Abstract
Studies of mammalian genes activated in response to an acute stimulus have suggested diverse mechanisms through which chromatin structure and nucleosome remodeling events contribute to inducible gene transcription. However, because of this diversity, the logical organization of the genome with respect to nucleosome remodeling and gene induction has remained obscure. Numerous proinflammatory genes are rapidly induced in macrophages in response to microbial infection. Here, we show that in lipopolysaccharide-stimulated macrophages, the catalytic BRG1/BRM subunits of the SWI/SNF class of ATP-dependent nucleosome remodeling complexes are consistently required for the activation of secondary response genes and primary response genes induced with delayed kinetics, but not for rapidly induced primary response genes. Surprisingly, a Mi-2β complex was selectively recruited along with the SWI/SNF complexes to the control regions of secondary response and delayed primary response genes, with the Mi-2β complex acting antagonistically to limit the induction of these gene classes. SWI/SNF and Mi-2β complexes influenced cell size in a similarly antagonistic manner. These results provide insight into the differential contributions of nucleosome remodeling complexes to the rapid induction of defined classes of mammalian genes and reveal a robust anti-inflammatory function of Mi-2β.
Keywords: Inflammation, cytokines, chromatin, SWI/SNF, NuRD
The ability of eukaryotic cells to respond to external stimuli depends on the coordinated activation of specific subsets of genes. To facilitate gene activation, repressive chromatin structures must be altered by histone-modifying complexes and ATP-dependent remodeling complexes (Roth et al. 2001; Peterson 2002; Carrozza et al. 2003; Lusser and Kadonaga 2003; Cairns 2005). In mammalian cells, two of the most widely studied families of ATP-dependent remodeling complexes are the SWI/SNF and Mi-2/NuRD complexes. SWI/SNF complexes contain either of two ATPase subunits, BRG1 and BRM, along with a number of BRG-associated factors (BAFs) (Becker and Horz 2002; Martens and Winston 2003). BRG1 and BRM are nearly 75% homologous and have partially redundant and specific roles during gene activation (Reyes et al. 1998; Bultman et al. 2000; Kadam and Emerson 2003). The Mi-2/NuRD complexes contain the Mi-2α or Mi-2β ATPase subunit, along with several associated factors that include histone deacetylases (Becker and Horz 2002; Feng and Zhang 2003).
Several transcription factors can interact with SWI/SNF complexes and recruit the complexes to specific genes (Peterson and Workman 2000; Kadam and Emerson 2002; Chi 2004). Furthermore, a large number of genes have been identified in yeast, fruit flies, and mammals that require SWI/SNF complexes for activation (Krebs et al. 2000; Sudarsanam et al. 2000; Liu et al. 2001; Armstrong et al. 2002; Ng et al. 2002). However, a broad understanding of the biological and mechanistic logic that distinguishes SWI/SNF-dependent from SWI/SNF-independent genes has not been obtained, particularly in mammalian cells. For example, the promoters of some genes activated in response to an acute stimulus, such as heat-shock genes, appear to exist in an open chromatin structure, sometimes with preinitiated and paused RNA polymerase II molecules, suggesting that these genes may not require nucleosome remodeling for transcriptional induction (Tsukiyama et al. 1994; Shopland et al. 1995; Herrera et al. 1997; Armstrong et al. 2002). Nevertheless, SWI/SNF complexes have been implicated in mouse hsp70 induction, with nucleosome remodeling linked to relief of the block in RNA polymerase II elongation (Corey et al. 2003). In contrast to this scenario, the promoter for another primary response gene, human IFNB1, contains a positioned nucleosome overlapping the TATA box, with a nucleosome-free distal promoter region that forms an enhanceosome upon binding of inducible transcription factors (Agalioti et al. 2000). Transcriptional induction of this gene requires SWI/SNF-dependent translocation of the positioned nucleosome. In a third classic scenario, the entire promoter within the mouse mammary tumor virus (MMTV) long terminal repeat (LTR) is contained within positioned nucleosomes, which must be remodeled by SWI/SNF complexes prior to transcription initiation (Hebbar and Archer 2003; Nagaich et al. 2004).
Mi-2/NuRD complexes have been shown to interact with DNA-binding proteins that have been implicated in transcriptional repression, such as Ikaros and BCL-6, consistent with the presence of histone deacetylases in the complexes (Kehle et al. 1998; Kim et al. 1999; Feng and Zhang 2003; Fujita et al. 2004; Liu and Bagchi 2004). Mi-2/NuRD complexes can be recruited to specific target genes by these proteins, although they can also be recruited to DNA through their association with methyl-CpG-binding proteins (Feng and Zhang 2003). Despite strong evidence that Mi-2/NuRD complexes contribute to transcriptional repression, the first mammalian loss-of-function study of Mi-2β demonstrated an essential role in activation of the T-cell-restricted Cd4 gene (Williams et al. 2004). In addition, at least two biochemical studies have revealed the existence of large multiprotein complexes that contain both BRG1 and Mi-2 (Nakamura et al. 2002; Shimono et al. 2003). However, the roles of these large complexes have not been explained by functional studies or current models.
The inflammatory response, an essential component of host defense against microbial infection, requires the rapid and selective activation of numerous proinflammatory genes in macrophages, dendritic cells, and other cells of the innate immune system (Janeway and Medzhitov 2002). Toll-like receptors (TLRs) play a critical role in responding to microbial components, such as lipopolysaccharide (LPS), by activating several common signal transduction pathways (Akira et al. 2001; Vaidya and Cheng 2003; Iwasaki and Medzhitov 2004; Jenner and Young 2005). In one example, nucleosome remodeling appears to contribute to the rapid induction of the p40 subunit of the proinflammatory cytokine interleukin-12 (IL-12) (Weinmann et al. 1999). In murine macrophages, the promoter of the Il12b gene, which encodes IL-12 p40, is contained within a positioned nucleosome, which upon LPS stimulation is rapidly and selectively remodeled (Weinmann et al. 1999). Nucleosome remodeling requires TLR4 signaling and new protein synthesis but is independent of the NF-κB subunit c-Rel, which is essential for transcription (Weinmann et al. 2001). Recently, an enhancer region located 10 kb upstream of the Il12b transcription start site was also found to exhibit increased restriction enzyme access in response to LPS signaling (Zhou et al. 2004; L. Zhou and S.T. Smale, unpubl.).
One challenge in defining the in vivo roles of nucleosome remodeling complexes is that their absence usually results in rapid cell death (Sumi-Ichinose et al. 1997). For this reason, in vivo studies of SWI/SNF functions in mammalian cells have relied more strongly on dominant-negative approaches and on the SW13 cell line, which has adapted to growth in the absence of SWI/SNF complexes, than on mice or cells with disrupted SWI/SNF alleles (de La Serna et al. 2000; Liu et al. 2001; Chi et al. 2003; Chi 2004). As an alternative strategy for performing loss-of-function studies, we disrupted expression of remodeling complexes by retroviral delivery of small interfering RNAs (siRNAs) and focused on gene activation in response to LPS, an acute extracellular stimulus. This strategy allowed us to study cells possessing greatly reduced concentrations of remodeling proteins before their absence resulted in a loss of viability. Our results reveal clear logic in LPS-stimulated macrophages, in that consistent requirements for SWI/SNF complexes were observed at secondary response genes and primary response genes induced with delayed kinetics but not at rapidly induced primary response genes. Furthermore, strong and consistent antagonism between SWI/SNF and Mi-2β complexes was observed, revealing that Mi-2β is a potent anti-inflammatory molecule.
Results
Kinetics of LPS-induced nucleosome remodeling at Il12b control regions
Previous studies revealed that the activation of primary and transformed macrophages by LPS results in increased restriction enzyme accessibility at a single positioned nucleosome located at the Il12b promoter, and at an enhancer located 10 kb upstream of the transcription start site (Weinmann et al. 1999; Zhou et al. 2004). In the assay used here, nuclei from unstimulated and LPS-stimulated J774 macrophages were treated with a restriction enzyme; after purification of genomic DNA and cleavage with a second enzyme, the efficiency of cleavage in the isolated nuclei was monitored by Southern blot (Zhou et al. 2004). The Il12b promoter and enhancer both contain SpeI restriction sites, which allow a direct comparison of accessibility at the two locations (Fig. 1A). At both the promoter and enhancer, a small increase in SpeI cleavage was detected 30 min after LPS stimulation, with more pronounced increases after 1 h (Fig. 1B). SpeI cleavage at the enhancer was much more efficient than at the promoter, with similar results obtained when another enzyme that cleaves at both locations was tested (data not shown). The reason for this difference is not known. Consistent with previous studies, the LPS-induced increases in restriction enzyme cleavage were dependent on new protein synthesis, as pretreatment of cells with the protein synthesis inhibitor cycloheximide (CHX) abrogated the increases (Fig. 1B; Weinmann et al. 1999; L. Zhou and S.T. Smale, unpubl.).
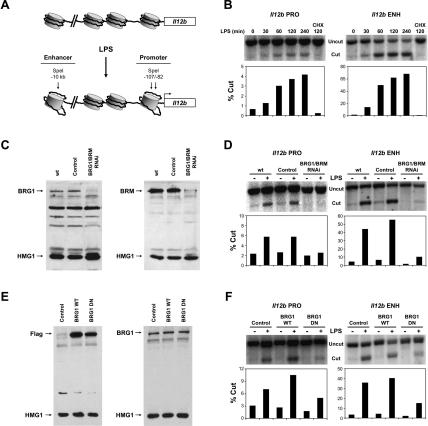
Figure 1.
SWI/SNF-dependent nucleosome remodeling at Il12b control regions. (A) The locations of SpeI restriction enzyme recognition sites used for the analysis of LPS-induced nucleosome remodeling at the Il12b promoter and enhancer are shown. (B) Nuclei from J774 macrophages stimulated with LPS for the indicated times were digested with SpeI for 15 min (Cut). The purified DNA was then digested with two reference enzymes (Uncut) and analyzed by Southern blot using promoter-specific (PRO) and enhancer-specific (ENH) 32P-labeled probes as described (Weinmann et al. 1999; Zhou et al. 2004). Nuclei from J774 cells pretreated with cycloheximide (CHX) for 15 min before LPS activation were also analyzed. The efficiency of SpeI cleavage was quantified by PhosphorImager analysis and plotted as the percentage of genomic DNA cleaved by SpeI (% Cut). The data are representative of three independent experiments. (C) BRG1 and BRM were simultaneously depleted in J774 macrophages using a retroviral RNAi strategy. The siRNA hairpin was designed to target a homologous sequence of the BRG1 and BRM mRNAs. The expression of BRG1 (left) and BRM (right) was monitored by Western blot in whole-cell extracts prepared from uninfected J774 macrophages (wt) or from J774 cells infected with an empty RNAi retroviral vector (control) or the BRG1/BRM RNAi vector. The DNA-binding protein HMG1 was analyzed as a loading control. (D) The restriction enzyme accessibility assay was used to monitor nucleosome remodeling at the Il12b promoter (PRO) and enhancer (ENH) in nuclei from uninfected cells (wt), empty vector (control) cells, and BRG1/BRM-depleted cells before (-) or after (+) LPS stimulation for 4 h. (E) Wild-type (WT) and dominant-negative (DN) BRG1 proteins were stably expressed in J774 macrophages. Expression of BRG1 WT and BRG1 DN was monitored by Western blot using Flag (left) and BRG1 (right) antibodies and whole-cell extracts from uninfected J774 cells (control) or J774 cell clones expressing Flag-tagged dominant-negative BRG1 or wild-type BRG1. (F) Restriction enzyme accessibility assays were performed with nuclei from uninfected J774 cells or cell clones expressing BRG1 WT or BRG1 DN, before (-) and after (+) LPS stimulation for 4 h.
SWI/SNF complexes are required for LPS-induced nucleosome remodeling at Il12b control regions
To identify the remodeling complexes required for LPS-induced nucleosome remodeling at the Il12b promoter and enhancer, the expression of BRG1 and BRM, the catalytic subunits of SWI/SNF remodeling complexes, was disrupted in J774 macrophages using a retroviral RNA interference (RNAi) strategy (Supplementary Fig. 1). This strategy results in the stable integration and expression of siRNA hairpins. The retroviral vector also includes a GFP reporter cassette and a puromycin selectable marker, which allow the measurement and enrichment of cell populations that express the hairpins. Because BRG1 and BRM are thought to be partially redundant and compensatory, an siRNA hairpin was designed to target a 19-base-pair (bp) homologous region of the BRG1 and BRM mRNAs. Retroviral delivery of the BRG1/BRM siRNA resulted in the simultaneous depletion of the BRG1 and BRM proteins (Fig. 1C). Substantial depletion of BRG1 and BRM was observed from 3 to 10 d after retroviral transduction (data in Fig. 1C obtained 5 d after transduction). However, after 10 d, BRG1/BRM depletion became highly toxic to the cells. Thus, in the following experiments, BRG1/BRM-depleted cells were assayed 5 d after retroviral transduction, before any noticeable cell death (Supplementary Fig. 1; data not shown).
To determine whether BRG1/BRM depletion affects LPS-induced remodeling at Il12b control regions, the LPS-induced increases in SpeI cleavage were monitored (Fig. 1D). The cleavage efficiencies at the Il12b promoter and enhancer were strongly reduced in nuclei from BRG1/BRM-depleted cells relative to nuclei from uninfected (wt) cells and cells transduced with the control virus.
In parallel experiments, a dominant-negative version of BRG1 was used to disrupt SWI/SNF function in J774 macrophages. The dominant-negative BRG1 contains a single point mutation in the ATP-binding domain that permits proper folding and complex assembly, but disrupts nucleosome remodeling (de La Serna et al. 2000). Flag-tagged wild-type and dominant-negative BRG1 (BRG1 WT and BRG1 DN) were stably expressed in J774 cells by retroviral transduction (Fig. 1E, left panel). There were no apparent differences in cell growth and proliferation between these cell lines, and the overall BRG1 levels increased less than twofold compared with uninfected (control) cells (Fig. 1E, right panel). Using the restriction enzyme accessibility assay, a decrease in SpeI cleavage efficiency was observed at both the Il12b promoter and enhancer in nuclei from LPS-stimulated BRG1 DN cells in comparison to control cells or BRG1 WT cells (Fig. 1F). Although the magnitudes of the dominant-negative effects are smaller than the magnitudes of effects observed following siRNA-mediated depletion, the results support the hypothesis that nucleosome remodeling at the Il12b promoter and enhancer requires the expression and function of the SWI/SNF catalytic subunits.
Selective functions of SWI/SNF complexes during LPS stimulation of J774 cells
We hypothesized that nucleosome remodeling at the Il12b promoter and enhancer is essential for Il12b expression (Weinmann et al. 1999). To determine whether BRG1/BRM depletion influences Il12b induction in response to LPS, the secretion of IL-12 p40 protein from J774 macrophages was measured by ELISA. Four hours after LPS treatment, BRG1/BRM-depleted cells produced sixfold less IL-12 p40 relative to wt or control cells (Fig. 2A). This decrease could be due to a direct requirement for BRG1/BRM at the Il12b locus or to indirect effects on cell viability or the expression of other genes. To begin to distinguish between these possibilities, we examined the expression of other LPS-induced genes, beginning with the Cxcl2 gene encoding macrophage inflammatory protein 2 (MIP-2), an inducible chemokine involved in attracting neutrophils to sites of infection. Interestingly, MIP-2 induction by ELISA was unaffected by BRG1/BRM depletion (Fig. 2A). Similar results were obtained with BRG1 DN macrophages (Fig. 2B). Thus, SWI/SNF ATPase subunits appear to be required for the activation of only a subset of LPS-induced genes.
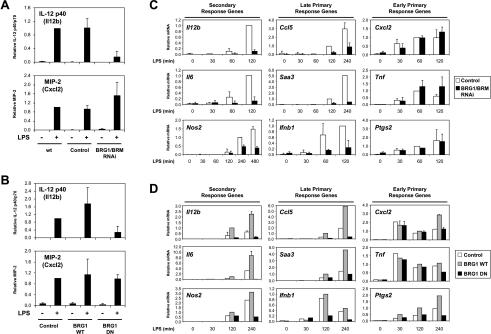
Figure 2.
Selective requirement for SWI/SNF at secondary response and late primary response genes. (A) IL-12 p40 (Il12b) and MIP-2 (Cxcl2) production were monitored by ELISA in supernatants from uninfected J774 cells (wt), cells infected with the empty retroviral vector (control), and BRG1/BRM-depleted cells, before (-) and after (+) LPS stimulation for 4 h. The amounts of secreted protein are plotted relative to uninfected J774 cells and include data from three independent experiments. (B) IL-12 p40 and MIP-2 production were monitored by ELISA in supernatants from uninfected J774 cells (control) or J774 clones expressing wild-type (BRG1 WT) or dominant-negative (BRG1 DN) BRG1 before (-) and after (+) LPS stimulation for 4 h. Data were quantified as in A. (C) Quantitative real-time RT–PCR was used to analyze RNA harvested from J774 cells infected with the empty retroviral vector (white bars) or BRG1/BRM RNAi-depleted (black bars) cells stimulated with LPS for the indicated times. Nine potent LPS-induced genes were grouped according to their sensitivity to CHX (primary vs. secondary response genes) and kinetics of induction (early vs. late). For each gene examined, mRNA levels were normalized to the constitutive Gapd mRNA. The mRNA levels were plotted relative to time points at which significant signals were obtained. This approach was necessary because fold inductions would otherwise be weighted relative to weak background signals obtained in the absence of LPS. For secondary and late primary response genes, mRNA levels were plotted relative to the level observed at 120 min of LPS stimulation in control cells, with the exception of Nos2, which was plotted relative to the 240-min time point. For early primary response genes, mRNA levels were plotted relative to the 60-min time point from control cells. The relative mRNA values were determined from three to five independent BRG1/BRM RNAi depletion experiments. (D) Quantitative real-time RT–PCR was used to analyze mRNA levels in uninfected J774 cells (white bars), BRG1 WT cells (gray bars), and BRG1 DN cells (black bars) stimulated with LPS for the indicated time points. mRNA levels were plotted relative to the 120-min time point from BRG1 WT cells.
SWI/SNF complexes are selectively required for the induction of secondary response and delayed primary response genes
To explore the logic underlying the selective requirement for SWI/SNF complexes at LPS-induced genes, the expression properties of Il12b and Cxcl2 were compared.
Consistent with previous reports (Saccani et al. 2001; Bradley et al. 2003), Cxcl2 mRNA levels increased substantially 30 min after LPS stimulation, whereas Il12b mRNA accumulated at later times, with high levels observed after 2 h (Fig. 2C). In addition, LPS-induced Il12b transcription is inhibited by CHX (Weinmann et al. 1999), whereas Cxcl2 transcription was enhanced in the presence of CHX (Supplementary Fig. 2). Consequently, in activated macrophages, Il12b can be classified as a secondary response gene and Cxcl2 as a primary response gene.
To determine whether SWI/SNF dependence is observed only at secondary response genes following LPS stimulation, mRNAs for nine LPS-induced genes were monitored by quantitative RT–PCR in BRG1/BRM-depleted and control macrophages (Fig. 2C). For this analysis, primary response genes were defined as those whose induced mRNA levels remained unchanged or increased in the presence of CHX, whereas secondary response genes were those whose induced mRNA levels were inhibited at least fivefold in the presence of CHX (Supplementary Fig. 2). The analysis was also restricted to genes that displayed potent LPS induction (at least 10-fold by 2 h after LPS stimulation) in J774 cells.
The data in Figure 2C show that BRG1/BRM depletion inhibited the LPS-induced expression of three secondary response genes (Il12b, Il6, and Nos2 [encoding iNOS]). However, the expression of only three of six primary response genes was significantly reduced following BRG1/BRM depletion. Interestingly, the three primary response genes that were sensitive to BRG1/BRM depletion (Ccl5 [RANTES], Saa3, and Ifnb1) were induced with delayed kinetics, whereas the three that were resistant to BRG1/BRM depletion (Cxcl2, Tnf, and Ptgs2 [Cox-2]) were induced more rapidly. In these experiments and those described below, early primary response genes were defined as those whose mRNA levels increased more than fivefold by 30 min after LPS treatment. In contrast, mRNA levels for the late primary response genes were either undetectable or induced by less than twofold at the 30-min time point.
To validate the RNAi results, the same group of genes was examined in BRG1 DN and BRG1 WT macrophages (Fig. 2D). The results were largely consistent with the RNAi results, although they are more difficult to interpret because ectopic expression of the BRG1 WT protein led to super-activation of some genes. When the effects of BRG1 WT and BRG1 DN are compared, the BRG1 DN protein strongly inhibited the three secondary response and three delayed primary response genes. However, when the BRG1 DN results were compared with the control results, the BRG1 DN protein had little effect on Saa3 expression. At the early primary response genes, the BRG1 DN protein had relatively little effect on the Cxcl2 and Tnf genes, but it resulted in substantially decreased expression of Ptgs2 (Fig. 2D). This latter effect may be related to prior evidence that the Ptgs2 gene is regulated in a biphasic manner (Caivano et al. 2001). Taken together, the RNAi and DN results suggest that SWI/SNF complexes may be critical for the LPS induction of secondary response and late primary response genes, but not for the induction of early primary response genes. This trend was upheld when many more LPS-induced genes were analyzed (see Fig. 4C, below).
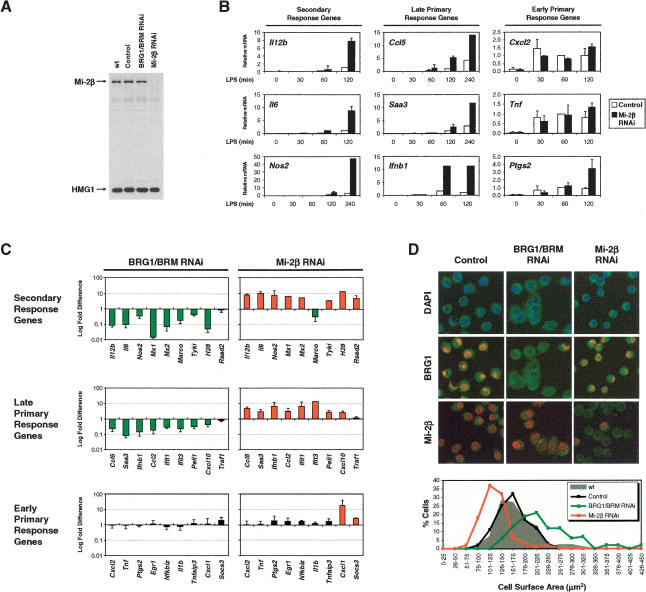
Figure 4.
Antagonistic functions of SWI/SNF and Mi-2β complexes. (A) Mi-2β was depleted from J774 cells using the retroviral RNAi strategy. Mi-2β expression was monitored by Western blot in whole-cell extracts prepared from uninfected J774 cells (wt), cells infected with the empty RNAi vector (control), or cells infected with RNAi vectors that target BRG1/BRM or Mi-2β. HMG1 expression was analyzed as a loading control. (B) The effect of Mi-2β depletion on gene expression was monitored by real-time RT–PCR, using RNA harvested from J774 cells infected with the empty retroviral vector (white bars) or Mi-2β-depleted (black bars) cells stimulated with LPS for the indicated time points. Relative mRNA values were calculated as in Figure 2C. (C) mRNA levels (fold differences in log scale) were compared between BRG1/BRM-depleted cells and cells infected with the empty retroviral vector (left column), or between Mi-2β-depleted cells and cells infected with the empty vector (right column). For each gene, the time-dependent induction in mRNA levels in response to LPS stimulation was measured by quantitative real-time RT–PCR. The effects on mRNA levels due to BRG1/BRM or Mi-2β depletion were pooled from different time points after LPS stimulation whenever the induction was at least fivefold in the control cells. Differences in mRNA levels greater than twofold are represented as green bars (decrease) or red bars (increase), whereas differences less than twofold are represented as black bars. (D) Confocal fluorescence microscopy was used to analyze J774 macrophages infected with the empty RNAi vector (control), as well as the BRG1/BRM and Mi-2β RNAi vectors, all of which express GFP (green). Cells were stained with DAPI (blue) and analyzed for protein depletion using BRG1 or Mi-2β antibodies (red). Digital images were used to measure cell surface area. The graph shows the distribution of surface area of uninfected cells (n = 83, gray area), empty vector-infected cells (n = 114, black plot), BRG1/BRM-depleted cells (n = 126, green plot), and Mi-2β-depleted cells (n = 111, red plot).
SWI/SNF and Mi-2β complexes are recruited to proinflammatory control regions
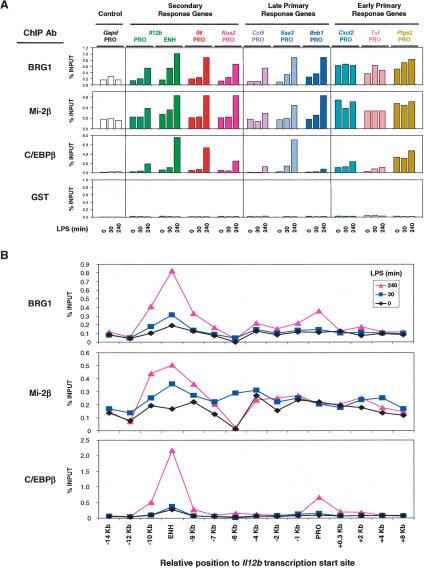
To determine whether SWI/SNF complexes associate directly with the control regions of secondary response genes and late primary response genes, chromatin immunoprecipitation (ChIP) experiments were performed using BRG1 antibodies and sheared cross-linked chromatin prepared from unstimulated or LPS-stimulated J774 cells (Fig. 3A, top). Quantitative real-time PCR analysis of DNA precipitated with BRG1 antibodies suggested that BRG1 associates in an LPS-induced fashion with the control regions of all six secondary response and late primary response genes examined. At these control regions, the real-time PCR signals increased fourfold to 10-fold by 240 min after LPS stimulation. These control regions were not enriched when a control GST antibody was used in the ChIP assay (Fig. 3A, bottom). At the Il12b locus, 15 different primer pairs were examined that amplify sequences between -14 kb and +8 kb relative to the transcription start site; the results demonstrate that inducible BRG1 association is concentrated at the enhancer and promoter (Fig. 3B). Interestingly, ChIP signals of similar magnitude were observed when the control regions of early primary response genes were examined, but at these control regions, BRG1 association appeared to be largely LPS independent, with modest induction observed in some experiments (Fig. 3A; see also Fig. 5A,B, below). The constitutive association of BRG1 with the promoters of early primary response genes suggests that SWI/SNF complexes might contribute to the initial establishment of “open” chromatin structures during macrophage development, or to the maintenance of open structures in mature macrophages.
Figure 3.
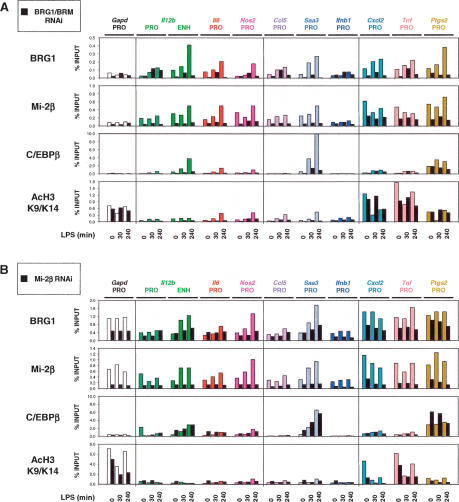
Association of BRG1 and Mi-2β with proinflammatory control regions. (A) A ChIP assay was employed using antibodies directed against BRG1, Mi-2β, C/EBPβ, and GST. Sheared, cross-linked chromatin was prepared from J774 cells treated with LPS for 0, 30, or 240 min. Precipitated DNA was quantified by real-time PCR using primers specific for the indicated control regions. The relative abundance of each control region was plotted relative to input DNA (% INPUT). The data are representative of experiments from three independent chromatin preparations. (B) ChIP analysis at the Il12b locus using chromatin prepared from J774 cells treated with LPS for 0, 30, and 240 min and precipitated with antibodies directed against BRG1, Mi-2β, and C/EBPβ. Precipitated DNA samples were amplified using primer pairs specific to the indicated regions relative to the Il12b transcriptional start site.
Figure 5.
ChIP analyses in BRG1/BRM- and Mi-2β-depleted cells (A) ChIP experiments were performed side-by-side with chromatin prepared from cells infected with the empty RNAi vector (color bars) and from BRG1/BRM-depleted cells (black bars) treated with LPS for 0, 30, and 240 min. Antibodies directed against BRG1, Mi-2β, C/EBPβ, and histone H3 acetylated at Lys 9 and Lys 14 (Ac-H3 K9/K14) were used. Precipitated DNA was quantified and plotted as in Figure 3A. (B) Parallel ChIP experiments with chromatin prepared from cells infected with the empty RNAi vector (color bars) and from Mi-2β-depleted cells (black bars) treated with LPS for 0, 30, and 240 min.
To document the level of reproducibility of ChIP results obtained with antibodies that yield relatively weak signals, it seems most informative to present the results obtained in multiple independent experiments. Independent experiments examining the association of BRG1 with these same control regions are shown in Figure5A and B (below) and Supplementary Figure 3A and B. The only difference in experimental design between these later experiments and the experiment in Figure 3A is that the later experiments made use of cells transduced with the control retrovirus (for comparison to the siRNA-expressing retroviruses examined in those experiments). Although some variability was observed, the strong trend was upheld toward LPS-induced association of BRG1 with the control regions of secondary response and late primary response genes. At the early primary response genes, no inducible association of BRG1 was observed in three of the five experiments shown (Fig. 3A; see also Fig. 5B, below; Supplementary Fig. 3B) and in four other independent experiments (at least one early primary response gene examined in each experiment). Although some induction of BRG1 association was observed in the remaining two experiments (see Fig. 5A, below; Supplementary Fig. 3A), BRG1 association with these genes in the uninduced cells was consistently higher than with the secondary response and late primary response genes, resulting in a relatively modest increase following stimulation.
The transcription factor C/EBPβ is involved in the transcriptional induction of several proinflammatory genes (Poli 1998). By ChIP, C/EBPβ association with the control regions of most secondary response genes and late primary response genes was greatly increased at late time points following LPS stimulation (Fig. 3A, third row). In contrast, C/EBPβ association with the control regions of early response genes was often enhanced in unstimulated cells and only moderately increased following LPS stimulation, similar to the results obtained with BRG1. Although C/EBPβ was previously reported to recruit SWI/SNF complexes to target genes through residues found in only one C/EBPβ isoform (Kowenz-Leutz and Leutz 1999), that isoform is not expressed in murine macrophages (Bradley et al. 2003). Thus, the relevance of C/EBPβ for SWI/SNF recruitment remains unknown.
As an additional control for the above experiments, ChIP assays were performed with antibodies against Mi-2β, a catalytic subunit of the NuRD remodeling complex. Surprisingly, the association of Mi-2β with the proinflammatory loci closely coincided with the association of BRG1 (Fig. 3A, second row; see also Fig. 5A,B, below; Supplementary Fig. 3A,B). That is, Mi-2β appeared to associate with the control regions of secondary response genes and late primary response genes in an LPS-dependent manner but associated constitutively with the promoters of early primary response genes.
Antagonistic functions of SWI/SNF and Mi-2β complexes
To determine the role of the Mi-2β remodeling complexes during LPS-induced gene expression, Mi-2β was depleted in J774 cells using the retroviral RNAi strategy. Five days after retroviral delivery of Mi-2β siRNAs, Mi-2β protein was nearly undetectable in Western blots, in comparison to wild-type cells or cells transduced with retroviruses lacking siRNA sequences (control) or encoding the BRG1/BRM siRNA (Fig. 4A). Surprisingly, an analysis of LPS-induced transcription revealed that the expression of secondary response and late primary response genes was greatly increased in Mi-2β-depleted macrophages relative to cells transduced with the control retrovirus (Fig. 4C; please note that the Y-axis scales differ from those used in earlier figures). In contrast, the expression of early primary response genes was influenced to a lesser extent by Mi-2β depletion. These results demonstrate that Mi-2β complexes temper the induction of secondary response and late primary response genes.
To further examine the differential effects of BRG1/BRM and Mi-2β, the analysis was expanded to include other LPS-induced genes, including genes described previously and genes identified in microarray experiments (V.R. Ramirez-Carrozzi and S.T. Smale, unpubl.). This analysis was restricted to genes whose mRNAs were induced by at least fivefold in LPS-stimulated J774 cells in quantitative RT–PCR experiments (see Supplementary Fig. 2). Figure 4C shows the fold difference in mRNA levels derived from a comparison of control cells to BRG1/BRM-depleted cells or Mi-2β-depleted cells. The data reveal strong reciprocal effects on most of the secondary response and late primary response genes studied, with BRG1/BRM depletion reducing expression and Mi-2β depletion enhancing expression. In contrast, with only a few exceptions, early primary response genes were unaffected by depletion of either BRG1/BRM or Mi-2β.
It is noteworthy that although we have been unable to achieve efficient RNAi-mediated depletion of BRG1/BRM or Mi-2β in primary macrophages, the kinetics of induction and CHX sensitivity of all 27 genes examined were similar in murine bone marrow-derived macrophages in comparison to J774 cells (data not shown). Thus, the roles of remodeling complexes in the J774 line are likely to reflect the requirements in primary macrophages.
Previous studies demonstrated that SWI/SNF complexes can regulate cell size and morphology (Hill et al. 2004; Medjkane et al. 2004). Specifically, cell lines that expressed dominant-negative BRG1 or were deficient in the SNF5/INI1 subunit of the SWI/SNF complex were larger than wild-type cells. To further characterize the antagonism between SWI/SNF and Mi-2β complexes, the size and morphology of BRG1/BRM-depleted and Mi-2β-depleted J774 macrophages was monitored by fluorescence microscopy (Fig. 4D). GFP expression from the integrated retroviruses enabled visualization of the cytoplasm of infected cells and a comparison of cell size. The fluorescent images revealed clear differences, with BRG1/BRM-depleted cells appearing larger and Mi-2β-depleted cells appearing smaller than control cells. DAPI staining revealed changes in nuclear size but not in the organization of pericentromeric heterochromatin. Immunostaining with BRG1 and Mi-2β antibodies confirmed that the targeted proteins were specifically depleted from most cells in a retrovirus-transduced population. The digital fluorescence images collected were used to measure the cell surface area using previously described methods (Hill et al. 2004). Consistent with the previous reports, BRG1/BRM depletion resulted in an overall increase in cell surface area relative to control cells. In contrast, Mi-2β depletion resulted in an overall decrease in surface area. The average surface areas of BRG1/BRM-depleted versus Mi-2β-depleted cells was 228 ± 59 μm2 versus 123 ± 30 μm2, which corresponds to a 2.5-fold difference in cell volume (assuming that cell shape remains unchanged, as documented in the previous studies of Hill et al. 2004). Opposing changes in the average size of cell nuclei were also observed when BRG1/BRM-depleted and Mi-2β-depleted cells were compared with control cells (data not shown). Polynucleated cells were frequently observed with BRG1/BRM-depleted macrophages but not with Mi-2β-depleted macrophages. These results suggest that the antagonistic functions of SWI/SNF and Mi-2β complexes extend beyond the expression of individual genes, to the control of cell size and morphology.
ChIP analyses in BRG1/BRM- and Mi-2β-depleted cells
One central question is whether the expression of early primary response genes is truly independent of BRG1/BRM and Mi-2β, especially since the ChIP data suggest that these factors are constitutively associated with these genes. One possibility is that the preassociated BRG1/BRM and Mi-2β complexes are highly stable and therefore resistant to RNAi-mediated depletion. To examine this possibility, ChIP experiments were performed in depleted cells. Following BRG1/BRM depletion, BRG1 association with all control regions examined was strongly reduced, including the control regions for early primary response genes (Fig. 5A; for a second independent experiment, see Supplementary Fig. 3A). This result supports the hypothesis that BRG1/BRM complexes are truly dispensable for expression of early primary response genes within the time-frame of the analysis (see Discussion). Interestingly, the association of Mi-2β with proinflammatory control regions was also substantially diminished in BRG1/BRM-depleted chromatin, although to a greater extent at secondary and late primary response genes than at early primary response genes (Fig. 5A; Supplementary Fig. 3A). These results suggest that Mi-2β association might require prior remodeling by SWI/SNF complexes or corecruitment with SWI/SNF as part of a larger multiprotein complex.
In the BRG1/BRM-depleted cells, the association of C/EBPβ with secondary response and late primary response genes was also greatly reduced, whereas modest decreases were observed at early primary response genes (Fig. 5A; Supplementary Fig. 3A). Finally, the acetylation of histone H3 on Lys 9 and Lys 14 was examined. In wild-type J774 cells, acetylation increased following LPS stimulation at most of the secondary response and late primary response genes examined, but appeared to be lower than the levels observed at early primary response genes and at the control Gapd gene. The low acetylation levels at the secondary response and late primary response genes were substantially reduced in the BRG1/BRM-depleted cells, whereas the higher levels observed at the early response genes and Gapd gene were unchanged or reduced only slightly (Fig. 5A).
In Mi-2β-depleted cells, Mi-2β association with all genes examined was greatly reduced (Fig. 5B; Supplementary Fig. 3B). In contrast, BRG1 association with the genes in the Mi-2β-depleted cells appeared to be reduced to a lesser extent. Thus, Mi-2β association is strongly dependent on BRG1/BRM (Fig. 5A), whereas BRG1 association appears to exhibit a somewhat weaker dependence on Mi-2β (Fig. 5B). Mi-2β depletion had relatively little effect on C/EBPβ association or on histone acetylation (Fig. 5B). However, small but consistent increases in C/EBPβ binding and histone acetylation were often observed in the unstimulated Mi-2β-depleted cells and at early time-points after LPS stimulation (Fig. 5B; Supplementary Fig. 3B). Although these ChIP signals are very weak and although the magnitudes of the increases are small, the results may be meaningful because they represent the only instances in which ChIP signals increased following depletion of either BRG1/BRM or Mi-2β. These results raise the possibility that the enhanced gene expression observed in Mi-2β-depleted cells might be due to changes in chromatin structure that result in increases in histone acetylation and the binding of C/EBPβ.
LPS-induced nucleosome remodeling at proinflammatory loci
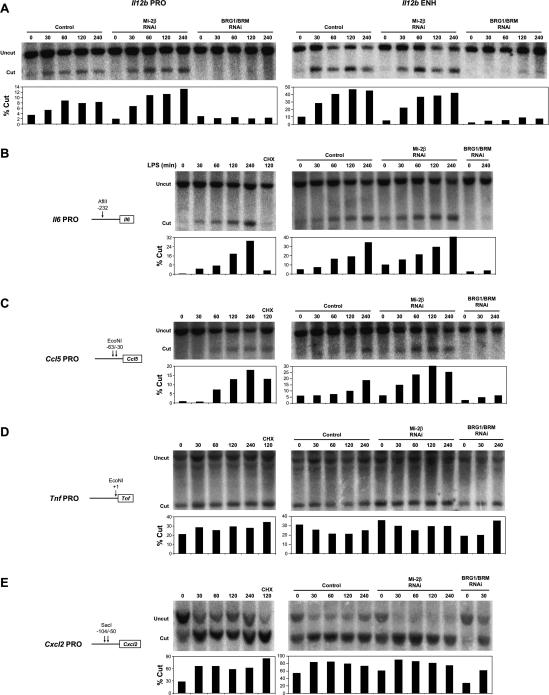
To gain further insight into the relationship between BRG1/BRM, Mi-2β, and LPS-induced transcription, the promoters of the Il6, Ccl5 (RANTES), Tnf, and Cxcl2 (MIP-2) promoters, as well as the Il12b promoter and enhancer, were examined using the restriction enzyme accessibility assay (Fig. 6). The Il6 secondary response promoter, like the Il12b promoter and enhancer, was cleaved at a low efficiency in unstimulated cells, with substantial time-dependent increases in cleavage efficiency following LPS stimulation (Fig. 6A,B). The promoter for the Ccl5 delayed primary response gene also exhibited LPS-induced cleavage (Fig. 6C). However, inducible restriction enzyme cleavage was resistant to CHX at the Ccl5 promoter, but sensitive to CHX at the Il6 promoter and at the Il12b promoter and enhancer (Figs. 1B, 6B,C). At all four of these control regions, restriction enzyme cleavage in nuclei from LPS-induced cells was strongly reduced in BRG1/BRM-depleted cells (Fig. 6A–C). These results are consistent with the hypothesis that BRG1/BRM complexes directly remodel nucleosomes at these control regions following LPS stimulation. Importantly, the results implicate at least two distinct nucleosome remodeling mechanisms at proinflammatory loci in response to LPS; remodeling at secondary response genes involves a mechanism that requires new protein synthesis, whereas remodeling at late primary response genes was independent of new protein synthesis (see Discussion).
Figure 6.
Restriction enzyme accessibility analysis of proinflammatory control regions. The restriction enzyme accessibility assay was used to monitor LPS-induced nucleosome remodeling at the Il12b promoter and enhancer (A), and at the promoter regions of the proinflammatory genes Il6 (B), Ccl5 (C), Tnf (D), and Cxcl2 (E). Uninfected J774 cells, empty vector (control) cells, Mi-2β-depleted cells, and BRG1/BRM-depleted cells were treated with LPS for the indicated times and pretreated with CHX where indicated. Isolated nuclei were digested with the restriction enzymes indicated in the figures, and the cleaved DNAs were analyzed by Southern blot as described in the legend for Figure 1.
In Mi-2β-depleted cells, cleavage efficiencies were generally comparable to control cells, although slight increases were observed at the Il12b, Il6, and Ccl5 promoters. Because the magnitudes of these increases are small, it is difficult to draw conclusions from these results regarding the mechanism by which Mi-2β tempers the induction of these genes. Nevertheless, the results raise the possibility that the enhanced expression in Mi-2β-depleted cells is due to subtle but enhanced remodeling at the control regions of secondary response and delayed primary response genes.
The promoters of the early primary response genes Tnf and Cxcl2 (MIP-2) were cleaved at a much higher efficiency in unstimulated cells (20%–30%) (Fig. 6D,E) than was observed at the secondary response and delayed primary response genes (1%–10%) (Fig. 6A–C). Although the precise cleavage efficiencies at the various primary response and secondary response genes are difficult to compare because different restriction enzymes were generally used, it is noteworthy that the Ccl5 and Tnf promoters were both analyzed in the same DNA preparation after nuclear cleavage with EcoN1. Following LPS stimulation, the cleavage efficiency increased only slightly at the Tnf promoter but to a much greater extent at the Cxcl2 promoter. The significance of these increases at early primary response promoters is not known. However, it is important to note that the increases were unaffected by BRG1/BRM depletion, suggesting that they are not SWI/SNF dependent (Fig. 6D,E). These increases following LPS stimulation could therefore be due to the activity of a different remodeling complex or could reflect an imperfect relationship between restriction enzyme accessibility and the activity of ATP-dependent nucleosome remodeling complexes.
Discussion
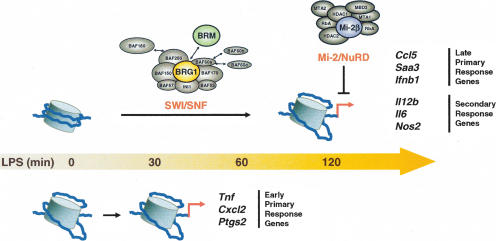
In this study, a retroviral RNAi strategy was used to study the functional roles of SWI/SNF and Mi-2β complexes during gene activation in macrophages in response to LPS stimulation. The results revealed considerable consistency in the SWI/SNF-dependence and SWI/SNF-independence of different classes of inducible genes (Fig. 7). The hypothesis that the SWI/SNF complexes directly regulate the activation of secondary response and late primary response genes was supported by the direct BRG1 association, by the LPS-induced increases in restriction enzyme access, and by the BRG1/BRM dependence of these increases. A second unexpected finding was that SWI/SNF and Mi-2β complexes acted antagonistically in LPS-stimulated macrophages (Fig. 7). Although the mechanism by which Mi-2β complexes limit gene activation remains unknown, three findings suggest that the mechanism may be direct. First, the ChIP results suggest that Mi-2β is recruited to secondary response and late primary response genes in a SWI/SNF-dependent manner. Second, in the functional siRNA experiments, the Mi-2β effect was limited to SWI/SNF-dependent genes. Third, the antagonism between SWI/SNF and Mi-2β was not restricted to LPS-induced genes, but was also apparent in their opposing effects on cell size.
Figure 7.
Differential contributions of nucleosome remodeling complexes during inflammatory gene induction. A summary of the results depicts the selective requirement for SWI/SNF complexes at secondary response and late primary response genes, with Mi-2β negatively influencing these same sets of genes. Early primary response genes do not appear to be regulated by either SWI/SNF or Mi-2β complexes.
The observation that SWI/SNF complexes regulate defined classes of genes following LPS stimulation of macrophages indicates that significant logic underlies the contributions of these complexes to gene regulation. The next challenge will be to examine in greater depth the properties and regulation of each of the three main classes of genes, to determine if additional consistent logic can be uncovered. It will be interesting to determine whether consistent features distinguish genes within one class from those in the other two classes. Genes in all three classes rely on transcription factors in the NF-κB, AP-1, and C/EBP families. However, the reasons the genes fall into different classes with respect to protein synthesis requirement, induction kinetics, and nucleosome remodeling requirements remain largely unknown.
At the early primary response genes, a key question is whether similar mechanisms are used in the various genes within this class to establish the open chromatin structure that appears to exist in unstimulated macrophages. A second question will be to understand the constitutive association of BRG1 with these promoters. Constitutive association of SWI/SNF complexes was also observed with IFN-α-inducible promoters in a recent study, which proposed that the SWI/SNF complexes might be poised to play a critical role in rapid transcriptional induction (Liu et al. 2002; Cui et al. 2004). However, in our study, the early primary response genes were induced normally in the BRG1/BRM-depleted cells, despite greatly reduced levels of BRG1 at the promoters. We propose that, at these genes, SWI/SNF complexes might be involved in the establishment of accessibility during macrophage development. The complexes may also help with the long-term maintenance of accessible chromatin structures in mature macrophages, but might be expendable during the relatively short time-frame of our RNAi experiments.
At secondary response genes, new protein synthesis was found to be required, not only for transcription but also for nucleosome remodeling, as monitored by restriction enzyme accessibility. These results suggest that (1) a primary response gene product may be consistently required for recruiting SWI/SNF complexes to secondary response genes, and (2) the protein synthesis requirement for induction of these genes may largely or solely be due to the requirement for this hypothetical SWI/SNF-recruiting factor. We speculate that one or more primary response gene products may be dedicated to the recruitment of SWI/SNF complexes to secondary response genes. One attractive candidate is IκBζ, which is essential for the induction of at least two secondary response genes, Il12b and Il6, but not for induction of one early primary response gene, Tnf (Yamamoto et al. 2004).
The SWI/SNF dependence of late primary response genes suggests that a mechanism must also exist for recruiting SWI/SNF complexes in the absence of new protein synthesis. The IFN-β enhanceosome may provide an example of this class of genes (Agalioti et al. 2000). In this case, cooperative binding of multiple factors to the enhanceosome appears to present a surface that promotes the subsequent recruitment of histone modifying activities and the SWI/SNF complex. Interestingly, several of the late primary response genes studied here are among a subset of LPS-induced genes that require the IRF3 signaling pathway for activation, in addition to the NF-κB and JNK pathways involved in the activation of early primary response genes (Doyle et al. 2002). Thus, IRF3 may contribute to the recruitment of nucleosome remodeling complexes at these genes, although it may consistently function in the form of an enhanceosome, as observed at the IFN-β promoter (Agalioti et al. 2000). One additional unanswered question is whether the delayed activation of genes within this class is primarily due to the requirement for nucleosome remodeling, which may delay the initiation of transcription.
The SWI/SNF requirement for the activation of secondary response and late primary response genes was demonstrated by the simultaneous depletion of BRG1 and BRM by RNAi. Similar results were obtained by expression of dominant-negative BRG1, which may inhibit the functions of both BRG1 and BRM complexes. We therefore do not know whether BRG1 and BRM complexes act redundantly during induction of these genes, or whether only BRG1 complexes are relevant. Unfortunately, attempts to independently and efficiently deplete either BRG1 or BRM using the retroviral RNAi strategy have been unsuccessful.
Previous work demonstrated that the p65 subunit of NF-κB is recruited to proinflammatory loci in two different waves after LPS treatment of RAW264.7 macrophages (Saccani et al. 2001). The Cxcl2 promoter was immediately accessible to p65, whereas the Il6, Ccl5, and Ccl2 (MCP-1) promoters displayed delayed association. The results described here suggest that differences in SWI/SNF-mediated remodeling are at least partially responsible for these differences in the time course of NF-κB binding.
It is not clear how Mi-2β inhibits the induction of secondary response and late primary response genes. The mechanism by which Mi-2β is recruited to the proinflammatory loci also remains unknown. The SWI/SNF dependence of recruitment suggests that SWI/SNF and Mi-2β complexes may be associated with each other in vivo, as has been suggested in previous biochemical studies (Nakamura et al. 2002; Shimono et al. 2003). Alternatively, Mi-2β may be recruited by a specific DNA-binding protein, but only after nucleosome remodeling by SWI/SNF complexes.
Finally, these results suggest that ATP-dependent nucleosome remodeling complexes may be attractive targets for modulating the strength of an inflammatory response or other inducible responses. It is noteworthy that preliminary microarray experiments with J774 cells depleted for BRG1/BRM or Mi-2β revealed that a surprisingly small number of constitutively expressed genes were misregulated in the absence of these complexes in the experimental time-frame used (5 d after retroviral transduction and 0–4 h after LPS stimulation). These results are consistent with studies performed in SW13 cells and cells expressing dominant-negative forms of BRG1 (Liu et al. 2001; de la Serna et al. 2005). Thus, the open chromatin structures that are likely to be required for the expression of housekeeping genes and uninduced macrophage-specific genes appear to be relatively stable, suggesting that antagonists of BRG1/BRM or Mi-2β could influence the expression of specific subsets of inducible genes without influencing constitutively expressed genes.
Materials and methods
Cell culture and reagents
The J774 murine macrophage cell line was maintained in DMEM medium containing 10% fetal bovine serum (FBS) (Omega Scientific) and penicillin/streptomycin. J774 cells were activated with LPS (10 μg/mL) and, where indicated, were pretreated with CHX (10 μg/mL) for 15 min. Sandwich ELISA kits were used to measure the production of IL-12 p40/p70 (PharMingen) and MIP-2 (R&D Systems). Antibodies against BRG1 (Santa Cruz, sc-10768), BRM (BD Pharmingen, 610390), and HMG1 (BD Pharmingen, 556528) and polyclonal antisera against GST-Mi-2β (residues 22–155) were used for Western blot analysis.
RT–PCR and real-time quantitative PCR
RNA was extracted using TRI-reagent, treated with RNase-free DNaseI, and purified using an RNeasy kit (Qiagen). Quantified RNA (2 μg) was reverse-transcribed using Omniscript RT Kit (Qiagen) and random hexamer primers. cDNA fragments were analyzed by real-time PCR using iQ SYBR Green Supermix (Bio-Rad) and the iCycler System (Bio-Rad). The PCR amplification conditions were 95°C (3 min) and 45 cycles of 95°C (15 sec), 60°C (30 sec), and 72°C (30 sec). Primer pairs were designed to amplify 80–120 bp mRNA-specific fragments, and unique products were tested by melt-curve analysis. Primer sequences are available upon request.
Retroviral RNAi
The retroviral RNAi vectors were based on the pQCXIP vector (Clontech). The destabilized GFP gene was first cloned into the multiple cloning site of pQCXIP. Mouse U6 or human H1 promoters were cloned into the NheI site within the 3′LTR (Barton and Medzhitov 2002). Oligonucleotides encoding siRNA hairpins against BRG1/BRM and Mi-2β were annealed and cloned after the U6 and H1 promoters, respectively. Hairpin sequences are available upon request. For retrovirus generation, 293T cells were grown in DMEM with 10% FBS and penicillin/streptomycin. Cells were grown to 90% confluency in 10-cm dishes and transfected with the retroviral vectors and the 10A1 packaging vector using lipofectamine 2000 (Invitrogen). The medium was replaced 24 h post-transfection. The virus supernatants were collected 36 and 48 h after transfection, filtered through a 0.22-μm syringe filter, and stored at 4°C. For retroviral infection, J774 cells (7.5 × 105/well) were seeded in six-well plates, and 2 mL of virus supernatant were supplemented with 8 μL 2 M HEPES (pH 7.5) and 8 μL polybrene (20 μg/mL final concentration) was added to the cells. Spin infections were performed at 2500 rpm for 1.5 h at 30°C and were repeated once the following day. One day after the second infection, puromycin (3 μg/mL) selection was started and GFP expression was monitored by flow cytometry. RNAi depletion was monitored by Western blotting.
Restriction enzyme accessibility
Experiments were performed as described previously (Weinmann et al. 1999; Zhou et al. 2004). Isolated cell nuclei and limiting amounts of restriction enzyme (100 U) were incubated for 15 min at 37°C, followed by genomic DNA isolation. Purified DNA (10–15 μg) was digested to completion to generate reference cleavage products using the following restriction enzymes: KpnI and SphI for the Il12b promoter and enhancer, XbaI and SpeI for Il6, EcoRI and HindIII for Ccl5, HindIII and XbaI for Tnf, and EcoRI and DraI for Cxcl2. Samples were analyzed by Southern blotting with32P-labeled gene-specific probes designed at the following regions: Il12b promoter (+64 to +437), Il12b enhancer (-8711 to -9113), Il6 promoter (-544 to -1043), Ccl5 promoter (-297 to -667), Tnf promoter (+14 to +504), and Cxcl2 promoter (+922 to +1409).
ChIP
J774 cells (8 × 107) were untreated or stimulated with LPS as required. Chromatin was cross-linked using 1% formaldehyde for 10 min at room temperature. The cells were collected after two PBS washes in cell lysis buffer (5 mM PIPES at pH 8.0, 85 mM KCl, 0.5% NP-40) and lysed for 10 min on ice. Nuclei were resuspended in nuclei lysis buffer (50 mM Tris-HCl at pH 8.1, 10 mM EDTA, 1% SDS) and sonicated six times with 15-sec pulses followed by 45-sec recovery periods at output 6.0 (Sonicator 3000, Misonix). The purified chromatin (100 μg) was diluted 2.5-fold with ChIP dilution buffer (16.7 mM Tris-HCl at pH 8.1, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100) supplemented with protease inhibitors. Diluted chromatin was immunoprecipitated overnight at 4°C using antibodies (2–10 μg) against BRG1 (Upstate, 07-478) and H3 K9/K14 (Upstate, 07-352, 07-353). Polyclonal antisera against GST-Mi-2β (residues 22–155), GST-C/EBPβ (residues 22–195), and GST were prepared by our laboratory. Immune complexes were recovered by a 1-h incubation at 4°C with 50 μL of Protein A agarose/salmon sperm DNA beads (Upstate, 16-157) and were washed four times with high-salt wash buffer (50 mM HEPES at pH 7.9, 0.5 M NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 0.1% deoxycholate) and two times with TE buffer. Each wash step was for 10 min at room temperature. The DNA was extracted in 300 μL of elution buffer (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, 1% SDS) supplemented with proteinase K (20 μg) for 2 h at 55°C, followed by reversal of DNA cross-links overnight at 65°C. The DNA was purified with QIAquick columns (Qiagen) and was detected by real-time quantitative PCR using 5% of each input as standards. The following regions (relative to the start of transcription) were amplified using specific primer pairs: Gapd PRO (-76 to -187), Il12b PRO (-46 to -150), Il12b ENH (-9834 to -9934), Il6 PRO (-4 to -96), Nos2 PRO (+5 to -111), Ccl5 PRO (+8 to -78), Saa3 PRO (-68 to -171), Ifnb1 PRO (-85 to -201) Cxcl2 PRO (-38 to -134), Tnf PRO (+30 to -89), and Ptgs2 PRO (-69 to -158).
Fluorescence microscopy
J774 cells (2 × 105) were attached to coverslips, washed once in PBS, fixed for 1 min in methanol on dry ice, and permeabilized for 10 min in blocking buffer (PBS with 0.1% saponin and 10% FBS). The cells were then incubated for 1 h with rabbit anti-BRG1 antibody (Upstate) or anti-Mi-2β antibody diluted 1:1000 in blocking buffer. The cells were washed three times (3 min each) in PBS 0.1% saponin, then incubated for 30 min with Texas Red-labeled goat anti-rabbit antibody (Jackson Immunoresearch) diluted 1:500 in blocking buffer. The cells were washed as above and mounted in Vectashield (Vector Labs) containing 0.1 μg/mL DAPI. Slides were analyzed on a Leica Inverted Confocal Microscope. Cell size measurements were performed using the Adobe Photoshop program, as described by Hill et al. (2004).
Acknowledgments
We thank Arnold Berk, Craig Peterson, and Gang Wang for helpful advice and discussions, and David Turner for providing a mouse U6 promoter plasmid. This work was supported by P.H.S. Training Grant CA009120 and a Giannini Family Fellowship (to V.R.C.). S.T.S. in an Investigator of the Howard Hughes Medical Institute.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1383206
References
- Agalioti T., Lomvardas, S., Parekh, B., Yie, J., Maniatis, T., and Thanos, D. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103: 667-678. [DOI] [PubMed] [Google Scholar]
- Akira S., Takeda, K., and Kaisho, T. 2001. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2: 675-680. [DOI] [PubMed] [Google Scholar]
- Armstrong J.A., Papoulas, O., Daubresse, G., Sperling, A.S., Lis, J.T., Scott, M.P., and Tamkum, J.W. 2002. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 21: 5245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton G.M. and Medzhitov, R. 2002. Retroviral delivery of small interfering RNA into primary cells. Proc. Natl. Acad. Sci. 99: 14943-14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P.B. and Horz, W. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71: 247-273. [DOI] [PubMed] [Google Scholar]
- Bradley M.N., Zhou, L., and Smale, S.T. 2003. C/EBPβ regulation in lipopolysaccharide-stimulated macrophages. Mol. Cell. Biol. 23: 4841-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S., Gebuhr, T., Yee, D., La Mantia, C., Nicholson, J., Gilliam, A., Randazzo, F., Metzger, D., Chambon, P., Crabtree, G., et al. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6: 1287-1295. [DOI] [PubMed] [Google Scholar]
- Cairns B.R. 2005. Chromatin remodeling complexes: Strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15: 185-190. [DOI] [PubMed] [Google Scholar]
- Caivano M., Gorgoni, B., Cohen, P., and Poli, V. 2001. The induction of cyclooxygenase-2 mRNA in macrophages is biphasic and requires both CCAAT enhancer-binding protein β (C/EBP β) and C/EBP δ transcription factors. J. Biol. Chem. 276: 48693-48701. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Utley, R.T., Workman, J.L., and Cote, J. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19: 321-329. [DOI] [PubMed] [Google Scholar]
- Chi T. 2004. A BAF-centred view of the immune system. Nat. Rev. Immunol. 4: 965-977. [DOI] [PubMed] [Google Scholar]
- Chi T.H., Wan, M., Lee, P.P., Akashi, K., Metzger, D., Chambon, P., Wilson, C.B., and Crabtree, G.R. 2003. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 19: 169-182. [DOI] [PubMed] [Google Scholar]
- Corey L.L., Weirich, C.S., Benjamin, I.J., and Kingston, R.E. 2003. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes & Dev. 17: 1392-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K., Tailor, P., Liu, H., Chen, X., Ozato, K., and Zhao, K. 2004. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol. Cell Biol. 24: 4476-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Serna I.L., Carlson, K.A., Hill, D.A., Guidi, C.J., Stephenson, R.O., Sif, S., Kingston, R.E., and Imbalzano, A.N. 2000. Mammalian SWI–SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 20: 2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna I.L., Ohkawa, Y., Berkes, C.A., Bergstrom, D.A., Dacwag, C.S., Tapscott, S.J., and Imbalzano, A.N. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25: 3997-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S., Vaidya, S., O'Connell, R., Dadgostar, H., Dempsey, P., Wu, T., Rao, G., Sun, R., Haberland, M., Modlin, R., et al. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17: 251-263. [DOI] [PubMed] [Google Scholar]
- Feng Q. and Zhang, Y. 2003. The NuRD complex: Linking histone modification to nucleosome remodeling. Curr. Top. Microbiol. Immunol. 274: 269-290. [DOI] [PubMed] [Google Scholar]
- Fujita N., Jaye, D.L., Geigerman, C., Akyildiz, A., Mooney, M.R., Boss, J.M, and Wade, P.A. 2004. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 119: 75-86. [DOI] [PubMed] [Google Scholar]
- Hebbar P.B. and Archer, T.K. 2003. Chromatin remodeling by nuclear receptors. Chromosoma 111: 495-504. [DOI] [PubMed] [Google Scholar]
- Herrera R.E., Nordheim, A., and Stewart, A.F. 1997. Chromatin structure analysis of the human c-fos promoter reveals a centrally positioned nucleosome. Chromosoma 106: 284-292. [DOI] [PubMed] [Google Scholar]
- Hill D.A., Chiosea, S., Jamaluddin, S., Roy, K., Fischer, A.H., Boyd, D.D., Nickerson, J.A., and Imbalzano, A.N. 2004. Inducible changes in cell size and attachment area due to expression of a mutant SWI/SNF chromatin remodeling enzyme. J. Cell Sci. 117: 5847-5854. [DOI] [PubMed] [Google Scholar]
- Iwasaki A. and Medzhitov, R. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5: 987-995. [DOI] [PubMed] [Google Scholar]
- Janeway C.A. and Medzhitov, R. 2002. Innate immune recognition. Annu. Rev. Immunol. 20: 197-216. [DOI] [PubMed] [Google Scholar]
- Jenner R.G. and Young, R.A. 2005. Insights into host responses against pathogens from transcriptional profiling. Nat. Rev. Microbiol. 3: 281-294. [DOI] [PubMed] [Google Scholar]
- Kadam S. and Emerson, B.M. 2002. Mechanisms of chromatin assembly and transcription. Curr. Opin. Cell Biol. 14: 262-268. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11: 377-389. [DOI] [PubMed] [Google Scholar]
- Kehle J., Beuchle, D., Treuheit, S., Christen, B., Kennison, J.A., Bienz, M., and Muller, J. 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282: 1897-1900. [DOI] [PubMed] [Google Scholar]
- Kim J., Sif, S., Jones, B., Jackson, A., Koipally, J., Heller, E., Winandy, S., Viel, A., Sawyer, A., Ikeda, T., et al. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10: 345-355. [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E. and Leutz, A. 1999. A C/EBP β isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4: 735-743. [DOI] [PubMed] [Google Scholar]
- Krebs J.E., Fry, C.J., Samuels, M.L., and Peterson, C.L. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102: 587-598. [DOI] [PubMed] [Google Scholar]
- Liu X.F. and Bagchi, M.K. 2004. Recruitment of distinct chromatin-modifying complexes by tamoxifen-complexed estrogen receptor at natural target gene promoters in vivo. J. Biol. Chem. 279: 15050-15058. [DOI] [PubMed] [Google Scholar]
- Liu R., Liu, H., Chen, X., Kirby, M., Brown, P.O., and Zhao, K. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106: 309-318. [DOI] [PubMed] [Google Scholar]
- Liu H., Kang, H., Liu, R., Chen, X., and Zhao, K. 2002. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol. Cell. Biol. 22: 6471-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A. and Kadonaga, J.T. 2003. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 25: 1192-1200. [DOI] [PubMed] [Google Scholar]
- Martens J.A. and Winston, F. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13: 136-142. [DOI] [PubMed] [Google Scholar]
- Medjkane S., Novikov, E., Versteege, I., and Delattre, O. 2004. The tumor suppressor hSNF5/INI1 modulates cell growth and actin cytoskeleton organization. Cancer Res. 64: 3406-3413. [DOI] [PubMed] [Google Scholar]
- Nagaich A.K., Walker, D.A., Wolford, R., and Hager, G.L. 2004. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell 14: 163-174. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Mori, T., Tada, S., Krajewski, W., Rozovskaia, T., Wassell, R., Dubois, G., Mazo, A., Croce, C.M., and Canaani, E. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10: 1119-1128. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Robert, F., Young, R.A., and Struhl, K. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes & Dev. 16: 806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L. 2002. Chromatin remodeling: Nucleosomes bulging at the seams. Curr. Biol. 12: R245-R247. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. and Workman, J.L. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10: 187-192. [DOI] [PubMed] [Google Scholar]
- Poli V. 1998. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 273: 29279-29282. [DOI] [PubMed] [Google Scholar]
- Reyes J.C., Barra, J., Muchardt, C., Camus, A., Babinet, C., and Yaniv, M. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 17: 6979-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.Y., Denu, J.M., and Allis, C.D. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70: 81-120. [DOI] [PubMed] [Google Scholar]
- Saccani S., Pantano, S., and Natoli, G. 2001. Two waves of nuclear factor κB recruitment to target promoters. J. Exp. Med. 193: 1351-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y., Murakami, H., Kawai, K., Wade, P.A., Shimokata, K., and Takahashi, M. 2003. Mi-2 β associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J. Biol. Chem. 278: 51638-51645. [DOI] [PubMed] [Google Scholar]
- Shopland L.S., Hirayoshi, K., Fernandes, M., and Lis, J.T. 1995. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes & Dev. 9: 2756-2769. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P., Iyer, V.R., Brown, P.O., and Winston, F. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 97: 3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi-Ichinose C., Ichinose, H., Metzger, D., and Chambon, P. 1997. SNF2β-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol. Cell. Biol. 17: 5976-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T., Becker, P.B., and Wu, C. 1994. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367: 525-532. [DOI] [PubMed] [Google Scholar]
- Vaidya S.A. and Cheng, G. 2003. Toll-like receptors and innate antiviral responses. Curr. Opin. Immunol. 15: 402-407. [DOI] [PubMed] [Google Scholar]
- Weinmann A.S., Plevy, S.E., and Smale, S.T. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11: 665-675. [DOI] [PubMed] [Google Scholar]
- Weinmann A.S., Mitchell, D.M., Sanjabi, S., Bradley, M.N., Hoffmann, A., Liou, H.C., and Smale, S.T. 2001. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat. Immunol. 2: 51-57. [DOI] [PubMed] [Google Scholar]
- Williams C.J., Naito, T., Arco, P.G., Seavitt, J.R., Cashman, S.M., De Souza, B., Qi, X., Keables, P., Von Andrian, U.H., and Georgopoulos, K. 2004. The chromatin remodeler Mi-2β is required for CD4 expression and T cell development. Immunity 20: 719-733. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Yamazaki, S., Uematsu, S., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Kuwata, H., Takeuchi, O., Takeshige, K., et al. 2004. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 430: 218-222. [DOI] [PubMed] [Google Scholar]
- Zhou L., Nazarian, A.A., and Smale, S.T. 2004. Interleukin-10 inhibits interleukin-12 p40 gene transcription by targeting a late event in the activation pathway. Mol. Cell. Biol. 24: 2385-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]