Abstract
Nonsense-mediated mRNA decay (NMD) is a surveillance mechanism that degrades mRNA containing premature termination codons (PTCs). In mammalian cells, recognition of PTCs requires translation and depends on the presence on the mRNA with the splicing-dependent exon junction complex (EJC). While it is known that a key event in the triggering of NMD is phosphorylation of the trans-acting factor, Upf1, by SMG-1, the relationship between Upf1 phosphorylation and PTC recognition remains undetermined. Here we show that SMG-1 binds to the mRNA-associated components of the EJC, Upf2, Upf3b, eIF4A3, Magoh, and Y14. Further, we describe a novel complex that contains the NMD factors SMG-1 and Upf1, and the translation termination release factors eRF1 and eRF3 (SURF). Importantly, an association between SURF and the EJC is required for SMG-1-mediated Upf1 phosphorylation and NMD. Thus, the SMG-1-mediated phosphorylation of Upf1 occurs on the association of SURF with EJC, which provides the link between the EJC and recognition of PTCs and triggers NMD.
Keywords: SMG-1, eRF, Upf, phosphorylation, EJC, NMD
Nonsense-mediated mRNA decay (NMD) is a surveillance mechanism that detects and degrades mRNA containing premature termination codons (PTCs) to eliminate potentially harmful C-terminally truncated proteins (Peltz et al. 1994; Wilkinson and Shyu 2001; Wilusz et al. 2001; Culbertson and Leeds 2003; Baker and Parker 2004; Maquat 2004; Conti and Izaurralde 2005; Yamashita et al. 2005). Aberrant proteins encoded by PTC-mRNA can have gain-of-function and dominant-negative effects that can result in genetic diseases (Frischmeyer and Dietz 1999; Holbrook et al. 2004). In some cases, truncated proteins retain at least some of their normal function, and the selective inhibition of NMD provides a novel strategy to rescue the mutant phenotype in PTC-related mutations (Cali and Anderson 1998; Usuki et al. 2004). NMD also degrades certain unspliced or unproductively spliced mRNAs, and upstream open reading frames (uORFs) containing mRNAs as natural targets (Mitrovich and Anderson 2000, 2005; He et al. 2003; Mendell et al. 2004), and, in the case of the vertebrate immune system, mRNAs emanating from out-of-frame gene rearrangements (Li and Wilkinson 1998; Frischmeyer and Dietz 1999).
In mammalian cells, translation termination codons and exon-exon junctions are cis-acting elements that allow recognition of PTCs. mRNA is subject to rapid decay when translation terminates more than ∼50-55 nucleotides upstream of the last exon-exon junction (Holbrook et al. 2004; Maquat 2004). The positional information that marks exon-exon junctions is provided by the components of the splicing-dependent exon junction complex (EJC) that persist during export and until the mRNA is translated (Kataoka et al. 2000; Le Hir et al. 2000; Zhou et al. 2000; Dreyfuss et al. 2002; Tange et al. 2004; Moore 2005). Indeed, some of the EJC proteins—notably Y14 and magoh—remain at the same position on the mRNA and translation is required to remove them (Dostie and Dreyfuss 2002).
Three UPF genes and seven smg genes have been genetically identified in Saccharomyces cerevisiae (Culbertson et al. 1980; Leeds et al. 1991) and Caenorhabditis elegans (Hodgkin et al. 1989; Pulak and Anderson 1993; Cali et al. 1999), respectively, as essential trans-acting factors for NMD. All these genes are conserved in Drosophila melanogaster and mammals and have essential roles in NMD (Sun et al. 1998; Yamashita et al. 2001; Mendell et al. 2002; Gatfield et al. 2003; Ohnishi et al. 2003; Kim et al. 2005). The three components Upf1/SMG-2, Upf2/SMG-3, and Upf3/SMG-4 are conserved in eukaryotes, and therefore, the three Upf/SMG proteins are core components of NMD (Culbertson and Leeds 2003). Upf1, an RNA helicase (Czaplinski et al. 1995), interacts with Upf2, and Upf2, in turn, binds directly to Upf3 in S. cerevisiae (Weng et al. 1996) and in mammals (Lykke-Andersen et al. 2000; Mendell et al. 2000; Serin et al. 2001). In addition, Upf3b is part of the EJC and directly interacts with Y14, a core component of the EJC, in mammals (Kim et al. 2001a; Le Hir et al. 2001; Lykke-Andersen et al. 2001). Moreover, in S. cerevisiae, these three Upf proteins and eukaryotic release factors (eRF1 and eRF3) have been proposed to function as part of a “surveillance complex” that recognizes PTCs and triggers NMD (Peltz et al. 1994; Culbertson and Leeds 2003), based on the genetic interactions of UPF genes for the recognition of translation termination codons and direct interactions between Upf proteins and eRF1 and eRF3 in vitro (Czaplinski et al. 1998; Wang et al. 2001) and in vivo (Kobayashi et al. 2004). However, in mammals, the in vivo formation of the Upf protein and eRF1-eRF3 complex, as well as the mechanism of the interactions between the Upf/SMG proteins and EJC, remains to be clarified.
Additional metazoan-specific SMG proteins SMG-1, SMG-5, SMG-6, and SMG-7 appear to regulate the sequential phosphorylation and dephosphorylation of Upf1/SMG-2 that is required for NMD. Genetic studies have determined that the six smg genes are regulators of the phosphorylation state of SMG-2. smg-1, smg-3, and smg-4 are required for the phosphorylation of SMG-2, whereas smg-5, smg-6, and smg-7 are required for dephosphorylation in C. elegans (Page et al. 1999). Recent studies have also provided mechanistic insights into the regulation of Upf1/SMG-2 phosphorylation: SMG-1 kinase activity is essential for Upf1/SMG-2 phosphorylation (Yamashita et al. 2001; Grimson et al. 2004), whereas SMG-5 and SMG-7 are part of the protein phosphatase 2A (PP2A) complex (Anders et al. 2003; Ohnishi et al. 2003) and selectively associate with phosphorylated Upf1 in vivo (Ohnishi et al. 2003). However, the relationship between the phosphorylation/dephosphorylation of Upf1 and the recognition of PTC and/or EJC by the surveillance complex remains to be clarified. In addition, the mechanism whereby Upf2/SMG-3 and Upf3/SMG-4 are involved in the phosphorylation of Upf1/SMG-2 is not well defined.
In the present study, we demonstrate that SMG-1 associates with the post-spliced mRNA through Upf2 and Y14, and that this association is essential for Upf1 phosphorylation during NMD. Together with the Upf2- and Y14-independent formation of the SMG-1-Upf1-eRF1-eRF3 complex (SURF), our results reveal the critical steps during recognition of PTC in mammalian cells.
Results
SMG-1 associates with the post-splicing mRNP complex through an EJC component, Y14
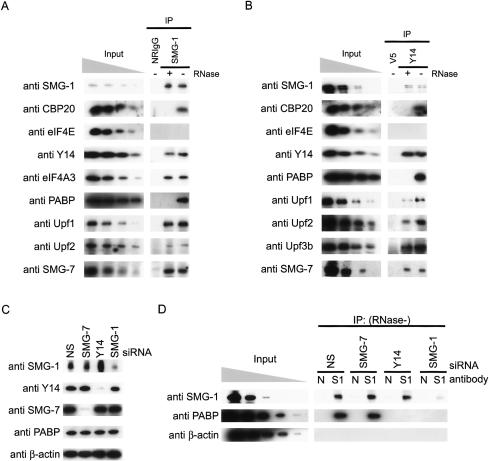
To explore the possibility that the phosphorylation of Upf1 by SMG-1 occurs on mRNAs, we analyzed SMG-1 immunoprecipitates (SMG-1-IP) for the presence of proteins that associate with mRNP. For this purpose, total HeLa TetOff cell lysates were treated with or without ribonuclease (RNase), immunoprecipitated with anti-SMG-1 antibodies or control immunoglobulin G (IgG), and analyzed by Western blotting for the presence of cap-binding proteins, EJC components, and poly(A)-binding protein (PABP), indicators of an mRNP complex (Dreyfuss et al. 2002; Moore 2005). As shown in Figure 1A, CBP20 (a component of nuclear cap-binding protein complex), Y14 (an EJC component) (Le Hir et al. 2000; Kim et al. 2001b), and cytoplasmic PABP, were present in SMG-1-IP in the absence of RNase. However, the coprecipitation of Y14 with SMG-1 is not abolished by RNase treatment, whereas precipitation of CBP20 and PABP was abolished (Fig. 1A). Similar experiments on Y14 immunoprecipitates showed that Y14 associates with SMG-1 in an RNase-insensitive manner, whereas the association of CBP20 and PABP is RNase sensitive (Fig. 1B). Note that we could not detect eIF4E in precipitates of SMG-1 and Y14 (Fig. 1A,B). In addition, SMG-1-IPs contain Upf1, Upf2, SMG-7, and eIF4A3 (a core component of EJC) (Chan et al. 2004; Ferraiuolo et al. 2004; Palacios et al. 2004; Shibuya et al. 2004) in the presence of RNase (Fig. 1A). Y14 immunoprecipitates contain Upf1, Upf2, Upf3b, and SMG-7 in RNase-insensitive manner (Fig. 1B). Because there is little supporting evidence for the complex formation of Upf2 and Y14, the simultaneous formation of the Upf2, Upf3b, and Y14 complex in vivo is also observed for Upf2 immunoprecipitation (Supplementary Fig. 1). Intriguingly, the RNase-sensitive coprecipitation of PABP with SMG-1 is greatly reduced by the knockdown of Y14 using short interfering RNA (siRNA), but is not reduced by the knockdown of SMG-7 (Fig. 1C,D). Because Y14 is a part of the post-splicing mRNP complex (Kataoka et al. 2000), we concluded that SMG-1 specifically associates with the post-splicing mRNP complex containing CBP20 and PABP through Y14. Additional coimmunoprecipitation experiments further revealed the association of HA-Upf3b, Magoh (a core component of EJC that stably binds Y14) (Kataoka et al. 2001), and Upf3aS (a component of the SMG-5-SMG-7 complex) (Ohnishi et al. 2003), with ectopically expressed wild-type Flag-SMG-1 in the presence of RNase (Fig. 2A-C). The RNase-resistant coprecipitation of Upf2, Upf3b, Y14, and Magoh with wild-type Flag-SMG-1 was observed in the cytoplasmic fraction (Fig. 2D). Both fractions contained a small amount of contamination, as judged by the presence of GAPDH (an indicator of the cytoplasmic fraction) and lamin C (an indicator of the nuclear fraction) (Fig. 2D).
Figure 1.
SMG-1 associates with the post-splicing mRNP through Y14. (A,B,D) HeLa TetOff cell lysates were immunoprecipitated with antibodies shown at the top of the figure. The immunoprecipitates (IPs), and fractionations of the cell lysates (input: 10%, 3.3%, 1.1%, and 0. 33% in A,B, and 0.11% in D of the amount immunoprecipitated) were analyzed by Western blotting with the indicated antibody probes. (C) Western blotting of siRNA-treated HeLa TetOff cell lysates. (D) N indicates the normal rabbit IgG, and S1 indicates anti-SMG-1 antibodies.
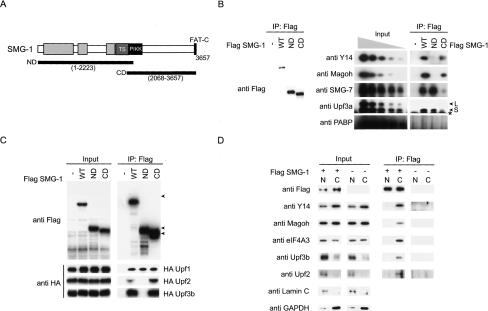
Figure 2.
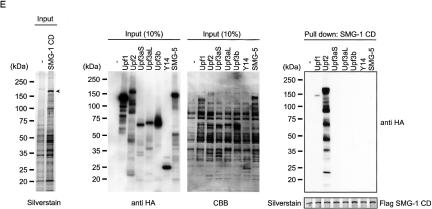
C-terminal half of SMG-1 directly binds to Upf2. (A) Schematic representation of SMG-1 mutants. In SMG-1-ND, the C-terminal 1589 amino acids are deleted. In SMG-1-CD, the N-terminal 2223 amino acids are deleted. The putative PIKK (PI3K-related protein kinase) and FAT-C domains are shown as black boxes. The TS (TOR-SMG-1) domains are shown as a dark-gray box. OCR (SMG-1 [one]-specific conserved region) domains are shown in light gray. (B,C) HeLa TetOff cells transfected with the plasmids shown above. A minus sign (-) indicates an empty vector; the blots were lysed and immunoprecipitated with the antibodies shown on the right (IP). IP or cell lysates (input; 10% in C, and 10%, 3.3%, 1.1%, 0. 33%, and 0.01% of the empty vector transfected in B of the amount immunoprecipitated) were then probed with the antibodies shown on the left. Arrowheads indicate the positions of SMG-1-WT, SMG-1-ND, and SMG-1-CD. SMG-1 mutants do not affect the expressions of probed proteins (data not shown). An asterisk indicates the IgG signals. (D) Immunoprecipitations and Western blotting were carried out on the nucleoplasm (N) and cytoplasm (C) of HeLa TetOff cells. Input represents 10% of the amount immunoprecipitated. (E) The SMG-1-CD region binds Upf2 in vitro. Details are described in Materials and Methods. A minus sign (-) indicates no template mRNA in an in vitro translation reaction. (Left panel) Input represents 5% of the in vitro translated reaction used for binding. Arrowheads indicate the positions of SMG-1-CD.
Upf2 directly binds SMG-1 and is essential for the association between SMG-1 and the post-splicing mRNP complex
We next determined the region of SMG-1 that associates with its binding partners. To do so, we expressed wild-type Flag-SMG-1, SMG-1-ND, and SMG-1-CD (Fig. 2A) and carried out immunoprecipitation with RNase-treated HeLa TetOff total cell extracts. These proteins can localize to the cytoplasm (Supplementary Fig. 2). HA-Upf1 and known EJC components Y14, Magoh, HA-Upf3b, and HA-Upf2 were coprecipitated by Flag-SMG-1 and SMG-1-CD. In addition, Flag-SMG-1-ND coprecipitated with SMG-7, Upf3aS, and HA-Upf1, although it failed to precipitate Y14, Magoh, HA-Upf3b, and HA-Upf2 (Fig. 2B,C). These results suggest that the SMG-1-CD is the region involved in the selective association with most EJC components. To identify the protein(s) that directly interact with SMG-1, we carried out pull-down analysis using purified recombinant Flag-SMG-1-CD, and in vitro translated proteins Y14, Upf2, Upf3b, Upf3aL, Upf3aS, SMG-5, and Upf1. As shown in Figure 2E, purified Flag-SMG-1-CD strongly interacted with in vitro translated Upf2 and weakly interacted with Upf1, but did not detectably interact with the other proteins. Similar experiments on Flag-SMG-1-ND revealed that it cannot pull down in vitro translated proteins except Upf1, indicating the specificity of the association (data not shown). In this assay, SMG-1 did not significantly interact with Y14, suggesting that SMG-1 associates with Y14 through Upf2, although we could not exclude the possibility that other EJC components, which we did not test for at this time, can also directly bind to SMG-1.
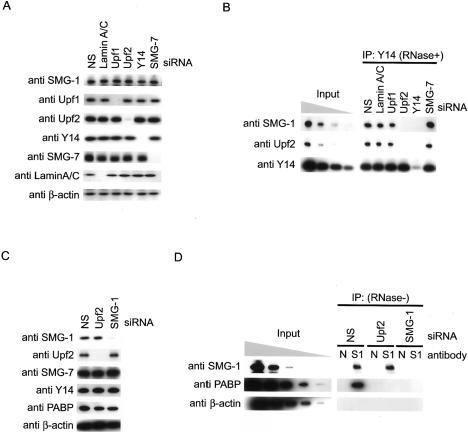
To further investigate the role of Upf2 in the complex formation between SMG-1 and Y14, we carried out immunoprecipitation experiments with anti-Y14 antibody under Upf2 knockdown conditions using siRNA. Depletion of Upf2 or Y14 strongly decreased the quantity of SMG-1 coimmunoprecipitating with Y14 in RNase-treated HeLa TetOff total cell extracts. On the other hand, knockdown of Upf1, SMG-7, or lamin A/C caused no significant change in the precipitation of SMG-1 with Y14 (Fig. 3B). In addition, depletion of Upf2 resulted in a decrease in the quantity of PABP that coimmunoprecipitated with SMG-1 using anti-SMG-1 antibodies in the absence of RNase (Fig. 3D). Considering these results together, we concluded that Upf2 is required for the association between SMG-1 and the Y14-post-splicing mRNP complex. All tested proteins were efficiently knocked down as assessed by Western blotting (Fig. 3A,C).
Figure 3.
Association between SMG-1 and mRNP requires Upf2. (A,C) HeLa TetOff cell lysates transfected with the siRNAs shown above the blots were analyzed by Western blotting with the indicated antibodies as probes. (B,D) HeLa TetOff cell lysates transfected with siRNAs were immunoprecipitated with anti-Y14 antibody in the presence of RNase (B), or with anti-SMG-1 or normal rabbit IgG in the absence of RNase (D; IP). IP or cell lysates (input; 10%, 3.3%, 1.1%, and 0. 33% in B, and 0.11% in D) were then probed with the antibodies shown on the left.
SMG-1-mediated phosphorylation of Upf1 requires Upf2, Upf3b, and Y14
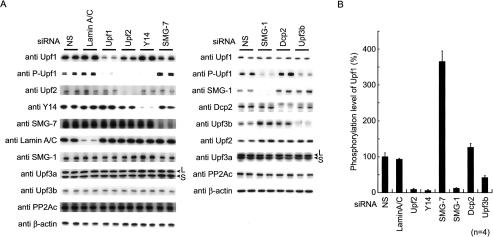
What does the interaction between SMG-1 and post-spliced mRNAs mean? One of the most plausible explanations is that the SMG-1-mediated phosphorylation of Upf1 occurs on the EJC. Therefore, impairing any of the adaptor proteins, Upf2, Y14, or Upf3b (a putative adaptor protein between Upf2 and Y14) of SMG-1 to the mRNP should result in a decrease of phosphorylated Upf1 (P-Upf1). In support of this, genetic analysis has indicated that smg-3 (Upf2 in mammals) and smg-4 (Upf3 in mammals) genes are required for SMG-2 (Upf1 in mammals) phosphorylation in C. elegans, although the mechanism is unknown (Page et al. 1999). To test this, we performed siRNA knockdown of a number of proteins (Fig. 4A) and looked at P-Upf1 levels. As shown in Figure 4A and B, P-Upf1, as detected by anti-P-Upf1 antibody (Ohnishi et al. 2003), was significantly reduced in cells with reduced Upf2, Upf3b, Y14, or SMG-1, whereas SMG-7 knockdown caused a significant increase in P-Upf1. On the other hand, knockdown of Lamin A/C and Dcp2 had no significant effect on Upf1 phosphorylation. These results show that Upf2, Y14, and Upf3b are required for the SMG-1-mediated phosphorylation of Upf1.
Figure 4.
SMG-1-mediated phosphorylation of Upf1 requires Upf2, Upf3b, and Y14 in vivo. (A) HeLa TetOff cell lysates transfected with the siRNAs shown above the blots were analyzed by Western blotting with the indicated antibody probes. Anti-P-Upf1 antibody recognizes phosphorylated S1078 and S1096 residues. Arrowheads indicate the positions of Upf3aL and Upf3aS. (B) Quantification of the phosphorylation level of Upf1 in vivo when NMD components were depleted.
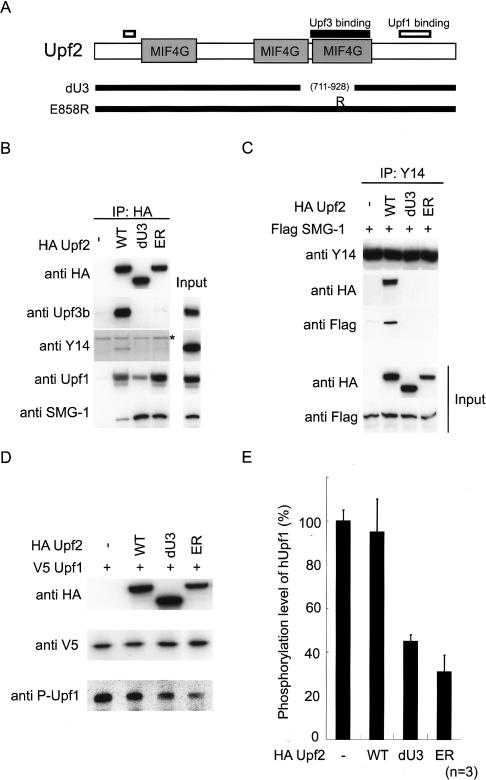
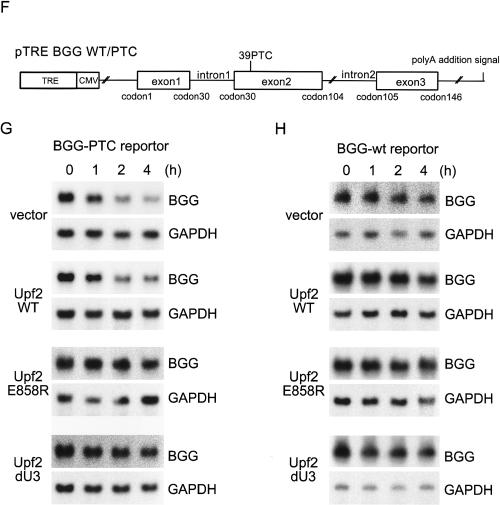
To further confirm the role of Upf2 and Upf3b in SMG-1-mediated phosphorylation of Upf1, we created two Upf2 mutants, which do not retain the ability to associate with Upf3b (Fig. 5A). The first was Upf2-dU3, which has a deletion between residues 711 and 928 (Serin et al. 2001), and the second was Upf2-E858R, which harbored a glutamic acid-to-arginine mutation at residue 858, which is an essential amino acid residue for Upf3b binding in vitro (Kadlec et al. 2004). As shown in Figure 5B, both mutants lost the ability to coprecipitate Upf3b and Y14 following overexpression of HA-tagged proteins and immunoprecipitation. Intriguingly, Upf2 mutants retained the ability to interact with Upf1 and SMG-1 (Fig. 5B). Based on these and the above results, we next tested whether the Upf2 mutants upon overexpression in cells could interrupt the association of SMG-1 and Y14. For this, we expressed either wild-type or mutant HA-Upf2 along with Flag-SMG-1 in HeLa TetOff cells, and subsequently immunoprecipitated endogenous Y14. As also shown in Figure 1B, overexpression of wild-type Upf2 had no effect on Y14's ability to associate with SMG-1. Upf2-dU3 and Upf2-E858R, on the other hand, completely disrupted the association between Y14 and Flag-SMG-1 (Fig. 5C). Further, when the mutants were overexpressed with V5-Upf1, both Upf2-dU3 and Upf2-E858R caused a decrease in phosphorylation of Upf1 whereas wild-type Upf2 had no effect (Fig. 5D,E). Furthermore, we looked at whether these mutants affect wild-type or PTC-containing β-globin mRNA degradation. Briefly, Hela TetOff cells were transfected with either wild-type or PTC-containing β-globin construct (Fig. 5F) along with wild-type or mutant Upf2. Cells were treated with doxycycline, and the half-life of the mRNAs was measured. As shown in Figure 5G and H, Upf2-dU3 and Upf2-E858R had no effect on the half-life of wild-type mRNA but did stabilize the PTC-bearing mRNA. Wild-type Upf2 had no effect on either transcript (see graphs in Supplementary Figure 5; PTC-bearing mRNA: T1/2 > 240 min for expression of mutant Upf2 vs. T1/2 = ∼180 min for expression of wild-type Upf2). Thus, most likely due to the inhibition of Upf1 phosphorylation, Upf2-dU3 and Upf2-E858R inhibit NMD. Since these mutants block association of Upf2 with Upf3b, this interaction is essential for the association of Y14 with SMG-1 and thus SMG-1-mediated phosphorylation of Upf1.
Figure 5.
Interaction between Upf2 and Upf3b is required for the association between SMG-1 and mRNP during NMD. (A) Schematic representation of Upf2 mutants. The solid line indicates the Upf3-binding region. The white line indicates the Upf1-binding region. MIF4G (mammalian eIF4G like) domains are shown as dark-gray boxes. (B) Upf2 mutants do not interact with Upf3b and Y14. Ectopic HA-Upf2-WT, HA-Upf2-dU3, or HA-Upf2-E858R expressed in HeLa TetOff cells was immunoprecipitated under the conditions of RNase-treatment and analyzed for the presence of endogenous proteins by Western blotting with the indicated antibody probes. A minus sign (-) indicates an empty vector. Upf2 mutants do not affect expressions of probed proteins (data not shown). Input represents 10% of the amount immunoprecipitated and of the empty vector transfected. An asterisk indicates the IgG signals. (C) HeLa TetOff cell lysates transfected with plasmids were immunoprecipitated with anti-Y14 antibody under conditions of RNase-treatment (IP). IP and the input fraction (10% of the amount immunoprecipitated) were analyzed by Western blotting with the indicated antibody probes. (D) Overexpression of Upf2 mutants inhibits the phosphorylation of Upf1. (E) Quantification of the phosphorylation level of Upf1 when Upf2 mutants were overexpressed. (F) Schematic representation of human β-globin gene (BGG) reporter constructs BGG-WT and BGG-PTC. The ORF is represented by boxes and introns and UTRs are represented by lines. (G,H) Overexpression of Upf2-dU3 and Upf2-E852R inhibit PTC-containing β-globin mRNA decay. HeLa TetOff cells were transfected with a reporter plasmid (BGG-WT or BGG-PTC). Cells were simultaneously transfected with the Upf2 plasmids, as indicated on the left of blots. Panels show typical examples of the results of Northern blotting. The quantities of BGG mRNA, normalized to GADPH signals, are graphed in Supplementary Figure 5.
SMG-1 forms a complex with Upf1, eRF1, and eRF3 in a Upf2- and Y14-independent manner
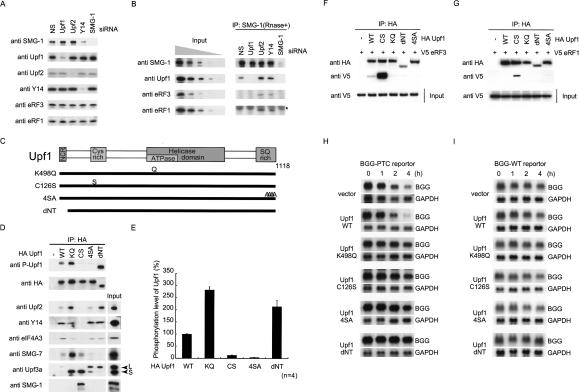
The requirements for EJC components for the phosphorylation of Upf1 strongly support the notion that SMG-1-mediated phosphorylation of Upf1 occurs on EJCs that are deposited on the post-spliced mRNA. To further evaluate the nature of the association between SMG-1 and EJC/mRNA, in vitro splicing coupling coimmunoprecipitation analysis on HEK293T cell extracts was performed. The results show that SMG-1 does not associate with spliced mRNA after in vitro splicing. Control Upf3b and Y14 specifically precipitated spliced mRNA (Supplementary Fig. 3). However, analysis of SMG-1-IPs from HeLa TetOff total cell extracts using anti-SMG-1 antibodies combined with knockdown of Upf2 or Y14 revealed that the depletion of Upf2 or Y14 stimulated the coprecipitation of Upf1 instead of inhibiting their interaction (Fig. 6B). These results not only indicated that SMG-1 is not a stable EJC component but also suggested that SMG-1 can form a complex with Upf1 in the absence of the association with Upf2 on EJC. The most intriguing possibility is that the association between SMG-1-Upf1 and the Upf2-EJC complexes follows after recognition of PTC on mRNA. If this was the case, the eukaryotic release factors (eRF1 and eRF3) should be involved in the SMG-1-Upf1 complex, although an in vivo association with eRF1 and eRF3 has not been previously detected in mammals. To test this possibility, we probed the SMG-1-IPs with antibodies against eRF1 or eRF3. Intriguingly, the knockdown of Upf2 or Y14 greatly enhanced the recovery of eRF1 or eRF3 in SMG-1-IPs (Fig. 6B). All tested proteins were efficiently depleted after siRNA transfection (Fig. 6A).
Figure 6.
The SMG-1-Upf1 complex with eRF1 and eRF3. (A) HeLa TetOff cell lysates transfected with the siRNAs shown above the blots were analyzed by Western blotting with the indicated antibody probes. (B) HeLa TetOff cell lysates transfected with siRNA were immunoprecipitated with anti-SMG-1 antibodies (IP). IP or cell lysates (input; 10%, 3.3%, 1.1%, 0. 33%, and 0.11% of the amount immunoprecipated) were then probed with the antibodies shown on the left. An asterisk indicates the IgG signals. (C) Schematic structures of human Upf1 mutants. The metazoan-specific N-terminal conserved region (NCR), cysteine-rich regions, helicase domains, and SQ-rich region of Upf1 are shown in gray boxes. (D,F,G) HeLa TetOff cells transfected with the plasmids indicated above each blot; a minus sign (-) indicates an empty vector. Cell lysates (input) were immunoprecipitated with the antibodies under the conditions of RNase-treatment shown on the top and probed with the antibodies shown on the left. Upf1 mutants do not affect expressions of probed proteins (data not shown). Input represents 10% of the amount immunoprecipitated. (D) Immunoprecipitations of Upf1 mutants. (E) Quantification of the phosphorylation level of Upf1 mutants. (F,G) Upf1-C126S strongly interact with eRF1 and eRF3. (H,I) Similar to Figure 5G, HeLa TetOff cells were transfected with a reporter plasmid (BGG-WT or BGG-PTC). Cells were simultaneously transfected with the Upf1 plasmids, as indicated on the left. Panels show typical examples of the results of Northern blotting. The quantities of BGG mRNA, normalized to GADPH signals, are graphed in Supplementary Figure 5.
To further confirm the above idea, we designed a series of Upf1 mutants that were expected to loose some of the functions of Upf1: C126S (C-S mutation at residue 126 whose analogous substitution in S. cerevisiae causes a loss of Upf2 binding) (Weng et al. 1996), K498Q (K498, a putative ATP-binding residue, is substituted with Q that abolishes the ATPase activity) (Supplementary Fig. 4), 4SA (four amino acid substitutions in Upf1 preferentially phosphorylated by SMG-1; S1073A, S1078A, S1096A, and S1116A), and dNT (a deletion of N-terminal 63 residues resulting in a reduction of the ability to associate with SMG-5) (Fig. 6C).
Wild-type or mutant Upf1 and V5-tagged eRF1 or eRF3 were expressed in HeLa TetOff cells. HA-Upf1 was immunopurified from HeLa TetOff total extracts using HA-matrix in the presence of RNase. Consistent with the Upf2-independent formation of the SMG-1, Upf1, eRF1, and eRF3 complex, Upf1-C126S coprecipitated more SMG-1, V5-eRF1, and V5-eRF3 than did wild type, whereas it did not coprecipitate detectable amounts of Upf2, Y14, or eIF4A3 (Fig. 6D,F,G). Next, we measured the phosphorylation state of Upf1 mutants using the anti-P-Upf1 antibody. Consistent with the results of the knockdown of Upf2, Upf3b, or Y14, the phosphorylation state of Upf1-C126S is greatly reduced compared with wild-type Upf1. On the other hand, other Upf1 mutants did not produce any significant change in SMG-1, eRF1, or eRF3 binding except for a modest decrease in eRF3 precipitation for Upf1-K498Q, Upf1-dNT, and Upf1-4SA mutants (Fig. 6D,F,G). In addition, Upf1-K498Q and Upf1-dNT show increases in Upf2 precipitation and are more phosphorylated compared with wild-type Upf1 (Fig. 6D; Ohnishi et al. 2003). Furthermore, compared with wild-type Upf1, the amount of coprecipitated SMG-7 was greatly reduced for Upf1-4SA and Upf1-dNT precipitates, increased for Upf1-K498Q, and modestly increased for Upf1-C126S (Fig. 6D). In addition, Upf1-K498Q and Upf1-C126S preferentially associated with the Upf3aS isoform, whereas Upf1-4SA and Upf1-dNT favored the Upf3aL isoform (Fig. 6D).
As shown Figure 6H and I, all Upf1 mutants have an ability to inhibit NMD when overexpressed in HeLa TetOff cells, as measured by the half-life of the β-globin-PTC reporter (see graphs in Supplementary Fig. 5; T1/2 > 240 min for expression of mutant Upf1 vs. T1/2 = ∼180 min for expression of wild-type Upf1).
To evaluate the consequences of the phosphorylation and eRF3 association of Upf1, we coexpressed V5-eRF3 and wild-type His-SMG-1 or His-SMG-1-DA, a kinase inactive mutant, and immunoprecipitated V5-eRF3 to analyze the coprecipitation of endogenous Upf1. As shown in Figure 7, a large increase in Upf1 coprecipitation was observed in V5-eRF3 precipitates when His-SMG-1-DA was coexpressed, suggesting that the SMG-1-mediated phosphorylation of Upf1 induces the dissociation of eRF3 from Upf1.
Figure 7.
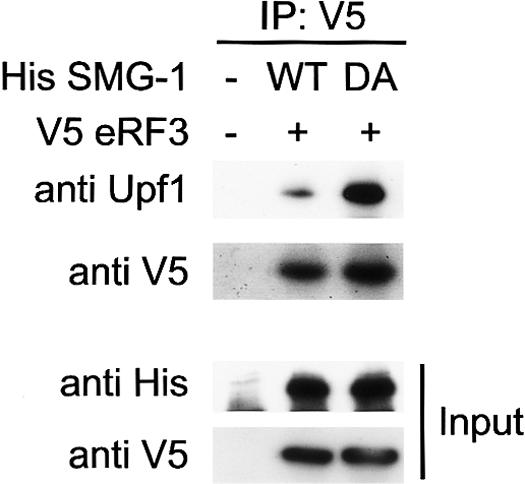
The interaction between Upf1 and eRF3 increased upon overexpression of His-SMG-1-DA. HeLa TetOff cell lysates transfected with the plasmids shown above. Cell lysates (input) were immunoprecipitated with the antibodies under the conditions of RNase-treatment as shown on the top and were probed with the antibodies shown on the left. Input represents 10% of the amount immunoprecipitated.
Discussion
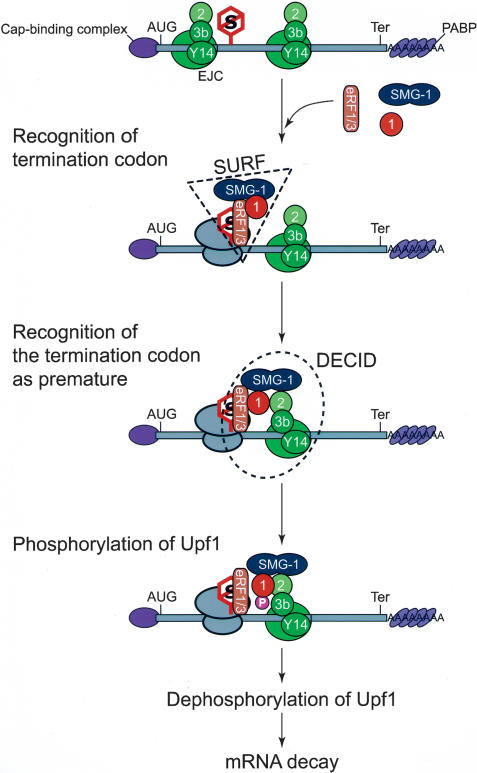
The SMG-1-mediated phosphorylation of Upf1 is a critical step in NMD in mammalian cells (Fig. 6H,I; Yamashita et al. 2001; Ohnishi et al. 2003). In the present study, we demonstrated that the SMG-1-mediated phosphorylation of Upf1 is an essential part of PTC recognition and occurs in a complex, the SURF associated with the EJC on mRNPs. Figure 8 illustrates our view of formation of the SURF-EJC complex, which we termed the decay-inducing complex (DECID), and the steps that lead up to the phosphorylation of Upf1 by SMG-1.
Figure 8.
A model illustrating the SMG-1-mediated phosphorylation of Upf1 that occurs on post-splicing mRNP after translation termination. Details are described in the text. (P) Phosphate group; (1) Upf1; (2) Upf2; (3b) Upf3b; (eRF1/3) eRF1-eRF3 complex; (EJC) exon junction complex; (AUG) start codon; (Ter) termination codon; (S) stop codon; (SURF) SMG-1-Upf1-eRF1-eRF3 complex; (DECID) decay-inducing complex.
SMG-1 forms a SURF
Several lines of evidence led us to hypothesize the presence of a SURF. First, Upf1 interacts with both SMG-1-ND and SMG-1-CD, whereas Upf2 and EJC components (Upf2-EJC) can only bind SMG-1-CD, suggesting that SMG-1 interacts with Upf1 and Upf2-EJC via different regions (Fig. 2C). Second, SMG-1 coprecipitates with Upf1 in Upf2- or Y14-depleted conditions (Fig. 6A,B). Third, Upf1-C126S, which cannot bind Upf2-EJC, associates with SMG-1 (Fig, 6D). Furthermore, human eRF1 and eRF3 were identified as additional SURF components (Fig, 6A,B,F,G). Intriguingly, the SURF accumulated greatly when Upf2 or Y14 was depleted (Fig. 6A,B) or the Upf2-binding region of Upf1 is mutated (Fig. 6D,F,G). These observations not only support Upf2-EJC-independent SURF formation but also indicate the transient nature of SURF. The SURF accumulated when the next step, the association of the SURF with Upf2-EJC (to be discussed later), was impaired. The presence of translation termination factors eRF1 and eRF3 suggested that transient SURF formation most likely occurs after recognition of the translation termination codon, and that the eRF1-eRF3 complex probably recruits Upf1 and SMG-1 to a PTC.
The SURF associates with the post-splicing mRNA through Upf2-EJC to form the DECID
The RNA- and Y14-dependent association of SMG-1 and PABP or CBP20 indicates that SMG-1 binds post-splicing mRNAs through EJC (Fig. 1A,D). The presence of CBP20, not eIF4E, in SMG-1-containing post-splicing mRNP complexes (Fig. 1A) suggests that SMG-1 primarily associates with a complex in an early phase, the so-called pioneer round of translation (Chiu et al. 2004; Maquat 2004). Interestingly, SMG-1 associates with the known EJC components Upf2, Y14, Magoh, Upf3b, and eIF4A3 through the CD region in the absence of RNA (Figs. 1A, 2B,C; data not shown). Furthermore, Upf2 (Fig. 3B,D) and its ability to bind Upf3b (Fig. 5B,C), but not Upf1 (Fig. 3B), is essential for SMG-1-Y14/EJC-mRNP complex formation, suggesting that SMG-1 associated with post-splicing mRNP complexes through Upf2 as an anchor protein. These observations are also supported by the in vitro direct interactions (most likely) of SMG-1-Upf2 (Fig. 2D), Upf2-Upf3b (Kadlec et al. 2004), and Upf3b-Y14 (Kim et al. 2001a). Like SMG-1, Upf1 binds EJC through Upf2 (Fig. 6D), which contributes to the RNA-dependent association between Upf1 and PABP (Schell et al. 2003; I. Kashima and S. Ohno unpubl.), suggesting that the Upf1-Upf2-Upf3-Y14/EJC protein interaction is also present on the post-spliced mRNA. It is also possible that the Upf1-C126S mutant loses the ability to interact with other Upf1-binding proteins that are involved in the mRNP complex formation of Upf1 (Hosoda et al. 2005; Kaygun and Marzluff 2005; Kim et al. 2005). Also, given the Upf2- or Y14-independent SURF formation of SMG-1, Upf1, eRF1, and eRF3 (Fig. 6B,D,F,G), we concluded that SURF binds EJC-mRNP through Upf2 after recognition of the translation termination codon, which seems to link the termination codon and EJC through protein-protein interactions. Based on the essential role of the complex formation of SURF-Upf2-EJC in NMD (Figs. 5G,H, 6H,I), we named this complex DECID.
SMG-1 phosphorylates Upf1 in DECID after recognition of premature translation termination codons
Four lines of evidence we provide here strongly support the notion that the SMG-1-mediated phosphorylation of Upf1 occurs on the post-spliced mRNA, and that the complex formation of DECID, namely, the SURF-Upf2-EJC complex, is required for inducing Upf1 phosphorylation. First, Upf2 mutants that do not bind Upf3b-Y14 cannot induce phosphorylation of Upf1, even though they make a complex with SMG-1 and Upf1 (Fig. 5D,E). Second, knockdown of essential components required for the association between SMG-1 and the post-splicing mRNP complex; namely Upf2, Upf3b, or Y14 strongly inhibit Upf1 phosphorylation (Fig. 4). Third, detectable phosphorylation is not observed in Upf1-C126S (Fig. 6D), and fourth, in both cases, Upf1 forms the SURF with SMG-1 (Fig. 6B,D,E). The hypothesis that Upf1 phosphorylation occurs on the post-spliced mRNA is also supported by the observation that the phosphorylated form of Upf1 primarily presents in the polysomal fraction in mammalian cells (Pal et al. 2001), although we do not know whether it is mainly restricted to the 80S mono-ribosome, as is Y14 (Dostie and Dreyfuss 2002). To further support the above observations, inhibition of translation initiation suppresses Upf1 phosphorylation (N. Izumi and S. Ohno, unpubl.).
We did not detect any difference between Upf2-knockdown and Upf3b- or Y14-knockown in Upf1 phosphorylation or SMG-1-mRNP complex formation, even though 3′ untranslated region (UTR) tethering experiments indicate a distinguishable difference in cytoplasmic 3′UTR-tethered mRNA abundance between Upf2-tethering and Upf3b- or Y14-tethering (Gehring et al. 2005). In addition, tethering assay indicated that the Upf2-binding ability of Upf3b is dispensable for tethering-inducing mRNA decay (Gehring et al. 2003). At the very least, there is a complex containing Upf2, Upf3, and Y14 (Fig. 1; Supplementary Fig. 1), and Upf2 and its Upf3b-binding ability are essential for the interaction between SMG-1 and Y14-mRNP, and Upf1 phosphorylation and NMD (Figs. 3, 5, 6). These differences might reflect the possibility that data obtained using tethering assay cannot directly reflect bona fide NMD. There is also a possibility that there is substrate specificity of EJC components for NMD target mRNAs.
The requirement for smg-3 and smg-4 for SMG-2 phosphorylation and NMD (Pulak and Anderson 1993; Page et al. 1999), and the smg-3-independent complex formation of SMG-1 and SMG-2 (Grimson et al. 2004) suggest that a similar mechanism to mammals exists in C. elegans. However, similar to S. cerevisiae (Leeds et al. 1991) and D. melanogaster (Gatfield et al. 2003), exon-exon junctions are most likely not required for PTC recognition in C. elegans. For example, exon-exon junctions do not affect PTC recognition in a C. elegans unc-54 mutant (Pulak and Anderson 1993). Based on the present results, the most probable model is that in these eukaryotes the SURF and the downstream sequence element (DSE; a cis-acting element of NMD)-binding protein complex containing Upf3/SMG-4, and probably Upf2/SMG-3, distinguish between PTC and normal termination codons, although no obvious DSE has been identified in C. elegans and D. melanogaster. A similar model was proposed for S. cerevisiae (Culbertson and Neeno-Eckwall 2005); nevertheless, the association between DSE and Upf3p and/or Upf2p needs to be clarified. Recently, phosphorylation of Upf1p was described in S. cerevisiae (de Pinto et al. 2004), although S. cerevisiae lacks an obvious SMG-1 homolog. In addition, Upf1p-C65S, which is a similar mutation to human Upf1-C126S, shows significant phosphorylation, but S. cerevisiae might have different means of Upf1 phosphorylation (de Pinto et al. 2004).
Role of the SMG-1-mediated phosphorylation of Upf1
What is the consequence of Upf1 phosphorylation during NMD? To date, we have not been able to detect any differences in the activity of the nonphosphorylated and phosphorylated forms of purified human Upf1 ATPase (Supplementary Fig. 4). However, there is evidence that phosphorylation induces remodeling of the surveillance complex containing Upf1 (Ohnishi et al. 2003). In addition to the previous findings, we have established that Upf1 phosphorylation causes dissociation of eRF3 from Upf1. The following observations further support this notion: (1) Overproduction of kinase inactive mutant of SMG-1 causes the accumulation of the eRF3 and Upf1 complex (Fig. 7); and (2) compared with the wild-type, Upf1-dNT and Upf1-K498Q have higher phosphorylation levels of Upf1, and cause a decrease in coprecipitated eRF3, together with an increase in coprecipitated Upf2 (Fig. 6D,F). On the other hand, wild-type and kinase-inactive mutant of SMG-1 did not show significant difference in coprecipitation of Upf1, Upf2, Upf3b, and Y14 (data not shown).
The phosphorylation-induced remodeling of the Upf1 complex might also regulate its ATPase/helicase activity. In support of this notion, eRF1 and eRF3 (components of SURF) are putative inhibitor proteins of Upf1 (Czaplinski et al. 1998) and probably suppress Upf1 activity in the SURF. In fact, the ATPase-inactive mutant of Upf1, Upf1-K498Q, can bind Upf2-EJC (Fig. 6D). This suggests that Upf1 ATPase activity is not required for the association between Upf1 and Upf2-EJC. We might therefore expect that Upf1 activity would increase after Upf1 phosphorylation, which mediates the dissociation of eRF1-eRF3 from Upf1 during the remodeling of the Upf1 complex. Further analysis—for example, an ATPase assay of immunocomplexes of Upf1 mutants— would be helpful for addressing this issue.
Furthermore, phosphorylated Upf1 recruits the SMG-5-SMG-7 complex to form the wee complex (WC) together with PP2A and Upf3aS (Ohnishi et al. 2003), which induces dephosphorylation of P-Upf1 (Ohnishi et al. 2003; Anders et al. 2003) and might regulate the cellular localization of Upf1 (Unterholzner and Izaurralde 2004). Consistently, the TPR containing the N-terminal region of SMG-7 is structurally related to 14-3-3 and can bind phosphorylated S1096 peptide in vitro (Fukuhara et al. 2005). Further support for the above notions is that Upf1-4SA reduces the affinity for SMG-7 (Fig. 6D). In addition, a specific association between the N-terminal half of SMG-1 and the WC components SMG-7 and Upf3aS (Fig. 2B), and the efficient association of Upf1-C126S and SMG-1, Upf3aS, or SMG-7 (Fig. 6D) probably suggests the involvement of SMG-1 in the recruitment of the SMG-5-SMG-7-Upf3aS complex to P-Upf1.
In conclusion, our data demonstrate that SMG-1 forms a SURF with Upf1, eRF1, and eRF3, most likely just after the recognition of the translation termination codon on post-spliced mRNAs. If the SURF can recognize downstream Upf2-EJC, the SURF associates with Upf2-EJC to form the DECID to induce Upf1 phosphorylation. Because Upf1 phosphorylation is the rate-limiting step in NMD (Yamashita et al. 2001; Ohnishi et al. 2003), we propose that the complex formation of DECID and the subsequent Upf1 phosphorylation represent the step that recognizes the termination codon as PTC in mammalian cells. This might imply that some regulatory mechanism of NMD via the SMG-1-mediated phosphorylation of Upf1 might also control the expression of natural NMD target genes.
Materials and methods
Plasmids and antibodies
Expression vectors for Flag-SMG-1-wild-type (WT), Flag-SMG-1-ND (amino acids 1-2223), Flag-SMG-1-CD (amino acids 2068-3657), and V5-Upf1-WT, HA-Upf2-WT were constructed in one of the mammalian expression vectors SR-Flag, SR-HA, or SR-V5, following standard procedures. V5-eRF1 and V5-eRF3 were created by modifying pFlagTB3-1 and pFlagGSPT2 (Hoshino et al. 1998). The HA-Upf1 mutants (HA-Upf1-K498Q, HA-Upf1-C126S, and HA-Upf1-4SA [S1073A S1078A S1096A S1116A]) and the HA-Upf2 mutants (HA-Upf2-dU3 and HA-Upf2-E858R) were constructed by site-directed mutagenesis of the HA-Upf1-WT and HA-Upf2-WT plasmids, respectively. HA-Upf1-WT, HA-Upf1-dNT, pTRE-BGG-WT, and pTRE-BGG-PTC reporter plasmids were constructed as previously described (Yamashita et al. 2001; Ohnishi et al. 2003). Anti-Upf3b and Dcp2 antibodies were generated against the peptide sequence (CAERKQESGISHRKEGGEE) in Upf3b and recombinant human Dcp2 (269-420 amino acids) protein fused to maltose-binding protein. Affinity purification of the antibody was performed following standard procedures. The anti-SMG-1, Upf1, P-Upf1 (clone 3B8), Upf2, Upf3a, SMG-7, eRF1, eRF3, Y14 (clone 4C4), and eIF4A3 (clone 3F1) antibodies were generated as described previously (Hoshino et al. 1998; Kataoka et al. 2000; Yamashita et al. 2001; Ohnishi et al. 2003; Chan et al. 2004). The anti-CBP20 antibody was a generous gift from Dr. Ian Mattaj (European Molecular Biology Laboratory, Heidelberg, Germany). Antibodies to eIF4E (Santa Cruz), PABP (Abcam), β-actin (Sigma), GAPDH (Abcam), and lamin A/C (Santa Cruz) were used. The anti-tag antibodies (HA, Flag, V5) were obtained commercially.
Transfection with plasmids and siRNA
HeLa TetOff cells (Clontech) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. Plasmid transfections were performed in six-well plates, 10-cm dishes, or 15-cm dishes using Polyfect (Qiagen) according to the manufacturer's recommendations. For the immunoprecipitation of HA- or Flag-tagged proteins, transfected cells were harvested 36 h later. Transient siRNA transfections were performed in 12-well plates or 15-cm dishes; cells were transfected with RNAifect (Qiagen) and 100 nM siRNA according to the manufacturer's protocol; and cells were harvested 72 h later. The following target sequences were used: SMG-1, AAGTGTATGTGCGCCAAAGTA; Upf1, AAGATGCAGTTCCGCTCCATT; Upf2, AACAACAGCCCTTCCAGAATC, AAGGCTTTTGTCCCAGCCATC; Y14, AACGCTCTGTTGAAGGCTGGA, AATTCGCAGAATATGGGGAAA; SMG-7, AACAGCACAGTCTACAAGCCA; lamin A/C, AACTGGACTTCCAGAAGAACA; nonsilencing (NS), AATTCTCCGAACGTGTCACGT; Upf3b, SMARTpool siRNA no. M-012871-00 (Dharmacan); and Dcp2, SMARTpool no. M-008425-00 (Dharmacan).
Immunoprecipitation and Western blotting
HeLa TetOff cells were transfected using Polyfect (Qiagen) or RNAifect (Qiagen) and incubated for 30 min at 4°C in a lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1% Nonidet-P40, 100 nM okadaic acid (Calbiochem), and protease inhibitor cocktail (Sigma). The precleared lysates were added to 200 μg/mL RNaseA (Qiagen) or 0.1 U/mL RNAsin Plus (Promega), then incubated with antibodies preadsorbed on 30 μL of protein G or protein A-Sepharose (Amersham Biosciences) for 2 h at 4°C with rotation. To immunoprecipitate, 25 μL of anti-HA-affinity matrix (Roche) or anti-Flag-M2 agarose (Sigma) or 15 μg of appropriate antibodies were used instead of protein G-Sepharose preadsorbed with antibody. The immunocomplexes were washed with a washing buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1% Nonidet-P40, and 100 nM okadaic acid; boiled in 100 μL of standard 1× SDS sample buffer; and then analyzed by Western blotting. In experiments where HA-affinity matrix or Flag-agarose was used, proteins recovered on the matrix were eluted by incubation for 15 min at 37°C in the presence of 50 μL of washing buffer containing 1 mg/mL HA-peptide or 0.5 mg/mL Flag × 3 peptide, boiled with 50 μL of 2× SDS sample buffer, and analyzed by Western blotting. All Western blot experiments were detected with an ECL Western blotting detection system (Amersham Biosciences) and quantified with a LuminoImager, LAS-3000, and Science Lab 2001 Image Gauge software (Fuji Photo Film). The quantities of P-Upf1, normalized to Upf1 signals, are graphed in Figures 4B and 6D. All immunoprecipitation experiments were performed three or four times, and typical results are shown.
Cell fractionation
For the preparation of cytoplasmic extracts, 1 × 109 cells were resuspended in an equal volume of hypotonic lysis buffer containing 10 mM Tris HCl (pH 8.0), 10 mM NaCl, 2 mM MgCl2, 0.2 mM DTT, 1× Complete Protease Inhibitor Cocktail (Roche), 50 nM okadaic acid, and 100 μg/mL RNaseA (Qiagen) and were subsequently lysed by 30 hand-strokes with a loose-fit Potter-Elvehjem homogenizer. The lysates were centrifuged at 500 × g for 10 min at 4°C to remove cell nuclei. Corrected supernatants were centrifuged at 15,000 × g for 20 min at 4°C to remove membranes and particulate material. Finally, the lysates were supplemented with NaCl to obtain a final concentration of 50 mM. For preparation of nuclear extracts, corrected cell nuclei were washed twice with 10 mL of hypotonic lysis buffer, then resuspended in an equal volume of nuclear extraction buffer containing 10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 0.1 mM EGTA, 0.1 mM EDTA, 420 mM NaCl, 25% glycerol, 0.5 mM DTT, and 1× Complete Protease Inhibitor Cocktail (Roche) and incubated for 30 min on ice. The lysates were centrifuged at 12,000 × g for 10 min at 4°C to remove the precipitate. The same amount of fractionated protein lysate was then immunoprecipitated by anti-Flag-M2 agarose (Sigma).
In vitro protein-binding experiment
HA- or Flag-tagged proteins were produced in vitro using a wheat germ cell-free system (Cell-Free Science). All HA-tagged proteins shown in Figure 2 were expressed, and Flag-SMG-1-CD was expressed and purified using anti-Flag-M2 agarose. In vitro translated products were added to 300 ng of Flag-SMG-1-CD coupled to anti-Flag-M2 agarose resin in binding buffer (20 mM Tris-HCl at pH 7.4, 2.5 mM MgCl2, 100 mM NaCl, 0.5 mM DTT, and 0.05% Tween20) and mixed for 1 h at 4°C. After binding, resins were washed five times with the same buffer, and the bound protein was eluted with an 1 × SDS sample buffer and then analyzed by Western blotting. Experiments were performed at least three times, and typical results are shown.
NMD analysis
NMD analysis was performed in essentially the same manner as described elsewhere (Yamashita et al. 2001; Ohnishi et al. 2003), except that HeLa TetOff cells (1.8 × 106) were transfected with 3 μg of effecter plasmids, which are indicated in Figures 5G and H and 6H and I, in combination with 3 μg of pTRE-BGG-WT or pTRE-BGG-PTC. Briefly, 24 h after transfection, doxycyclin was added at time zero to inhibit de novo transcription, cells were harvested at the indicated time points, and total RNAs were analyzed by Northern blotting using either a β-globin or GAPDH (control) probe. The quantities of BGG mRNA, normalized to GADPH signals, are graphed in Supplementary Figure 5. Values in Supplementary Figure 5A and C represent mean ± SE for three independent experiments. Data in Supplementary Figure 5B and D are the average of two independent experiments.
Acknowledgments
We thank Dr. Iain Mattaj (European Molecular Biology Laboratory) for the kind gift of the antibody against CBP20. We also thank Yuri Kikuchi, Makiko Ono, Tomoko Morita, Yukiko Yamashita, and Dr. Tetsuo Ohnishi for technical support, and Michael Diem for critical reading of the manuscript. This work was supported in part by grants to S.O. from the Japan Society for the Promotion of Science; from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and from the Japan Science and Technology Corporation. I.K. is a Research Fellow of the Japan Society for the Promotion of Sciences. A.Y. received support from the Yokohama Foundation for Advancement of Medical Science.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1389006.
References
- Anders K.R., Grimson, A., and Anderson, P. 2003. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 22: 641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K.E. and Parker, R. 2004. Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr. Opin. Cell Biol. 16: 293-299. [DOI] [PubMed] [Google Scholar]
- Cali B.M. and Anderson, P. 1998. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol. Gen. Genet. 260: 176-184. [DOI] [PubMed] [Google Scholar]
- Cali B.M., Kuchma, S.L., Latham, J., and Anderson, P. 1999. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 151: 605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.C., Dostie, J., Diem, M.D., Feng, W., Mann, M., Rappsilber, J., and Dreyfuss, G. 2004. eIF4A3 is a novel component of the exon junction complex. RNA 10: 200-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S.Y., Lejeune, F., Ranganathan, A.C., and Maquat, L.E. 2004. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes & Dev. 18: 745-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E. and Izaurralde, E. 2005. Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17: 316-325. [DOI] [PubMed] [Google Scholar]
- Culbertson M.R. and Leeds, P.F. 2003. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13: 207-214. [DOI] [PubMed] [Google Scholar]
- Culbertson M.R. and Neeno-Eckwall, E. 2005. Transcript selection and the recruitment of mRNA decay factors for NMD in Saccharomyces cerevisiae. RNA 11: 1333-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M.R., Underbrink, K.M., and Fink, G.R. 1980. Frameshift suppression Saccharomyces cerevisiae, II: Genetic properties of group II suppressors. Genetics 95: 833-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Weng, Y., Hagan, K.W., and Peltz, S.W. 1995. Purification and characterization of the Upf1 protein: A factor involved in translation and mRNA degradation. RNA 1: 610-623. [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria, M.J., Paushkin, S.V., Han, X., Weng, Y., Perlick, H.A., Dietz, H.C., Ter-Avanesyan, M.D., and Peltz, S.W. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes & Dev. 12: 1665-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pinto B., Lippolis, R., Castaldo, R., and Altamura, N. 2004. Overexpression of Upf1p compensates for mitochondrial splicing deficiency independently of its role in mRNA surveillance. Mol. Microbiol. 51: 1129-1142. [DOI] [PubMed] [Google Scholar]
- Dostie J. and Dreyfuss, G. 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 12: 1060-1067. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim, V.N., and Kataoka, N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3: 195-205. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo M.A., Lee, C.S., Ler, L.W., Hsu, J.L., Costa-Mattioli, M., Luo, M.J., Reed, R., and Sonenberg, N. 2004. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl. Acad. Sci. 101: 4118-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer P.A. and Dietz, H.C. 1999. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 8: 1893-1900. [DOI] [PubMed] [Google Scholar]
- Fukuhara N., Ebert, J., Unterholzner, L., Lindner, D., Izaurralde, E., and Conti, E. 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell 17: 537-547. [DOI] [PubMed] [Google Scholar]
- Gatfield D., Unterholzner, L., Ciccarelli, F.D., Bork, P., and Izaurralde, E. 2003. Nonsense-mediated mRNA decay in Drosophila: At the intersection of the yeast and mammalian pathways. EMBO J. 22: 3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring N.H., Neu-Yilik, G., Schell, T., Hentze, M.W., and Kulozik, A.E. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11: 939-949. [DOI] [PubMed] [Google Scholar]
- Gehring N.H., Kunz, J.B., Neu-Yilik, G., Breit, S., Viegas, M.H., Hentze, M.W., and Kulozik, A.E. 2005. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 20: 65-75. [DOI] [PubMed] [Google Scholar]
- Grimson A., O'Connor, S., Newman, C.L., and Anderson, P. 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol. Cell Biol. 24: 7483-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Li, X., Spatrick, P., Casillo, R., Dong, S., and Jacobson, A. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12: 1439-1452. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Papp, A., Pulak, R., Ambros, V., and Anderson, P. 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J.A., Neu-Yilik, G., Hentze, M.W., and Kulozik, A.E. 2004. Nonsense-mediated decay approaches the clinic. Nat. Genet. 36: 801-808. [DOI] [PubMed] [Google Scholar]
- Hoshino S., Imai, M., Mizutani, M., Kikuchi, Y., Hanaoka, F., Ui, M., and Katada, T. 1998. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF): Its identification as eRF3 interacting with eRF1. J. Biol. Chem. 273: 22254-22259. [DOI] [PubMed] [Google Scholar]
- Hosoda N., Kim, Y.K., Lejeune, F., and Maquat, L.E. 2005. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 12: 893-901 [DOI] [PubMed] [Google Scholar]
- Kadlec J., Izaurralde, E., and Cusack, S. 2004. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat. Struct. Mol. Biol. 11: 330-337. [DOI] [PubMed] [Google Scholar]
- Kataoka N., Yong, J., Kim, V.N., Velazquez, F., Perkinson, R.A., Wang, F., and Dreyfuss, G. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6: 673-682. [DOI] [PubMed] [Google Scholar]
- Kataoka N., Diem, M.D., Kim, V.N., Yong, J., and Dreyfuss, G. 2001. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 20: 6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H. and Marzluff, W.F. 2005. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12: 794-800. [DOI] [PubMed] [Google Scholar]
- Kim V.N., Kataoka, N., and Dreyfuss, G. 2001a. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293: 1832-1836. [DOI] [PubMed] [Google Scholar]
- Kim V.N., Yong, J., Kataoka, N., Abel, L., Diem, M.D., and Dreyfuss, G. 2001b. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 20: 2062-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Furic, L., Desgroseillers, L., and Maquat, L.E. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195-208. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Funakoshi, Y., Hoshino, S., and Katada, T. 2004. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J. Biol. Chem. 279: 45693-45700. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Izaurralde, E., Maquat, L.E., and Moore, M.J. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19: 6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P., Peltz, S.W., Jacobson, A., and Culbertson, M.R. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes & Dev. 5: 2303-2314. [DOI] [PubMed] [Google Scholar]
- Li S. and Wilkinson, M.F. 1998. Nonsense surveillance in lymphocytes? Immunity 8: 135-141. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu, M.D., and Steitz, J.A. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121-1131. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836-1839. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. 2004. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 5: 89-99. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Medghalchi, S.M., Lake, R.G., Noensie, E.N., and Dietz, H.C. 2000. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell Biol. 20: 8944-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J.T., ap Rhys, C.M., and Dietz, H.C. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298: 419-422. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Sharifi, N.A., Meyers, J.L., Martinez-Murillo, F., and Dietz, H.C. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36: 1073-1078. [DOI] [PubMed] [Google Scholar]
- Mitrovich Q.M. and Anderson, P. 2000. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes & Dev. 14: 2173-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2005. mRNA surveillance of expressed pseudogenes in C. elegans. Curr. Biol. 15: 963-967. [DOI] [PubMed] [Google Scholar]
- Moore M.J. 2005. From birth to death: The complex lives of eukaryotic mRNAs. Science 309: 1514-1518. [DOI] [PubMed] [Google Scholar]
- Ohnishi T., Yamashita, A., Kashima, I., Schell, T., Anders, K.R., Grimson, A., Hachiya, T., Hentze, M.W., Anderson, P., and Ohno, S. 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12: 1187-1200. [DOI] [PubMed] [Google Scholar]
- Page M.F., Carr, B., Anders, K.R., Grimson, A., and Anderson, P. 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell Biol. 19: 5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M., Ishigaki, Y., Nagy, E., and Maquat, L.E. 2001. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7: 5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I.M., Gatfield, D., St Johnston, D., and Izaurralde, E. 2004. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753-757. [DOI] [PubMed] [Google Scholar]
- Peltz S.W., He, F., Welch, E., and Jacobson, A. 1994. Nonsense-mediated mRNA decay in yeast. Prog. Nucleic Acid Res. Mol. Biol. 47: 271-298. [DOI] [PubMed] [Google Scholar]
- Pulak R. and Anderson, P. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes & Dev. 7: 1885-1897. [DOI] [PubMed] [Google Scholar]
- Schell T., Kocher, T., Wilm, M., Seraphin, B., Kulozik, A.E., and Hentze, M.W. 2003. Complexes between the nonsense-mediated mRNA decay pathway factor human upf1 (upframeshift protein 1) and essential nonsense-mediated mRNA decay factors in HeLa cells. Biochem. J. 373: 775-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin G., Gersappe, A., Black, J.D., Aronoff, R., and Maquat, L.E. 2001. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell Biol. 21: 209-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T., Tange, T.O., Sonenberg, N., and Moore, M.J. 2004. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 11: 346-351. [DOI] [PubMed] [Google Scholar]
- Sun X., Perlick, H.A., Dietz, H.C., and Maquat, L.E. 1998. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. 95: 10009-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange T.O., Nott, A., and Moore, M.J. 2004. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 16: 279-284. [DOI] [PubMed] [Google Scholar]
- Unterholzner L. and Izaurralde, E. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 16: 587-596. [DOI] [PubMed] [Google Scholar]
- Usuki F., Yamashita, A., Higuchi, I., Ohnishi, T., Shiraishi, T., Osame, M., and Ohno, S. 2004. Inhibition of nonsense-mediated mRNA decay rescues the phenotype in Ullrich's disease. Ann. Neurol. 55: 740-744. [DOI] [PubMed] [Google Scholar]
- Wang W., Czaplinski, K., Rao, Y., and Peltz, S.W. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20: 880-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y., Czaplinski, K., and Peltz, S.W. 1996. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell Biol. 16: 5491-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.F. and Shyu, A.B. 2001. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays 23: 775-787. [DOI] [PubMed] [Google Scholar]
- Wilusz C.J., Wang, W., and Peltz, S.W. 2001. Curbing the nonsense: The activation and regulation of mRNA surveillance. Genes & Dev. 15: 2781-2785. [DOI] [PubMed] [Google Scholar]
- Yamashita A., Ohnishi, T., Kashima, I., Taya, Y., and Ohno, S. 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes & Dev. 15: 2215-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Kashima, I., and Ohno, S. 2005. The role of SMG-1 in nonsense-mediated mRNA decay. Biochim. Biophys. Acta 1754: 305-315. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Luo, M.J., Straesser, K., Katahira, J., Hurt, E., and Reed, R. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401-405. [DOI] [PubMed] [Google Scholar]