material turned out to be the leader sequence, which had been inserted into the genome in a manner that appeared to result from nonhomologous recombination with subgenomic RNA7 (sgRNA7), the mRNA for the N protein. In revertants 3E and 1H, both the stop codon and codon 228 point mutations were present. In revertants 1A and 1M, only the codon 228 point mutation occurred. For the latter four revertants these events were not relevant to the MΔ2 ORF, but for revertant 1I, we were subsequently able to conclude that the 9-amino-acid substitution was responsible for reversion. The composition of these revertants is consistent with their having been formed by a disruption of discontinuous negative-strand sgRNA7 synthesis (see Discussion).
FIG. 4.
Recombination of part of the MHV leader RNA into the 3′ terminus of the M gene. (A) Nucleotide sequence of the M gene-N gene region of revertant 1I aligned with the corresponding regions of the MΔ2 sequence and the leader-N gene region of sgRNA7. The broken line indicates the locus of the apparent crossover between MΔ2 and sgRNA7. Revertants 1H and 3E, which contain mutations in both the MΔ2 stop codon and codon 228 (underlined), and revertants 1A and 1M, which contain just the codon 228 mutation, also appear to result from more-limited crossovers between MΔ2 and sgRNA7. TRS7 denotes the TRS governing synthesis of sgRNA7 (N mRNA). (B) Potential mechanism of formation of the substitution in revertant 1I via a nonhomologous recombination event between sgRNA7 and genomic RNA. (C) Proposed mechanism of formation of the substitution in revertant 1I through an aberrant shift from synthesis of negative-strand sgRNA7 to synthesis of the full-length antigenome, as detailed in Discussion. In panels B and C, note that the leader region is not drawn to scale. UTR, untranslated region.
Genetic reconstruction of the MΔ2 revertants.
Because almost every revertant contained at least two mutations (Fig. 3), we decided to examine a specific subset of these for the ability to suppress the MΔ2 lesion. For this purpose, we chose to focus on the changes T185I, D195G, D195N, G196S, and S206F in the M protein, because mutations at each of these positions arose multiple times independently. For the N protein, we concentrated on S98L, Q437L, and Q437MMA for the same reason. Each of these selected mutations was introduced individually into the MHV genome in conjunction with the MΔ2 mutation in order to assess its phenotype in the absence of extraneous amino acid changes. To accomplish this, we generated RT-PCR fragments encompassing the specific mutations of interest from the appropriate revertants and these were introduced into the pLK61 plasmid. Recombinant viruses were then generated with the fMHV-based targeted RNA recombination procedure described above.
At least two independent recombinants containing each mutation were examined for the presence of both the MΔ2 mutation and the second-site mutation by restriction digestion and direct sequencing of analytic RT-PCR products.
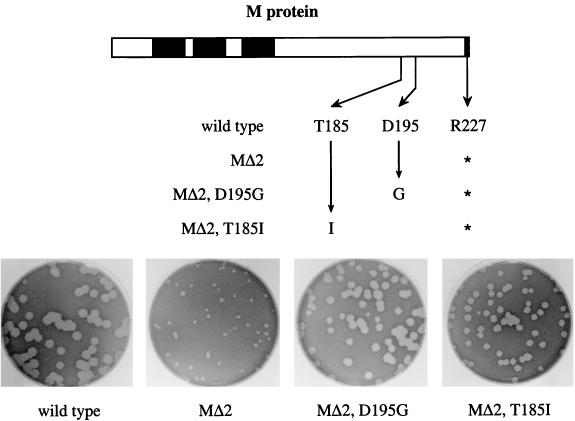
Every one of the selected mutations in the M protein was able to compensate for the MΔ2 defect, as judged by the plaque size and viral titers of the five reconstructed revertant candidates. Figure 5 shows examples of two of these reconstructed revertants. Plaques of both recombinants, harboring either the T185I or D195G mutation in an MΔ2 background, were significantly larger than those of the MΔ2 mutant, and only slightly smaller than those of the wild type. This was also true for the other three reconstructed revertants containing either D195N, G196S, or S206F (data not shown). For the latter candidate, the plaque sizes were somewhat smaller than for the other four compensating mutations but were still clearly much larger than those of the MΔ2 recombinant. All five reconstructed revertants grew more rapidly and had much higher titers than did the original MΔ2 mutant. Based on this genetic evidence, we concluded that each of these five changes in the M protein was individually able to compensate for the MΔ2 defect.
FIG. 5.
Reconstruction of viruses containing potential M gene reverting mutations of the MΔ2 lesion. Derivative plasmids were constructed from pLK61, containing the MΔ2 stop codon at codon 227 in addition to a single candidate compensating mutation elsewhere in the M gene. Donor RNAs transcribed from these vectors were used to construct recombinants from fMHV, and the plaque sizes of the resulting double mutants were assessed at 68 h postinfection.
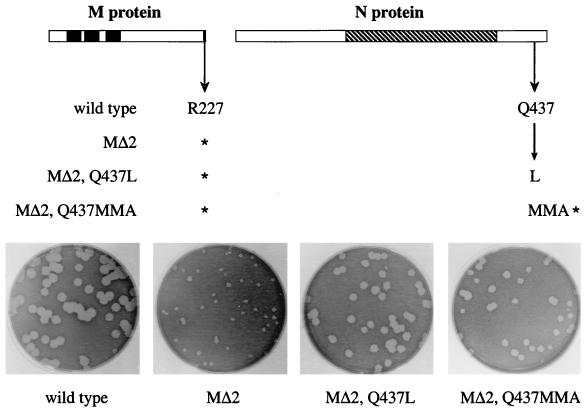
For the selected mutations in the N protein, those located at the carboxy terminus were found to almost completely compensate for the MΔ2 lesion. Reconstructed revertant candidates containing either the point mutation Q437L or the deletion frameshift resulting in Q437MMA, in the presence of the MΔ2 mutation, gave rise to plaques only slightly smaller than the wild-type plaques (Fig. 6). Additionally, these viruses produced much faster-spreading infections in cell monolayers and had much higher titers than did the MΔ2 mutant. These results provide the first genetic evidence for a direct interaction between the N and M proteins of MHV. It is noteworthy that in the Q437MMA revertant, the loss of 15 amino acids at the carboxy terminus of the N protein does not seriously impair the function of this viral component and it indeed overcomes the deficiency due to the deletion of the two final residues of the M protein. Consistent with this, the Q437MMA mutation had only a minor impact on the virus when constructed in a wild-type background (i.e., in the absence of the MΔ2 mutation) (data not shown). At this time, it is not clear what role is being played by the MMA substitution at residues 437 to 439 of the N protein in this altered N-M interaction.
FIG. 6.
Reconstruction of viruses containing potential N gene reverting mutations of the MΔ2 lesion. Derivative plasmids were constructed from pLK61, containing the MΔ2 stop codon at codon 227 in addition to a single candidate compensating mutation in the N gene. Donor RNAs transcribed from these vectors were used to construct recombinants from fMHV, and the plaque sizes of the resulting double mutants were assessed at 68 h postinfection.
The remaining candidate reverting mutation in the N protein, S98L, was the only one not able to compensate for the MΔ2 defect. Reconstructed recombinants containing the S98L mutation in conjunction with the MΔ2 mutation produced plaques indistinguishable from those of the original MΔ2 mutant and grew just as poorly. This result was not unexpected in light of the finding that, in most of the revertants in which the S98L mutation appeared (revertants 1A, 1C, 1H, 1M, 1G, 1L, 1B, 1D, and 3F), the revertant phenotype was accounted for by the compensating mutations D195G, T185I, or G196S in the M protein or Q437L or Q437MMA in the N protein (Fig. 3). We concluded, therefore, that the S98L mutation in N was irrelevant to the revertant phenotype. We currently do not have an explanation for why this extraneous mutation independently arose so frequently in two separate searches.
DISCUSSION
Power of fMHV as a tool in reverse genetics.
In recent years, significant progress in the study of coronavirus assembly has led to an increased understanding of the roles played by individual structural proteins and has begun to define the basic requirements for virion formation. Much of this new information has been provided by investigations of the VLP assembly process due to its sensitivity and accessibility to manipulation (2-4, 6-8, 11, 52). Based on predictions from the VLP system, some of the same mutations have also been incorporated into MHV recombinants by using targeted RNA recombination (6, 18). These constructed mutant viruses enabled us to look at the coronavirus assembly process in the environment of an authentic MHV infection, in which all participants were present, including nucleocapsids and the S protein. The mutual reinforcement of results from the VLP and targeted recombination systems was restricted, however, by the limitations of recombinant selection based on the thermolability of a parental recipient virus (6, 10, 16, 22).
In the present study, a much more powerful mutant selection was enabled by the use of the MHV-FIPV chimeric virus fMHV (18) as the recipient in targeted RNA recombination. The stringent selection afforded by fMHV, based on host cell species specificity, allowed us to straightforwardly obtain a highly defective MHV recombinant, MΔ2. In previous attempts it had been impossible to isolate this mutant, in which the carboxy-terminal two amino acids of the M protein were truncated (6).
Very recently, infectious cDNA clones of three coronaviruses, TGEV, human coronavirus 229E, and avian infectious bronchitis virus, have been developed based on different platforms (1, 5, 48, 53). A similar construct for MHV is likely to become available in the future. The development of infectious cDNA clones provides some obvious advantages to coronavirus reverse genetics, especially the ability to introduce mutations into the replicase gene that occupies some 20 to 22 kb at the 5′ end of the genome. At the present time, however, the efficiency and manageability of the fMHV-based targeted RNA recombination system still make it the method of choice for the manipulation of the structural genes at the 3′ end of the genome of MHV. Besides the MΔ2 mutant, we have obtained other defective recombinant viruses mutated in different structural genes of MHV in the same manner. The versatility of this technique is further supported by its recent application to a strain of MHV other than A59, MHV-JHM (29). More broadly, the host range-based selection provides a general blueprint for the genetic manipulation of any coronavirus.
The MΔ2 mutant is the most defective MHV mutant that we have ever encountered. This recombinant yielded extremely small plaques and had a viral titer that was 3 to 4 orders of magnitude lower than the infectious titers of the wild-type controls. The devastating effect of the MΔ2 lesion was not unexpected, given the lethality of this mutation in VLP formation. It is highly likely that the MΔ2 viral recombinant is viable because the presence of nucleocapsids provides a stabilizing force helping to drive the assembly process.
Genetic evidence for an interaction between the M and N proteins.
Although the severity of the MΔ2 phenotype was a hindrance to working with this virus, it did provide the distinct advantage of a strong selection for the isolation of revertants to study second-site compensating mutations. This allowed, for the first time, a systematic in vivo genetic approach to map protein-protein interactions involving the carboxy terminus of the M protein. Sequence analysis of the entire E, M, and N genes of all 22 independent revertants pointed to a number of mutations potentially capable of compensating for the MΔ2 lesion. All identified changes were in the M protein itself or in the N protein; none was found in the E protein (Fig. 3). This latter result may indicate that the small, highly conserved E protein either cannot tolerate or cannot provide changes that would compensate for the truncation of the M protein. This possibly mirrors results from a previous study in which the revertants of deleterious E protein mutants were isolated and second-site compensating mutations mapping in the M protein were not found (10).
Reconstruction of recombinant viruses containing individual potential reverting mutations provided solid evidence that any one of the five carboxy-terminal mutations, T185I, D195G, D195N, G196S, or S206F, of the M protein was capable of compensating for the MΔ2 lesion (Fig. 5). Four of these mutations independently arose multiple times, and collectively they account for almost all of the revertants that map in the M protein. However, we did not comprehensively examine the remaining mutations in M, and it remains possible that some of the additional mutations make minor contributions to the phenotypes of the above revertants. Also, for some revertants, e.g., 3G, the relevant compensating change can be inferred, although it has not yet been rigorously proven.
Similarly, a reconstruction of candidate reverting mutations in the N protein revealed only one locus, near the carboxy terminus, that was sufficient for reversion of the MΔ2 defect (Fig. 6). Remarkably, mutations in four independent revertants converged at two possible changes near the same residue, Q437, of the N protein. The more dramatic mutation was Q437MMA, which resulted from a 10-nt deletion in the 3′-terminal region of the N gene. This deletion frameshifted and truncated the N ORF, creating a protein 15 amino acids shorter than wild-type N. The same mutation, paired with a full-length M protein, resulted in a virus that formed plaques only slightly smaller than those of the wild type. This indicated that the Q437MMA mutation specifically compensates for the MΔ2 mutation and is not merely a general growth-enhancing mutation for MHV.
The N protein Q437L and Q437MMA reverting mutations argue strongly that the carboxy-terminal region of the N protein directly cooperates with the carboxy-terminal region of the M protein during virion formation (Fig. 7). This finding now provides us with an avenue for probing the involvement and functions of the N protein in the coronavirus assembly process. The interpretation of the MΔ2 revertants that map in the M protein is less straightforward. All of these reverting mutations fall into a relatively narrow window some 21 to 42 amino acids distant from the site of the primary lesion (Fig. 3 and 7). It is possible that they individually enhance the interaction of the MΔ2 protein with the N protein, or alternatively, they might augment interactions between molecules of the MΔ2 protein. It will be necessary to further test all of these conclusions by biochemical methods.
FIG. 7.
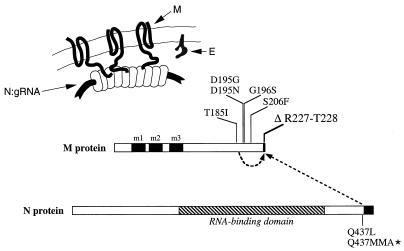
Summary of intramolecular interactions within the M protein and intermolecular interactions between the M and N proteins suggested by this study. The schematic at the top shows the relative topologies and interfaces of the M, N, and E proteins at or within the virion envelope. The linear representation of the M and N proteins depicts the genetic interactions revealed by the MΔ2 revertant analysis. Solid rectangles indicate the three transmembrane domains of the M protein (39); the hatched rectangle indicates the RNA-binding domain of the N protein (20, 27). The carboxy-terminal truncations of the M and N proteins introduced, respectively, by the MΔ2 mutation and the Q437MMA mutation are also indicated by solid rectangles. gRNA, genomic RNA.
In both classes of revertants, the second-site mutation may compensate for the loss of an interaction between the R227 residue of the M protein and the nucleocapsid either by increasing the affinity of an adjacent interaction or by providing a new contact point between M and N to stabilize the virion. In particular, we hypothesize that the R227 residue of the wild-type M protein is electrostatically linked with one or more of the nine aspartate or glutamate residues in the carboxy terminus of the N protein. Overall, the MHV N protein is highly basic, but there is a concentration of acidic residues in its carboxy-terminal domain (33); this general pattern is common to all coronavirus N proteins (19). It is conceivable that, in the absence of the M protein R227 residue, the lost electrostatic interaction can be replaced by a hydrophobic interaction between residues L225 and L226 of the M protein and residue L437 or residues M437 and M438 of the revertant N proteins. To test this, we are currently carrying out a comprehensive mutagenesis of the charged residues of the carboxy terminus of the N protein.
We do not yet know how the intermolecular interaction that we have uncovered by genetic criteria may be related to coronavirus M protein-N protein interactions that three groups have defined previously by biochemical approaches. An early study by Sturman et al. (46) was the first to show a direct and temperature-dependent association between the M protein and nucleocapsids purified from MHV. This elegant work provided the first glimpse of the relationship between the two major constituents of virions. Nearly two decades later, Narayanan et al. (24) demonstrated an M protein-N protein interaction by reciprocal coimmunoprecipitation analyses of proteins from MHV-infected cells. More importantly, they found that M protein only interacted with N protein that was bound to genomic RNA, thereby providing an explanation for the highly selective incorporation of the nucleocapsid into budding virions. Recently, this selective association has been shown to depend absolutely on the presence of the MHV genomic RNA packaging signal, the transfer of which can confer selective packaging to nonviral RNAs (25).
For the porcine coronavirus TGEV, Escors et al. (9) defined an interaction between the carboxy terminus of in vitro-translated M protein and purified viral nucleocapsid. Using a combination of M protein mutagenesis, inhibition by antibodies of defined specificity, and peptide competition, they were able to map this interaction to residues 233 to 257 of the TGEV M protein. This corresponds to a roughly homologous segment of residues 201 to 224 of the MHV M protein, a region that coincides neither with the MΔ2 deletion nor with the major population of reverting mutations in the M protein (Fig. 3 and 7) but rather is situated between these two loci. Because of the proximity of the TGEV and MHV M protein nucleocapsid-interacting regions, we think that they are likely to be contributory parts of the same M protein-N protein interface. However, further work, corroborated by both genetic and biochemical approaches, will be needed to more precisely define the relationship between the two.
Implications for the mechanism of MHV transcription.
In addition to information about structural protein interactions, the sequences of five of the MΔ2 revertants provided an unexpected insight into MHV RNA synthesis. One of the characteristic features of the coronavirus and arterivirus families, which together make up the order Nidovirales, is their strategy of gene expression by synthesis of a nested set of sgRNAs that serve as the mRNAs for all genes downstream of the replicase gene (gene 1). Structurally, each of the positive-strand sgRNA species has a 5′ leader sequence, corresponding to the 5′ end of the genome, fused to a body sequence corresponding to the 3′ end of the genome (50). The points of fusion, which exhibit various levels of homology to the 3′ end of the leader sequence, are designated TRSs. Each positive-strand sgRNA has a negative-strand counterpart, present in much lower abundance (44, 45), and the negative-strand sgRNAs are contained within double-stranded RNA structures thought by some to represent replicative intermediates (42, 43). It is generally agreed that leader-body fusion is brought about by a discontinuous transcription mechanism, but it is currently a matter of considerable controversy whether this occurs during negative- or positive-strand synthesis (see reference 32 and references therein). Although the debate is by no means resolved, work with the arterivirus equine arteritis virus has provided some of the most elegant and convincing evidence for the case that the discontinuous step during nidovirus transcription occurs during negative-strand synthesis (34, 51).
In revertant 1I we observed the replacement of the 3′ end of the MΔ2 ORF by a sequence of nearly equal size that originated from the 3′ end of the MHV leader (Fig. 4A). It is conceivable that this replacement came about by a nonhomologous recombination between sgRNA7 (the N mRNA) and genomic RNA (Fig. 4B). However, such a potential mechanism offers no explanation for why the two templates would have been nearly perfectly aligned. We propose instead that the nonhomologous recombination event that gave rise to this revertant genome took place as the result of a disruption of negative-strand discontinuous transcription (Fig. 4C). The unusual structure of the M gene of revertant 1I is consistent with its having been formed during synthesis of negative-strand sgRNA7 from a full-length, positive-strand genomic template. During this process, the discontinuous transcription complex collapsed, resulting in resumption of synthesis of a full-length antigenome containing the partial leader insertion. This insertion fortuitously altered the MΔ2 ORF to a variant that was able to compensate for the truncation of R227. In four other revertants, 3E, 1A, 1H, and 1M, a less extensive occurrence of the same mechanism appears to have given rise to extraneous mutations in the MΔ2 stop codon and the next codon downstream (equivalent to codon 228 in the wild-type M ORF) (Fig. 3 and 4A). We can find no aberrant version of positive-strand discontinuous RNA synthesis that similarly gives rise to the observed products. Therefore, we think that the sequences of all of these five revertants support a model of negative-strand discontinuous transcription. It is not clear how often, and at how many TRSs, such an event occurs during wild-type MHV RNA synthesis. Generally, it would be expected to disadvantageously alter the upstream gene, owing to the close spacing of MHV ORFs. In the case of revertant 1I, there was a strong selection favoring recovery of the longest leader insertion. However, for the other four revertants, the acquired mutations were extraneous with respect to the reverting mutation. Thus, transcriptional collapse may be more frequent than previously appreciated. In this regard, we note that the leader RNA remnants found in the genomes of wild-type human coronavirus OC43 (23) and a mutant of MHV strain S (47) may have been generated by variations of the same mechanism.
Acknowledgments
We thank Matthew Shudt, Heather Berry, and Tim Moran of the Molecular Genetics Core Facility of the Wadsworth Center for the synthesis of oligonucleotides and for DNA sequencing.
This work was supported in part by Public Health Service grant AI 39544 from the National Institutes of Health.
REFERENCES
- 1.Almazan, F., J. M. Gonzalez, Z. Penzes, A. Izeta, E. Calvo, J. Plana-Duran, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos, E. C. W., W. Luytjes, H. van der Meulen, H. K. Koerten, and W. J. M. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos, E. C. W., W. Luytjes, and W. J. M. Spaan. 1997. The function of the spike protein of mouse hepatitis virus strain A59 can be studied on virus-like particles: cleavage is not required for infectivity. J. Virol. 71:9427-9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Haan, C. A. M., L. Kuo, P. S. Masters, H. Vennema, and P. J. M. Rottier. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72:6838-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Haan, C. A. M., M. Smeets, F. Vernooij, H. Vennema, and P. J. M. Rottier. 1999. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J. Virol. 73:7441-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Haan, C. A. M., H. Vennema, and P. J. M. Rottier. 2000. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 74:4967-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godeke, G. J., C. A. de Haan, J. W. Rossen, H. Vennema, and P. J. M. Rottier. 2000. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 74:1566-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes, K. V., E. W. Dollar, and L. S. Sturman. 1981. Tunicamycin resistant glycosylation of a coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology 115:334-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsue, B., T. Hartshorne, and P. S. Masters. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 74:6911-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsue, B., and P. S. Masters. 1997. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 71:7567-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. M. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koetzner, C. A., M. M. Parker, C. S. Ricard, L. S. Sturman, and P. S. Masters. 1992. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 66:1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krijnse-Locker, J., M. Ericsson, P. J. M. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo, L., G.-J. Godeke, M. J. B. Raamsman, P. S. Masters, and P. J. M. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laude, H., and P. S. Masters. 1995. The coronavirus nucleocapsid protein, p. 141-163. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 20.Masters, P. S. 1992. Localization of an RNA-binding domain in the nucleocapsid protein of the coronavirus mouse hepatitis virus. Arch. Virol. 125:141-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masters, P. S. 1999. Reverse genetics of the largest RNA viruses. Adv. Virus Res. 53:245-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masters, P. S., C. A. Koetzner, C. A. Kerr, and Y. Heo. 1994. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J. Virol. 68:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mounir, S., and P. J. Talbot. 1993. Human coronavirus OC43 RNA4 lacks two open reading frames located downstream of the S gene of bovine coronavirus. Virology 192:355-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narayanan, K., and S. Makino. 2001. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 75:9059-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navas, S., S.-H. Seo, M. M. Chua, J. Das Sarma, E. Lavi, S. T. Hingley, and S. R. Weiss. 2001. Murine coronavirus spike protein determines the ability of the virus to replicate in the liver and cause hepatitis. J. Virol. 75:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson, G. W., and S. A. Stohlman. 1993. Localization of the RNA-binding domain of mouse hepatitis virus nucleocapsid protein. J. Gen. Virol. 74:1975-1979. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen, V.-P., and B. Hogue. 1997. Protein interactions during coronavirus assembly. J. Virol. 71:9278-9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ontiveros, E., L. Kuo, P. S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230-238. [DOI] [PubMed] [Google Scholar]
- 30.Opstelten, D.-J. E., P. de Groote, M. C. Horzinek, H. Vennema, and P. J. M. Rottier. 1993. Disulfide bonds in folding and transport of mouse hepatitis coronavirus glycoproteins. J. Virol. 67:7394-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Opstelten, D.-J. E., M. J. B. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. M. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker, M. M., and P. S. Masters. 1990. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology 179:463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasternak, A. O., E. van den Born, W. J. M. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips, J. J., M. M. Chua, E. Lavi, and S. R. Weiss. 1999. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 73:7752-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricard, C. S., C. A. Koetzner, L. S. Sturman, and P. S. Masters. 1995. A conditional-lethal murine coronavirus mutant that fails to incorporate the spike glycoprotein into assembled virions. Virus Res. 39:261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Risco, C., I. M. Antón, L. Enjuanes, and J. L. Carrascosa. 1996. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 70:4773-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Risco, C., M. Muntión, L. Enjuanes, and J. L. Carrascosa. 1998. Two types of virus-related particles are found during transmissible gastroenteritis virus morphogenesis. J. Virol. 72:4022-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rottier, P. J. M. 1995. The coronavirus membrane glycoprotein, p. 115-139. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 40.Rottier, P. J. M., M. C. Horzinek, and B. A. M. van der Zeijst. 1981. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effects of tunicamycin. J. Virol. 40:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription, p. 215-219. In L. Enjuanes, S. G. Siddell, and W. Spaan (ed.), Advances in experimental medicine and biology, vol. 440. Coronaviruses and arteriviruses. Plenum Press, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 44.Sethna, P. B., M. A. Hofmann, and D. A. Brian. 1991. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J. Virol. 65:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sethna, P. B., S.-L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sturman, L. S., K. V. Holmes, and J. Behnke. 1980. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 33:449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taguchi, F., T. Ikeda, S. Makino, and H. Yoshikura. 1994. A murine coronavirus MHV-S isolate from persistently infected cells has a leader and two consensus sequences between the M and N genes. Virology 198:355-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 49.Tooze, S. A., J. Tooze, and G. Warren. 1988. Site of addition of N-acetyl-galactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J. Cell Biol. 106:1475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Most, R. G., and W. J. M. Spaan. 1995. Coronavirus replication, transcription, and RNA recombination, p. 11-31. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 51.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vennema, H., G.-J. Godeke, J. W. A. Rossen, W. F. Voorhout, M. C. Horzinek, D.-J. E. Opstelten, and P. J. M. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]