Abstract
To identify cell proteins regulated by the Epstein-Barr virus (EBV) transcription factor EBNA-2, we analyzed a cell line with conditional EBNA-2 activity by using microarray expression profiling. This led to the identification of two novel target genes induced by EBNA-2. The first of these, interleukin-16, is an immunomodulatory cytokine involved in the regulation of CD4 T cells. The second, AML-2, is a member of the Runt domain family of transcription factors. Quiescent B cells initially expressed AML-1 but, 48 h after virus infection, the levels of AML-1 decreased dramatically, whereas the amount of AML-2 protein increased. Analysis of a panel of B-cell lines indicated that AML-2 expression is normally predominant in EBV latency III, whereas AML-1 is associated with EBV latency I or EBV-negative cells. The AML genes are the first example of cell transcription factors whose expression correlates with the latency I/III phenotype.
EBNA-2-mediated transactivation of cell and viral genes plays a critical role in the generation and maintenance of continuously proliferating cells after infection with Epstein-Barr virus (EBV). Mutant viruses (e.g., P3HR1) which contain deletions or truncations of the EBNA-2 gene are unable to immortalize primary B cells after infection (7, 40, 41), but this function can be restored by complementation of the EBNA-2 deletion. Lymphoblastoid cell lines (LCLs) generated by EBV infection have a defined pattern of latent viral gene expression that, in addition to EBNA-2, includes the EBV nuclear antigens EBNA-1, EBNA-3A, EBNA-3B, and EBNA-3C and the latent membrane proteins LMP-1, LMP-2A, and LMP-2B. LCLs also express two nonpolyadenylated viral RNA transcripts, EBER1 and EBER2, and low levels of the BamHI-A rightward transcripts (BARTs). This pattern of latency is also characteristic of EBV-positive Burkitt's lymphoma (BL) cell lines with what is termed a group III phenotype. BL cell lines with a group I phenotype have a much more restricted expression pattern that is generally confined to EBNA-1 and the EBERs, whereas group II BL cells express a variable pattern of viral genes intermediate between group I and group III. Group I BL cells can sometimes drift in culture to express the full range of latent genes, acquiring a group III phenotype (14, 43).
Since EBNA-2 is one of the first viral genes expressed upon infection and is absolutely essential for immortalization, many studies have been carried out to identify both viral and cellular genes that are regulated by EBNA-2. Among the direct targets of EBNA-2 transcriptional activation are the viral Cp promoter, which controls the expression of the EBNA genes, the latent membrane proteins LMP-1, LMP-2A, and LMP-2B, the cellular proteins CD21, CD23, and c-fgr (reviewed in reference 24), and the proto-oncogene c-myc (21, 49). The activation of the LMP-1 promoter and the LMP-1/LMP-2B bidirectional regulatory element by EBNA-2 is enhanced by functional cooperation between EBNA-2 and EBNA-LP (16, 38). In primary B cells, transfection of both EBNA-LP and EBNA-2 expression plasmids into cells activated by ligation of CD21 with the virus gp350 protein results in the induction of cyclin D2 mRNA (47). As well as its role in activating cellular genes, EBNA-2 is also able to suppress the expression of the immunoglobulin mu gene (19).
EBNA-2 has no intrinsic DNA-binding activity, but it can be tethered to EBNA-2-responsive promoters through interactions with cellular factors. The recruitment of EBNA-2 to DNA can occur via interactions with RBP-Jκ (17, 32, 59, 68), Spi-1/PU.1 (20, 27), and ATF/CRE (48). EBNA-2 itself can also recruit the CREB-binding protein CBP (60) and the SWI/SNF chromatin remodeling complex (62, 63) and can interact via its acidic transactivation domain with components of the basal transcription machinery (55-57) to regulate promoter activity.
We and others have described a number of genes including cyclin D2, cdk-4 and the cytokines tumor necrosis factor alpha (TNF-α), granulocyte colony-stimulating factor (G-CSF), and lymphotoxin (LT) (21, 50) that are induced rapidly after EBNA-2 activation but are not regulated directly by EBNA-2. These genes require the synthesis of other intermediary factors for their transcription, since their activation is completely inhibited by the addition of protein synthesis inhibitors prior to the activation of EBNA-2. In this study we used microarray expression profiling to identify novel cellular genes involved in the cascade of events regulated by EBNA-2. By this process we identified several genes transcriptionally regulated after EBNA-2 activation. One of the genes regulated is the Runt domain transcription factor AML-2 (RUNX3). We have also found a direct association between AML-2 expression and the EBV group III latency phenotype in BL cell lines and LCLs and an inverse correlation between AML-2 expression and that of another Runt domain family member (AML-1). AML-1 expression was associated with the group I latency phenotype in Burkitt lymphomas. This is the first example of a cell transcription factor whose expression distinguishes between the two phenotypes.
MATERIALS AND METHODS
Cell lines.
DG75 (2) BL2, BL41, and Akata 31 are EBV-negative BL cell lines. Karpas 620 is an EBV-negative plasma cell leukemia kindly provided by Abraham Karpas (Department of Haematology, University of Cambridge) (36). BJAB is an EBV-negative B-cell lymphoma line. IB4, LCL-3, and LCL-C are EBV-immortalized LCLs generated by the infection of B cells with B95-8 EBV. Mak 1, Mutu cl216, Akata 2000, Mutu cl179, Elijah, Wewack, and Rael are all EBV positive and have been described as having a group I phenotype. BL37, P3HR1, Namalwa, and Raji are EBV positive with a group II phenotype, whereas Jijoye and Mutu III cl148 express a group III latency phenotype (43). BL41/P3HR1 and BL41/B95.8 are BL41 cells infected with the P3HR1 and B95.8 viruses, respectively. The cell lines were maintained in RPMI 1640 medium (Gibco-BRL) supplemented with 10 to 20% (vol/vol) heat-inactivated fetal calf serum and antibiotics. EREB2.5 cells (23) contain a conditional EBNA-2 regulated by estrogen and were maintained in RPMI 1640 without phenol red (Gibco-BRL) supplemented with 10% heat-inactivated fetal calf serum, antibiotics, and 1 μM β-estradiol. SV LMP-1 11c and SV LMP-1 13c are derived from EREB2.5 cells and constitutively express LMP-1. pHEBo1A is a control cell line stably transfected with empty vector pHEBo (67). These cells were maintained as for EREB2.5 but SV LMP-1 11c, SV LMP-1 13c, and pHEBo1A also had hygromycin B added (75, 75, and 150 μg/ml, respectively). For estrogen withdrawal experiments, cells were washed twice in serum-free medium before being resuspended at 5 × 105/ml in RPMI 1640 medium without β-estradiol. Cells were then incubated for 5 days. In experiments to identify direct targets of EBNA-2 transcription, protein synthesis was inhibited by pretreating cells for 2 h with 50 μg of cycloheximide/ml and 100 μM anisomycin (Sigma) prior to the addition of estrogen.
Purification of B cells, and EBV and virus infections.
Primary B cells from peripheral blood were isolated as described previously (4, 47). Buffy coats were centrifuged over Ficoll-Paque (Pharmacia LKB) gradients, and CD19-positive lymphocytes were immunoselected by using pan-B Dynabeads M450 (Dynal). Beads were removed by competition with Detachabeads (Dynal), and the cells were resuspended at 106/ml in RPMI 1640 supplemented with penicillin, streptomycin, and 15% heat-inactivated fetal calf serum. The cells were incubated for 40 h prior to infection with EBV. The isolated cells were analyzed by flow cytometry for purity by using fluorescein isothiocyanate-conjugated anti-CD20. Cells were infected with the B95-8 strain of EBV as described previously (18).
Microarray analysis.
Estrogen-starved cells were treated with protein synthesis inhibitors for 2 h and either left unstimulated or stimulated with estrogen for 4 h. Total cell RNA was extracted by using RNAzol B. Samples of the RNA were analyzed by RNase protection assay (RPA) for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) content to ensure that the RNA was intact. Then, 50-μg samples from unstimulated and stimulated cells were then labeled in reverse transcription (RT) reactions with dCTP-Cy3 and dCTP-Cy5. Unstimulated RNA labeled with Cy3 was mixed with stimulated RNA labeled with Cy5 and vice versa. Both mixes were used in overnight hybridization reactions with Ludwig/Sanger/ICRF Consortium microarray chips (5K1 and 5K2). The chips contained 9,932 sequence-verified probes representing ca. 5,300 different genes (see online site [http://www.sanger.ac.uk/Projects/Microarrays/] for details of the arrays). Labeling both samples with Cy3 and Cy5 enabled us to minimize the variation within the experiment caused by differential labeling with two different dyes. The slides were washed and then scanned by using a GSI Lumonics ScanArray 4000 and analyzed by using QuantArray software. Six data sets from three independent experiments were collated by using GeneSpring analysis software. Genes induced more than twofold were analyzed further by independent RPA or RT-PCR assays.
Immunoblotting and antibodies.
Radioimmunoprecipitation assay (RIPA) lysates were prepared and quantitated, and immunoblots were performed as described previously (3). Proteins were fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. After the membranes were blocked with 10% milk powder in phosphate-buffered saline-0.05% Tween 20, the membranes were probed with the following antibodies: a 1/500 dilution of anti-pleckstrin mouse monoclonal (clone 25; Transduction Laboratories), a 1/500 dilution of anti-EBNA-2 mouse monoclonal (PE-2; Dako), and a 1/10 dilution of anti-LMP-1 mouse monoclonal S12 (33), a 1/1,000 dilution of anti-cleaved caspase 3 (D175; Cell Signaling Technology, Beverly, Mass.), 1/40 dilutions of rabbit polyclonal anti-AML-1 (Ab-2) and AML-2 (Ab-1; Oncogene Research Products), a 1/1,000 dilution of anti-interleukin-16 (anti-IL-16) (14.1) (PharMingen, B.D.), and 1 μg of anti-PI3KR1 p85α (AB6; Calbiochem)/ml. The secondary antibodies were horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (Dako) and sheep anti-mouse immunoglobulin (Amersham). Bound immunocomplexes were detected by enhanced chemiluminescence (Amersham).
RPA.
Total cellular RNA was extracted by using RNAzol B and then quantified by measuring its absorbance at 260 nm. Probes for use in RPAs were generated by in vitro transcription of linearized plasmids. The plasmids were as follows: pleckstrin, Image clone 299599 linearized with the SpeI (T3 transcript) 367-bp probe; GADD45B, Image clone 772288 linearized with the AvaI (T3 transcript) 316-bp probe; and IL-16, Image clone 809776 linearized with the BstXI (T3 transcript) 300-bp probe. The c-myc and cyclin D2 probes were included in a RiboQuant custom-made template (PharMingen, BD Bioscience); PI3KR1 p85α, a 522-bp HincII/PstI fragment, was subcloned from Image Clone 34184 into pBluescript II SK and linearized with the HincII (T3 transcript) 522-bp probe; and PTK2B was prepared from Image Clone 163468 linearized with the HincII (T3 transcript) 300-bp probe. All probes for the candidate genes were resequenced to confirm their identities. RPAs were performed as recommended by the manufacturers of the RPA III kit (Ambion). Briefly, 1 μg of linearized plasmid was used to generate 32P-labeled antisense RNA probes. Cellular RNA was hybridized overnight at 42°C with 50,000 cpm of the probe. An equivalent amount of yeast RNA was included in a hybridization reaction as a negative control. Single-stranded RNA was digested with an RNase A-T1 mixture for 30 min at 37°C. Protected fragments were precipitated and separated on an acrylamide gel, and the gel was analyzed on a phosphorimager.
cDNA synthesis and RT-PCR.
Total cell RNA was extracted with RNAzol B, and 5 μg of the RNA was included in a cDNA synthesis reaction by using a ProStar first-strand RT-PCR kit (Stratagene) as recommended by the manufacturer. PCR for AML-2 was carried out with the forward primer 5′-ATT GCT CTT CCT ACC CCA TCC CCC-3′ and the reverse primer 5′-CGT GCT TCC TAC ATC AGT GTG TTT G-3′. PCR conditions were 94°C for 5 min, followed by 22 cycles of 94°C/30 s, 56°C/30 s, and 72°C/30 s, with a final incubation at 72°C for 7 min. PCR for GAPDH was carried out with the forward primer 5′-TGA AGG TCG GAG TCA ACG GAT TG-3′ and the reverse primer 5′-GCC ATG GAA TTT GCC ATG CCA TGG GTG G-3′. PCR products were analyzed by gel electrophoresis. GAPDH PCR conditions were 95°C for 5 min, followed by 35 cycles of 95°C/60 s, 59°C/55 s, and 72°C/45 s, and a final incubation at 72°C for 7 min.
RESULTS
EBNA-2 transcription targets identified by using the EBNA-2 conditional EREB2.5 cell line.
Our aim was to identify novel targets of EBNA-2 transcription. We have successfully used this system in a previous study to distinguish between genes regulated directly (e.g., c-myc) and indirectly (e.g., cyclin D2, TNF-α, and LT) by EBNA-2 (50). In this study we analyzed ca. 5,300 microarrayed cellular genes.
We compared RNA extracted from EREB2.5 cells that were either untreated or treated with estrogen for 4 h in the presence of protein synthesis inhibitors. From three independent array experiments (two determinations per experiment) we identified 13 genes that were, on average, regulated by our criterion of more than a twofold induction. Two of the genes identified were c-myc (Fig. 1, 5.2-fold induction) and CD21 (2.8-fold induction). Both of these genes have been identified as direct targets of EBNA-2 and therefore provided useful internal positive controls to confirm that our assay successfully detected EBNA-2-regulated genes.
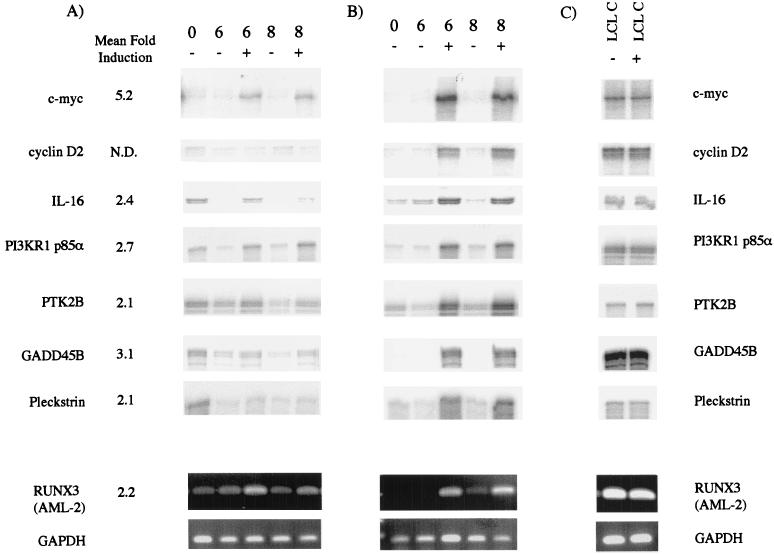
FIG. 1.
(A) Validation of the microarray data (shown as arithmetic mean fold induction) by using independent RPA and RT-PCR. To confirm genes as direct targets of EBNA-2 activity, EREB2.5 cells were washed and starved of estrogen for 5 days. The protein synthesis inhibitors cycloheximide and anisomycin were added for 2 h prior to the start of the experiment, and the cells were then either left untreated (−) or were treated with 1 μM estrogen (+) for 6 or 8 h as indicated. Total cellular RNA was extracted and analyzed by RPA or RT-PCR. (B) To confirm that genes were regulated during normal cell cycle progression after EBNA-2 activation, a similar time course was performed in the absence of protein synthesis inhibitors. EREB2.5 cells were washed and starved of estrogen for 5 days as in panel A and were then either left untreated or treated with estrogen for 6 or 8 h. Total cellular RNA was analyzed by RPA (5 μg) or RT-PCR for AML-2 and GAPDH. (C) As a control for the possible effects of addition of estrogen to B cells, LCL cells were treated with protein synthesis inhibitors and were then either left untreated or treated with estrogen for 4 h. RNA was extracted from the cells and assayed as described for panels A and B.
For a more complete analysis of the regulation of the candidate genes, we subsequently analyzed RNA levels (using RT-PCR or RPA) immediately after protein synthesis inhibition and after 6 or 8 h in the absence or presence of estrogen (Fig. 1A). We also compared RNA levels at the same time points and conditions but in the absence of protein synthesis inhibitors to establish that the genes were regulated during the normal course of EBNA-2-mediated proliferation (Fig. 1B).
Of the 13 initial targets, 5 were increased by estrogen stimulation of cells treated with protein synthesis inhibitors, but there was no clear induction in cells that had not been treated with the inhibitors (data not shown). The Image clones forming these probes and corresponding genes were 26418 (EDG1), 1335534 (BLR1), 810881 (EHD1), 302539 (caspase 4), and 469768 (BRF2). Presumably, the effect on these genes was specific to the experimental system, and they are not normally regulated during cell proliferation driven by EBNA-2. These genes were therefore excluded from further analysis.
The RPA and RT-PCR analyses of the remaining seven genes is shown in Fig. 1. c-myc was clearly induced at 6 and 8 h after the addition of estrogen in the treated cells (Fig. 1A) and to an even greater level in the untreated cells (Fig. 1B), confirming previous results (50). Cyclin D2 was included as an example of an indirect target of EBNA-2 activity and, as expected, was only induced in the absence of protein synthesis inhibitors (Fig. 1B). Figure 1C shows the levels of RNA detected in an LCL, LCL C, treated for 2 h with protein synthesis inhibitors and then either left unstimulated or stimulated with estrogen for 4 h (comparable to the experimental conditions used in the microarray experiment). This demonstrated that estrogen treatment itself did not affect the levels of any of the RNAs analyzed in our system and that changes in expression levels were due to the presence of the ER-EBNA-2 fusion protein.
The other genes investigated included those encoding a cytokine (IL-16), a transcription factor (RUNX3), components of cell signaling pathways (pleckstrin, PI3KR1 p85α, and PTK 2B), and a gene involved in the stress response (GADD45B). The RNA for GADD45B was upregulated after EBNA-2 activation by a predominantly indirect mechanism (Fig. 1B). This result is consistent with the regulation of GADD45B by NF-κB (9), whose activity is induced by LMP-1 (a known EBNA-2 target). It might additionally respond to activation of the TNF signaling pathway after TNF-α induction by EBNA-2 (50).
The RNA for PTK2B, PI3KR1 p85α subunit, and pleckstrin was upregulated by estrogen addition to a far higher degree in the absence of protein synthesis inhibitors than in the presence of inhibitors, thus demonstrating that all three genes are predominantly induced indirectly. In the case of pleckstrin the induction might be due to the activity of the EBNA-3 family of viral proteins (25). Despite large increases in the RNA levels, no increases in protein levels were detected by Western blotting (data not shown).
The regulation of the immunomodulatory cytokine IL-16 was analyzed by RPA. IL-16 RNA was present after the 2-h treatment of estrogen-starved EREB2.5 cells with protein synthesis inhibitors (Fig. 1A, T0). After a further 6 h of incubation without estrogen, this basal level could no longer be detected, but we could still detect IL-16 RNA in cells stimulated with estrogen during the same 6-h period. The relative levels of RNA detected in the 6− and 6+ samples were consistent with the fold induction detected by microarray, thus confirming our initial observation. IL-16 RNA was also clearly induced in estrogen-stimulated cells in the absence of protein synthesis inhibitors, indicating that IL-16 is a genuine target for regulation after EBNA-2 activation (Fig. 1B).
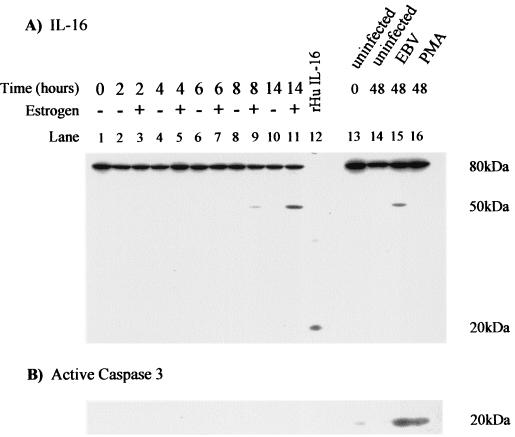
Figure 2A shows the result of a Western blot for IL-16 with the 14.1 monoclonal antibody which recognizes an epitope within the C-terminal 130 amino acids of the protein. IL-16 is predominantly expressed as an 80-kDa precursor protein (pro-IL-16) that is cleaved at the C terminus to release the mature 20-kDa secreted form (1, 6). The antibody therefore recognizes both the precursor and mature forms of IL-16. A time course of estrogen stimulation of EREB2.5 cells is shown in lanes 1 to 11. Lane 12 was loaded with 10 ng of recombinant human IL-16 as a positive control for the secreted mature form of the protein. After estrogen addition, there was little observable effect on the level of the 80-kDa precursor protein during the 14-h time course. However, we detected a smaller protein of ca. 50 kDa in cells stimulated with estrogen for 8 and 14 h and in primary B-cell infection. Since this protein was recognized by the 14.1 antibody, it represents a truncated form including the C terminus and is therefore a potential donor of mature IL-16. The 50-kDa protein was not induced in B cells mitotically stimulated by phorbol myristate acetate (PMA) in the absence of EBV infection (Fig. 2A). Caspase 3, which mediates processing of the IL-16 precursor to the active form (44, 65), was induced by both EBV infection and PMA treatment (Fig. 2B). The kinetics of induction of the 50-kDa IL-16 protein indicate that it is induced in B cells activated by functional EBNA-2 but that it is unlikely to be a direct target of EBNA-2.
FIG. 2.
Analysis of IL-16 protein expression. (A) EREB2.5 cells were starved of estrogen for 5 days and were then either left unstimulated (−) or were activated by the addition of estrogen (+) for the times indicated. RIPA extracts were prepared, and 50 μg of protein was analyzed by SDS-PAGE (lanes 1 to 11). Then, 10 ng of human recombinant IL-16 was included to ensure detection of the 20-kDa mature secreted form of IL-16 (lane 12). Primary B cells were isolated by CD19-positive selection and either left uninfected, infected with EBV, or stimulated with PMA for 48 h (lanes 13 to 16). Then, 50 μg of protein extract was analyzed for the presence of IL-16 with the mouse monoclonal antibody 14.1. (B) The membrane was reprobed for the expression of the 20-kDa cleaved active form of caspase 3.
EBNA-2-mediated regulation of AML-2 (RUNX3).
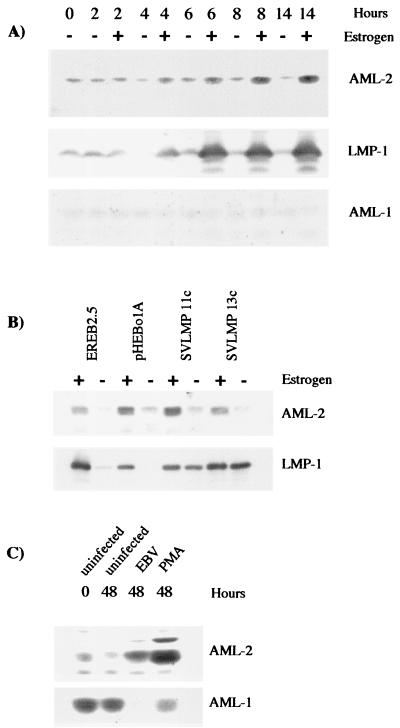
The array experiment also identified AML-2 as being a potential target for regulation by EBNA-2. AML-2 RNA was induced 2.2-fold after EBNA-2 activation by estrogen addition. Validation of our array experiment by RT-PCR with AML-2-specific primers showed that AML-2 RNA was induced after estrogen activation in cells both in the absence and in the presence of protein synthesis inhibitors (Fig. 1A and B), even though there was a slight stabilization of AML-2 RNA by the protein synthesis inhibitors. Subsequent analysis of protein expression in growth-arrested EREB2.5 cells during a time course of estrogen refeeding showed increased AML-2 protein levels detectable 4 h after estrogen addition (Fig. 3A). Accumulation of AML-2 then continued throughout the 14-h experiment. The kinetics of AML-2 induction were very similar to the expression of LMP-1 (a direct target of EBNA-2) during the same experiment (Fig. 3A). AML-2 (also known as RUNX3, CBFα-3, or PEBP2α-C) is a member of the mammalian Runt domain family of transcription factors, which includes two other proteins, AML-1 and AML-3. All three members of the family can bind the same consensus DNA sequence, but AML-3 is not usually expressed in B-cell lines (34). The same Western blot was therefore stripped and reprobed with an antibody specific for the other family member, AML-1. AML-1 showed no regulation in response to EBNA-2 activity in the EREB2.5 cells (Fig. 3A).
FIG. 3.
AML-2 protein levels increase after activation of EBNA-2 in EREB2.5 cells and after EBV infection of primary B cells. (A) EREB2.5 cells were treated as described in Fig. 2 and analyzed for the expression of AML-2 and LMP-1. The blot was then stripped and reprobed with an anti-AML-1 antibody. (B) The indicated cell lines were maintained in the presence or absence of estrogen for 5 days. Extracts were analyzed by Western blotting for AML-2 or LMP-1 expression. (C) Purified B cells were isolated by CD19-positive selection and were left uninfected, were infected with EBV, or were stimulated with PMA for 48 h. RIPA lysates were prepared, and 50 μg of protein was analyzed by SDS-PAGE.
The expression of AML-2 was not maintained in estrogen-starved EREB2.5 cells transfected with a plasmid constitutively expressing LMP-1 (SVLMP-1 11c and 13c). These cells express LMP-1 even when EBNA-2 is no longer functional after the withdrawal of estrogen. The expression of AML-2 was identical in parental EREB2.5 cells, in EREB2.5 cells stably transfected with the control plasmid pHEBo1A, and in the 11c and 13c cell lines (Fig. 3B). The expression of AML-2 therefore depended upon the activity of EBNA-2 rather than on expression of LMP-1. The RT-PCR and the protein expression data taken together indicate that AML-2 can be a direct target of EBNA-2 regulation.
Western blotting showed the expression of AML-2 and AML-1 protein in primary B cells undergoing infection with EBV or activation by treatment with PMA (Fig. 3C). As in growth-arrested EREB2.5 cells, the amount of AML-2 expressed in quiescent primary B cells was low and remained low during the 48-h experiment in uninfected cells. After virus infection, AML-2 expression levels increased and were induced to an even higher degree in cells activated by PMA. This may reflect the higher proportion of cells that we are able to activate by using PMA compared with the 25% of cells we routinely infect with EBV (49).
The expression of AML-1 in primary B cells contrasted significantly with the expression seen in growth arrested EREB2.5 cells. AML-1 was highly expressed in primary B cells, but there was no evidence that AML-1 was expressed in estrogen-starved EREB2.5 cells. Upon both EBV infection and PMA activation of primary B cells, the amount of AML-1 detected was greatly reduced. The difference between primary B cells and EREB2.5 cells starved of estrogen in their AML-1 protein levels is even greater than what we observed previously for p27 (50). The estrogen-starved EREB2.5 cells thus only partially mimic the conditions found in primary B cells.
AML-2 and AML-1 expression distinguishes between group III and group I phenotypes.
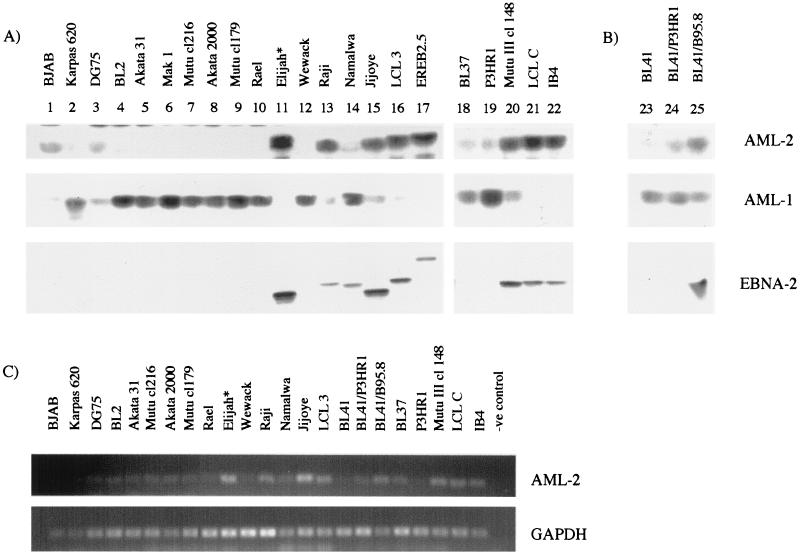
AML expression patterns were compared in LCLs and lymphoma cell lines that were either EBV negative or expressed different EBV proteins ranging from group I to group III latency phenotypes (Fig. 4A). EBNA-2 is characteristically absent in latency I but is expressed in latency II and III. The variation in size of EBNA-2 protein (Fig. 4A) is a result of repeat copy number variation in different EBV strains, except in the EREB2.5 cells which contain an EBNA-2 estrogen receptor fusion protein. There was a remarkable correlation between a group III or LCL phenotype and expression of AML-2. AML-2 protein levels were high in all five LCLs tested, as well as in the group III BL cell line Mutu III cl148 and in the EBNA-2 expressing group II BL Raji. Our observation that AML-2 is highly expressed in Raji cells agrees with previous published data (28). The Elijah cells were originally group I but have drifted in culture to express EBNA-2, and these cells also expressed AML-2. The BL41 converts (BL41 cells infected with either the P3HR1 or B95-8 viruses) also showed a similar correlation between EBNA-2 expression and higher AML-2 expression (Fig. 4B). The increased expression of AML-2 protein in certain B-cell lines was reflected by an increase in the level of RNA transcribed (Fig. 4C), indicating that the control of expression is likely to be at the RNA level. One exception was Namalwa, which did not express AML-2 despite having detectable EBNA-2, although it is not known whether the EBNA-2 in Namalwa EBV is functional.
FIG. 4.
AML family expression in B cell lines. (A and B) RIPA lysates were prepared from a range of B-cell lines, and 50 μg of protein extract was analyzed by SDS-PAGE for expression of the proteins indicated. The lines used included an EBV-negative B-cell lymphoma (lane 1), an EBV-negative plasma cell leukemia (lane 2), EBV-negative BL cell lines (lanes 3 to 5 and lane 23), group I BL cells (lanes 6 to 10 and lane 12), group II BL cells (lanes 13, 14, 18, 19, and 24), group III BL cells (lanes 15, 20, and 25), and LCLs (lanes 16, 17, 21, and 22). ∗, Elijah (lane 11) normally has a group I phenotype, but these cells drifted in culture so that they then expressed EBNA-2. (C) Analysis of AML-2 RNA levels in BL and LCL cell lines by RT-PCR. RT-PCR for GAPDH is shown as a control for cDNA synthesis.
The cell lines that did not express AML-2 expressed high levels of AML-1 instead. These cell lines included a plasmacytoma (Karpas 620), EBV-negative BL (BL2 and Akata 31), group I BL cell lines, and the BL cells of intermediate phenotype that lacked EBNA-2. In agreement with our data, BL2 cells have previously been shown to contain predominantly AML-1 DNA-binding activity (34). Previous work on AML-2 in Mutu cells did not distinguish whether the Mutu line was group I or group III phenotype (34), but our data would predict that the line was latency group III. Although there were some B-cell lines where AML-1 and AML-2 were coexpressed (e.g., in DG75 cells where both proteins were present at low level), usually one of the two AML family members dominated.
DISCUSSION
We have used microarray technology to identify novel cellular genes that are induced in response to the activation of EBNA-2. The candidate genes identified in our initial array analysis covered a range of proteins, including a cytokine, transcription factors, cell signaling molecules, and a gene involved in cell stress. For several of these genes we did detected changes in protein level in response to EBNA-2 activation, but the RNA levels were increased. Our analysis concentrated on IL-16 and the AML genes, for which the effects were observed at the protein level.
IL-16.
The cytokine identified as a candidate gene was IL-16, which is an addition to the list of cytokines identified previously as being regulated by EBNA-2 (i.e., TNF-α, LTβ, LT, and G-CSF [50]). The evidence presented here suggests that IL-16 is an indirect target of EBNA-2, although the RNA analysis in cells treated with protein synthesis inhibitors is often difficult to interpret due to possible effects of cycloheximide and anisomycin on RNA stability (13). No effect was observed on the 80-kDa precursor protein of IL-16 after EBNA-2 activation; however, we identified the induction of a 50-kDa IL-16 product. Cloned cDNA fragments encoding IL-16 proteins of this size have been identified previously and are capable of acting as a donor of mature IL-16 (65). In addition, multiple proteins of <80 kDa (including a major band at 50 kDa) have been detected after activation of CD4+ and CD8+ T cells by treatment with anti-CD3 antibodies (6). It is possible therefore that EBV enhances the release of mature IL-16 in vivo via induction of the 50-kDa form of the protein.
IL-16 has previously been shown to be secreted by both EBV-negative and EBV-positive B-cell lines, including BJAB, Raji, Namalwa, and Daudi cells (45). We tested IL-16 secretion upon infection of purified B cells (data not shown) but were unable to detect an enhancement of release of active IL-16 because of a high background of IL-16 release from the uninfected cell preparation, perhaps reflecting the high endogenous levels of the 80-kDa precursor in the purified B cells. We did, however, observe induction of activated caspase 3 upon EBV infection, this being the caspase involved in IL-16 maturation (44, 65). It thus remains possible that IL-16 release is stimulated during primary EBV infection in vivo. IL-16 is a cytokine involved in the modulation of CD4+ T-cell activity. It directly binds to CD4, effects migration of CD4+ T cells and can induce a loss of responsiveness via the T-cell receptor (8, 54). CD4+ effector T cells can inhibit the proliferation of newly infected B cells in vitro (65) and may therefore play an important role in the immune regulation and prevention of the early phases of EBV infection (37). Any perturbation of the immune effector response after infection caused by release of IL-16 might be beneficial to the establishment of the EBV infection.
An alternative potential role for IL-16 in the context of EBV infection could involve the superantigen encoded by an endogenous human retrovirus whose expression is induced by EBV infection (51). This activates T-cell proliferation, perhaps providing T-cell help for B-cell outgrowth, and may be responsible for some of the T-cell proliferation observed in infectious mononucleosis. It is possible that IL-16 secreted in response to EBV infection of B cells might serve as the attractant that recruits T cells to the infected B lymphocytes (22). The potential role of IL-16 in these processes should be assessed in in vivo models.
AML transcription factors.
We found a clear general correlation between AML-1 expression and the group I phenotype in BL and between AML-2 expression and the latency III phenotype. Some BL cell lines that have been in culture for many years, such as Daudi and Namalwa, have acquired intermediate phenotypes in culture that do not correspond well to the group I to III paradigms. Thus, Namalwa was exceptional in having EBNA-2 expression, but no AML-2 and Daudi cells have been reported to have high AML-2 RNA expression despite having a deletion of the EBNA-2 gene (30). Namalwa and Daudi seem to be exceptions to the general relationship between the group I and III phenotype and AML expression. AML-2 (also known as RUNX3, CBFα-3, or PEBP2α-C) is a member of the “Runt domain” family of transcription factors which are important regulators of hematopoiesis and osteogenesis. The family, which includes AML-1, AML-2, and AML-3, has a highly conserved 100- to 120-amino-acid Runt homology domain (RHD) that mediates binding to the DNA motif PyGPyGGT (called the Runt domain-binding element). The activity of the AML proteins is enhanced by the formation of the core binding factor (CBF) heterodimer containing one of the AML proteins and CBFβ. This CBF transcription factor complex may repress or activate genes depending on the promoter (29) and the cell type. The Runt domain binding element is frequently found in the promoters of hematopoietic genes, and the known target genes of AML-1 include the cytokines IL-3 and G-CSF (52, 58), cytokine receptors (64), subunits of the T- and B-cell receptors (10, 39), and the B-cell-specific protein BLK (31).
The AML transcription factors are already known to be important in human leukemia. The CBF complex is one of the most frequent targets of chromosomal translocation in acute leukemia (reviewed in reference 61). The translocations occur primarily between the AML-1 gene locus on chromosome 21 and up to 10 other loci, often resulting in the generation of fusion proteins between AML-1 and transcriptional repressors, thus altering the normal function of the AML protein (12, 35, 42). Although its function is frequently disrupted in leukemias, AML-1 also acts as an oncogene in its wild-type form (26, 53). Similar experiments with AML-2 did not reveal any transforming activity. Transformation of the group I BL cells that express high levels of AML-1 occurs through translocation of the c-myc gene so that it is constitutively expressed under the control of the immunoglobulin enhancer element. There is therefore no evidence yet to suggest that either AML-1 or AML-2 have an oncogenic function in EBV-infected B cells, but the presence of either AML-1 or AML-2 in the group I or group II BL cell lines may contribute to other phenotypic differences.
Our observation that both AML-2 and AML-1 are regulated within 48 h of primary B-cell infection suggests that the AML transcription factors may have a role in EBV induced proliferation. AML-1 has previously been reported to bind the EBV LMP-1 promoter, although this binding was not thought to contribute to the activation of the LMP-1 promoter caused by EBNA-2 (20). That study was completed prior to the identification of two other members of the AML family, AML-2 and AML-3. Our data suggest that where EBNA-2 and LMP-1 are both active, then the AML family member most likely to be present and binding the LMP-1 promoter is AML-2. However, the promoter mutation analysis concluding that the AML site does not contribute to EBNA-2 responsiveness was carried out in BJAB cells, which themselves contain much more AML-2 than AML-1, so it is not likely that AML-2 is a significant regulator of LMP-1.
We have shown that AML-1 is the dominant AML family member present in primary B cells prior to its rapid downregulation after EBV infection. Three major isoforms of AML-1 are generated by alternate splicing and the activity of two distinct promoters: the proximal promoter driving expression of AML-1A and AML-1B and the distal promoter generating the AML-1C isoform (11). Primary B cells have previously been reported to lack AML-1A and AML-1B transcripts but contain AML-1 transcripts generated from the distal promoter lacking exons 1 to 4 and exons 2 to 4 (5). The products generated from these transcripts would not contain an RHD region and were therefore predicted to interfere with growth factor signaling in normal circulating B cells. In our study, we have detected AML-1 by Western blotting with an AML-1-specific antibody generated against the region corresponding to residues 50 to 177. This antibody therefore recognizes the RHD, and we would predict that the AML-1 protein detected in peripheral blood B cells would be functional. It was notable that the expression of AML-1 was rapidly downregulated after EBV infection and PMA stimulation, whereas the levels of AML-2 simultaneously increased. Both the distal and proximal promoter regions of AML-1 have CBF binding sites, and it has been suggested previously that AML-1 expression may be regulated by one of the AML family members (11). We could speculate therefore that the increased AML-2 levels induced by EBV infection could have a direct effect on the activity of one or both of the AML-1 promoters.
Relatively little is known about the function of AML-2, which maps to the short arm of chromosome 1 at p36 (30). As well as being induced by EBV infection in our system, AML-2 is induced in hematopoietic cells through the retinoic acid receptor α-dependent signaling pathway (28). In the mouse system, AML-2 is induced by TGFβ1 (46) and both binds and functionally cooperates with SMAD3 to stimulate transcription of the germ line immunoglobulin Cα promoter (15, 66). It therefore appears to have an important function in class switching to produce immunoglobulin A. Since all of the family members bind to the same consensus DNA sequence, there is likely to be highly complex transcriptional regulation of responsive genes depending on the AML family members and cofactors present. We are carrying out further investigations into the possible effects of induction of AML-2 in B cells and whether AML-2 might contribute to downstream effects ascribed indirectly to EBNA-2. The data we have presented provides evidence that AML-2 is a direct target of EBNA-2 transcription but, because EBNA-2 has wide-ranging effects on many viral genes, we cannot rule out the possibility that other EBV genes are involved in its regulation.
To our knowledge, this is the first cell transcription factor identified whose expression distinguishes between phenotypically group I and group III BL cells and may play an important role in contributing toward phenotypic differences between these two cell types.
Acknowledgments
We thank Abraham Karpas and Bettina Kempkes for cell lines, Martin Allday for comments on the manuscript, and the Sanger Centre microarray consortium.
REFERENCES
- 1.Baier, M., N. Bannert, A. Werner, K. Lang, and R. Kurth. 1997. Molecular cloning, sequence, expression, and processing of the interleukin 16 precursor. Proc. Natl. Acad. Sci. USA 94:5273-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Bassat, H., N. Goldblum, S. Mitrani, T. Goldblum, J. M. Yoffey, M. M. Cohen, Z. Bentwich, B. Ramot, E. Klein, and G. Klein. 1977. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D.G.-75). Int. J. Cancer 19:27-33. [DOI] [PubMed] [Google Scholar]
- 3.Brimmell, M., R. Mendiola, J. Mangion, and G. Packham. 1998. BAX frameshift mutations in cell lines derived from human haemopoetic malignancies are associated with resistance to apoptosis and microsatellite instability. Oncogene 16:1803-1812. [DOI] [PubMed] [Google Scholar]
- 4.Cannell, E., P. Farrell, and A. Sinclair. 1996. Epstein-Barr virus exploits the normal cell pathway to regulate Rb activity during the immortalisation of primary B cells. Oncogene 13:1413-1421. [PubMed] [Google Scholar]
- 5.Chimienti, G., M. Alaibac, F. Marzullo, A. Carbone, and G. Pepe. 2000. The expression pattern of the AML1 gene in non-Hodgkin's B-cell lymphomas and normal B lymphocytes. Blood Cell Mol. Dis. 26:186-192. [DOI] [PubMed] [Google Scholar]
- 6.Chupp, G. L., E. A. Wright, D. Wu, M. Vallen-Mashikian, W. W. Cruikshank, D. M. Center, H. Kornfeld, and J. S. Berman. 1998. Tissue and T-cell distribution of precursor and mature IL-16. J. Immunol. 161:3114-3119. [PubMed] [Google Scholar]
- 7.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruikshank, W. W., K. Lim, A. C. Theodore, J. Cook, G. Fine, P. F. Weller, and D. M. Center. 1996. IL-16 inhibition of CD3-dependent lymphocyte activation and proliferation. J. Immunol. 157:5240-5248. [PubMed] [Google Scholar]
- 9.De Smaele, E., F. Zazzeroni, F. Papa, D. Nguyen, R. Jin, J. Jones, R. Cong, and G. Franzoso. 2001. Induction of gadd45β by NF-κB downregulates pro-apoptotic JNK signalling. Nature 414:308-313. [DOI] [PubMed] [Google Scholar]
- 10.Erman, B., M. Cortes, B. S. Nikolajczyk, N. A. Speck, and R. Sen. 1998. ETS-core binding factor: a common composite motif in antigen receptor gene enhancers. Mol. Cell. Biol. 18:1322-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghozi, M. C., Y. Bernstein, V. Negreanu, D. Levanon, and Y. Groner. 1996. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc. Natl. Acad. Sci. USA 93:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golub, T. R., G. F. Barker, S. K. Bohlander, S. W. Hiebert, D. C. Ward, P. Bray-Ward, E. Morgan, S. C. Raimondi, J. D. Rowley, and D. G. Gilliland. 1995. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 92:4917-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg, M. E., A. L. Hermanowski, and E. B. Ziff. 1986. Effect of protein synthesis inhibitors on growth factor activation of c-fos, c-myc, and actin gene transcription. Mol. Cell. Biol. 6:1050-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory, C. D., T. Tursz, C. F. Edwards, C. Tetaud, M. Talbot, B. Caillou, A. B. Rickinson, and M. Lipinski. 1987. Identification of a subset of normal B cells with a Burkitt's lymphoma (BL)-like phenotype. J. Immunol. 139:313-318. [PubMed] [Google Scholar]
- 15.Hanai, J., L. F. Chen, T. Kanno, N. Ohtani-Fujita, W. Y. Kim, W. H. Guo, T. Imamura, Y. Ishidou, M. Fukuchi, M. J. Shi, J. Stavnezer, M. Kawabata, K. Miyazono, and Y. Ito. 1999. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Cα promoter. J. Biol. Chem. 274:31577-31582. [DOI] [PubMed] [Google Scholar]
- 16.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP (EBNA-LP) stimulates EBNA-2 acidic domain mediated transcriptional activation. J. Virol. 71:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 18.Hollyoake, M., A. Stuhler, P. Farrell, J. Gordon, and A. Sinclair. 1995. The normal cell cycle activation program is exploited during the infection of quiescent B lymphocytes by Epstein-Barr virus. Cancer Res. 55:4784-4787. [PubMed] [Google Scholar]
- 19.Jochner, N., D. Eick, U. Zimber-Strobl, M. Pawlita, G. W. Bornkamm, and B. Kempkes. 1996. Epstein-Barr virus nuclear antigen 2 is a transcriptional suppressor of the immunoglobulin mu gene: implications for the expression of the translocated c-myc gene in Burkitt's lymphoma cells. EMBO J. 15:375-382. [PMC free article] [PubMed] [Google Scholar]
- 20.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaser, A., S. Dunzendorfer, F. A. Offner, O. Ludwiczek, B. Enrich, R. O. Koch, W. W. Cruikshank, C. J. Wiedermann, and H. Tilg. 2000. B lymphocyte-derived IL-16 attracts dendritic cells and Th cells. J. Immunol. 165:2474-2480. [DOI] [PubMed] [Google Scholar]
- 23.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieff, E., and A. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Philadelphia, Pa.
- 25.Kienzle, N., D. Young, S. L. Silins, and T. B. Sculley. 1996. Induction of pleckstrin by the Epstein-Barr virus nuclear antigen 3 family. Virology 224:167-174. [DOI] [PubMed] [Google Scholar]
- 26.Kurokawa, M., T. Tanaka, K. Tanaka, S. Ogawa, K. Mitani, Y. Yazaki, and H. Hirai. 1996. Overexpression of the AML1 proto-oncoprotein in NIH 3T3 cells leads to neoplastic transformation depending on the DNA-binding and transactivational potencies. Oncogene 12:883-892. [PubMed] [Google Scholar]
- 27.Laux, G., B. Adam, L. J. Strobl, and F. Moreau-Gachelin. 1994. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-Jκ interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 13:5624-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le, X. F., Y. Groner, S. M. Kornblau, Y. Gu, W. N. Hittelman, D. Levanon, K. Mehta, R. B. Arlinghaus, and K. S. Chang. 1999. Regulation of AML2/CBFA3 in hematopoietic cells through the retinoic acid receptor α-dependent signaling pathway. J. Biol. Chem. 274:21651-21658. [DOI] [PubMed] [Google Scholar]
- 29.Levanon, D., R. E. Goldstein, Y. Bernstein, H. Tang, D. Goldenberg, S. Stifani, Z. Paroush, and Y. Groner. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA 95:11590-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levanon, D., V. Negreanu, Y. Bernstein, I. Bar-Am, L. Avivi, and Y. Groner. 1994. AML1, AML2, and AML3, the human members of the Runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics 23:425-432. [DOI] [PubMed] [Google Scholar]
- 31.Libermann, T. A., Z. Pan, Y. Akbarali, C. J. Hetherington, J. Boltax, D. A. Yergeau, and D. E. Zhang. 1999. AML1 (CBFα2) cooperates with B cell-specific activating protein (BSAP/PAX5) in activation of the B cell-specific BLK gene promoter. J. Biol. Chem. 274:24671-24676. [DOI] [PubMed] [Google Scholar]
- 32.Ling, P. D., J. J. Hsieh, I. K. Ruf, D. R. Rawlins, and S. D. Hayward. 1994. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J. Virol. 68:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mann, K. P., D. Staunton, and D. A. Thorley-Lawson. 1985. Epstein-Barr virus-encoded protein found in plasma membrane in transformed cells. J. Virol. 55:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyers, S., N. Lenny, W. Sun, and S. W. Hiebert. 1996. AML-2 is a potential target for transcriptional regulation by the t(8;21) and t(12;21) fusion proteins in acute leukemia. Oncogene 13:303-312. [PubMed] [Google Scholar]
- 35.Miyoshi, H., T. Kozu, K. Shimizu, K. Enomoto, N. Maseki, Y. Kaneko, N. Kamada, and M. Ohki. 1993. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 12:2715-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nacheva, E., P. E. Fischer, P. D. Sherrington, W. Labastide, E. Lawlor, E. Conneally, C. Blaney, F. G. Hayhoe, and A. Karpas. 1990. A new human plasma cell line, Karpas 620, with translocations involving chromosomes 1,11 and 14. Br. J. Haematol. 74:70-76. [DOI] [PubMed] [Google Scholar]
- 37.Nikiforow, S., K. Bottomly, and G. Miller. 2001. CD4+ T-cell effectors inhibit Epstein-Barr virus-induced B-cell proliferation. J. Virol. 75:3740-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein (EBNA-LP) enhances EBNA-2 mediated transactivation of latent membrane protein 1 expression; a role for the W1W2 repeat domain. J. Virol. 71:6619-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prosser, H. M., D. Wotton, A. Gegonne, J. Ghysdael, S. Wang, N. A. Speck, and M. J. Owen. 1992. A phorbol ester response element within the human T-cell receptor beta-chain enhancer. Proc. Natl. Acad. Sci. USA 89:9934-9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabson, M., L. Gradoville, L. Heston, and G. Miller. 1982. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J. Virol. 44:834-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rickinson, A. B., L. S. Young, and M. Rowe. 1987. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J. Virol. 61:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romana, S. P., M. Mauchauffe, M. Le Coniat, I. Chumakov, D. Le Paslier, R. Berger, and O. A. Bernard. 1995. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood 85:3662-3670. [PubMed] [Google Scholar]
- 43.Rowe, M., D. T. Rowe, C. D. Gregory, L. S. Young, P. J. Farrell, H. Rupani, and A. B. Rickinson. 1987. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J. 6:2743-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sciaky, D., W. Brazer, D. M. Center, W. W. Cruikshank, and T. J. Smith. 2000. Cultured human fibroblasts express constitutive IL-16 mRNA: cytokine induction of active IL-16 protein synthesis through a caspase-3-dependent mechanism. J. Immunol. 164:3806-3814. [DOI] [PubMed] [Google Scholar]
- 45.Sharma, V., J. L. Sparks, and J. D. Vail. 2000. Human B-cell lines constitutively express and secrete interleukin-16. Immunology 99:266-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, M. J., and J. Stavnezer. 1998. CBF α3 (AML2) is induced by TGF-β1 to bind and activate the mouse germline Ig α promoter. J. Immunol. 161:6751-6760. [PubMed] [Google Scholar]
- 47.Sinclair, A. J., I. Palmero, G. Peters, and P. J. Farrell. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 13:3321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sjoblom, A., W. Yang, L. Palmqvist, A. Jansson, and L. Rymo. 1998. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J. Virol. 72:1365-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spender, L., E. Cannell, M. Hollyoake, B. Wensing, J. Gawn, M. Brimmell, G. Packham, and P. Farrell. 1999. Control of cell cycle entry and apoptosis in B lymphocytes infected by Epstein-Barr virus. J. Virol. 73:4678-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spender, L. C., G. H. Cornish, B. Rowland, B. Kempkes, and P. J. Farrell. 2001. Direct and indirect regulation of cytokine and cell cycle proteins by EBNA-2 during Epstein-Barr virus infection. J. Virol. 75:3537-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sutkowski, N., B. Conrad, D. A. Thorley-Lawson, and B. T. Huber. 2001. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 15:579-589. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi, A., M. Satake, Y. Yamaguchi-Iwai, S. C. Bae, J. Lu, M. Maruyama, Y. W. Zhang, H. Oka, N. Arai, K. Arai, et al. 1995. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood 86:607-616. [PubMed] [Google Scholar]
- 53.Tanaka, T., M. Kurokawa, K. Ueki, K. Tanaka, Y. Imai, K. Mitani, K. Okazaki, N. Sagata, Y. Yazaki, Y. Shibata, T. Kadowaki, and H. Hirai. 1996. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol. Cell. Biol. 16:3967-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theodore, A. C., D. M. Center, J. Nicoll, G. Fine, H. Kornfeld, and W. W. Cruikshank. 1996. CD4 ligand IL-16 inhibits the mixed lymphocyte reaction. J. Immunol. 157:1958-1964. [PubMed] [Google Scholar]
- 55.Tong, X., R. Drapkin, D. Reinberg, and E. Kieff. 1995. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc. Natl. Acad. Sci. USA 92:3259-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong, X., R. Drapkin, R. Yalamanchili, G. Mosialos, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 15:4735-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong, X., F. Wang, C. J. Thut, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J. Virol. 69:585-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uchida, H., J. Zhang, and S. D. Nimer. 1997. AML1A and AML1B can transactivate the human IL-3 promoter. J. Immunol. 158:2251-2258. [PubMed] [Google Scholar]
- 59.Waltzer, L., F. Logeat, C. Brou, A. Israel, A. Sergeant, and E. Manet. 1994. The human Jκ recombination signal sequence binding protein (RBP-J kappa) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 13:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westendorf, J. J., and S. W. Hiebert. 1999. Mammalian Runt-domain proteins and their roles in hematopoiesis, osteogenesis, and leukemia. J. Cell Biochem. 1999(Suppl. 32-33):51-58. [DOI] [PubMed] [Google Scholar]
- 62.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, D. Y., A. Krumm, and W. H. Schubach. 2000. Promoter-specific targeting of human SWI-SNF complex by Epstein-Barr virus nuclear protein 2. J. Virol. 74:8893-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, D. E., K. Fujioka, C. J. Hetherington, L. H. Shapiro, H. M. Chen, A. T. Look, and D. G. Tenen. 1994. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1). Mol. Cell. Biol. 14:8085-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Y., D. M. Center, D. M. Wu, W. W. Cruikshank, J. Yuan, D. W. Andrews, and H. Kornfeld. 1998. Processing and activation of pro-interleukin-16 by caspase-3. J. Biol. Chem. 273:1144-1149. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, Y., and R. Derynck. 2000. Transcriptional regulation of the transforming growth factor-beta-inducible mouse germ line Ig alpha constant region gene by functional cooperation of Smad, CREB, and AML family members. J. Biol. Chem. 275:16979-16985. [DOI] [PubMed] [Google Scholar]
- 67.Zimber-Strobl, U., B. Kempkes, G. Marschall, R. Zeidler, C. Van Kooten, J. Banchereau, G. Bornkamm, and W. Hammerschmidt. 1996. Epstein-Barr virus latent membrane protein (LMP1) is not sufficient to maintain proliferation of B cells but both it and activated CD40 can prolong their survival. EMBO J. 15:7070-7078. [PMC free article] [PubMed] [Google Scholar]
- 68.Zimber-Strobl, U., E. Kremmer, F. Grasser, G. Marschall, G. Laux, and G. W. Bornkamm. 1993. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 12:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]