Abstract
The lipid-containing bacteriophage PRD1 infects a variety of gram-negative cells by injecting its linear double-stranded DNA genome into the host cell cytoplasm, while the protein capsid is left outside. The virus membrane and several structural proteins are involved in phage DNA entry. In this work we identified a new infectivity protein of PRD1. Disruption of gene XXXII resulted in a mutant phenotype defective in phage reproduction. The absence of the protein P32 did not compromise the particle assembly but led to a defect in phage DNA injection. In P32-deficient particles the phage membrane is unable to undergo a structural transformation from a spherical to a tubular form. Since P32− particles are able to increase the permeability of the host cell envelope to a degree comparable to that found with wild-type particles, we suggest that the tail-tube formation is needed to eject the DNA from the phage particle rather than to reach the host cell interior.
PRD1, the type species of the Tectiviridae family, infects gram-negative bacterial hosts harboring an IncP, IncN, or IncW conjugative plasmid. The plasmid functions are involved only in the initial stage of infection, as the plasmid encodes the cell envelope bridging phage receptor complex (21, 24, 30). This broad-host-range double-stranded DNA phage is composed of an outer protein capsid and an internal membrane (1, 5, 40). The proteinaceous membrane vesicle encloses the 14,925-bp-long linear doubled-stranded-DNA genome (8, 15). The viral genome encodes some 25 structural proteins (8).
The icosahedral protein coat of the PRD1 particle is composed of the major capsid protein P3 organized on a pseudo T=25 lattice, similarly to that of the human adenovirus coat (10, 15). A small cementing protein, P30, stabilizes the capsid structure (45). The fivefold positions are built up by an adenovirus-like spike-penton complex composed of proteins P2, P5, and P31 (7, 16, 23, 46). Each of the vertices is a metastable structure (46). PRD1 infection starts with the specific recognition of the receptor by the adsorption protein P2. This interaction probably results in a conformational change and detachment of P2 from the particle (23). This is considered to be a signal for the rest of the vertex proteins and the peripentonal capsid protein trimers to be released. The phage membrane can undergo a structural transformation from a spherical vesicle to a tubular form, and the vertex opening enables the membranous tube to protrude from the particle. The tail-tube structure is seen only in particles that have ejected their DNA, and consequently the tube formation has been considered to be involved in the PRD1 DNA injection (1, 29). Interestingly, two murolytic phage-specific enzymes have been shown to be membrane associated (43, 44). The enzymatic activity of these proteins, P7 and P15, is thought to assist in the phage genome entry.
The understanding of the considerably elaborated PRD1 system has relied on the isolation and analysis of phage mutants. In spite of extensive attempts to saturate the genome with nonsense mutations (32, 46), we still lack mutations in several genes. Conventional mutagenesis systems based on in vitro manipulation of the isolated DNA molecule are not easily applicable to the PRD1 genome. The linear genome contains covalently attached priming proteins at both 5′ termini (3). These hydrophobic proteins complicate the handling of the genome in vitro. Recently a targeted mutagenesis system based on combined in vitro manipulation and in vivo recombination for PRD1 genome was developed (7).
Very little is known about the mechanisms by which DNA crosses the bacterial cell envelope in transformation, conjugation, or virus infection. These phenomena can be elucidated by studying alterations in the cellular energetics, which take place during these processes. The envelope of a gram-negative bacterium forms a protective barrier to the cell, which only allows the passage of very small molecules by simple diffusion. Hydrophilic ions and ATP can cross the outer membrane (OM) through porins; however, lipophilic ions and ionophoric antibiotics cannot (for reviews, see references 37 and 38). Thus, the distribution of lipophilic ions (such as the membrane voltage indicator tetraphenylphosphonium [TPP+]) between the cell and the medium can be used to determine the energy state of the cytoplasmic membrane (CM) as well as the OM permeability. The CM is rather impermeable to inorganic ions such as potassium (K+); thus, the efflux of intracellular K+ is an indication of an increased CM permeability. Ion fluxes across the cell envelope can be measured by selective electrodes, which monitor changes of the indicator ion concentration in the medium.
Using the mutagenesis-recombination system, we generated a lacZα insertion mutation in the PRD1 gene XXXII. The gene XXXII mutation does not interfere with the particle formation or DNA packaging. P32− particles are able to bind to host cells and induce alterations in the cell envelope permeability comparable to those induced by wild-type (wt) viruses. However, the phage genome delivery to the host cell cytoplasm is compromised.
MATERIALS AND METHODS
Bacteria, plasmids, and phages.
The bacterial strains used in this study are listed in Table 1, and plasmids and phages are listed in Table 2. Cells were grown in Luria-Bertani (LB) medium (47). When appropriate, ampicillin (100 μg/ml), chloramphenicol (10 μg/ml), kanamycin (30 μg/ml), or tetracycline (20 μg/ml) was added. Phage PRD1 (40) was propagated on Salmonella enterica serovar Typhimurium DS88, and its mutant derivatives sus1 (32) and sus539 (46) were propagated on the suppressor strains PSA(pLM2) and DB7156(pLM2), respectively. The recombinant PRD1 [lacZα]-9 was propagated on a complementation strain, HMS174(pLM2, pPR18). For the production of wt and mutant phage particles, DS88 cells were infected at a multiplicity of infection (MOI) of about 10. After lysis, the particles were concentrated and purified by rate zonal sucrose gradient centrifugation, as previously described (6). The virus zones from these sucrose gradients were further purified by ion-exchange chromatography on Sartorius D100 anion exchange cartridges (Sartorius), essentially as described by Walin et al. (50). Purified particles were collected by differential centrifugation, and the pellets were dissolved in 20 mM sodium phosphate buffer, pH 7.4.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype and/or phenotype | Reference |
|---|---|---|
| E. coli K12 | ||
| DH5α | F−supE44 Δ(lacZYA-argF) U169 (φ80 lacZΔM15) hsdR17 recA1 endA gyrA96 thi1 relA1 | 47 |
| HB101 | recA1 endA hsdS thi1 ara14 leuB6 proA2 lacY1 galK xyl mtl1 supE44 | 11 |
| HMS174 | F−recA1 hsdR | 17 |
| JE2571 | leu thr thi lacY thy pil | 13 |
| S. enterica sv. Typhi- murium LT2 | ||
| DS88 | SL5676 ΔH2 H1-i::Tn10 (Tcs) non rev (pLM2) | 6 |
| DB7156 | DB700 leuA141(Am) hisC527(Am) supF30 | 51 |
| PSA | supE | 32 |
TABLE 2.
Plasmids and phages used in this study
| Plasmid or phage | Descriptiona | Relevant property | Selective marker(s)b | Reference |
|---|---|---|---|---|
| pALH21 | pSU19Δ[EcoRI-HindIII]Ω[PRD1 9937-11910] | PRD1 gene XVIII-XI+ | Cmr | 8 |
| pJB15 | Encodes PRD1 receptor | Tcr | 25 | |
| pLM2 | Encodes PRD1 receptor | Kmr | 34 | |
| pMG69 | pALH21Ω[ApaI at position PRD1 10442]c | Cmr | This study | |
| pMG73 | pMG69Δ[ApaI]Ω[lacZα at position PRD1 10442]d | Insertion mutant of gene XXXII | Cmr | This study |
| pPR18 | pSU18Δ[BamHI-PstI]Ω[PRD1 10440-10604]e | PRD1 gene XXXII+ | Cmr | This study |
| pSU18 | Cloning vector | Cmr | 9 | |
| pSU19 | Cloning vector | Cmr | 9 | |
| RP4 | Encodes PRD1 receptor | Apr, Kmr, Tcr | 18 | |
| PRD1 wt | Wild type | 40 | ||
| PRD1 sus1 | Amber mutation in gene IX | Defect in DNA packaging | 32 | |
| PRD1 sus539 | Amber mutation in gene II | Defect in adsorption | 46 | |
| PRD1 [lacZα]-9 | lacZα fragment in gene XXXII | Defect in DNA entry | α-complementation | This study |
The cloning vector is indicated, followed by the restriction enzymes used and the origin of the insert in brackets. PRD1 sequence coordinates were taken from GenBank/EMBL (accession number M69077).
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance.
Mutagenesis primers for generating the ApaI site were OP302 (GGCAGCTAATGGGCCCGTTTGGCAAAACCC) and OP303 (GGGTTTTGCCAAACGGGCCCATTAGCTGCC). The nucleotides differing from PRD1 sequence are shown in bold.
PCR primers used for amplification of the lacZα fragment were FOR (AAAAAGGGCCCCCAATACGCAAACCGCCTCTCCCC) and REV (AAAAAGGGCCCGCGCCATTCGCCATTCAGGCTGCG).
PCR primers used for amplification of gene XXXII were OP154 (TATATGGATCCGAGGAGAAATTAACATATGGGCGAGTTTGGCAAAAC) and OP155 (TATATCTGCAGTTAAATGATTGGCGCAACGG).
DNA techniques.
Standard molecular cloning techniques were performed as described by Sambrook and Russell (47), and the Escherichia coli K12 strain HB101 (11) was used as the host for plasmids. The coding sequence for gene XXXII was amplified by PCR using viral DNA as a template and inserted into plasmid pSU18, resulting in plasmid pPR18. For the cloning strategy, see Table 2. For mutagenesis of gene XXXII, the plasmid pALH21 was used as a template. An ApaI restriction site was generated at the beginning of the gene XXXII using the QuickChange site-directed mutagenesis kit (Stratagene), resulting in the plasmid pMG69. Chromosomal DNA from E. coli K12 JE2571 (13), carrying the wild-type β-galactosidase gene, was isolated and used as a template to amplify the lacZα fragment. The fragment was inserted into the ApaI site of plasmid pMG69, and a clone was selected where the fragment was oriented in the opposite direction in respect to the PRD1 genes. The resulting plasmid was named pMG73.
Construction of the recombinant phage.
The lacZα fragment was inserted into the virus genome by recombination between the wt genome and plasmid pMG73, essentially as described by Bamford and Bamford (7). To obtain the recombinant virus, strain JE2571(RP4, pMG73) was infected with wt PRD1. The resulting lysate was plated on a complementation strain, DH5α(pJB15, pALH21), in the presence of 30 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside per ml and 0.2 mM isopropyl-β-d-thiogalactopyranoside to select for blue plaques. One plaque was obtained and further purified on the same strain and named PRD1 [lacZα]-9.
Electron microscopy.
For thin-section electron microscopy, S. enterica serovar Typhimurium DS88 cells were grown to a density of 109 CFU/ml and infected with PRD1 [lacZα]-9 [grown on the complementation strain HMS174(pLM2, pPR18)] at a MOI of about 10. Samples were taken at 20, 40, and 55 min postinfection and fixed with 3% (vol/vol) glutaraldehyde for 20 min at 22°C. The cells were collected by centrifugation, washed twice with 20 mM potassium phosphate buffer, pH 7.4, and prepared for transmission electron microscopy as described elsewhere (4). The micrographs were taken with a JEOL 1200 EX electron microscope operating at 60 kV (electron microscopy unit, Institute of Biotechnology, University of Helsinki).
In another experiment, DS88 cells were grown to the optimal adsorption phase (approximately 2 × 109 CFU/ml [28]), and purified mutant phage particles were adsorbed to these cells at 22°C. Samples were taken after 5, 15, and 50 min postinfection, washed twice with LB medium, and prepared for thin-section electron microscopy as described above.
Assay for phage membrane aggregation and tail-tube formation.
Purified mutant phage particles were disrupted by 2.5 M guanidine hydrochloride (GuHCl) treatment for 45 min at 37°C (1). Samples were diluted 1:1 in 20 mM sodium phosphate buffer, pH 7.4, and analyzed by rate zonal centrifugation in 10 to 40% (wt/vol) sucrose gradient (Sorvall TH 641 rotor; 30,000 rpm, 45 min, 20°C). The protein concentration in gradient fractions was determined as described below.
For the analysis of tail-tube formation, purified wt and mutant particles were treated either with 2 M GuHCl for 30 min at 37°C or 20 mM Tris-HCl, pH 7.4, for 48 h at 22°C. Samples were stained with either 1% phosphotungstic acid, pH 6.5, or 1% ammonium molybdate, pH 6.5, as described by Bamford and Mindich (4). Electron micrographs were taken as described above.
DNA replication assay.
The labeling of replicating PRD1 DNA with [3H]thymidine was accomplished according to the method of Mosig and Colowick (36), except that nalidixic acid (40 μg/ml) was added to inhibit host replication (20).
Measurements of ion fluxes and determination of membrane voltage.
The ion flux experiments were performed as described by Daugelavičius et al. (19). In short, S. enterica serovar Typhimurium DS88 cells were grown to the optimal adsorption phase, collected by centrifugation, and dissolved in 50 mM Tris-HCl, pH 7.5, to a 1:100 proportion of the original volume. When appropriate, the OM permeabilization was achieved by treatment with 100 mM Tris-HCl [pH 8.0]-10 mM EDTA [pH 8.0] at 22°C for 10 min (27). The concentrated cell suspension was added to an appropriate buffer in a 5-ml thermostated reaction vessel to the final concentration of 3 × 109 CFU/ml. The concentration of TPP+ and K+ ions in the medium was monitored by selective electrodes. The K+ content of the cells and the nonspecific binding of TPP+ were measured by adding gramicidin D (GD) and polymyxin B (PMB) to the cell suspension in the reaction vessel. The membrane voltage values were calculated as described by Daugelavičius et al. (19).
Analytical methods.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis was performed either as described by Olkkonen and Bamford (39) or as described by Schägger and von Jagow (49). The proteins were either stained with Coomassie brilliant blue or transferred from the gel onto a polyvinylidene difluoride membrane (Millipore) and visualized with the ECL detection system (Pierce) using horseradish peroxidase-conjugated swine anti-rabbit immunoglobulin Gs (IgGs) (DAKO) or peroxidase-conjugated horse anti-mouse IgGs (Vector) as secondary antibodies. Polyclonal antiserum recognizing protein P31 (46) and a monoclonal antibody recognizing protein P7 and P14 (7N41) (25) were used as primary antibodies for protein composition analysis of mutant phage particles. The protein concentration of the purified viruses was determined with Coomassie brilliant blue G250 using bovine serum albumin as a standard (12). The zymogram assay was performed as described by Rydman and Bamford (43).
RESULTS
A new gene, XXXII, is essential for viral growth.
Early genetic and biochemical analysis of PRD1 nonsense mutants revealed that at least 25 gene products are encoded by the phage genome (31, 32, 35). The sequencing of the genome led to the firm assignment of 33 genes and open reading frames (ORF) (8, 41, 42, 48). A second attempt to isolate phage nonsense mutations in the uncharacterized ORFs, and analysis of these mutants, has identified three additional genes (43, 45, 46). There are still some 10 ORFs with unknown function, for which no mutants are available. Using the PRD1 site-directed mutagenesis system (7), a new gene essential for PRD1 propagation was identified.
A small ORF, ORFn, was disrupted by the insertion of the lacZα fragment. The fragment was first cloned to the beginning of ORFn in a plasmid construct, pALH21, containing the PRD1 genes from XVIII to XI (to enable recombination). The disrupted ORFn was then introduced into the virus genome by in vivo recombination between the linear virus DNA and the circular plasmid (see Materials and Methods). In a mixture of wt and recombinant phages, those containing the lacZα fragment were selected by plating viruses on an E. coli strain, DH5α(pJB15, pALH21), producing the ω fragment of the LacZ protein. In this strain only recombinant phages expressing the α fragment of the LacZ protein give blue plaques. After three single plaque isolations, the presence of the lacZα fragment in ORFn was verified by restriction enzyme analysis of the viral DNA isolated from the recombinant phage PRD1 [lacZα]-9. The mutant phenotype could be rescued when ORFn was provided in trans in a plasmid replicon (pPR18), the determined titers of purified concentrated mutant viruses being 3 × 105 and 4 × 1011 PFU/ml on the noncomplementing DH5α(pJB15) and complementing DH5α(pJB15, pPR18) strains, respectively. This finding confirmed that the defect is in ORFn and that ORFn is essential for PRD1 propagation. Accordingly, this ORF was designated gene XXXII, encoding protein P32 (for the PRD1 gene and protein nomenclature see reference 8).
Protein P32-deficient mutant forms stable DNA-containing particles.
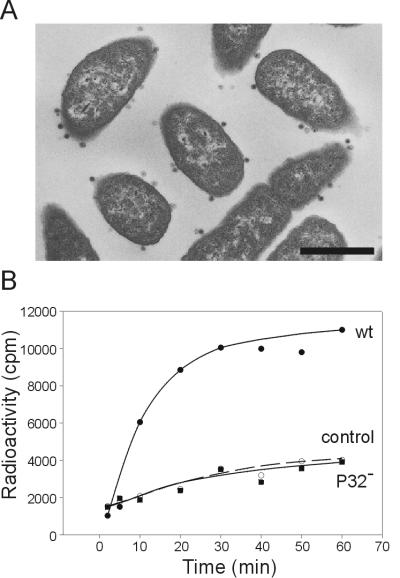
To analyze the phenotypic defect resulting from the disruption of the gene XXXII, DS88 cells were infected with PRD1 [lacZα]-9 grown on a complementation strain, HMS174(pLM2, pPR18). Samples were taken for thin-section electron microscopy at different time points postinfection. The mutant infection followed that of the wt one, yielding a large number of DNA-containing particles (Fig. 1A). The integrity of the P32-deficient particles was comparable to that of the wt phages. In both cases, 80% of the particles, after purification by rate zonal sucrose gradient centrifugation, were filled DNA-containing viruses. The amount of these particles was estimated from the intensities of the slower- and faster-sedimenting light-scattering zones in sucrose gradients, respectively (data not shown).
FIG. 1.
(A) An electron micrograph of thin-sectioned DS88 cells infected with PRD1 [lacZα]-9. The image, obtained 55 min postinfection, shows a large number of peripheral filled phage particles (bar, 500 nm). (B) Protein composition of PRD1 wt and P32-deficient mutant particles is shown in a Coomassie blue-stained Tricine SDS-polyacrylamide gel. In the mutant phage preparation the only protein missing is the 5.4-kDa P32 (marked by an arrow). Molecular weight markers and some PRD1 proteins are indicated on the left and right, respectively.
The protein composition of the purified mutant particles was determined by SDS-polyacrylamide gel electrophoresis followed by Coomassie blue staining (Fig. 1B) or immunological detection of proteins using specific antisera (data not shown). The presence of a lytic muramidase, protein P15, which is not detectable by Coomassie blue staining, was verified by a zymogram analysis (data not shown). In accordance with the complementation analysis of the P32-deficient mutant, no other phage structural protein that was detectable using these methods was missing.
Gene XXXII mutation interferes with phage membrane tube formation.
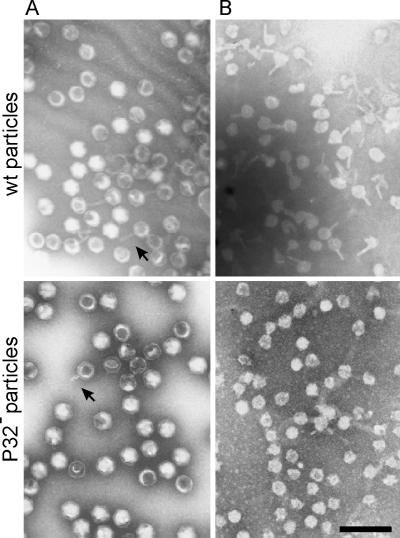
One specific feature of purified PRD1 virions is the presence of a tail-tube structure, which is an extended, protein-rich, membrane tube of approximately 14 nm in diameter and 60 to 65 nm in length (1). Particles with these tubes occur with a frequency of about 10% in the wt preparation (46). The formation of these structures can be induced by treating the virus with Tris buffer at room temperature (Fig. 2A). In P32-deficient mutant particles, tail-tube structures were observed at a much lower frequency (Fig. 2A). In addition, these structures were shorter, approximately one-third of the wt tube length. Also, disrupting the phage particles with 2.5 M guanidine hydrochloride results in vesicles, most of which are empty with protruding tubular structures (1) (Fig. 2B). When P32− particles were treated with guanidine hydrochloride, practically all membrane vesicles obtained were spherical, unlike the case in a wt preparation, where in almost every vesicle a tubular structure was observed (Fig. 2B).
FIG. 2.
Electron micrographs of negatively stained wt and P32-deficient particles showing extended or short tail-tube structures, respectively. The arrow points to the tail appendage. (A) Phage particles treated with 20 mM Tris-HCl, pH 7.4, for 48 h at 22°C. (B) Disruption of the wt or P32− particles with 2 M GuHCl treatment reveals membrane vesicles with or without tail-tubes, respectively (bar, 200 nm).
Another feature described for the PRD1 membrane is that isolated membrane vesicles readily aggregate. This phenotype is caused by the phage structural protein P11 (1). Purified P32− particles behaved like wt ones in respect to this phenotype. Disruption of the particles with guanidine hydrochloride and subsequent separation of the membrane vesicles by sucrose gradient centrifugation revealed that in the absence of P32, phage membrane proteins are found in the pellet fraction (data not shown). In these conditions aggregating membranes sediment to the bottom of the gradient and nonaggregating membranes form a slower sedimenting peak (1).
Protein P32 is needed for phage DNA entry but not for receptor recognition.
The first step in PRD1 infection is the recognition and attachment to the receptor on the host cell surface. This binding is dependent on protein P2 residing in the capsid vertices (23, 33). The ability of P32− particles to bind to cells was studied by thin-section electron microscopy of infected DS88 cells. The electron micrograph shows that mutant phage particles are attached directly to the cell surface, similar to the wt particles (Fig. 3A). However, no progeny particles were observed inside the cell, even after incubation time (70 min postinfection) where the wt infection led to extensive phage propagation (data not shown). Since the receptor recognition property of the P32− particles is unaltered, the DNA entry step most probably is compromised. This was confirmed by a DNA synthesis assay where [3H]thymidine was incorporated into replicating DNA, only after the genome had entered the cell. It has been previously shown that newly synthesized viral DNA appears approximately 20 min after infection (20). Figure 3B shows that in the wt infection, saturation levels of phage DNA were detected at 20 to 30 min after infection, while P32− mutant infection behaved like the uninfected control cells.
FIG. 3.
(A) An electron micrograph of thin-sectioned DS88 cells infected with P32-deficient mutant particles. The image, obtained 50 min postinfection, shows several phage particles bound on the cell surface (bar, 500 nm). (B) DNA synthesis, determined by [3H]thymidine incorporation into replicating DNA, in DS88 cells treated with nalidixic acid (open circles) and cells infected with PRD1 wt (filled circles) or P32− (squares) particles. The DNA gyrase inhibitor does not affect PRD1 DNA replication, unlike the case with host replication.
Deficiency in the infectivity protein P32 does not prevent PRD1-induced changes in cell envelope permeability.
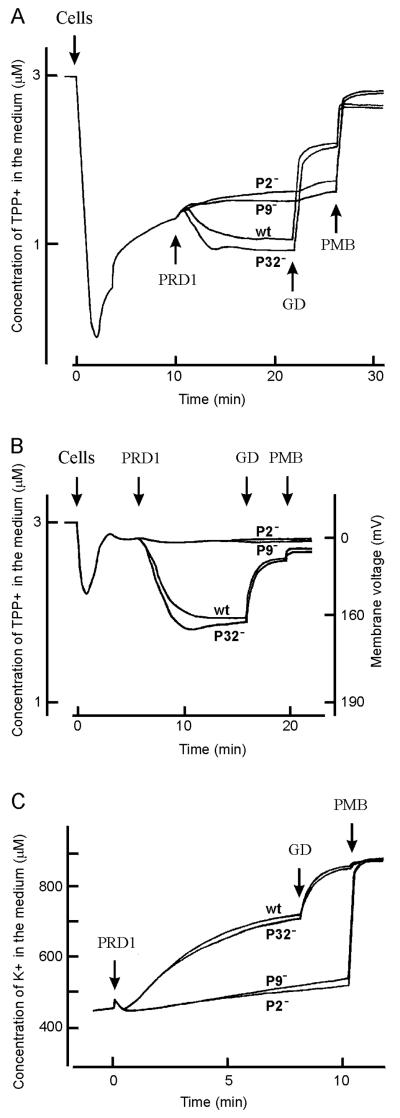
After binding to the receptor, the phage nucleic acid has to traverse all layers of the cell envelope to reach the cytoplasm. Measurements of ion fluxes during phage infection have been used to study the DNA entry process. It has been previously shown that DNA containing PRD1 particles, but not adsorbing empty particles, is able to increase the permeability of both the cytoplasmic and outer membranes (19). The effects of P32− particles on the envelope permeability of DS88 cells were studied by monitoring TPP+ accumulation in the cell cytosol and by determining the efflux of intracellular potassium ions.
The OM of gram-negative cells is rather impermeable to TPP+ and the channel-forming antibiotic GD; however, these molecules freely penetrate the cytoplasmic membrane. Tris-EDTA-treated cells are highly permeable to TPP+ but not to the rather bulky GD molecules (see Fig. 4A). Thus, initially cells rapidly accumulate TPP+, which is followed by an equilibration of the TPP+ concentration between the cell interior and the external milieu. Infection of Tris-EDTA-treated cells with either wt or P32− phages induced additional TPP+ uptake by DS88 cells, indicating an increase in the membrane voltage (see also reference 19). In control infections with empty particles (P9−) or particles unable to bind to cells (P2−), no additional phage-induced TPP+ accumulation was observed (Fig. 4A). The addition of GD induced the depolarization of the plasma membrane and, because of that, the release of accumulated TPP+ only from cells infected with wt or P32− particles. The polycationic antibiotic PMB, which does not require the increased permeability of the OM for its depolarizing action, induced efflux of accumulated TPP+ in all infections tested.
FIG. 4.
Electrochemical measurements of PRD1-infected cells. Accumulation of TPP+ by EDTA-treated (A) or intact (B) DS88 cells, and efflux of K+ (from intact cells) followed by phage infection (C). The experiments were performed at 37°C in 50 mM Tris-HCl, pH 7.5. Cells, phages, GD, and PMB were added to the final concentration of 3 × 109 CFU/ml, 24 μg of protein/ml (that is, MOI of ∼100), 5 μg/ml, and 100 μg/ml, respectively. Ion-selective electrodes were used to monitor the concentration of the indicator ions in the external medium. For interpretation of the experiment, see the text.
When the OM was intact, the cells bound a small amount of TPP+ before infection. The addition of wt or P32− particles induced a considerable uptake of TPP+ by DS88 cells (Fig. 4B). In the control P2− or P9− infections no such effects were observed. The P32− mutant-induced TPP+ uptake indicates that these particles are able to increase the permeability of the OM to lipophilic compounds to a level comparable to that caused by wt viruses, whereas the control phage particles did not.
In addition to the effects on accumulation of TPP+, the P32− mutant induced an efflux of K+ ions from the cytosol (Fig. 4C). The rate of efflux and the amount of K+ remaining in the infected cells equaled those of the wt infection, indicating that P32− particles also increase the permeability of the host CM.
Sequence alignment reveals a close similarity of P32 to PRD1 protein P34.
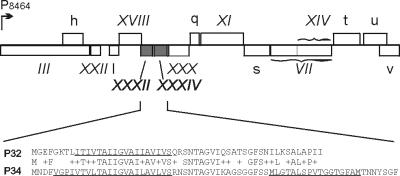
The sequence analysis of the protein P32 for potential secondary structure elements revealed one possible membrane-spanning α-helix near the N terminus of this 54-amino-acid-long polypeptide, suggesting membrane association. Homology search revealed a close similarity to another small gene product of PRD1, encoded by ORFo. These polypeptides share a high local similarity (84%), the overall similarity being 66% (Fig. 5). However, ORFo is incapable of complementing the defect in the gene XXXII (data not shown). So far, our attempts to isolate mutations in ORFo have been unsuccessful. N-terminal amino acid sequencing of a peptide fragment (8) has confirmed that ORFo encodes a structural PRD1 protein, and accordingly ORFo is designated gene XXXIV, encoding the protein P34.
FIG. 5.
Organization of the late operon OL3 of phage PRD1 (nucleotides 8464 to 13886) (8, 22). Roman numerals and lowercase letters identify genes and open reading frames, respectively. The inset shows the alignment of proteins P32 and P34, sharing an overall similarity of 66%. The regions predicted (26) to form a membrane-spanning α-helix are underlined.
DISCUSSION
Recombinant DNA techniques enable targeted mutagenesis to introduce base substitutions, deletions, and insertions. This powerful technology, however, is not easily applicable to all genetic systems, such as linear genomes with terminal proteins. Using the site-directed mutagenesis method designed to manipulate the PRD1 genome (7), we were able to create a lacZα insertion into gene XXXII. Even though this methodology was successfully used to introduce nonsense mutations in gene V (7), our attempts to exchange the lacZα fragment in gene XXXII to an amber mutation failed. One reason for this could be that the recombination frequency differs in different genomic regions. Gene XXXII is some 4 kb downstream of gene V. Also, the phenotype of the resulting amber mutant might affect the outcome. The enrichment of the mutant viruses during recombination is influenced by the selective advantage of the suppressed phenotype over the mutant one.
Our current understanding of PRD1 DNA delivery relies on genetic, biochemical, and structural data (7, 16, 23, 32, 43, 44, 46). This multistep process seems to be dependent on several phage-encoded proteins. Here we described the identification and analysis of a new component, P32, involved in phage DNA delivery. Host recognition and receptor binding properties of the P32-deficient particles were unaltered, but no progeny viruses were obtained in the mutant infection. A DNA replication assay confirmed that these particles are unable to inject the genome into the host cell cytoplasm. Why is the genome release compromised in the absence of P32?
Observations of PRD1 particles forming tail-like appendages (but with the membrane still inside the particle) have been reported several times (14, 15, 29). The tail-tube formation is dependent on at least protein P18, and it has been suggested to be involved in PRD1 DNA delivery (1, 29). Also, mutations in the vertex proteins P5 and P31 are associated with the deficiency in the formation of this tubular structure (7, 46). The phage membrane transformation from a spherical to a tubular form seems to play an important role in the PRD1 DNA entry process, since also in the new P32− mutant the tube formation is compromised. P32− particles, however, were able to induce efflux of K+ ions equal to that induced by wt infection, suggesting that P32 deficiency does not prevent PRD1-induced effects at the CM. These new findings suggest that the tail-tube per se is not needed to reach the CM, but rather in the translocation of the DNA from the virus head to the cell interior. Preliminary data (to be published) from our laboratory suggest that the formation of the tail-tube extension is dependent on several additional phage structural proteins and that in all cases the tail-tube formation is compromised.
The amino acid sequence of P32 shows characteristic elements for integral membrane proteins. According to the orientation of the predicted α-helix, the topology of protein P32 would be such that the N-terminal part spans the membrane, leaving the bulk of the protein (C terminus) in the cytoplasmic side of the CM. If our model of a clathrin-like engulfment of the phage membrane from the host cytoplasmic membrane during particle assembly (2) is valid, the C terminus of P32 would face outward on the virus membrane surface.
Sequence analysis revealed another interesting feature, a close similarity of P32 to another small phage membrane protein, P34 (Fig. 5). This is a second example of a high level of sequence similarity between two PRD1 gene products, suggesting a single gene origin for the two elements. Also, the multimeric vertex proteins P5 and P31 (6, 16, 46) are encoded from genome regions sharing considerable sequence homology. It is intriguing to propose that gene multiplication is used to conserve interactions when building increasingly complex structures. Obviously we are in the process of obtaining mutations in gene XXXIV to verify its predicted role in DNA delivery.
Acknowledgments
We are grateful to Anna Rantala for skillful technical assistance. Pia Rydman is warmly thanked for providing plasmid pPR18 and for performing the zymogram assay.
Research grants 162993, 164298, and 172621 (Finnish Center of Excellence Programme [2000-2005]) from the Academy of Finland supported this study.
REFERENCES
- 1.Bamford, D., and L. Mindich. 1982. Structure of the lipid-containing bacteriophage PRD1: disruption of wild-type and nonsense mutant phage particles with guanidine hydrochloride. J. Virol. 44:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamford, D. H., J. Caldentey, and J. K. H. Bamford. 1995. Bacteriophage PRD1: a broad host range dsDNA tectivirus with an internal membrane. Adv. Virus Res. 45:281-319. [DOI] [PubMed] [Google Scholar]
- 3.Bamford, D. H., and L. Mindich. 1984. Characterization of the DNA-protein complex at the termini of the bacteriophage PRD1 genome. J. Virol. 50:309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamford, D. H., and L. Mindich. 1982. Electron microscopy of cells infected with nonsense mutants of bacteriophage Φ6. Virology 710:222-228. [DOI] [PubMed] [Google Scholar]
- 5.Bamford, D. H., L. Rouhiainen, K. Takkinen, and H. Söderlund. 1981. Comparison of the lipid-containing bacteriophages PRD1, PR3, PR4, PR5 and L17. J. Gen. Virol. 57:365-373. [DOI] [PubMed] [Google Scholar]
- 6.Bamford, J. K. H., and D. H. Bamford. 1990. Capsomer proteins of bacteriophage PRD1, a bacterial virus with a membrane. Virology 177:445-451. [DOI] [PubMed] [Google Scholar]
- 7.Bamford, J. K. H., and D. H. Bamford. 2000. A new mutant class, made by targeted mutagenesis, of phage PRD1 reveals that protein P5 connects the receptor binding protein to vertex. J. Virol. 74:7781-7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bamford, J. K. H., A.-L. Hänninen, T. M. Pakula, P. M. Ojala, N. Kalkkinen, M. Frilander, and D. H. Bamford. 1991. Genome organization of membrane-containing bacteriophage PRD1. Virology 183:658-676. [DOI] [PubMed] [Google Scholar]
- 9.Bartolomé, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 10.Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 11.Bouyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 12.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 13.Bradley, D. E. 1980. Morphological and serological relationship of conjugative pili. Plasmid 4:155-169. [DOI] [PubMed] [Google Scholar]
- 14.Bradley, D. E., and E. L. Rutherford. 1975. Basic characterization of lipid-containing bacteriophage specific for plasmids of the P, N, and W incompatibility groups. Can. J. Microbiol. 21:152-163. [DOI] [PubMed] [Google Scholar]
- 15.Butcher, S. J., D. H. Bamford, and S. D. Fuller. 1995. DNA packaging orders the membrane of bacteriophage PRD1. EMBO J. 14:6078-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caldentey, J., R. Tuma, and D. H. Bamford. 2000. Assembly of bacteriophage PRD1 spike complex: the role of the multidomain protein P5. Biochemistry 39:10566-10573. [DOI] [PubMed] [Google Scholar]
- 17.Campbell, J. L., C. C. Richardson, and F. W. Studier. 1978. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc. Natl. Acad. Sci. USA 75:2276-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datta, N., R. W. Hedges, E. J. Shaw, R. B. Sykes, and M. H. Richmond. 1971. Properties of an R factor from Pseudomonas aeruginosa. J. Bacteriol. 108:1244-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daugelavičius, R., J. K. H. Bamford, and D. H. Bamford. 1997. Changes in host cell energetics in response to bacteriophage PRD1 DNA entry. J. Bacteriol. 179:5203-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis, T. N., and J. E. J. Cronan. 1983. Nonsense mutants of the lipid-containing bacteriophage PR4. Virology 126:600-613. [DOI] [PubMed] [Google Scholar]
- 21.Davis, T. N., E. D. Muller, and J. E. Cronan, Jr. 1982. The virion of the lipid-containing bacteriophage PR4. Virology 120:287-306. [DOI] [PubMed] [Google Scholar]
- 22.Grahn, A. M., J. K. H. Bamford, M. C. O'Neill, and D. H. Bamford. 1994. Functional organization of the bacteriophage PRD1 genome. J. Bacteriol. 176:3062-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grahn, A. M., J. Caldentey, J. K. H. Bamford, and D. H. Bamford. 1999. Stable packaging of phage PRD1 DNA requires adsorption protein P2, which binds to the IncP plasmid-encoded conjugative transfer complex. J. Bacteriol. 181:6689-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grahn, A. M., J. Haase, D. H. Bamford, and E. Lanka. 2000. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transfer systems. J. Bacteriol. 182:1564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hänninen, A.-L., D. H. Bamford, and J. K. H. Bamford. 1997. Probing the phage PRD1 specific proteins during infection by monoclonal and polyclonal antibodies. Virology 227:198-206. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann, K., and W. Stoffel. 1993. TMbase—A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 27.Kalasauskaitė, E. V., D. L. Kadišaite̊, R. J. Daugelavičius, L. L. Grinius, and A. A. Jasaitis. 1983. Studies on energy supply for genetic processes. Requirement for membrane potential in Escherichia coli infection by T4. Eur. J. Biochem. 130:123-130. [PubMed] [Google Scholar]
- 28.Kotilainen, M. M., A. M. Grahn, J. K. H. Bamford, and D. H. Bamford. 1993. Binding of an Escherichia coli double-stranded DNA virus PRD1 to a receptor coded by an IncP-type plasmid. J. Bacteriol. 175:3089-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundström, K. H., D. H. Bamford, E. T. Palva, and K. Lounatmaa. 1979. Lipid-containing bacteriophage PR4: structure and life cycle. J. Gen. Virol. 43:583-592. [DOI] [PubMed] [Google Scholar]
- 30.Lyra, C., H. Savilahti, and D. H. Bamford. 1991. High-frequency transfer of linear DNA containing 5′-covalently linked terminal proteins: electroporation of bacteriophage PRD1 genome into Escherichia coli. Mol. Gen. Genet. 228:65-69. [DOI] [PubMed] [Google Scholar]
- 31.McGraw, T., H.-L. Yang, and L. Mindich. 1983. Establishment of a physical and genetic map for bacteriophage PRD1. Mol. Gen. Genet. 190:237-244. [DOI] [PubMed] [Google Scholar]
- 32.Mindich, L., D. Bamford, C. Goldthwaite, M. Laverty, and G. Mackenzie. 1982. Isolation of nonsense mutants of lipid-containing bacteriophage PRD1. J. Virol. 44:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mindich, L., D. Bamford, T. McGraw, and G. Mackenzie. 1982. Assembly of bacteriophage PRD1: particle formation with wild-type and mutant viruses. J. Virol. 44:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mindich, L., J. Cohen, and M. Weisburd. 1976. Isolation of nonsense suppressor mutants in Pseudomonas. J. Bacteriol. 126:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mindich, L., and T. McGraw. 1983. Molecular cloning of bacteriophage PRD1 genomic fragments. Mol. Gen. Genet. 190:233-236. [DOI] [PubMed] [Google Scholar]
- 36.Mosig, G., and N. Colowick. 1995. DNA replication of bacteriophage T4 in vivo. Methods Enzymol. 262:587-604. [DOI] [PubMed] [Google Scholar]
- 37.Nikaido, H. 1993. Transport across the bacterial outer membrane. J. Bioenerg. Biomembr. 25:581-589. [DOI] [PubMed] [Google Scholar]
- 38.Nikaido, H., and M. Vaara. 1985. Molecular basis of outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olkkonen, V. M., and D. H. Bamford. 1989. Quantitation of the adsorption and penetration stages of bacteriophage Φ6 infection. Virology 171:229-238. [DOI] [PubMed] [Google Scholar]
- 40.Olsen, R. H., J.-S. Siak, and R. H. Gray. 1974. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J. Virol. 14:689-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pakula, T. M., H. Savilahti, and D. H. Bamford. 1989. Comparison of the amino acid sequence of the lytic enzyme from broad-host-range bacteriophage PRD1 with sequences of other cell wall lytic enzymes. J. Eur. Biochem. 180:149-152. [DOI] [PubMed] [Google Scholar]
- 42.Pakula, T. M., H. Savilahti, and D. H. Bamford. 1989. The organization of the right-end early region of bacteriophage PRD1 genome. Gene 85:53-58. [DOI] [PubMed] [Google Scholar]
- 43.Rydman, P. S., and D. H. Bamford. 2000. Bacteriophage PRD1 DNA entry uses a viral membrane-associated transglycosylase activity. Mol. Microbiol. 37:356-363. [DOI] [PubMed] [Google Scholar]
- 44.Rydman, P. S., and D. H. Bamford. 2002. The lytic enzyme of bacteriophage PRD1 is associated with the viral membrane. J. Bacteriol. 184:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rydman, P. S., J. K. H. Bamford, and D. H. Bamford. 2001. A minor capsid protein P30 is essential for bacteriophage PRD1 assembly. J. Mol. Biol. 313:785-795. [DOI] [PubMed] [Google Scholar]
- 46.Rydman, P. S., J. Caldentey, S. J. Butcher, S. D. Fuller, T. Rutten, and D. H. Bamford. 1999. Bacteriophage PRD1 contains a labile receptor-binding structure at each vertex. J. Mol. Biol. 291:575-587. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Savilahti, H., and D. H. Bamford. 1987. The complete nucleotide sequence of the left very early region of Escherichia coli bacteriophage PRD1 coding for the terminal protein and the DNA polymerase. Gene 57:121-130. [DOI] [PubMed] [Google Scholar]
- 49.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 50.Walin, L., R. Tuma, G. J. Thomas, and D. H. Bamford. 1994. Purification of viruses and macromolecular assemblies for structural investigations using a novel ion-exchange method. Virology 201:1-7. [DOI] [PubMed] [Google Scholar]
- 51.Winston, F., D. Botstein, and J. H. Miller. 1979. Characterization of amber and ochre suppressors in Salmonella typhimurium. J. Bacteriol. 137:433-439. [DOI] [PMC free article] [PubMed] [Google Scholar]