Abstract
Tertiary interactions are crucial in maintaining the tRNA structure and functionality. We used a combined sequence analysis and quantum mechanics approach to calculate accurate energies of the most frequent tRNA tertiary base pairing interactions. Our analysis indicates that six out of the nine classical tertiary interactions are held in place mainly by H-bonds between the bases. In the remaining three cases other effects have to be considered. Tertiary base pairing interaction energies range from −8 to −38 kcal/mol in yeast tRNAPhe and are estimated to contribute roughly 25% of the overall tRNA base pairing interaction energy. Six analyzed posttranslational chemical modifications were shown to have minor effect on the geometry of the tertiary interactions. Modifications that introduce a positive charge strongly stabilize the corresponding tertiary interactions. Non-additive effects contribute to the stability of base triplets.
INTRODUCTION
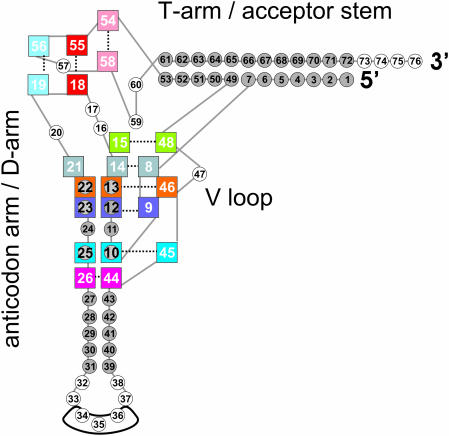
The canonical tRNA conformation, characterized by two helical domains perpendicularly oriented, has already been known for 30 years (1–5). Besides the regular Watson–Crick pairs enclosed in the tRNA stems, specific base pairing interactions have been observed in crystal and NMR structures of tRNA molecules, which are known as tRNA tertiary interactions (Figure 1).
Figure 1.
A schematic representation of the tRNA standard structure. Nucleotides enclosed into the tRNA stems are represented as gray circles. Nucleotides involved in tertiary interactions are enclosed in colored squares. Tertiary base pairing bonding interactions are indicated as dashed lines. The anticodon triplet is also highlighted.
The major interactions maintaining the well-known tRNA L-shape are generally assumed to occur at the corner of the molecule. This region, where the D and T-loops meet, contains several unique elements, including the T54–A58 reverse-Hoogsteen (RH) base pair, the G18–Ψ55 and G19–C56 inter-loop base pairs and stack of the four mutually intercalated A58–G18–A/G57–G19 purine bases. All these interactions are highly conserved in cytosolic tRNAs, indicating their importance for the tRNA function. Available experimental data also support the functional importance of such a region. Mutants of the Escherichia coli tRNAAla(CUA) obtained by random mutagenesis and having at least one of the above-mentioned interactions disrupted were shown to be non-functional (6), and insertion of additional nucleotides into the T-loop or deletion of G19 from the D-loop affected the accuracy of the codon–anticodon recognition, resulting in a frameshift (7,8). It is also known that modification of the D- and T-loops region can influence the processing of the 3′ and 5′ termini, as well as the CCA-addition. For instance, disruption of the base pairs between the D and T-loop had a negative effect on the 3′ and 5′ processing of tRNAHis from Drosophila (9), while positions 57 and 58 of the T-loop were involved in the recognition by the CCA-adding enzyme (10).
Other interactions at the elbow of the structure given by conserved and semiconserved nucleotides of the D and V arms define the core of tRNA molecules. These are the G15–C48 reverse Watson–Crick (RWC) base pair; and the 10–25–45, 9–12–23 and 13–22–46 base triplets, where a canonical WC base pair of the D-stem interacts in the wide groove with a third base from the D or V arm. In the D arm, nucleotide U8 also gives a RH base pair with A14, which in turn, in several canonical tRNAs as yeast tRNAAsp, H-bonds to A21. A purine–purine 26–44 bp, causing a kink between the anticodon and D stems, completes the picture of the tertiary interactions in canonical tRNA. Experimental studies support the importance of the core structure in aminoacylation of several tRNAs, such as E.coli tRNACys (11–13), yeast tRNAAsp (14)and E.coli tRNAGln (15). To quote some examples, the substitution of the 15–48 bp in E.coli tRNACys has been shown to decrease the catalytic efficiency of aminoacylation by two orders of magnitude (13), whereas mutations designed to disrupt the tertiary structure of yeast tRNAAsp, involving the D-stem or D-loop, have been shown to decrease the specificity constant for aspartylation up to three–four orders of magnitude (14). Moreover, recent studies on mitochondrial tRNAs exhibiting a canonical cytosolic-like structure have shown that the identity of nt 48 governs the human tRNALeu(CUN) accepting activity (16) and mutation of G10 in yeast tRNAGln and deletion of position 45 in yeast tRNAArg2 determine defective acylation (17). The 10–25 bp has been also shown to be one of the major binding sites of E.coli tRNAGln to the parent synthetase (15).
Many of the above-mentioned studies showed that the functional importance of non-standard interactions could be ascribed to their role in maintaining the tRNA tertiary structure (14,16–21).
Recently, structural elements resembling the D and/or T-loop region of tRNAs have been identified in RNase P and in the ribosome (22,23), as well as in RNA viruses genome (24,25). This finding suggests that larger RNAs may use conformation-stabilizing strategies analogous to those found in the small tRNA molecule.
Altogether such data call for a thorough analysis of the particular role played by each tRNA element in its structure and function. The systematic elucidation of strategies specific to different RNAs may indeed constitute a very important step toward the understanding of how RNA tertiary structure forms and how it defines the function of the molecule. In order to contribute to the on-going systematic investigation of the different forces that determine RNAs structure, here we focus on the interaction energy of H-bonded nucleic acid base pairs and triplets in tRNA tertiary interactions.
It is important to note that the structure of RNAs is determined by the interplay of several forces with H-bonds interaction between bases being one of the most relevant. Other important forces are due to base–base stacking interactions, base–backbone and backbone–backbone interactions as well as effects due to the ‘environment’ such as RNA interaction with solvent, metal ions, small-molecules and proteins. Several studies were performed to shed light on the different forces that determine nucleic acids structure. In particular advanced quantum mechanics methods are especially suitable for the evaluation of the strength of H-bonded bases interaction (26–41).
In this context, Šponer et al. (33) demonstrated recently that computationally expensive second order Møller–Plesset (MP2) technique (42), in conjugation with rather extended basis sets, is probably the method of choice for the evaluation of base pairing interactions. Unfortunately, routine calculations at this level are unfeasible even for nucleic acid pairs. The situation has improved with the implementation of the resolution of identity MP2 (RIMP2) procedure which provides essentially identical results as the MP2 method with a fraction of computational resources (43–45). This opened the route to large-scale MP2-level calculations of base pairing. Nevertheless, when the complexity (and the size) of the problem raises from base pairs to base triplets computer power remains a limitation. Density functional theory (DFT) based methods are the most reasonable alternative approaches for the calculation of geometry and energy of H-bonded base clusters. Comparison of the hybrid B3LYP (46–49) and of the pure PW91 (50,51) functionals with RIMP2 results on an extended set of base pairs showed that geometries are much less sensitive to the level of calculations than base pairing energies (33). In fact, B3LYP optimizations were shown to provide extremely similar donor–acceptor distances as the RIMP2/cc-pVTZ method, with a typical elongation around 0.03 Å. As far as energies are concerned, the B3LYP method systematically underestimates the reference RIMP2 data by 1.4–3.6 kcal/mol. The largest discrepancy in the relative order of stability between two bp is 2.2 kcal/mol. The PW91 functional, suggested by Müller et al. (52) and Tsuzuki and Lüthi (53) to perform better than the B3LYP functional, improves the agreement with the reference RIMP2 values to −0.8/+1.8 kcal/mol (33). However, the relative stability of two bp is not improved compared with the B3LYP method (largest error is 2.6 kcal/mol). Moreover, the PW91 functional slightly exaggerates monomer deformation energies. Incidentally, DFT methods completely fail to describe base stacking (29,32).
Since we are interested here in base pairs and triplets RIMP2 cannot be the workhorse. We thus decided to evaluate geometries using the B3LYP approach and then calculate interaction energies at the RIMP2 level of theory. Considering that our recipe is extremely close to the approach used by Šponer et al. to obtain highly accurate interaction energies (33), it is not surprising that our computed interaction energies almost match the values they reported. In the few cases where comparison is possible, the stability of the base pairs we calculated is on the average 0.3 kcal/mol from the stability they obtained using the full RIMP2/aug-cc-pVTZ method. This validates the approach we used and, consequently, makes the comparison with their results straightforward. Finally, since accurate evaluation of H-bonded bases energy is a rather demanding task, other forces will be not discussed here.
Optimal geometries and accurate interaction energies have been evaluated for all the H-bonded bases, including ribose C1′ atoms, in the nine classical tertiary interactions (schematic diagram in Figure 1), as extracted from the best-resolution tRNA structure now available. This is the yeast tRNAPhe X-ray structure solved at 1.93 Å (PDB code: 1ehz) (54).
Yeast tRNAPhe was the first tRNA molecule to have its structure solved and its tertiary H-bonding interactions described and characterized (55–57). A statistical analysis on the available tRNA sequences, here performed to identify the most frequently occurring nucleic acid base combinations for each classical tertiary interaction, showed that yeast tRNAPhe is also a very good representative for class I cytolosic tRNAs. However, when the most frequent combinations did not correspond to those present in yeast tRNAPhe (2 out of the 9 analyzed cases), their geometry and interaction energy were also computed. In all cases, when the tertiary interaction included chemically modified bases, we also considered the corresponding interaction with the unmodified bases, in order to allow a comparison.
This study thus provides the first accurate estimate of the interaction energy of H-bonded bases in tRNA tertiary interactions. It also gives insight into the effect of chemical modifications of the bases on the structure and stability of the interaction. Comparison of the energy of the different interactions outlines which of them contribute most to the tRNA structure stability, and possibly constitutes a valuable basis for the rational design of combinatorial tRNA gene libraries and tRNA site-directed mutants. The latter aspect is especially interesting as it has been shown that affinity of mutant tRNA for protein targets can arise from stabilization of RNA tertiary interactions rather than optimization of RNA–protein contacts (58). The calculated energies may also be useful for implementing algorithms for RNA structure prediction.
MATERIALS AND METHODS
tRNA sequence and structure analysis
We analyzed all the 466 tRNA cytoplasmic sequences, 369 of class I and 97 of class II, including posttranslational modified bases downloaded from the ‘Compilation of tRNA sequences (September 2004 edition)’ (59,60). For each sequence we recorded the occurring nucleic acid base combination in the positions relative to each classical tertiary interaction. Twenty-one non-redundant structures of free tRNAs (PDB codes: 1ehz, 2tra, 1fir), tRNA-synthetase complexes (PDB codes: 1f7u, 1asz, 1il2, 1c0a, 1o0c, 1n78, 1ffy, 1eiy, 1h4s, 1qf6, 1h3e, 1ser, 1j1u, 1ivs, 1u0b, 1wz2) and tRNA-elongation factor Tu complexes (PDB codes: 1ttt, 1b23) determined by X-ray crystallography at a resolution of 3.3 Å or better were collected from the PDB data archive (61). Structure analysis was performed using the InsightII software (62). Using the set of tools available in the package, we analyzed H-bond patterns of observed tertiary interactions and, when of interest, performed structural superimpositions.
Computational details
The TURBOMOLE package was used for all the calculations discussed in the present paper (63). The hybrid B3LYP functional was used for all geometry optimizations unless otherwise specified. The cc-pVTZ basis set (64) was used for geometry optimizations. Interaction energies were calculated on the B3LYP/cc-pVTZ geometries at the RIMP2 level of theory using the cc-pVTZ basis set on C atoms and on H atoms bonded to C atoms, while the more extended aug-cc-pVTZ basis sets (64) was used for N, O and polar H atoms. The extended aug-cc-pVTZ basis sets could not be used for all the atoms due to severe linear dependency of the resulting molecular orbitals in the triplets. All interaction energies were corrected for basis set superimposition error (BSSE) using the counterpoise procedure of Boys and Bernardi (65).
Two different sets of total interaction energies were evaluated. In the first approach we optimized the position of all the atoms in the base pair/triplet, and the total interaction energy was calculated relative to the fully optimized and isolated bases. This is the standard approach used in the literature, and a detailed discussion on this approach can be found in previous papers (33,66). We refer to these total interaction energies as ΔEOpt. In the case of a base pair ΔEOpt is calculated as shown in the following equation.
| 1 |
where is the energy of the optimized X–Y base pair, and and are the energies of the isolated and optimized X and Y bases. Within this approach the deformation energy EDef, which is the energy required to deform the isolated and relaxed bases to the geometry they assume in the base pair/triplet, can be also obtained. In the case of a base pair EDef is calculated as shown in the following equation.
| 2 |
where and are the energies of the isolated X and Y bases with the geometry frozen to that in the fully optimized X–Y pair. By definition EDef always destabilizes the bases interaction. Finally, the interaction energy EInter, which is the interaction energy between the bases in the geometry they assume in the base pair/triplet, can also be obtained. In the case of a base pair EInter is calculated as shown in the following equation.
| 3 |
Equations 1–3 can be combined as ΔEOpt = EInter + EDef.
In the second approach we optimized the position of the H atoms only, while all the heavy atoms in the base pair/triplet were frozen as in the X-ray structure. The interaction energy was calculated relative to the unrelaxed isolated bases using a rigid body approximation, i.e. the base pair/triplet was rigidly fragmented into the bases, which implies that in the rigid body approximation EDef is not defined. We refer to these rigid body interaction energies as ΔERBI. In the case of a base pair the ΔERBI is calculated as shown in the following equation.
| 4 |
where is the energy of the X–Y base pair with the heavy atoms frozen to the X-ray geometry, and are the energies of the isolated X and Y bases with the geometry frozen to that in the X–Y pair.
Extension of ΔEOpt, EDef and ΔERBI to a base triplet or to the decomposition of a X–Y–Z triplet into the X–Y and Z fragments is straightforward. Furthermore, as indicated by Burda et al. (66) the interaction energy of base triplets can be decomposed into 3 bp pairwise interactions and a three-body contribution E3 as shown in the following equation.
| 5 |
where , and are the X–Y, X–Z and Y–Z pairs interaction energy with the bases fixed to the geometry they have in the optimized triplet. The three-body contribution E3 is a measure of the non-additivity of the base pair interactions (66).
RESULTS
tRNA sequence and structure analysis
Some tRNA tertiary interactions, such as 8–14–21, 18–55, 19–56 and 54–58, involve the highly conserved nucleotides U8, A14, G18, G19, A21, T54, Ψ55, C56 and A58, whereas others allow a certain nucleotide variability. For each classical tertiary interaction, we recorded all the possible nucleic acid base combinations occurring in the available cytoplasmic tRNA sequences (59,60). Interactions corresponding to the most frequent base combinations in class I cytosolic tRNAs have been considered here and are reported in Table 1 along with relative occurrences. They have been visually verified in available tRNA structures. PDB codes of the best resolution structures exhibiting each specific interaction are also reported in Table 1.
Table 1.
All computed interactions are reported with the relative occurrence in cytosolic tRNA sequences from the Sprinzl database (60)
| Interaction | Occurrence | PDB | Res (Å) | Base pair | Burkard et al. | L&W | |
|---|---|---|---|---|---|---|---|
| 8–14–21 | U–A–A | 301 (82%) | 1ehz | 1.93 | uracil8…adenine14 | Reverse hoogsten (RH) | 4 |
| adenine14…adenine21 | – | – | |||||
| 9–12–23 | A–U–A | 170 (46%) | 1ehz | 1.93 | uracil12…adenine23 | Watson–Crick (WC) | 1 |
| adenine9…adenine23 | N7-amino,symmetric (N7as) | 4 | |||||
| 10–25–45 | G–C–G | 128 (35%) | 1ffy | 2.10 | guanine10…cytosine25 | Watson–Crick (WC) | 1 |
| guanine10…guanine45 | – | – | |||||
| m2G-C-G | 94 (25%) | 1ehz | 1.93 | N2-methylguanine10…cytosine25 | Watson–Crick (WC) | 1 | |
| N2-methylguanine10…guanine45 | – | – | |||||
| 13–22–46 | C–G–m7G | 179 (49%) | 1ehz | 1.93 | cytosine13…guanine22 | Watson–Crick (WC) | 1 |
| guanine22…7-methylguanine46 | N7-imino (N7i) | 4 | |||||
| C–G–G | 35 (9%) | 1j1u | 1.95 | C13…G22 | Watson–Crick | 1 | |
| guanine22…guanine46 | N7-imino (N7i) | 4 | |||||
| 15–48 | G–C | 186 (50%) | 1ehz | 1.93 | guanine15…cytosine48 | Reverse Watson–Crick (RWC) | 2 |
| 18–55 | G–Ψ | 349 (94%)a | 1ehz | 1.93 | guanine18…pseudouracil55 | imino-2-carbonyl, amino-2-carbonyl, bifurcatedb | 2 |
| G–U | 17 (19%) | 1j1u | 1.95 | guanine18…uracil55c | imino:amino-2-carbonyl, bifurcatedb | 2 | |
| 19–56 | G–C | 365 (99%)d | 1ehz | 1.93 | guanine19…cytosine56 | Watson–Crick (WC) | 1 |
| 26–44 | m22G–A | 94 (25%) | 1ehz | 1.93 | N2,N2-dimethylguanine26…adenine44 | Imino | 1 |
| G–A | 46 (12%) | 1j1u | 1.95 | guanine26…adenine44 | Imino | 1 | |
| 54–58 | T–A | 114 (31%) | 1c0a | 2.40 | thymine54…adenine58 | Reverse hoogsten (RH) | 4 |
| T–m1A | 84 (23%) | 1ehz | 1.93 | thymine54…1-methyladenine58 | Reverse hoogsten (RH) | 4 | |
| U–Ae | 34 (9%) | 1j1u | 1.95 | uracil54…adenine58 | Reverse hoogsten (RH) | 4 | |
| U–m1Ae | 23 (6%) | – | – | uracil54…1-methyladenine58 | Reverse hoogsten (RH) | 4 | |
PDB codes of the best resolution structure where each specific interaction is observed are also reported. For all computed base pairs with at least two hydrogen bonds, classification is given according to Burkard et al. (73) and Leontis and Westhof (74) schemes; in (74) ‘1’ stays for Cis Watson–Crick/Watson–Crick Antiparallel, ‘2’ for Trans Watson–Crick/Watson–Crick Parallel and ‘4’ for Trans Watson–Crick/Hoogsteen Antiparallel. Bases nomenclature is according to Sprinzl et al. (59).
a71 sequences bear an additional methyl group on the ribose O2′ atom of guanosine18.
bNomenclature from the NCIR database, http://prion.bchs.uh.edu/bp_type/ (75).
cComputed starting from the 1j1u coordinates.
d51 sequences bear an additional methyl group on the ribose O2′ atom of guanosine19 or cytidine56.
eThese are the fourth and fifth most populated nucleic acid base combinations, after 1-methylpseudouracil54-adenine 58 (m1Ψ-A), present in39 sequences (11%).
The most represented combination corresponds to that present in the yeast tRNAPhe, which has also the best resolution structure solved to date (Pdb code: 1ehz, resolution 1.93 Å), for seven of the nine interactions. The only exceptions are represented by the interactions at positions 10–25–45 and 54–58, where yeast tRNAPhe nucleotide combinations are N2-methylguanine-cytosine-guanine (m2G-C-G) and thymine-1-methyladenine (T-m1A). m2G10-C25-G45 and T54-m1A58 are the second most populated combinations with an occurrence of 25 and 23%, respectively, after the corresponding interactions (G10–C25–G45, T54–A58) lacking a posttranslational chemical modification.
On these grounds, we chose the structure of tRNAPhe (Pdb code: 1ehz) as the reference system for our calculation of interaction energy of tRNA tertiary base pairing interactions.
Molecular structures and interaction energies
Optimized geometries and relative interaction energies (ΔEOpt) were computed for the base pairs and triplets reported in Table 1. These are the ideal geometries and interaction energies that would be obtained in the gas phase in the absence of any other effect. The relative H-bond distances are reported in Table 2. Additionally, interaction energies were also computed for the tertiary base pairs and triplets with the geometry fixed to the yeast tRNAPhe 1ehz X-ray structure (ΔERBI). The interaction energy ΔERBI is an estimate of the contribution of each base pairing interaction to the overall stability of the tRNA. The base pairs interaction energy is calculated relative to the two separated bases, while in the case of base triplets two different interaction energies are reported. In fact, besides the interaction energy relative to the three separated bases, we also calculated the interaction energy between the main base pair and the third interacting base. This provides an estimate of the tertiary interaction energy for triplets where a WC pair enclosed into the D stem interacts with a third base. All the calculated ΔEOpt and ΔERBI are reported in Table 3.
Table 2.
H-bond lengths of the optimized base pairs and triplets obtained at the B3LYP level and r.m.s.d. values for the heavy atoms superimposition on the corresponding 1ehz structures (see also Figures 2 and 3)
| Interaction | Base pair | H-bond length | H-bond length | H-bond length | RMSD (Å) |
|---|---|---|---|---|---|
| 8–14–21 | U8…A14 | O2(U)–N6(A) 3.04 | N3(U)–N7(A) 2.86 | 1.38 | |
| A14…A21 | N6(A)–N3(A) 3.06 | 0.21 | |||
| 9–12–23 | U12…A23 | O2(U)–N6(A) 2.96 | N3(U)–N1(A) 2.88 | 0.27 | |
| A9…A23 | N6(A)–N7(A) 3.08 | N7(A)–N6(A) 3.00 | 0.19 | ||
| 10–25–45 | m2G10…C25 | N2(m2G)–O2(C) 2.91 | N1(m2G)–N3(C) 2.96 | O6(m2G)–N4(C) 2.82 | 2.11 |
| m2G10…G45 | O6(m2G)–N2(G) 2.88 | 0.38 | |||
| C25…G45 | N4(C)–N3(G) 3.23 | ||||
| G10…C25 | N2(G)–O2(C) 2.92 | N1(G)–N3(C) 2.96 | 06(G)–N4(C) 2.82 | ||
| G10…G45 | O6(G)–N2(G) 2.89 | ||||
| C25…G45 | N4(C)–N3(G) 3.29 | ||||
| 13–22–46 | C13…G22 | O2(C)–N2(G) 2.83 | N3(C)–N1(G) 2.95 | N4(C)–06(G) 2.94 | 0.17 |
| G22…m7G46 | O6(G)–N1(m7G) 2.84 | N7(G)–N1(m7G) 2.78 | 0.12 | ||
| C13…G22 | O2(C)–N2(G) 2.90 | N3(C)–N1(G) 2.95 | N4(C)–06(G) 2.85 | ||
| G22…G46 | O6(G)–N1(G) 3.13 | N7(G)–N1(G) 2.88 | |||
| 15–48 | G15…C48 | N1(G)–O2(C) 2.80 | N6(G)–O2(C) 2.99 | 1.15 | |
| 18–55 | G18…Ψ55 | N1(G)–O2(Ψ) 2.95 | N2(G)–O2(Ψ) 3.13 | 0.30 | |
| G18…U55a | N1(G)–O2(U) 2.95 | N2(G)–O2(U) 3.19 | |||
| 19–56 | G19…C56 | N2(G)–O2(C) 2.94 | N1(G)–N3(C) 2.95 | 06(G)–N4(C) 2.80 | 0.45 |
| 26–44 | m22G26…A44 | N1(m22G)–N1(A) 3.03 | O6(m22G)–N6(A) 2.84 | 0.25 | |
| G26…A44 | N1(G)–N1(A) 2.97 | O6(G)–N6(A) 2.86 | |||
| 54–58 | T54…A58 | O2(T)–N6(A) 2.97 | N3(T)–N7(A) 2.86 | ||
| T54…m1A58 | O2(T)–N6(m1A) 2.72 | N3(T)–N7(m1A) 2.92 | 0.21 | ||
| U54…A58 | O2(U)–N6(A) 2.99 | N3(U)–N7(A) 2.85 | |||
| U54…m1A58 | O2(U)–N6(m1A) 2.73 | N3(U)–N7(m1A) 2.92 |
aComputed starting from the 1j1u coordinates.
Table 3.
Interaction energies, in kcal/mol, of the tRNA nine tertiary interactions
| Base complex | Interaction | EDef | EInter | ΔEOpt | E3(Opt) | ΔERBI | E3(RBI) |
|---|---|---|---|---|---|---|---|
| U8–A14–A21 | U…A…A | 1.7 | −25.7 | −24.0 | −1.5 | −19.3 | 0.1 |
| (U–A)…A | 0.3 | −9.3 | −9.0 | −3.5 | |||
| A9–U12–A23 | A…U…A | 2.8 | −28.9 | −26.1 | −1.3 | −24.0 | −0.4 |
| A…(U–A) | 1.1 | −12.8 | −11.7 | −9.7 | |||
| G10–C25–G45 | G…C…G | 5.6 | −44.6 | −39.0 | |||
| (G-C)…G | 2.0 | −14.4 | −12.4 | ||||
| m2G10–C25–G45 | m2G…C…G | 4.9 | −44.8 | −39.9 | −6.4 | −38.7 | −0.2 |
| (m2G–C)…G | 2.3 | −14.6 | −12.3 | −8.4 | |||
| C13–G22–m7G46 | C…G…m7G | 6.8 | −70.5 | −63.6 | −7.0 | −68.7 | −2.8 |
| (C–G)…m7G | 0.6 | −38.6 | −38.0 | −38.1 | |||
| C13–G22–G46 | C…G…G | 5.8 | −52.0 | −46.2 | |||
| (C–G)…G | 1.8 | −21.2 | −19.4 | ||||
| G15–C48 | 1.5 | −16.1 | −14.6 | −13.7 | |||
| G18–Ψ55 | 0.9 | −13.4 | −12.5 | −13.8 | |||
| G18–U55 | 1.7 | −14.2 | −12.6 | ||||
| G19–C56 | 3.4 | −30.2 | −26.8 | −29.5 | |||
| m22G26–A44 | 1.9 | −18.1 | −16.2 | −13.9 | |||
| G26–A44 | 1.5 | −18.5 | −17.0 | ||||
| T54–A58 | 1.1 | −16.8 | −15.7 | ||||
| T54–m1A58 | 2.1 | −24.6 | −22.5 | −20.7 | |||
| U54–A58 | 1.7 | −16.7 | −15.0 | ||||
| U54–m1A58 | 2.5 | −23.7 | −21.2 |
ΔEOpt is the stabilization energy of the base pair/triplet starting from the optimized and isolated bases. ΔEOpt can be decomposed as ΔEOpt = EDef + EInter, where EDef is the energy required to deform the isolated bases to the geometry they assume in the base pair/triplet and EInter is the interaction energy between the bases in the geometry they assume in the base pair/triplet. ΔERBI is the interaction energy of the base pair/triplet with the geometry fixed to the yeast tRNAPhe 1ehz X-ray structure. For triplets two energy values are reported. In the first approach the X–Y–Z triplet is fragmented into the three bases, X, Y and Z, and the interaction energy reported reflects the X…Y…Z interaction. In the second approach the X–Y–Z triplet is fragmented into the base pair (X–Y) and the base Z, and the interaction energy reported reflects the (X–Y)…Z interaction. E3 is the three-body contribution to the total interaction energy of triplets; it is a measure of the non-additivity of the pairwise base interactions. Further details can be found in Computational details section.
Furthermore, in the case of base triplets we also calculated the interaction energy for the three possible base pairs that can be obtained from fragmentation of the triplet. In these calculations we fixed the base pairs at the geometry they have in the optimized or X-ray triplets. For base pairs giving at least two H-bonds, ideal geometry and energy were also computed in the absence of the third interacting base, to investigate the effect of the tertiary interaction on the base pair geometry and stability. All the pairwise interaction energies are reported in Table 4. The difference between the triplet ΔEOpt and the sum of the three pairwise interaction energies is a measure of non-additive effects (E3). These non-additive effects cannot be recovered by empirical force fields, which are based on two-body interaction terms, and their magnitude underlines the necessity of using a quantum mechanics approach in the calculation of base triplets and beyond.
Table 4.
Interaction energies, in kcal/mol, of the base pairs X–Y, X–Z and Y–Z that can be obtained by fragmentation of the base triplets X–Y–Z
| Triplet | Pair | Isolated pair | Pair in the optimized triplet | Pair in the X-ray triplet |
|---|---|---|---|---|
| U8–A14–A21 | 8–14 | −15.0 | −14.6 | −15.8 |
| 8–21 | 0.6 | −2.4 | ||
| 14–21 | −8.5 | −1.2 | ||
| A9–U12–A23 | 9–12 | −0.3 | −1.3 | |
| 9–23 | −11.1 | −10.7 | −8.8 | |
| 12–23 | −13.8 | −13.8 | −14.3 | |
| m2G10–C25–G45 | 10–25 | −27.1 | −26.7 | −30.8 |
| 10–45 | −5.8 | −7.3 | ||
| 25–45 | −1.0 | −0.4 | ||
| G13–G22–m7G46 | 13–22 | −26.8 | −25.6 | −30.7 |
| 13–46 | 4.0 | 1.5 | ||
| 22–46 | −34.2 | −32.5 | −36.8 |
The ‘Isolated Pair’ column reports the interaction energy of the isolated and optimized base pair. These calculations have been performed for base pairs with at least two H-bonds. The ‘Optimized Triplet’ column reports the interaction energy between base pairs in the geometry they have in the optimized triplet. The ‘X-ray Triplet’ column reports the interaction energy between base pairs in the geometry they have in the X-ray structure.
The superimposition of the optimized and X-ray base triplets and pairs are shown in Figures 2 and 3, respectively; root mean square deviation (r.m.s.d.) values are reported in Table 2.
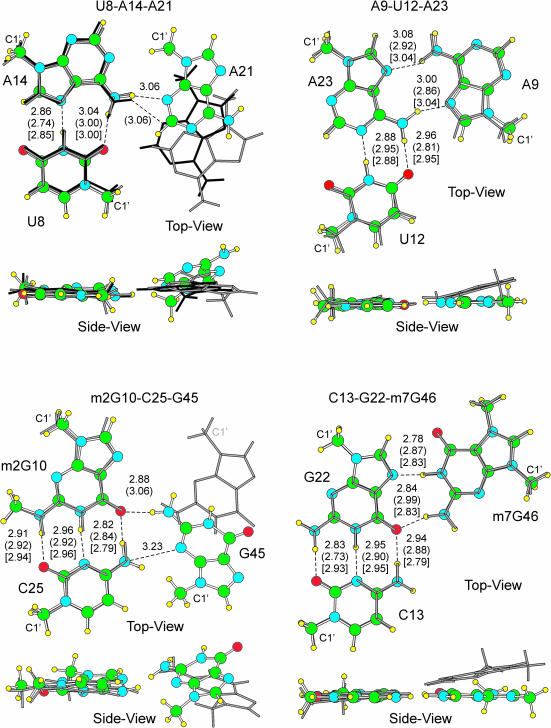
Figure 2.
Superimposition of the optimized (balls and sticks) and 1ehz X-ray (gray sticks) base triplets. For the U8–A14–A21 triplet, superimposition with the 1j1u X-ray triplet (black sticks) is also reported. Superimposition has been performed on the two bases on the left. H-bonds distances between heavy atoms are reported in Å. Out of parentheses the optimized values, in parentheses the 1ehz X-ray values, in square brackets values for the isolated base pair. The C1′ ribose atom is also indicated.
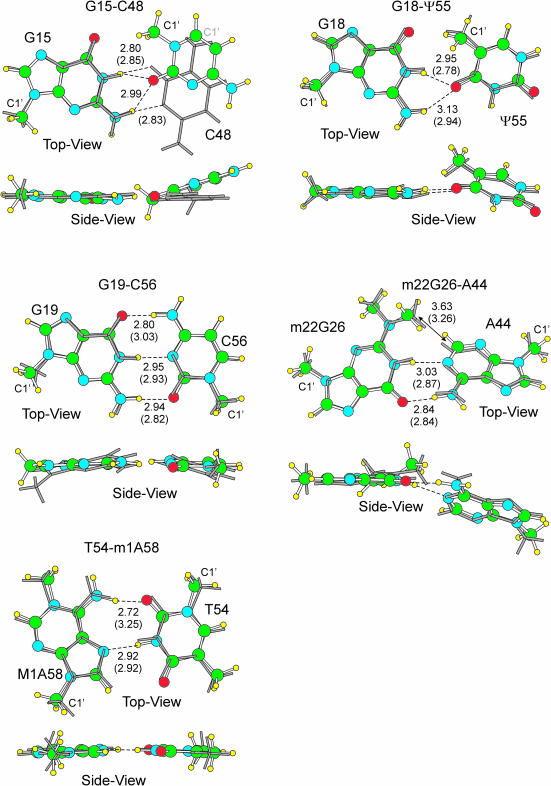
Figure 3.
Superimposition of the optimized (balls and sticks) and 1ehz X-ray (gray sticks) base pairs. H-bonds distances between heavy atoms are reported in Å. Out of parentheses the optimized values, in parentheses the 1ehz X-ray values. The C1′ ribose atom is also indicated.
The U8–A14–A21 triplet
The geometry of the U8–A14 RH base pair in the optimized triplet is quite similar to the U8-A14 pair in 1ehz (r.m.s.d. = 0.21 Å, H-bonds within 0.12 Å) (Figure 2 and Table 2) as well as to an isolated U–A RH base pair (r.m.s.d. = 0.03 Å, H-bonds within 0.01 Å). This implies that the presence of the A21 base does not influence the geometry of the slightly non-planar U8–A14 pair. As regards the A14–A21 interaction, in the optimized triplet the N6(A14):N3(A21) H-bond is formed. In 1ehz this H-bond is missing, while a H-bond instead occurs between N1(A21) and ribose O2′(U8). Nevertheless, the N6(A14):N3(A21) H-bond is present in other tRNA structures such as Methanococcus jannaschii tRNALys (PDB code: 1j1u) (67), which also presents the N1(A21):O2′(U8) H-bond as 1ehz, and yeast tRNAAsp (PDB code: 2tra) (68), which instead presents a N1(A14):O2′(A21) H-bond. The skeleton of 1j1u is reported in black in Figure 2. Visual inspection clearly indicates that the same A14–A21 H-bond is present in the optimized and in the 1j1u triplets. However, in the optimized triplet A21 rotates remarkably as compared with A21 in 2tra.
The U8–A14 interaction ΔEOpt in the optimized triplet is only 0.4 kcal/mol weaker than the interaction energy in the isolated optimized U–A RH pair (Table 4), which further supports that the U8–A14 pair is very little perturbed in the triplet. Overall, the stability of the optimized base triplet starting from the three isolated bases (Table 3) is very close to the sum of the three possible base pairs (U8–A14, U8–A21 and A14–A21) pairwise interaction energies, −22.5 kcal/mol. This implies that non-additive effects, E3 = −1.5 kcal/mol only slightly stabilize the triplet.
The U8–A14 and U8–A21 rigid body interaction energies ΔERBI in the 1ehz structure are rather similar to those obtained for the optimized base triplet, although heavy atoms were not relaxed (Table 4). Instead, the A14–A21 interaction is quite weaker in the 1ehz triplet relative to the optimized structure, consequence of the missing A41–A21 H-bond in the X-ray structure. The overall stability of the 1ehz base triplet is thus consistently reduced as compared with the optimized triplet. As final control, we calculated the rigid body A14–A21 interaction in 1j1u and 2tra, which both present the N6(A14):N3(A21) H-bond. In 1j1u the A14–A21 interaction amounts to ΔERBI = −6.2 kcal/mol, while in 2tra it amounts to ΔERBI = −5.9 kcal/mol. These values are much more similar to the A14–A21 interaction in the optimized triplet, and consistent with the presence of a H-bond between the bases.
The A9–U12–A23 triplet
The geometry of the canonical U12–A23 WC base pair in the optimized triplet is very similar to the geometry in 1ehz (r.m.s.d. = 0.19 Å, H-bonds within 0.15 Å), as well as to the optimized geometry of an isolated U–A WC base pair (r.m.s.d. = 0.02 Å, H-bonds within 0.01 Å). In all the three cases the U–A WC pair is planar. These results imply that the presence of the third A9 base has minor influence on the geometry adopted by the U12–A23 pair. On the contrary, the A9–A23 N7-amino symmetric (N7as) pair in the optimized triplet as well as the isolated N7as A–A pair is planar, whereas this pair is slightly buckled in 1ehz. Despite of the planarity in the optimized triplet a rather good superimposition between the optimized and the X-ray triplets is obtained (r.m.s.d. = 0.27 Å, H-bonds within 0.16 Å).
The U12–A23 as well as the A9–A23 interaction energies in the optimized triplet are very close to interaction energies in the isolated U–A WC and A–A N7as pairs, respectively. This further supports that the two pairs are very little perturbed in the optimized triplet. Overall, the stability of the optimized base triplet starting from the three isolated bases is very close to the sum of the three pairwise interaction energies, −24.8 kcal/mol. This again indicates that non-additive effects, E3 = −1.3 kcal/mol, slightly stabilize the triplet.
The rigid body interaction energies of the three possible base pairs in the 1ehz structure are rather similar (within 1.9 kcal/mol) to the interaction energies obtained from the optimized base triplet (Table 4). The overall stability of the 1ehz base triplet is only 2.1 kcal/mol lower than the optimized value. Considering the rigid body approximation used the rather good agreement between ΔERBI and ΔEOpt is quite remarkable.
The m2G10–C25–G45 triplet
The m2G10–C25 WC base pair in the optimized triplet is very similar to that in 1ehz (r.m.s.d. = 0.38 Å, H-bonds within 0.04 Å). Conversely, the m2G10–C25 WC pair in the optimized triplet is somewhat different from the isolated m2G–C WC base pair (r.m.s.d. = 0.17 Å), although H-bonds are within 0.03 Å. The main difference lies in the planarity of the isolated m2G–C WC pair, whereas in both the optimized and X-ray triplets the m2G10–C25 pair is not planar.
Moreover, visual inspection of Figure 2 clearly shows that G45 assumes a different orientation after geometry optimization. Specifically, the G45 base rotates by roughly 180° around the O6(m2G10):N2(G45) H-bond, and in the optimized geometry the G45(C1′) atom is near to the C25 base. This remarkable geometrical rearrangement of G45 allows the formation of a weak H-bond like interaction between N4(C25) and N3(G45). To test whether this rearrangement of G45 is dependent on the theoretical approach used we performed a further optimization of the triplet at the more accurate (but much more computationally expensive) RIMP2/cc-pVTZ level. Also at this level of theory G45 rearranges as shown in Figure 2.
The main difference between the rigid body and optimized interaction energies is the higher stability of the interaction between the m2G10–C25 pair and the G45 base in the optimized structure. The overall stability of the X-ray triplet is only 0.2 kcal/mol higher than the sum of the 3 bp pairwise interaction energies, which indicates that non-additive effects are substantially negligible in the X-ray geometry.
The C13–G22–m7G46 triplet
The geometry of the C13–G22 WC base pair in the optimized triplet is very similar to the geometry in 1ehz (r.m.s.d. = 0.12 Å, H-bonds within 0.10 Å), but it is somewhat different from the geometry of an isolated C–G WC base pair. In fact, in the optimized triplet the H-bonds are even 0.15 Å different from the isolated pair. A possible explanation is that the positively charged m7G base electrostatically repels the positively charged N4 amino group of C13.
As regards the G22–m7G46 N7-imino (N7i) pair, the geometry in the optimized triplet is planar, whereas it is slightly non-planar in 1ehz. The G22–m7G46 H-bonds in the optimized triplet are roughly 0.10–0.15 Å shorter than in the X-ray structure while they are rather close to the H-bonds in the isolated G–m7G N7i pair. This suggests that in the optimized triplet the interaction between the charged m7G46 base and G22 is overestimated. It is likely that in the complete tRNAPhe structure the m7G46 and G22 interaction is damped by surrounding phosphate groups.
The C13–G22 and G22–m7G interactions in the fully optimized triplet are 1.2 and 1.7 kcal/mol weaker than in the isolated C–G WC and G–m7G N7i pairs, respectively, which further supports that the triplet is somewhat perturbed by the repulsive C13 and m7G46 interaction. Remarkably, in this triplet the non-canonical G22–m7G46 interaction is roughly 7 kcal/mol stronger than the canonical WC C13–G22 interaction. Overall, the stability of the fully optimized base triplet starting from the three isolated bases is remarkably higher than the sum of the three pairwise interaction energies, −54.1 kcal/mol. This indicates that non-additive effects, E3 = −7.0 kcal/mol, are particularly strong.
In the rigid body approximation, the stability of the three possible base pairs is 3–5 kcal/mol higher than those obtained for the optimized base triplet. This is due to the quite high deformation energy we calculated for this triplet, EDef = 5.8 kcal/mol, that reduces the ΔEOpt value and is not considered in the rigid body approximation (see Computational details). The overall stability of the base triplet is roughly 5 kcal/mol higher than that calculated for the optimized triplet. Considering the rigid body approximation used, and the remarkably high stability of this triplet, we believe there is a reasonable agreement between the ΔEOpt and ΔERBI values also in this case.
The G15–C48 pair
This pair presents an RWC geometry in the 1ehz structure. However, previous quantum mechanics studies evidenced that the isolated G–C RWC pair is not a stable geometry due to repulsive amino–amino and carbonyl–carbonyl contacts (35). Consistently, in our geometry optimization of the G15–C48 1ehz pair the RWC geometry is not retained and the optimized G15–C48 pair presents the bifurcated H-bond pattern shown in Figure 3. The severe geometrical rearrangement indeed alleviates the amino–amino repulsion. However, it also breaks the N2(G15):N3(C48) H-bond of the X-ray structure. The O2(C48):N1(G15) H-bond is retained after optimization, but C48 shifts in such a way that the O2(C48):N2(G15) H-bond is formed.
The interaction between G15 and C48 in the optimized pair is quite strong. Interestingly, the rigid body interaction clearly indicates that the G15–C48 RWC pair contributes noticeably to the 1ehz stability, although the isolated GC RWC pair is not a stable structure.
The G18–Ψ55 pair
The optimized base pair is quite similar to the geometry in 1ehz (r.m.s.d. = 0.30 Å, H-bonds within 0.19 Å). Both the X-ray and optimized pairs are substantially twisted. The main difference is in the stretch of the H-bonds. The optimized H-bonds are 0.17/0.19 Å longer than in the X-ray structure. The G18–Ψ55 interaction in the optimized pair is rather close to the rigid body interaction energy, which again indicates an overall similarity between the optimized and the X-ray pairs.
The G19–C56 pair
The optimized base pair is relatively similar to the geometry in 1ehz (r.m.s.d. = 0.45 Å). However, the X-ray pair is substantially non-planar whereas the optimized pair is only slightly twisted. Consistently the optimized H-bonds are somewhat different from the X-ray values. In particular the optimized O6(G19):N4(G56) H-bond is 0.23 Å shorter, and the optimized N2(G19):O2(C56) H-bond is 0.12 Å longer than in the X-ray structure.
The G19–C56 interaction in the optimized pair is 2.7 kcal/mol weaker than in 1ehz. While the difference between the ΔEOpt and ΔERBI could be somewhat surprising, the ΔERBI is only 0.7 kcal/mol away from the Einter calculated for the optimized pair. In short, the G–C WC pair is destabilized by a sizeable deformation energy, EDef = 3.4 kcal/mol.
The m22G26–A44 pair
The optimized base pair is very similar to the geometry in 1ehz (r.m.s.d. = 0.25 Å, H-bonds within 0.16 Å). Both the optimized and the X-ray pairs are substantially non-planar.
The m22G22–A44 interaction in the optimized structure is 2.3 kcal/mol stronger than that in 1ehz. The poor stability of the X-ray pair is probably due to the substantial planarity around the N2(m22G26) atom, which results in a rather short distance, 3.26 Å, and consequently repulsive interaction between the CM1(m22G26) and C2(A44) atoms (Figure 3). In the optimized pair the N2(m22G26) atom is substantially pyramidal, and this pyramidalization results in a much longer distance, 3.63 Å, between the CM1(m22G26) and C2(A44) atoms.
The T54–m1A58 pair
The geometry of the optimized base pair is quite similar to the geometry in 1ehz (r.m.s.d. = 0.21 Å). However, the optimized structure presents the O2(T54):N6(m1A58) H-bond, whereas the distance between these two atoms is 0.53 Å longer in the X-ray structure. Both the optimized and the X-ray pairs are substantially planar. The T54–m1A58 interaction in the optimized pair is 1.8 kcal/mol stronger than in 1ehz. The higher stability of the optimized pair is a consequence of the additional O2(T54):N6(m1A58) H-bond.
Comparison with corresponding interactions lacking posttranslational chemical modifications
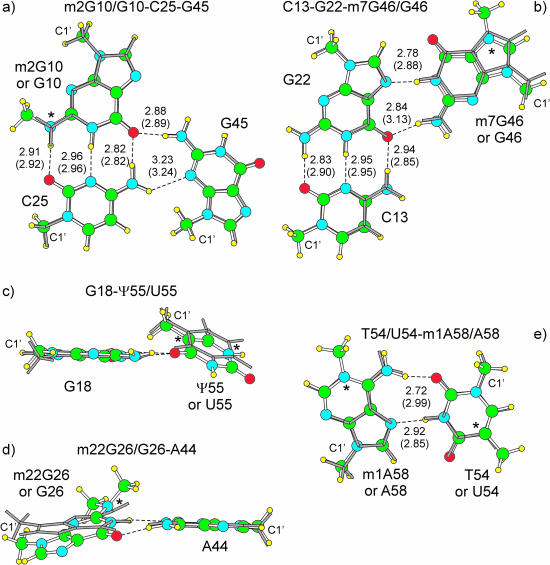
To get insight on the role of the chemical modification on the geometry and stability of the tertiary interactions, we also examined the G10–C25–G45 and C13–G22–G46 base triplets, as well as the G18–U55, G26–A44 and T54–A58/U54–m1A58/U54–A58 base pairs. They are all represented in available tRNA sequences and, but for the U54–m1A58 pair, observed in solved tRNA structures (Table 1). With the exception of U54–m1A58/U54–A58, they all correspond to the first or second most populated nucleic acid bases combination for each specific interaction. Superimposition between the modified and unmodified base pairs and triplets is reported in Figure 4.
Figure 4.
Superimposition of optimized yeast tRNAPhe interactions presenting posttranslational modifications (ball and sticks) on the corresponding unmodified interactions (gray sticks). H-bonds distances between heavy atoms are reported in Å. Out of parentheses values for the modified structures, in parentheses those for the unmodified structures. A star indicates where the chemical modification occurs. The C1′ ribose atom is also indicated. In (a) and (b) superimposition has been performed on the base pair on the left; (c) and (d) on the G18 and A44 pair, respectively; (e) on corresponding atoms of both the bases.
The optimized G10–C25–G45 triplet is extremely similar to the m2G10–C25–G45 triplet (r.m.s.d. = 0.04 Å). Consistently, the total interaction energy of the unmodified triplet is only 1.8 kcal/mol away from the ΔEOpt of the modified triplet.
Conversely, the optimized geometry of the C13–G22–G46 triplet is somewhat different from that of the C13–G22-m7G46 triplet, with the G13–G22 WC pair in the unmodified triplet more similar to an isolated G–C WC pair. However, the main difference is the remarkable elongation, 0.29 Å, of the N2(G46):O6(G13) H-bond in the unmodified triplet. Of course, these geometrical rearrangements are consequence of the positive charge on the m7G46 base of the modified triplet (69). Similar effects are observed when comparing the charged T54–m1A58/U54–m1A58 pairs with the neutral T54–A58/U54–A58 pairs (69). The O2(T54/U54):N6(A58) H-bonds are remarkably longer than in the modified T54–m1A58/U54–m1A58 pair. As expected, the stability of the unmodified and neutral C13–G22–G46 triplet and T54–A58/U54–A58 pairs is remarkably lower than the stability of the positively charged modified base triplet and pair, by 17.4 and 6.8/6.2 kcal/mol, respectively.
The unmodified G18–U55 pair is rather similar to the G18–Ψ55 pair, although the modified pair is slightly more planar. Quite different, instead, is the geometry of the unmodified G26–A44 pair relative to the modified m22G26–A44 pair. The unmodified pair is quite more planar than the modified pair, although still propeller twisted. The reason is steric in nature. In fact, the two additional methyl groups on the N2 atom of m22G26 base force the modified base pair to assume a more pronounced non-planar geometry to avoid steric interactions between the CM1(m22G26) and the C2(A44) atoms. Consequence of the reduced steric repulsion between the two bases is the shrink of the N1(G26):N1(A44) H-bond. Nevertheless, the unmodified G26–A44 base pair is only 0.8 kcal/mol more stable than the modified base.
DISCUSSION
Geometries of six out of the nine optimized yeast tRNAPhe tertiary interactions: A9–U12–A23, C13–G22–m7G46, G18–Ψ55, G19–C56, T54–m1A58 and m22G26–A44, reproduce very well those observed in the best resolution structure 1ehz (Figures 2 and 3). The r.m.s.d. values for the bases heavy atoms superimposition are indeed within 0.45 Å and H-bond lengths differ for <0.23 Å, except for one H-bond in the T54–m1A58 pair. Consequently, the computed optimized and rigid body interaction energies ΔEOpt and ΔERBI are very similar. Considering that the ΔERBI does not take into account the unfavorable deformation energy term EDef, a more accurate comparison can be made with the EInter term for optimized structures. The slight geometrical differences result here in a change in the energy values that reaches a maximum of 4.9 kcal/mol for the A9–U12–A23 triplet but can be as little as 0.4 kcal/mol for the G18–Ψ55 pair.
These results strongly suggest that the above-mentioned interactions are held in place mainly by H-bond interactions between the bases; other forces such as base–backbone interaction, base–base stacking and environment effects seem to play a minor role. Note that this finding holds true in the light of the analysis of 20 lower resolution cytosolic tRNA structures (for the Pdb codes list see Materials and Methods).
Optimized structures of G15–C48, U8–A14–A21 and m2G10–C25–G45 instead do not reproduce those observed in 1ehz (Figures 2 and 3). As concerns the U8–A14–A21 triplet, the U8–A14 RH pair geometry is very well reproduced in the optimized structure, whereas the third interacting base A21 approaches A14 to give an N3-amino H-bond interaction. The optimized triplet is thus more stable than in 1ehz by ∼5 kcal/mol. Stabilization given by the additional A14–A21 N3-amino H-bond can be thus quite easily overcome with other effects, such as the interaction of bases with backbone, base–base stacking (A21 participates into a six purine bases stacking interaction with m7G46, A9, G45, A44 and G43, Figure 5) or contribution from ions and solvent molecules or other molecular partners. This is in good agreement with the observation that, of the available standard tRNA structures, some present such A14–A21 interaction while others do not. For example, structure of yeast tRNAAsp (PDB code 2tra) and M.jannaschii tRNALys (PDB code: 1j1u) are quite similar to our optimized structure presenting the A14-A21 N3-amino H-bond interaction. 1j1u is shown superimposed to both the optimized and RB 1ehz structure in Figure 2. The computed ΔERBI for the A14–A21 interaction in the 1j1u and 2tra structures is about −6 kcal/mol, which is intermediate between the 1ehz ΔERBI and ΔEOpt values.
Figure 5.
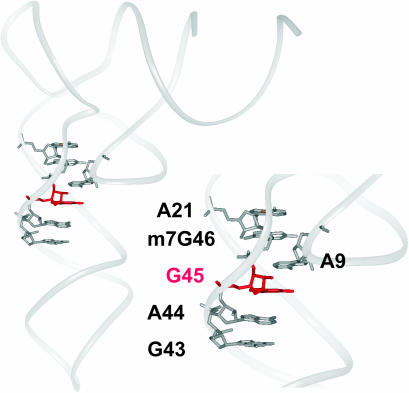
Detail of yeast tRNAPhe X-ray structure at 1.93 Å resolution [PDB code: 1ehz (54)]. Ribose-phosphate backbone is shown as grey solid oval ribbon. Six staggered purine bases of the D and V arms are shown in a stick representation and labeled. The central G45 is colored in red.
The cases of G15–C48 pair and m2G10–C25–G45 triplet are probably the most intriguing. The G15–C48 pair presents an RWC geometry in the 1ehz structure. However, it is well known that the isolated G–C RWC pair is not a stable geometry due to repulsive amino–amino and carbonyl–carbonyl contacts (35). Indeed, in the fully optimized structure, we observe a severe geometrical rearrangement which decreases the amino–amino repulsion, but also breaks the N2(G15):N3(C48) H-bond of the X-ray structure, dealing to a bifurcated H-bond pattern (in Figure 3).
The r.m.s.d. for the optimized and X-ray heavy atoms m2G10–C25–G45 structures superimposition is the highest observed here, 2.11 Å. Also in this case, as for 8–14–21, the main interacting base pair, the m2G10–C25 WC, is well superimposable in the two structures. However the third interacting base G45 assumes a remarkably different orientation after geometry optimization (Figure 2).
Our results show that the 10–25–45 interaction, as well as 15–48, although stable (−13.8 kcal/mol for the RWC G15–C48 pair, −8.4 kcal/mol for the interaction of G45 with m2G10–C25), does not represent an energy minimum when extracted from 1ehz. For both the interactions, geometries corresponding to the global minima are not compatible with the overall tRNA structure. However in 1ehz other forces can contribute to stabilize such geometries. In particular the effect of stacking interactions, of H-bonded water molecules and neighbor metal cations, as well as of base–backbone and backbone–backbone interactions need to be investigated, in order to explain the observed geometries. That is besides the purpose of this paper.
Stacking effects seem to play a particularly important role in maintaining the observed 1ehz G10–C25–G45 geometry. G45 is indeed closely stacked between nucleotides A9 and A44, participating in a series of five staggered purine bases (Figure 5). The nt 9 is known to be a conserved purine in cytosolic tRNAs (95% of all analyzed cytosolic sequences). Interestingly, nt 44 is a conserved purine (79%) in class I tRNA, where nt 45 gives tertiary interaction with the D stem, whereas it is a pyrimidine (91%) in class II tRNA where nt 45 instead participates into the additional V stem.
It is commonly accepted that tRNA tertiary interactions substantially contribute to the tRNA fold stabilization. But, how much does this contribution amount to? An approximate estimate for the tertiary base pairing contribution can be obtained using the rigid body interaction energies (ΔERBI) we calculated. By summing the energy contribution from each tertiary interaction in the yeast tRNAPhe structure, we obtain a total contribution in terms of base pairing interactions of −167 kcal/mol (only the stabilization given to the triplet from the third bases is counted for the 9–12–23, 10–25–45 and 13–22–46 triplets, for instance in the case of A9–U12–A23 only the interaction energy between A9 and the U12–A23 base pair is considered). This roughly contributes for a remarkable 25% to the total base pairing interaction energies of the overall yeast tRNAPhe structure, by considering a contribution of about −450 kcal/mol from the base pairing interactions in the 1ehz stems [12 G–C WC, 8 A–U WC and 1 G–U Wobble base pair, our values and Ref. (33)]. Of course, this estimate can only be considered an approximation since deformation energies are not included in the ΔERBI values.
Our calculations indicate that the most stable base pairing tertiary interaction occurs between the charged m7G46 with the WC base pair C13–G22. Note that such modification is observed in 240 out of the 369 analyzed class I tRNA sequences (65%), while it is totally absent in class II tRNAs where nt 46 is enclosed in the additional V arm. This interaction is estimated to contribute −38.0 kcal/mol to the tRNA structure stabilization. This value can be overestimated, owing to the presence of a positive charge not compensated in the computation by counter ions, solvent molecules and the ribose-phosphate backbone. However, the stability of the corresponding unmodified interaction is still as high as −19.4 kcal/mol.
The second and third most stable interactions are the G19–C56 WC and T54–m1A58 RH base pairs, with an energy of −26.8 kcal/mol and −22.5 kcal/mol, respectively. For the latter a little overestimation due to an uncompensated charge on the nt 58, analogously to the case of 7mG46, cannot be excluded. This finding is in good agreement with all available data. Such interactions indeed, localized at the corner of the molecule and involving highly conserved nucleotides, are very well conserved in cytosolic tRNAs, indicating their importance for the tRNA function. Note also that, among yeast tRNAPhe tertiary interactions, the G19–C56 pair is the only canonical WC and that the U54–m1A58 RH pair flanks the very well conserved U turn motif in the T loop. The functional and structural relevance of such interactions is also well documented in literature (9,18,20,21,70). It is worth noting that the stability of each remaining tertiary base pairing interaction, except for A9–U12–A23 and m2G10–C25–G45, is always comparable with or higher than that of a canonical WC A–T base pair.
As the effect of chemical modification is concerned, we investigated six modifications on the five interactions: m2G10–C25–G45 and C13–G22–m7G46 base triplets, and G18–Ψ55, m22G26–A44 and T54–m1A58 base pairs. Superimposition between the modified and unmodified base pairs/triplets is reported in Figure 4.
The geometrical effect of the additional methyl on G10 amino group, as well as on U54 C4 atom, is practically undetectable. On the contrary, the other investigated chemical modifications introduce a modest geometrical distortion in the computed interactions. In particular, the addition of a methyl group on N7 of m7G46 and on N1 of m1A58, both introducing a positive charge on the corresponding interaction, particularly affect the inter-base distances (H-bonds lengths change up to 0.29 Å). The C13–G22–m7G46 is indeed the only triplet where the C13–G22 WC pair is substantially deformed and destabilized in the triplet as compared with an isolated C–G WC base pair (Table 4). This can be explained with the electrostatic repulsion between the positively charged m7G46 base and the partial positive charge on the C13 amino group.
The additional double methyl group on N2 of m22G26 and the isomerization from U55 to Ψ55 instead both affect the deviation from planarity of the corresponding G26–A44 and G18–U55 unmodified interactions, enhancing and decreasing it, respectively. Note that m22G26–A44 is a Cis WC base pair. Cis A–G WC base-pairs have been widely investigated in a computational and statistical study (32), where they were found to be very flexible, being able to adopt a wide range of low-energy geometries, although with a clear preference for the propeller twisted conformation around the O6(G)–N6(A) axis. They occur mostly at the ends of canonical helices, where they serve as interface with other motifs introducing conformational flexibility. The G26–A44 interaction thus can introduce structural flexibility in tRNA at the interface between the AC and D helix, where the pattern of tertiary base pairing interactions begins. However we showed that the chemical modification on N2(G26) introduces a structural constraint which blocks the base pair in a high propeller twisted conformation, limiting the possible conformational flexibility.
The stabilizing effect of the charged m7G46 and m1A58 nucleotides on the C13–G22–m7G46 base triplet and T54–m1A58/U54–m1A58 pair, as compared with the unmodified and neutral C13–G22–G46 triplet and T54–A58/U54–A58 pair, is remarkable (17.4 and 6.8/6.2 kcal/mol, respectively) although possibly overestimated here, as already discussed. Conversely, the effect of the remaining four investigated chemical modifications, m2G10, m22G26, T54 and Ψ55, on the base pairing interactions stability, is not very significant (despite of the slight planarity distortion caused by m22G26 and Ψ55 on the corresponding interactions). In particular modifications on nucleotides m2G10 and T54 contribute to the stabilization of the G10–C25–G45 and U54–m1A58/U54–A58 interactions by a slight −0.9 kcal/mol and −0.7 kcal/mol, respectively. The effect of m22G26 and Ψ55 instead goes in the direction of a destabilization of the modified interaction by 0.8 and 0.1 kcal/mol for m22G26–A44 and G18–Ψ55, respectively.
Therefore, of the three investigated modifications occurring in the T loop, only m1A58 substantially contributes to the stability of the corresponding base pairing interaction, whereas T54 does not have a very significant stabilizing effect (−0.7) and Ψ55 has a negligible destabilizing effect (+0.1). However, Ψ55 has been experimentally shown to slightly enhance the stability of the interaction between the T and D domains from −6.2 ± 1.6 kcal/mol of the unmodified T loop to −6.9 ± 2.5 kcal/mol (71). Although results of our calculations lie within the experimental error, possible stabilizing contributions of such modified nucleotide could be ascribed to other factors determining the RNA structure, different from the base pairing interactions considered here, for instance the demonstrated base-stacking contribution of Ψ in a tRNA base-paired region (72). In the same study a stabilization effect from −6.2 ± 1.6 to −7.4 ± 2.0 kcal/mol has been measured for T54, that is in qualitative agreement with our results. However, also in this case additional contributions from other structural factors cannot be excluded.
CONCLUDING REMARKS
In this paper we systematically investigated the stability of tRNA tertiary interactions focusing on the base pairing contribution. To this aim we employed advanced quantum mechanics computational techniques to calculate geometries and interaction energies for H-bonded base pairs and triplets corresponding to the nine classical tertiary interactions in the yeast tRNAPhe model system.
What we learned from this approach is that tRNA tertiary interactions are mostly inter-base H-bonds driven. Indeed in only three of the nine analyzed interactions, G15–C48, U8–A14–A21 and m2G10–C25–G45, other effects from the context of the overall tRNA structure are needed to explain the observed geometries. Furthermore, we estimate here that the tertiary contribution to the overall base pairing tRNA interaction energy is as high as 25%, with most of the tertiary base pairing interactions contributing with an energy comparable with or higher than that of a canonical WC A–T base pair. Finally, the effect of chemical modifications on the tertiary base pairing stability in the analyzed cases has been shown to range from important for the positively charged m7G46 and m1A58, to moderate for the additionally methylated neutral m2G10, m22G26 and T54 and negligible for the isomerized Ψ55. With the exception of Ψ55 and of m22G, they all stabilize the corresponding base pairing interaction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by University ‘La Sapienza’ of Rome (C26A039249) and by the Biosapiens Network of Excellence, European Commission FP6 Programme, contract number LHSG-CT-203-503265. R.O. is recipient of a post-doctoral grant from Centro Linceo Interdisciplinare ‘Beniamino Segre’, Accademia dei Lincei. Funding to pay the Open Access publication charges for this article was provided by EU.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kim S.H., Suddath F.L., Quigley G.J., McPherson A., Sussman J.L., Wang A.H., Seeman N.C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- 2.Robertus J.D., Ladner J.E., Finch J.T., Rhodes D., Brown R.S., Clark B.F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 3.Hingerty B., Brown R.S., Jack A. Further refinement of the structure of yeast tRNAPhe. J. Mol. Biol. 1978;124:523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- 4.Rich A., Kim S.H. The three-dimensional structure of transfer RNA. Sci. Am. 1978;238:52–62. doi: 10.1038/scientificamerican0178-52. [DOI] [PubMed] [Google Scholar]
- 5.Sussman J.L., Holbrook S.R., Warrant R.W., Church G.M., Kim S.H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J. Mol. Biol. 1978;123:607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- 6.Hou Y.M., Schimmel P. Novel transfer RNAs that are active in Escherichia coli. Biochemistry. 1992;31:4157–4160. doi: 10.1021/bi00132a001. [DOI] [PubMed] [Google Scholar]
- 7.O'Mahony D.J., Mims B.H., Thompson S., Murgola E.J., Atkins J.F. Glycine tRNA mutants with normal anticodon loop size cause -1 frameshifting. Proc. Natl Acad. Sci. USA. 1989;86:7979–7983. doi: 10.1073/pnas.86.20.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuohy T.M., Thompson S., Gesteland R.F., Atkins J.F. Seven, eight and nine-membered anticodon loop mutants of tRNA(2Arg) which cause +1 frameshifting. Tolerance of DHU arm and other secondary mutations. J. Mol. Biol. 1992;228:1042–1054. doi: 10.1016/0022-2836(92)90313-9. [DOI] [PubMed] [Google Scholar]
- 9.Levinger L., Bourne R., Kolla S., Cylin E., Russell K., Wang X., Mohan A. Matrices of paired substitutions show the effects of tRNA D/T loop sequence on Drosophila RNase P and 3′-tRNase processing. J. Biol. Chem. 1998;273:1015–1025. doi: 10.1074/jbc.273.2.1015. [DOI] [PubMed] [Google Scholar]
- 10.Li Z., Gillis K.A., Hegg L.A., Zhang J., Thurlow D.L. Effects of nucleotide substitutions within the T-loop of precursor tRNAs on interaction with ATP/CTP:tRNA nucleotidyltransferases from Escherichia coli and yeast. Biochem. J. 1996;314:49–53. doi: 10.1042/bj3140049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou Y.M., Motegi H., Lipman R.S., Hamann C.S., Shiba K. Conservation of a tRNA core for aminoacylation. Nucleic Acids Res. 1999;27:4743–4750. doi: 10.1093/nar/27.24.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamann C.S., Hou Y.M. Probing a tRNA core that contributes to aminoacylation. J. Mol. Biol. 2000;295:777–789. doi: 10.1006/jmbi.1999.3353. [DOI] [PubMed] [Google Scholar]
- 13.Christian T., Lipman R.S., Evilia C., Hou Y.M. Alternative design of a tRNA core for aminoacylation. J. Mol. Biol. 2000;303:503–514. doi: 10.1006/jmbi.2000.4169. [DOI] [PubMed] [Google Scholar]
- 14.Puglisi J.D., Putz J., Florentz C., Giege R. Influence of tRNA tertiary structure and stability on aminoacylation by yeast aspartyl-tRNA synthetase. Nucleic Acids Res. 1993;21:41–49. doi: 10.1093/nar/21.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibba M., Hong K.W., Sherman J.M., Sever S., Soll D. Interactions between tRNA identity nucleotides and their recognition sites in glutaminyl-tRNA synthetase determine the cognate amino acid affinity of the enzyme. Proc. Natl Acad. Sci. USA. 1996;93:6953–6958. doi: 10.1073/pnas.93.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao R., Zhao M.W., Hao Z.X., Yao Y.N., Wang E.D. A T-stem slip in human mitochondrial tRNALeu(CUN) governs its charging capacity. Nucleic Acids Res. 2005;33:3606–3613. doi: 10.1093/nar/gki677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francisci S., De Luca C., Oliva R., Morea V., Tramontano A., Frontali L. Aminoacylation and conformational properties of yeast mitochondrial tRNA mutants with respiratory deficiency. RNA. 2005;11:914–927. doi: 10.1261/rna.2260305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zagryadskaya E.I., Kotlova N., Steinberg S.V. Key elements in maintenance of the tRNA L-shape. J. Mol. Biol. 2004;340:435–444. doi: 10.1016/j.jmb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Doyon F.R., Zagryadskaya E.I., Chen J., Steinberg S.V. Specific and non-specific purine trap in the T-loop of normal and suppressor tRNAs. J. Mol. Biol. 2004;343:55–69. doi: 10.1016/j.jmb.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 20.Zagryadskaya E.I., Doyon F.R., Steinberg S.V. Importance of the reverse Hoogsteen base pair 54–58 for tRNA function. Nucleic Acids Res. 2003;31:3946–3953. doi: 10.1093/nar/gkg448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du X., Wang E.D. Tertiary structure base pairs between D- and TΨC-loops of Escherichia coli tRNA(Leu) play important roles in both aminoacylation and editing. Nucleic Acids Res. 2003;31:2865–2872. doi: 10.1093/nar/gkg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J.C., Cannone J.J., Gutell R.R. The lonepair triloop: a new motif in RNA structure. J. Mol. Biol. 2003;325:65–83. doi: 10.1016/s0022-2836(02)01106-3. [DOI] [PubMed] [Google Scholar]
- 23.Krasilnikov A.S., Mondragon A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Smit M.H., Gultyaev A.P., Hilge M., Bink H.H., Barends S., Kraal B., Pleij C.W. Structural variation and functional importance of a D-loop-T-loop interaction in valine-accepting tRNA-like structures of plant viral RNAs. Nucleic Acids Res. 2002;30:4232–4240. doi: 10.1093/nar/gkf539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piron M., Beguiristain N., Nadal A., Martinez-Salas E., Gomez J. Characterizing the function and structural organization of the 5′ tRNA-like motif within the hepatitis C virus quasispecies. Nucleic Acids Res. 2005;33:1487–1502. doi: 10.1093/nar/gki290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Šponer J., Leszczynski J., Hobza P. Hydrogen bonding and stacking of DNA bases: a review of quantum-chemical ab initio studies. J. Biomol. Struct. Dyn. 1996;14:117–135. doi: 10.1080/07391102.1996.10508935. [DOI] [PubMed] [Google Scholar]
- 27.Šponer J., Florian J., Hobza P., Leszczynski J. Nonplanar DNA base pairs. J. Biomol. Struct. Dyn. 1996;13:827–833. doi: 10.1080/07391102.1996.10508895. [DOI] [PubMed] [Google Scholar]
- 28.Šponer J., Burda J.V., Mejzlik P., Leszczynski J., Hobza P. Hydrogen-bonded trimers of DNA bases and their interaction with metal cations: ab initio quantum-chemical and empirical potential study. J. Biomol. Struct. Dyn. 1997;14:613–628. doi: 10.1080/07391102.1997.10508161. [DOI] [PubMed] [Google Scholar]
- 29.Hobza P., Šponer J. Structure, energetics, and dynamics of the nucleic Acid base pairs: nonempirical ab initio calculations. Chem. Rev. 1999;99:3247–3276. doi: 10.1021/cr9800255. [DOI] [PubMed] [Google Scholar]
- 30.Šponer J., Leszczynski J., Hobza P. Electronic properties, hydrogen bonding, stacking, and cation binding of DNA and RNA bases. Biopolymers. 2001;61:3–31. doi: 10.1002/1097-0282(2001)61:1<3::AID-BIP10048>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Hobza P., Šponer J. Toward true DNA base-stacking energies: MP2, CCSD(T), and complete basis set calculations. J. Am. Chem. Soc. 2002;124:11802–11808. doi: 10.1021/ja026759n. [DOI] [PubMed] [Google Scholar]
- 32.Šponer J., Hobza P. Molecular interactions of nucleic acid bases. A review of quantum-chemical studies. Collect. Czech. Chem. Commun. 2003;68:2231–2282. [Google Scholar]
- 33.Šponer J., Jurecka P., Hobza P. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J. Am. Chem. Soc. 2004;126:10142–10151. doi: 10.1021/ja048436s. [DOI] [PubMed] [Google Scholar]
- 34.Perez A., Šponer J., Jurecka P., Hobza P., Luque F.J., Orozco M. Are the hydrogen bonds of RNA (AU) stronger than those of DNA (AT)? A quantum mechanics study. Chemistry. 2005;11:5062–5066. doi: 10.1002/chem.200500255. [DOI] [PubMed] [Google Scholar]
- 35.Zhanpeisov N.U., Šponer J., Leszczynski J. Reverse Watson–Crick Isocytosine–Cytosine and Guanine–Cytosine base pairs stabilized by the formation of the minor tautomers of bases. An ab initio study in the gas phase and in a water cluster. J. Phys. Chem. A. 1998;102:10374. [Google Scholar]
- 36.Swart M., Guerra C.F., Bickelhaupt F.M. Hydrogen bonds of RNA are stronger than those of DNA, but NMR monitors only presence of methyl substituent in uracil/thymine. J. Am. Chem. Soc. 2004;126:16718–16719. doi: 10.1021/ja045276b. [DOI] [PubMed] [Google Scholar]
- 37.Guerra C.F., Bickelhaupt F.M., Baerends E.J. Orbital interactions in hydrogen bonds important for cohesion in molecular crystals and mismatched pairs of DNA bases. Cryst. Growth Des. 2002;2:239–245. [Google Scholar]
- 38.Guerra C.F., Bickelhaupt F.M., Baerends E.J. Hydrogen bonding in mimics of Watson–Crick base pairs involving C-H proton donor and F proton acceptor groups: a theoretical study. ChemPhysChem. 5:481–487. doi: 10.1002/cphc.200301069. [DOI] [PubMed] [Google Scholar]
- 39.Guerra C.F., Bickelhaupt F.M. Orbital interactions in strong and weak hydrogen bonds are essential for DNA replication. Angew. Chem. Int. Ed. Engl. 2002;41:2092–2095. [PubMed] [Google Scholar]
- 40.Rueda M., Luque F.J., Lopez J.M., Orozco M. Amino–Imino Tautomerism in derivatives of cytosine: effect on hydrogen-bonding and stacking properties. J. Phys. Chem. A. 2001;105:6575–6580. [Google Scholar]
- 41.Rueda M., Kalko S.G., Luque F.J., Orozco M. The structure and dynamics of DNA in the gas phase. J. Am. Chem. Soc. 2003;125:8007–8014. doi: 10.1021/ja0300564. [DOI] [PubMed] [Google Scholar]
- 42.Møller C., Plesset M.S. Note on an approximation treatment for many-electron systems. Phys. Rev. 1934;46:618–622. [Google Scholar]
- 43.Feyereisen M., Fitzgerald G., Komornicki A. Use of approximate integrals in ab initio theory. An application in MP2 energy calculations. Chem. Phys. Lett. 1993;208:359. [Google Scholar]
- 44.Weigend F., Häser M. RI-MP2: first derivatives and global consistency. Theor. Chem. Acc. 1997;97:331. [Google Scholar]
- 45.Jurečka P., Nachtigall P., Hobza P. RI-MP2 calculations with extended basis sets—a promising tool for study of H-bonded and stacked DNA base pairs. Phys. Chem. Chem. Phys. 2001;3:4578–4582. [Google Scholar]
- 46.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 47.Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 48.Becke A.D. Density-functional thermochemistry. IV. A new dynamical correlation functional and implications for exact-exchange mixing. J. Chem. Phys. 1996;104:1040–1046. [Google Scholar]
- 49.Stephens P.J., Devlin F.J., Chabalowski C.F., Frisch M.J. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 1994;98:11623–11627. [Google Scholar]
- 50.Perdew J.P., Burke K., Wang Y. Generalized gradient approximation for the exchange-correlation hole of a many-electron system. Phys. Rev. B Condens Matter. 1996;54:16533–16539. doi: 10.1103/physrevb.54.16533. [DOI] [PubMed] [Google Scholar]
- 51.Perdew J.P., Wang Y. Accurate and simple analytic representation of the electron-gas correlation energy. Phys. Rev. B Condens Matter. 1992;45:13244–13249. doi: 10.1103/physrevb.45.13244. [DOI] [PubMed] [Google Scholar]
- 52.Tsuzuki S., Lüthi H.P. Interaction energies of van der Waals and hydrogen bonded systems calculated using density functional theory: Assessing the PW91 model. J. Chem. Phys. 2001;114:3949–3957. [Google Scholar]
- 53.Müller A., Losada M., Leutwyler S. Ab initio benchmark study of (2-Pyridone)2, a strongly bound doubly hydrogen-bonded dimer. J. Phys. Chem. A. 2004;108:157–165. [Google Scholar]
- 54.Shi H., Moore P.B. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quigley G.J., Wang A.H., Seeman N.C., Suddath F.L., Rich A., Sussman J.L., Kim S.H. Hydrogen bonding in yeast phenylalanine transfer RNA. Proc. Natl Acad. Sci. USA. 1975;72:4866–4870. doi: 10.1073/pnas.72.12.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salemink P.J., Yamane T., Hilbers C.W. Demonstration of a tertiary interaction in solution between the extra arm and the D-stem in two different transfer RNA's by NMR. Nucleic Acids Res. 1977;4:3727–3741. doi: 10.1093/nar/4.11.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agris P.F., Sierzputowska-Gracz H., Smith C. Transfer RNA contains sites of localized positive charge: carbon NMR studies of [13C]methyl-enriched Escherichia coli and yeast tRNAPhe. Biochemistry. 1986;25:5126–5131. doi: 10.1021/bi00366a022. [DOI] [PubMed] [Google Scholar]
- 58.Bullock T.L., Sherlin L.D., Perona J.J. Tertiary core rearrangements in a tight binding transfer RNA aptamer. Nature Struct. Biol. 2000;7:497–504. doi: 10.1038/75910. [DOI] [PubMed] [Google Scholar]
- 59.Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sprinzl M., Vassilenko K.S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berman H.M., Bhat T.N., Bourne P.E., Feng Z., Gilliland G., Weissig H., Westbrook J. The Protein Data Bank and the challenge of structural genomics. Nature Struct. Biol. 2000;7:957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- 62.Dayringer H.E., Tramontano A., Sprang S.R., Fletterick R.J. Interactive program for visualization and modeling of protein, nucleic acids and small molecules. J. Mol. Graph. 1986;4:82–87. [Google Scholar]
- 63.Ahlrichs R., Bar M., Haser M., Horn H., Kolmel C. Electronic structure calculations on workstation computers: The program system turbomole. Chem. Phys. Lett. 1989;162:165–169. [Google Scholar]
- 64.Dunning T.H., Jr Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. J. Chem. Phys. 1989;90:1007–1023. [Google Scholar]
- 65.Boys S.F., Bernardi F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Phys. 1970;19:553–566. [Google Scholar]
- 66.Burda J.V., Šponer J., Leszczynski J., Hobza P. Interaction of DNA base pairs with various metal cations (Mg2+, Ca2+, Sr2+, Ba2+, Cu+, Ag+, Au+, Zn2+, Cd2+, and Hg2+): nonempirical ab initio calculations on structures, energies, and nonadditivity of the interaction. J. Phys. Chem. B. 1997;101:9670–9677. [Google Scholar]
- 67.Kobayashi T., Nureki O., Ishitani R., Yaremchuk A., Tukalo M., Cusack S., Sakamoto K., Yokoyama S. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nature Struct. Biol. 2003;10:425–432. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- 68.Westhof E., Dumas P., Moras D. Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Crystallogr. A. 1988;44:112–123. [PubMed] [Google Scholar]
- 69.Sierzputowska-Gracz H., Gopal H.D., Agris P.F. Comparative structural analysis of 1-methyladenosine, 7-methylguanosine, ethenoadenosine and their protonated salts IV: 1H, 13C, and 15N NMR studies at natural isotope abundance. Nucleic Acids Res. 1986;14:7783–7801. doi: 10.1093/nar/14.19.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fukai S., Nureki O., Sekine S., Shimada A., Vassylyev D.G., Yokoyama S. Mechanism of molecular interactions for tRNA(Val) recognition by valyl-tRNA synthetase. RNA. 2003;9:100. doi: 10.1261/rna.2760703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nobles K.N., Yarian C.S., Liu G., Guenther R.H., Agris P.F. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 2002;30:4751–4760. doi: 10.1093/nar/gkf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis D.R., Veltri C.A., Nielsen L. An RNA model system for investigation of pseudouridine stabilization of the codon–anticodon interaction in tRNALys, tRNAHis and tRNATyr. J. Biomol. Struct. Dyn. 1998;15:1121. doi: 10.1080/07391102.1998.10509006. [DOI] [PubMed] [Google Scholar]
- 73.Burkard M.E., Turner D.H., Tinoco I., Jr . Structures of base pairs involving at least two hydrogen bonds. In: Gesteland R.F., Cech T.R., Atkins J.F., editors. The RNA World, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 675–680. [Google Scholar]
- 74.Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nagaswamy U., Larios-Sanz M., Hury J., Collins S., Zhang Z., Zhao Q., Fox G.E. NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res. 2002;30:395–397. doi: 10.1093/nar/30.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.