Abstract
Eukaryotic transcription activation domains (ADs) are not well defined on the proteome scale. We systematicallly tested ∼6000 yeast proteins for transcriptional activity using a yeast one-hybrid system and identified 451 transcriptional activators. We then determined their transcription activation strength using fusions to the Gal4 DNA-binding domain and a His3 reporter gene which contained a promoter with a Gal4-binding site. Among the 132 strongest activators 32 are known transcription factors while another 35 have no known function. Although zinc fingers, helix–loop–helix domains and several other domains are highly overrepresented among the activators, only few contain characterized ADs. We also found some striking correlations: the stronger the activation activity, the more acidic, glutamine-rich, proline-rich or asparagine-rich the activators were. About 29% of the activators have been found previously to specifically interact with the transcription machinery, while 10% are known to be components of transcription regulatory complexes. Based on their transcriptional activity, localization and interaction patterns, at least six previously uncharacterized proteins are suggested to be bona fide transcriptional regulators (namely YFL049W, YJR070C, YDR520C, YGL066W/Sgf73, YKR064W and YCR082W/Ahc2).
INTRODUCTION
Transcriptional regulators (TRs) that activate transcription are usually composed of a DNA-binding domain (DBD) and an activation domain (AD). The DBD targets these proteins to a specific binding site in the promoter or enhancer region of a gene and the AD mediates transcription initiation (1).
Transcriptional activators in yeast were among the first to be studied in detail. In all known cases they recruit additional proteins or whole complexes to the pertinent promoters, eventually leading to the binding of one of the three RNA polymerases. For example, the yeast transcription regulator Gal4 is involved in regulation of galactose metabolism and activates transcription by recruitment of the basal transcription machinery (2–5).
The Gal4–AD can recruit Tra1, a component of the SAGA complex, to the upstream activating sequence (UAS), where Gal4 itself is bound. SAGA, in turn, recruits the mediator complex to the UAS. The Gal4–AD can also directly recruit the mediator complex. In any case, the UAS bound mediator is required for recruitment of general transcription factors to the core promoter and assembly of the pre-initiation complex.
While DBDs are extremely well characterized both functionally and structurally, ADs do not share easily recognizable motifs or structures (6). Accordingly, no specific pattern or motif for the identification of an AD has been defined in pattern/domain databases such as Prosite (7) or SMART (8). In contrast, >50 patterns for DBDs have been documented in the SMART database.
Because the activation properties of transcriptional activators cannot easily be recognized by sequence homology, several studies tried to identify more general sequence features resulting in a number of different AD classes (9), e.g. acidic activators (10), glutamine-rich activators (11) and proline-rich activators (12). In addition, a few rather unspecific properties like hydrophobic patches interspersed with hydrophilic residues (13) or amphipatic alpha helices (14) have been identified. These analyses culminated in the finding that even small chemical compounds with a certain pattern of hydrophobic and hydrophilic residues are sufficient for transcriptional activation (15).
Only a few proteins have been identified as specific interaction partners of transcriptional activators, including TATA-box binding protein, TFIIB, TFIIH (4,16–20) and several others [reviewed in (21)].
Although previous studies have attempted to screen for transcriptional activators [e.g. (22)], we do not know of such a systematic screen in yeast. Incidentally, we approached the problem indirectly by doing systematic yeast-two-hybrid (Y2H) screens (23) which are based on a split transcription regulator, e.g. Gal4. In these screens all yeast open reading frames (ORFs) have been fused to the DBD of yeast Gal4 and tested for interaction with every other yeast protein. Unfortunately, in many cases no interactions could be detected because the Gal4–DBD fusion protein was activating transcription without requiring a second fusion protein, that is, the Gal4–AD fusion, also known as prey.
Gal4–DBD fusion proteins that activate transcription of a reporter gene without requirement of a prey protein are called ‘auto-activators’ (or activators for short) and thereby possess properties of an AD. Our genome-wide two-hybrid screen thus provided data about activation properties of nearly all yeast proteins. Up to now this information has not been used for the understanding of transcription in yeast. However, similar assays have been used previously by Wiesner et al. (24) and Ma and Ptashne (25) for screening human and Escherichia coli proteins/peptides for their transcriptional activation properties. By design such screens do not necessarily identify physiological activators as the fusion partners are artificially targeted to the promoter of a single, arbitrarily selected reporter gene.
Bearing these caveats in mind, we not only selected Y2H auto-activators but also measured their activation strength. In a second step we analyzed these auto-activators for their domain content and other sequence properties. We found that auto-activators possess specific physicochemical properties, like certain amino acid clusters, and can often be found in known complexes of the transcription machinery. Many of the activators identified in this study are known physiological activators. However, many do not appear to be localized to the nucleus under standard laboratory conditions and thus may not act as bona fide transcription factors.
Nevertheless, we believe that even such cases can shed light on the mechanisms of transcriptional activation similar to heterologous proteins such as VP16 which helped to uncover many mechanistic details of transcriptional activation.
MATERIALS AND METHODS
Selection of transcriptional activators
Transcriptional activators were identified in two genome-wide Y2H studies of yeast proteins (26,27). The ‘Ito screen’ resulted in 392 auto-activators which were identified by testing essentially all yeast proteins for transcription activation when fused to the DBD of Gal4 as full-length ORFs (27). The ‘Uetz set’ contains 68 auto-activators and was identified in two-hybrid screens of ∼600 yeast proteins (26).
For analysis of transcription activator properties activators identified by Ito et al. were selected from the Uetz bait library (26) and combined with the Uetz activators to form a set of 451 proteins. These two independent selection steps ensured correct identification of activators. For further quality control, we sequenced 48 samples from the Ito collection and checked another 29 from the Uetz lab by colony PCR for correct insert size. Among these 77 clones only 1 (out of the 29 mentioned) did not match the expected identity and was thus excluded from further analysis. The activators of the ‘Ito set’ were then divided into activation strength groups (see below). However, 79 strains did not grow well enough to be quantified, although they were reported to be activators. Their activation strength has been annotated as ‘not available’ (NA) in Supplementary Table 1.
Yeast strains
The activators were selected from the bait proteins described by Uetz et al. (26) and Hazbun et al. (28), and consisted of full-length ORFs fused to the Gal4–DBD in the CEN4 plasmid pOBD2 (29). These DBD-activator gene constructs were expressed in the strain YULH (MATa, ura3-52, trp1, lys2, his3, leu2, gal4Δ, gal80Δ, GAL1-URA3, GAL1-lacZ). This ‘activator strain’ was re-arrayed onto 96-well plates and mated with yeast cells of the opposite mating type [namely PJ69-4α: MAT α, trp1-901, leu2–3, ura3–52, his3–200, gal4Δ, gal80Δ, GAL2-ADE2, LYS::GAL1-HIS3, met2::GAL7-lacZ (26,30)] carrying an empty prey vector pLP-GADT7 (Clontech). This mating was necessary in order to introduce the HIS3 reporter gene of the PJ69-4 α strain.
The handling of yeast colonies was done by automated robotic procedures employing a Biomek 2000 robotic workstation (Beckman Coulter).
Measurements of transcriptional activation strength
The activation strength was measured by two different assays which measure the expression of two reporter genes: His3, which encodes imidazoleglycerol-phosphate dehydratase and catalyzes the sixth step in histidine biosynthesis (LTH assay) and beta-galactosidase (bGal assay).
His3 gene expression was measured (LTH assay) by growing yeast cells on selective media lacking leucine (L), tryptophane (T) and histidine (H) with the former two corresponding to markers on the two vectors pOBD2 and pLP-GADT7. Activity of the HIS3 reporter was quantified by increasing amounts of 3-aminotriazole (3-AT), a competitive inhibitor of His3. The lowest concentration of 3-AT that inhibited growth was considered as the activation strength of the gene construct (29). This assay was done in quadruplicate in order to ensure reproducibility (for detailed results see Supplementary Table 1).
The bGal assay was performed in 96-well plates using ONPG as a substrate (31). Briefly, diploid yeast cells were grown overnight at 30°C in 100 µl selective media (leucine and tryptophane deficient), the media was replaced by 50 µl Z-buffer [1.6% (w/v) Na2HPO4, 0.55% (w/v) NaH2PO4, 0.075% (w/v) KCl and 0.025% (w/v) MgSO4, pH 7] and the cells lysed by two freeze-thaw cycles. For normalization of the cell density OD600 was measured using a microplate reader (Elx808, Bio-Tek Instruments, VT). The assay was started by adding 50 µl of 1.5 mg/ml ONPG in Z-buffer and the initial OD405i was measured immediately. The reaction was incubated at 37°C and OD405 measured at different time points. Time points still in the linear range were considered for calculation of the activation strength.
Finally, the mean bGal activity of a randomly chosen set of genes not found to possess auto-activation properties was subtracted. This assay was done in triplicates and mean and SEM were calculated for each activator (Supplementary Table 1).
Data sources and analysis
General yeast protein properties, e.g. molecular mass and pI, protein annotations and gene ontology (GO) classification were downloaded from SGD (32) as of June 2005. Protein localization data were from Huh et al. (33), which was assumed to be less biased compared with curated data (e.g. GO component data). The abundance of proteins was compared using the genome-wide measurements by Ghaemmaghami et al. (34). Protein interaction data were downloaded from the MIPS database (35).
Overrepresentation of certain GO terms was assessed using the program FuncAssociate (36). For in detail analysis of known TR function and definition of the set of known TRs the GO slim term ‘transcription regulator activity (F-GO 30528)’ was used. Table 1 summarizes the protein sets that were used for analysis. Proteins annotated with the F-GO term ‘Transcriptional activator activity’ (TermID 16563) from the YPD database (Biobase, Germany) served as an additional reference set (37).
Table 1.
Protein sets
| Protein set | Description |
|---|---|
| All yeast | All yeast proteins |
| Non acts | All yeast proteins not in the activator list |
| TR | Known transcriptional activators/regulators (GO slim term ‘transcription activator activity’) |
| All acts | All Y2H activators |
| Nucleus | Nuclear proteins [Huh et al. (33)] |
| Acts nucleus | Y2H activators localized to the nucleus |
| LTHw, LTHm, LTHs | Weak (LTHw), medium (LTHm), and strong (LTHs) activators in LTH assay |
| TR+1, nucleus+1, acts nucleus+1 | Protein sets as indicated above but including directly interacting proteins in the analysis of binary protein interactions. |
Physicochemical properties
General physicochemical properties, e.g. molecular mass and isoelectric point, were taken directly from the datasets of the SGD (32). Calculation of amino acid groups and segments/clusters with specific physicochemical properties, e.g. charge, were done employing the SAPS program (38). Frequencies of amino acids were directly calculated from SGD sequence data. The minimal and maximal pI of a protein was calculated as the lowest and highest pI of a 20 amino acid window, respectively. Amino acid clusters in a protein were defined as the maximal count of a specific amino acid in a 20 amino acid window of the protein. GRAVY (grand average of Hydropathicity) values were calculated as the sum of hydropathy values for all of the amino acids, divided by the number of residues in the sequence (39). Aromaticity is the relative frequency of aromatic amino acids (39) and the codon adaptation index (CAI) is an empirical measure of synonymous codon usage bias, which is positively correlated with the expression level of genes (40) (see also ‘http://www.yeastgenome.org/help/protein_page.html’).
Protein domains
The occurrence of protein domains in the set of Y2H activators was assessed using the SMART domain database (8). The number of proteins with each domain in the Y2H activator set and the whole genome was counted and the enrichment in the former set was calculated. Significance was tested using Fisher's exact test and Holm's procedure for multiple testing correction (41).
Protein interactions
A protein–protein interaction map of transcription activators was generated using Cytoscape (42). In the Cytoscape map only physical binary protein interactions from the MIPS database were considered. Only activators and their direct interaction partners, bridging at least two activators, were selected.
The percentage of activators that interacted with other transcription factors (i.e. proteins with the GO slim term ‘transcription [P-GO 6350])’ was calculated using the set of physical binary protein interactions from the MIPS database. Similarly, we counted with how many transcription complexes our activators interacted. A ‘transcription complex’ was defined as a protein complex with at least 50% of the proteins annotated as being involved in transcription. The protein interactions with these complexes were assessed by using the MIPS data of protein complexes filtered to contain only high-throughput data (for reduction of bias of well-characterized proteins).
Ranking of interaction partners was done by counting with how many proteins of a specific protein set a given yeast protein interacted (using high-throughput protein complex data from the MIPS database).
Statistical analysis
Data processing was done with PERL (www.perl.org) and statistical calculations with R (43). Correction for multiple testing was done with Holm's procedure using the multtest package of R (41,44).
RESULTS
Y2H activators and their activation strength
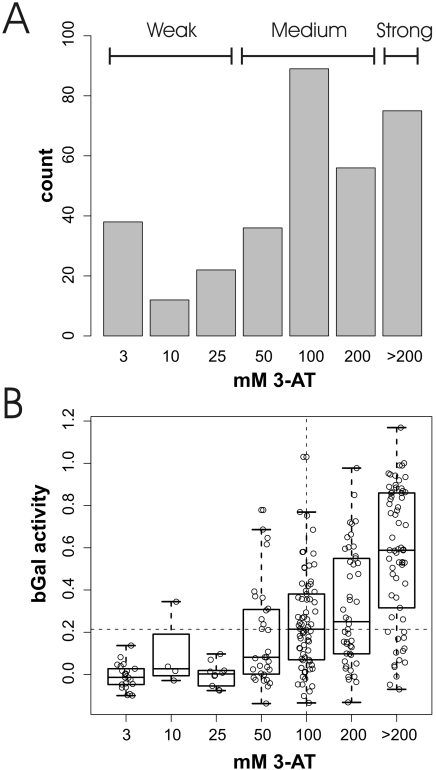
About 450 Y2H activators were identified previously in two genome-wide Y2H screens (Supplementary Table 1) (26,27). However, these proteins were not listed in the original publication nor did these studies provide any quantitative data for the activation strength. Therefore, we measured the activation strength of the activators using two different reporter genes whose expression depends on their level of transcription: His3 and β-galactosidase (bGal assay) (Figure 1, see also Materials and Methods and Supplementary Table 1). According to the activation strength in the LTH assay the activators were grouped into weak (3–25 mM 3-AT), medium (50–200 mM 3-AT) and strong (>200 mM 3-AT) activators. These groups were termed LTHw, LTHm and LTHs, respectively. The bGal assay distribution was divided into strong activators (bGals) which activate above the median and weak activators that activate below the median (bGalW).
Figure 1.
Activation strength in Y2H activators. (A) Number of activators showing the indicated activation strength in the LTH assay. They were divided into weak (LTHw), medium (LTHm) and strong (LTHs) activators as indicated. (B) Correlation of activation strength in LTH and bGal assays. The actually measured values are shown as dots, the median and two quartiles by an overlayed boxplot. Dotted lines indicate the median of bGal activity and LTH activity, respectively. For an explanation of datasets see Table 1.
A comparison of activation strength in both assays revealed an intermediate (Pearson's correlation coefficient of 0.58) but highly significant correlation (P < 2.2e−16) of activation values (Figure 1B). Although both reporters are driven by the Gal1 promoter, the lacZ reporter is actually present in two copies with two different promoters (a GAL7 promoter in PJ69-4a and a GAL1 promoter in YULH). We have not tried to determine the contribution of each of the two lacZ genes and restricted the following analysis to the His3 data.
In summary, a total of 72 weak, 179 medium and 75 strong transcriptional activators were identified in these experiments (Table 2 and Supplementary Table 1).
Table 2.
Strong transcriptional activators when fused C-terminally to a Gal4–DBD and expressed in yeast strain with Gal4-binding sites in the His3 reporter gene
| Name | ORF | Process/function | Loc. |
|---|---|---|---|
| ACE2 | YLR131C | Transcription | N |
| ACF2 | YLR144C | Cytoskeleton | S |
| ADR1 | YDR216W | Transcription | N |
| AOS1 | YPR180W | Protein modification | N |
| APC4 | YDR118W | Protein catabolism | N |
| APL2 | YKL135C | Vesicular transport | C |
| ASM4 | YDL088C | Nuclear organization | N,ER |
| ATC1 | YDR184C | Response to stress# | N |
| BCK2 | YER167W | Cell cycle# | C,N |
| BFR2 | YDR299W | Vesicular transport# | N |
| CAK1 | YFL029C | Meiosis,cell cycle | C |
| CHA4 | YLR098C | Transcription | N |
| CRZ1 | YNL027W | Transcription | C,N |
| CSE2 | YNR010W | Transcription | N |
| CUP2 | YGL166W | Transcription | N |
| ECM22 | YLR228C | Transcription | N |
| FCP1 | YMR277W | Transcription | N |
| GAC1 | YOR178C | Meiosis | |
| GAL11 | YOL051W | Transcription | N |
| GLN3 | YER040W | Transcription | N |
| GYP1 | YOR070C | Vesicular transport | G |
| HAP4 | YKL109W | Transcription | N |
| HSF1 | YGL073W | Transcription | N |
| IDS2 | YJL146W | Meiosis# | C,N |
| IKI1 | YHR187W | Transcription | N |
| IME1 | YJR094C | Transcription | N |
| ISF1 | YMR081C | Cellular respiration# | ? |
| LTV1 | YKL143W | Unknown# | C |
| MAS6 | YNR017W | Transport | M |
| MBR1 | YKL093W | Cellular respiration# | ? |
| MCD4 | YKL165C | Protein modification# | S |
| MED4 | YOR174W | Transcription | N |
| MED8 | YBR193C | Transcription | N |
| MET4 | YNL103W | Transcription | N |
| MRS6 | YOR370C | Transport | C |
| MSB4 | YOL112W | Cytoskeleton | S |
| MSN2 | YMR037C | Response to stress | N |
| MVP1 | YMR004W | Transport# | C |
| NAM7 | YMR080C | RNA metabolism | C |
| NAP1 | YKR048C | Budding | C,N |
| NCA3 | YJL116C | Organelle organization# | ? |
| NUP100 | YKL068W | Nuclear organization | N,ER |
| PAT1 | YCR077C | Cell cycle# | C |
| PCL6 | YER059W | Carbohydrate metabolism | |
| PDC2 | YDR081C | Transcription | N |
| PEX19 | YDL065C | Organelle organization# | C |
| PFK27 | YOL136C | Carbohydrate metabolism | C |
| PHO4 | YFR034C | Transcription | C,N |
| PPG1 | YNR032W | Protein modification | C,N |
| PUP2 | YGR253C | Response to stress | N,ER |
| PUT3 | YKL015W | Transcription | N |
| REC114 | YMR133W | DNA metabolism# | N |
| RIM15 | YFL033C | Response to stress | C |
| RLM1 | YPL089C | Cell wall | N |
| ROX3 | YBL093C | Transcription | N |
| RPA12 | YJR063W | Transcription | N |
| RPB3 | YIL021W | Transcription | N |
| RPI1 | YIL119C | Signal transduction | N |
| RPP1A | YDL081C | Protein biosynthesis | C |
| RSP5 | YER125W | Nuclear organization | N |
| RTG1 | YOL067C | Transcription | C,N |
| SCD5 | YOR329C | Vesicular transport | V |
| SCH9 | YHR205W | Protein modification | C |
| SIN4 | YNL236W | Transcription | N |
| SIW14 | YNL032W | Response to stress | C |
| SLA1 | YBL007C | Cell wall | S |
| SNF2 | YOR290C | Transcription | N |
| SPT21 | YMR179W | Transcription# | N |
| SSN2 | YDR443C | Transcription | N |
| STE5 | YDR103W | Signal transduction | V,C,N |
| SWI3 | YJL176C | Transcription | N |
| SWI5 | YDR146C | Transcription | C,N |
| TOA1 | YOR194C | Transcription | N |
| UBP3 | YER151C | Protein modification | C |
| YAL014C | YAL014C | Transport | C |
| YAP1802 | YGR241C | Vesicular transport | S |
| YBL081W | YBL081W | Unknown# | ? |
| YBR061C | YBR061C | Protein biosynthesis | C |
| YBR271W | YBR271W | Unknown | C |
| YCR082W | YCR082W | Unknown# | C,N |
| YDL161w | YDL161W | Vesicular transport | S |
| YDR031w | YDR031W | Unknown# | C,N |
| YDR213W | YDR213W | Transcription | C,N |
| YDR223W | YDR223W | Unknown# | ? |
| YDR260C | YDR260C | Cell cycle# | N |
| YDR291W | YDR291W | Unknown | N |
| YDR320C | YDR320C | Organelle organization# | ER |
| YDR330W | YDR330W | Protein catabolism# | C,N |
| YDR489W | YDR489W | DNA metabolism | N |
| YDR520C | YDR520C | Unknown# | C,N |
| YER045c | YER045C | Transcription | N |
| YER051W | YER051W | Unknown# | ? |
| YGL036W | YGL036W | Unknown# | C |
| YGL079W | YGL079W | Unknown# | C |
| YGL223C | YGL223C | Vesicular transport# | G |
| YGR120C | YGR120C | Vesicular transport | G |
| YGR130C | YGR130C | Unknown# | C |
| YHL012W | YHL012W | Unknown# | ? |
| YHR160C | YHR160C | Transport | P |
| YIL019W | YIL019W | Unknown# | C,N |
| YIL079C | YIL079C | Transport# | N |
| YJL029C | YJL029C | Vesicular transport# | G,C |
| YJL181W | YJL181W | Unknown# | ? |
| YJR070C | YJR070C | Unknown | C,N |
| YJR119C | YJR119C | Unknown# | C,N |
| YKL059C | YKL059C | RNA metabolism# | N |
| YKL171W | YKL171W | Protein catabolism | C |
| YKR060W | YKR060W | Ribosome biogenesis | N |
| YKR064W | YKR064W | Unknown# | C,N |
| YKR077W | YKR077W | Unknown# | C,N |
| YLL013C | YLL013C | RNA metabolism | C |
| YLR135W | YLR135W | DNA metabolism | N |
| YLR192C | YLR192C | Protein biosynthesis | C |
| YLR285W | YLR285W | Vitamin metabolism | C |
| YLR445W | YLR445W | Unknown# | ? |
| YML037C | YML037C | Unknown# | V |
| YMR030W | YMR030W | Cellular respiration# | M,N |
| YMR048W | YMR048W | Cell cycle# | N |
| YMR181C | YMR181C | Unknown# | ? |
| YMR195W | YMR195W | Unknown# | V |
| YMR295C | YMR295C | Unknown# | S |
| YMR299C | YMR299C | Unknown# | C |
| YNR069C | YNR069C | Unknown# | ? |
| YOR066W | YOR066W | Unknown# | ? |
| YOR166C | YOR166C | Unknown# | ? |
| YOR262W | YOR262W | Unknown# | C |
| YPL105C | YPL105C | Unknown# | C |
| YPL229W | YPL229W | Unknown# | C |
| YPL233W | YPL233W | Cell cycle# | N |
| YPL250C | YPL250C | Unknown# | ? |
| YPR008W | YPR008W | Transcription | N |
| YPR179C | YPR179C | Transcription | N |
3AT concentrations of 200 mM or more are required to suppress the amount of His3 expressed in these strains. Process/function abbreviated after GO annotation in SGD, # = molecular function unknown. Localization (Loc.) encoded as nuclear (N), cytoplasmic (C), cytoskeletal/cell wall (S), Golgi (G), endoplasmic reticulum (ER), peroxisome (P), vesicle/membranes (V), mitochondrion (M) or unknown (?).
Function
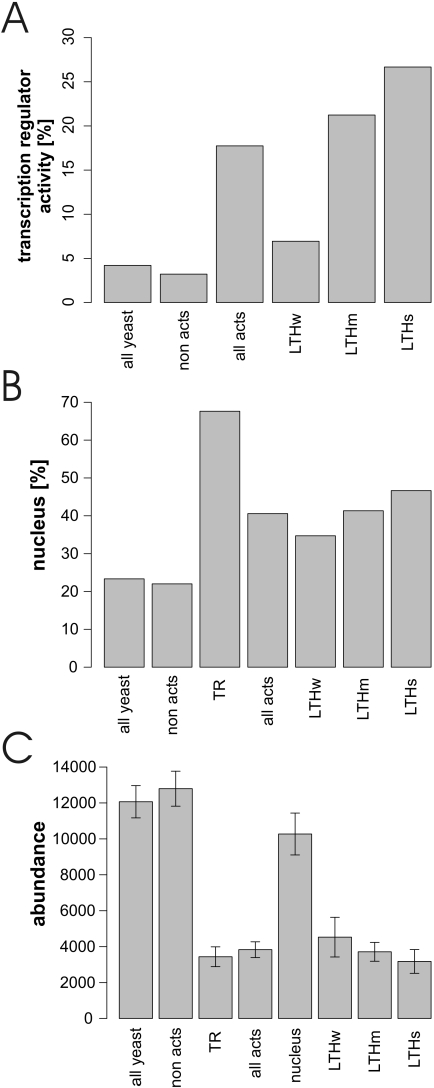
First, we wondered if Y2H activators also act in vivo as transcriptional activators. Surprisingly, only 92 (or 20%) of the 451 activators had the GO attribute ‘transcription regulator activity’ (GO 0030528). Transcription activity was significantly correlated with activation strength: 27% of all strong activators are annotated as having ‘transcription regulatory activity’ (Figure 2A) whereas this annotation drops to 21 and 7% of the medium and weak activators, respectively. These numbers are significantly higher than for the Y2H non-activators (∼3%). As an additional reference set we selected 138 proteins annotated to possess ‘transcriptional activator activity’ in the YPD database. Only 55 (or 12%) of the Y2H activators were annotated as transcriptional activators in YPD. This annotation correlated with the activational strength: 19% of strong, 16% of medium and 4% of weak activators were annotated with this term (Supplementary Table 1). This suggests that many more proteins may act as transcriptional activators than currently known.
Figure 2.
Function, localization and abundance of yeast activators. (A) Percentage of proteins annotated to have ‘TR activity’ (GO term, see also Supplementary Table 1). (B) Percentage of protein localized to the nucleus as analyzed by Huh et al. (33). (C) Abundance of proteins as analyzed by Ghaemmaghami et al. (34).
Localization
Transcription takes place in the nucleus. We wondered if Y2H activators were also localized in the nucleus and analyzed their localization using the large-scale localization study by Huh et al. (33). While 22% of the Y2H non-activators were localized to the nucleus, 41% of the activators were nuclear (Figure 2B). The activation strength was also correlated with nuclear localization: e.g. the percentage of nuclear localization grew from 35% for weak activators to almost 50% in strong activators. However, the percentage of strong activators in the nucleus was still lower than the value of 67% found for known transcriptional activators (GO annotation ‘transcription regulator activity’).
Correlating GO function and localization revealed a significant overrepresentation of known TRs among nuclear activators and among nuclear-cytoplasmic proteins, but not in the set of exclusively cytoplasmic proteins (Supplementary Figure 1). Strikingly, 47% of nuclear Y2H activators are known TRs. This number indicates that a large fraction of these activators may act as transcription factors in vivo although this activity has not been recognized previously. As a large fraction of activator proteins have no assigned function (∼35%) this finding can contribute to the functional annotation of these proteins (see below) (Table 3).
Table 3.
List of Y2H activators of unknown function predicted to be TRs
| ORF | Name | Activation strength (in mM 3AT) | Biological process (GO) | Localization | Interaction with transcription machinery | Additional evidence |
|---|---|---|---|---|---|---|
| YFL049W | YFL049W | 100 | Unknown | Nucleus | Yes | Yes |
| YJR070C | YJR070C | 200 | Unknown | Cytoplasm, Nucleus | No | Yes |
| YDR520C | YDR520C | >200 | Unknown | Cytoplasm, Nucleus | Yes | Yes |
| YGL066W | SGF73 | 50 | Protein modification | Nucleus | Yes | No |
| YKR064W | YKR064W | 200 | Unknown | Cytoplasm, Nucleus | No | Yes |
| YCR082W | YCR082W | 200 | Unknown | Cytoplasm, Nucleus | Yes | No |
Abundance
It has been known for a long time that transcriptional activators are often expressed at low levels. This notion is also reflected in the lower concentration of known transcription regulators (Figure 2C). With an average copy number of ∼4000 proteins per cell the Y2H activators confirmed this finding and even showed a trend towards lower abundance the stronger the activation was (Figure 2C). However, this trend was not significant.
General physicochemical properties of Y2H activators
Although characteristics such as abundance and localization allow us to classify activators, they do not explain their behavior. In fact, it has been unclear which properties turn a protein into a transcriptional activator even though certain physicochemical properties such as acidic stretches have been identified (10). We therefore revisited the influence of physicochemical properties on the propensity of a protein to activate transcription.
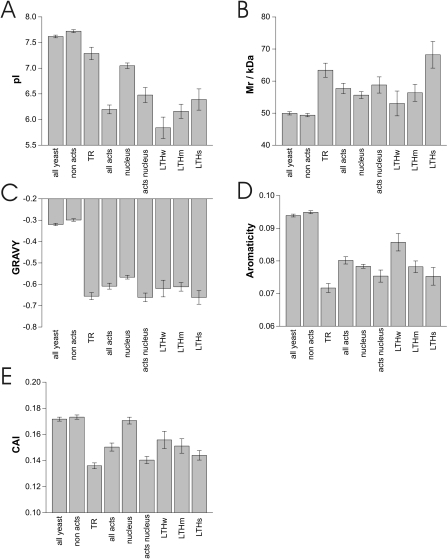
Well-defined general properties were analyzed first and included the isoelectric point (Figure 3A), molecular mass (Figure 3B), overall hydrophobicity (GRAVY score) (Figure 3C) and aromaticity (Figure 3D). The most pronounced effect was found for the isoelectic point: the mean pI for activators was found to be ∼1.5 pH units below the value for non-activators. This effect was also observed for activators in the nucleus, although with ∼0.5 pH units the difference between nuclear localized activators and nuclear proteins in general was lower. Interestingly, known TRs had only a slightly reduced mean pI, which was even higher than for nuclear proteins in general. However, proteins annotated as ‘TRs’ may include non-activating regulators too. The influence of activation strength onto the mean pI was not significant (judged by Student's t-test).
Figure 3.
General physicochemical properties of activators. Mean and SEM of physicochemical properties is shown for several protein sets. (A) Isoelectric point (pI), (B) molecular mass (Mr), (C) GRAVY score (overall hydrophobicity), (D) aromaticity and (E) Codon adaptation index (CAI). For details see text.
The mean molecular weight of activators was found to be ∼9 kDa higher than for non-activators. This tendency is also reflected by the higher mean molecular weight of known TRs. Activators in the nucleus were not significantly larger than nuclear proteins in general but we found that the mean molecular weight increased with activation strength (Figure 3B).
The overall hydrophobicity (GRAVY score) and the aromaticity showed a similar pattern. Both properties were reduced in activators as well as in known TRs. The effect was much smaller for the GRAVY score but highly significant for nuclear localized proteins and activators (Students't t-test P = 2.4e−5).
In addition, the CAI was analyzed (Figure 3E). As lower codon scores are associated with lower protein expression levels (34), the lower CAI for activators as well as for known TRs supports the previous finding of lower protein levels for these two protein sets (Figure 2C).
As expected, known TRs generally showed similar properties as the Y2H activators except for the isoelectric point.
Amino acid clusters
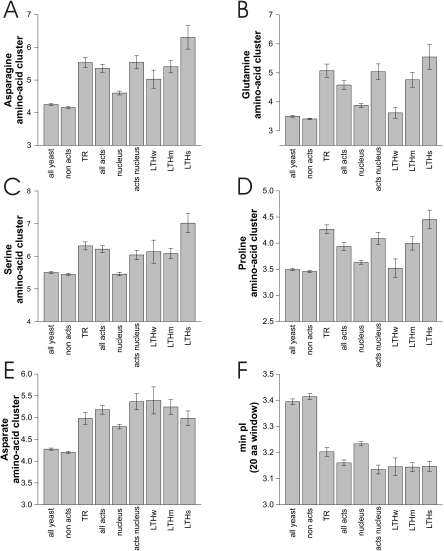
It has been known for a long time that certain amino acids are overrepresented in transcriptional activators, in particular acidic and basic residues and certain other amino acids such as glutamine. We wondered if this is true for our activator set as well. As basic amino acid clusters are also acting as nuclear localization signals, we only took activators into account that are known to be localized to the nucleus and compared them with the remaining nuclear proteins. After correction for multiple testing using Holm's procedure (41) 15 parameters involving certain amino acids remained highly significant (Supplementary Table 2). These included the minimum pI in a 20 amino acid window, the overall percentage of several amino acids (Ala, Gly, Ser, Val, Lys, Asn, Gln) and amino acid clusters (Ser, Asp, Pro, Asn, Gln) and, in addition, the amino acids Lys and Arg and the net charge (amino acids [Lys + Arg] − [Glu + Asp] = KR − ED). A detailed analysis of the amino acid clusters and the minimum pI is shown in Figure 4. A detailed analysis of the remaining properties is shown in Supplementary Figure 2.
Figure 4.
Amino acid clusters of activators. (A–E) Amino acid clusters indicating the maximum number of the respective amino acid in a window of 20 amino acids of the protein (For the remaining amino acid clusters see Supplementary Figure 2). (F) Minimum isoelectric point (pI) in a 20 amino acids window, summarizing the effect of charged amino acids (glutamate, aspartate, arginine, lysine).
Taken together, the analysis of physicochemical properties showed that Y2H activators tend to possess a lower isoelectric point, have a lower hydrophobicity, higher molecular weight, lower CAI and show specific properties like enrichment of asparagine clusters.
Protein domains
Transcriptional ADs are still not well defined structurally. This is also reflected in specialized domain databases such as SMART (8) or Interpro (45) which have hardly any defined entries for transcriptional ADs. Therefore we have analyzed the Y2H activators for enrichment of known domains. Not unexpectedly, certain DBDs such as helix–loop–helix motifs or zinc fingers are indeed significantly overrepresented among the activators (Supplementary Table 5).
Certain domains are also significantly underrepresented such as the AAA domain. However, these domains are not further considered here.
Binary protein–protein interactions
Specific transcription activation is mediated by physical contact with the transcriptional machinery or other factors necessary for transcription, like chromatin remodeling proteins (4,5,16,18–20).
Activators form a tightly connected protein interaction network (Figure 5A). Therefore, we wondered if the Y2H activators had any known interactions with the transcription machinery or other proteins (Figure 5B).
Figure 5.
Protein–protein interactions of transcription activators. (A) This protein interaction network shows the activators (red) and the proteins interacting with at least two activators. Proteins involved in transcription are indicated by a diamond shape and the activation strength (bGal and LTH combined) is reflected by the node size. Individual protein names can be identified in the online version of this figure. (B) Protein–protein interactions of activators and control groups with other proteins involved in transcription (P-GO term ‘transcription’). Physical, binary protein interaction data from the MIPS database (35) were used. The right three columns include indirect interactions involving a bridging protein (indicated by ‘+1’) between the activator and the transcription protein. (C) Percentage of proteins interacting with components of protein complexes involved in transcription. Only high-throughput derived protein complex data from the MIPS database were considered.
Significantly more nuclear Y2H activators (∼36%) interacted with components of the transcription machinery compared with all nuclear proteins (∼23%). The same is true for known transcription regulators which show an even higher fraction of proteins interacting with the transcription machinery (∼46%). Taking bridging proteins into account (i.e. transcription proteins that can be reached over two protein interaction edges), the number of proteins interacting with transcription factors even increases to ∼57% (nuclear Y2H activators) and ∼64% (transcription regulators), respectively. All mentioned differences are highly significant judged by Fisher's exact test (P < 0.01). Remarkably, activation strength showed no effect on the number of proteins interacting with the transcription machinery.
Protein complexes
Because transcription is carried out by several large protein complexes such as RNA polymerase, TAFs, mediator and the like, we wondered with which of these complexes the Y2H activators interacted. Therefore we analyzed complexes from large scale MS studies (35,46,47) which had at least 50% of their proteins annotated as ‘transcription’-related for the presence of Y2H activators (Figure 5C). Of nuclear activators 19% interacted with transcription complexes compared with 8% of all nuclear proteins. An even higher fraction (27%) of known TRs interacted with complexes involved in transcription. Strikingly, Y2H activators in the nucleus that were not annotated as TRs showed a significant higher number of interactions with the transcription machinery compared with other nuclear non-TR proteins. This might indicate that many of these proteins are also bona fide TRs (see below). All these differences are highly significant judged by Fisher's exact test (P < 0.01).
To reveal the nature of proteins interacting with transcriptional activators we compiled the most frequent interactors of our Y2H activators. The top 15 interacting proteins of different protein query sets are given in Supplementary Table 4. This analysis reveals that a high percentage of activators, as well as known TRs interact with components required for transcription. These were components of the RNA-polymerase II holoenzyme and proteins involved in chromosome remodeling, like the SAGA complex. Nuclear proteins in general do not show this tendency with only one interactor (YDL014W) shared between Y2H activators and nuclear proteins in general (Supplementary Table 4). Interestingly, both nuclear activators and transcription regulators interacted with the metabolic enzyme alcohol dehydrogenase. With 21 and 19%, respectively, the fractions were much higher than in the set of nuclear proteins (∼10%, not shown).
In summary, Y2H activators as well as known TRs tend to specifically interact with components of the transcription machinery even though such interactions have not been shown for the majority of either group.
Y2H activators as bona fide transcriptional regulators
Many of the Y2H activators described here may be bona fide TRs even if they have not been annotated as such. We therefore may have identified Y2H activators of yet unknown function given their ability to efficiently activate transcription, their nuclear localization and their protein interaction pattern with other components of the transcription machinery. The following Y2H activators of yet unknown function are predicted to be physiological activators.
YFL049W, a protein with no assigned function, was co-purified with Rtt102p, Snf2p and Snf5p indicating that it is a real Swi/Snf component (48). The yeast SWI/SNF complex is required for transcription of several yeast genes and has been shown to alter nucleosome structure in an ATP-dependent reaction. Transcription stimulation by SWI/SNF requires an AD with which it directly interacts. Strikingly, the acidic ADs of VP16, Gcn4, Swi5 and Hap4 interacted directly with the purified SWI/SNF complex and with the SWI/SNF complex in whole-cell extracts (16). A physical interaction of YFL049W with the SWI/SNF complex together with the strong transcription activation properties (strong Y2H activator) strongly supports the possible transcription stimulation function involving the SWI/SNF complex.
YJR070C is both localized to the nucleus and the cytoplasm. Nucleic acid binding proteins are overrepresented in its set of genetic and physical interaction partners. As it is known to bind eIF5 (49) it might have regulatory functions involving translation in the cytoplasm and transcription in the nucleus.
YDR520C, also localized to the cytoplasm and the nucleus, was shown to bind a zinc finger-containing transcriptional repressor, Dal80 (27), suggesting the interesting hypothesis that Dal80 is repressing the activator properties of YDR520C.
YGL066W/SGF73, a protein with unknown molecular function, was identified as a novel subunit of the SAGA (Spt/Ada/Gcn5 acetylase) multisubunit complex (50). Ataxin-7 is the human ortholog of the yeast SAGA SGF73 subunit and is a bona fide subunit of the human TFTC-like transcriptional complexes (TATA-binding protein-free TAF-containing complex) (51). The physical association of YGL0066W with the TFTC transcription complex and its activation properties supports a role in transcriptional regulation function.
A profiles/patterns analysis of YKR064W revealed the presence of a Zn[2]-Cys[6] fungal-type binuclear cluster domain in the N-terminal region. This domain binds to DNA and is also found in ArgR2p, a component of the ARGR transcription regulatory complex (ArgR1p, ArgR2p, ArgR3p, Mcm1p) (this domain already lead to the annotation as transcription factor in the YPD database) (52). A search in the eMOTIF database (53) reveals a ‘fungal transcriptional regulatory protein’ motif (the eMotif search tool is also available in SGD). Together with the strong transcriptional activation properties of YKR064W, this finding indicates a true transcriptional regulation and activator function of YKR064W relying on its DNA binding as well as its AD, probably involving the ARGR transcription regulatory complex.
YCR082w (=Ahc2) is a protein localized in the nucleus as well as in the cytoplasm without known function. It was identified as a strong activator in this study and is known to interact with chromatin reorganization components like a histone acetyltransferase complex (Ahc1) (26) and Abf1, a DNA-binding protein involved in chromatin reorganization (54). In addition, YCR082w interacts with a transcription factor, Srb4, a subunit of the RNA polymerase II mediator complex (27).
DISCUSSION
Transcriptional activation is the basis of the Y2H system (26,27). However, some bait proteins activate transcription without requiring an interacting protein that bears a separate AD. In fact, this property prevents the study of many TRs by means of the two-hybrid system. Nevertheless, we realized that this observation can extend our knowledge about the properties of transcriptional ADs.
In the course of this study we took previously identified (but mostly unpublished) activators from several Y2H screens (26,27) and measured their activation strength. In addition, we looked for properties distinguishing this set from non-activator proteins. Although many well-characterized TRs and nuclear proteins are highly overrepresented in the set of Y2H activators, we identified many which have not been associated with transcription previously. Irrespective of their physiological role, these activators must be able to interact with and recruit the transcriptional machinery.
Which features or sequences mediate the recruitment of the general transcriptional machinery to the activator? Although there is no single feature several rather general properties of transcription activators have been defined previously. These included the protein sequence composition as in acidic activators (25) or more specific features such as defined interactions between activators and the basal transcription machinery [e.g. (21)].
Acidic activators, i.e. activators with stretches of acidic amino acids, were the first class to be identified in yeast and the ones studied most extensively (10,55). Their importance was emphasized by screens for random activating fragments of the E.coli genome or activating peptides, both of which mainly identified acidic activators (56). The finding was confirmed in this study by the lower isoelectric point of activators (Figure 3A), more acidic stretches (Figure 4F) and by the increased clustering of aspartate (Figure 4E).
Other previously defined activator classes were also mirrored by this study: glutamine-rich activators (Figure 4B, Supplementary Figure 2) (11), proline-rich activators (Figure 4D) (12) and serine-rich activators (Figure 4C) (57). Elevated asparagine levels and cluster values (Figure 4A and Supplementary Figure 2) of activators may indicate a role of an additional amino acid in transcription activation, possibly similar to the closely related amino-acid glutamine in glutamine-rich activators.
It is generally accepted that ADs mediate their function by specific interactions with the basal transcription machinery (5) or additional factors involved in transcription, like chromosome remodeling complexes (58). Transcription activators analyzed in this study form a highly interconnected protein interaction network involving transcription related proteins (Figure 5A). Protein interactions with transcription-related proteins are highly overrepresented, even in the set of activators which have not been described as TRs (Figure 5B and C).
A significant fraction of nuclear Y2H activators as well as known TRs interact with the basal transcription machinery (like RNA polymerase II) and chromosome remodeling complexes (like the SAGA complex) (Supplementary Table 4). These interactions possibly explain their activation properties. Strikingly, the top ranking interaction partner of nuclear activators not known to regulate transcription was also RNA polymerase II. This indicates that genuine transcription regulators are contained in this set and form the basis for annotating them as TRs (see Results). However, for a large fraction of the Y2H activators no interactions are known and thus their mode of action remains unclear.
A surprising finding is the frequent interaction of activators with alcohol dehydrogenase. This enzyme is found with ∼10% of nuclear proteins (data not shown) but with ∼20% in both the nuclear activator and known transcription regulator dataset. Thus, alcohol dehydrogenase might be more a specific component of activation complexes than an unspecific contamination. Such moonlighting functions (59) have been found in other components of various transcription factor complexes but also among other proteins such as actin which acts both as a cytoskeletal protein and a transcription factor (60).
Screening of protein fragments fused to the DBD of Gal4 has been used before to identify transcriptional activators. Ruden et al. (56) screened fragments of the E.coli genome and mainly identified acidic stretches that activate transcription in yeast. Wiesner et al. (24) established a screening system based on a GFP reporter gene in a murine cell line and identified human transcription factors using a cDNA library. In contrast to this study we could make use of extensive large-scale studies which not only identified a set of transcription activators (26,27), but also localized proteins (33), measured protein concentrations (34) and protein interactions (26,27,46,47). The measurement of the activation strength let us focus on sets of proteins highly enriched in known TRs. Combining several data sources, for six uncharacterized yeast proteins we could identify additional evidence for their genuine role in transcriptional regulation.
We are aware that we may have missed a number of activators: of 138 proteins annotated with the GO-term ‘transcriptional activator activity’ in the YPD database 55 (40%) were detected as activators in this study (Supplementary Table 1), suggesting a false negative rate of ∼60%. This discrepancy may be explained by the fact that some TRs are annotated as activators although they do not act as such in our assay. Other proteins may require specific promoters, cofactors or conditions to exhibit activating activity. For example, several activators are only active when yeast is grown in glucose-free media. Interestingly, Gal4 itself becomes a very weak activator when fused to another Gal4–DBD, i.e. the DBD appears to inhibit the activating properties of some proteins. Thus, our studies need to be extended in order to get a more complete picture of all transcriptional activators in yeast, e.g. by using different conditions or fusing the DBD C-terminally.
Outlook
Although we have identified a large number of transcriptional activators in yeast and semi-quantitatively measured their activation strength, we have not measured activation in a truly quantitative way. New reporter strains with luciferase or some other enzyme need to be used in the future for more precise measurements.
In addition, we have not determined the actual ADs. Given the poor definition of ADs, it remains an important challenge to map these domains by fragmenting the proteins described here. As ADs do not appear to be well-defined structural domains it appears to be likely that they do not require a defined 3D structure. Instead, short linear peptides may contain the activation activity proper.
Once the ADs have been mapped, their interactions with the transcriptional machinery have to be identified. Unfortunately, the classical two-hybrid system cannot be used for this purpose. Thus either classical biochemistry or alternative systems have to be used such as the split-Ubiquitin system (61).
Finally, activators can differ in their ability to activate transcription dependent on the promoters to which they are bound (62,63). Hence all activators need to be tested in different promoter contexts and their transcriptional activity quantified.
Once their interactions have been mapped on a proteome-wide scale and their promoter context evaluated, it may be possible to generalize these findings and predict the transcriptional activity of every protein, including their activation strength. This would be a major step towards quantitative and thus systems biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Olivier Kassel and Ivan Sadowski for helpful comments on the manuscript. Ed Hurt and Jochen Baßler kindly provided help with the YPD database. This work was supported by DFG grant Ue 50/2-1. B.T. is a fellow of the Studienstiftung des deutschen Volkes. Funding to pay the Open Access publication charges for this article was provided by the Forschungszentrum Karlsruhe GmbH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kadonaga J.T. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 2.Bhaumik S.R., Green M.R. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaumik S.R., Green M.R. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 2003;370:445–454. doi: 10.1016/S0076-6879(03)70038-X. [DOI] [PubMed] [Google Scholar]
- 4.Koh S.S., Ansari A.Z., Ptashne M., Young R.A. An activator target in the RNA polymerase II holoenzyme. Mol. Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 5.Ptashne M., Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 6.Triezenberg S.J. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 7.Hulo N., Sigrist C.J., Le Saux V., Langendijk-Genevaux P.S., Bordoli L., Gattiker A., De Castro E., Bucher P., Bairoch A. Recent improvements to the PROSITE database. Nucleic Acids Res. 2004;32:D134–D137. doi: 10.1093/nar/gkh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell P.J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 10.Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 11.Courey A.J., Holtzman D.A., Jackson S.P., Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 12.Mermod N., O'Neill E.A., Kelly T.J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989;58:741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 13.Regier J.L., Shen F., Triezenberg S.J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc. Natl Acad. Sci. USA. 1993;90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giniger E., Ptashne M. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature. 1987;330:670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 15.Minter A.R., Brennan B.B., Mapp A.K. A small molecule transcriptional activation domain. J. Am. Chem. Soc. 2004;126:10504–10505. doi: 10.1021/ja0473889. [DOI] [PubMed] [Google Scholar]
- 16.Neely K.E., Hassan A.H., Wallberg A.E., Steger D.J., Cairns B.R., Wright A.P., Workman J.L. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 17.Melcher K., Johnston S.A. GAL4 interacts with TATA-binding protein and coactivators. Mol. Cell. Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodrich J.A., Hoey T., Thut C.J., Admon A., Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 19.Stringer K.F., Ingles C.J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 20.Ingles C.J., Shales M., Cress W.D., Triezenberg S.J., Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 21.Green M.R. Eukaryotic transcription activation: right on target. Mol. Cell. 2005;18:399–402. doi: 10.1016/j.molcel.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 22.Escher D., Schaffner W. Improved ‘activator trap’ method for the isolation of transcriptional activation domains from random DNA fragments. Biotechniques. 1996;21:848–854. doi: 10.2144/96215st02. [DOI] [PubMed] [Google Scholar]
- 23.Fields S., Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 24.Wiesner C., Hoeth M., Binder B.R., de Martin R. A functional screening assay for the isolation of transcription factors. Nucleic Acids Res. 2002;30:e80. doi: 10.1093/nar/gnf079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 26.Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 27.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazbun T.R., Malmstrom L., Anderson S., Graczyk B.J., Fox B., Riffle M., Sundin B.A., Aranda J.D., McDonald W.H., Chiu C.H., et al. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- 29.Cagney G., Uetz P., Fields S. High-throughput screening for protein–protein interactions using two-hybrid assay. Methods Enzymol. 2000;328:3–14. doi: 10.1016/s0076-6879(00)28386-9. [DOI] [PubMed] [Google Scholar]
- 30.James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serebriiskii I.G., Toby G.G., Golemis E.A. Streamlined yeast colorimetric reporter activity assays using scanners and plate readers. Biotechniques. 2000;29:278–279. doi: 10.2144/00292st03. 282–274, 286–278. [DOI] [PubMed] [Google Scholar]
- 32.Christie K.R., Weng S., Balakrishnan R., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 34.Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 35.Mewes H.W., Amid C., Arnold R., Frishman D., Guldener U., Mannhaupt G., Munsterkotter M., Pagel P., Strack N., Stumpflen V., et al. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 2004;32:D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berriz G.F., King O.D., Bryant B., Sander C., Roth F.P. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- 37.Costanzo M.C., Crawford M.E., Hirschman J.E., Kranz J.E., Olsen P., Robertson L.S., Skrzypek M.S., Braun B.R., Hopkins K.L., Kondu P., et al. YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 2001;29:75–79. doi: 10.1093/nar/29.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brendel V., Bucher P., Nourbakhsh I.R., Blaisdell B.E., Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc. Natl Acad. Sci. USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lobry J.R., Gautier C. Hydrophobicity, expressivity and aromaticity are the major trends of amino-acid usage in 999 Escherichia coli chromosome-encoded genes. Nucleic Acids Res. 1994;22:3174–3180. doi: 10.1093/nar/22.15.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharp P.M., Li W.H. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Statist. 1979;6:65–70. [Google Scholar]
- 42.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R-Development-Core-Team. 2004. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 44.Pollard K.S., Dudoit S., van der Laan M.J. Multiple testing procedures: the multtest package and applications to genomics. In: Gentleman R.C., Carey V.J., Huber W., Irizarry R., Dudoit S., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer-Verlag; 2005. pp. 249–271. Chapter 15. [Google Scholar]
- 45.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 47.Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 48.Graumann J., Dunipace L.A., Seol J.H., McDonald W.H., Yates J.R., III, Wold B.J., Deshaies R.J. Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol. Cell. Proteomics. 2004;3:226–237. doi: 10.1074/mcp.M300099-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Thompson G.M., Cano V.S., Valentini S.R. Mapping eIF5A binding sites for Dys1 and Lia1: in vivo evidence for regulation of eIF5A hypusination. FEBS Lett. 2003;555:464–468. doi: 10.1016/s0014-5793(03)01305-x. [DOI] [PubMed] [Google Scholar]
- 50.Sanders S.L., Jennings J., Canutescu A., Link A.J., Weil P.A. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helmlinger D., Hardy S., Sasorith S., Klein F., Robert F., Weber C., Miguet L., Potier N., Van-Dorsselaer A., Wurtz J.M., et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum. Mol. Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- 52.Amar N., Messenguy F., El Bakkoury M., Dubois E. ArgRII, a component of the ArgR–Mcm1 complex involved in the control of arginine metabolism in Saccharomyces cerevisiae, is the sensor of arginine. Mol. Cell. Biol. 2000;20:2087–2097. doi: 10.1128/mcb.20.6.2087-2097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J.Y., Brutlag D.L. The EMOTIF database. Nucleic Acids Res. 2001;29:202–204. doi: 10.1093/nar/29.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venditti P., Costanzo G., Negri R., Camilloni G. ABFI contributes to the chromatin organization of Saccharomyces cerevisiae ARS1 B-domain. Biochim. Biophys. Acta. 1994;1219:677–689. doi: 10.1016/0167-4781(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 55.Ptashne M., Gann A.A. Activators and targets. Nature. 1990;346:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 56.Ruden D.M., Ma J., Li Y., Wood K., Ptashne M. Generating yeast transcriptional activators containing no yeast protein sequences. Nature. 1991;350:250–252. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- 57.van de Wetering M., Oosterwegel M., van Norren K., Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Utley R.T., Ikeda K., Grant P.A., Cote J., Steger D.J., Eberharter A., John S., Workman J.L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 59.Jeffery C.J. Moonlighting proteins. Trends Biochem. Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 60.Hofmann W.A., Stojiljkovic L., Fuchsova B., Vargas G.M., Mavrommatis E., Philimonenko V., Kysela K., Goodrich J.A., Lessard J.L., Hope T.J., et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nature Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 61.Johnsson N., Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl Acad. Sci. USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harbury P.A., Struhl K. Functional distinctions between yeast TATA elements. Mol. Cell. Biol. 1989;9:5298–5304. doi: 10.1128/mcb.9.12.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen W.C., Bhaumik S.R., Causton H.C., Simon I., Zhu X., Jennings E.G., Wang T.H., Young R.A., Green M.R. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.