Abstract
Frequency (FRQ) is a central component of interconnected negative and positive limbs of feedback loops of the circadian clock of Neurospora. In the negative limb, FRQ inhibits its transcriptional activator White Collar Complex (WCC) and in the positive limb, FRQ supports accumulation of WCC. We show that these conflicting functions are confined to distinct subcellular compartments and coordinated in temporal fashion. Inactivation of the transcriptional activator WCC requires nuclear FRQ and occurs early after the onset of FRQ expression. Support of WCC accumulation requires cytosolic FRQ and occurs on a post-translational level, when high amounts of FRQ have accumulated. The transcriptional function of FRQ in the negative loop and its post-translational function in the positive loop are independent and associated with distinct regions of FRQ. Phosphorylation of FRQ at the PEST-2 region triggers its maturation from a nuclear repressor toward a cytoplasmic activator.
Keywords: Circadian, Neurospora, frq, PEST, post-translational
Circadian rhythms are evolutionary widespread mechanisms that provide the means for temporal organization of physiology and behavior in many organisms. They are driven by molecular clocks, which oscillate in self-sustained fashion with an endogenous (free-running) period of ∼1 d. The free-running oscillation is synchronized to the 24-h geophysical rhythm of earth rotation via perception and integration of environmental stimuli (socalled Zeitgebers) such as light and ambient temperature. Interconnected negative and positive transcriptional/translational feedback loops are critical for robust circadian rhythmicity in many systems (Roenneberg and Merrow 2003; Dunlap and Loros 2004; Gachon et al. 2004; Hardin 2004).
In mice, Period1/2 and Chryptochrome1/2 genes (mPer1/2 and Cry1/2) are rhythmically expressed under control of their transcriptional activator CLOCK/BMAL1. mPER/CRY heterodimers inhibit CLOCK/BMAL1 and thereby feedback-regulate transcription of their own genes. CLOCK/BMAL1 is also an activator of Rev-erbα. REV-ERBα is a transcriptional repressor of Bmal1 (Preitner et al. 2002), which competes with the transcriptional activator RORα (Sato et al. 2004). Hence, Bmal1 transcription is indirectly activated by mPER/CRY, leading to rhythmic expression of Bmal1 (Reppert and Weaver 2002; Gachon et al. 2004; Akashi and Takumi 2005).
In the Drosophila system, dPer and Timeless (Tim) genes are rhythmically transcribed. dPER/TIM inhibits its activator dCLOCK/CYCLE, which is also a transcriptional activator of Vrille and Pdp1. Since VRILLE is a repressor and PDP1 is an activator of dClock (Cyran et al. 2003), dPER/TIM indirectly drives rhythmic transcription of dClock (Stanewsky 2003; Hardin 2004).
Thus, mPER/CRY and dPER/TIM both act on a transcriptional level in the nucleus. Furthermore, they act by the same molecular mechanism in the positive and in the negative limb of the circadian feedback loops; they inhibit their transcriptional activator. This leads to repression of transcription of genes in the negative limb and to indirect activation of transcription of genes in the positive limb via repression of a repressor gene (Gachon et al. 2004; Hardin 2004).
Clock components are regulated by phosphorylation and dephosphorylation (Lee et al. 1998, 2001; Kim et al. 2002; Nawathean and Rosbash 2004). These post-translational mechanisms are critical for circadian rhythmicity in flies and mammals (Reppert and Weaver 2002; Stanewsky 2003; Gachon et al. 2004; Hardin 2004). Recent findings suggest that in cyanobacteria, even the core of the circadian pacemaker is based on autoregulatory rhythmic phosphorylation of the circadian clock protein KaiC rather than on a transcriptional/translational feedback loop (Tomita et al. 2005). KaiC, when incubated in vitro with KaiB and C, undergoes robust cycles of autophosphorylation and dephosphorylation (Nakajima et al. 2005).
In Neurospora, the frequency gene (frq) is transcribed in rhythmic fashion and FRQ protein acts in a temperature-compensated manner in the negative and positive feedback loop of the circadian clock (Roenneberg and Merrow 2003; Dunlap and Loros 2004; Diernfellner et al. 2005). In the negative limb, FRQ inhibits its transcriptional activator White Collar Complex (WCC) by facilitating its phosphorylation (Schafmeier et al. 2005), and thus, FRQ represses its own gene on the level of transcription (Aronson et al. 1994a; Froehlich et al. 2003). In the course of a circadian period, FRQ is progressively phosphorylated by several kinases, in particular CK-1a, a homolog of mammalian CKIε/δ, CKII and CAMK-1 (Görl et al. 2001; Yang et al. 2001, 2002, 2003). The protein phosphatases PP1 and PP2A dephosphorylate FRQ and antagonize the effects of the kinases (Yang et al. 2004). The PEST-1 sequence in the central portion of FRQ and nearby sites are targets for phosphorylation (Liu et al. 2000; Görl et al. 2001; Yang et al. 2002, 2003, 2004). Hyperphosphorylated FRQ is recognized by FWD-1, an F-box protein of an SCF-type ubiquitin ligase (He et al. 2003). Phosphorylation, ubiquitination, and subsequent degradation of FRQ are major determinants of circadian period length.
The negative feedback loop is connected with a positive loop. In this loop, FRQ facilitates accumulation of high levels of WCC, which is essential to generate high-amplitude self-sustained circadian rhythmicity (Lee et al. 2000; Cheng et al. 2001). FRQ affects wc-2 transcription and to some extent also trenscription of wc-1, but neither gene is transcribed in rhythmic fashion (Lee et al. 2000; Cheng et al. 2001, 2003; Dragovic et al. 2002). FRQ controls expression of WC-1 and probably also WC-2 on a post-transcriptional level (Cheng et al. 2001). WC-2 protein levels are constant throughout a circadian period, whereas WC-1 abundance oscillates with a low amplitude ∼6 h delayed compared with the FRQ abundance rhythm (Lee et al. 2000; Cheng et al. 2001).
FRQ contains a functionally essential nuclear localization sequence, NLS, (Luo et al. 1998). Yet, the majority of FRQ is located in the cytosol (Cheng et al. 2005; Schafmeier et al. 2005), suggesting a regulation of its subcellular localization.
We show here that FRQ, in contrast to mPER/CRY and dPER/TIM, acts in distinct subcellular compartments and via distinct mechanisms in the negative and positive limb. Thereby, apparently contradicting functions of FRQ are separated in terms of space and time. Negative feedback is carried out by nuclear FRQ early after FRQ expression, whereas support of WC-1 expression, which is limiting for WCC formation, requires progressive accumulation of FRQ in the cytosol. Distinct populations of FRQ, distinguished by phosphorylation status, mediate nuclear and cytosolic functions. In particular, phosphorylation of serine residues within the PEST-2 region of FRQ appears to be crucial for the ability of FRQ to support WC-1 expression, but has no affect on the negative feedback capacity of FRQ. We suggest that progressive phosphorylation triggers maturation of FRQ from a nuclear repressor toward a cytoplasmic activator.
Results
Temporal coordination of FRQ functions
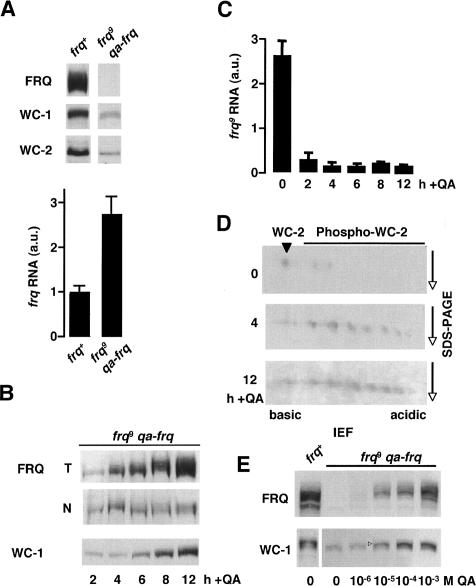
FRQ inhibits synthesis of its own RNA by promoting phosphorylation-dependent inactivation of its transcriptional activator WCC (Schafmeier et al. 2005) and it supports WC-1 and WC-2 expression (Lee et al. 2000; Cheng et al. 2001). To analyze the kinetics of these apparently contradicting FRQ functions, we expressed in a frq-deficient strain (frq9) a functional copy of the frq ORF under control of the quinic acid (QA) inducible qa-2 promoter. The frq9 allele encodes, due to a frame-shift mutation, a C-terminally truncated protein that is defective in negative feedback (Aronson et al. 1994b; Merrow et al. 1997) and in its ability to support WC-1 and WC-2 accumulation (Cheng et al. 2001; Schafmeier et al. 2005). In the absence of QA in the growth medium the frq9;qa-frq strain expressed, like the parental frq9 strain, reduced levels of WC-1 and WC-2, but elevated levels of frq9 RNA (Fig. 1A). Functional FRQ was then induced with QA and the kinetics of accumulation of FRQ in total cell extracts and in nuclei was analyzed over a time course of 12 h (Fig. 1B). Low levels of newly synthesized, hypophosphorylated FRQ were detected in total cell extracts and in nuclei 2 h after addition of QA. Levels and phosphorylation status of total FRQ increased and reached levels similar to those in frq+ after 8–12 h of induction (Fig. 1B; Supplementary Fig. S1). In contrast, nuclear FRQ reached a plateau value already after 4 h, suggesting that its accumulation in the nucleus is limited. Analysis of WC-1 revealed that the newly induced FRQ was inefficient in supporting accumulation of WC-1. WC-1 levels increased significantly 8 h after FRQ induction and reached frq+ levels after 12 h, confirming that FRQ supports expression of WC-1 with a substantial time lag (Lee et al. 2000). The data suggest that high levels of FRQ and/or hyperphosphorylated FRQ are required to support accumulation of WC-1.
Figure 1.
Contradicting FRQ functions are coordinated in temporal fashion. (A) Expression levels of clock proteins and frq mRNA in wild-type (frq+) and frq9 qa-frq. Total protein extracts and RNA were prepared and analyzed by Western blotting (top) and quantitative RT-PCR (bottom), respectively. Blots were decorated with the indicated antibodies. (B) Kinetics of accumulation of FRQ and WC-1. frq9;qa-frq was induced with 10-2 M QA and samples were harvested at the indicated time points after QA addition. Expression of FRQ in total protein extracts (T) and nuclei (N) and of WC-1 (accumulates in nuclei only) was analyzed by Western blotting. Two-hundred micrograms of protein were loaded per lane. (C) Kinetics of repression of the in frq9 locus in frq9;qa-frq cells after induction of FRQ expression. RNA prepared at the indicated time points was analyzed by RT-PCR with primers and probes specific to the 5′-UTR of frq9, which is absent in qa-frq. (D) WC-2 is hyperphosphorylated 4 h after qa-frq induction. Samples from B were subjected to two-dimensional gel electrophoresis—isoelectric focusing (IEF) and subsequent SDS-PAGE (Schafmeier et al. 2005)—and immunobloting with αWC-2 antiserum. (E) frq9;qa-frq cultures were grown in the presence of the indicated concentrations of QA to induce different steady-state levels of FRQ. Samples were analyzed by Western blotting with antibodies against FRQ and WC-1. The open arrowhead indicates hyperphosphorylated WC-1. (A,C) Values are mean ± SD.
frq9 RNA was then measured by quantitative real-time PCR (RT-PCR) using a probe specific to the 5′-UTR of the frq9 allele, which is absent in the chimaeric qa-frq gene. frq9 RNA was almost fully repressed 2 h after induction of qa-frq and reached its minimum after 4 h (Fig. 1C), supporting that the kinetics of negative feedback is fast (Merrow et al. 1997). Two-dimensional gel analysis revealed that WC-2 was hyperphosphorylated 4 h after qa-frq induction (Fig. 1D), indicating that the WCC was inactivated by FRQ-dependent phosphorylation (Schafmeier et al. 2005). This demonstrates that low amounts of hypophosphorylated FRQ were efficient in negative feedback and rapid repression of the frq9 allele. Furthermore, frq9 RNA levels remained low throughout the time course despite accumulation of substantial amounts of WC-1. This demonstrates that FRQ promotes expression of transcriptionally inactive WCC. Interestingly, the kinetics of WCC phosphorylation and negative feedback on frq9 RNA correlated well with the kinetics of FRQ accumulation in the nucleus, while the kinetics of WC-1 accumulation correlated with accumulation of total FRQ levels, which reflect primarily accumulation of FRQ in the cytosol (Schafmeier et al. 2005).
To discriminate whether high levels of FRQ are required to support WC-1 accumulation or whether low levels of newly synthesized FRQ would also directly or indirectly support WC-1 expression with a time lag, we induced different FRQ levels with increasing QA concentrations and analyzed steady-state levels of WC-1. As shown in Figure 1E, accumulation of high steady-state levels of WC-1 required high levels of FRQ.
Together, the data suggest that negative feedback on one hand, and support of WC-1 expression on the other hand, display distinct requirements for FRQ abundance and phosphorylation. Low levels of hypophosphorylated FRQ are immediately active in negative feedback, while support of WC-1 expression occurs with a time lag, when high levels and hyperphosphorylated FRQ have accumulated. This allows temporal coordination of apparently contradicting FRQ functions.
Spatial coordination of FRQ functions
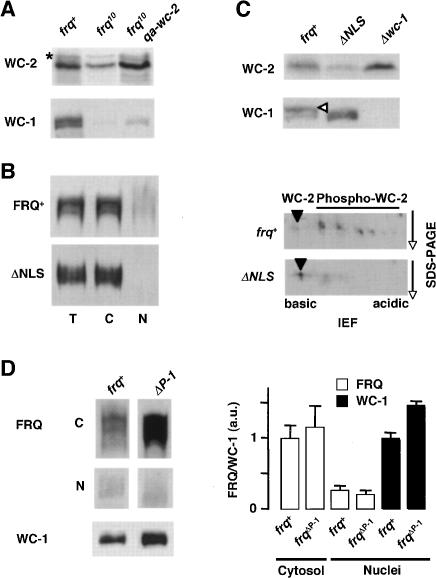
FRQ supports wc-2, and to some extent, also wc-1 transcription and accumulation of WC-1 on a post-transcriptional level (Lee et al. 2000; Cheng et al. 2001; Dragovic et al. 2002; Káldi et al. 2005). Since WC-1 is not stable in the absence of WC-2 (Cheng et al. 2001), the observed post-transcriptional support of WC-1 accumulation could be an indirect effect of FRQ supporting WC-2 expression on the transcriptional level. Thus, in frq-deficient strains the concentration of free WC-2 could become rate limiting for the kinetics of WCC formation. This would lead to low WC-1 levels, since unassembled WC-1 is efficiently degraded. To test this hypothesis, we expressed in frq10 (frq deletion) an inducible qa-wc-2 gene. When transcription of qa-wc-2 was induced, WC-2 accumulated to wild-type levels (Fig. 2A). Yet, levels of WC-1 increased only marginally, demonstrating that WC-2 concentration is not rate limiting for WCC formation in frq10.
Figure 2.
Support of WC-1 expression is a cytosolic function of FRQ. (A) Overexpression of WC-2 does not significantly elevate WC-1 levels in frq10. An inducible extra-copy of wc-2 (qa-wc-2) was expressed in frq10 background. Indicated strains were grown in LL, and total extracts were analyzed by Western blotting with antibodies against WC-2 and WC-1. (⋆) Unspecific band. (B) Subcellular distribution of FRQ and FRQΔNLS. (T) Total extract, (C) cytosol, (N) nuclei. Two-hundred micrograms of protein were loaded per lane. (C) Expression of WCC in frqΔNLS. (Top) Total cell extracts were subjected to SDS-PAGE and immunodecoration. (Open arrowhead) Hyperphosphorylated WC-1. (Bottom) Extracts were analyzed by two-dimensional gel electrophoresis and immunoblotting with αWC-2. (Arrowhead) Unphosphorylated WC-2. (D) FRQ and WC-1 accumulation in a PEST-1 deletion strain. (Left) frq+ and frqΔP-1 were grown in LL. Samples were harvested and fractionated into cytosol (C) and nuclei (N), and analyzed by Western blotting. (Right) Quantification of FRQ and WC-1 from Western blots. Mean ± SD (n = 3).
We then asked whether the post-transcriptional support of WC-1 accumulation is associated with a function of FRQ in the cytosol. FRQ contains a nuclear localization signal, which is essential for its function (Luo et al. 1998). Yet, only a small fraction of FRQ is localized in the nucleus and the majority of the protein accumulates in the cytosol (Cheng et al. 2005; Schafmeier et al. 2005). We used the strain frqΔNLS that encodes a FRQ version where the NLS was deleted (Luo et al. 1998). As reported previously (Luo et al. 1998), FRQΔNLS levels in the nucleus were substantially reduced (Fig. 2B). WC-2 levels were low in frqΔNLS, demonstrating that nuclear functions of FRQ were compromised (Fig. 2C, top). WC-1 levels, however, were not reduced, suggesting that its accumulation was supported by cytoplasmic FRQΔNLS. WC-1 and WC-2 (Fig. 2C) were hypophosphorylated in frqΔNLS, indicating that hyperphosphorylation of WCC requires nuclear FRQ.
We have recently shown that deletion of the PEST-1 sequence of FRQ leads to accumulation of elevated levels of FRQΔP-1 and of WC-1 (Görl et al. 2001). As shown in Figure 2D, the cytosolic levels of FRQΔP-1 are elevated in comparison with FRQ levels in frq+, while nuclear levels of FRQΔP-1 and FRQ are similar. Thus, the increase in WC-1 amount (Fig. 2D) correlates with the amount of cytosolic FRQΔP-1.
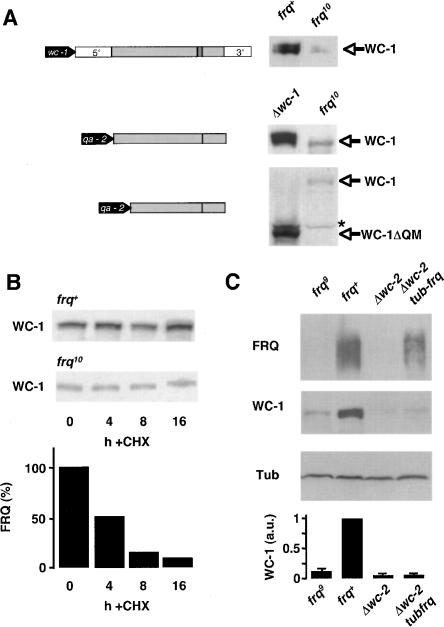
Together, the data suggest that accumulation of WC-1 is supported by cytoplasmic FRQ. By what mechanism does cytoplasmic FRQ support WC-1 expression? Signals for translational regulation are often located in the 5′- and 3′-untranslated region (UTR) of mRNAs (Gebauer and Hentze 2004). To exclude such potential regulation, we expressed the wc-1 ORF (spliced c-DNA without UTRs) under control of the qa-2 promoter in Δwc-1 and in frq10. We also expressed an N-terminally truncated version of the ORF (wc-1ΔQM). The deleted region of wc-1 encodes the N-terminal polyglutamine stretch and a methionine-rich region of predicted low complexity (data not shown), and we reasoned that this region could potentially contain sequences regulating mRNA translation. Following induction of the qa-2 driven wc-1 gene, WC-1 and WC-1ΔQM were efficiently expressed in Δwc-1, while expression levels of both proteins were low in frq10 (Fig. 3A), in agreement with similar results by Cheng et al. (2003). As measured by RT-PCR, wc-1 RNA levels were 2.3 ± 0.4 (n = 4) times higher in wc-1 frq10 qawc-1 than in frq+ (data not shown). The data suggest that FRQ supports WC-1 accumulation on a post-translational level. However, we cannot rule out that signals for translational control are located in the wc-1ΔQM ORF encoding the evolutionary conserved LOV and PAS domains and the GATA-like zinc-finger of WC-1.
Figure 3.
FRQ supports WC-1 expression in a post-translational fashion. (A, left) The wc-1 gene (top), qa-2 promoter-driven cDNA copies of the spliced full-size wc-1 ORF (middle), and the truncated wc-1ΔQM ORF (bottom) are schematically outlined. The wc-1ΔQM ORF lacks the first 860 base pairs (bp) of wc-1 and the truncated protein starts presumably at methionine 297. (Filled block arrows) Promoters; 5′ and 3′ untranslated regions. (Gray and hatched boxes) Exons and introns. (Right) Expression levels of full-length and truncated WC-1 forms are shown by representative Western blots. (Top) WC-1 expression in frq+ and frq10. (Middle) WC-1 expression in Δwc-1;qa-wc-1 and frq10;qa-wc-1. (Bottom) Expression of WC-1ΔQM in Δwc-1;qa-wc-1ΔQM and frq10;qa-wc-1ΔQM. (⋆) Unspecific band. (B) Stability of WC-1 in complex with WC-2. Indicated strains were grown in DD and harvested at the indicated time points after addition of 10 μg/mL cycloheximide (CHX). (Top) Total extracts were analyzed by Western blotting and decoration with αWC-1 antiserum. (Bottom) Quantification of FRQ degradation kinetics is shown for control. (C) FRQ does not support WC-1 accumulation in the absence of WC-2. The indicated strains were grown in LL. Extracts were prepared and analyzed by Western blotting. FRQ, WC-1, and β-tubulin (Tub) are shown. (Bottom) Expression levels of WC-1 relative to β-tubulin. The ratio of WC-1/Tub in frq+ was set to 1 (n = 2).
We then measured whether the turnover kinetics of WC-1 is dependent on FRQ (Fig. 3B). When protein synthesis was inhibited with cycloheximide, WC-1 was equally stable in frq+ and frq10. Since WC-1 does not accumulate to significant levels in the absence of WC-2, (Cheng et al. 2002) the experiment measures only turnover of assembled WC-1. Accordingly, we concluded that FRQ does not affect stability of WC-1 once the protein is assembled in the WCC.
We then asked whether FRQ could stabilize newly synthesized WC-1 prior to its assembly with WC-2; i.e., on the level of translation or folding. For this purpose, we expressed in a Δwc-2 strain FRQ under control of the β-tubulin promoter (tub-frq), since the endogenous frq is not transcribed in wc-2-deficient strains. Levels of FRQ were lower in tub-frq than in frq+, but much higher than in Δwc-2 (Fig. 3C). Yet, WC-1 levels remained unaffected, suggesting that FRQ may not facilitate translation of wc-1 RNA or folding of the polypeptide. Rather, FRQ promotes accumulation of WC-1 only in presence of WC-2.
Since FRQ does not apparently control translation and folding of WC-1 nor turnover of the WCC complex, it seems plausible that cytosolic FRQ assists the process of assembly of WC-1 with WC-2 (see Discussion).
The PEST-2 sequence in FRQ is required for support of WC-1 expression
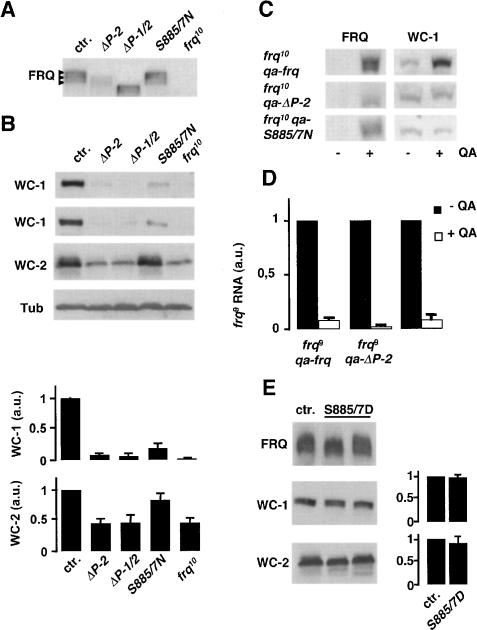
FRQ9 is defective in negative feedback and does not support WC-1 accumulation, indicating that the C-terminal portion of FRQ is necessary for both functions. The C-terminal portion of FRQ contains the PEST-2 sequence (Merrow and Dunlap 1994). PEST-2 is phosphorylated in vitro by CK-1a (Görl et al. 2001). Analysis of GST-PEST-2 by mass spectrometry revealed that PEST-2 is phosphorylated at two serine residues, S885 and S887, and a PEST-2-derived peptide, containing S885 and S887, was phosphorylated by CK-1a (Supplementary Fig. S2). To access whether PEST-2 has a function in negative feedback or in support of WC-1 accumulation, we constructed the mutant alleles frqΔP-2 and frq-S885/7N, in which PEST-2 was deleted or the serine (S) codons 885 and 887 were changed to asparagine (N) codons. Since we had previously shown that deletion of PEST-1 results in stabilization of FRQ (Görl et al. 2001), we also deleted PEST-1 in addition to PEST-2 (frqΔP-1/2). The mutant frq alleles and a wild-type copy (control) were expressed in frq10, and expression levels (LL) of the FRQ variants were analyzed (Fig. 4A). FRQΔP-2 levels were lower, while FRQΔP-1/2 levels were similar to control levels of FRQ. This suggests that PEST-1 and PEST-2 have different functions. Deletion of PEST-2 leads to a reduced expression of FRQ, which can be compensated for by the stabilizing effect of the PEST-1 deletion. PEST-1 is phosphorylated by CK-1a (Görl et al. 2001). PEST-2, which is phosphorylated in vitro by CK-1a/b (Görl et al. 2001), may also be a target for phoshorylation in vivo, since hyperphosphorylated species of FRQΔP-2 appear to be underrepresented in comparison with FRQ (Fig. 4A; Supplementary Fig. S3). FRQ-S885/7N was expressed at almost control levels and its phosphorylation status was similar to that of FRQ (Fig. 4A).
Figure 4.
PEST-2 of FRQ is crucial for positive but not for negative feedback. (A) Expression of PEST-2 mutant versions of FRQ. frq10, his-3 was transformed with the wild-type frq gene (ctr.) and the indicated PEST-2 mutant alleles. Arrowheads indicate three major FRQ species of different electrophoetic mobility reflecting heterogeneous phosphorylation status. frq10 is shown for control. (B) Expression levels of WC-1 (two different experiments are shown) and WC-2 in the indicated strains. β-Tubulin (Tub) is shown for control. (Bottom) Quantification of WC-1 and WC-2 relative expression levels. Mean ± SD; n = 3. (C) Expression levels of WC-1 after induction of wild-type and mutated frq alleles under control of the qa-2 promoter. Cells were harvested 16 h after induction and total extracts of uninduced and induced cells were subjected to Western analysis. (D) Repression of the endogenous frq9 allele by QA-induced wild-type FRQ, FRQΔP-2, and FRQ-S885/7N. Mean values of two independent RT-PCR determinations with triplicate reactions (±SD). RNA levels before QA-induction were set equal to 1. (E) FRQS885/7D supports expression of WC-1 and WC-2. frq10, his-3 was transformed with frq+ (ctr.) and frqS885/7D alleles (two transformants are shown). (Left) Expression levels of WC-1 and WC-2 were analyzed by Western blotting. (Right) Quantification of WC-1 and WC-2 levels in the indicated strains (mean ± SD, n = 4).
To characterize the PEST-2 mutant proteins, we determined their stability and subcellular distribution. Turnover kinetics of FRQΔP-2 and FRQ-S885/7N were not significantly different from those of FRQ (Supplementary Fig. S4) and their subcellular distribution was not affected (data not shown). As expected, FRQΔP-1/2 was stabilized due to the deletion of PEST-1 (Supplementary Fig. S4).
We then determined expression levels of WC-1 in the transformed strains (Fig. 4B,C). WC-1 was inefficiently expressed in frqΔP-2, frqΔP-1/2, and frq-S885/7N. In contrast, efficient WC-1 accumulation was supported by authentic FRQ (Fig. 4B,C). PEST-1 had no direct effect on WC-1 expression (see Fig. 2D); the elevated levels of FRQΔP-1 supported accumulation of correspondingly elevated amounts of WC-1. WC-2 levels were reduced in frqΔP-2 and frqΔP-1/2, but not in frq-S885/7N (Fig. 4B). For further analysis, we expressed inducible copies of frq, frqΔP-2, and frq-S885/7N under control of the qa-2 promoter in an frq10 strain and analyzed WC-1 expression (Fig. 4C). Without induction of the qa-2 promoter, WC-1 levels were low in all three strains. WC-1 level increased when expression of wild-type frq was induced, but not upon induction of frqΔP-2 and frq-S885/7N. The data indicate that expression of the mutant FRQ proteins did not affect WC-1 levels. Rather, PEST-2 is required for the support of WC-1 expression. S885 and S887 of FRQ appear to be crucial for this function.
A key function of FRQ is its ability to inhibit synthesis of its own mRNA. To measure whether PEST-2 affects negative feedback, we expressed qa-frq, qa-frqΔP-2, and qa-frqS885/7N in a frq9 strain. Prior to addition of QA, the frq9 allele was efficiently transcribed, since FRQ9 protein is defective in negative feedback (Fig. 4D). When QA was added (DD), frq9 RNA was efficiently repressed by FRQ as well as by the induced FRQΔP2 and FRQS885/7N, demonstrating that PEST-2 is not required for negative feedback. This indicates that negative feedback and support of WC-1 accumulation are two mechanistically distinct functions that are associated with distinct regions of FRQ.
Support of WC-1 accumulation by FRQ appears to be dependent on the putative phosphorylation sites S885 and/or S887. To mimic phosphorylation, we constructed frq alleles, where the codons S885 and S887 were changed by site directed mutagenesis to aspartate (D) codons. When expressed in a frq10 background, the mutant protein, FRQS885/7D, supported accumulation of high levels of WC-1 (Fig. 4E), suggesting that WC-1 accumulation is regulated by phosphorylation of FRQ at S885 and S887. As in frqS885/7N, WC-2 levels were not affected in frqS885/7D (Fig. 4E).
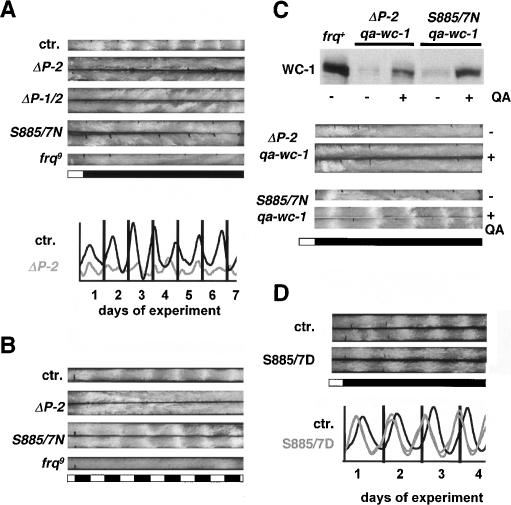
frqΔP-2 and frq-S885/7N strains did not show a molecular rhythm of frq RNA (data not shown) and FRQ protein abundance (Supplementary Fig. S5) and displayed no conidial banding when grown in constant darkness (DD) on race-tubes (Fig. 5A). This demonstrates that the positive limb of the interconnected feedback loops is crucial for free-running rhyhtmicity. To assess whether the strains had generally lost the ability to form conidial bands, race tubes were exposed to 12 h light/12 h dark (LD) cycles (Fig. 5B). frq9 was arrhythmic under such regime, since light-driven conidial banding requires FRQ (Chang and Nakashima 1997; Merrow et al. 1999). Interestingly, frqS885/7N showed conidial banding in LD cycles, while frqΔP-2 did not. This suggests that deletion of PEST-2 causes a more severe functional defect than exchange of S885 and S887.
Figure 5.
Conidiation rhythm in wild-type and PEST-2 mutant strains. (A) Racetube assay showing arrhythmic behavior of PEST-2 mutants under free-running conditions (DD). White bars indicate light periods and black bars indicate darkness. (Bottom) Densitometric traces of ctr. and frqΔP2. (B) frq-S885/7N shows rhythmic conidiation in LD cycles. Indicated strains were exposed to 12 h light/12 h dark cycles. (C, top) Induced expression of WC-1 from an additional gene copy under qa-2 promoter control. Cells were grown in the absence of QA (-) or wc-1 was induced for 16 h (+). (Bottom) Rescue of overt rhythmicity in frq-S885/7N by QA-induced expression of WC-1. Racetubes without (-) or with (+) QA were inoculated with the indicated strains and incubated in DD. (D) Ser-to-Asp exchange of residues 885 and 887 of FRQ result in a 3-h phase advance of the conidiation rhythm. Racetube analysis of frqS885/7D and frq+ (ctr.) transformed frq10 strains.
To analyze whether the failure to support accumulation of WC-1 is the sole defect of the PEST-2 mutant strains, frqΔP-2 and frq-S885/7N were transformed with a qa-wc-1 gene. In the absence of QA, low levels of WC-1 were expressed in both mutant strains (Fig. 5C). When wc-1 RNA synthesis was induced, increased expression of WC-1 was detected. However, despite the presence of a highly transcribed extra copy of the wc-1 gene (data not shown), WC-1 levels remained markedy lower than in frq+ (Fig. 5C), demonstrating once more that functional FRQ is required for WC-1 accumulation. WC-1 levels were higher in frq-S885/7N;qa-wc-1 than in frqΔP-2;qa-wc-1, supporting the notion that deletion of PEST-2 causes a more severe defect than exchange of S885 and S887. The data demonstrate that the PEST-2-associated function of FRQ is required to support expression of WC-1, even when efficient wc-1 transcription is driven by a heterologous promoter.
Race tubes were inoculated with frq-S885/7N;qa-wc-1 and frqΔP-2;qa-wc-1 and incubated in DD (Fig. 5D). In the absence of QA in the growth medium, both strains did not show rhythmic conidiation. In the presence of QA, frqΔP-2;qa-wc-1 remained arrhythmic, while frq-S885/7N;qa-wc-1 displayed rhythmic circadian conidiation.
Finally, frq-S885/7D was rhythmic on race tubes in DD (Fig. 5E). The free-running period (21.52 ± 0.74x h, n = 8) was similar to that of the frq+-transformed control strain (21.59 ± 0.5 h, n = 2), indicating that the principle functions of FRQ were not compromised. However, the phase of conidiation of frq-S885/7D (DD 8 ± 0.55) was advanced by 3 h compared with the control strain (DD 11.1 ± 0.4) (Fig. 5E).
Together, the data demonstrate that nuclear and cytoplasmic functions of FRQ are distinct and associated with distinct portions of FRQ. Overt rhythmicity is lost when the cytoplasmic function of FRQ is compromised. PEST-2 is crucial for the support of WC-1 accumulation, but is neither required for negative feedback nor rate limiting for FRQ turnover. The kinetics of FRQ phosphorylation at S885 and S887 appear to determine WC-1 accumulation and the phase of conidiation.
Discussion
FRQ is part of interlocked positive and negative feedback loops that are crucial for robust circadian rhythmicity of Neurospora. In these loops, FRQ exerts apparently conflicting functions. In the negative limb of the interlocked loops, FRQ inhibits its own expression by phosphorylation-dependent inactivation of its transcriptional activator WCC (Schafmeier et al. 2005). In the positive limb, FRQ supports accumulation of WCC (Lee et al. 2000; Cheng et al. 2001).
We present evidence that these conflicting functions of FRQ are confined to distinct subcellular compartments, coordinated in temporal fashion and regulated by phosphorylation of FRQ. Phosphorylation-dependent inactivation of WCC requires nuclear FRQ. Low levels of FRQ are sufficient and hypophosphorylated FRQ appears to be particularly active in negative feedback. Thus, negative feedback sets in early after FRQ expression. Support of WC-1 accumulation, in contrast, is mediated by cytosolic FRQ. It requires higher levels of FRQ than negative feedback and it requires phosphorylation of FRQ. Accordingly, WCC accumulation is supported late after FRQ expression, when significant levels of cytoplasmic FRQ have built up and when FRQ is hyperphosphorylated.
A function of FRQ in the cytosol was surprising and unexpected, since cytoplasmic sequestration of clock proteins in mammals and flies is generally assumed to provide the means for accumulation of inactive protein that becomes activated upon regulated nuclear import. When imported into the nucleus, mPER/CRY and dPER/TIM act by a single mechanism; they inactivate CLOCK/BMAL1 and dCLOCK/CYCLE, rerspectively, and thus repress genes that are activated by these transcription factors (Reppert and Weaver 2002; Stanewsky 2003; Gachon et al. 2004; Hardin 2004). This leads to direct repression of genes in the negative limb and activation of genes in the positive limb via repression of a repressor gene.
In addition, post-transcriptional mechanisms contribute to the circadian loops in flies and mammals. In Drosophila, dCLOCK/CYCLE activates transcription of Vrille and Pdp1, encoding a repressor and a transcriptional activator of dClock, respectively. Yet, VRILLE levels peak 3–6 h before PDP1 (Cyran et al. 2003), suggesting a considerably complex regulation beyond the level of rhythmic transcription. Furthermore, flies remain rhythmic when dClock is expressed in antiphase under control of the per promoter (Kim et al. 2002), even when dPER and Tim are constitutively expressed (Yang and Sehgal 2001), suggesting that transcriptional control of the positive limb and the negative limb is not absolutely essential for clock function. In the mouse system, mPer1/2, mCry1/2, Rev-erbα, and RORα transcript levels peak at different circadian times, which cannot be explained by a simple transcriptional control of these genes by CLOCK/BMAL1 (Gachon et al. 2004; Sato et al. 2004) Futhermore, in a rev-erbα knock-out mouse, Bmal1 is constitutively transcribed at elevated levels; yet mPer2 and mCry1/2 are rhythmically transcribed and the circadian clock is running (Preitner et al. 2002). This demonstrates that transcriptional control of the positive limb is not strictly required for rhythmicity in mice. Interestingly, dimerization of BMAL1 with the CLOCK paralog NPAS2 is regulated on a metabolic level through a heme-binding PAS-domain (Dioum et al. 2002), suggesting that also the interaction of BMAL1 with CLOCK may be regulated by a post-translational mechanism. There is, however, no evidence that PERs, CRYs, or TIMs are directly involved in post-translational regulation of the circadian clock in mammals and flies.
In Neurospora, FRQ activates wc-2 transcription by an unknown mechanism (Cheng et al. 2003). This function of FRQ could be indirect and analogous to the function of mPER/CRY and dPER/TIM in mammals and flies; i.e., via transcriptional repression of a WCC-controlled putative repressor of wc-2. FRQ also affects transcription of wc-1 (Dragovic et al. 2002; Káldi et al. 2005). However, levels of WCC are dependent on FRQ even when wc-1 or wc-2 are constitutively transcribed from a heterologous promoter, demonstrating that transcriptional control of the positive limb is not required. Thus, FRQ acts on a transcriptional level in the negative limb of the circadian feedback loop; the critical function of FRQ in the positive limb appears to be associated with post-translational regulation. How does FRQ exert apparently diverse functions such as inactivation of WCC in the nucleus and support of WCC accumulation in the cytosol? We have shown recently that FRQ feeds back on the transcription of its own gene by promoting phosphorylation of WCC, which leads to inactivation of the transcription factor (Schafmeier et al. 2005). Here, we present evidence that support of WCC accumulation is a post-translational process supported by FRQ in the cytosol. FRQ appears to act on the level of WCC assembly. Although it seems unlikely that FRQ is a general chaperone, it may specifically facilitate biogenesis of WCC. It is tempting to speculate that FRQ supports WCC accumulation by a molecular mechanism similar to the mode by which it inhibits WCC in the negative limb. Thus, FRQ may facilitate phosphorylation of cytosolic WC-1 or WC-2. Phosphorylation could enhance assembly competence of WC-1 with WC-2, either directly or indirectly, by promoting their accumulation in the nucleus. Recently, five adjacent phosphorylation sites in WC-1 have been identified that affect expression levels of WCC (He et al. 2005). However, it was not investigated whether phosphorylation of WC-1 at these sites is facilitated by FRQ.
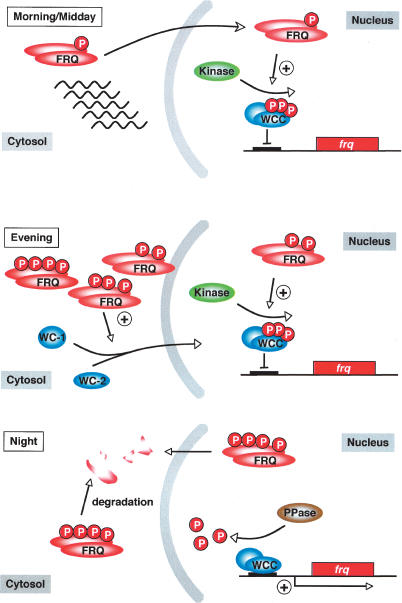
In the course of a circadian period, FRQ is progressively phosphorylated and degraded. The phosphorylation targets identified hitherto, in particular PEST-1 and nearby sites, regulate turnover of FRQ and are major determinants of period length (Görl et al. 2001; Yang et al. 2001, 2002, 2003). We show here that PEST-2 has no rate-limiting function in FRQ turnover. Rather, PEST-2 is required to support accumulation of WCC. In particular, the putative CK-1a phosphorylation sites S885 and S887 are critical for this function. PEST-2 is not required for negative feedback. The functions of FRQ in the negative and positive limbs of the circadian feedback loops are therefore associated with distinct regions or domains of FRQ. We present evidence that hyperphosphorylated FRQ is significantly less efficient in negative feedback than the hypophosphorylated protein. Thus, not only FRQ stability, but also its functions in the negative and positive limps of the feedback loop appear to be regulated by phosphorylation. Given the lack of homology of FRQ with evolutionary conserved protein domains of known function, FRQ could provide a platform for association of kinases and phosphatases modifying WC-1, WC-2, and also FRQ itself. Phosphorylation of FRQ could modulate its affinity for its interaction partners and thus regulate specific FRQ functions. We propose that progressive phosphorylation of FRQ in the course of a circadian period triggers its gradual maturation to a cytoplasmic activator (Fig. 6). Early after expression, hypophosphorylated FRQ acts as a repressor by promoting phosphorylation of WCC in the nucleus. Later, when hypo- and hyperphosphorylated forms of FRQ have accumulated, both functions of FRQ are active; cytoplasmic FRQ supports accumulation of WCC, which is efficiently inactivated by nuclear FRQ. Thereby, FRQ supports accumulation of high levels of its inactive transcription factor, and finally, when FRQ is predominantly hyperphosphorylated and on the pathway to degradation, negative feedback is relieved and the accumulated inactive WCC becomes progressively activated by PP2A-dependent dephosphorylation (Schafmeier et al. 2005), which then dominates kinetically over FRQ dependent WCC phoshorylation.
Figure 6.
Model for FRQ function in nucleus and cytosol. (Morning/Midday) Newly synthesized FRQ is active as a repressor of WCC in the nucleus. Low levels of hypophosphorylated FRQ are sufficient to promote phosphorylation and inactivation of WCC. (Evening) FRQ supports accumulation of inactive WCC. High levels of hyperphosphorylated cytosolic FRQ are required to support WC-1 accumulation. Cytosolic FRQ may act on the level of WCC assembly. Newly assembled WCC accumulates in the nucleus and is inactivated via FRQ-dependent phosphorylation. frq transcription remains repressed. (Night) FRQ is progressively phosphorylated and degraded. As the FRQ concentration decreases, PP2A dependent dephosphorylation of WCC dominates over phosphorylation (Schafmeier et al. 2005). Dephosphorylated WCC binds to the frq promoter and activates transcription.
Materials and methods
Strains and growth conditions
Neurospora strains used in this study (frq+, frq9, frq10, frqΔNLS, Δwc-1, and Δwc-2) carried the bd mutation (Loros et al. 1986). For transformations, bd; frq10; his-3 and bd; Δwc-1; his-3 were used. Standard medium contained 2% glucose, 0.5% L-arginine, 1× Vogel's, and 10 ng/mL biotin, solid medium contained an additional 2% agar. For induction of genes under control of the qa-2 promoter, Neurospora was transferred to induction medium (2% sucrose, 0.17% L-arginine, 1× Vogel's, and 10 ng/mL biotin) containing 0.3% QA. In race-tube medium, glucose content was reduced to 0.1%, and arginine was reduced to 0.17%. Analysis of race tubes was performed using the ChronoII program (T. Roenneberg, Ludwig-Maximilians-Universitat, Munich, Germany).
Plasmid constructs and Neurospora transformation
The frq ORF was cloned into a pGEM4 plasmid, resulting in pG4-ORF (Görl et al. 2001). In pG4-ORFΔP2, nucleotides 2535–2661 of frq-ORF (encoding PEST-2) were replaced with CGCGGCCGC, creating a NotI site. In pG4-ORF-S885/7N, the codons for the serine residues 885 and 887 were exchanged to AAT by site-directed mutagenesis (QuikChange XL, Stratagene) resulting in asparagine codons. The SfiI–XbaI fragment of pBM61frq was replaced by the corresponding fragment of pG4-ORFΔP2 and pG4-ORF-S885/7N, resulting in pBM61frqΔPEST-2 and pBM61frq S885/7N, respectively. pBM61frqΔPEST-1/2 was constructed correspondingly using the vector pBM61frqΔPEST-1 (Görl et al. 2001). pBM61frq S885/7D was obtained by site-directed mutagenesis.
For transformation, Neurospora cells were grown on solid medium for 5–7 d, and conidia were harvested by shaking in 1 M sorbitol. Conidia were washed in 1 M sobitol several times and concentrated to an optical density of 300–600/mL suspension. Fifty microliters of the suspension were incubated with linearized plasmid DNA (derivatives of pBM60 for targeted insertion into the his-3 locus) for 5 min on ice. Cells were transformated by electroporation in a 0.2-cm cuvette (voltage gradient: 1.5 kV/cm; capacitance: 25 μ FD; resistance: 600 ohms; Gene Pulser; Bio-Rad). One milliliter of 1 M sorbitol was added to each cuvette, the suspension was mixed by gently pipetting up and down, and 200–800 μL were added to 10 mL of top-agar, and plated on medium. Transformants were picked after 2–3 d of incubation at 30°C and transformed into standard liquid medium. Insertion of the target gene was verified by PCR. Homocaryotic strains were generated by purification by several plating steps and checked by quantitative PCR.
Protein analysis
Extraction of Neurospora protein and subcellular fractionation was performed as described (Luo et al. 1998; Görl et al. 2001). Nuclear extracts were prepared by incubating nuclei at 4°C under shaking for 30 min in protein extraction buffer (50 mM HEPES at pH 7.4, 10% glycerol, 137 mM NaCl, 5 mM EDTA, 1 mM phenylmethyl sulfonyl fluoride, 1 μg/mL leupeptin, 1 μg/mL pepstatin A). Two-dimensional gel analysis was performed as described in Schafmeier et al. (2005). Samples contained the following phosphatase inhibitors: 30 mM Na-pyrophosphate, 20 mM Na phosphate, 5 mM EDTA, 400 nM ocadaic acid, 20 mM caliculin, and 2 mM vanadate. Western blotting was performed as described (Görl et al. 2001). Enhanced chemiluminescence signals were detected with X-ray films. Series of exposures in the range of 5 sec to 30 min were generated. Quantification was performed using the QuantityOne software (Bio-Rad).
GST-PEST-2 and a PEST-2-derived peptide were phosphorylated in vitro using recombinant CK1a (Görl et al. 2001). Phosphorylation sites were determined by mass spectrometry by F. Lottspeich, Max-Planck-Institut, Martinsried, Germany.
RNA analysis
RNA was prepared and analyzed by quantitative real-time PCR (RT-PCR) essentially as described (Görl et al. 2001; Merrow et al. 2001); cDNA was synthesized from 2 μg of DNase-treated total RNA using the SuperScriptII RT-system (Invitrogen) and random hexamer primers. frq and actin cDNA was detected by RT-PCR (ABI-Prism 7000, Applied Biosystems) using TaqMan probes (Görl et al. 2001). Triplicate reactions (25 μL) containing cDNA equivalent to 0.1 μg RNA were analyzed.
Acknowledgments
We thank J. Dunlap and J. Loros for Δwc-1 and Δwc-2 strains and J. Payk and J. Scholz for excellent technical assistance. This work was supported by grants from Deutsche Forschungsgemeinschaft (BR 1375-1 and SFB 638), by Fonds der Chemischen Industrie and from Alexander von Humboldt-Stiftung to K.K.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.360906.
References
- Akashi M. and Takumi, T. 2005. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 12: 441-448. [DOI] [PubMed] [Google Scholar]
- Aronson B.D., Johnson, K.A., Loros, J.J., and Dunlap, J.C. 1994a. Negative feedback defining a circadian clock: Autoregulation of the clock gene frequency. Science 263: 1578-1584. [DOI] [PubMed] [Google Scholar]
- Aronson B.D., Johnson, K.A., and Dunlap, J.C. 1994b. Circadian clock locus frequency: Protein encoded by a single open reading frame defines period length and temperature compensation. Proc. Natl. Acad. Sci. 91: 7683-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B. and Nakashima, H. 1997. Effects of light–dark cycles on the circadian conidiation rhythm in Neurospora crassa. J. Plant Res. 110: 449-453. [Google Scholar]
- Cheng P., Yang, Y., and Liu, Y. 2001. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc. Natl. Acad. Sci. 98: 7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Yang, Y., Gardner, K.H., and Liu, Y. 2002. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol. Cell. Biol. 22: 517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Yang, Y., Wang, L., He, Q., and Liu, Y. 2003. WHITE COLLAR-1, a multifunctional Neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. J. Biol. Chem. 278: 3801-3808. [DOI] [PubMed] [Google Scholar]
- Cheng P., He, Q., He, Q., Wang, L., and Liu, Y. 2005. Regulation of the Neurospora circadian clock by an RNA helicase. Genes & Dev. 19: 234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran S.A., Buchsbaum, A.M., Reddy, K.L., Glossop, N.R., Hardin, P.E., Young, M.W., Storti, R.V., and Blau, J. 2003. vrille, Pdp1 and dClock form a second loop in the Drosophila circadian clock. Cell 112: 329-341. [DOI] [PubMed] [Google Scholar]
- Diernfellner A.C.R., Schafmeier, T., Merrow, M.W., and Brunner, M. 2005. Molecular mechanism of temperature-sensing by the circadian clock of Neurospora crassa. Genes & Dev. 19: 1968-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum E.M., Rutter, J., Tuckerman, J.R., Gonzalez, G., Gillez-Gonzalez, M.A., and McKnight, S.L. 2002. NPAS2: A gas-responsive transcription factor. Science 298: 2385-2387. [DOI] [PubMed] [Google Scholar]
- Dragovic Z., Tan, Y., Gorl, M., Roenneberg, T., and Merrow, M. 2002. Light reception and circadian behavior in `blind' and `clock-less' mutants of Neurospora crassa. EMBO J. 21: 3643-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J.C. and Loros, J.J. 2004. The Neurospora circadian system. J. Biol. Rhythms 19: 414-424. [DOI] [PubMed] [Google Scholar]
- Froehlich A.C., Loros, J.J., and Dunlap, J.C. 2003. Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc. Natl. Acad. Sci. 100: 5914-5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F., Nagoshi, E., Brown, S.A., Ripperger, J., and Schibler, U. 2004. The mammalian circadian timing system: From gene expression to physiology. Chromosoma 113: 103-112. [DOI] [PubMed] [Google Scholar]
- Gebauer F. and Hentze, M.W. 2004. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell. Biol. 5: 827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görl M., Merrow, M., Huttner, B., Johnson, J., Roenneberg, T., and Brunner, M. 2001. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J. 20: 7074-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin P.E. 2004. Transcription regulation within the circadian clock: The E-box and beyond. J. Biol. Rhythms 19: 348-360. [DOI] [PubMed] [Google Scholar]
- He Q., Cheng, P., Yang, Y., He, Q., Yu, H., and Liu, Y. 2003. FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. EMBO J. 22: 4421-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Shu, H., Cheng, P., Chen, S., Wang, L., and Liu, Y. 2005. Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J. Biol. Chem. 280: 17526-17532. [DOI] [PubMed] [Google Scholar]
- Káldi K., Herreros González, B., and Brunner M. 2005. Transcriptional regulation of the Neurospora circadian clock gene wc-1 affects the phase of circadian output. EMBO reports [Epub December 23, 2005]. [DOI] [PMC free article] [PubMed]
- Kim E.Y. Bae, K., Ng, F.S., Glossop, N.R.J., Hardin, P.E., and Edery, I. 2002. Drosophila Clock protein is under posttranscriptional control and influences light-induced activity. Neuron 34: 69-81. [DOI] [PubMed] [Google Scholar]
- Lee C., Bae, K., and Edery, I. 1998. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER–TIM complex. Neuron 21: 857-867. [DOI] [PubMed] [Google Scholar]
- Lee K., Loros, J.J., and Dunlap, J.C. 2000. Interconnected feedback loops in the Neurospora circadian system. Science 289: 107-110. [DOI] [PubMed] [Google Scholar]
- Lee C., Etchegaray, J.P., Cagampang, F.R., Loudon, A.S., and Reppert, S.M. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107: 855-867. [DOI] [PubMed] [Google Scholar]
- Liu Y., Loros, J.J., and Dunlap, J. 2000. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc. Natl. Acad. Sci. 97: 234-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros J.J., Richman, A., and Feldman J.F. 1986. A recessive circadian clock mutation at the frq locus of Neurospora crassa. Genetics 114: 1095-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Loros, J.J., and Dunlap, J.C. 1998. Nuclear localization is required for function of the essential clock protein FRQ. EMBO J. 17: 1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M.W. and Dunlap, J.C. 1994. Intergeneric complementation of a circadian rhythmicity defect: Phylogenetic conservation of structure and function of the clock gene frequency. EMBO J. 13: 2257-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M.W., Garceau, N., and Dunlap, J. 1997. Dissection of a circadian oscillation into discrete domains. Proc. Natl. Acad. Sci. 94: 3877-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M.W., Brunner, M., and Roenneberg, T. 1999. Assignment of circadian function for the Neurospora clock gene frequency. Nature 399: 584-586. [DOI] [PubMed] [Google Scholar]
- Merrow M., Franchi, L., Dragovic, Z., Görl, M., Johnson, J., Brunner, M., Macino, G., and Roenneberg, T. 2001. Circadian regulation of the light input pathway in Neurospora crassa. EMBO J. 20: 307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M., Imai, K., Ito, H., Nishiwaki, T., Murayama, Y., Iwasaki, H., Oyama, T., and Kondo, T. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308: 414-415. [DOI] [PubMed] [Google Scholar]
- Nawathean P. and Rosbash, M. 2004. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol. Cell 13: 213-223. [DOI] [PubMed] [Google Scholar]
- Preitner N., Damiola, F., Lopez-Molina, L., Zakany, J., Duboule, D., Albrecht, U., and Schibler, U. 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251-260. [DOI] [PubMed] [Google Scholar]
- Reppert S.M. and Weaver, D.R. 2002. Coordination of circadian timing in mammals. Nature 418: 935-941. [DOI] [PubMed] [Google Scholar]
- Roenneberg T. and Merrow, M. 2003. The network of time: Understanding the molecular circadian system. Curr. Biol. 13: R198-R207. [DOI] [PubMed] [Google Scholar]
- Sato T.K., Panda, S., Miraglia, L.J., Reyes, T.M., Rudic, R.D., McNamara, P., Naik, K.A., FitzGerald, G.A., Kay, S.A., and Hogenesch, J.B. 2004. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43: 527-537. [DOI] [PubMed] [Google Scholar]
- Schafmeier T., Haase, A., Káldi, K., Scholz, J., Fuchs, M., and Brunner, M. 2005. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122: 1-12. [DOI] [PubMed] [Google Scholar]
- Stanewsky R. 2003. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J. Neurobiol. 54: 111-147. [DOI] [PubMed] [Google Scholar]
- Tomita J., Nakajima, M., Kondo, T., and Iwasaki, H. 2005. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307: 251-254. [DOI] [PubMed] [Google Scholar]
- Yang Z. and Sehgal, A. 2001. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron 29: 453-467. [DOI] [PubMed] [Google Scholar]
- Yang Y., Cheng, P., Zhi, G., and Liu, Y. 2001. Identification of a calcium/calmodulin-dependent protein kinase that phosphorylates the Neurospora circadian clock protein FREQUENCY. J. Biol. Chem. 276: 41064-41072. [DOI] [PubMed] [Google Scholar]
- Yang Y., Cheng, P., and Liu, Y. 2002. Regulation of the Neurospora circadian clock by casein kinase II. Genes & Dev. 16: 994-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Cheng, P., He, Q., Wang, L., and Liu, Y. 2003. Phosphorylation of FREQUENCY protein by casein kinase II is necessary for the function of the Neurospora circadian clock. Mol. Cell. Biol. 23: 6221-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., He, Q., Cheng, P., Wrage, P., Yarden, O., and Liu, Y. 2004. Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes & Dev. 18: 255-260. [DOI] [PMC free article] [PubMed] [Google Scholar]