Abstract
A common feature of all the known cancer genetic syndromes is that they predispose only to selective types of malignancy. However, many of the genes mutated in these syndromes are ubiquitously expressed, and influence seemingly universal processes such as DNA repair or cell cycle control. The tissue specificity of cancers that arise from malfunction of these apparently universal traits remains a key puzzle in cancer genetics. Mutations in DNA mismatch repair (MMR) genes cause the most common known cancer genetic syndrome, hereditary non-polyposis colorectal cancer, and the fundamental biology of MMR is one of the most intensively studied processes in laboratories all around the world. This review uses MMR as a model system to understand mechanisms that may explain the selective development of tumors in particular cell types despite the universal nature of this process. We evaluate recent data giving insights into the specific tumor types that are attributable to defective MMR in humans and mice under different modes of inheritance, and propose models that may explain the spectrum of cancer types observed.
INTRODUCTION
DNA mismatch repair (MMR) and mechanisms of cancer prevention
More than 1 billion years old, MMR plays critical roles in the maintenance of genomic stability in prokaryotes, simple eukaryotes and metazoan organisms such as humans and rodents (1–3). Interest in DNA mismatch repair and its mechanisms of action exploded in the early 1990s with the observation that germline mutations in MMR genes cause hereditary non-polyposis colorectal cancer (HNPCC) (4–7). The biology of MMR has been extensively characterized in model organisms, and these studies have given tremendous insights into the function of specific proteins in humans. The model organisms in which MMR has been most extensively studied are Escherichia coli and Saccharomyces cerevisiae. Other systems in which MMR genes have been rigorously characterized include Caenorhabditis elegans and Arabidopsis Thaliana. The precise mechanistic function that each MMR protein performs in these model organisms, as well as humans and mice, has been covered in detail in numerous excellent reviews (3,8–15). Therefore, this subject will be summarized only succinctly here, focusing on mechanisms most likely to be relevant to the discussion of tissue specific carcinogenesis.
Briefly, there are nine mammalian MMR genes (MLH1, MLH3, PMS1-2 and MSH2-6) (3,8). The MMR proteins interact with each other to create a combinatorial code of complexes that mediate distinct functions. The mammalian E.coli MutS homologs (MSH proteins) are thought to directly contact double-stranded DNA, scanning along the genomic DNA for mismatches analogous to a ‘sliding clamp’ until they encounter a base pair containing a mismatch (16,17). The MSH proteins interact with multiple proteins including the mammalian E.coli MutL homologs (MLH) and yeast post-meiotic segregation (PMS) homolog proteins (which have significant amino acid identify and structural similarity to the MLH proteins), as well as RPA, EXO1, RFC, possibly HMGB1, and other less well-characterized proteins [reviews (3,8) as well as the new primary research papers (18–20) discuss this topic in excellent detail]. With respect to the mutator function, the MSH2–MSH6 heterodimer is thought primarily to repair single-base substitutions and 1 bp insertion–deletion mutations, while MSH2–MSH3 is thought primarily to repair 1–4 bp insertion–deletion mutations (3,8). The E.coli MutL homologs (MLH) and yeast PMS proteins interact with heterodimers of MSH proteins to help catalyze their different functions. MLH1–PMS2 is the primary MutL complex that interacts with both MSH2/6 and MSH3 complexes in mechanisms thought to be relevant to cancer prevention. Recent studies, including those from our laboratory, suggest mammalian MLH1–MLH3 also contributes to some of these processes as well, but in all mechanisms tested to a lesser degree than MLH1–PMS2 (21–24). MLH1–PMS1 clearly exists in mammalian cells but it currently has no clearly defined role in processes relevant to cancer susceptibility (25,26). MSH4 and MSH5 are thought to play roles exclusively in meiosis and are currently not thought to contribute to mechanisms of cancer prevention (27).
A mutator phenotype causing increased mutation rates for single-base substitution and insertion/deletion mismatches have traditionally been thought to be the main functions in MMR cancer prevention. Defects in the insertion/deletion repair function create a phenotype that is largely thought to be unique to MMR. Since this function was originally characterized on short repetitive DNA microsatellite sequences, it is often referred to as Microsatellite Instability or MSI (28–30). MSI is divided into MSI-Low and MSI-High (MSI-H) subtypes depending on the percentage of microsatellite markers that are mutated, but only the MSI-High subtype is clearly associated with defective MMR. Tumors with no MSI are referred to as Microsatellite Stable (MSS). [Consensus thinking on the topic of MSI was recently covered in detail by Umar et al. (31)]. The precise mechanisms whereby MSI causes cancer are hypothesized to include a genome wide increased mutation rate that causes mutations not only in microsatellites, but also in exonic coding sequences of genes important in cancer suppression. MSI mutation rates in these short repetitive sequences in MLH1- or MSH2-deficient cells are estimated to be more than 100 times higher than that seen in MMR-proficient cells and used to test directly whether MMR deficiency is involved in specific tumor types. Almost all described MLH1/MSH2 mutations cause MSI-H tumors, although recently MSS MLH1 non-synonymous amino acid substitutions causing deleterious mutations have been described (32,33). Current thinking is that MMR mutations are recessive (i.e. require two hits under the Knudson hypothesis) before cells become susceptible to cancer. However, it has been suggested that cells from subjects carrying heterozygous germline mutations may have detectable MSI using a more sensitive assay referred to as single-molecule or small pool PCR (34,35) that can detect approximately 5- to 10-fold elevations in MSI rates. These new findings are intriguing, but require further investigation before the role of MMR haploinsufficiency in cancer susceptibility is clearly established.
In addition to its essential roles in insertion/deletion and single-base substitution repair, MMR proteins also participate in additional mechanisms that could contribute to carcinogenesis, most notably initiation of apoptosis in response to DNA damage (16,36). Recent studies using mouse models of single-amino acid substitutions causing separation of function mutations have demonstrated clearly that decreased apoptosis plays an important role in MMR-deficient tumorgenesis (37,38). The precise mechanism by which MMR contributes to the initiation of programmed cell death remains unclear, with both futile cycles of repair causing high levels of double-strand breaks and direct signaling proposed (39,40). MMR mutant cells fail to activate a p73-dependent cell death activation pathway (41), which may explain the absence of a critical requirement for p53-mediated apoptosis (42). This failure impairs cell cycle and DNA damage checkpoint recognition (3).
In summary, the current models suggest that MMR mutations cause cancer primarily through the contribution of both mechanisms: (i) cells acquire mutations in components of critical tumor suppressor gene pathways that allow them to proliferate and (ii) cells do not initiate apoptosis appropriately. It is unclear at present whether these mechanisms operate sequentially or concurrently in different cell types. MMR proteins also contribute to suppress homeologous recombination (39,43), which could in principle contribute as an important mechanism of carcinogenesis. However, the contribution of increased rates of homeologous recombination from MMR mutations to carcinogenesis is not well defined at this point in time.
The spectrum of cancer susceptibility attributable to MMR deficiency
Mutations in the MMR genes MLH1 and MSH2 were first identified because of their association with colorectal cancer in HNPCC families (4–7). Approximately 70–85% of HNPCC families have identifiable mutations in these two genes (9). Subsequently, HNPCC families with reduced penetrance and identifiable MSH6 and PMS2 mutations were identified (44,45). Further studies demonstrated that somatically acquired defects in MMR mutations are attributable for ∼17 000 sporadic colorectal cancer (CRC) (46,47) and 10 000 endometrial cancer (48) new diagnoses in the United States each year. Germline MSH6 mutations were also shown to be responsible for an appreciable proportion of familial colorectal cancer and hereditary uterine cancer families (49–51). More recently, pediatric patients harboring homozygous recessive MLH1 or MSH2 germline mutations have also been identified. In these individuals, the most notable susceptibility is to hematopoietic malignancies (which are not generally present in HNPCC) and colorectal cancer (52–56).
Numerous studies have attempted to characterize the tumor spectrum and age of onset of HNPCC-related tumors in affected individuals, but these studies have been fraught with challenges given the relative rarity of the syndrome and difficulty in gathering prospective data. Most studies have focused on identifying high-risk families of those that meet clinical criteria for HNPCC and comparing incidence data with that of the general population. However, this approach leads to biases in the data toward higher than actual rates of tumor development since the most strongly affected families tend to be included while those with a less severe phenotype are less frequently recognized (referral bias). HNPCC-Lynch Syndrome was originally described as a familial syndrome manifested by elevated incidence of colorectal and endometrial cancer and further studies have related this to MMR defects as well to numerous other cancer susceptibilities (4–7). Colorectal cancer has been reported in men with HNPCC with a cumulative lifetime risk upwards of 74% (57–59), and standardized incidence ratio of 9.6 (95% CI 7.5–12.3) in a clinic-based study of individuals with MSI-H tumors and as low as 2.0 (1.3–2.7) in the same study for population-based data in MSS individuals (60). In women, the risk of CRC is lower and lifetime risk of endometrial cancer has been reported as low as 30% (61) in a study based on registry data from Finland, Holland and the United States. Other studies have placed the lifetime risks of endometrial cancer as much higher: 42% for women with a mutation in MLH1, 61% for MSH2 (58). While the elevated risk for both CRC and endometrial cancer in these patients is undisputed, it soon became clear upon observing these patients and their families that cancers at other specific sites seemed to appear far more frequently than one would see in the general population. Most reliable studies have relied on the calculation of standard incidence rates to evaluate these observations, i.e. the ratio of observed to expected cancer rates compared with population based information. The first study on this subject demonstrated an elevated SIR for gastric cancer (SIR = 4.1), hepatobiliary (SIR = 4.9), urologic tract (SIR = 2.5), ovarian (SIR = 3.6), and small bowel cancer (SIR = 25), as well and an increased but not statistically significant SIR of 1.6 for brain tumors (62) which has not been substantiated by further studies. Further work in this area has verified these results, and has repeatedly shown increased risk of small bowel, hepatobiliary, gastric, skin keratoacanothoma, ureter/kidney and ovarian tumors. Anecdotal reports of tumors such as leukemia, lymphoma, pancreas, larynx and breast tumors have not been substantiated by larger scale observational studies (60,63).
Recent large-scale population and clinic-based analyses have been performed that have less overall bias than single institution case series (60,64–66). These studies use MSI or loss of immunohistochemical staining as the screening methodology, and therefore identify both germline and sporadic MMR defects. In these studies, the greatest cancer susceptibility is manifest as colorectal cancer [odds ratio (OR) 4.3–9.6], endometrial cancer (OR 3.4–5.4), stomach cancer (OR 3.3–7.1), kidney (OR 1.8–4.1), ovary (OR 1.6–2.6), small intestine (OR 3.6–9.7) and ureter (OR 6.8–10.0) (Table 1) (60). Surprisingly, significantly lower cancer rates are also observed, most convincingly and repeatedly seen in prostate, breast and lung cancers (60,63,67). Environmental factors have also been demonstrated to play a role in the affecting the tumor spectrum. Recent data from HNPCC mutation carriers support the assertion that smoking significantly increases CRC risk in HNPCC mutation carriers (hazard ratio of 1.43, P < 0.04) (68).
Table 1.
Standardized incidence ratios (SIRs) of cancer risk by site in population-based and clinic-based ascertainment of MMR defective cancers
| Tumor site | All MSI-H SIR (60) | Population-based SIR (60) | Clinic-based SIR (60) | Incidence per 100 000 (60) | Clinic-based SIR (63) | Incidence per 100 000 (63) | Combined incidence per 100 000 (60,63) |
|---|---|---|---|---|---|---|---|
| Colon | 6.1 (5.2–7.2) | 4.3 (3.4–5.3) | 9.6 (7.5–12.3) | 5319 | N/Aa | ||
| Gastric | 4.6 (2.7–6.6) | 3.3 (1.4–5.4) | 7.1 (3.1–11.7) | 614 | 4 | 1291 | 802 |
| Uterus | 4.1 (2.9–5.6) | 3.4 (1.9–4.8) | 5.4 (3.1–7.9) | 2440 | N/Aa | 8049 | 4020 |
| Kidney | 2.6 (1.4–4.0) | 1.8 (0.5–3.3) | 4.1 (1.5–7.1) | 438 | 3 | 759 | 528 |
| Lung | 0.3 (0.2–0.5) | 0.2 (0.0–0.4) | 0.6 (0.2–1.1) | 351 | |||
| Ovary | 2.0 (1.0–3.2) | 1.6 (0.5–3.1) | 2.6 (0.8–5.1) | 714 | 3.5 | 1974b | 1069b |
| Breast | 0.5 (0.3–0.7) | 0.4 (0.2–0.6) | 0.6 (0.3–1.0) | 1250 | <1 | ||
| Hematopoietic | 0.5 (0.2–0.8) | 0.7 (0.2–1.1) | 0.3 (0.0–0.7) | 321 | 1 | ||
| Pancreas | 1.7 (0.7–2.8) | 2.1 (0.8–3.8) | 1.0 (0.0–2.7) | 292 | 1 | ||
| Prostate | 0.3 (0.1–0.5) | 0.3 (0.1–0.6) | 0.3 (0.0–0.7) | 292 | |||
| Small bowel | 7.6 (2.5–13.9) | 9.7 (1.9–19.3) | 3.6 (0.0–12.0) | 175 | 25 | 759 | 338 |
| Cervix | 0.3 (0.1–0.5) | 0.2 (0.0–0.4) | 0.5 (0.0–0.9) | 357 | 1 | 1063b | 556b |
| Bladder | 0.4 (0.1–0.9) | 0.3 (0.0–0.7) | 0.8 (0.0–1.8) | 146 | 1 | 380 | 211 |
| Hepatobiliary | 2.4 (0.6–5.0) | 0.9 (0.0–3.2) | 5.3 (0.0–11.7) | 117 | 5 | 532 | 232 |
| Ureter | 9.0 (2.0–18.3) | 10.0 (0.0–22.9) | 6.8 (0.0–21.8) | 117 | 22 | 380 | 190 |
| Brain | 0.7 (0.0–1.7) | 0.7 (0.0–1.9) | 0.6 (0.0–2.1) | 88 | 1 | ||
| Head/neck | 0.3 (0.0–0.6) | 0.4 (0.0–0.9) | N/A | 88 | 1 | ||
| Melanoma | 0.3 (0.0–0.4) | 0.2 (0.0–0.6) | N/A | 58 |
The spectrum of cancer susceptibility in germline homozygous MMR mutation mouse models
Mouse models for all nine MMR genes, as well as numerous combinations of these genes, have been created and have given significant insights into the mechanisms and cancer susceptibility of specific cell types (25,39,69–77). Msh2−/− and Mlh1−/− mice have a highly penetrant cancer susceptibility, which manifests primary as lymphomas, GI epithelial adenomas and basal cell carcinomas starting at ∼6 months of age (25,39,76,78). Msh6−/− mice display essentially the same tumor spectrum, but with the same tumor types appearing around 9–12 months of age (72). Pms2−/− mice develop lymphomas and sarcomas but not GI tumors, starting around 9–12 months of age (25,79,80). Mlh3−/− mice have lower penetrance GI cancer susceptibility and develop GI cancers with a mean onset of 12 months of age, as well as lymphomas and skin cancers (23). Pms1−/−, Msh3−/−, Msh4−/− and Msh5−/− mice have to date demonstrated no clear susceptibility to cancer (25,71,81). In summary, the tissue specificity of all MMR mouse knock out models is very similar, but with varying penetrance. In mice, the main tissues affected are GI epithelial tumors and mixed B and T cell lineage lymphomas. It is not well understood at present why endometrial, ovarian and renal cancers do not occur with appreciable frequency as they do in humans. It has been speculated that this difference might reflect the shorter lifespan of mice, the lower penetrance of these other cancer types, differences in hormonal exposure of the estrous versus menstrual cycles or environmental exposures.
Contribution of the mutator phenotype: distinct sequences have higher mutation rates in genes critical for different cell types
Since different cell types have distinct critical tumor suppressor and tumor promoter gene requirements, the precise sequences of these genes in part may help define the tissue specificity of MMR defects as mutation targets. It is well appreciated that different cell types have distinct patterns of gene expression/splicing (82,83) and are dependent on distinct tumor suppressor and tumor promoter genes [for recent review, see (84)]. Therefore, it is not surprising to accept the premise that specific cell types are more susceptible to perturbations in specific signaling pathways than others. It is clear that defective MMR preferentially causes specific sequences to mutate at faster rates than other sequences. The best-characterized highly specific preference is for mutation of short pair repetitive sequences, a phenomenon originally described by Perucho, Shibata, Thibodeau and colleagues (28,29). Current thinking is that insertion/deletion mutations of most non-coding genomic microsatellites do not contribute directly to cancer susceptibility, but that these mutations do play a role when they occur in coding or proximal promoter regulatory sequences. Perhaps the best-characterized target is the transforming growth factor β type II receptor, which in humans has a 10 Adenine repeat in its coding region that is frequently mutated in MSI-H CRC tumors (85). Since TGF Beta signaling is particularly significant for GI epithelial cells, and repression of this pathway's signaling is relieved by transforming growth factor beta type II receptor (TGFβR2) adenine repeat frameshift mutations that occur at high rates in MMR defective cells, the combination of these two events may explain in part why GI epithelial cells are preferentially transformed in HNPCC. However, because this 10 adenine coding repeat is not present in mouse Tgfbr2 and the MMR knockout models develop GI carcinomas, it cannot be the only explanation for the preference of tumors to develop from this cell type. Recent genome wide surveys have cataloged genes whose coding sequence contains repetitive mono- and dinucleotide repeat sequences (86–88) that are likely candidates for frameshift mutations in MMR-deficient cells. Because nonsense-mediated decay (NMD) causes the degradation of many mRNAs with frameshift mutations causing premature stop codons, cross comparisons of different gene expression patterns in MSI-H cancers versus normal mucosa with the list of genes encoding longer repetitive mono- and dinucleotide repeat sequences are a well focused strategy to identify selective targets in different tissues that explain the predilection of MMR defects to cause specific cancer types. Experiments in cell lines using this approach and selective stabilization of NMD targeted mRNA transcripts with chemical or genetic approaches has suggested UVRAG, p300 and GRK as potential important targets in MSI-H CRCs (89,90). It will be interesting to see how the targets identified by this approach when tested in other cell types such as uterine epithelial cells, or use primary cancer cells.
There is also evidence that even mildly repetitive sequences in genes whose function is critical are selected with higher frequency by MMR deficiency as common mutation targets. For example, the spectrum of Apc mutations in MMR-deficient tumors has been exhaustively characterized in mouse models. In mouse GI epithelial tumors, Apc deletions of a single ‘CA’ in TCC CAC ACA CA or a single ‘GA’ in AAG AGA GAG AG, both of which are in the Apc coding region, cause frameshift truncation mutations have each been observed more than 20 times and occur significantly more frequently than any base-substitution mutation (91–93). Similar observations have been made in human MSI-H colorectal tumors (94). Therefore, the contribution of the MMR mutator function to cancer cell type specificity is likely to involve elements of (i) the presence of repetitive coding sequences in specific genes, (ii) the expression pattern of those genes and (iii) the functional importance of those genes so that even rare mutations can be selected for if they confer a proliferative advantage.
How do targets of the MMR mutator phenotype influence tumor specificity?
The observation that human and mouse null mutations for critical MMR genes allow essentially normal development demonstrate clearly that the loss of MMR alone is insufficient to initiation a carcinogenic transforming event, but that additional genetic changes are also required for the transition to frank malignancy. The genes that are inactivated by frameshift mutations in MMR-deficient cancers have been evaluated by several groups (95,96). Of all the genes, TGFBR2, BAX and CASP-5 stand out as the most consistently mutated in MMR defective tissues (95,96). TGFBR2 is consistently mutated in most MSI-H cell lines and CRC tumors with 1 or 2 bp deletions in a coding poly(A)10 tract causing a frameshift mutation (85) and this finding has been consistently replicated by multiple groups (95–97). Subsequently, multiple other genes with putative tumor suppressor functions and containing coding microsatellites have shown to be mutated at various frequencies in sporadic MSI-H and HNPCC associated colorectal, gastric and endometrial cancers. These include additional genes such as IGFIIR, PTEN (98) involved in signal transduction, apoptosis (BAX, Caspase 5) (95,99), DNA repair, MSH3, MSH6, MBD4 (100,101), as well as genes that play roles in transcriptional regulation, protein stability and immune surveillance. It is thought that by analyzing different cell lines, coding repetitive sequences that are not under growth selection (found in either tissue or temporally-specific non-transcriptionally active genes) or which are essential to cell survival would only appear with a low frequency proportional to that of the mutation rate of the MMR-deficient cells. These genes are sometimes referred to as ‘bystander’ or ‘survivor’ genes' (102). One would expect a bias toward decreased mutation frequencies in these coding MS (cMS). Finally, that genes in which frameshift mutations would convey a growth or selective advantage to the cell, presumably by abrogation of some tumor suppressor function, can be identified by a higher than expect frequency of mutation in MSI-H tumors. Duval et al. (103) used the maximum likelihood statistical analysis to search for new genes in which coding repetitive sequence mutations would provide a selective advantage to growth or tumor progression in a group of 9 known target genes and 19 others with an expected role in carcinogenesis. They also analyzed a series of 22 non-coding versus 25 coding short mononucleotide repeats (≤10 bp) where 7/25 coding repeats (in CASP5, MSH6, IGFIIR, GRB14, BAX, TCF-4 and TGFβ2R) showed a significantly higher mutation frequency; however, a great amount of variability was seen with mutation frequencies ranging from 2 to 92% in coding, and 0 to 40% in non-coding sequences (104). Other approaches have (102) used comprehensive literature search to identify 169 coding and 25 non-coding repeat sequences in colon, gastric and endometrial cancer and a cumulative mutation model to predict the genes with the strongest evidence for involvement in MMR defective mechanisms of carcinogenesis in the colon. These genes included TGFβ2R, BAX, TCF-4, MSH3, ACVR2, PTHL3, HT001, AC1, SLC23A1 MACS, NDUFC2 and TAF1B (88,102). Two other genes, WAF1 and BCL2, were found with mutation frequencies below the lower prediction limit arguing for negative selection in these cases. If this prediction is correct, it could be tested by creating models of MMR deficiency combined with Waf1 or Bcl2 null mice. It follows that observation of decreased tumor incidence or a shift in tumor spectrum would provide further evidence for a negative selection bias against these cMS. Similar techniques have also been used to discern a temporal relationship between target genes in the progression from adenoma to full blown malignancy emphasizing the likelihood that loss of BAX, TGFβ2R, MACS, NDUFC2, and TAF1B as early events and potential therapeutic targets (88).
Multiple studies have continued to identify a small subset of genes with coding repetitive sequences whose loss of function mutations likely contribute to tissue specificity of carcinogenesis. More problematic, has been the question of tissue specificity of these genes and pathways since there are obvious differences in target genes according to tissue type. This has been most frequently studied with regard to attempts to distinguish genes specific for either colorectum stomach or endometrium carcinomas. The spectrum of target genes in colon and gastric cancers appears very similar (97,105) with the exception of two genes, TGFβ2R (P = 0.0004) and MSH6 (P < 0.0001). Overall, endometrial cancers showed far less instability with mutation frequencies of TGFβ2R, MSH6 and MSH3 were significantly lower than in gastrointestinal than endometrial tumors (P < 0.01). As predicted, in all three cancer types the distribution of mutation frequencies in putative target genes was bimodal suggesting a peak for genuine target genes and ‘bystanders’ at lower frequencies (102).
Why GI epithelial cells and where along the proximal-distal axis?
Because mutation rates are significantly higher in MMR defective versus proficient cells, current thinking is that the cell types with the highest overall proliferation rate (and therefore the total number of genomic replication cycles) would be most susceptible to cancer susceptibility attributable from the MMR mutator phenotype (3,101,106,107). This observation is likely to contribute to the spectrum of MMR cancers. In humans, the GI epithelium has the highest known proliferation rate of all cell types that have been measured and is thought to turn over approximately every 3–5 days (108). Other cell types that cycle rapidly include bone marrow, skin and epithelial cells of the upper GI tract. The in vivo ‘experiment’ of which cells have the most rapid proliferation rate is repeated daily in patients with cancer who are treated with standard cytotoxic chemotherapy agents. These drugs target universal processes in proliferation such as genome replication by DNA polymerases or unwinding by topoisomerases and kill the cells with the highest proliferation rates. Acutely, patients develop diarrhea, which reflects cell death of lower GI epithelium. In the next few days later, red, white and platelet blood cells nadir, hair falls out, and mucositis develops reflecting the slightly less rapid turnover rates of bone marrow progenitor, hair/skin and upper GI epithelial cells. It is likely that these high proliferation rates explain in part the high rates of lower GI epithelial tumors that results from MMR mutations, although there is a paucity of direct experimental evidence to support the assertion. However, it is clear that ranking cell types by proliferation rates alone does not explain why other tissues, such as the genitourinary tract or ovarian epithelium, are affected. Therefore, additional causes must exist as well.
MMR-environment interactions in the gastrointestinal and genitourinary epithelium
MSH2–MSH6 can specifically recognize distinct types of damaged DNA (such as those caused by oxidative or alkylating agents). It has previously been proposed that the tissues susceptible to malignancy in the context of defective MMR could be explained by a specific type of DNA damage occurring primarily at these sites (16). The affected cells would not correctly initiate apoptosis to this damage, thereby leading to a selective advantage and transformation (16,109–111). While a specific type of DNA damage has not to date been shown to be unique to the cell types affected by MMR deficiency (and absent in the unaffected cell types), environmental insults have been demonstrated to play a role in the affecting the tumor spectrum in both patients and mouse models. Recent data from HNPCC mutation carriers support the assertion that smoking significantly increases CRC risk in HNPCC mutation carriers (hazard ratio of 1.43, P < 0.04) (68). Ethnic background also affects tumor spectrum (112–114), and marked inter-family variation in risk of HNPCC spectrum cancer is clearly documented (58,67,115–120). The variance is significant in the expressivity of the syndrome (i.e. the distribution of different tumor types that develops in different HNPCC families) (115). However, the precise environmental (or other genetic) modifiers that influence these phenotypic variances are incompletely characterized.
It is notable that changes in relative tumor susceptibility of the stomach, small intestine and colorectum are observed to vary with both the temporal era as well as geography. The first HNPCC family identified was described by Aldred Warthin, a pathologist in 1913, in Europe and is referred to as Family G (121,122). Members of the original family are still alive today, and descendents have been followed in Europe and the United States. Similarly, many other HNPCC kindreds have family histories that extend over a century (48,122). In the early 20th century, the GI tumors in Family G and other similar kindreds were distinguished largely by an excess of stomach adenocarcinoma (123). In mid and late 20th and early 21st centuries, the GI cancer susceptibility has shifted to include mostly colorectal cancers. There are also notable differences between the cancer spectrum of HNPCC families that have been identified in Asia, Europe, Australia, North and South America and Africa. For example, it is of note that the GI cancer susceptibility of Asian families generally has greater incidence of stomach cancer compared to North America, South America and Europe, where colorectal cancer predominates (118,119,122). It has been speculated that the shift in the GI cancer susceptibility rostral-caudal axis has reflected changes in food preparation and content. In particular, the decreased use of ‘smoked’ and cured meats resulting from refrigeration, and the high fat content of the modern European–American diet have been proposed as important factors (122). The smoking process of meats is thought to introduce heterocyclic amine byproducts in the food, and the curing process involved nitrate salts that cause nitroso-compounds, which are thought to act as potential mutagens. Epidemiologic and animal model studies provide evidence for this model (68,124). Direct experimental evidence supports the epidemiologic hypothesis that specific environmental mutagens play important roles. Cell culture experiments using Msh2+/− cells in culture demonstrate that treatments with the genotoxic agents such as alkylators (ethylnitrosourea) or DNA cross-likers (mitomycin C) selects for Msh2−/− cells that increase in relation to the number of Msh2+/− heterozygous cells (125). Part of this selection may reflect the observation that MMR cells appear to have higher overall proliferation rates compared to MMR-deficient cells, which would give them a growth advantage.
In MMR mouse models, the relative susceptibility of GI epithelium along the rostral-caudal axis (stomach, small bowel and colorectum) to form tumors is modulated by dietary fat and pharmaceuticals. Diets that are high in fat and low in calcium and folate (commonly referred to as ‘Western Diets’) increase most significantly the incidence of colorectal tumors compared with small intestine or stomach tumors (126), which parallels the higher colorectal cancer rates observed in HNPCC families in North America and Europe. Non-steroidal anti-inflammatory agents (NSAIDs) and PPAR gamma agonists can decrease small intestine tumors, while increasing stomach and colon tumors in MMR-deficient mouse models (127,128). Two randomized prospective human chemoprevention trials involving NSAIDs and HNPCC family members are ongoing, and more precise data as to how well the mouse model findings can be extended to humans will be available in the next several years (129,130). CDX2 is a transcription fact that helps specify GI epithelial cell fate through a rostral-caudal expression gradient. Mutations in Cdx2 are implicated with carcinogenesis in GI epithelium (131,132). As CDX2 down-regulation is frequently observed in colorectal cancer and mutations and lower expression are strongly associated with MSI-H CRCs (133), it would be interesting to test directly, perhaps using mouse models, whether dietary modulation of Cdx2 function might be an important mechanism that underlies the shifts along the different epithelia along the lower GI tract for MMR defective tumors.
While no precise environmental exposures or dietary or pharmaceutical agents have been specifically implicated in the non-GI MMR tumor spectrum sites, it is proposed that such agents concentrated in the urine may contribute to the higher rates of renal and ureteral cancers seen in HNPCC (134). Since cadmium is associated with both inactivation of MMR (135) and sporadic renal carcinoma (RR 2.0) (136,137), testing the hypothesis that this agent induces MMR-deficient renal cancers could be carried out by exposing MMR defective animal models, or measurements of this element in renal tissues from affected patients as cadmium should not be affected by formalin fixation. Other potential agents to test prospectively in these models associated with renal cell cancer risk include asbestos, trichloroethylene and other petroleum by-products that are concentrated in urine.
Target genes in tumors from germline recessive versus sporadically acquired MMR deficiency
Patients with germline recessive MMR mutations (two mutant alleles) have a different spectrum of tumor types that that develops with dominant MMR (one mutant germline allele and somatic loss of the second allele, or sporadic functional loss of both alleles). Compared with HNPCC, recessive MLH1, MSH2, MSH6 and PMS2 mutations cause GI epithelial cancers as well as acute and chronic myeloid leukemias, Non-Hodgkin's lymphomas, dermal neurofibromas characteristic of Neurofibromatosis Type I and perhaps glioblastomas (52–56,138).
The observation that the cancer susceptibility of recessive versus dominant MMR mutations is different may give clues as to potential target genes. In particular, the leukemia susceptibility of patients with germline recessive MMR mutations suggests an interesting experimental model. This model would predict that there exists a primitive myeloid progenitor cell type during early development (i.e. present in individuals who inherit two MMR mutant alleles) not present in adults (such as in patients who inherit one mutant allele and have somatic loss of the second allele) that contains (i) target genes particularly sensitive to MMR-deficient mutation, or (ii) critical dependence on MMR mediated apoptosis. Furthermore, this model would also predict that this primitive myeloid progenitor cell type is rare or absent during adult life, and therefore not susceptible to transformation by dominant MMR mutations with subsequent loss of the second allele. In support of the first prediction by this model, mechanistic studies in mouse models looked at MMR germline defects combined with mice that are mutant for Nf1, the gene mutated in Neurofibromatosis Type I. Mlh1−/−;Nf1+/− mice develop myeloid leukemias, similar to the hematopoietic malignancies observed in patients with recessive MMR mutations (139). The tumors had significant MSI-H mutations, and loss of neurofibromin expression in all tumors analyzed. These results suggest that MMR deficiency can accelerate myeloid leukemogenesis in Nf1+/− mice, presumably by inactivating Nf1 gene expression as a myeloid lineage tumor suppressor gene. As there is substantial evidence that inherited versus sporadically acquired mutations significantly influence the types of tumors that develop [reviewed in (140)], and all the current described MMR mouse models use germline mutations, future experiments involving inducible Mlh1 or Msh2 mutations using Cre-Lox technology in ovary, kidney or uterus will be important to test whether these models develop cancers in those tissues.
Immune surveillance and tissue specificity of cancer susceptibility
The insertion/deletion frameshift mutation phenotype of MMR deficiency is predicted not only to cause gene mutations, but also lead to the creation of novel protein antigens that are generated by the alternate reading frames and expressed in cells. It has been suggested that additional tumor types may develop in MMR null patients, but that they are suppressed by the immune system. In support of this concept, MSI-H CRCs are associated with a higher concentration of infiltrating lymphocytes than are MSS CRCs, and this pathological finding is included as one of the features to help physicians screen for MSI-H tumors (141).
Because MMR null mouse models develop NHL while HNPCC patients generally do not, a recent study screened a large series human NHLs for MSI. The MSI-H phenotype was present only in patients with HIV or iatrogenic (post bone marrow transplant) induced immunodeficiency (142). These data are consistent with a model whereby helper T-cell mediated immune surveillance does play a role in suppressing NHLs that develop in the context of MMR deficiency (presumably in these cases due to homozygous somatic inactivation), and thereby contributes to limiting the clinically apparent cancers in patients. The observation is supported by mouse model studies that show combining MMR deficiency with immunodeficiency (Msh2−/−;Foxn1−/− and Msh6−/−;Foxn1−/−) significantly increases NHL susceptibility (143). Why immunocompetant MMR homozygous mutant pediatric patients and mouse models develop NHLs is unclear, but the parallel findings suggest the same mutator or anti-apoptotic targets may be affected in both species. The identification of frameshift-inactivating mutations of pro-apoptotic factors BAX and Caspase-5 in the NHLs from immunodeficient patients predicts they may be affected in the mouse models. However, since mouse Bax lacks the polyguanine repeat present in human BAX and the mouse ortholog of Casp5 is controversial, these experiments would require ‘humanizing’ these genes in mice before analyzing mouse lymphomagenesis.
Tissue specificity of apoptosis mechanisms in MMR defective cancers
Failure of apoptosis is considered to be one of the six fundamental hallmarks of malignant disease (144) and clearly plays a major physiologic role in MMR-deficient tumors (145,146). These observations provide a preliminary basis for the examination of the role of apoptosis in MMR defective cell type specific cancer susceptibility. There are three reasons for thinking that initiation of apoptosis may contribute to MMR defective tumor specificity. First, current quantitative biological models of colorectal cancer emphasize abrogation of apoptosis as a very early stage in progression (147,148). These integrative biology models provide evidence that defects in apoptosis precede increases in mutation rates or genomic instability, which are thought to affect progression. Second, different cell types have distinct critical requirement for different genes in apoptotic signal transduction pathways. These cell type specific requirements have been characterized in different tissues, with the best-characterized example the somatic selection for BCL2 expression in B-cell lymphomas (149). Other mutations in apoptotic genes, in contrast, are critical for other cell types. For example, Caspase-9, Cytochrome C and Bax models demonstrate critical roles for these genes in neural apoptosis, while caspase-8 and FADD are critical in cardiomyocyte apoptosis (150). In wild-type and MMR defective cells, there is preliminary evidence that apoptosis may be mediated through a PARP-1 dependent but caspase independent mechanism (93,151). Since there exist selective blockers of PARP-1 (DHIQ and DPQ) and caspase inhibitors (BAF and Z-VAD.fmk), studies to investigate the precise contribution of these different apoptotic pathways in different cell types that are either susceptible or resistant to MMR defective carcinogenesis may be informative whether critical signaling defects correlate with cell types susceptible to defective MMR-induced carcinogenesis. It is notable that BAX, which helps mediate Caspase-3 inducible apoptosis, is a common target of MMR mutations. It is underappreciated that mathematical and computer models of multistage theories of progression can be used to predict cancer rates in different cell types from quantitative aspects of apoptosis and DNA repair (152). If precise quantitative data on cell proliferation and mouse model tumor incidence data can be collected from tissues that are susceptible and resistant to MMR defective carcinogenesis (colorectal and breast epithelium for example), specific predictions of the relative contributions of apoptosis can be used to predict which additional tumor types would be susceptible to MMR inactivation by using age-specific penetrance, stage and metachronicity parameters. Third, apoptosis is particularly critical for all the cell types that are susceptible to cell autonomous MMR deficiency, including gastrointestinal, ovarian and endometrial epithelium and white blood cells. As discussed, many of the most crucial members of apoptotic pathways for these organ sites can be targeted MMR-deficient mutation, most notably BAX, APAF-1, BCL10, CASP-5, FAS and others (105).
In particular, the role that apoptosis plays in uterine endometrium and ovarian epithelium as part of normal tissue physiology is underappreciated. In response to hormonal stimulation, the uterine endometrium undergoes monthly cycling including a proliferative or follicular (pre-ovulatory), secretory or luteal (post-ovulatory) and menstrual phases. During each of these phases, there is marked change in the quantity of and quality of the endometrial cells. The proliferative phase is marked by high mitotic index, and Ki-67 indicative of rapid cellular proliferation, as well as elevated levels of BCL2, an anti-apoptotic factor, presumably involved in stabilizing the cells during this period and allowing for normal physiologic growth in preparation for implantation and pregnancy (153). During the luteal phase, BCL2 expression falls, consistent with the appearance of apoptotic cells, and with higher expression of Fas, Fas-L and elevated activity of caspases 3, 8 and 9 from the luteal phase to menstruation (154). These cycle-related changes in expression are most prominent in the glandular epithelial cells where menstrual-related endometrial thickening and sloughing occurs, as well as the most common site of endometrial carcinogenesis in MMR deficiency. More consistent basal levels of BCL2 expression are maintained in basal laminal, stromal and epithelial cells, where escape of apoptotic death is necessary for these cells to rebuild the functional endometrium with the next hormonal cycle. Similar studies in the ovary demonstrate the importance of programmed cell death in this organ. During the menstrual cycle, a single mature follicle is released by rupture of the ovarian epithelium. Multiple studies have implicated apoptosis and related signaling pathways as the mechanism of ovarian cell death (155,156). BCL2 expression is markedly elevated during the luteal phase and early pregnancy, but notably absent in the regressing corpus luteum (157). Assays of DNA fragmentation as a marker for apoptotic cell death showed high molecular weight DNA during early luteal phases, while mid-luteal phase DNA demonstrates cleavage into low molecular weight ladders (158).
The critical role of apoptosis in the susceptibility of ureter and urologic system, however, is less clear. Urothelial cells have a relatively low basal rate of proliferation and apoptosis in order to maintain tissue homeostasis (159); there is some evidence that this rate changes over time with exposure to toxins and other environmental exposures in the urine. This may also lead to potential apoptotic cell death and entry into wound healing pathways; however, the evidence for apoptotic signaling pathways contributing to GU cancers is less clear.
Acceleration and deceleration of proliferation: are the rates of change of proliferation rates important?
Although less intensively studied than the GI tract, it is clear that uterine endometrial and ovarian epithelial cancers are susceptible to defective MMR (Table 1). A study recently compared on a large-scale GI epithelial and uterine endometrial cancers as to which MSI susceptible genes contain frameshift mutations (97). This study suggested that the profile of target gene mutations demonstrates both qualitative and quantitative differences between gastrointestinal and endometrial MSI-H cancers. For colorectal and gastric MSI-H tumors, the genes affected were essentially identical. Mutations in the DNA-dependent protein kinase catalytic subunit, Glnac, CCAAT-box binding factor and CtBP-interacting protein coding regions appeared to be specific for endometrial but not GI MSI-H tumors (97). However, they do not clearly establish cell type specific GI-uterine targets that would not be expected to play roles in other tissues as well. In MMR-deficient mouse model experiments, Pten, a gene mutated also in human MMR defective cancers (103), was shown to be an important target whose mutation increases endometrial cancer rates (160). Since PTEN mutations are implicated in many non-MMR spectrum malignancies (such as thyroid), other factors must be included as well to explain why the high penetrance of endometrial cancer in HNPCC, which in women can exceed that of CRC (161,162).
What might explain the susceptibility of uterine endometrium and ovarian epithelium? Recently, it has been proposed that the rate of change of cell proliferation, and not just the rate of proliferation, may play an important role in tumor initiation (163), and both these tissues experience hormonally induced rapid stop/start changes in proliferation rates rather than gradual changes. The endometrium experiences a monthly cycle of estrogen and progesterone responsive abrupt rapid proliferation, followed by abrupt apoptosis and regression. The ovarian epithelium has a similar monthly cycle of hormonally induced rupture to release follicles, and then abrupt rapid proliferation to repair the ruptured epithelium. In summary, these tissues are characterized not only by relatively high proliferation rates but particularly by rapid acceleration and deceleration in the rates of proliferation, which can be characterized by the first derivative of the cell proliferation velocity.
It is noteworthy that stomach and colorectum epithelial cells also are characterized by unusually variable changes in proliferation rate velocity. The classic studies of cell kinetic studies using thymidine labeling of epithelial cells from human stomach, small intestine and colorectum both in vivo (164) and in vitro short term tissue culture (108,165) show similar overall rates of proliferation in human small intestine, but significantly higher changes in proliferation rate, in human stomach or colorectum epithelium compared with the small intestine (108,164,165). Since the penetrance of stomach and colorectal cancer is consistently much higher than small bowel cancer in HNPCC regardless of geography or diet, these studies provide evidence for a potential link between proliferation rate acceleration/deceleration and the cell types most susceptible to MMR defective mechanisms of carcinogenesis. Furthermore, hematopoietic white blood cell and non-melanotic dermal lineages are susceptible to immunological and (in the latter case) sunlight induced abrupt cycles of stop/start rapid proliferation.
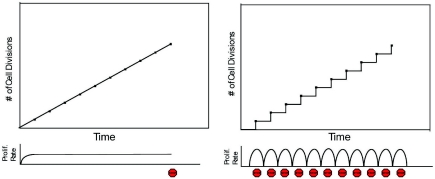
Based on the available data, we propose a model that the hormonally induced cycle of monthly rapid acceleration and deceleration of proliferation rate may play a significant role in uterine endometrial and ovarian epithelial cells to MMR defective cancer susceptibility. If correct, this model would suggest several testable features. First, that endometrial/ovarian cancers rates would be higher with inducible rapid bursts of stop/start proliferation compared to the same number of cell division cycles induced more gradually. Similarly, the model would predict that abruptly blocking endometrial/ovarian epithelial cell hormonal dependent proliferation while controlling for the overall number of cell division cycles would make cells more susceptible to carcinogenesis. Importantly, the model would also predict mechanistically that genes critical for terminating the cell cycle and initiating apoptosis or senescence (stepping on the brakes of proliferation) would be mutated at higher rates in the tissues with rapid bursts of stop/start proliferation compared to those with high proliferation rates but more gradual changes in rates (Figure 1).
Figure 1.
Model of cell proliferation at constant and varying rates of cell division. The left panel shows a conceptual model of cells dividing at a constant rate. The number of cell divisions is plotted on the y-axis and time on the x-axis. The proliferation rate is constant until the tenth cell division, when proliferation stops (as denoted by a Stop sign). The right panel shows a model of cells dividing at varying rates. There are repeated episodes of sharp acceleration and deceleration (as denoted by a Stop sign) of the proliferation rate, starting at time point zero. The proliferation rate is constantly changing and is not constant. The potential selective advantages for growth and increased overall mutations in critical tumor suppressor genes for cells experiencing cycles of abrupt stop/start cell division is discussed in the section ‘ACCELERATION and deceleration of proliferation: are the rates of change of proliferation rates important?’
This model could first be tested in synchronized cell culture directly using MMR defective cell lines, where mutation rates can be quantified. If there is a proliferative advantage for MMR defective cells in an environment of fast acceleration/deceleration, it would also predict that, analogous to experiments using DNA damaging agents (125), MMR second hit mutations would be selected for by this treatment. Also, it could be tested using MMR-deficient mice and tissue specific inducible oncogenes such as Myc that are active only in the presence of specific ligands. The same models could be used to evaluate the affect of rapid acceleration/deceleration of proliferation in tissues with consistently have lower cancer rates in MMR defective individuals such as prostate, breast (60,63,67,117). It is important to note that while hormonally responsive cycles of stop/start proliferation are also seen in breast epithelium, it is distinctive from that seen in the endometrium and ovary. The primary rapid proliferative responses of human breast epithelium are during puberty and pregnancy, not during menses, while the primary hormonally induced response during menses is largely hypertrophic, not hyperproliferative (166,167). If correct, our model would predict in the context of MMR deficiency, pulsatile induction of Myc or other oncogenes would have a much greater affect than continuous Myc expression.
CONCLUSION
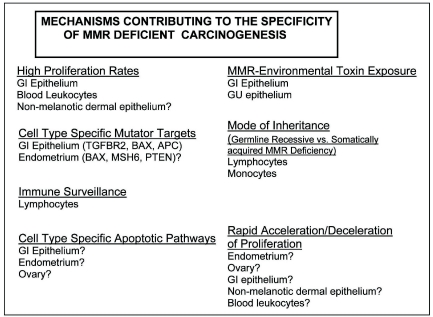
The tissue specificity of cancers that arise from malfunction of ubiquitously expressed processes such as MMR remains a key puzzle in cancer genetics. Based on the accumulated data, we propose there is likely no single mechanism that can explain fully the cell type specificity observed in the spectrum of MMR defective cancers. A more likely explanation is that several distinct mechanisms contribute to shape the observed tissue specificity, and contribute in different ways to different tissues (Figure 2). For the GI epithelium, important facts include particularly rapid proliferation rates, exposure to mutagens from the diet, the dependence on critical genes whose sequences contain short repetitive sequences. For the endometrium, cell type specific targets of the mutator and anti-apoptosis functions of MMR are likely important, but endometrium specific targets need to be more precisely characterized. For the renal tract, concentration of environmental mutagens in the urine are likely important, but these mutagens need to be studied in more detail. For the ovary, cell type specific targets are likely to be important, but they too need to be understood more clearly. For lymphocytes, both the mode of inheritance (Germline Recessive versus Somatically acquired MMR Deficiency) and immune surveillance play important roles. Finally, we propose that not only high proliferation rates, but the rapid acceleration and deceleration of proliferation rates may play a significant role in the specific cell types affected by MMR deficiency, and suggest experiments to test this hypothesis directly.
Figure 2.
Summary of different mechanisms contributing to the specificity of cell types that are susceptible to MMR deficiency.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the American Cancer Society RSG-CCE-02-153-01 and NCI 1R03CA10835.
Conflict of interest statement. None declared.
REFERENCES
- 1.Eisen J.A. A phylogenomic study of the MutS family of proteins. Nucleic Acids Res. 1998;26:4291–4300. doi: 10.1093/nar/26.18.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolodner R.D., Marsischky G.T. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 3.Muller A., Fishel R. Mismatch repair and the hereditary non-polyposis colorectal cancer syndrome (HNPCC) Cancer Invest. 2002;20:102–109. doi: 10.1081/cnv-120000371. [DOI] [PubMed] [Google Scholar]
- 4.Fishel R., Lescoe M.K., Rao M.R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer [Erratum (1994) Cell, 77, 167.] Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 5.Leach F.S., Nicolaides N.C., Papadopoulos N., Liu B., Jen J., Parsons R., Peltomaki P., Sistonen P., Aaltonen L.A., Nystrom-Lahti M., et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 6.Bronner C.E., Baker S.M., Morrison P.T., Warren G., Smith L.G., Lescoe M.K., Kane M., Earabino C., Lipford J., Lindblom A., et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 7.Papadopoulos N., Nicolaides N.C., Wei Y.F., Ruben S.M., Carter K.C., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D., et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel T.A., Erie D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 9.de la Chapelle A. Genetic predisposition to colorectal cancer. Nature Rev. Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 10.Edelmann L., Edelmann W. Loss of DNA mismatch repair function and cancer predisposition in the mouse: animal models for human hereditary nonpolyposis colorectal cancer. Am. J. Med. Genet. C. Semin. Med. Genet. 2004;129:91–99. doi: 10.1002/ajmg.c.30021. [DOI] [PubMed] [Google Scholar]
- 11.Marcon E., Moens P.B. The evolution of meiosis: recruitment and modification of somatic DNA-repair proteins. Bioessays. 2005;27:795–808. doi: 10.1002/bies.20264. [DOI] [PubMed] [Google Scholar]
- 12.Neuberger M.S., Di Noia J.M., Beale R.C., Williams G.T., Yang Z., Rada C. Somatic hypermutation at A.T pairs: polymerase error versus dUTP incorporation. Nature Rev. Immunol. 2005;5:171–178. doi: 10.1038/nri1553. [DOI] [PubMed] [Google Scholar]
- 13.Kunz C., Schar P. Meiotic recombination: sealing the partnership at the junction. Curr. Biol. 2004;14:R962–964. doi: 10.1016/j.cub.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 14.Stojic L., Brun R., Jiricny J. Mismatch repair and DNA damage signaling. DNA Repair (Amst.) 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Kolodner R.D., Putnam C.D., Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 16.Fishel R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: revising the mutator hypothesis. Cancer Res. 2001;61:7369–7374. [PubMed] [Google Scholar]
- 17.Acharya S., Foster P.L., Brooks P., Fishel R. The coordinated functions of the E.coli MutS and MutL proteins in mismatch repair. Mol. Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y., Yuan F., Presnell S.R., Tian K., Gao Y., Tomkinson A.E., Gu L., Li G.M. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 19.Constantin N., Dzantiev L., Kadyrov F.A., Modrich P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 2005;48:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dzantiev L., Constantin N., Genschel J., Iyer R.R., Burgers P.M., Modrich P. A defined human system that supports bidirectional mismatch-provoked excision. Mol. Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y., Berends M.J., Sijmons R.H., Mensink R.G., Verlind E., Kooi K.A., van der Sluis T., Kempinga C., van dDer Zee A.G., Hollema H., et al. A role for MLH3 in hereditary nonpolyposis colorectal cancer. Nature Genet. 2001;29:137–138. doi: 10.1038/ng1001-137. [DOI] [PubMed] [Google Scholar]
- 22.Liu H.X., Zhou X.L., Liu T., Werelius B., Lindmark G., Dahl N., Lindblom A. The role of hMLH3 in familial colorectal cancer. Cancer Res. 2003;63:1894–1899. [PubMed] [Google Scholar]
- 23.Chen P., Dudley S., Hagen W., Dizon D., Paxton L., Reichow D., Yoon S., Yang K., Arnheim N., Liskay M., Lipkin S.M. Contributions by MutL homologs Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005;65:8662–8670. doi: 10.1158/0008-5472.CAN-05-0742. [DOI] [PubMed] [Google Scholar]
- 24.Cannavo E., Marra G., Sabatés-Bellver J., Menigatti M., Lipkin S.M., Fischer S., Cejka P., Jiricny J. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65:10759–10766. doi: 10.1158/0008-5472.CAN-05-2528. [DOI] [PubMed] [Google Scholar]
- 25.Prolla T.A., Baker S.M., Harris A.C., Tsao J.L., Yao X., Bronner C.E., Zheng B., Gordon M., Reneker J., Arnheim N., et al. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nature Genet. 1998;18:276–279. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- 26.Raschle M., Marra G., Nystrom-Lahti M., Schar P., Jiricny J. Identification of hMutLbeta, a heterodimer of hMLH1 and hPMS1. J. Biol. Chem. 1999;274:32368–32375. doi: 10.1074/jbc.274.45.32368. [DOI] [PubMed] [Google Scholar]
- 27.Svetlanov A., Cohen P.E. Mismatch repair proteins, meiosis, and mice: understanding the complexities of mammalian meiosis. Exp. Cell Res. 2004;296:71–79. doi: 10.1016/j.yexcr.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Thibodeau S.N., Bren G., Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 29.Shibata D., Peinado M.A., Ionov Y., Malkhosyan S., Perucho M. Genomic instability in repeated sequences is an early somatic event in colorectal tumorigenesis that persists after transformation. Nature Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- 30.Dietmaier W., Wallinger S., Bocker T., Kullmann F., Fishel R., Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 31.Umar A., Boland C.R., Terdiman J.P., Syngal S., de la Chapelle A., Ruschoff J., Fishel R., Lindor N.M., Burgart L.J., Hamelin R., et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu T., Tannergard P., Hackman P., Rubio C., Kressner U., Lindmark G., Hellgren D., Lambert B., Lindblom A. Missense mutations in hMLH1 associated with colorectal cancer. Hum. Genet. 1999;105:437–441. doi: 10.1007/s004390051127. [DOI] [PubMed] [Google Scholar]
- 33.Lipkin S.M., Rozek L.S., Rennert G., Yang W., Chen P.C., Hacia J., Hunt N., Shin B., Fodor S., Kokoris M., et al. The MLH1 D132H variant is associated with susceptibility to sporadic colorectal cancer. Nature Genet. 2004;36:694–699. doi: 10.1038/ng1374. [DOI] [PubMed] [Google Scholar]
- 34.Coolbaugh-Murphy M., Maleki A., Ramagli L., Frazier M., Lichtiger B., Monckton D.G., Siciliano M.J., Brown B.W. Estimating mutant microsatellite allele frequencies in somatic cells by small-pool PCR. Genomics. 2004;84:419–430. doi: 10.1016/j.ygeno.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Alazzouzi H., Domingo E., Gonzalez S., Blanco I., Armengol M., Espin E., Plaja A., Schwartz S., Capella G., Schwartz S., Jr Low levels of microsatellite instability characterize MLH1 and MSH2 HNPCC carriers before tumor diagnosis. Hum. Mol. Genet. 2005;14:235–239. doi: 10.1093/hmg/ddi021. [DOI] [PubMed] [Google Scholar]
- 36.Li G.M. The role of mismatch repair in DNA damage-induced apoptosis. Oncol Res. 1999;11:393–400. [PubMed] [Google Scholar]
- 37.Lin D.P., Wang Y., Scherer S.J., Clark A.B., Yang K., Avdievich E., Jin B., Werling U., Parris T., Kurihara N., et al. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;64:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- 38.Yang G., Scherer S.J., Shell S.S., Yang K., Kim M., Lipkin M., Kucherlapati R., Kolodner R.D., Edelmann W. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 39.de Wind N., Dekker M., Berns A., Radman M., te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 40.Fishel R. Signaling mismatch repair in cancer. Nature Med. 1999;5:1239–1241. doi: 10.1038/15191. [DOI] [PubMed] [Google Scholar]
- 41.Gong J.G., Costanzo A., Yang H.Q., Melino G., Kaelin W.G., Jr, Levrero M., Wang J.Y. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 42.Zabkiewicz J., Clarke A.R. DNA damage-induced apoptosis: insights from the mouse. Biochim. Biophys. Acta. 2004;1705:17–25. doi: 10.1016/j.bbcan.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 43.de Wind N., Dekker M., Claij N., Jansen L., van Klink L., Radman M., Riggins G., van der Valk M., van't Wout M., te Riele H. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nature Genet. 1999;23:359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- 44.Akiyama Y., Sato H., Yamada T., Nagasaki H., Tsuchiya A., Abe R., Yuasa Y. Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res. 1997;57:3920–3923. [PubMed] [Google Scholar]
- 45.Nicolaides N.C., Carter K.C., Shell B.K., Papadopoulos N., Vogelstein B., Kinzler K.W. Genomic organization of the human PMS2 gene family. Genomics. 1995;30:195–206. doi: 10.1006/geno.1995.9885. [DOI] [PubMed] [Google Scholar]
- 46.Gryfe R., Kim H., Hsieh E.T., Aronson M.D., Holowaty E.J., Bull S.B., Redston M., Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N. Engl. J. Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 47.Gryfe R., Gallinger S. Microsatellite instability, mismatch repair deficiency, and colorectal cancer. Surgery. 2001;130:17–20. doi: 10.1067/msy.2001.112738. [DOI] [PubMed] [Google Scholar]
- 48.Lynch H.T. Inherited susceptibility to cancer: clinical, predictive and ethical perspectives [In Process Citation] Gut. 1999;44:765B–7765. doi: 10.1136/bmj.318.7197.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolodner R.D.e.a. Germline MSH6 mutation in colorectal cancer families. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
- 50.Goodfellow P.J., Buttin B.M., Herzog T.J., Rader J.S., Gibb R.K., Swisher E., Look K., Walls K.C., Fan M.Y., Mutch D.G. Prevalence of defective DNA mismatch repair and MSH6 mutation in an unselected series of endometrial cancers. Proc. Natl Acad. Sci. USA. 2003;100:5908–5913. doi: 10.1073/pnas.1030231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ollikainen M., Abdel-Rahman W.M., Moisio A.L., Lindroos A., Kariola R., Jarvela I., Poyhonen M., Butzow R., Peltomaki P. Molecular analysis of familial endometrial carcinoma: a manifestation of hereditary nonpolyposis colorectal cancer or a separate syndrome? J. Clin. Oncol. 2005;23:4609–4616. doi: 10.1200/JCO.2005.06.055. [DOI] [PubMed] [Google Scholar]
- 52.Wang Q., Lasset C., Desseigne F., Frappaz D., Bergeron C., Navarro C., Ruano E., Puisieux A. Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res. 1999;59:294–297. [PubMed] [Google Scholar]
- 53.Ricciardone M.D., Ozcelik T., Cevher B., Ozdag H., Tuncer M., Gurgey A., Uzunalimoglu O., Cetinkaya H., Tanyeli A., Erken E., et al. Human MLH1 deficiency predisposes to hematological malignancy and neurofibromatosis type 1. Cancer Res. 1999;59:290–293. [PubMed] [Google Scholar]
- 54.Vilkki S., Tsao J.L., Loukola A., Poyhonen M., Vierimaa O., Herva R., Aaltonen L.A., Shibata D. Extensive somatic microsatellite mutations in normal human tissue. Cancer Res. 2001;61:4541–4544. [PubMed] [Google Scholar]
- 55.Whiteside D., McLeod R., Graham G., Steckley J.L., Booth K., Somerville M.J., Andrew S.E. A homozygous germ-line mutation in the human MSH2 gene predisposes to hematological malignancy and multiple cafe-au-lait spots. Cancer Res. 2002;62:359–362. [PubMed] [Google Scholar]
- 56.Gallinger S., Aronson M., Shayan K., Ratcliffe E.M., Gerstle J.T., Parkin P.C., Rothenmund H., Croitoru M., Baumann E., Durie P.R., et al. Gastrointestinal cancers and neurofibromatosis type 1 features in children with a germline homozygous MLH1 mutation. Gastroenterology. 2004;126:576–585. doi: 10.1053/j.gastro.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 57.Aarnio M., Sankila R., Pukkala E., Salovaara R., Aaltonen L.A., de la Chapelle A., Peltomaki P., Mecklin J.P., Jarvinen H.J. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int. J. Cancer. 1999;81:214–218. doi: 10.1002/(sici)1097-0215(19990412)81:2<214::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 58.Vasen H.F., Wijnen J.T., Menko F.H., Kleibeuker J.H., Taal B.G., Griffioen G., Nagengast F.M., Meijers-Heijboer E.H., Bertario L., Varesco L., et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology. 1996;110:1020–1027. doi: 10.1053/gast.1996.v110.pm8612988. [DOI] [PubMed] [Google Scholar]
- 59.Dunlop M.G., Farrington S.M., Carothers A.D., Wyllie A.H., Sharp L., Burn J., Liu B., Kinzler K.W., Vogelstein B. Cancer risk associated with germline DNA mismatch repair gene mutations. Hum. Mol. Genet. 1997;6:105–110. doi: 10.1093/hmg/6.1.105. [DOI] [PubMed] [Google Scholar]
- 60.Lindor N.M., Rabe K., Petersen G.M., Haile R., Casey G., Baron J., Gallinger S., Bapat B., Aronson M., Hopper J., et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA. 2005;293:1979–1985. doi: 10.1001/jama.293.16.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watson P., Vasen H.F., Mecklin J.P., Jarvinen H., Lynch H.T. The risk of endometrial cancer in hereditary nonpolyposis colorectal cancer. Am. J. Med. 1994;96:516–520. doi: 10.1016/0002-9343(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 62.Watson P., Lynch H.T. Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer. 1993;71:677–685. doi: 10.1002/1097-0142(19930201)71:3<677::aid-cncr2820710305>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 63.Watson P., Lynch H.T. The tumor spectrum in HNPCC. AntiCancer Res. 1994;14:1635–1639. [PubMed] [Google Scholar]
- 64.Pinol V., Castells A., Andreu M., Castellvi-Bel S., Alenda C., Llor X., Xicola R.M., Rodriguez-Moranta F., Paya A., Jover R., et al. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 65.Stormorken A.T., Bowitz-Lothe I.M., Noren T., Kure E., Aase S., Wijnen J., Apold J., Heimdal K., Moller P. Immunohistochemistry identifies carriers of mismatch repair gene defects causing hereditary nonpolyposis colorectal cancer. J. Clin. Oncol. 2005;23:4705–4712. doi: 10.1200/JCO.2005.05.180. [DOI] [PubMed] [Google Scholar]
- 66.Hampel H., Frankel W.L., Martin E., Arnold M., Khanduja K., Kuebler P., Nakagawa H., Sotamaa K., Prior T.W., Westman J., et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N. Engl. J. Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 67.Watson P., Lynch H.T. Cancer risk in mismatch repair gene mutation carriers. Fam. Cancer. 2001;1:57–60. doi: 10.1023/a:1011590617833. [DOI] [PubMed] [Google Scholar]
- 68.Watson P., Ashwathnarayan R., Lynch H.T., Roy H.K. Tobacco use and increased colorectal cancer risk in patients with hereditary nonpolyposis colorectal cancer (Lynch syndrome) Arch. Intern. Med. 2004;164:2429–2431. doi: 10.1001/archinte.164.22.2429. [DOI] [PubMed] [Google Scholar]
- 69.Baker S.M., Bronner C.E., Zhang L., Plug A.W., Robatzek M., Warren G., Elliott E.A., Yu J., Ashley T., Arnheim N., et al. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 70.Baker S.M., Plug A.W., Prolla T.A., Bronner C.E., Harris A.C., Yao X., Christie D.M., Monell C., Arnheim N., Bradley A., et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nature Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 71.de Wind N., Dekker M., Claij N., Jansen L., van Klink Y., Radman M., Riggins G., van der Valk M., van't Wout K., te Riele H. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nature Genet. 1999;23:359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- 72.Edelmann W., Cohen P., Kane P., Lau K., Morrow B., Bennett S., Umar A., Kunkel T., Cattoretti G., Chaganti R., et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 73.Edelmann W., Cohen P.E., Kneitz B., Winand N., Lia M., Heyer J., Kolodner R., Pollard J.W., Kucherlapati R. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nature Genet. 1999;21:123–127. doi: 10.1038/5075. [DOI] [PubMed] [Google Scholar]
- 74.Edelmann W., Yang K., Umar A., Heyer J., Lau K., Fan K., Liedtke W., Cohen P.E., Kane M.F., Lipford J.R., et al. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- 75.Kneitz B., Cohen P.E., Avdievich E., Zhu L., Kane M.F., Hou H., Jr, Kolodner R.D., Kucherlapati R., Pollard J.W., Edelmann W. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 76.Reitmair A.H., Schmits R., Ewel A., Bapat B., Redston M., Mitri A., Waterhouse P., Mittrucker H.W., Wakeham A., Liu B. MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nature Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- 77.Lipkin S.M., Wang V., Jacoby R., Banerjee-Basu S., Baxevanis A.D., Lynch H.T., Elliott R.M., Collins F.S. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nature Genet. 2000;24:27–35. doi: 10.1038/71643. [DOI] [PubMed] [Google Scholar]
- 78.Heyer J., Yang K., Lipkin M., Edelmann W., Kucherlapati R. Mouse models for colorectal cancer. Oncogene. 1999;18:5325–5333. doi: 10.1038/sj.onc.1203036. [DOI] [PubMed] [Google Scholar]
- 79.Yao X., Buermeyer A.B., Narayanan L., Tran D., Baker S.M., Prolla T.A., Glazer P.M., Liskay R.M., Arnheim N. Different mutator phenotypes in Mlh1- versus Pms2-deficient mice. Proc. Natl Acad. Sci. USA. 1999;96:6850–6855. doi: 10.1073/pnas.96.12.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Narayanan L., Fritzell J.A., Baker S.M., Liskay R.M., Glazer P.M. Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene Pms2. Proc. Natl Acad. Sci. USA. 1997;94:3122–3127. doi: 10.1073/pnas.94.7.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edelmann W., Umar A., Yang K., Heyer J., Kucherlapati M., Lia M., Kneitz B., Avdievich E., Fan K., Wong E., et al. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer Res. 2000;60:803–807. [PubMed] [Google Scholar]
- 82.Shmueli O., Horn-Saban S., Chalifa-Caspi V., Shmoish M., Ophir R., Benjamin-Rodrig H., Safran M., Domany E., Lancet D. GeneNote: whole genome expression profiles in normal human tissues. C R Biol. 2003;326:1067–1072. doi: 10.1016/j.crvi.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 83.Okumura M., Kondo S., Ogata M., Kanemoto S., Murakami T., Yanagida K., Saito A., Imaizumi K. Candidates for tumor-specific alternative splicing. Biochem. Biophys. Res. Commun. 2005;334:23–29. doi: 10.1016/j.bbrc.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 84.Vogelstein B., Kinzler K.W. Cancer genes and the pathways they control. Nature Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 85.Markowitz S., Wang J., Myeroff L., Parsons R., Sun L., Lutterbaugh J., Fan R.S., Zborowska E., Kinzler K.W., Vogelstein B., et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 86.Woerner S.M., Gebert J., Yuan Y.P., Sutter C., Ridder R., Bork P., von Knebel Doeberitz M. Systematic identification of genes with coding microsatellites mutated in DNA mismatch repair-deficient cancer cells. Int. J. Cancer. 2001;93:12–19. doi: 10.1002/ijc.1299. [DOI] [PubMed] [Google Scholar]
- 87.Park J., Betel D., Gryfe R., Michalickova K., Di Nicola N., Gallinger S., Hogue C.W., Redston M. Mutation profiling of mismatch repair-deficient colorectal cncers using an in silico genome scan to identify coding microsatellites. Cancer Res. 2002;62:1284–1288. [PubMed] [Google Scholar]
- 88.Woerner S.M., Kloor M., Mueller A., Rueschoff J., Friedrichs N., Buettner R., Buzello M., Kienle P., Knaebel H.P., Kunstmann E., et al. Microsatellite instability of selective target genes in HNPCC-associated colon adenomas. Oncogene. 2005;24:2525–2535. doi: 10.1038/sj.onc.1208456. [DOI] [PubMed] [Google Scholar]
- 89.Ionov Y., Nowak N., Perucho M., Markowitz S., Cowell J.K. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene. 2004;23:639–645. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- 90.El-Bchiri J., Buhard O., Penard-Lacronique V., Thomas G., Hamelin R., Duval A. Differential nonsense mediated decay of mutated mRNAs in mismatch repair deficient colorectal cancers. Hum. Mol. Genet. 2005;14:2435–2442. doi: 10.1093/hmg/ddi245. [DOI] [PubMed] [Google Scholar]
- 91.Kuraguchi M., Edelmann W., Yang K., Lipkin M., Kucherlapati R., Brown A.M. Tumor-associated Apc mutations in Mlh1−/− Apc1638N mice reveal a mutational signature of Mlh1 deficiency. Oncogene. 2000;19:5755–5763. doi: 10.1038/sj.onc.1203962. [DOI] [PubMed] [Google Scholar]
- 92.Kuraguchi M., Yang K., Wong E., Avdievich E., Fan K., Kolodner R.D., Lipkin M., Brown A.M., Kucherlapati R., Edelmann W. The distinct spectra of tumor-associated Apc mutations in mismatch repair-deficient Apc1638N mice define the roles of MSH3 and MSH6 in DNA repair and intestinal tumorigenesis. Cancer Res. 2001;61:7934–7942. [PubMed] [Google Scholar]
- 93.Edelmann W., Yang K., Kuraguchi M., Heyer J., Lia M., Kneitz B., Fan K., Brown A.M., Lipkin M., Kucherlapati R. Tumorigenesis in Mlh1 and Mlh1/Apc1638N mutant mice. Cancer Res. 1999;59:1301–1307. [PubMed] [Google Scholar]
- 94.Huang J., Papadopoulos N., McKinley A.J., Farrington S.M., Curtis L.J., Wyllie A.H., Zheng S., Willson J.K., Markowitz S.D., Morin P., et al. APC mutations in colorectal tumors with mismatch repair deficiency. Proc. Natl Acad. Sci. USA. 1996;93:9049–9054. doi: 10.1073/pnas.93.17.9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schwartz S., Jr, Yamamoto H., Navarro M., Maestro M., Reventos J., Perucho M. Frameshift mutations at mononucleotide repeats in caspase-5 and other target genes in endometrial and gastrointestinal cancer of the microsatellite mutator phenotype. Cancer Res. 1999;59:2995–3002. [PubMed] [Google Scholar]
- 96.Yamamoto H., Gil J., Schwartz S., Jr, Perucho M. Frameshift mutations in Fas, Apaf-1, and Bcl-10 in gastro-intestinal cancer of the microsatellite mutator phenotype. Cell Death Differ. 2000;7:238–239. doi: 10.1038/sj.cdd.4400651. [DOI] [PubMed] [Google Scholar]
- 97.Duval A., Reperant M., Compoint A., Seruca R., Ranzani G.N., Iacopetta B., Hamelin R. Target gene mutation profile differs between gastrointestinal and endometrial tumors with mismatch repair deficiency. Cancer Res. 2002;62:1609–1612. [PubMed] [Google Scholar]
- 98.Souza R.F., Appel R., Yin J., Wang S., Smolinski K.N., Abraham J.M., Zou T.T., Shi Y.Q., Lei J., Cottrell J., et al. Microsatellite instability in the insulin-like growth factor II receptor gene in gastrointestinal tumours [letter] [Erratum (1996) Nature Genet., 14, 488.] Nature Genet. 1996;14:255–257. doi: 10.1038/ng1196-255. [DOI] [PubMed] [Google Scholar]
- 99.Rampino N., Yamamoto H., Ionov Y., Li Y., Sawai H., Reed J.C., Perucho M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 100.Baranovskaya S., Soto J.L., Perucho M., Malkhosyan S.R. Functional significance of concomitant inactivation of hMLH1 and hMSH6 in tumor cells of the microsatellite mutator phenotype. Proc. Natl Acad. Sci. USA. 2001;98:15107–15112. doi: 10.1073/pnas.251234498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Malkhosyan S., Rampino N., Yamamoto H., Perucho M. Frameshift mutator mutations. Nature. 1996;382:499–500. doi: 10.1038/382499a0. [DOI] [PubMed] [Google Scholar]
- 102.Woerner S.M., Benner A., Sutter C., Schiller M., Yuan Y.P., Keller G., Bork P., Doeberitz M.K., Gebert J.F. Pathogenesis of DNA repair-deficient cancers: a statistical meta-analysis of putative real common target genes. Oncogene. 2003;22:2226–2235. doi: 10.1038/sj.onc.1206421. [DOI] [PubMed] [Google Scholar]
- 103.Duval A., Rolland S., Compoint A., Tubacher E., Iacopetta B., Thomas G., Hamelin R. Evolution of instability at coding and non-coding repeat sequences in human MSI-H colorectal cancers. Hum. Mol. Genet. 2001;10:513–518. doi: 10.1093/hmg/10.5.513. [DOI] [PubMed] [Google Scholar]
- 104.Duval A., Reperant M., Hamelin R. Comparative analysis of mutation frequency of coding and non coding short mononucleotide repeats in mismatch repair deficient colorectal cancers. Oncogene. 2002;21:8062–8066. doi: 10.1038/sj.onc.1206013. [DOI] [PubMed] [Google Scholar]
- 105.Duval A., Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62:2447–2454. [PubMed] [Google Scholar]
- 106.Knudson A. Hereditary predisposition to cancer. Ann. N Y Acad. Sci. 1997;29:58–67. doi: 10.1111/j.1749-6632.1997.tb48593.x. [DOI] [PubMed] [Google Scholar]
- 107.Mao E.F., Lane L., Lee J., Miller J.H. Proliferation of mutators in A cell population. J. Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lipkin M. Proliferation and differentiation of gastrointestinal cells. Physiol. Rev. 1973;53:891–910. doi: 10.1152/physrev.1973.53.4.891. [DOI] [PubMed] [Google Scholar]
- 109.Peto R. Epidemiology, multistage models and short-term mutagenicity tests. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1977. [DOI] [PubMed] [Google Scholar]
- 110.Tomlinson I.P., Bodmer W.F. Failure of programmed cell death and differentiation as causes of tumors: some simple mathematical models. Proc. Natl Acad. Sci. USA. 1995;92:11130–11134. doi: 10.1073/pnas.92.24.11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cairns J. Mutation and cancer: the antecedents to our studies of adaptive mutation. Genetics. 1998;148:1433–1440. doi: 10.1093/genetics/148.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee S.C., Guo J.Y., Lim R., Soo R., Koay E., Salto-Tellez M., Leong A., Goh B.C. Clinical and molecular characteristics of hereditary non-polyposis colorectal cancer families in Southeast Asia. Clin. Genet. 2005;68:137–145. doi: 10.1111/j.1399-0004.2005.00469.x. [DOI] [PubMed] [Google Scholar]
- 113.Liu S.R., Zhao B., Wang Z.J., Wan Y.L., Huang Y.T. Clinical features and mismatch repair gene mutation screening in Chinese patients with hereditary nonpolyposis colorectal carcinoma. World J. Gastroenterol. 2004;10:2647–2651. doi: 10.3748/wjg.v10.i18.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Y.Z., Sheng J.Q., Li S.R., Zhang H. Clinical phenotype and prevalence of hereditary nonpolyposis colorectal cancer syndrome in Chinese population. World J. Gastroenterol. 2005;11:1481–1488. doi: 10.3748/wjg.v11.i10.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lynch H.T., de la Chapelle A. Hereditary colorectal cancer. N. Engl. J. Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 116.Neugut A.I., Terry M.B. Cigarette smoking and microsatellite instability: causal pathway or marker-defined subset of colon tumors? J. Natl Cancer Inst. 2000;92:1791–1793. doi: 10.1093/jnci/92.22.1791. [DOI] [PubMed] [Google Scholar]
- 117.Watson P., Lynch H.T. Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer. 1993;71:677–685. doi: 10.1002/1097-0142(19930201)71:3<677::aid-cncr2820710305>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 118.Lynch H.T., Watson P., Lanspa S., Marcus J., Smyrk T., Fitzgibbons R., Jr, Cristofaro G., Kriegler M., Lynch J. Clinical nuances of Lynch syndromes I and II. Prog. Clin. Biol. Res. 1988;279:177–188. [PubMed] [Google Scholar]
- 119.Lynch H.T., Watson P., Lanspa S.J., Marcus J., Smyrk T., Fitzgibbons R.J., Jr, Kriegler M., Lynch J.F. Natural history of colorectal cancer in hereditary nonpolyposis colorectal cancer (Lynch syndromes I and II) Dis. Colon Rectum. 1988;31:439–444. doi: 10.1007/BF02552613. [DOI] [PubMed] [Google Scholar]
- 120.Aarnio M., Sankila R., Pukkala E., Salovaara R., Aaltonen L.A., de la Chapelle A., Peltomaki P., Mecklin J.P., Jarvinen H.J. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int. J. Cancer. 1999;81:214–218. doi: 10.1002/(sici)1097-0215(19990412)81:2<214::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 121.Lynch H.T., Smyrk T.C. Identifying hereditary nonpolyposis colorectal cancer. N. Engl. J. Med. 1998;338:1537–1538. doi: 10.1056/NEJM199805213382109. [DOI] [PubMed] [Google Scholar]
- 122.Lynch H.T., Smyrk T., Lynch J.F. Molecular genetics and clinical-pathology features of hereditary nonpolyposis colorectal carcinoma (Lynch syndrome): historical journey from pedigree anecdote to molecular genetic confirmation. Oncology. 1998;55:103–108. doi: 10.1159/000011843. [DOI] [PubMed] [Google Scholar]
- 123.Douglas J.A., Gruber S.B., Meister K.A., Bonner J., Watson P., Krush A.J., Lynch H.T. History and molecular genetics of Lynch syndrome in family G: a century later. JAMA. 2005;294:2195–2202. doi: 10.1001/jama.294.17.2195. [DOI] [PubMed] [Google Scholar]
- 124.Reddy B.S. The Fourth DeWitt S. Goodman lecture. Novel approaches to the prevention of colon cancer by nutritional manipulation and chemoprevention. Cancer Epidemiol. Biomarkers Prev. 2000;9:239–247. [PubMed] [Google Scholar]
- 125.Borgdorff V., van Hees-Stuivenberg S., Meijers C.M., de Wind N. Spontaneous and mutagen-induced loss of DNA mismatch repair in Msh2-heterozygous mammalian cells. Mutat. Res. 2005;574:50–57. doi: 10.1016/j.mrfmmm.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 126.Lamprecht S.A., Lipkin M. Chemoprevention of colon cancer by calcium, vitamin D and folate: molecular mechanisms. Nature Rev. Cancer. 2003;3:601–614. doi: 10.1038/nrc1144. [DOI] [PubMed] [Google Scholar]
- 127.Yang K., Fan K., Kurihara N., Shinozaki H., Rigas B., Augenlicht L., Kopelovich L., Edelmann W., Kucherlapati R., Lipkin M. Regional response leading to tumorigenesis after sulindac in small and large intestine of mice with Apc mutations. Carcinogenesis. 2003;24:605–611. doi: 10.1093/carcin/24.3.605. [DOI] [PubMed] [Google Scholar]
- 128.Yang K., Fan K.H., Lamprecht S.A., Edelmann W., Kopelovich L., Kucherlapati R., Lipkin M. Peroxisome proliferator-activated receptor gamma agonist troglitazone induces colon tumors in normal C57BL/6J mice and enhances colonic carcinogenesis in Apc(1638 N/+) Mlh1(+/−) double mutant mice. Int. J. Cancer. 2005;116:495–499. doi: 10.1002/ijc.21018. [DOI] [PubMed] [Google Scholar]
- 129.Annie Yu H.J., Lin K.M., Ota D.M., Lynch H.T. Hereditary nonpolyposis colorectal cancer: preventive management. Cancer Treat Rev. 2003;29:461–470. doi: 10.1016/s0305-7372(03)00084-7. [DOI] [PubMed] [Google Scholar]
- 130.Velasquez J.L., Lipkin S.M. What Are SNPs and haplotypes and how will they help us manage the prevention of adult cancer? Curr. Oncol. Rep. 2005;7:475–479. doi: 10.1007/s11912-005-0013-1. [DOI] [PubMed] [Google Scholar]
- 131.Chawengsaksophak K., James R., Hammond V.E., Kontgen F., Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- 132.Hinoi T., Tani M., Lucas P.C., Caca K., Dunn R.L., Macri E., Loda M., Appelman H.D., Cho K.R., Fearon E.R. Loss of CDX2 expression and microsatellite instability are prominent features of large cell minimally differentiated carcinomas of the colon. Am. J. Pathol. 2001;159:2239–2248. doi: 10.1016/S0002-9440(10)63074-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rozek L.S., Lipkin S.M., Fearon E.R., Hanash S., Giordano T.J., Greenson J.K., Kuick R., Misek D.E., Taylor J.M., Douglas J.A., et al. CDX2 polymorphisms, RNA expression, and risk of colorectal cancer. Cancer Res. 2005;65:5488–5492. doi: 10.1158/0008-5472.CAN-04-3645. [DOI] [PubMed] [Google Scholar]
- 134.Lynch H.T., Smyrk T.C. Identifying hereditary nonpolyposis colorectal cancer. New Engl. J. Med. 1998;338:1537–1538. doi: 10.1056/NEJM199805213382109. [DOI] [PubMed] [Google Scholar]
- 135.Jin Y.H., Clark A.B., Slebos R.J., Al-Refai H., Taylor J.A., Kunkel T.A., Resnick M.A., Gordenin D.A. Cadmium is a mutagen that acts by inhibiting mismatch repair. Nature Genet. 2003;34:326–329. doi: 10.1038/ng1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kolonel L.N. Association of cadmium with renal cancer. Cancer. 1976;37:1782–1787. doi: 10.1002/1097-0142(197604)37:4<1782::aid-cncr2820370424>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 137.Pesch B., Haerting J., Ranft U., Klimpel A., Oelschlagel B., Schill W. Occupational risk factors for renal cell carcinoma: agent-specific results from a case-control study in Germany. MURC Study Group. Multicenter urothelial and renal cancer study. Int. J. Epidemiol. 2000;29:1014–1024. doi: 10.1093/ije/29.6.1014. [DOI] [PubMed] [Google Scholar]
- 138.Ostergaard J., Sunde L., Okkels H. Neurofibromatosis von Recklinghausen type I phenotype and early onset of cancers in siblings compound heterozygous for mutations in MSH6. Am. J. Med. Genet. 2005;139:96–105. doi: 10.1002/ajmg.a.30998. [DOI] [PubMed] [Google Scholar]
- 139.Gutmann D.H., Winkeler E., Kabbarah O., Hedrick N., Dudley S., Goodfellow P.J., Liskay R.M. Mlh1 deficiency accelerates myeloid leukemogenesis in neurofibromatosis 1 (Nf1) heterozygous mice. Oncogene. 2003;22:4581–4585. doi: 10.1038/sj.onc.1206768. [DOI] [PubMed] [Google Scholar]
- 140.Tuveson D.A., Jacks T. Technologically advanced cancer modeling in mice. Curr. Opin. Genet. Dev. 2002;12:105–110. doi: 10.1016/s0959-437x(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 141.Umar A., Risinger J.I., Hawk E.T., Barrett J.C. Testing guidelines for hereditary non-polyposis colorectal cancer. Nature Rev. Cancer. 2004;4:153–158. doi: 10.1038/nrc1278. [DOI] [PubMed] [Google Scholar]
- 142.Duval A., Raphael M., Brennetot C., Poirel H., Buhard O., Aubry A., Martin A., Krimi A., Leblond V., Gabarre J., et al. The mutator pathway is a feature of immunodeficiency-related lymphomas. PNAS. 2004;101:5002–5007. doi: 10.1073/pnas.0400945101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Campbell M.R., Nation P.N., Andrew S.E. A lack of DNA mismatch repair on an athymic murine background predisposes to hematologic malignancy. Cancer Res. 2005;65:2626–2635. doi: 10.1158/0008-5472.CAN-04-3158. [DOI] [PubMed] [Google Scholar]
- 144.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 145.Zhang H., Richards B., Wilson T., Lloyd M., Cranston A., Thorburn A., Fishel R., Meuth M. Apoptosis induced by overexpression of hMSH2 or hMLH1. Cancer Res. 1999;59:3021–3027. [PubMed] [Google Scholar]
- 146.Hickman M.J., Samson L.D. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc. Natl Acad. Sci. USA. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Frank S.A., Nowak M.A. Problems of somatic mutation and cancer. Bioessays. 2004;26:291–299. doi: 10.1002/bies.20000. [DOI] [PubMed] [Google Scholar]
- 148.Michor F., Frank S.A., May R.M., Iwasa Y., Nowak M.A. Somatic selection for and against cancer. J. Theor. Biol. 2003;225:377–382. doi: 10.1016/s0022-5193(03)00267-4. [DOI] [PubMed] [Google Scholar]
- 149.Yang E., Korsmeyer S.J. Molecular thanatopsis: a discourse on the BCL2 family and cell death. Blood. 1996;88:386–401. [PubMed] [Google Scholar]
- 150.Reed J.C., Doctor K.S., Godzik A. The domains of apoptosis: a genomics perspective. Science STKE. 2004;239:re9. doi: 10.1126/stke.2392004re9. [DOI] [PubMed] [Google Scholar]
- 151.Halappanavar S.S., Rhun Y.L., Mounir S., Martins L.M., Huot J., Earnshaw W.C., Shah G.M. Survival and proliferation of cells expressing caspase-uncleavable Poly(ADP-ribose) polymerase in response to death-inducing DNA damage by an alkylating agent. J. Biol. Chem. 1999;274:37097–37104. doi: 10.1074/jbc.274.52.37097. [DOI] [PubMed] [Google Scholar]
- 152.Michor F., Hughes T.P., Iwasa Y., Branford S., Shah N.P., Sawyers C.L., Nowak M.A. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–1270. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 153.Mertens H.J., Heineman M.J., Evers J.L. The expression of apoptosis-related proteins Bcl-2 and Ki67 in endometrium of ovulatory menstrual cycles. Gynecol. Obstet. Invest. 2002;53:224–230. doi: 10.1159/000064569. [DOI] [PubMed] [Google Scholar]
- 154.Otsuki Y. Apoptosis in human endometrium: apoptotic detection methods and signaling. Med. Electron. Microsc. 2001;34:166–173. doi: 10.1007/s007950100011. [DOI] [PubMed] [Google Scholar]
- 155.Vaskivuo T.E., Tapanainen J.S. Apoptosis in the human ovary. Reprod. Biomed. Online. 2003;6:24–35. doi: 10.1016/s1472-6483(10)62052-4. [DOI] [PubMed] [Google Scholar]
- 156.Nnene I.O., Nieto J.J., Crow J.C., Sundaresan M., MacLean A.B., Perrett C.W., Hardiman P. Cell cycle and apoptotic proteins in relation to ovarian epithelial morphology. Gynecol Oncol. 2004;92:247–251. doi: 10.1016/j.ygyno.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 157.Sugino N., Suzuki T., Kashida S., Karube A., Takiguchi S., Kato H. Expression of Bcl-2 and Bax in the human corpus luteum during the menstrual cycle and in early pregnancy: regulation by human chorionic gonadotropin. J. Clin. Endocrinol. Metab. 2000;85:4379–4386. doi: 10.1210/jcem.85.11.6944. [DOI] [PubMed] [Google Scholar]
- 158.Shikone T., Yamoto M., Kokawa K., Yamashita K., Nishimori K., Nakano R. Apoptosis of human corpora lutea during cyclic luteal regression and early pregnancy. J. Clin. Endocrinol. Metab. 1996;81:2376–2380. doi: 10.1210/jcem.81.6.8964880. [DOI] [PubMed] [Google Scholar]
- 159.King E.D., Matteson J., Jacobs S.C., Kyprianou N. Incidence of apoptosis, cell proliferation and bcl-2 expression in transitional cell carcinoma of the bladder: association with tumor progression. J. Urol. 1996;155:316–320. [PubMed] [Google Scholar]
- 160.Wang H., Douglas W., Lia M., Edelmann W., Kucherlapati R., Podsypanina K., Parsons R., Ellenson L.H. DNA mismatch repair deficiency accelerates endometrial tumorigenesis in Pten heterozygous mice. Am. J. Pathol. 2002;160:1481–1486. doi: 10.1016/S0002-9440(10)62573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wijnen J., de Leeuw W., Vasen H., van der Klift H., Moller P., Stormorken A., Meijers-Heijboer H., Lindhout D., Menko F., Vossen S., et al. Familial endometrial cancer in female carriers of MSH6 germline mutations. Nature Genet. 1999;23:142–144. doi: 10.1038/13773. [DOI] [PubMed] [Google Scholar]
- 162.Wijnen J.T., Vasen H.F., Khan P.M., Zwinderman A.H., van der Klift H., Mulder A., Tops C., Moller P., Fodde R. Clinical findings with implications for genetic testing in families with clustering of colorectal cancer. N. Engl. J. Med. 1998;339:511–518. doi: 10.1056/NEJM199808203390804. [DOI] [PubMed] [Google Scholar]
- 163.Frank S.A. Genetic predisposition to cancer - insights from population genetics. Nature Rev. Genet. 2004;5:764–772. doi: 10.1038/nrg1450. [DOI] [PubMed] [Google Scholar]
- 164.Lipkin M., Sherlock P., Bell B. Generation time of epithelial cells in the human colon. Nature. 1962;195:175–177. doi: 10.1038/195175b0. [DOI] [PubMed] [Google Scholar]
- 165.Deschner E., Lewis C., Lipkin M. In vitro study of human rectal epithelial cells. Atypical zoe of thymidine incorporation in mucosa of multiple polyposis. J. Clin. Invest. 1963;42:1922–1928. doi: 10.1172/JCI104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Scully R., Puget N. Hereditary breast and ovarian cancer genes. Methods Mol. Biol. 2003;222:41–57. doi: 10.1385/1-59259-328-3:041. [DOI] [PubMed] [Google Scholar]
- 167.Rosfjord E.C., Dickson R.B. Growth factors, apoptosis, and survival of mammary epithelial cells. J. Mammary Gland Biol. Neoplasia. 1999;4:229–237. doi: 10.1023/a:1018789527533. [DOI] [PubMed] [Google Scholar]