Abstract
The formation of G-quadruplex and i-motif structures in the 5′ end of the retinoblastoma (Rb) gene was examined using chemical modifications, circular dichroism (CD) and fluorescence spectroscopy. It was found that substitutions of 8-methylguanine at positions that show syn conformations in antiparallel G-quadruplexes stabilize the structure in the G-rich strand. The complementary C-rich 18mer forms an i-motif structure, as suggested by CD spectroscopy. Based on the C to T mutation experiments, C bases participated in the C–C+ base pair of the i-motif structure were determined. Experiments of 2-aminopurine (2-AP) substitution reveal that an increase of fluorescence in the G-quadruplex relative to duplex is attributed to unstacked 2-AP within the loop of G-quadruplex. The fluorescence experiments suggest that formation of the G-quadruplex and i-motif can compete with duplex formation. Furthermore, a polymerase arrest assay indicated that formation the G-quadruplex structure in the Rb gene acts as a barrier in DNA synthesis.
INTRODUCTION
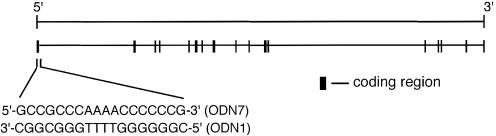
The retinoblastoma (Rb) gene encodes a nuclear phosphoprotein that acts as a tumor suppressor by affecting the cell cycle (1,2). The Rb/E2F pathway, regulating the E2F transcription factor (1,2), is critical in regulating the initiation of DNA replication, and the control of this pathway is disrupted in virtually all human cancers (2). Examination of the Rb gene sequences reveals that regions at the 5′ termini are extremely rich in G and C residues (Figure 1) (3,4). G-rich sequences have been proposed to have important biological roles through the formation of G-quadruplex structures (5–10). For example, telomeric DNA is fundamental in protecting the cell from recombination and degradation through the formation of G-quadruplex structures (11). It has been demonstrated that a G-quadruplex structure formed in the c-myc promoter region functions as a transcriptional repressor element (12). We recently demonstrated, using a photochemical method, that the sequence d(CGGGGGGTTTTGGGCGGC) (ODN 1)at the 5′ terminus of the Rb gene can form an antiparallel G-quadruplex (13).
Figure 1.
Structure of the human retinoblastoma gene. The inset shows the DNA base sequences used in this study.
It is well known that C-rich DNA strands adopt an i-motif structure, whose building block is hemiprotonated C-C+-base pairs at slightly acidic pH (14). Sequences with the potential to form i-motifs are frequent in the human genome, as exemplified by its presence in the regulatory region of myeloid specific genes and human c-ki-ras (15,16). However, the C-rich strand from the Rb gene has received relatively less attention. Here, we propose the formation of G-quadruplex and i-motif structures at the 5′ end of Rb gene, examined by chemical modifications, circular dichroism (CD) and fluorescence spectroscopy experiments. One possible biologically relevant role for the G-quartet is control of DNA synthesis. This hypothesis was tested using a rapid and simple polymerase arrest assay carried out in physiological salt conditions.
MATERIALS AND METHODS
Synthesis of oligonucleotides
Oligonucleotides containing 8-methylguanine (m8G) were prepared by the phosphoramidite method on controlled pore glass supports (1 µmol) using a Beckman OLIGO1000 and Applied Biosystems 3400 DNA synthesizer (17,18). After automated synthesis, the oligomers were detached from the support, deprotected and purified by high-performance liquid chromatography. The oligomers were identified by electrospray ionization mass spectrometry on a Perkin Elmer SCIEX API 165 mass spectrometer (negative mode). The purity and concentrations of these oligomers were determined by complete digestion of the oligomers with alkaline phosphatase and P1 nuclease to 2′-deoxymononucleosides.
CD measurements and analysis of CD melting profiles
CD spectra were measured using an AVIV MODEL 62 DS/202 CD spectrophotometer. CD spectra were recorded using a 1 cm path-length cell. In CD melting studies, diluted samples were equilibrated at room temperature for several hours to obtain equilibrium spectra. Solutions for CD spectra were prepared as 1 ml samples at 0.1 mM (base concentration) in the presence of 100 mM KCl at 25°C. The sample pH was adjusted for each experiment with HCl and KOH, and using a glass microelectrode.
Fluorescence measurements
The fluorescence spectra were measured using a JASCO FP-6300 spectrofluorometer. The samples were excited at 317 nm and the fluorescence emission spectra were collected in the wavelength range 335–500 nm. Solutions for fluorescence spectra were prepared as 1 ml samples at 0.1 mM (base concentration) in the presence of 100 mM KCl at 25°C. In the G-quartet to duplex experiments, 1.5 equiv. of the cytosine-rich complementary strand and TMPyP was added.
Polymerase stop assay
Labeled DNA primer d(TAATCAGCACTACATATG) (5′-labeled with Texas red) (24 nM), template Temp[Rb] d[CCTAATCTATCTTAC(CGGGGGGTTTTGGGCGGC)TTACGCACTCGAATGCATATGTAGTGCTGATTA] (12 nM) and mutated template Temp [Rb mutation] containing the mutant sequence d[CCTAATCTATCTTAC(CAGGGAGTTTTAGGCAGC)TTACGCACTCGAATGCATATGTAGTGCTGATTA] (12 nM) were annealed in buffer [10 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA and 1.5 µg/µl BSA] with 0.1 mM dNTP by heating to 95°C and slow cooling to room temperature. Taq DNA polymerase (5 U) was added and the mixture was incubated for 20 min at different temperatures. The polymerase extension was stopped by adding two times stop buffer [10 mM EDTA, 10 mM NaOH, 0.1% xylene cyanole and 0.1% bromophenol blue in formamide solution], and the solution was loaded onto a 12% denaturing polyacrylamide gel.
RESULTS AND DISCUSSION
Structure characterization of G-rich sequence
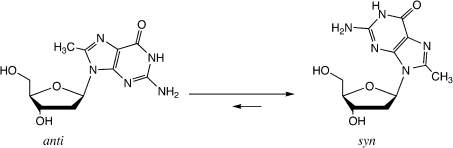
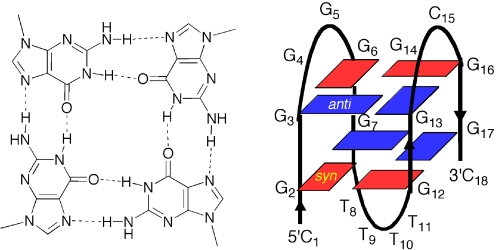
The chemical and biochemical properties of G-quadruplexes could be better understood by controlling G-quartet-folding topologies in a specified manner (19–22). The G-rich strand may adopt a G-quartet structure depending on the orientation of the strands and the syn/anti conformations of guanines (23–29). In anti-parallel G-quadruplex structures, G residues adapt syn/anti conformations around individual G-tetrads (23–29). For example, analogous determination of the arrangement of anti/syn conformations of the G residues by using 8-bromoguanine substituted G-quadruplexes has been reported (19). Based on NMR analysis, we have demonstrated that m8G adopts a syn conformation (Figure 2) (18). It is reasonable to believe that substitutions of m8G at positions that show syn conformations in antiparallel G-quadruplexes might stabilize the structure, whereas substitution at positions required to be anti might destabilize the structures. Based on the efficient deoxyribonolactone formation by the uracil-5-yl radical in the diagonal loop, we recently proposed that the G-rich sequence ODN 1 at the 5′ end of the Rb gene forms an antiparallel G-quadruplex (13). To understand the syn/anti conformations of guanine in the antiparallel G-quadruplex of the Rb gene, the thermal stability of the G-quadruplex formed by ODN 1 with specifically positioned m8G residues were examined using CD melting experiments. Substitution at the first dG of each GG step results in an increase in stability. The melting temperature (Tm) values of ODN 2, ODN 4, ODN 5 and ODN 6 were increased by 3–5°C, whereas substitution at the second dG of GG step results in a decrease in stability (ODN 3) (Table 1 and Supplementary Figure 1S). The m8G substitution at syn positions in the antiparallel G-quadruplex contributes to reducing the energetic penalty for folding, whereas the substitution at anti positions results in an energetic penalty for flipping the base from syn to anti. The CD spectra of each oligonucleotide exhibit a positive band at 295 nm and a negative band around 265 nm in the presence of 100 mM K+ ions, which is characteristic of an antiparallel G-quadruplex structure (Supplementary Figure 2S). Based on the predicted effect of m8G, these results suggest that the guanine syn/anti patterns are along individual strands and around individual G-tetrads in the antiparallel structure (Figure 3).
Figure 2.
Structure of m8G. Incorporation of a methyl group markedly stabilizes the syn conformation (17–18).
Table 1.
Thermal stability of m8G-containing G-quadruplexes
| ODN | Tm (°C) | |
|---|---|---|
| d(CGGGGGGTTTTGGGCGGC) | (ODN 1) | 51 |
| d(C(m8G)GGGGGTTTTGGGCGGC) | (ODN 2) | 55 |
| d(CG(m8G)GGGGTTTTGGGCGGC) | (ODN 3) | 45 |
| d(CGGGG(m8G)GTTTTGGGCGGC) | (ODN 4) | 56 |
| d(CGGGGGGTTTT(m8G)GGCGGC) | (ODN 5) | 54 |
| d(CGGGGGGTTTTGGGC(m8G)GC) | (ODN 6) | 54 |
aThe Tms of the oligonucleotides were determined in 2 mM sodium cacodylate buffer (pH 7.0) and 150 mM KCl at 2.5 µM with each strand. SEs were <0.5°C in duplicate determinations.
bThe position of the m8G in ODN 1, ODN 2, ODN 4, ODN 5 and ODN 6 for the syn position; and ODN 3 for the anti position.
Figure 3.
Four guanines assemble in a planar ring to form a G-quartet (left panel). Schematic representation of the G-quadruplex structure adopted by d(CGGGGGGTTTTGGGCGGC) (ODN 1) (right panel), guanine bases in the syn and anti conformations are represented, respectively.
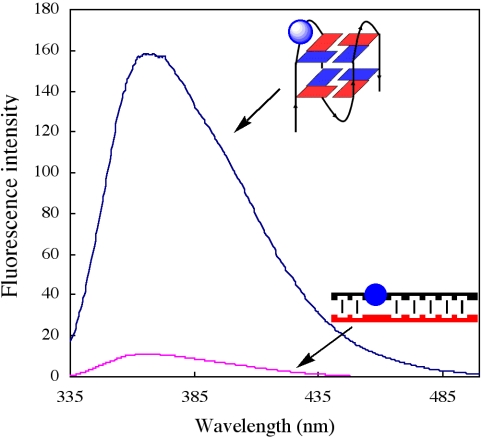
To further understand the molecular basis of the structure, quadruplex-to-duplex conversion was monitored through changes in fluorescence intensity of 2-aminopurine (2-AP), a highly structure-sensitive fluorescent probe, which was incorporated into the loop of the G-quadruplex. An important observation was that the fluorescence intensity of the quadruplex was 10 times higher than that of the duplex (Figure 4). Disrupting the G-quadruplex by hybridization of a cytosine-rich complementary strand to the quadruplex resulted in a marked decrease in fluorescence intensity. The reduction is attributed to different stacking interactions of the nucleobases in the quadruplex and duplex structures (30,31). In the quadruplex, the nucleobase segment in the loop is highly distorted and π-stacking is disrupted. The short decay time observed for 2-AP in a duplex is attributed to the fully stacked state, whereas longer lifetimes come from unstacked 2-AP in the G-quadruplex (30).
Figure 4.
Fluorescence changes accompanying quadruplex to duplex transition from addition of the cytosine-rich complementary strand. Quadruplex, d(CGGG(2-AP)GGTTTTGGGCGGC); 2-AP was incorporated into the loop position. Complementary strand, d(GCCGC CCAAAACCTCCCG) (red).
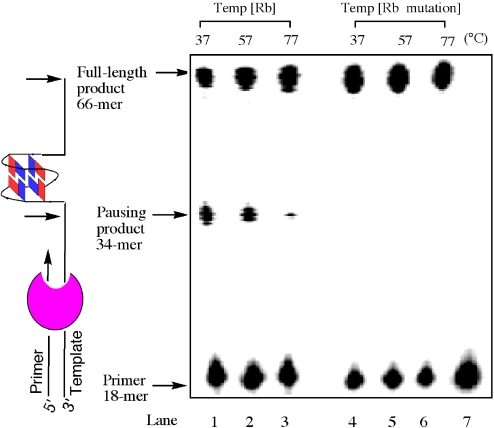
In theory, the structures we have described could form in vivo any time that the duplex region containing the sequence becomes unpaired. Replication or transcription would provide such an opportunity, allowing the G-quadruplex to form upon exposure of the single-stranded G-rich region. The Rb sequence, which contains a G-rich segment, might block DNA synthesis by forming such a G-quadruplex that is sufficient to impede progress of the polymerase. To test this possibility, we studied the ability of the G-quadruplex to arrest DNA synthesis. A 66mer DNA template containing the ODN 1 sequence was incubated in the presence of Taq DNA polymerase with a primer with a complementary sequence to the 3′ end of the 66mer template. The principle of the assay is shown to the left of the gel in Figure 5. If complex extension of the primer occurs, a full-length 66mer product is formed. However, factors that promote and stabilize G-quadruplex formation will lead to a specific pause site on the template, resulting in the formation of a truncated 34mer product. The polymerase primer extension reactions were carried out at three different temperatures. A significant pause is observed for primer extension under physiological salt conditions at 37°C, and there is less 34mer product with increasing temperature. The polymerase pausing is less apparent at 77°C, which is higher than the melting point of the G-quadruplex structure formed in this G-rich region of the template DNA. To further confirm that the pause results from the formation of a G-quadruplex structure in the template DNA, a mutated template DNA that contains the G to A mutations G2, G6, G12 and G16 within individual G-tetrads was used in a parallel experiment. The primer extension results with the mutated template indicate that no significant pause occurs at all three temperatures tested. The results suggest that an intramolecular G-quadruplex can be formed in the G-rich region of the Rb gene, leading to pronounced DNA synthesis arrest in the original G-rich template. Non-B DNA conformations were also associated with genomic rearrangement breakpoints (32,33), and such a G-quadruplex element in Rb gene could contribute to double-strand breaks, resulting in destabilization of the gene.
Figure 5.
Temperature-dependent block of DNA synthesis by the G-quadruplex structure formed in the DNA template containing the Rb sequence. Temp [Rb] contains sequence 5′-d(CGGGGGGTTTTGGGCGGC)-3′ (ODN 1) (lanes 1–3); Temp [Rb mutation] contains the mutant sequence 5′-d(CAGGGAGTTTTAGGCAGC)-3′ (lanes 4–6); lane 7 is primer alone. The primer was labeled at the 5′ end with Texas Red. Arrows indicate the positions of the full-length DNA synthesis product, the G-quadruplex pause site and the free primer.
Structure characterization of C-rich sequence
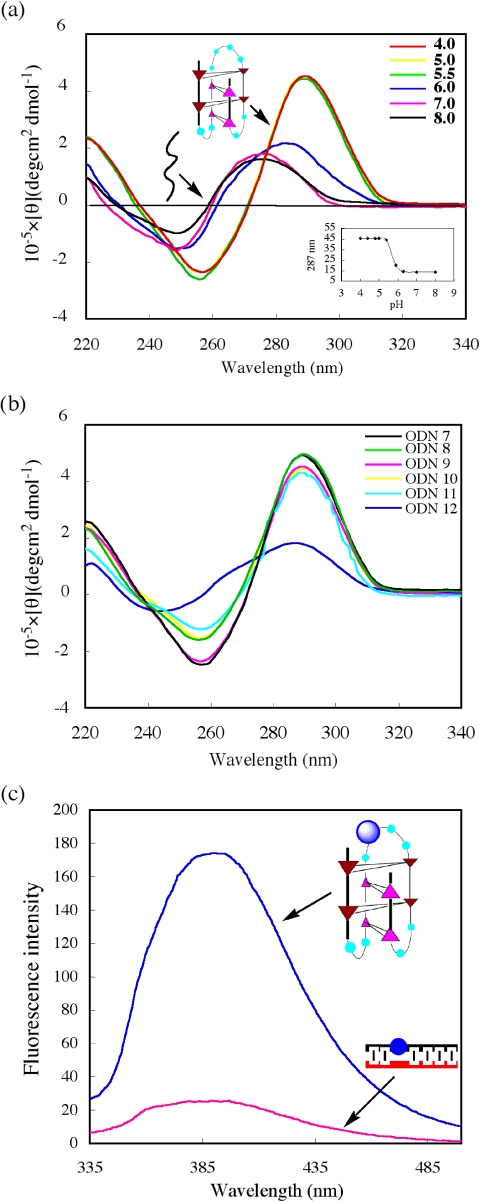
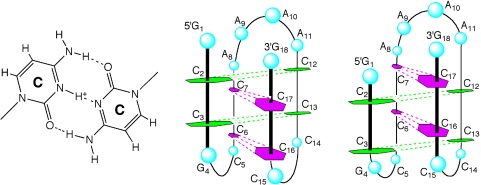
C-rich strands are known to fold into i-motif structures with hemiprotonated C-C+-base pairs in acidic conditions (34,35). To investigate whether the i-motif structure was formed by the C-rich complementary strand of the 5′ end of the Rb gene, CD spectra of ODN 7 were measured at pHs ranging from 4.0 to 8.0 (Figure 6a). For the C-rich strand, measurement at pH < 5.5 revealed a maximum at 285–290 nm, which is characteristic of i-motif structures (36,37). At pH > 7, the maximum shifted towards 275 nm, consistent with the unstructured signal-strand DNA. A pH profile plotted by measuring changes in CD Cotton effect of the positive band at 287 nm shows conformational integrity in the range pH 4.0–5.5, further increases in pH resulted in unfolding to a single strand. A transition mid-point at pH 5.9 ± 0.2 was determined from this plot (Figure 6a, inset). To identify further the nature of the i-motif structure, five different C-to-T mutations in ODN 7 were designed and prepared (Figure 6b). If these Cs are not involved in the C–C+ base pairs, then mutation to T should not significantly affect i-motif stabilization. CD experiments showed that C5, C14 and C15 are not required for formation of a stable i-motif structure. However, C7 is clearly critical for i-motif stability, showing a decrease in CD intensity at 287 nm. Based on these results, two types of i-motif structures can be formed by the four C–C+ intercalated units, as shown in Figure 7. Our observations suggest the possibility of the formation of intramolecular i-motif in the Rb sequence. Although acidic pH favors the formation of i-motif structures and there is no conclusive proof that they exist in vivo, the potential sequences to form i-motifs are frequent in the human genome. To date, i-motif structures have been proposed to form in both centromeric and telomeric regions of human chromosomes, as well as in the promoter region of the human c-myc gene (38–41).
Figure 6.
(A) CD spectra of the C-rich complementary strand d(G1C2C3G4C5C6C7A8A9A10A11C12C13C14C15C16C17G18) (ODN 7) at various pHs. All spectra were recorded at pHs ranging from 4.0 to 8.0 at 25°C, Inset, profile of ellipticity at 287 nm versus pH. (B) CD spectra of ODN7 and mutated oilgonucleotides. ODN8, C5→T, d(GCCGTCC AAAACCCCCCG); ODN9, C14→T, d(GCCGCCCAAAACCTCCCG); ODN 10, C15→T, d(GCCGCCCAAAACCCTCCG); ODN 11, dual mutants C14, C15→T, d(GCCGCCCAAAACCTTCCG); ODN 12, C7→T, d(GCCGCCTAAAACCCCCCG). (C) Fluorescence changes accompanying i-motif to duplex conversion by addition of the G-rich complementary strand. i-motif, d(GCCGCCCA(2-AP)AACCCCCCG); 2-AP was incorporated into the loop of i-motif.
Figure 7.
C–C+ base pairs (left panel) and schematic structures of the i-motif topology from the C-rich complementary strand d(G1C2C3G4C5C6C7A8A9A10A11C12C13C14C15C16C17G18) (middle and right panels).
To further understand the molecular basis of the i-motif structure, the fluorescence intensity of 2-AP at the loop of i-motif was investigated. A marked increase in fluorescence intensity in i-motif compared with duplex was observed (Figure 6c). The different stacking interactions of the nucleobases in the i-motif and duplex structures may be attributed to the differences of fluorescence. It is similar to the observation on G-quadruplex and duplex.
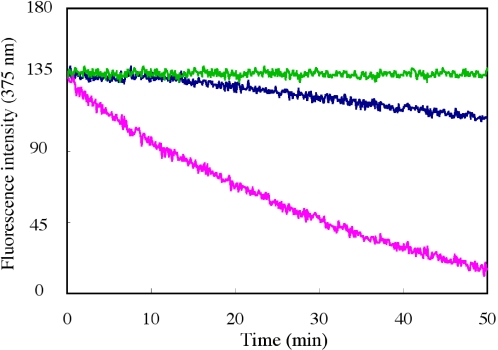
To elucidate the effect of i-motif and G-quadruplex structure on quadruplex-to-duplex conversion, the ratio of duplex formation was examined by fluorescence experiments. We found that quadruplex-to-duplex conversion occurs more slowly at lower pH conditions in comparison with neutral pH (Figure 8). It is believed that low pH stabilizes the folded i-motif structure of the C-rich strand, thus providing a competitor for duplex formation relative to the linear C-rich strand at neutral pH, that results in slow hybridization occurring and the formation of duplex (34,42). Furthermore, when the G-quartet-binding ligand TMPyP was added to the 2-AP-contaning 18mer solution, almost no fluorescence change was observed. This suggests that TMPyP stabilizes the G-quadruplex structure and prevents the duplex formation reaction.
Figure 8.
Effect of i-motif and G-quadruplex structure on quadruplex-to-duplex conversion. 2-AP-containg sequence d(CGGG(2AP)GGT TTTGGGCGGC) was used to monitor fluorescence change. Fluorescence traces at 375 nm of the complementary strand-induced quadruplex-to-duplex conversion are shown for addition of the normal C-rich single-strand (pH 7.0) (cyan) and an i-motif C-rich strand (pH 5.5) (blue) in the presence of TMPyP (green).
In conclusion, we found that methylation of dG at positions that exist in syn conformations in antiparallel G-quartets can stabilize the quadruplex structure in the G-rich strand of Rb gene. The quadruplex-to-duplex conformational change was monitored using the structure-sensitive fluorescence probe 2-AP. Importantly, we found that formation of such a G-quadruplex in the Rb gene impedes the progress of DNA polymerase, indicating a possible biological role for the quadruplex structure in Rb gene. Our experiments suggest that i-motif formation occurs in the C-rich complementary strand in vitro. Although the double-helix form was predominant at neutral pH, the G-quadruplex and i-motif efficiently competed with the duplex at lower pH values. These results suggest that the G-strand and the C-strand at the 5′ terminus of Rb gene are capable of forming the G-quadruplex and i-motif structures in vitro.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This study was partly supported by a Grant-in-Aid for Priority Research from the Ministry of Education, Science, Sports, and Culture, Japan; and SORST of Japan Science and Technology (JST). Funding to pay the Open Access publication charges for this article was provided by JST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Derbyshire K.M., Willetts N.S. Mobilization of the non-conjugative plasmid RSF1010: a genetic analysis of its origin of transfer. Mol. Gen. Genet. 1987;206:154–160. doi: 10.1007/BF00326551. [DOI] [PubMed] [Google Scholar]
- 2.Nevins J.R. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 2001;10:699–7032. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 3.Friend S.H., Bernards R., Rogeli S., Weinberg R.A., Rapaport J.M., Alberts D.M., Dryja T.P. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 4.Bernards R., Schacleford G.M., Gerber M.R., Horowitz J.M., Friend S.H., Schartl M., Bogenmann E., Rapaport J.M., McGee T., Dryja T.P., et al. Structure and expression of the murine retinoblastoma gene and characterization of its encoded protein. Proc. Natl Acad. Sci. USA. 1989;86:6474–6478. doi: 10.1073/pnas.86.17.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson J.R., Raghuraman M.K., Cech T.R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 6.Sen D., Gilbert W. A sodium–potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 7.Kang C., Zhang X., Ratliff R., Moyzis R., Rich A. Crystal structure of four-stranded oxytricha telomeric DNA. Nature. 1992;356:126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- 8.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murchie A.I., Lilly D.M. Retinoblastoma susceptibility genes contain 5′ sequences with a high propensity to form guanine-tetrad structures. Nucleic Acids Res. 1992;20:49–53. doi: 10.1093/nar/20.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hackett J.A., Feldser D.M., Greider C.W. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- 12.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y., Sugiyama H. Highly efficient photochemical 2′-deoxyribonolactone formation at the diagonal loop of a 5-iodouracil-containing antiparallel G-quartet. J. Am. Chem. Soc. 2004;126:6274–6279. doi: 10.1021/ja031942h. [DOI] [PubMed] [Google Scholar]
- 14.Gehring K., Leroy J.L., Gueron M. A tetrameric DNA structure with protonated cytosine–cytosine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 15.Postel E.H., Berberich S.J., Rooney J.W., Kaetzel D.M. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J. Bioenerg. Biomembr. 2000;32:277–284. doi: 10.1023/a:1005541114029. [DOI] [PubMed] [Google Scholar]
- 16.Manzini G., Yathindra N., Xodo L.E. Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic Acids Res. 1994;22:4634–4640. doi: 10.1093/nar/22.22.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugiyama H., Kawai K., Matsunaga A., Fujimoto K., Saito I., Robinson H., Wang A.H.-J. Synthesis, structure and thermodynamic properties of 8-methylguanine-containing oligonucleotides: Z-DNA under physiological salt conditions. Nucleic Acids Res. 1996;24:1272–1278. doi: 10.1093/nar/24.7.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y., Ikeda R., Sugiyama H. 8-Methylguanosine: a powerful Z-DNA stabilizer. J. Am. Chem. Soc. 2003;125:13519. doi: 10.1021/ja036233i. [DOI] [PubMed] [Google Scholar]
- 19.Dias E., Battiste J.L., Williamson J.R. Chemical probe for glycosidic conformation in telomeric DNAs. J. Am. Chem. Soc. 1994;116:4479–4480. [Google Scholar]
- 20.Dominick P.K., Jarstfer M.B. A conformationally constrained nucleotide analogue controls the folding topology of a DNA G-quadruplex. J. Am. Chem. Soc. 2004;126:5050–5051. doi: 10.1021/ja039192z. [DOI] [PubMed] [Google Scholar]
- 21.Davis J.T. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 22.Sacca B., Lacroix L., Mergny J.-L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 24.Rujan I.N., Meleney J.C., Bolton P.H. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazel P., Huppert J., Balasubramanian S., Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2005;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 26.Parkinson G.N., Lee M.P., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 27.Crnugelj M., Sket P., Plavec J. Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003;125:7866–7871. doi: 10.1021/ja0348694. [DOI] [PubMed] [Google Scholar]
- 28.Rezler E.M., Seenisamy J., Bashyam S., Kim M.-Y., White E., Wilson W.D., Hurley L.H. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 29.Phan A.T., Kuryavyi V., Gaw H.Y., Patel D. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nature Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jean J.M., Hall K.B. 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc. Natl Acad. Sci. USA. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura T., Kawai K., Majima T. Fluorescence properties of 2-aminopurine in human telomeric DNA. Chem. Comm. 2004;12:1338–1439. doi: 10.1039/b403913k. [DOI] [PubMed] [Google Scholar]
- 32.Baclla A., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinhe S.S., O'Connell C.D., Cooper D.N., Wells R.D. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl Acad. Sci. USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bacolla A., Wells R.D. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2005;280:941–95. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 34.Phan A.T., Mergny J.L. Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phan A.T., Gueron M., Leroy J.L. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000;299:123–144. doi: 10.1006/jmbi.2000.3613. [DOI] [PubMed] [Google Scholar]
- 36.Pataskar S.S., Dash D., Brahmachari S.K. Intramolecular i-motif structure at acidic pH for progressive myoclonus epilepsy (EPM1) repeat d(CCCCGCCCCGCG)n. J. Biomol. Struct. Dyn. 2001;19:307–313. doi: 10.1080/07391102.2001.10506741. [DOI] [PubMed] [Google Scholar]
- 37.Mathur V., Verma A., Maiti S. Thermodynamics of i-tetraplex formation in the nuclease hypersensitive element of human c-myc promoter. Biochem. Biophys. Res. Commun. 2004;320:1220–1227. doi: 10.1016/j.bbrc.2004.06.074. [DOI] [PubMed] [Google Scholar]
- 38.Gallego J., Chou S.H., Reid B.R. Centromeric pyrimidine strands fold into an intercalated motif by forming a double hairpin with a novel T:G:G:T tetrad: solution structure of the d(TCCCGTTTCCA) dimer. J. Mol. Biol. 1997;273:840–856. doi: 10.1006/jmbi.1997.1361. [DOI] [PubMed] [Google Scholar]
- 39.Ahmed S., Kintanar A., Henderson E. Human telomeric C-strand tetraplexes. Nature Struct. Biol. 1994;1:83–88. doi: 10.1038/nsb0294-83. [DOI] [PubMed] [Google Scholar]
- 40.Leroy J.L., Gueron M., Mergny J.L., Helene C. Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res. 1994;22:1600–1606. doi: 10.1093/nar/22.9.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simonsson T., Pribylova M., Vorlickova M. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun. 2000;278:158–166. doi: 10.1006/bbrc.2000.3783. [DOI] [PubMed] [Google Scholar]
- 42.Li W., Wu P., Ohmichi T., Sugimoto N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002;526:77–81. doi: 10.1016/s0014-5793(02)03118-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.