Abstract
The importance of certain structural features of the 5′ untranslated region of classical swine fever virus (CSFV) RNA for the function of the internal ribosome entry site (IRES) was investigated by mutagenesis followed by in vitro transcription and translation. Deletions made from the 5′ end of the CSFV genome sequence showed that the IRES boundary was close to nucleotide 65: thus, the IRES includes the whole of domain II but no sequences upstream of this domain. Deletions which invaded domain II even to a small extent reduced activity to about 20% that of the full-length structure, and this 20% residual activity persisted with more extensive deletions until the whole of domain II had been removed and the deletions invaded the pseudoknot, whereupon IRES activity fell to zero. The importance of both stems of the pseudoknot was verified by making mutations in both sides of each stem; this severely reduced IRES activity, but the compensating mutations which restored base pairing caused almost full IRES function to be regained. The importance of the length of the loop linking the two stems of the pseudoknot was demonstrated by the finding that a reduction in length from the wild-type AUAAAAUU to AUU almost completely abrogated IRES activity. Random A→U substitutions in the wild-type sequence showed that IRES activity was fairly proportional to the number of A residues retained in this pseudoknot loop, with a preference for clustered neighboring A residues rather than dispersed As. Finally, it was found that the sequence of the highly conserved domain IIIa loop is, rather surprisingly, not important for the maintenance of full IRES activity, although amputation of the entire domain IIIa stem and loop was highly debilitating. These results are interpreted in the light of recent models, derived from cryo-electron microscopy, of the interaction of the closely related hepatitis C virus IRES with 40S ribosomal subunits.
Hepatitis C virus (HCV) RNA is translated by a mechanism of internal ribosome entry that is distinctly different from the picornavirus precedent. Small (40S) ribosomal subunits can bind directly to the HCV internal ribosome entry site (IRES) at the correct site even in the absence of any translation initiation factors (19, 25), whereas the presence of factors is needed for small ribosomal subunit binding to picornavirus IRESs (23, 24). Consequently, initiation on the HCV IRES does not require eukaryotic initiation factor 4A (eIF4A), -4B, -4E, or -4G or ATP hydrolysis (25), in contrast to the picornavirus IRESs, which require eIF4A and -4B and at least the central domain of eIF4G as well as ATP (23, 24). Thus, at the superficial operational level, initiation on the HCV IRES is similar to initiation of translation of prokaryotic mRNAs, but with a complex 330-nucleotide (nt) IRES apparently playing a role analogous to the prokaryotic Shine-Dalgarno sequence. Recent results obtained with cryo-electron microscopy of IRES-40S subunit complexes, however, show that the two mechanisms are very different in detail. In the prokaryotic system the Shine-Dalgarno motif pairs with the 16S rRNA in the platform region of the small subunit, which places the mRNA in the appropriate channel, with the initiation codon in the active site (36). In contrast, the bulk of the HCV IRES binds to the back, or solvent side, of the 40S subunit, behind the platform region (33). Only a small part of the IRES lies in the vicinity of the subunit interface and the active sites of the subunit.
Among other viral IRESs, the HCV IRES is most closely similar to the pestivirus IRESs. Originally there were three recognized species of pestivirus, differing in their host animal: border disease virus (BDV), which infects sheep; bovine viral diarrhea virus (BVDV); and classical swine fever virus (CSFV), formerly known as hog cholera virus. Subsequently, it has been determined that there are two types of BVDV (BVDV-1 and -2), which are classified as two different species rather than different strains (14). In addition, it has recently been shown that the pestiviruses isolated from reindeer and giraffes are each a novel and distinct species (1), and a variety of yet to be classified viruses have been isolated from other mammalian species (2).
Although a great deal of work has been devoted to the HCV IRES, relatively little attention has been given to those of the pestiviruses (6, 21, 25, 27, 30), yet the picornavirus precedent has shown that a comparative study of closely similar IRESs can be particularly informative. There are strong similarities between the structure of the pestivirus IRES (Fig. 1) and the revised HCV IRES structure of Honda et al. (16). In terms of gross features, the main differences are that domain II is slightly longer in HCV than in the pestiviruses; stem 1 of the pseudoknot is bipartite in the pestiviruses and is longer than in the HCV IRES; the pseudoknot loop linking stem 1 to stem 2 is longer in pestiviruses; and whereas there are two side stem-loops, IIId1 and IIId2, in the pestivirus IRES, there is just a single IIId domain in the HCV IRES. On the other hand, there are remarkable similarities between the HCV and pestivirus IRESs in the sequence of the unpaired regions at the top of domain II and in the loops of domains IIIa, IIIc, and IIIe (in contrast to considerable differences in the sequences at the top of domain IIIb).
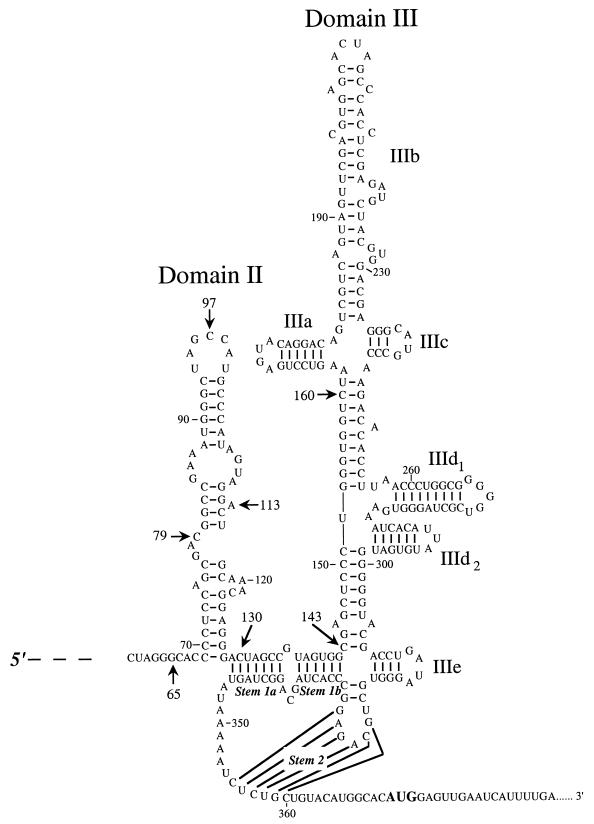
FIG. 1.
Secondary structure of the CSFV IRES. The structure shown is based on previously published structures (4, 9) plus our own structure probing data and phylogenetic comparisons, with the various domains designated according to the nomenclature in current usage. Arrows show the end points of the various 5′ deletions that were obtained for the experiments of Fig. 7 and 8; the designated residue is the extreme 5′ end of the CSFV sequences retained in the deletion mutant.
Here we describe the results of a mutagenesis approach to explore the significance of some features of the CSFV IRES as follows: the position of the 5′ boundary relative to domain II, the importance of domain II, the importance of base pairing in both stems of the pseudoknot, the significance of the length and composition of the A-rich pseudoknot loop, and the role of the highly conserved domain IIIa loop. While our work was in progress, some (but not all) of these questions have been addressed by others (19, 20, 21), but as these published results are rather contradictory, a third opinion is called for, which is why we have presented our data relating to these questions in addition to our results on completely novel aspects.
MATERIALS AND METHODS
Plasmid constructs.
The control dicistronic construct, pXLJ′OS, used in this work was identical to pXLJO described by Reynolds et al. (28, 29) except that the Asp718 site in the intercistronic cloning cassette was filled in to create a SnaBI site. Downstream of a bacteriophage T7 promoter, it has a Xenopus laevis 5′ untranslated region (UTR), coding region, and 3′ UTR and then a short cloning cassette with sites for SalI, SacI, SnaBI, and NcoI (in that order), followed by sequences coding for what is known as NS′, a slightly truncated form of the influenza virus (strain A/PR/34) NS1 protein described previously (3), and finally the complete NS1 3′ UTR terminating in an EcoRI site. The CSFV sequences used in this study were originally obtained as construct pCSFV/5B (generously donated by G. Meyers and H.-J. Thiel), which has the first 1,248 nt of the CSFV Alfort Tübingen genome (accession number JO4358). PCR was used to amplify nt 1 to 826 of this fragment, with the forward primer designed to change the extreme 5′-terminal sequence from GTATAG to GTCGAC in order to introduce a SalI site. Following restriction digestion with SalI and HindIII, the resulting product, which spanned nt 1 to 755 of the CSFV genome, was inserted into the corresponding sites of pGEM-1 (Promega). This was cut with AgeI (between nt 438 and 439 of the CSFV sequence) and the overhangs were filled in. The CSFV sequences were released by cutting with SalI and were cloned between the SalI and SnaBI sites of pXLJ′OS. The resulting dicistronic construct, pXLCSFV 1-442.NS′, has the complete CSFV 5′ UTR as an intercistronic spacer, and the downstream cistron consists of the first 67 nt of the CSFV coding sequences downstream of the AUG fused to the NS′ reading frame via a GUACC linker. For some experiments, pXLCSFV 1-423.NS′ was used. This was generated by making deletions from the AgeI site by the method of Henikoff (15): it has the first 48 nt of CSFV coding sequences fused to the NS′ reading frame via an ACC linker.
To prepare a nested set of 5′ deletions, pXLCSFV 1-423.NS′ was cut at the SalI site at the start of the CSFV 5′ UTR sequences, and the DNA was digested with exonuclease III for various times, followed by exonuclease VII, as described by Henikoff (15). The truncated CSFV sequences plus the NS′ coding sequences and 3′ UTR were released by cutting with EcoRI, and the fragments were cloned between the SnaBI and EcoRI sites of pXLJ′OS.
Point mutations and small internal deletions were generated by the PCR method of Picard et al. (26), incorporating modifications to the protocol described previously (17). All constructs were verified by dideoxynucleotide sequencing, and all plasmids were propagated by standard methods in Escherichia coli TG1 by using ampicillin selection (31).
In vitro transcription and translation assays.
All plasmids were linearized by digestion with EcoRI prior to transcription. The generation of capped or uncapped RNAs by transcription with bacteriophage T7 RNA polymerase was performed as described previously (18, 28). Translation assays were carried out as described previously (12), with added KCl at 100 mM and Mg2+ at 0.5 mM and with [35S]methionine as the radiolabeled amino acid. Aliquots of the translation assay samples were analyzed by gel electrophoresis on 20% polyacrylamide gels as described previously (8). Stained, dried gels were exposed to Hyperfilm β-Max (Amersham International). Quantitation was done by scanning densitometry of the dried films using Phoretix software; a range of different exposures was used in order to ensure that the determinations were made under conditions in which the response of the film was linear.
The coupled transcription-translation assays were carried out using the in-house system described by Craig et al. (7), but with added KCl at a final concentration of 100 mM, the optimum for translation dependent on the CSFV IRES, instead of the 40 to 60 mM concentration generally used for uncapped RNA products that would be translated by the conventional scanning mechanism (7).
Transfection assays.
BHK-21 cells were prepared at 50 to 80% confluence in six-well Costar tissue culture dishes and were then infected with recombinant vaccinia virus vTF7-3 (11). After 2.5 h at 37°C, the medium was removed and the cell monolayer was washed with Glasgow minimum essential medium and then OPTIMEM (Gibco-BRL). DNA transfection was carried out using 5 μg of plasmid DNA mixed with OPTIMEM and LIPOFECTIN (Gibco-BRL) as described in the supplier's protocol. At 20 h posttransfection the medium was replaced with methionine-free Eagle's medium. After a further 60 min, 30 μCi of [35S]methionine (Amersham International) was added per well and the plates were incubated for another 2 h. Cells were subsequently scraped from the plates, pelleted by centrifugation, and processed for gel electrophoresis. Following autoradiography of the dried gel, NS′ expression was quantitated by scanning densitometry of the autoradiograph.
To detect and quantify the yield of cyclin, samples of cell extracts were subjected to Western blotting essentially as described by Harlow and Lane (13), using a 1:100 dilution of a mouse monoclonal antibody raised against X. laevis cyclin B2 (a gift of J. Gannon and T. Hunt) and a 1:5,000 dilution of alkaline phosphatase-conjugated goat anti-mouse secondary antibody. Alkaline phosphatase activity was detected using 5-bromo-4-chloro-3-indolyl phosphate and Nitro Blue Tetrazolium as described by Harlow and Lane (13), and the staining was quantitated by scanning densitometry in reflectance mode.
RESULTS
Importance of both stems of the pseudoknot structure.
In HCV, stem 1 of the pseudoknot consists of nine contiguous base pairs (eight of them are G-C pairs) which are followed by just a single unpaired U residue before the 3′ part of stem 2 (16). In all the pestiviruses, in contrast, stem 1 is bipartite, consisting of two base-paired stems, referred to here as stem 1A and 1B (Fig. 1), with an intervening loop, or “bubble.” In the 5′ UTR of all pestiviruses, the top strand of stem 1 (as shown in Fig. 1) comprises 14 nt and the bottom strand comprises 16 residues, with the excess of 2 nt being absorbed in the unpaired bubble. There are differences between the three main species with respect to the lengths of the two parts of stem 1, with corresponding differences in the number of residues in the unpaired central bubble: in BDV, stem 1A is 7 bp and stem 1B is 5 bp; in BVDV the corresponding numbers are 8 and 6 bp, respectively; and in CSFV the corresponding numbers are 7 and 6 bp, respectively (Fig. 1).
The importance of both paired stems of the pseudoknot for HCV IRES function, and of just stem 2 in the CSFV IRES, has been well documented through mutational analysis (30, 34, 35). However, in a more recent publication doubts have been raised as to whether stem 2 of the HCV pseudoknot really forms as drawn or whether what is more important is the primary sequence of stem 2 rather than just base pairing (19). In addition, although these stems have been found to be susceptible to RNase V1, cleavage by single-strand specific nucleases has been reported to occur in stem 1B of the CSFV pseudoknot and in the 5′ side of stem 2 of both the CSFV and HCV IRESs (20, 21). We have also observed hits in the 5′ side of stem 2 of the CSFV IRES pseudoknot by reagents specific for single-stranded residues (kethoxal and RNases A and T1), as well as at the extremities of the bottom strand of stem 1A (data not shown). Given the doubts raised by Kieft et al. (19), the somewhat ambiguous structure probing data, and the fact that the CSFV pseudoknot is a much less formidable barrier to reverse transcription than the HCV pseudoknot (25), and thus is possibly less “tight,” the existence of the base-paired structure in the CSFV IRES and its significance for IRES activity cannot be taken for granted.
We therefore used a mutagenesis approach to address the issue of whether base pairing in stem 1A is necessary for IRES activity. Unless otherwise stated, the parent construct used throughout this work was pXLCSFV 1-442.NS′, a dicistronic construct with X. laevis cyclin B2 cDNA as the upstream cistron, followed by nt 1 to 442 of the CSFV genome, joined via a GUACC linker to what we refer to as the NS′ cistron, which consists of the coding sequences of a slightly truncated form of the influenza virus NS1 gene, plus the complete 3′ UTR of the NS1 cDNA (3, 28). Since the CSFV initiation codon is at nt 373 to 375, the first 67 nt of viral coding sequences following the AUG initiation codon are retained in this construct, and the downstream reading frame codes for a fusion protein consisting of the first part of viral Npro linked to NS′. The reason for adopting this approach is that we find that, as in the case of the HCV IRES (28, 29), maximum IRES activity with NS′ as a reporter requires retention of the 5′-proximal viral coding sequences (S. P. Fletcher and R. J. Jackson, unpublished observations).
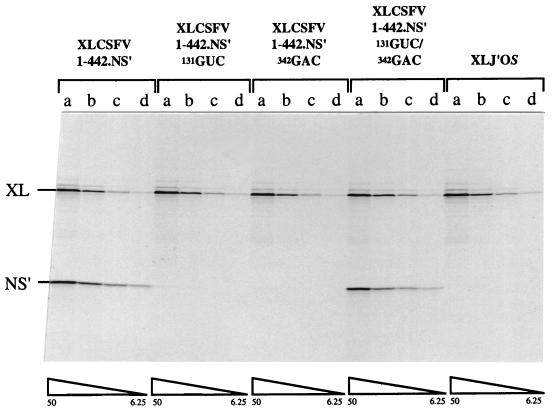
Three contiguous residues in the top strand of stem 1A were mutated to generate pXLCSFV 1-442.NS′ 131GUC, in which the disruption of 3 bp of stem 1A would probably be sufficient to destabilize the whole of stem 1A (Fig. 1). In a separate mutagenesis, three contiguous residues of the bottom strand of stem 1A were mutated to generate pXLCSFV 1-442.NS′ 342GAC, with the same likely outcome. Each of these mutations effectively inactivated internal initiation completely (Fig. 2). However, when the two mutations were incorporated into the same construct, which would allow the (re)formation of a 7-bp stem 1A, a high level of IRES activity was recovered (Fig. 2). For quantitation throughout this work we have translated all RNAs at four (occasionally three) different RNA concentrations and then used densitometry of the autoradiographs to determine the ratio of the yields of NS′ to cyclin, which is taken as a measure of IRES activity, for each assay. At each RNA concentration this yield ratio determined for the mutant was expressed as a percentage of the same ratio determined for the wild-type control construct, and then the four (or three) values so calculated were averaged. (In general, the defectiveness of debilitating mutations was actually greatest at low RNA concentrations and lowest at high concentrations.) By these criteria the double 131GUC/342GAC mutant recovered 75% (average value) of the wild-type activity, with individual values ranging from 56% at the lowest RNA concentration to 90% at the highest. This result provides compelling evidence that the base-paired stem 1A really does exist and is critical for IRES activity.
FIG. 2.
CSFV IRES activity requires base pairing in stem 1a of the pseudoknot. Capped dicistronic transcripts of the parent construct pXLCSFV 1-442.NS′, the negative control pXLJ′OS, the two individual mutants pXLCSFV 1-442.NS′ 131GUC and pXLCSFV 1-442.NS′ 342GAC, and the double (compensatory) mutant, pXLCSFV 1-442.NS′ 131GUC/342GAC, were translated at final concentrations of 50 (a), 25 (b), 12.5 (c), and 6.25 (d) μg/ml. The translation products were resolved on a 20% acrylamide gel, and the resulting autoradiograph is shown. XL, the cyclin B2 cistron translation product; NS′, the product derived from the downstream cistron.
We have also carried out similar mutagenesis on stem 2, which we consider to be 6 bp in length. It is true that there is a potential seventh base pair (Fig. 1), but we doubt whether this would exist in reality since it would be a terminal G-U pair and because a potential base pair in this position is not absolutely conserved in all pestiviruses. Evidence for the importance of stem 2 has been provided by Rijnbrand et al. (30), who generated a pair of mutants, each of which abolished the complementarity throughout the whole of stem 2 apart from one central position. Our mutations of stem 2 have been less drastic, being limited to two contiguous residues. Two pairs of such mutants were generated. In one of these pairs, the 325UC and 357GA mutants, each individual mutation would disrupt 2 bp in the center of stem 2, thus generating a bubble with, in theory, just 2 bp on either side (Fig. 1), which would probably destabilize the whole of stem 2. Each of these mutations severely compromised IRES activity (Table 1), but when both mutations were incorporated in the same construct, thus restoring six contiguous base pairs in stem 2, the recovery of IRES activity was complete (Table 1).
TABLE 1.
IRES activity of pseudoknot stem 2 mutantsa
| Mutated residues of wild type (XLCSFV 1-442.NS′)b | % IRES activity (relative to wild type) |
|---|---|
| 325AG/357CU; 327AG/355CU | 100 |
| 325UC/357CU | 16 |
| 325AG/357GA | 15 |
| 325UC/357GA | 99 |
| 327UC/355CU | 52 |
| 327AG/355GA | 15 |
| 327UC/355GA | 91 |
Capped transcripts of the designated mutants were translated at RNA concentrations of 50, 25, 12.5, and 6.25 μg/ml. Quantitative densitometry of the resulting autoradiographs was used to calculate the ratio of the yields of NS′ and cyclin at each RNA concentration, and this ratio was expressed as a percentage of the ratio observed with the wild-type construct. The averages of the four values of relative efficiency thus obtained for each mutant are shown.
Wild-type sequence is shown in bold.
The other pair of mutations, 327UC and 355GA, would each, individually, disrupt 2 bp of stem 2 but would leave four contiguous base pairs intact. Thus, these mutations might have less of a destabilizing effect on stem 2 than the pair discussed previously. In fact, the effect of these mutations, when tested individually, was asymmetric (Table 1). The 355GA mutation was almost as detrimental to IRES activity as the mutations described above which were located in the center of stem 2. However, an IRES with the 327UC mutation retained about half the activity of the wild-type control (Table 1). In the 327UC mutant there is a possibility of U-U pairing between positions 327 and 356, adjacent to four canonical base pairs, whereas in the 355GA mutant the equivalent positions are both occupied by A residues. It is possible that this provides an explanation for the much higher activity of the 327UC mutant than of the 355GA mutant, since U-U pairing adjacent to two (or more) canonical base pairs is found in other situations, notably in the wobble position of codon-anticodon interactions involving some mitochondrial tRNAs.
Nevertheless, despite the unexpectedly high activity of the IRES with the 327UC mutation, what is noteworthy is that incorporation of the two mutations (327UC and 355GA) into the same construct restored full IRES activity. Thus, the existence of stem 2 and its importance for CSFV IRES function are supported by direct experimentation.
Mutational analysis of the pseudoknot loop.
We next examined the loop joining stem 1A and stem 2. In BDV and CSFV strains this almost invariably has the sequence AUAAAAAU (on the assumption discussed above that the 3′-terminal U of this motif is not involved as a seventh base pair in stem 2 of the pseudoknot). In most BVDV strains it is simply AAAAA. In contrast, in HCV there is just a single invariant U residue between the two stems, which may correlate with the fact that stem 1 is shorter in HCV than in the pestiviruses. The questions we wished to ask were whether the length of the loop in the CSFV pseudoknot is critical for IRES activity, and if so, whether the nucleotide composition of the loop segment is important. Accordingly, the five consecutive A residues of this loop were either deleted or replaced by five U residues. Figure 3 shows the results of in vitro translation assays of capped dicistronic transcripts of these mutants over a range of RNA concentrations. The relative IRES efficiency, averaged over the range of RNA concentrations as described previously, was calculated to be about 6% in the case of the deletion mutant and 25% for the substitution mutant (Table 2). As in many assays of IRESs with debilitating mutations, the efficiency relative to the wild type was lower at low RNA concentrations than at high RNA input.
FIG. 3.
CSFV IRES activity is strongly influenced by the size and composition of the pseudoknot loop. Capped dicistronic transcripts of the parent construct pXLCSFV 1-442.NS′, the negative control pXLJ′OS, and the pseudoknot loop mutants pXLCSFV 1-442.NS′ Δ349-353 and pXLCSFV 1-442.NS′ 349A5→U5 were translated at final concentrations of 50 (a), 25 (b), 12.5 (c), and 6.25 (d) μg/ml. The translation products were resolved on a 20% acrylamide gel, and the resulting autoradiograph is shown. XL, the cyclin B2 cistron translation product; NS′, the product derived from the downstream cistron.
TABLE 2.
IRES activity of pseudoknot loop mutations
| Sequence of mutation | % IRES activity (relative to wild type)a |
|---|---|
| AU AAAAA U (wild type [XLCSFV 1-442.NS′]) | 100 |
| AU AAAAU U | 113 |
| AU AAAUU U | 94 |
| AU AAAUA U | 92 |
| AU AAUUU U | 53 |
| AU UUUAA U | 52 |
| AU UAAUA U | 41 |
| AU UUAAU U | 37 |
| AU UAUUA U | 29 |
| AU AUUUU U | 28 |
| AU UUUUU U | 25 |
| AU U (5-nt deletion) | 6 |
IRES activity relative to the wild-type control was calculated as described in Table 1.
Clearly, the length of the loop is quite critical for IRES activity, but the nucleotide composition is not unimportant, which suggests that this segment serves as more than just a spacer between the two stems of the pseudoknot. In a further examination of this aspect, we tested the IRES activity of a variety of mutants which retained a loop of wild-type length but of a varying ratio of A and U residues. The quantitative results are summarized in Table 2. In general, it appeared that the higher the number of A residues, the more efficient was the IRES. However, it also seemed that contiguous A residues allowed for higher IRES activity than dispersed As. Interestingly, sequences with potentially self-complementary ends did not give unusually high or unexpectedly low IRES activity, though it is of course possible that topological constraints imposed by the two stems of the pseudoknot might have prevented any base pairing between the two complementary ends.
As mentioned above, the equivalent loop in the HCV IRES consists of just a single U residue, which may be explained by the fact that stem 1 is considerably shorter in the HCV IRES than in the CSFV IRES and thus a single residue in the loop is sufficient to span between stem 1 and stem 2. In order to test whether there were any constraints on maintaining a small loop, we inserted five A residues into this site to generate a loop with the sequence UAAAAA. The results of translation assays of dicistronic mRNAs with this insertion are shown in Fig. 4. Visual inspection suggests that this insertion had no significant influence on IRES activity, and this was confirmed by densitometry and quantitation. The efficiency relative to that of the wild type was determined at each of the four RNA concentrations, and the average value was calculated to be 84%.
FIG. 4.
Expansion of the pseudoknot loop of the HCV IRES has no effect on IRES activity. (A) Structure and primary sequence of the HCV IRES pseudoknot, showing the position of insertion of five A residues in pXLHCV 40-372.NS′ 324insA5. (B) Capped dicistronic transcripts of the parent pXLHCV 40-372.NS′ and a low activity deletion mutant pXLHCV 40-339.NS′ (29), the negative control pXLJ′OS, and the insertion mutant pXLHCV 40-372.NS′ 324insA5 were translated at final concentrations of 50 (a), 25 (b), 12.5 (c), and 6.25 (d) μg/ml. The translation products were resolved on a 20% acrylamide gel, and the resulting autoradiograph is shown. XL, the cyclin B2 cistron translation product; NS′, the product derived from the downstream cistron. The smaller size of the downstream cistron product encoded by pXLHCV 40-339.NS′ is due to the fact that this construct has no viral coding sequences, whereas in pXLHCV 40-372.NS′ and its derivatives, the NS′ coding sequence is fused to the first 30 nt of viral coding sequences, and thus the product is NS′ with an N-terminal extension.
The highly conserved IIIa loop sequence may not be important for IRES function.
We next turned our attention to stem-loop IIIa, since this is not only very highly conserved among the pestiviruses but is also strongly conserved between pestiviruses and HCV. The stem is invariably 6 bp long, though in some species this includes a single G-U pair at or near the base of the stem. Among the ∼150 different pestivirus 5′ UTR sequences in the database at the time of writing, the AGUA loop sequence is absolutely invariant. Among the 314 different strains and isolates of HCV examined by Smith et al. (32), only 5 isolates (all of them type 4 strains) showed any variation from AGUA, and in all five cases it was a substitution of a pyrimidine for the last A of the loop sequence. Thus, on phylogenetic grounds there is good reason to believe that the loop sequence must be important. Accordingly, we introduced random mutations into either the central two positions of this loop sequence or the two flanking positions and recovered 11 different sequences, which taken together have all four possible nucleotides at each position (Fig. 5). Surprisingly, when these were tested in the coupled transcription-translation system, they were all highly (and equally) active, with no significant difference from the wild type.
FIG. 5.
Silent mutations in the domain IIIa loop. The figure lists all the mutants of the IIIa loop that were isolated, all of which were found to have high IRES activity that was comparable to that of the wild type. Mutated residues are indicated with lowercase letters.
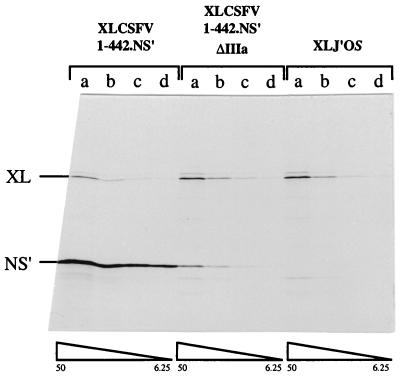
In view of this surprisingly high level of activity, no further experiments were conducted with these mutants, and instead we took the more drastic step of amputating all the base-paired residues and the loop of domain IIIa and replacing these 16 nt with a single C residue. Capped transcripts of this deletion mutant construct were translated (in the uncoupled system) over a range of RNA concentrations, and the outcome is shown in Fig. 6. Clearly, the complete amputation of the IIIA stem-loop seriously compromised IRES activity, although there was a small residual activity which, averaged over the four RNA concentrations, was 6% that of the wild type.
FIG. 6.
Amputation of domain IIIa stem and loop abolishes CSFV IRES activity. Capped dicistronic transcripts of the parent construct pXLCSFV 1-442.NS′, the negative control pXLJ′OS, and the domain IIIa deletion mutant pXLCSFV 1-442.NS′ ΔIIIa were translated at final concentrations of 50 (a), 25 (b), 12.5 (c), and 6.25 (d) μg/ml. The translation products were resolved on a 20% acrylamide gel, and the resulting autoradiograph is shown. XL, the cyclin B2 cistron translation product; NS′, the product derived from the downstream cistron.
The significance of domain II for IRES activity.
We made a series of deletions from the 5′ end of the CSFV sequence in order to map the 5′ boundary of the IRES and to assess the contribution of domain II to IRES activity. These deletions were made in the background of a construct (pXLCSFV 1-423.NS′) which retains fewer viral coding sequences than pXLCSFV 1-442.NS′ but nevertheless exhibits comparable IRES activity (S. P. Fletcher and R. J. Jackson, unpublished observations). The results obtained with a representative subset of these deletion mutants are shown in Fig. 7. The mutants were found to fall into three groups with respect to IRES activity. Those which included an intact domain II (Fig. 1), such as XLCSFV 38-423.NS′ and XLCSFV 65-423.NS′, retained a high level of IRES activity (Fig. 7). We also obtained deletion mutants with end points at nt 2 and 14, both of which showed similar activity to XLCSFV 38-423.NS′ and XLCSFV 65-423.NS′, with no perceptible gradient of IRES efficiency through this series (data not shown). Deletions which extend into domain II or remove it in its entirety fall into a second group, represented in Fig. 7 by pXLCSFV 130-423.NS′, which is missing the whole of domain II and also 1 bp of the pseudoknot stem 1A. The IRES activity of this mutant was very severely compromised but it was not zero (Fig. 7); the relative efficiency averaged over the four RNA concentrations was 19%. The deletions we obtained with end points at nt 79, 97, and 113, removing just part of domain II, were very similar in activity to pXLCSFV 130-423.NS′, which is missing the whole of this structural domain (data not shown). Finally, deletions which remove not only the whole of domain II but also invade stems 1A and 1B of the pseudoknot (e.g., pXLCSFV 143-423.NS′ and pXLCSFV 160-423.NS′) constitute a third group in which the IRES activity was effectively zero (Fig. 7). These results place the 5′ boundary such that the whole domain II, but very little sequence upstream of it, lies within the IRES. Deletions which enter and partially disrupt domain II or totally remove it (pXLCSFV130-423.NS′) reduce IRES activity to about 20% that of the control but do not totally abrogate it.
FIG. 7.
The position of the 5′ boundary of the CSFV IRES and the importance of domain II. Capped (+) or uncapped (−) transcripts, as indicated, of the designated 5′-deletion mutant constructs and the negative control pXLJ′OS were translated at final concentrations of 50 (a), 25 (b), 12.5 (c), and 6.25 (d) μg/ml. The translation products were resolved on a 20% acrylamide gel, and the resulting autoradiograph is shown. XL, the cyclin B2 cistron translation product; NS′, the product derived from the downstream cistron.
These conclusions were confirmed in transfection assays using selected deletion mutants from this series. The yield of NS′ from the IRES-dependent cistron was quantitated by metabolic labeling using [35S]methionine, followed by gel electrophoresis, autoradiography, and densitometry. The yield of cyclin from the upstream cistron was determined by Western blotting using an antibody that is specific for X. laevis cyclin B2 and does not cross-react with hamster cyclin. Data from a representative experiment are presented in Fig. 8, which confirms the results of the in vitro translation assays: the deletion mutant pXLCSFV 65-423.NS′, which has a complete domain II, retained full IRES activity; pXLCSFV130-423.NS′, which has lost the whole of domain II, was seriously debilitated but nevertheless retained about 22% of wild-type IRES activity; and more extensive deletions resulted in complete loss of activity.
FIG. 8.
Mapping the 5′ boundary of the CSFV IRES by in vivo transfection assays. BHK-21 cells were infected with recombinant vaccinia virus (vTF7-3) which expresses bacteriophage T7 RNA polymerase and were then transiently transfected with 5 μg of the designated dicistronic plasmid DNAs bearing 5′ deletions of the CSFV sequences. IRES activity was calculated as the ratio between expression of the downstream (IRES-dependent) cistron product (NS′) and the upstream cyclin B2 product, determined as described in Materials and Methods. Results from a representative experiment are shown.
DISCUSSION
There is some minor controversy in the literature with respect to where the 5′ boundary of the pestivirus IRES lies and more serious controversy over the significance of domain II for IRES activity. Rijnbrand et al. (30) found that the deletion of 26 nt from the 5′ end actually increased IRES activity by about 60%; a deletion to nt 66 reduced the activity to about two-thirds that of the full-length construct; and an internal deletion of nt 28 to 66 decreased activity still further. Consequently, they placed the 5′ boundary as lying within what was then called stem-loop B, between nt 28 and 66, whereas we would argue that all sequences up to the 5′ side of domain II do not participate directly in IRES function. In our experiments we did not observe any stimulation of IRES activity with short 5′ deletions (though admittedly our deletion series did not include any mutants with end points very near nt 28), and we found that a deletion to nt 65 retained full activity. We would argue that our position for the 5′ boundary is more consistent with the phylogenetic data: our analysis of covariances between BDV, BVDV, and CSFV 5′ UTR sequences shows strong conservation of structure downstream of the 5′ end of domain II (Fig. 1), but there is much less conservation upstream of this point. Moreover, a boundary for the CSFV IRES at the 5′ residue of domain II would be absolutely consistent with the position of the 5′ boundary of the HCV IRES (29) in the recently revised secondary structure model of the HCV 5′ UTR (16) and is also consistent with the position of the 5′ boundary of the BVDV IRES mapped by Chon et al. (6).
As for the significance of domain II in IRES function, Rijnbrand et al. (30) found that deletions which invaded domain II were essentially inactive in transfected Hep2 cells, whereas in our hands they retain about 20% of the activity of the full-length IRES both in vitro and in transfected BHK cells. This is very similar to the results we obtained with 5′ deletions of the HCV IRES (29). It is only when the deletions not only remove the whole of domain II but invade stem 1 of the pseudoknot that IRES activity falls to zero in our hands. In contrast, Kolupaeva et al. (20, 21) found that deletion of domain II of either the CSFV or HCV IRES reduced translation efficiency only mildly, to 60 to 65% of the control, although it did subtly alter the characteristics of 40S subunit binding to the IRES and was highly debilitating when combined with other mutations which on their own had only a mild phenotype.
The recent analysis by cryo-electron microscopy of the interaction of the 40S ribosomal subunit with the HCV IRES shows that the bulk of the IRES, domain III and the pseudoknot, binds to the solvent side of the 40S subunit, behind the platform (33). Domain II largely loops away from the ribosomal subunit, with the only contact being between the top of this domain and the E-site neighborhood of the 40S subunit. Deletion of domain II has no effect on the affinity of the IRES for the 40S subunit or on the actual contact sites between the rest of the IRES and the 40S subunit, but it does abolish the conformational change in the 40S subunit which occurs when the full-length IRES binds. It has been suggested that interaction between the top of domain II and the mRNA coding region may guide the RNA into the mRNA binding cleft on the 40S subunit (33). Alternatively, the conformational change in the 40S subunit may open up the mRNA binding cleft. Given that a domain II deletion doesn't affect the initial binding of the IRES to the ribosome, and so the initiation codon will be tethered tightly to the 40S subunit, it would seem surprising if the deletion completely abolished all IRES activity, as was reported by Rijnbrand et al. (30). On the other hand, retention of as much as 60 to 65% of the activity of the full-length IRES, as reported by Kolupaeva et al. (20, 21), would also seem surprising in view of the fact that the conformational changes induced in the 40S subunit are dependent on domain II.
The importance of both base-paired stems of the pseudoknot structure for the activity of the HCV IRES has previously been demonstrated by making compensatory mutations (34, 35). A similar approach was used by Rijnbrand et al. (30) to demonstrate that stem 2 of the CSFV IRES needs to be base paired, as has been confirmed here (Table 1). A further development described here and not addressed by Rijnbrand et al. (30) is the use of the same approach to demonstrate that stem 1, or at least stem 1A, is equally important (Fig. 2). In comparison with HCV, in which stem 1 of the pseudoknot is nine contiguous base pairs (eight of them being G-C pairs), the equivalent stem in the pestiviruses consists of two parts (stem 1A and stem 1B) separated by a bubble. Stem 1B, which is in the equivalent position to the HCV stem 1, is five or six contiguous base pairs (depending on the species of virus), and stem 1A is seven or eight contiguous base pairs. We have shown that base pairing in stem 1A is essential for CSFV IRES activity (Fig. 2), and although we have not directly examined stem 1B, topological constraints dictate that this, too, must surely be base paired as shown in Fig. 1. That base pairing in stem 1B is most likely to be critical for IRES activity is shown by the fact that disruption of 3 bp in this stem severely impaired IRES activity (21), although compensatory mutations to restore base pairing were not tested. In view of this overwhelming evidence in favor of the base pairing, but not the primary sequence, of the pseudoknot being critical for IRES activity, it is very puzzling that Kieft et al. (19) could not rescue HCV IRES activity by making compensating mutations in stem 2. The explanation for this unique negative result is not immediately self-evident, though there is one obvious difference in the technical details of these experiments. Kieft et al. (19) assayed IRES activity using the coupled transcription-translation system. Because this system generates uncapped transcripts and so has been optimized for translation (via the scanning mechanism) of uncapped mRNA, it is usually operated at a monovalent cation concentration that is considerably lower than that which is optimal for translation of capped mRNAs or translation dependent on the HCV and pestivirus IRESs.
Whereas there is just a single U residue between stem 1 and stem 2 in the HCV pseudoknot, in pestiviruses there is a longer loop varying from five consecutive A residues in BVDV strains to 8 nt (AUAAAAAU) in other species, including CSFV. This longer loop in CSFV than in HCV may be necessary for topological reasons related to the greater length of stem 1A plus 1B of the pestivirus pseudoknot. Certainly, reducing the length of the pseudoknot loop in the CSFV IRES by 5 nt very severely decreased IRES activity (Fig. 3). If our suggestion of a topological constraint is correct, this shortening would either distort the relative positions of the two stems of the pseudoknot or possibly cause the two stems to become partly unpaired.
However, our mutational analysis showed that for maximum IRES activity it is not just the length of the pseudoknot loop that is important; it also has to be A-rich. A-rich bulges like this are found in the group I intron ribozyme, where they are likewise essential for activity (5). The IRESs of cardioviruses and aphthoviruses also have an A-rich bulge, and mutations in this bulge influence not so much the activity of the IRES as its dependency on polypyrimidine tract binding protein for activity (17). X-ray crystallography of the group I intron ribozyme has shown that most of the residues of the A-rich bulge are involved in tertiary interactions which depend on such residues being specifically adenosines, a conclusion consistent with the influence of mutations of these residues on ribozyme activity (5). A common type of tertiary structure interaction, found in both the group I intron and in 23S rRNA, is the insertion of these adenosine residues into the minor groove of neighboring helices (5, 22). It would be intriguing if the A-rich pseudoknot bulge in the CSFV IRES were involved in similar tertiary interactions. On the whole, the influence of mutations of the A residues in the CSFV IRES pseudoknot loop is much less deleterious than in the case of the group I intron A-rich bulge, which would argue against the idea that they are involved in critical tertiary interactions. However, in structure probing experiments we find that even though this A-tract is not cleaved by cobra venom nuclease, the A residues are only moderately susceptible to reaction with dimethyl sulfate, indeed rather less reactive than nearby A residues such as the two between stem 2 and the initiation codon or the A residue in the bubble between stem 1A and stem 1B (S. P. Fletcher and R. J. Jackson, unpublished results).
Finally, the lack of any effect of mutation of the very highly conserved loop IIIa sequence was surprising. However, our results closely parallel those of Kolupaeva et al. (21), who found that mutation of the loop sequence from the wild-type AGUA to AAAA had very little effect on translation efficiency, but mutations which would have altered the length of the stem and/or the size of the loop were deleterious. In addition, these authors observed that the binding of 40S ribosomal subunits to the IRES afforded no protection of domain IIIa of either the CSFV or HCV IRESs (20, 21).
However, almost diametrically opposite results have been reported for the HCV IRES (19). Not only were residues in the IIIa loop protected when 40S subunits bound to the HCV IRES, but modification interference experiments strongly implicated the two A residues as critical for 40S subunit binding. In addition, mutation of the loop sequence to UCAU reduced translation efficiency and 40S subunit binding affinity about 10-fold. Nevertheless, the cryo-electron microscopy analysis actually suggests that domain IIIa, together with domains IIIb and IIIc, extends away from the surface of the 40S subunit (33), which is hard to reconcile with the mutagenesis and structure probing data published by the same group (19).
Since only two positions of this tetraloop were changed in any one of the many mutants we generated, it could conceivably be argued that the sequence of the loop is necessary but has very considerable inherent redundancy, with either just the two central residues or the two flanking A residues being sufficient. Although this type of explanation would be formally consistent with all the results of the point mutations made in the IIIa loop by Kolupaeva et al. (21), Kieft et al. (19), and ourselves, it seems highly implausible and without precedent in any other system. Another explanation that is formally consistent with all the data is that the CSFV and HCV IRESs differ with respect to the importance of the loop IIIa sequence, but this too seems highly improbable. It thus seems possible that despite its high degree of conservation, the sequence of the loop is not important for IRES activity, even though the length of the stem and the existence of the four-way junction probably is critical. This in turn raises the question of whether there may not be signals embedded within the IRES for other functions necessary for the viral life cycle apart from internal initiation of translation. Indeed, the results of recent experiments with the newly developed HCV replicon system indicate that there are cis-acting signals within the IRES which are critical for efficient RNA replication (10).
Acknowledgments
We thank G. Meyers and H.-J. Thiel for the original gift of the CSFV 5′ UTR plasmid and for advice, Tim Hunt and Julian Gannon for the gift of a highly specific antiserum raised against X. laevis cyclin B2, Paul Digard and Konrad Bishop for help with the transfection assays, and Catherine Gibbs for technical support.
The costs of this work were supported mainly by a grant from the Wellcome Trust, with additional support from the European Commission Biotechnology Program (BIO4-CT95-0045). S.P.F. gratefully acknowledges the support of a research studentship awarded by the Medical Research Council.
REFERENCES
- 1.Avalos-Ramirez, R., M. Orlich, H.-J. Thiel, and P. Becher. 2001. Evidence for the presence of two novel pestivirus species. Virology 286:456-465. [DOI] [PubMed] [Google Scholar]
- 2.Becher, P., M. Orlich, A. D. Shannon, G. Horner, M. Konig, and H.-J. Thiel. 1997. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J. Gen. Virol. 78:1357-1366. [DOI] [PubMed] [Google Scholar]
- 3.Borman, A., and R. J. Jackson. 1992. Initiation of translation of human rhinovirus RNA: mapping the internal ribosome entry site. Virology 188:685-696. [DOI] [PubMed] [Google Scholar]
- 4.Brown, E. A., H. Zhang, L. Ping, and S. M. Lemon. 1992. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 20:5041-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cate, J. H., A. R. Gooding, E. Podell, K. Zhou, B. L. Golden, C. E. Kundrot, T. R. Cech, and J. A. Doudna. 1996. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273:1678-1685. [DOI] [PubMed] [Google Scholar]
- 6.Chon, S. K., D. R. Perez, and R. O. Donis. 1998. Genetic analysis of the internal ribosome entry segment of bovine viral diarrhoea virus. Virology 251:370-382. [DOI] [PubMed] [Google Scholar]
- 7.Craig, D., M. T. Howell, C. L. Gibbs, T. Hunt, and R. J. Jackson. 1992. Plasmid cDNA-directed protein synthesis in a coupled eukaryotic in vitro transcription-translation system. Nucleic Acids Res. 20:4987-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dasso, M. C., and R. J. Jackson. 1989. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res. 17:3129-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, R., and K. V. Brock. 1993. 5′ and 3′ untranslated regions of pestivirus genome: primary and secondary structure analysis. Nucleic Acids Res. 21:1949-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesises bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grünert, S., and R. J. Jackson. 1994. The immediate downstream codon strongly influences the efficiency of utilization of eukaryotic translation initiation sites. EMBO J. 13:3618-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Heinz, F. X., M. S. Collett, R. H. Purcell, E. A. Gould, C. R. Howard, M. Houghton, R. J. M. Moorman, C. M. Rice, and H.-J. Thiel. 2000. Family Flaviviridae, p. 859-878. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 15.Henikoff, S. 1987. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 155:156-165. [DOI] [PubMed] [Google Scholar]
- 16.Honda, M., M. R. Beard, L.-H. Ping, and S. M. Lemon. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski, A., and R. J. Jackson. 1998. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4:626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminski, A., G. J. Belsham, and R. J. Jackson. 1994. Translation of encephalomyocarditis virus RNA: parameters influencing the selection of the internal initiation site. EMBO J. 13:1673-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kieft, J. S., K. Zhou, R. Jubin, and J. A. Doudna. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7:194-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolupaeva, V. G., T. V. Pestova, and C. U. T. Hellen. 2000. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 74:6242-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolupaeva, V. G., T. V. Pestova, and C. U. T. Hellen. 2000. Ribosomal binding to the internal ribosome entry site of classical swine fever virus. RNA 6:1791-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nissen, P., J. A. Ippolito, N. Ban, P. B. Moore, and T. A. Steitz. 2001. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. USA 98:4899-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestova, T. V., C. U. T. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pestova, T. V., I. N. Shatsky, and C. U. T. Hellen. 1996. Functional dissection of eukaryotic initiation factor eIF4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. T. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picard, V., E. Ersdalbadju, A. Q. Lu, and S. C. Bock. 1994. A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 22:2587-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole, T. L., C. Wang, R. A. Popp, L. N. D. Potgieter, A. Siddiqui, and M. S. Collett. 1995. Pestivirus translation initiation is by internal ribosome entry. Virology 206:750-754. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, J. E., A. Kaminski, A. R. Carroll, B. E. Clarke, D. J. Rowlands, and R. J. Jackson. 1996. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA 2:867-878. [PMC free article] [PubMed] [Google Scholar]
- 30.Rijnbrand, R., T. van der Straten, P. A. van Rijn, W. J. M. Spaan, and P. J. Bredenbeek. 1997. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J. Virol. 71:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Smith, D. B., J. Mellor, L. M. Jarvis, F. Davidson, J. Kolberg, M. Urdea, P. L.Yap, P. Simmonds, J. D. Conradie, A. G. S. Neill, G. M. Dusheiko, M. C. Kew, R. Crookes, A. Koshy, C. K. Lin, C. Lai, I. M. Murray-Lyon, A. Elguneid, A. A. Gunaid, T. Yemen, S. Yemen, D. Mutimer, M. Ahmed, C. Nuchprayoon, S. Tanprasert, F. E. Preston, M. Makris, A. Chuansumrit, C. Mahasandana, D. Pritchard, E. Riley, B. M. Greenwood, A. A. Saeed, A. M. Alrasheed, M. G. Saleh, I. McFarlane, C. Tibbs, R. Williams, J. Power, E. Lawlor, and H. Kiyokawa. 1995. Variations in the hepatitis C virus noncoding region—implications for secondary structure, virus detection and typing. J. Gen. Virol. 76:1749-1761. [DOI] [PubMed] [Google Scholar]
- 33.Spahn, C. M., J. S. Kieft, R. A. Grassucci, P. A. Penczek, K. Zhou, J. A. Doudna, and J. Frank. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291:1959-1962. [DOI] [PubMed] [Google Scholar]
- 34.Wang, C., P. Sarnow, and A. Siddiqui. 1994. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J. Virol. 68:7301-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, C., S.-Y. Le, N. Ali, and A. Siddiqui. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1:526-537. [PMC free article] [PubMed] [Google Scholar]
- 36.Yusupova, G. Z., M. M. Yusupov, J. H. D. Cate, and H. F. Noller. 2001. The path of messenger RNA through the ribosome. Cell 106:233-241. [DOI] [PubMed] [Google Scholar]